Abstract
Two genes, nahG and nahW, encoding two independent salicylate 1-hydroxylases have been identified in the naphthalene-degrading strain Pseudomonas stutzeri AN10. While nahG resides in the same transcriptional unit as the meta-cleavage pathway genes, forming the naphthalene degradation lower pathway, nahW is situated outside but in close proximity to this transcriptional unit. The nahG and nahW genes of P. stutzeri AN10 are induced and expressed upon incubation with salicylate, and the enzymes that are encoded, NahG and NahW, are involved in naphthalene and salicylate metabolism. Both genes, nahG and nahW, have been cloned in Escherichia coli JM109. The overexpression of these genes yields peptides with apparent molecular masses of 46 kDa (NahG) and 43 kDa (NahW), respectively. Both enzymes exhibit broad substrate specificities and metabolize salicylate, methylsalicylates, and chlorosalicylates. However, the relative rates by which the substituted analogs are transformed differ considerably.
Pseudomonas stutzeri AN10 is a naphthalene-degrading bacterium able to dissimilate naphthalene, 2-methylnaphthalene, and salicylate as sole carbon and energy sources (25). In contrast to the usual location on a plasmid (49), the genes for the naphthalene-catabolic pathway of P. stutzeri AN10 have been located in the chromosome (12, 25). Recently, the entire naphthalene degradation pathway of P. stutzeri AN10 has been cloned and sequenced (4). As with other extensively studied naphthalene-degrading strains, such as the archetype Pseudomonas putida G7, possessing the plasmid NAH7 (15, 29, 30, 52), and P. putida NCIB9816, possessing the NAH plasmid pWW60 (6, 23), the naphthalene-dissimilatory genes of P. stutzeri AN10 are organized in three operons: one coding for the enzymes involved in the conversion of naphthalene to salicylate (nahAaAbAcAdBFCED, naphthalene degradation upper pathway), the second coding for the conversion of salicylate to tricarboxylic acid cycle intermediates through the meta-cleavage pathway enzymes (nahGTHINLOMKJ; naphthalene degradation lower pathway), and the third coding for the regulatory gene (nahR).
The nahG gene, the gene most proximal to the naphthalene degradation lower pathway, codes for salicylate hydroxylase and has recently been cloned and sequenced in several naphthalene-degrading strains, such as P. putida G7 (53), P. putida S1 (34), P. putida KF715 (20), and P. stutzeri AN10 (4).
Salicylate hydroxylase is a flavoprotein monooxygenase that catalyzes the conversion of salicylate to catechol. The enzyme was first purified from P. putida S1 (50) and later from Pseudomonas cepacia (42), P. putida G7 (54), and other soil microorganisms (46). Mechanistic-kinetic properties of the salicylate hydroxylase have been studied extensively (10, 32, 36–40, 43, 44, 46, 47). Briefly, the enzyme binds salicylate and an external reductant (NADH or NADPH) to form a reduced enzyme-substrate complex. Subsequently, molecular oxygen binds to the complex for production of catechol, CO2, and H2O.
In this study, we have demonstrated that the salicylate hydroxylase activity in P. stutzeri AN10 is catalyzed by two isofunctional and inducible enzymes: NahG, the “classic” salicylate hydroxylase, the gene for which (nahG) resides in the same transcriptional unit as the meta-cleavage pathway genes (4), and NahW, a novel salicylate hydroxylase, whose encoding gene (nahW) is situated outside this transcriptional unit.
MATERIALS AND METHODS
Bacterial strains, plasmids, chemicals, media, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli and P. stutzeri strains were cultured in Luria-Bertani medium at 30°C (27) unless otherwise indicated. Ampicillin, tetracycline, and chloramphenicol were added at final concentrations of 100, 30, and 15 μg · ml−1, respectively, to select for the presence of plasmids. Chemicals and media were obtained from ADSA-Micro (Barcelona, Spain), Fluka Química (Madrid, Spain), Panreac Química S. A. (Barcelona, Spain), and Sigma-Aldrich Química (Madrid, Spain).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or characteristics | Source and/or reference |
|---|---|---|
| Escherichia coli | ||
| DH5α | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lacZYA-argF)U196 λ− (Φ80lacZΔM15) | 13 |
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB) λ− F′ [traD36 proAB lacIqlacZΔM15] | 51 |
| Pseudomonas stutzeri | ||
| AN10 | nah+a; genomovar 3 | Marine isolate, 24 |
| Plasmids | ||
| pSK(−) | Ampicillin-resistant cloning vector | Stratagene |
| pKS(+) | Ampicillin-resistant cloning vector | Stratagene |
| pBCSK(−) | Chloramphenicol-resistant cloning vector | Stratagene |
| pPA50C | pLFR3 with DNA insert of PsAN10 | 1 |
| pRAF104.2 | pBCSK(−) with PstI insert of pRAF104 | 4 |
| pRAF111 | pSK(−) with nahW under control of Plac | This study |
| pRAF127 | pKS(+) with nahG under control of Plac | This study |
nah+ indicates the ability to grow on naphthalene as a sole source of carbon and energy.
Standard DNA manipulations.
Standard molecular biology procedures were used throughout (27). Genomic DNA preparations were done as described previously (8). Digoxygenin DNA labelling, hybridization, and detection conditions were those recommended by the manufacturer (Boehringer Mannheim).
Plasmid construction.
Table 1 summarizes the plasmids constructed in this study. Plasmids pRAF127 and pRAF111 were constructed for overexpression of NahG and NahW, respectively, under the control of the promoter Plac. For pRAF127, a 1,751-bp EcoRV-PstI fragment from pRAF104.2 was inserted in pBluescript KS(+). Plasmid pRAF111 is a pBluescript SK(−) derivative containing the 1,433-bp XhoI-EcoRI fragment, generated by PCR from pPA50C using primers 5′-TGACTCGAGACGAATCGCCGCTTTTA-3′ and 5′-TGAGAATTCGCTGCTCCGCTTAGGTGA-3′. The fidelity of cloning was checked by DNA sequencing.
Overexpression and identification of NahG and NahW in E. coli.
NahG and NahW were overproduced by expression of the respective genes on plasmids pRAF127 and pRAF111, respectively, in E. coli JM109 by IPTG (isopropyl-β-d-thiogalactopyranoside) induction, as previously described (27). Cell extracts were obtained by sonication and clarified by centrifugation, and soluble proteins were separated by electrophoresis in sodium dodecyl sulfate-polyacrylamide (7% [wt/vol]) gels. The resolved proteins were stained with Coomassie blue (17).
