Abstract
Purpose of Review
In this review, we summarize the biological roles of methionine, methionine adenosyl transferase 2A (MAT2A) and S-adenosyl methionine (SAM) in methylation reactions during tumorigenesis. Newly emerged inhibitors targeting the Methionine-MAT2A-SAM axis will be discussed.
Recent Findings
SAM is the critical and global methyl-donor for methylation reactions regulating gene expression, and in mammalian cells, it is synthesized by MAT2A using methionine. Recent studies have validated methionine and MAT2A as metabolic dependencies of cancer cells because of their essential roles in SAM biosynthesis. MAT2A inhibition leads to synthetic lethality in methylthioadenosine-phosphorylase (MTAP)-deleted cancers, which accounts for 15% of all cancer types. Of note, remarkable progress has been made in developing inhibitors targeting the Methionine-MAT2A-SAM axis, as the first-in-class MAT2A inhibitors AG-270 and IDE397 enter clinical trials to treat cancer.
Summary
The Methionine-MAT2A-SAM axis plays an important role in tumorigenesis by providing SAM as a critical substrate for abnormal protein as well as DNA and RNA methylation in cancer cells. Targeting SAM biosynthesis through MAT2A inhibition has emerged as a novel and promising strategy for cancer therapy.
Keywords: methionine, MAT2A, SAM, methylation, MTAP-deleted cancer
Introduction
Although all cells in an organism carry the identical genetic information, the fate of an individual cell is determined by its unique gene expression program [1]. Under certain environmental conditions, the expression of specific genes is activated in a spatial and temporal dependent manner to achieve the predetermined differentiation and development, and to maintain biological functions of different tissues and organs [2]. While several chemical modifications of biomolecules, including proteins and nucleic acids, are involved in regulation of gene expression, methylation processes have been most extensively studied [3*]. Dysregulated methylation patterns have been reported to be associated with tumorigenesis. For example, DNA hypermethylation of tumor suppressor genes frequently leads to gene silencing and promotes tumorigenesis [4, 5]. Targeting of dysregulated methylation has been a rising focus in the development of cancer therapy [6, 7]. There are two main strategies to target abnormal methylation in cancers: Firstly, targeting the methylation modulators, such as inhibition of RNA methyltransferase METTL3 to reduce leukemogenic gene expression in acute myeloid leukemia [8*]; secondly, targeting the “Methionine-MAT2A-SAM” biosynthesis axis to starve cancer cells of the methylation substrate SAM. The latter strategy is a newly emerged field and will be discussed here. In this article, we summarize the roles of methionine, MAT2A and SAM in methylation during tumorigenesis, and highlight the most recent progresses in the development of pharmaceutical inhibitors targeting this pathway for anticancer therapies.
Methionine-MAT2A-SAM Axis
SAM biosynthesis is catalyzed by methionine adenosyl transferase (MAT) from methionine and ATP. There are two MAT isozymes in mammals, MAT1 and MAT2. MAT1 is predominantly expressed in the liver where it regulates the differentiation of hepatocytes and bile duct epithelial cells. MAT2 is ubiquitously expressed in normal cells and cancer cells [9, 10]. MAT2 contains two subunits, MAT2A and MAT2B, serving as catalytic and regulatory subunits, respectively [11]. To maintain SAM homeostasis, the expression of MAT2A is elegantly regulated by the dwell-time of RNA methyltransferase METTL16 on the hairpin structure of the retained intron between exons 8 and 9 of the MAT2A RNA [12, 13]: When adequate SAM levels are present, METTL16 utilizes SAM to methylate the hairpin structure of MAT2A RNA in a brief time and then leaves intron-retained MAT2A RNA alone for degradation; when the SAM level is depleted, this methylation step is impaired due to insufficient enzymatic turnover, prolonging the occupancy of METTL16 on the hairpin structure, which then drives the splicing of MAT2A RNA to produce functional MAT2A protein for increased SAM biosynthesis. Additionally, a recent study has revealed that the mechanistic target of rapamycin complex 1 (mTORC1) can promote MAT2A expression through downstream c-MYC, which is a transcription factor that binds directly to intron 1 of MAT2A, thereby promoting its expression [14*].
