Abstract
Type 1 diabetes (T1D4) in both humans and NOD mice is caused by T-cell mediated autoimmune destruction of pancreatic β-cells. Increased frequency or activity of autoreactive T-cells and failures of regulatory T-cells (Tregs) to control these pathogenic effectors have both been implicated in T1D etiology. Due to the expression of MHC-I molecules on β-cells, CD8 T-cells represent the ultimate effector population mediating T1D. Developing autoreactive CD8 T-cells normally undergo extensive thymic negative selection, but this process is impaired in NOD mice, and also likely T1D patients. Previous studies identified an allelic variant of Nfkbid, a NF-κB signal modulator, as a gene strongly contributing to defective thymic deletion of autoreactive CD8 T-cells in NOD mice. These previous studies found ablation of Nfkbid in NOD mice using the clustered regularly interspaced short palindromic repeats (CRISPR)/ CRISPR associated protein 9 (Cas9) system resulted in greater thymic deletion of pathogenic CD8 AI4 and NY8.3 TCR transgenic T-cells, but an unexpected acceleration of T1D onset. This acceleration was associated with reductions in the frequency of peripheral Tregs. Here we report transgenic overexpression of Nfkbid in NOD mice also paradoxically results in enhanced thymic deletion of autoreactive CD8 AI4 T-cells. However, transgenic elevation of Nfkbid expression also increased the frequency and functional capacity of peripheral Tregs in part contributing to the induction of complete T1D resistance. Thus, future identification of a pharmaceutical means to enhance Nfkbid expression might ultimately provide an effective T1D intervention approach.
Introduction
Genetic background is a key contributor to the development of autoimmune type 1 diabetes (T1D). Studies in the NOD mouse model and humans have linked over 60 genetic loci (designated Idd in mice and IDDM in humans) contributing to T1D, but with certain MHC (designated HLA in humans) haplotypes being the primary disease risk factor (1, 2). Although other leukocyte populations participate in β-cell destruction, T1D is largely considered a T-cell mediated disease (3, 4). It has long been known that autoreactive CD4 T-cell responses mediated by particular MHC/HLA class II variants play an important role in T1D development. However, since pancreatic β-cells express MHC class I, but not class II molecules, β-cell destruction in T1D is likely ultimately mediated by autoreactive CD8 T-cells (4–7). Defects in the negative selection of autoreactive T-cells during thymic development and failures of regulatory T-cells (Tregs) to control such pathogenic effectors are major features of T1D etiology (4, 8).
We previously identified NOD and C57BL/6 (B6) allelic variants of the NF-κB activity modulating Nfkbid gene, residing on a proximal region Chromosome (Chr.) 7 overlapping the Idd7 locus, as a major contributor exerting an intrinsic effect respectively inhibiting and promoting thymic negative selection of both AI4 and NY8.3 diabetogenic TCR transgenic CD8 T-cells (9). The protein coding regions of the NOD and B6 Nfkbid alleles are identical. However, the NOD Nfkbid allele is expressed at a higher level in thymocytes than the B6 variant (9). Thus, we originally used a CRISPR/Cas9 approach to ablate Nfkbid expression in NOD mice to assess if this enhanced thymic negative selection of AI4 CD8 T-cells. This was indeed found to be the case, but unexpectedly T1D development was accelerated in these resultant NOD-Nfkbid−/− mice. Such disease acceleration was concomitant with a significantly reduced number of Tregs in NOD-Nfkbid−/− mice, as well as the proportion of such cells expressing IL-10 (9). A similar effect on thymic derived Tregs was previously observed when Nfkbid expression was ablated in B6 background mice (10).
Nfkbid (a.k.a. IκBNS) is a nuclear IκB family protein that binds NF-κB subunit proteins through its ankyrin repeat domains (11). Such interactions with Nfkbid refine NF-κB pathway signaling outcomes downstream of TCR stimulation (12, 13). Depending on cell type and physiological context Nfkbid can serve as either a negative or positive modulator of NF-κB activity (14–17). Our previous studies found Nfkbid can enhance at least some NF-κB activities in thymocytes (9). Low to moderate NF-κB activity (primarily the p50/p65 isoform) reportedly inhibits thymic negative selection of CD8 lineage destined T-cells (18). However, high NF-κB activity can elicit thymic deletion of CD8 T-cells (18). Furthermore, NF-κB activity influences both Treg development and peripheral survival (10, 19–22). Thus, our original goal was to determine if NOD mice expressing the B6 Nfkbid variant in lieu of their endogenous allele were characterized by changes in NF-κB activity influencing the thymic deletion of diabetogenic CD8 T-cells and/or development of Tregs.