Salicylate hydroxylase induction in P. stutzeri AN10.
For induction of salicylate hydroxylase, P. stutzeri AN10 cells were grown overnight in minimal medium (2) containing 5 mM succinate. Cultures were harvested, washed, suspended in fresh minimal medium (A600, 0.8) supplemented with 5 mM succinate, and then incubated for 2 h at 30°C. After this, cells were harvested again, washed, suspended in fresh minimal medium (A600, 1.0), and incubated for 4.5 h in the presence of 2 mM salicylate (induction conditions) or 5 mM succinate (noninduction conditions).
Preparation of cell extracts.
Cells from 500 ml of E. coli or P. stutzeri cultures were collected by centrifugation (13,000 × g, 10 min at 4°C), washed twice with 25 ml of 50 mM Tris-HCl (pH 8.0) buffer (TB), and resuspended in 1 ml of TB. Cell extracts were obtained by two passages through a chilled French pressure cell at 18,000 lb · in−2. DNase I was added between the French press passages. Whole cells and cell debris were removed by centrifugation at 13,000 × g for 30 min at 4°C. Ultracentrifugation was carried out at 140,000 × g for 1 h at 4°C. The clear supernatant solution was kept on ice and used for assays of salicylate 1-hydroxylase activity.
Enzyme assays.
Salicylate 1-hydroxylase activities were measured according to reported procedures (3), following NADH-oxidation activity as a decrease in the absorbance at 340 nm (ɛNADH = 6,200 M−1 · cm−1), using a Pharmacia LKB Ultrospec III spectrophotometer. The reaction mixture (1 ml) contained 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 200 μM NADH, 150 μM salicylate (or derivatives), and 5 or 50 μl of cell extracts (E. coli or P. stutzeri, respectively). Protein concentrations were measured by the bicinchoninic acid method (31) with bovine serum albumin as the standard. Enzymatic conversions of salicylate to catechol by NahG and NahW were followed by high-performance liquid chromatography (HPLC) and changes in absorption spectra. E. coli JM109 (pRAF111) or E. coli JM109 (pRAF127) cells were washed twice in TB, and suspended in TB (A600, 1.0) supplemented with 100 μM salicylate. Cells were incubated at 30°C for 2 h. Aliquots were removed for measurement every 15 min. UV-visible spectra of cell supernatants were recorded with a Pharmacia Ultrospec III spectrophotometer. Reverse-phase HPLC was performed on a Beckman model 125 chromatograph equipped with a Beckman model 166 diode array UV-visible detector using a Ultrasphere C8 column (5 μm; 125 by 4.6 mm) (Beckman, Fullerton, Calif.), with methanol:water:H3PO4 (45:55:0.1) as the mobile phase, with a flow rate of 2.5 ml · min−1. The injection volume was 10 μl of cell supernatant. Commercial salicylic acid (absorbance peak at 296 nm; retention volume, 8.25 ml) and catechol (absorbance peak at 275 nm; retention volume, 1.00 ml) were used as standards.
Resolution of salicylate hydroxylases by fast protein liquid chromatography.
Cell extract (0.5 ml, 2.1 mg of protein) was applied to a MonoQ HR 10/10 ion exchange column. Elution was performed at a flow rate of 0.4 ml · min−1, and two linear gradients of increasing NaCl concentrations were used: 0 to 0.2 M over 5 min; 0.2 M to 0.35 M over 20 min. Fractions (0.5 ml) were collected, and salicylate hydroxylase activity was assayed as described above.
Gene probes.
Specific DNA probes for the respective salicylate hydroxylase genes were prepared from plasmids (indicated in Table 1) by PCR amplification, followed by restriction digestion and extraction of the linear fragment: nahG, a 1,322-bp fragment obtained from plasmid pRAF104.2 by PCR amplification using 5′-ATGAACGACATGAACGCT-3′ and 5′-ACGGCCTCTTACCCTTGA-3′ as primers; nahW, a 657-bp fragment obtained after SalI restriction of the 1,087-bp DNA fragment generated by PCR amplification from pPA50C using 5′-ATGCGCCACCACGGTATC-3′ and 5′-CAATCGAGGTGATGCACC-3′ as primers.
Nucleotide sequence determination and sequence analysis.
The nucleotide sequence of the salicylate hydroxylase nahW gene and its flanking regions was determined directly from plasmid pPA50C by using standard protocols of the manufacturers for Taq DNA polymerase-initiated cycle sequencing reactions with fluorescent-labelled dideoxynucleotide terminators and a 373A automated DNA sequencer (Perkin-Elmer, Applied Biosystems Inc.). Sequences were extended by using a primer walking strategy (27), designing new primers based on determined sequences. Analyses of sequence data were carried out by using the DNA-Strider 1.2 program (CEA, Cedex, France), the GeneWorks program (IntelliGenetics, Montana View, Calif.), and the Genetics Computer Group sequence analysis package (GCG Inc., Madison, Wis.) (7).
Nucleotide sequence accession number.
The nucleotide sequence reported in this study has been submitted to the GenBank/EMBL data bank (accession no. AF039534).
RESULTS
Nucleotide sequence of the nahW gene.
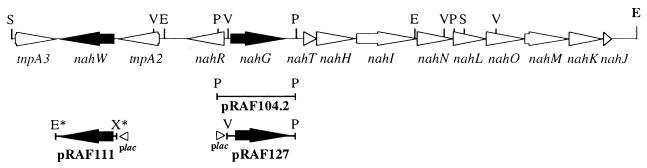
The nucleotide sequences (13,507 bp) for the entire naphthalene degradation lower pathway and for the regulatory nahR gene have been determined previously (4), showing an organization analogous to that found in other well-characterized naphthalene-degrading bacteria (Fig. 1) (6, 15, 23, 52). A tnpA-like gene (tnpA2), whose putative gene product, TnpA2, possesses 53.8% amino acid identity to transposase TnpA of the bacteriophage lambda KH100 is5 element (19), was detected downstream from the nahR gene (Fig. 1) (4).
FIG. 1.