SAM provides the aminopropyl group in aminopropylation to synthesize polyamines, while it is mainly consumed during methylation reactions to add methyl groups to the amino acid side chain of proteins, to nucleotide bases of DNA and RNA, and to many other acceptor molecules [15]. These processes are catalyzed by respective methyltransferases and are accompanied by the accumulation of the by-product S-adenosylhomocysteine, which is reversibly converted to homocysteine to synthesize cysteine through transsulfuration or to regenerate methionine (Figure. 1) [16].
Figure 1.
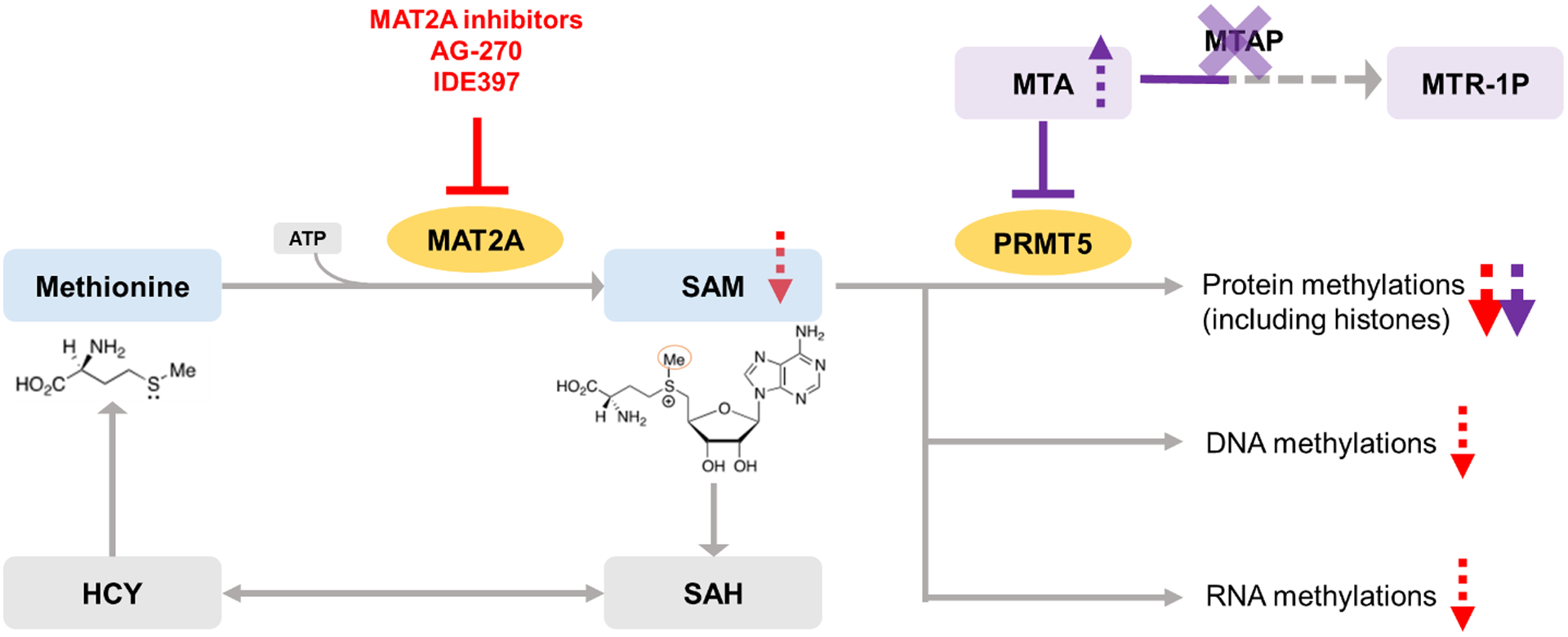
Targeting the Methionine-MAT2A-SAM axis in MTAP-deleted cancers. SAM is synthesized by MAT2A using methionine and ATP and consumed mainly for methylation reactions, in which, the by-product is S-adenosylhomocysteine (SAH). SAH is reversibly converted to homocysteine (HCY) contributing to methionine recycle. MTAP cleaves MTA into MTR-1P, and when MTAP is deleted, MTA accumulates and partially inhibits PRMT5. This partial inhibition results an increased dependence of PRMT5 on the concentration of its own substrate SAM. MAT2A inhibitors block the enzymatic activity of MAT2A, inhibit the nascent biosynthesis of SAM, and thereby suppress the activity of PRMT5 in MTAP-deleted cancer cells and lead to the synthetic lethality.