To pursue the above goal, we isolated a genomic fragment encompassing the B6 Nfkbid gene and transgenically introduced it into our NOD stock lacking this strain’s endogenous variant. Nfkbid expression in this new NOD transgenic stock surpassed that in either wild type NOD or B6 mice. Surprisingly, NOD mice with increased levels of Nfkbid expression also showed increased thymic negative selection of autoreactive CD8 T-cells. This was accompanied by a striking increase in the numbers and activity of Tregs and complete T1D resistance. Thus, this study evaluated how the varying extents of thymic deletion of autoreactive CD8 T-cells, coupled with the development and activity of Tregs elicited by Nfkbid ablation or overexpression respectively contributes to the acceleration or inhibition of T1D development in NOD mice. T1D protection resulting from elevating Nfkbid expression in NOD mice resulted at least in part from both increased thymic deletion of pathogenic CD8 T-cells and enhanced Treg numbers and/or activity.
Materials and Methods
Mice
C57BL/6J (B6), NOD/ShiLtDvs (NOD) (23), NOD.Cg-Emv30b Prkdcscid/Dvs (NOD.scid) (24) mice as well as an NOD stock carrying a bicistronic Foxp3-EGFP transgene (NOD.129X1(Cg)-Foxp3tm2Tch/DvsJ, hereafter NOD.Foxp3-GFP) (25, 26) were maintained in a specific pathogen-free research colony at The Jackson Laboratory. NOD/ShiLtDvs-Nfkbidem3Dvs/Dvs (hereafter NOD-Nfkbid−/−) mice have been previously described and carry a CRISPR/Cas9 generated mutation ablating functional Nfkbid mRNA expression (9). NOD and NOD-Nfkbid−/− mice expressing a transgenic Vα8.3+Vβ2+ TCR derived from the diabetogenic AI4 CD8 T-cell clone (officially designated NOD/ShiLtDvs-Tg(TcraAI4)1Dvs Tg(TcrbAI4)1Dvs/Dvs and NOD/ShiLtDvs-Nfkbidem3Dvs Tg(TcraAI4)1Dvs Tg(TcrbAI4)1Dvs/Dvs, and hereafter NOD-AI4 and NOD-AI4-Nfkbid−/−) have been described previously (9, 27). An NOD stock carrying an inactivated Rag1 gene as well as the AI4 transgenic TCR (NOD.Cg-Rag1tmMom Tg(TcraAI4)1Dvs Tg(TcrbAI4)1Dvs/Dvs, hereafter NOD.Rag1null. AI4) has also been previously described (28).
Generation of Nfkbid-overexpressing NOD mice
A genomic region encompassing the B6 Nfkbid allele was isolated from a BAC clone and cloned into a pUC57 Simple plasmid containing chicken HS4 insulator and retrieval sequences. A 303bp intergenic segment downstream of Nfkbid was removed for genotyping purposes. Nfkbid transgenic mice were generated by pronuclear injection of the excised B6 segment into NOD embryos. Transgene carriers were identified by PCR using a primer set that spans the omitted sequence (Nfkbid-g1F: CAA GCC TGA GCT ACC GAC AA; Nfkbid-g1R: GCC TTA CAC AAG CAG GGC TA). Founder mice were then crossed with the NOD-Nfkbid−/− strain to generate NOD/ShiLtDvs-Nfkbidem3Dvs Tg(Nfkbid)60–2Dvs/Dvs (hereafter NOD-60A) mice. The Nfkbid transgene is maintained in a hemizygous state. For experiments evaluating thymic negative selection of diabetogenic CD8 T-cells the 60A transgene was subsequently crossed into the Nfkbid-ablated NOD-AI4 line (9) to generate the NOD-AI4-60A strain (officially NOD/ShiLtDvs-Nfkbidem3Dvs Tg(Nfkbid)60–2Dvs Tg(TcraAI4)1Dvs Tg(TcrbAI4)1Dvs/Dvs). For studies employing fluorescently marked Foxp3+ Tregs, NOD-60A mice were bred with the NOD.Foxp3-GFP strain to yield both the NOD.Cg-Nfkbidem3Dvs Foxp3tm2Tch/Dvs (hereafter NOD-Nfkbid−/−.Foxp3-GFP) and NOD.Cg-Nfkbidem3Dvs Foxp3tm2Tch Tg(Nfkbid)60–2Dvs/Dvs (hereafter NOD-60A.Foxp3-GFP) stocks.