The genetic organization (physical and genetic maps) of the chromosomal naphthalene degradation lower pathway and the nahW gene of P. stutzeri AN10. The map of DNA fragments that have been subcloned and relevant restriction endonuclease sites are shown. E, EcoRI; P, PstI; S, SacI; V, EcoRV. E∗ and X∗ denote EcoRI and XhoI restriction sites, respectively, that were generated by PCR. Arrows indicate the directions of gene transcription. The genes and their gene products are as follows: nahG and nahW, salicylate hydroxylases; nahH, catechol 2,3-dioxygenase; nahI, hydroxymuconic semialdehyde dehydrogenase; nahJ, 4-oxalocrotonate isomerase; nahK, 4-oxalocrotonate decarboxylase; nahL, 2-oxopent-4-enoate hydratase; nahM, 2-oxo-4-hydroxypentanoate aldolase; nahN, hydroxymuconic semialdehyde hydrolase; nahO, acetaldehyde dehydrogenase; nahR, NahR regulatory protein; nahT, chloroplast ferredoxin-like protein.
Nucleotide sequence analyses of the tnpA2 downstream region revealed the presence of two new open reading frames (ORF) (Fig. 1). The ORF most distant from tnpA2, tnpA3, encodes a putative protein of 255 amino acids (molecular mass, 29,165 Da) that possesses 96% identity to the amino-terminal half (171 amino acids) of a putative ORF1 from the phenol-catabolic plasmid pEST1226 (18), and 97% identity to the first 174 amino-terminal residues of the putative TnpA1 and TnpA2 from P. stutzeri AN10 (4).
Between tnpA2 and tnpA3 another ORF, nahW, was detected. Its putative gene product, NahW (43,562 Da), possesses 41% identity to the putative salicylate hydroxylase (NahG) of Sphingomonas sp. AJ1 (GenBank accession no. AB000564), and 23 to 25% identity to other salicylate hydroxylases (20, 34, 53). Pairwise alignment (Fig. 2) indicated that P. stutzeri AN10 could possess two genes coding for salicylate hydroxylases, nahG and nahW. Electrophoresis of restriction enzyme-digested genomic DNA, followed by Southern blot hybridization, was used to investigate the presence of nahG and nahW homologs in different naphthalene-degrading P. stutzeri genomovars (25). All analyzed naphthalene-degrading strains of P. stutzeri were observed to possess both nahG and nahW (data not shown).
FIG. 2.
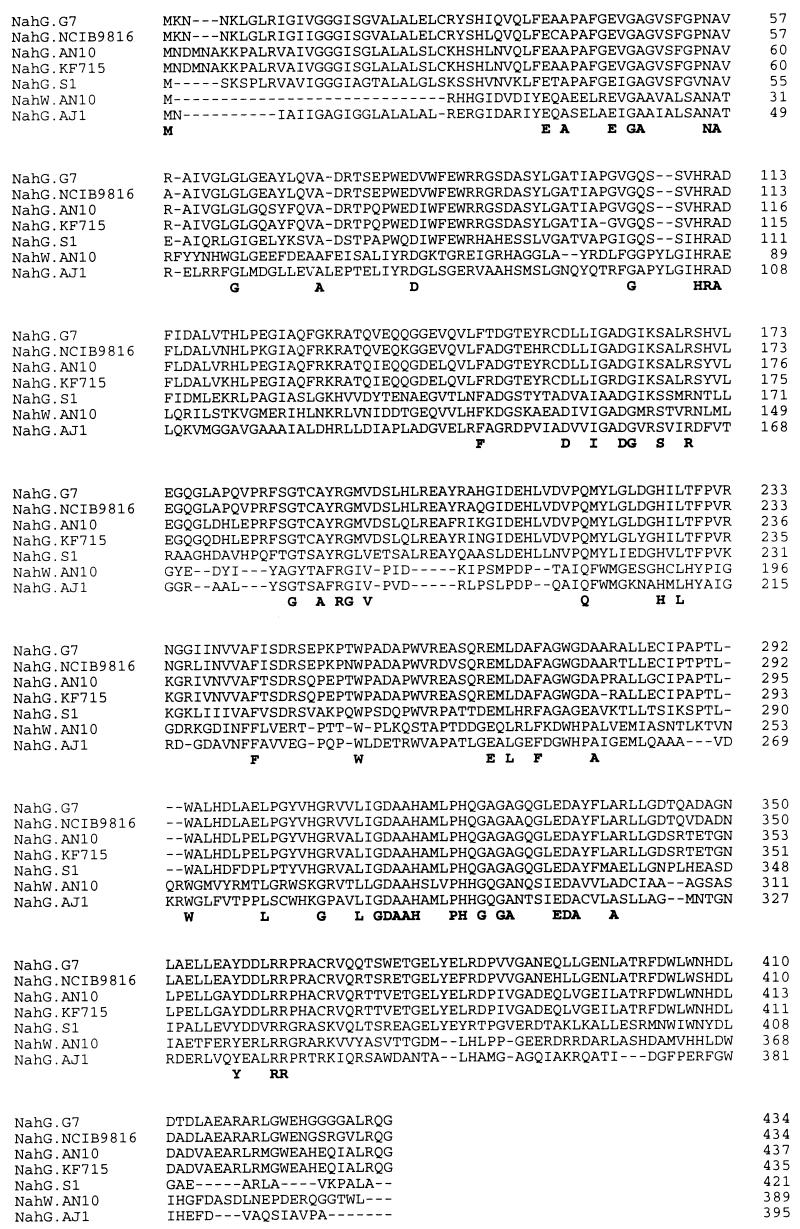
Amino acid sequence alignment of the P. stutzeri AN10 NahW protein and six different salicylate hydroxylase sequences available from the EMBL/GenBank database. The genetic sources are as follows: P. stutzeri AN10 (4), Sphingomonas sp. strain AJ1 (EMBL/GenBank accession no. AB000564), P. putida KF715 (20), P. putida G7 (53), P. putida NCIB9816 (EMBL/GenBank accession no. X83926), P. putida S1 (34). Dashes (–) indicate gaps that were introduced to optimize the alignments. Residues conserved in all sequences are shown in boldface below the alignment.