In the human genome, DNA that encodes more than 20,000 genes is precisely and tightly wound around histones into 23 pairs of chromosomes [17]. The density of compaction to regulate chromatin accessibility significantly relies on histone modifications, including phosphorylation, acetylation, crotonylation, ubiquitination, glycosylation, ADP ribosylation and principally methylation [18]. Histone methylation is catalyzed by histone methyltransferases (HMTs), which transfer one, two, or three methyl groups from SAM onto either lysine (K) or arginine (R) residues of histones H3 and H4. The sites and amounts of H3 and H4 methylation are of great significance for the regulation of gene expression. For example, while H3K9me3 mediates transcriptional repression, H3K4me3 mediates transcriptional activation [19, 20]. In addition to histones, many other proteins can be methylated. Protein methylation may affect protein-protein, protein-DNA or protein-RNA interactions, as well as protein stability, protein subcellular localization or enzymatic activity [21]. Moreover, in DNA methylation, DNA methyltransferases (DNMTs) recognize the 5’CG3’ sequence (CpG) of DNA and transfer the methyl group of SAM mainly to the C-5 position of cytosine (5mC), which regulates chromatin structure and DNA stability, thereby controlling gene expression [22]. Additionally, in RNA methylation, methyl groups from SAM can be added to RNA under the catalysis of RNA methyltransferases to mainly form N6-methyladenosine (m6A) [23]. m6A is closely related to human development, immunity, tumorigenesis and metastasis, stem cell renewal and differentiation [24]. In summary, SAM as the principal methyl-group donor in methylation reactions plays a critical role in many biological processes through regulating gene expression.
Methionine-MAT2A-SAM Axis in Tumorigenesis
Methionine
The essential amino acid methionine has been found critical for the maintenance of the epigenome of embryonic stem cells and induced pluripotent stem cells [25]. Depletion of methionine leads to decreased SAM levels, thereby impairing H3K4me3 histone methylation, which is conserved and transcriptionally regulates the signaling pathways involved in pluripotency [25]. This explains how the nutrient methionine can determine cell fates metabolically and epigenetically. Interestingly, an essential role of methionine has also been revealed in cancer stem cells (CSCs), contributing to tumorigenesis. Depleting methionine for a short period, but not other amino acids, dramatically inhibits the proliferation and tumorigenicity of CSCs in human non-small-cell lung cancer (NSCLC), accompanied with suppressed histone methylation levels [26]. These affects can be rescued by adding SAM but not other methionine metabolites, demonstrating the dependency of CSCs on methionine is attributed to the need for SAM biosynthesis [26]. Consistent findings on the role of methionine as a metabolic dependency were also discovered in human triple-negative breast cancer (TNBC) and leukemia [27, 28].