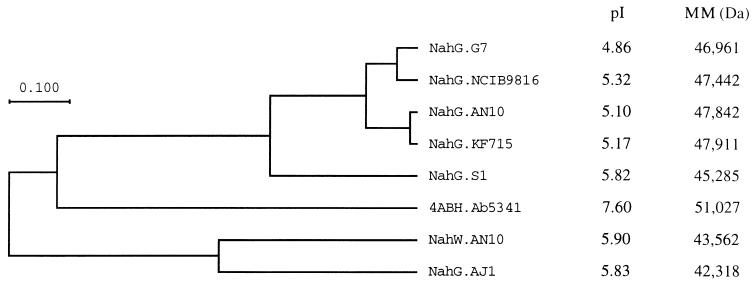
The putative NahG of P. stutzeri AN10 belongs to the same group of salicylate hydroxylases as the well-characterized NahG of P. putida G7 (53), while the putative NahW from P. stutzeri AN10 does not seem to belong to this major family of salicylate hydroxylases (Fig. 3). Interestingly, the well-conserved amino terminal FAD-binding site (GxGxxG) (9, 48), present in all salicylate hydroxylases described previously, is missing in the putative second salicylate hydroxylase (NahW) of this strain (Fig. 2).
FIG. 3.
UPGMA (unweighted-pair group method using arithmetic averages) dendrogram of similarities between bacterial salicylate hydroxylases. Genetic sources are given in the legend for Fig. 2. The amino acid sequence of the 4-aminobenzoate hydroxylase (4ABH) of Agaricus bisporus Ab5341 (41) was used as an outgroup. The bar indicates 10% estimated sequence divergence. Isoelectric points (pI) and molecular masses (MM) are based on the sequence data.
The nahW gene of P. stutzeri AN10 possesses 58.1% total G+C content, similar to the 59.5% G+C content of nahG and the 63.4% G+C content detected experimentally in P. stutzeri AN10 (24). However, when the third base usage was analyzed separately, the nahW and nahG genes of P. stutzeri AN10 demonstrated only 58.6 and 61.6% G or C preference, respectively, values that are quite different from the observed 80% overall preference for G or C in P. stutzeri (60.1% total G+C content) genes listed in databases. Despite this similarity between nahG and nahW of P. stutzeri AN10, different codon usage patterns were observed. While P. stutzeri genes in the databases and the nahW gene of P. stutzeri AN10 show strong G or C preferences in the third base of the codons for Ile (ATT, ATC, and ATA), His (CAT and CAC), and Gln (CAA and CAG), the nahG gene shows a preference for the A- or T-ended codons. In contrast, the nahW gene of P. stutzeri AN10 shows an A or T preference in the third base of codons for Phe (TTT and TTC), Asn (AAT and AAC), and Tyr (TAT and TAC), whereas P. stutzeri genes in databases and the nahG gene of P. stutzeri AN10 show a strong G or C preference in their third bases.
NahG and NahW: the two inducible salicylate hydroxylases of P. stutzeri AN10.
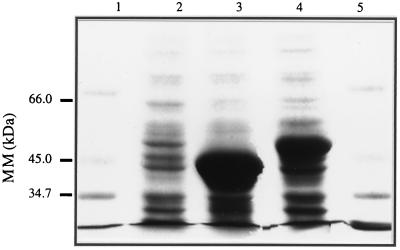
Both nahG and nahW were cloned separately under the transcriptional regulation of the Plac promoter (plasmids pRAF127 and pRAF111, respectively; Fig. 1) to test whether they encode for functional salicylate hydroxylases. Overexpression of nahG and nahW in E. coli JM109 yielded peptides with apparent molecular masses of 46 and 43 kDa (Fig. 4), respectively, similar to the predicted molecular masses of these peptides (Fig. 3). Both, E. coli JM109 (pRAF111) and E. coli JM109 (pRAF127), transformed salicylate to a product with spectral properties identical to those of catechol (data not shown). The identities of the reaction products with catechol were confirmed by HPLC analysis. Thus, both NahG and NahW were functional salicylate 1-hydroxylases.
FIG. 4.
Expression of the nahW and nahG gene products of P. stutzeri AN10 by recombinant E. coli strains using Plac induction. Coomassie-blue staining of a sodium dodecyl sulfate-polyacrylamide gel of IPTG-induced cell extracts of E. coli JM109 harboring plasmids pBluescript SK without insert (lane 2), pRAF111 (NahW; lane 3), or pRAF127 (NahG; lane 4). Lanes 1 and 5 show molecular mass (MM) markers.
Both NahG and NahW exhibited broad substrate specificities and metabolized salicylate, methylsalicylates, and chlorosalicylates (Table 2). However, the relative rates by which the substituted analogs were transformed differed considerably (Table 2). Whereas 3-methylsalicylate was transformed by NahG at a rate similar to that of salicylate, it was a poor substrate for NahW. Similarly, 5-methylsalicylate and 4,5-dimethylsalicylate were transformed at high rates by NahG, although at only approximately 10% of the rate observed for the transformation of salicylate by NahW. Whereas NahG seems to be a very efficient catalyst for the transformation of methylsalicylates, NahW appeared to be more efficient than NahG in the transformation of chlorosalicylates.
TABLE 2.
Salicylate hydroxylase activitya
| Substrate | Strains and produced proteins
|
||
|---|---|---|---|
| E. coli JM109 (pRAF127) NahG | E coli JM109 (pRAF111) NahW | PsAN10 NahG + NahW | |
| Salicylate | 1,961 ± 57 (100) | 1,460 ± 52 (100) | 18.0 ± 1.1 (100) |
| 3-Methylsalicylate | 1,612 ± 65 (82) | 6 ± 1 (<1) | 6.3 ± 0.2 (35) |
| 4-Methylsalicylate | 4,389 ± 201 (224) | 2,551 ± 107 (175) | 30.1 ± 1.3 (167) |
| 5-Methylsalicylate | 1,933 ± 54 (98) | 232 ± 8 (15) | 8.6 ± 0.2 (48) |
| 3-Chlorosalicylate | 20 ± 2 (1) | 118 ± 6 (8) | 2.0 ± 0.2 (11) |
| 4-Chlorosalicylate | 1,220 ± 35 (62) | 1,234 ± 19 (84) | 8.9 ± 0.3 (50) |
| 5-Chlorosalicylate | 130 ± 5 (6) | 192 ± 8 (13) | 1.8 ± 0.1 (10) |
| 4,5-Dimethylsalicylate | 2,503 ± 124 (127) | 121 ± 5 (8) | 6.4 ± 0.2 (35) |
| 3,5-Dichlorosalicylate | 131 ± 4 (6) | 90 ± 4 (6) | 1.1 ± 0.1 (6) |
Salicylate hydroxylase activities were obtained with extracts of induced cells. Activities, reported as averages ± standard deviations, are measured in nanomoles of NADH · min−1 · mg−1 of protein. Numbers in parenthesis are relative activities (%).
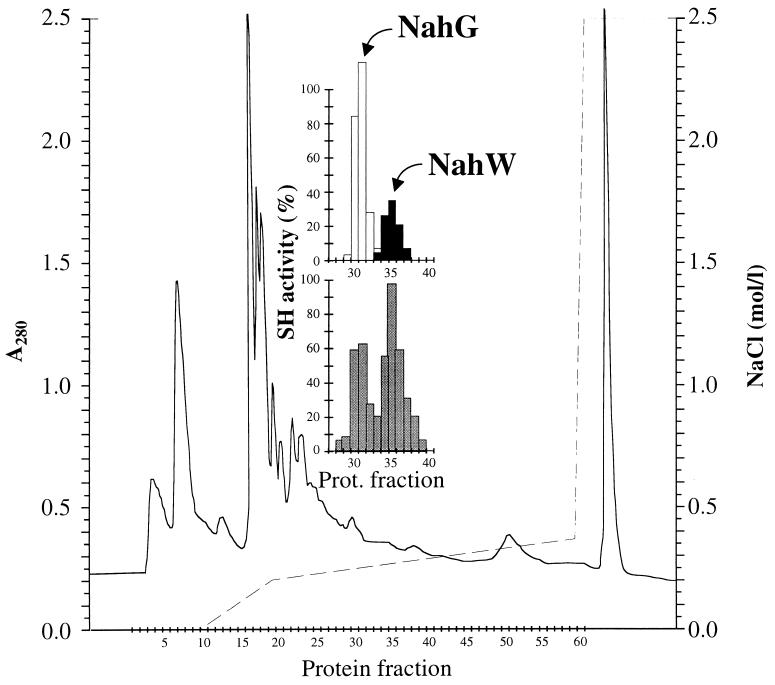
The transformation of salicylate and substituted salicylates by succinate-grown cells and cells incubated with the putative inducer salicylate was compared to analyze if both salicylate hydroxylases were expressed in the wild-type strain and if expression was inducible. Activities were detected only in extracts prepared from cells induced with salicylate. The relative rates for the transformations of methyl- and chlorosalicylates were between the rates observed for the individual enzymes prepared from E. coli cells (Table 2). Actually, two peaks of activity could be separated by ion-exchange chromatography of a cell extract from salicylate-induced P. stutzeri AN10 cells (Fig. 5). Whereas the protein eluting at 0.24 M NaCl, like NahG, showed high levels of activity with several methylsalicylates, such as 3-methylsalicylate, and a low level of activity with 3-chlorosalicylate (Fig. 5), the protein eluting at 0.26 M NaCl, like NahW, showed activity with 3-chlorosalicylate but not with 3-methylsalicylate (Fig. 5). Both peaks were also separated from cell extracts of naphthalene- and salicylate-grown P. stutzeri AN10 cells. Thus, both NahG and NahW appear to be induced by salicylate, and they seem to be involved in the metabolization of naphthalene and salicylate in P. stutzeri AN10.
FIG. 5.
Anion-exchange chromatography of extracts of salicylate-induced P. stutzeri AN10 cells. Protein was monitored as A280 (solid line). Gradient elution with NaCl (broken line) was used. (Inset) Salicylate hydroxylase activity was determined as NADH oxidation, as described in Materials and Methods, using salicylate (grey bars), 3-methylsalicylate (white bars), and 3-chlorosalicylate (black bars) as substrates.
DISCUSSION
In this study, we have demonstrated that conversion of salicylate to catechol in P. stutzeri AN10 is mediated by two isofunctional, induced, chromosomally encoded enzymes, NahG and NahW. NahG is encoded in the same transcriptional unit that contains the genes for the meta-cleavage pathway enzymes (4), forming the naphthalene degradation lower pathway, similar to the genetic organization of the well-characterized naphthalene degradation pathways encoded on plasmid NAH7 from P. putida G7 (15) and plasmid pWW60 from P. putida NCIB9816 (6). The structural gene encoding NahW is situated outside but in close proximity to (less than 3 kb) this transcriptional unit (Fig. 1). Thus, P. stutzeri AN10 represents the first example of a bacterium possessing two isofunctional salicylate hydroxylases. The genetic evidence (i.e., Southern blot analysis) demonstrated that both genes are present in all P. stutzeri naphthalene-degrading strains analyzed. Experiments to confirm that both genes are ubiquitous, being present in all known naphthalene-degrading strains, are now under way.
Both NahG and NahW exhibit a broad range of substrate specificity and metabolize salicylate, methylsalicylates, and chlorosalicylates (Table 2), as has been reported for the salicylate hydroxylase encoded by the nahG gene of NAH7 plasmid (21, 22). However, 3-chlorosalicylate was better converted by NahW, whereas NahG of P. stutzeri AN10 was more efficient metabolizing methylsalicylates. Relative consumption rates of salicylate and its derivatives by P. stutzeri AN10 were between the values obtained for each individual salicylate hydroxylase overproduced in E. coli. Thus, the consumption rates of salicylate in P. stutzeri AN10 appear to be due to the expression of both salicylate hydroxylases, NahG and NahW, being similar to the values obtained for NahG of plasmid NAH7 (21, 22). However, NahG of P. stutzeri AN10 converts 3-methylsalicylate at high rates (nearly 100% of the rate observed for salicylate) while NahG of P. putida G7 catalyzes its conversion at only 20% of the rate observed for salicylate (21).
The nahW and nahG genes of P. stutzeri AN10 are both induced and expressed with similar rates upon incubation with salicylate (Fig. 5). Additionally, a copy of a gene encoding the NahR-type protein, the putative LysR-type transcriptional activator of the entire naphthalene degradation pathway (28), was located between the nahG and nahW genes (Fig. 1) in P. stutzeri AN10 (4). Thus, it can be suggested that the regulation of both genes (nahG and nahW) is under the control of a LysR-type regulator, the nahR gene product, which probably activates their expression in the presence of the inducer salicylate. However, the ratio of the expression of both enzymes varies when different substituted salicylates are added as inducers (data not shown). Experiments to identify promoter regions and to clarify the regulation of these two transcriptional units of P. stutzeri AN10 are now under way.