MAT2A
MAT2A is highly expressed in various human cancer types, including colon, hepatoma, and breast cancer, and higher MAT2A expression in cancer patients is usually correlated with a worse prognosis [27, 29, 30*]. Of note, compared to other cancer types, leukemias have the highest MAT2A expression and leukemias carrying the Mixed Lineage Leukemia gene rearrangements (MLL-r), a characteristic related to dismal prognosis, have the highest MAT2A expression levels [28]. Additionally, a recent study revealed that the higher ratio of cytoplasmic versus nuclear (C/N) MAT2A expression in breast cancer correlates with lower 5-year overall survival rates of patients (C/N<1: 93.7%; C/N≥1: 83.3%), and the increased C/N ratio in breast cancer cells is accompanied with increased invasiveness, suggesting the C/N expression pattern of MAT2A as a biomarker in breast cancer prognosis [30*]. Moreover, microRNA-203 has been found to suppress hepatocellular carcinoma partially through inhibiting MAT2A expression [31].
Multiple studies have validated MAT2A as a key anti-cancer target [26–28]. To target the methionine metabolism in NSCLC CSCs, researchers evaluated the tumorigenic role of key enzymes involved, and found that rather than other enzymes in the methionine metabolism pathway, MAT2A expression was found strongly correlated with cancer progression and metastases [26]. Additionally, inhibition of MAT2A eliminated histone methylations and almost completely inhibited the tumorigenicity of NSCLC CSCs while sparing normal cells, suggesting targeting of MAT2A can selectively target cancer cells through disrupting methionine metabolism [26].
Methionine-MAT2A-SAM Axis in Immune Dysfunction
CD8+ T cells mediate anti-tumor immunity, and often become dysfunctional when infiltrated in tumor microenvironment [32]. This dysfunction is associated with dysregulated epigenetic modifications, and the underlying mechanism has been uncovered in a recent study showing that cancer cells can outcompete T cells for methionine consumption and disrupt the normal epigenetic pattern in T cells [33**]. Methionine is transported into cells through the solute carrier family proteins (SLC) [34]. SLC7A5 is similarly expressed in T cells and cancer cells, while SLC43A2 is selectively overexpressed in cancer cells, providing cancer cells an advantage in capturing methionine over CD8+ T cells in the cancer microenvironment [33**]. This results in low methionine levels in T cells, followed by decreased SAM levels, impairment of H3K79me2 methylation and suppression of the signal transducer and activator of transcription 5 (STAT5) signaling pathway [33**]. Suppression of STAT5 in T cells leads to T cell dysfunction and T cell death. Additionally, when SLC43A2 was knocked out, the growth of cancer cell was inhibited [33**]. Consistently, another recent study also revealed that the dysregulation of methionine metabolism in hepatocellular carcinoma promotes the exhaustion of CD8+ T cell [35*]. Therefore, targeting cancer methionine metabolism could be a promising immunotherapeutic strategy.
Targeting the Methionine-MAT2A-SAM Axis in Cancer
Combining Depletion of Methionine and Inhibition of MAT2A
Due to the increased addiction of cancer cells to methionine for SAM biosynthesis, depleting methionine through either dietary restriction or methionine degrading enzymes could be a promising strategy to target cancer cells [36]. However, since MAT2A is commonly upregulated after methionine depletion, a combination of methionine depletion and MAT2A inhibition is often used to efficiently inhibit SAM biosynthesis in cancer cells. Multiple studies have shown that this combination erases histone methylations and selectively eliminates cancer cells [26–28]. Moreover, in a recent study using a sarcoma patient-derived xenograft mouse model, a triple drug combination existing of an oral-recombinant methioninase (o-rMETase), MAT2A inhibitor cycloleucine and DNA methylation inhibitor decitabine, caused significant suppression of tumor burden and induction of tumor necrosis [37**].
MAT2A Inhibition in MTAP-deleted Cancers
MTAP locates on chromosome 9p21, approximately 100 kb telomeric to the common tumor suppressor gene cyclin-dependent kinase inhibitory protein 2A (CDKN2A), and the co-deletion of MTAP with CDKN2A happens in up to 15% of all cancers [9]. MTAP-deleted cancer cells lack the ability to metabolize 5’ methylthioadenosine (MTA) into 5-methylthioribose-1-phosphate (MTR-1P), and the resulting accumulation of MTA partially inhibits Protein Arginine Methyltransferase (PRMT) 5 [9, 27, 28, 38]. PRMT family methylates a variety of proteins and plays an important role in biological processes such as gene expression, splicing and DNA damage repair [39]. The affinity of PRMT5 for MTA is much higher than its own substrate SAM [40], and the partial inhibition of PRMT5 caused by MTA accumulation in MTAP-deleted cancers makes the catalytic activity of PRMT5 more dependent on the level of SAM, which is synthesized by MAT2A. Recent studies have shown that inhibition of MAT2A results in selective antiproliferative effects in MTAP-deleted cancers (Figure. 1) [41**, 42**, 43**]. Inhibition of MAT2A effectively reduces SAM levels in cancer cells by blocking the nascent biosynthesis of SAM, while concurrently PRMT5, which uses SAM as a substrate in a strong concentration-dependent manner, is subsequently inhibited [42**]. It then impairs splicing processes, induces DNA damage, defects mitosis and ultimately leads to inhibition of cell proliferation [42**].
Because of the enhanced dependence of PRMT5 on MAT2A in MTAP-deleted cancers, pharmacological inhibition of MAT2A has become a promising strategy to induce synthetic cell death in MTAP-deleted cancers.
Development of MAT2A Inhibitors
In 1983, cycloleucine was the first reported compound showing inhibitory effect on MAT [44]. However, the binding affinity of cycloleucine to MAT2A is weak, and the IC50 is blunt (140–190 uM). Because cycloleucine participates in other signaling pathways and its functional mechanisms remain unclear, it wasn’t further developed as an anti-cancer drug. In 2013, a family of fluorinated N,N-dialkylaminostilbene (FIDAS) agents were developed as a highly active class of MAT2A inhibitors to target liver and colorectal cancers, in which MAT2A is abnormally overexpressed [45]. Though FIDAS inhibits the survival and growth of cancer cells at low nanomolar concentrations, their C=C structure may participate in redox reactions without the specificity to only inhibit MAT2A. In 2017, PF-9366 was discovered by Pfizer as a small molecule allosteric MAT2A inhibitor, which retains the amino groups from cycloleucine and FIDAS but with a new core design to enhance the specificity [46]. However, since PF-9366 works through binding to MAT2B, the regulatory unit of MAT2A, it consequently increases the expression of MAT2A in vivo and shows insufficient cellular potency. In 2018, MAT2A inhibitor AGI-24512 (enzymatic IC50 ~8 nM) was structured by Agios from screening a library of more than 2000 small molecules and by analyzing the crystal structure of MAT2A/SAM binding [47]. Compared to the effects of PF-9366 treatment, MAT2A is also upregulated in cancer cells after AGI-24512 treatment, but this increase does not affect the antiproliferative activity. However, AGI-24512 shows poor oral absorption and short half-life. After further optimization, AG-270 was discovered which showed enhanced oral availability and better pharmacokinetic behavior [41**, 42**]. AG-270 reduced intracellular SAM levels more efficiently when compared to cycloleucine, PF-9366 and AG-24512, and showed selective antiproliferative activity in MTAP-deleted cancer cells [42**]. Importantly, after oral treatment with AG-270, mice bearing human colon cancer xenografts showed significantly reduced SAM levels and diminished tumor burden. Recently, the MAT2A inhibitor AG-270 has been approved by the FDA for a clinical trial to treat patients with advanced solid tumors or lymphoma with MTAP loss (NCT03435250). IDE397 is another MAT2A inhibitor discovered by IDEAYA and approved by the FDA to enter clinical trials based on its significant single-agent anti-tumor activity in 45 MTAP-deleted xenograft models (NCT04794699). Moreover, benefiting from current advanced drug screening tools, new MAT2A inhibitors keep emerging from preclinical studies, such as newly discovered Compound 28, which also shows promising in vivo anticancer effects in an MTAP-deleted xenograft model [43**].