Since both genes, nahG and nahW, in P. stutzeri AN10 encode physiologically active enzymes, one can speculate that the presence of two salicylate hydroxylases is advantageous to the host. It has been suggested that standard gene regulatory mechanisms allow cells to adjust their metabolisms to the range of conditions encountered most frequently. When extreme conditions cannot be accommodated by gene regulatory mechanisms, selection is imposed for increasing the copy number of a gene or set of linked genes that can improve growth (26). Thus, the variation in copy number (i.e., the presence of nahG and nahW) could provide an increased expression of the encoded enzymes (i.e., salicylate hydroxylases). Giving support to this plausible advantage of possessing two salicylate hydroxylases is the fact that P. putida PpG1064, carrying the nahG gene on a multicopy plasmid (high expression of NahG), demonstrated rates of salicylate degradation and growth rates higher than those in wild-type NAH-carrying P. putida PpG1064 (low expression of NahG) (11). In addition to this plausible physiological advantage, an evolutionary advantage of having two genes for salicylate hydroxylases can be suggested. Gene amplification as a stress response provides a plausible mechanism whereby bacteria might appear to be able to direct mutability to base pairs whose alteration improves growth (5, 26). In this sense, the presence of two salicylate hydroxylase-encoding genes (nahG and nahW) would provide a dispensable gene copy which permits the new activity of one of them to be enhanced (i.e., transformation of chlorosalicylates). Thus, a gene with a new function is formed (nahW) under continuous selection. In fact, divergent evolution of NahG and NahW of P. stutzeri AN10 from a common ancestor can be suggested, because three of the four criteria considered for assuming common ancestries for two proteins are met: NahG and NahW catalyze similar reactions, both proteins have similar subunit molecular masses, and their amino acid sequences are aligned without the introduction of multiple gaps (16). In any case, experiments are necessary to clarify plausible advantages for an organism possessing two salicylate hydroxylases.
According to the molecular masses, both salicylate hydroxylases of P. stutzeri AN10, NahG and NahW, belong, like NahG of P. putida G7 (53) and 4-hydroxybenzoate hydroxylase of P. fluorescens (45), to the subgroup of low-molecular-mass flavin-containing monooxygenases, which are approximately 45 kDa in size (14). All flavin-containing monooxygenases possess approximately 20% overall amino acid identity, with the strongest sequence conservation being in and adjacent to the flavin-binding regions. Structural comparisons between amino acid residues of resolved 4-hydroxybenzoate hydroxylase of P. fluorescens (45) and previously sequenced salicylate hydroxylases (20, 34, 53) indicate conserved residues which are important for the function of salicylate hydroxylases. The alignment of the putative primary amino acid sequence of the NahW protein of P. stutzeri AN10 with other salicylate hydroxylases (Fig. 2) could allow a better evaluation of the functional significance of conserved residues. Amino acid residues 152-TADVAIAADGIKSMR-167 have been designated the putative NADH-binding site in P. putida S1 salicylate hydroxylase, being Lys163 and Arg167, which are suggested to be involved in the formation of salt bridges with the oxygen atoms of pyrophosphate of NADH (33–35). Both amino acid residues are conserved in all salicylate hydroxylases, but Lys163 is substituted by another basic amino acid (Arg) in NahW (Fig. 2). Thus, we can assume the consensus NADH-binding domain of salicylate hydroxylases to be DXXIXXDGX[K,R]SXXR.
FAD has been described as the prosthetic group of salicylate hydroxylase (44). Interestingly, of the two salicylate hydroxylases of P. stutzeri AN10, only NahG, but not NahW, contains the well-conserved amino-terminal FAD-binding site (Fig. 2). In contrast, the putative second FAD-binding site of NahG of P. putida G7 (53), including two hydrophobic residues (positions 311 and 312 of P. putida G7 NahG sequence) and Gly and Asp in positions 313 and 314 of the same protein, respectively, seems to be conserved in all salicylate hydroxylases. The presence of this conserved domain suggests that both salicylate hydroxylases of P. stutzeri AN10, as the other known isofunctional enzymes, are flavin-dependent enzymes. Further preliminary biochemical data recently obtained in our laboratory give support to this hypothesis (data not shown).
The substrate catalytic active site of salicylate hydroxylases still remains to be elucidated. Following the putative second FAD-binding site, a cluster of 13 highly conserved amino acid residues was observed possessing the consensus sequence AAH[A,S][M,L][L,V]PH[Q,H]G[Q,A]GA (Fig. 2). Residues 319-MLPHQGA-325 of NahG of P. putida G7 (53) are similar to those in one of the two regions of 4-hydroxybenzoate hydroxylase shown to be involved in positioning the substrate 4-hydroxybenzoate properly with respect to the carboxyl group of the substrate and the isoalloxazine ring of FAD (45). The second region mentioned above corresponds to residues 48-AGV-50 of NahG of P. putida G7 (53). Ala48 and Val50 are conserved in all salicylate hydroxylases except for NahG of Sphingomonas sp. strain AJ1 (GenBank accession no. AB000564). Thus, these two conserved domains could be proposed to be the putative substrate active site of salicylate hydroxylases.
However, detailed biochemical and genetic studies of putative catalytically and structurally important amino acid residues and the purification and crystallography of the enzymes will be needed to clarify the exact enzymatic mechanism of salicylate hydroxylation.
ACKNOWLEDGMENTS
We are grateful to J. Lalucat for critical reading of the manuscript; J. Armengaud for advice on fast protein liquid chromatography procedures; and A. Krüger, A. Peterseim, and C. Strömpl for technical support with the nucleic acid sequencing. R.B. thanks K. N. Timmis for his encouragement and inspiration.
This work was supported by grants AMB94-1038 and BIO97-0639 (Spanish CICYT) and by grant 0319-433B (German Ministry of Education and Research). R.B. was the recipient of a short-term fellowship from the European Environmental Research Organization (EERO) for travel and work at the GBF. Part of this work was carried out in the framework of the European Community Human Capital and Mobility Network grant CHRX CT93-0194.