Conclusion
The Methionine-MAT2A-SAM pathway has emerged as a druggable target in cancer therapy due to its critical role in the regulation of gene expression through methylation. Recent studies revealed essential roles of this pathway in tumorigenesis, especially in MTAP-deleted cancers. The first-in-class MAT2A inhibitors AG-270 and IDE397 were recently enrolled in clinical trials based on their remarkable potencies in MTAP-deleted cancers, suggesting MAT2A inhibitors as promising novel anti-cancer therapeutics, especially for the treatment of a large number of patients with MTAP deletion. Future studies are expected to better utilize targeting this axis, such as investigating the application of MAT2A inhibitors in other cancer types and the combined effect of MAT2A inhibitors with other approved anti-cancer drugs.
Key points.
Methylation reactions play an important role in tumorigenesis through abnormal regulation of gene expression.
As the principal methyl-group donor in methylation reactions, SAM is biosynthesized by MAT2A from methionine and ATP.
Targeting SAM biosynthesis through MAT2A inhibition has emerged as a novel strategy for cancer therapy.
Financial support and sponsorship
This work was supported by National Institute of Health grants R01 CA201496 (National Cancer Institute), R01 GM133850 (National Institute of General Medical Sciences) and Held Foundation Fellowship (City of Hope).
Footnotes
Conflicts of interest
There are no conflicts of interest.
References and Recommended Reading
Papers of particular interest
* of special interest
** of outstanding interest
- 1.Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity. 2010;105(1):4–13. [DOI] [PubMed] [Google Scholar]
- 2.Chandrasekaran S, Zhang J, Sun Z, et al. Comprehensive Mapping of Pluripotent Stem Cell Metabolism Using Dynamic Genome-Scale Network Modeling. Cell Rep. 2017;21(10):2965–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3*.Altundag O, Canpinar H, Celebi-Saltik B. Methionine affects the expression of pluripotency genes and protein levels associated with methionine metabolism in adult, fetal, and cancer stem cells. J Cell Biochem. 2021. [DOI] [PubMed] [Google Scholar]; This paper compared the different effects of methionine in metabolic and epigenetic regulations in bone marrow-mesenchymal stem cells (MSCs), umbilical cord blood MSCs, and breast cancer stem cells (CSCs).
- 4.Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 2012;31(13):1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garinis GA, Patrinos GP, Spanakis NE, et al. DNA hypermethylation: when tumour suppressor genes go silent. Hum Genet. 2002;111(2):115–127. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Y, He C, Wang M, et al. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther. 2019;4:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nepali K, Liou JP. Recent developments in epigenetic cancer therapeutics: clinical advancement and emerging trends. J Biomed Sci. 2021;28(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Yankova E, Blackaby W, Albertella M, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593(7860):597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper identified and characterized the first-in-class inhibitor of METTL3 STM2457, which inhibits the expression of leukemogenic genes by reducing the m6A modifications on their mRNA in acute myeloid leukemia cells
- 9.Marjon K, Kalev P, Marks K. Cancer Dependencies: PRMT5 and MAT2A in MTAP/p16-Deleted Cancers. Annual Review of Cancer Biology, Vol 5, 2021. 2021;5:371–390. [Google Scholar]
- 10.Kotb M, Mudd SH, Mato JM, et al. Consensus nomenclature for the mammalian methionine adenosyltransferase genes and gene products. Trends in Genetics. 1997;13(2):51–52. [DOI] [PubMed] [Google Scholar]
- 11.Murray B, Antonyuk SV, Marina A, et al. Crystallography captures catalytic steps in human methionine adenosyltransferase enzymes. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(8):2104–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pendleton KE, Chen B, Liu K, et al. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017;169(5):824–835 e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pendleton KE, Park SK, Hunter OV, et al. Balance between MAT2A intron detention and splicing is determined cotranscriptionally. Rna. 2018;24(6):778–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Villa E, Sahu U, O’Hara BP, et al. mTORC1 stimulates cell growth through SAM synthesis and m(6)A mRNA-dependent control of protein synthesis. Mol Cell. 2021;81(10):2076–2093 e2079. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrated that, c-MYC, which is the downstream of mTOCR1, directly binds to the intro 1 of MAT2A RNA, and the activatiion of mTORC1 stimulates SAM biosynthesis by promoting the expression of MAT2A.