REFERENCES
- 1.Amengual J F. Metabolismo de derivados halogenados y metilados del naftaleno por Pseudomonas stutzeri AN10. Ph.D. thesis. Palma de Mallorca, Spain: Universitat de les Illes Balears; 1992. [Google Scholar]
- 2.Aragno M, Schlegel H G. The hydrogen-oxidizing bacteria. In: Starr M P, et al., editors. The prokaryotes. A handbook on habitats, isolation, and identification of bacteria. Berlin and Heildelberg, Germany: Springer-Verlag; 1981. pp. 865–893. [Google Scholar]
- 3.Barnsley E A. The induction of enzymes of naphthalene metabolism in pseudomonads by salicylate and 2-aminobenzoate. J Gen Microbiol. 1975;88:193–196. doi: 10.1099/00221287-88-1-193. [DOI] [PubMed] [Google Scholar]
- 4.Bosch, R., E. García-Valdés, and E. R. B. Moore. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation pathway from Pseudomonas stutzeri AN10. Submitted for publication. [DOI] [PubMed]
- 5.Cairns J, Overbaugh J, Miller S. The origin of mutants. Nature. 1988;335:142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- 6.Cane P A, Williams P A. A restriction map of naphthalene catabolic plasmid pWW60-1 and the location of some of its catabolic genes. J Gen Microbiol. 1986;132:2919–2929. [Google Scholar]
- 7.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhaese P, De Greve H, Decraemer H, Schell J, Van Mongatu M. Rapid mapping of transposon insertion and deletion mutations in the large Ti-plasmids of Agrobacterium tumefaciens. Nucleic Acids Res. 1979;7:1837–1849. doi: 10.1093/nar/7.7.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eggink G, Engel H, Vriend G, Terpstra P, Witholt B. Rubredoxin reductase of Pseudomonas oleovorans. Structural relationship to other flavoprotein oxidoreductases based on one NAD and two FAD fingerprints. J Mol Biol. 1990;212:135–142. doi: 10.1016/0022-2836(90)90310-I. [DOI] [PubMed] [Google Scholar]
- 10.Einarsdottir G H, Stankovich M T, Tu S-C. Studies of electron transfer properties of salicylate hydroxylase from Pseudomonas cepacia and effects of salicylate and benzoate binding. Biochemistry. 1988;27:3277–3285. doi: 10.1021/bi00409a023. [DOI] [PubMed] [Google Scholar]
- 11.Fujita M, Hashimoto S, Takeo M, Hagino K. Plasmid-coded degradation of salicylate using the catechol cleavage pathway of the host. J Ferment Bioeng. 1989;67:286–290. [Google Scholar]
- 12.Ginard M, Römling U, Tümmler B, Lalucat J. Abstracts of 1996 Biodegradation of Organic Pollutants Symposium. Palma de Mallorca, Spain. 1996. Naphthalene degradation is chromosomally encoded in Pseudomonas stutzeri; p. 183. [Google Scholar]
- 13.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 14.Harayama S, Kok M, Neidle E L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 15.Harayama S, Rekik M, Wasserfallen A, Bairoch A. Evolutionary relationships between catabolic pathways for aromatics: conservation of gene order and nucleotide sequences of catechol oxidation genes of pWW0 and NAH7 plasmids. Mol Gen Genet. 1987;210:241–247. doi: 10.1007/BF00325689. [DOI] [PubMed] [Google Scholar]
- 16.Harayama S, Timmis K N. Aerobic biodegradation of aromatic hydrocarbons by bacteria. In: Sigel H, Sigel A, editors. Degradation of environmental pollutants by microorganisms and their metalloenzymes. New York, N.Y: Marcel Dekker Inc.; 1992. pp. 99–156. [Google Scholar]
- 17.Harlow E, Lane E. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 18.Kasak L, Horak R, Nurk A, Talvik K, Kivisaar M. Regulation of the catechol 1,2-dioxygenase- and phenol monooxygenase-encoding pheBA operon in Pseudomonas putida PaW85. J Bacteriol. 1993;175:8038–8042. doi: 10.1128/jb.175.24.8038-8042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kröger M, Hobom G. Structural analysis of insertion sequence IS5. Nature. 1982;297:159–162. doi: 10.1038/297159a0. [DOI] [PubMed] [Google Scholar]
- 20.Lee J, Oh J, Min K R, Kim Y. Nucleotide sequence of salicylate hydroxylase gene and its 5′-flanking region of Pseudomonas putida KF715. Biochem Biophys Res Commun. 1996;218:544–548. doi: 10.1006/bbrc.1996.0097. [DOI] [PubMed] [Google Scholar]
- 21.Lehrbach P R, Zeyer J, Reineke W, Knackmuss H-J, Timmis K N. Enzyme recruitment in vitro: use of cloned genes to extend the range of haloaromatics degraded by Pseudomonas sp. strain B13. J Bacteriol. 1984;158:1025–1032. doi: 10.1128/jb.158.3.1025-1032.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris C M, Barnsley E A. The cometabolism of 1- and 2-chloronaphthalene by pseudomonads. Can J Microbiol. 1982;28:73–79. [Google Scholar]
- 23.Platt A, Shingler V, Taylor S C, Williams P A. The 4-hydroxy-2-oxovalerate aldolase and acetaldehyde dehydrogenase (acylating) encoded by the nahM and nahO genes of the naphthalene catabolic plasmid pWW60-22 provide further evidence of conservation of meta-cleavage pathway gene sequences. Microbiology. 1995;141:2223–2233. doi: 10.1099/13500872-141-9-2223. [DOI] [PubMed] [Google Scholar]
- 24.Rossello R, García-Valdés E, Lalucat J, Ursing J. Genotypic and phenotypic diversity of Pseudomonas stutzeri. Syst Appl Microbiol. 1991;14:150–157. [Google Scholar]
- 25.Rosselló-Mora R A, Lalucat J, García-Valdés E. Comparative biochemical and genetic analysis of naphthalene degradation among Pseudomonas stutzeri strains. Appl Environ Microbiol. 1994;60:966–972. doi: 10.1128/aem.60.3.966-972.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roth J R, Benson N, Galitski T, Haack K, Lawrence J G, Miesel L. Rearrangements of the bacterial chromosome: formation and applications. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2256–2276. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schell M A. Transcriptional control of the nah and sal hydrocarbon-degradation operons by the nahR gene product. Gene. 1985;36:301–309. doi: 10.1016/0378-1119(85)90185-4. [DOI] [PubMed] [Google Scholar]