- 15.Roje S S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry. 2006;67(15):1686–1698. [DOI] [PubMed] [Google Scholar]
- 16.Finkelstein JD, Martin JJ. Homocysteine. Int J Biochem Cell Biol. 2000;32(4):385–389. [DOI] [PubMed] [Google Scholar]
- 17.Simpson B, Tupper C, Al Aboud NM. Genetics, DNA Packaging. StatPearls. Treasure Island (FL); 2022. [PubMed] [Google Scholar]
- 18.Sadakierska-Chudy A, Filip M. A comprehensive view of the epigenetic landscape. Part II: Histone post-translational modification, nucleosome level, and chromatin regulation by ncRNAs. Neurotox Res. 2015;27(2):172–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Wang C, Liu W, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537(7621):558–562. [DOI] [PubMed] [Google Scholar]
- 20.Becker JS, Nicetto D, Zaret KS. H3K9me3-Dependent Heterochromatin: Barrier to Cell Fate Changes. Trends Genet. 2016;32(1):29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YH, Stallcup MR. Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol. 2009;23(4):425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oerum S, Meynier V, Catala M, et al. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021;49(13):7239–7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang X, Liu B, Nie Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiraki N, Shiraki Y, Tsuyama T, et al. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014;19(5):780–794. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Yip LY, Lee JHJ, et al. Methionine is a metabolic dependency of tumor-initiating cells. Nat Med. 2019;25(5):825–837. [DOI] [PubMed] [Google Scholar]
- 27.Strekalova E, Malin D, Weisenhorn EMM, et al. S-adenosylmethionine biosynthesis is a targetable metabolic vulnerability of cancer stem cells. Breast Cancer Res Treat. 2019;175(1):39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Secker KA, Bloechl B, Keppeler H, et al. MAT2A as Key Regulator and Therapeutic Target in MLLr Leukemogenesis. Cancers (Basel). 2020;12(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai J, Sun WM, Hwang JJ, et al. Changes in S-adenosylmethionine synthetase in human liver cancer: molecular characterization and significance. Hepatology. 1996;24(5):1090–1097. [DOI] [PubMed] [Google Scholar]
- 30*.Chu PY, Wu HJ, Wang SM, et al. MAT2A Localization and Its Independently Prognostic Relevance in Breast Cancer Patients. Int J Mol Sci. 2021;22(10). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates the higher MAT2A level in breast cancer patients is associated with worse survival rate, and the subcellular localization of MAT2A has a great significance on the prognosis of breast cancer patients.
- 31.Simile MM, Peitta G, Tomasi ML, et al. MicroRNA-203 impacts on the growth, aggressiveness and prognosis of hepatocellular carcinoma by targeting MAT2A and MAT2B genes. Oncotarget. 2019;10(29):2835–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Leun AM, Thommen DS, Schumacher TN. CD8(+) T cell states in human cancer: insights from single-cell analysis. Nat Rev Cancer. 2020;20(4):218–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33**.Bian Y, Li W, Kremer DM, et al. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature. 2020;585(7824):277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper revealed that cancer cells compete with T cells for methionine through upregulated SLC43A2, which starves T cells of methionine to produce SAM for histone methylation and leads to T cell dysfunction.
- 34.Pizzagalli MD, Bensimon A, Superti-Furga G. A guide to plasma membrane solute carrier proteins. FEBS J. 2021;288(9):2784–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35*.Hung MH, Lee JS, Ma C, et al. Tumor methionine metabolism drives T-cell exhaustion in hepatocellular carcinoma. Nat Commun. 2021;12(1):1455. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrated the dysregulated methionine metabolism promotes CD8+ T cell dysfunction in hepatocellular carcinoma, suggesting targeting tumor methionine metabolism pathway is a promising strategy in cancer immunotherapy.
- 36.Lu WC, Saha A, Yan W, et al. Enzyme-mediated depletion of serum l-Met abrogates prostate cancer growth via multiple mechanisms without evidence of systemic toxicity. Proc Natl Acad Sci U S A. 2020;117(23):13000–13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37**.Higuchi T, Han Q, Sugisawa N, et al. Combination Methionine-methylation-axis Blockade: A Novel Approach to Target the Methionine Addiction of Cancer. Cancer Genomics Proteomics. 2021;18(2):113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates the combination of degrading methionine through methioninase, inhibiting MAT2A through cycloleucine and impairing DNA methylation through decitabine effectively impairs methionine-methylation-axis and is a promising cancer chemotherapy.
- 38.Wang ZX, Yip LY, Lee JHJ, et al. Methionine is a metabolic dependency of tumor-initiating cells. Nature Medicine. 2019;25(5):825-+. [DOI] [PubMed] [Google Scholar]
- 39.Kim H, Ronai ZA. PRMT5 function and targeting in cancer. Cell Stress. 2020;4(8):199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marjon K, Cameron MJ, Quang P, et al. MTAP Deletions in Cancer Create Vulnerability to Targeting of the MAT2A/PRMT5/RIOK1 Axis. Cell Rep. 2016;15(3):574–587. [DOI] [PubMed] [Google Scholar]
- 41**.Konteatis Z, Travins J, Gross S, et al. Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion. J Med Chem. 2021;64(8):4430–4449. [DOI] [PubMed] [Google Scholar]; This paper reported the discovery of the first-in-class MAT2A inhibitor AG-270 via the fragment screening followed by an iterative structure-guided design.
- 42**.Kalev P, Hyer ML, Gross S, et al. MAT2A Inhibition Blocks the Growth of MTAP-Deleted Cancer Cells by Reducing PRMT5-Dependent mRNA Splicing and Inducing DNA Damage. Cancer Cell. 2021;39(2):209–224 e211. [DOI] [PubMed] [Google Scholar]; This paper characterized the selective antiproliferative effects of first-in-class MAT2A inhibitor AG-270 in MTAP-depleted cancers.
- 43**.De Fusco C, Schimpl M, Borjesson U, et al. Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model. J Med Chem. 2021;64(10):6814–6826. [DOI] [PubMed] [Google Scholar]; This paper reported a potent MAT2A inhibitor Compound 28 and evaluated its effects on methylation reactions and tumor proliferation in MTAP-deleted cancers.
- 44.Lombardini JB, Sufrin JR. Chemotherapeutic potential of methionine analogue inhibitors of tumor-derived methionine adenosyltransferases. Biochem Pharmacol. 1983;32(3):489–495. [DOI] [PubMed] [Google Scholar]
- 45.Sviripa VM, Zhang W, Balia AG, et al. 2’,6’-Dihalostyrylanilines, pyridines, and pyrimidines for the inhibition of the catalytic subunit of methionine S-adenosyltransferase-2. J Med Chem. 2014;57(14):6083–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan CL, Kaiser SE, Bolanos B, et al. Targeting S-adenosylmethionine biosynthesis with a novel allosteric inhibitor of Mat2A. Nat Chem Biol. 2017;13(7):785–792. [DOI] [PubMed] [Google Scholar]
- 47.Marjon K, Kalev P, Hyer M, et al. Abstract 2714: Targeting MAT2A in CDKN2A/MTAP-deleted cancers. Cancer Research. 2019;79(13_Supplement):2714–2714. [Google Scholar]