- 29.Schell M A, Wender P E. Identification of the nahR gene product and nucleotide sequences required for its activation of the sal operon. J Bacteriol. 1986;166:9–14. doi: 10.1128/jb.166.1.9-14.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon M J, Osslund T D, Saunders R, Ensley B D, Suggs S, Harcourt A, Suen W, Cruden D L, Gibson D T, Zylstra G J. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB9816-4. Gene. 1993;127:31–37. doi: 10.1016/0378-1119(93)90613-8. [DOI] [PubMed] [Google Scholar]
- 31.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki K, Katagiri M. Mechanism of salicylate hydroxylase-catalyzed decarboxylation. Biochim Biophys Acta. 1981;657:530–534. doi: 10.1016/0005-2744(81)90337-5. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki K, Mizuguchi M, Gomi T, Itagaki E. Identification of a lysine residue in the NADH-binding site of salicylate hydroxylase from Pseudomonas putida S-1. J Biochem. 1995;117:579–585. doi: 10.1093/oxfordjournals.jbchem.a124747. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki K, Mizuguchi M, Ohnishi K, Itagaki E. Structure of chromosomal DNA coding for Pseudomonas putida S-1 salicylate hydroxylase. Biochim Biophys Acta. 1996;1275:154–156. doi: 10.1016/0005-2728(96)00069-2. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki K, Ohnishi K. Functional modification of an arginine residue on salicylate hydroxylase. Biochim Biophys Acta. 1990;1040:327–336. doi: 10.1016/0167-4838(90)90130-8. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki K, Takemori S, Katagiri M. Mechanism of the salicylate hydroxylase reaction. IV. Fluorometric analysis of the complex formation. Biochim Biophys Acta. 1969;191:77–85. doi: 10.1016/0005-2744(69)90316-7. [DOI] [PubMed] [Google Scholar]
- 37.Takemori S, Hon-nami K, Kawahara F, Katahiri M. Mechanism of the salicylate hydroxylase reaction. VI. The monomeric nature of the enzyme. Biochim Biophys Acta. 1974;342:137–144. doi: 10.1016/0005-2795(74)90115-9. [DOI] [PubMed] [Google Scholar]
- 38.Takemori S, Nakamura M, Suzuki K, Katagiri M, Nakamura T. Mechanism of salicylate hydroxylase reaction. V. Kinetic analyses. Biochim Biophys Acta. 1972;284:382–393. doi: 10.1016/0005-2744(72)90134-9. [DOI] [PubMed] [Google Scholar]
- 39.Takemori S, Yasuda H, Mihara K, Suzuki K, Katagiri M. Mechanism of the salicylate hydroxylase reaction. II. The enzyme-substrate complex. Biochim Biophys Acta. 1969;191:58–68. doi: 10.1016/0005-2744(69)90314-3. [DOI] [PubMed] [Google Scholar]
- 40.Takemori S, Yasuda H, Mihara K, Suzuki K, Katagiri M. Mechanism of the salicylate hydroxylase reaction. III. Characterization and reactivity of chemically or photochemically reduced enzyme-flavin. Biochim Biophys Acta. 1969;191:69–76. doi: 10.1016/0005-2744(69)90315-5. [DOI] [PubMed] [Google Scholar]
- 41.Tsuji H, Oka T, Kimoto M, Hong Y M, Natori Y, Ogawa T. Cloning and sequencing of cDNA encoding 4-aminobenzoate hydroxylase from Agaricus bisporus. Biochim Biophys Acta. 1996;1309:31–36. doi: 10.1016/s0167-4781(96)00131-5. [DOI] [PubMed] [Google Scholar]
- 42.Tu S-C, Romero F A, Wang L-H. Uncoupling of the substrate monooxygenation and reduced pyridine nucleotide oxidation activities of salicylate hydroxylase by flavins. Arch Biochem Biophys. 1981;209:423–432. doi: 10.1016/0003-9861(81)90299-x. [DOI] [PubMed] [Google Scholar]
- 43.Wang L-H, Tu S-C. The kinetic mechanism of salicylate hydroxylase as studied by initial rate measurement, rapid reaction kinetics, and isotope effects. J Biol Chem. 1984;259:10682–10688. [PubMed] [Google Scholar]
- 44.Wang L-H, Tu S-C, Lusk R C. Apoenzyme of Pseudomonas cepacia salicylate hydroxylase. Preparation, fluorescence property, and nature of flavin binding. J Biol Chem. 1984;259:1136–1142. [PubMed] [Google Scholar]
- 45.Weijer W J, Hofsteenge J, Vereijken J M, Jekel P A, Beintema J J. Primary structure of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens. Biochim Biophys Acta. 1982;704:385–388. doi: 10.1016/0167-4838(82)90170-4. [DOI] [PubMed] [Google Scholar]
- 46.White-Stevens R H, Kamin H. Studies of a flavoprotein, salicylate hydroxylase. I. Preparation, properties, and the uncoupling of oxygen reduction from hydroxylation. J Biol Chem. 1972;247:2358–2370. [PubMed] [Google Scholar]
- 47.White-Stevens R H, Kamin H, Gibson Q H. Studies of a flavoprotein, salicylate hydroxylase. II. Enzyme mechanism. J Biol Chem. 1972;247:2371–2381. [PubMed] [Google Scholar]
- 48.Wierenga R K, Terpstra P, Hol W G J. Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 49.Williams P A, Sayers J R. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation. 1994;5:195–217. doi: 10.1007/BF00696460. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto S, Katagiri M, Maeno H, Hayaishi O. Salicylate hydroxylase, a monooxygenase requiring flavin adenine dinucleotide. I. Purification and general properties. J Biol Chem. 1965;240:3408–3413. [PubMed] [Google Scholar]
- 51.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 52.Yen K, Gunsalus I C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci USA. 1982;79:874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You I, Ghosal D, Gunsalus I C. Nucleotide sequence analysis of the Pseudomonas putida PpG7 salicylate hydroxylase gene (nahG) and its 3′-flanking region. Biochemistry. 1991;30:1635–1641. doi: 10.1021/bi00220a028. [DOI] [PubMed] [Google Scholar]
- 54.You I-S, Murray R I, Jollic D, Gunsalus I C. Purification and characterization of salicylate hydroxylase from Pseudomonas putida PpG7. Biochem Biophys Res Commun. 1990;169:1049–1054. doi: 10.1016/0006-291x(90)92000-p. [DOI] [PubMed] [Google Scholar]