Flow Cytometry
Single cell suspensions were prepared from spleen, pancreatic lymph node (Panc LN) and thymus by passing cells through nytex membranes. For spleen samples, red blood cells were lysed using Gey’s buffer (29). Aliquots of the respective cells were prepared and labeled with the following fluorescently labeled antibodies obtained from Biolegend (San Diego, CA), BD Biosciences (San Jose, CA), eBioscience or Invitrogen at Thermo Fisher Scientific or TONBO Biosciences (San Diego, CA): CD45.1 (A201.7 or A20), CD4 (GK1.5 or RM4–5; BV450, BV785, FITC), CD8 (53–6.72; APC, BV480 or PE-CY7), TcrVα8.3 (B21.14; FITC), TcrVβ2 (B20.6; AlexaFluor647), TCRβ (H57–597; BV711); CD25 (PC61; BV605), CD44 (IM7; RF700); CD62L (MEL-14; BV650); CD39 (24DMS1; SB436); CD73 (TY/11.8; APC); GITR (DTA 1; PE-Cy7); ICOS (C398.4A; BV605); PD-1 (29F.1A12; BV711); TIGIT (1G9; APC); CTLA-4 (UC10–4B9; PE). Staining with AI4 tetramer (Db/MimA2.YAIENYLEL, PE; provided by the NIH Tetramer Core Facility) has been described previously (9). In some experiments, following cell surface and viability staining (Fixable Viability Dye eFlour 780, Invitrogen Thermo Fisher Scientific) cells were fixed and permeabilized with Foxp3 Fix/Perm Buffer Set (Biolegend) according to maker’s protocols. Intracellular staining was then performed for Foxp3 (MF23; AlexaFluor 488) (BD Biosciences) and/or IL-10 (JESS-16E3; APC) (Biolegend). For the detection of IL-10 in splenocytes, cells were first plated in RPMI 1640 supplemented with penicillin/streptomycin, 5mM NEAA, 2mM L-glutamate, 5mM sodium pyruvate (all from Life Technologies, Grand Island, NY), 10% FBS (Hyclone, Logan, UT) and 50uM β-mercaptoethanol (Thermo Fisher Scientific). Cell stimulation cocktail [50ng/ml phorbol myristate acetate (PMA), 500ng/ml ionomycin, 10ng/ml lipopolysaccharide (LPS) with 2μM monensin] was added for 5 hours prior to IL-10 and Foxp3 staining for flow cytometry analyses as indicated above. Data acquisition was performed using a BD FACS Symphony A5 flow cytometer (BD Biosciences), an Attune NXT Acoustic Focusing Cytometer (Thermo Fisher) or a FACSCalibur (BD Biosciences). Data were analyzed using FlowJo software version 10 (BD Life Sciences). Cell sorting experiments were performed using a FACS Aria II sorter (BD Biosciences).
Gene expression
Total RNA was prepared from whole thymus or sorted splenic CD4+CD25+ or CD4+Foxp3-GFP+ Tregs. Thymic samples were resuspended in TRIzol (Thermo Fisher Scientific, Waltham, MA) and frozen at −80°C until use. For sorted splenic Tregs, we initially performed negative enrichment of CD4 T-cells. Briefly single cell suspensions were labeled with biotin-conjugated antibodies (anti-B220, anti-CD11c, anti-CD8, anti-CD11b and anti-TER-119; (TONBO Biosciences and BD Biosciences)) then incubated with Streptavidin MicroBeads (Miltenyi Biotec; North Rhine-Westphalia, Germany) and passed over LD Columns (Miltenyi Biotec). Flowthrough containing enriched CD4 T-cells was collected and sorted for the CD25+or Foxp3-GFP+ fraction into FBS, resuspended in TRIzol reagent and frozen. Phenol-chloroform extracted total RNA was reverse transcribed using SuperScript IV VILO Master Mix (Thermo Fisher Scientific) according to the manufacturer’s directions. qPCR was performed using SYBR Green PCR Master Mix (Applied Biosystems, Thermo Fisher Scientific) on a ViiA 7 Real Time PCR system (Applied Biosystems). The CRISPR/Cas9-generated mutation in NOD.Nfkbid−/− mice yields an mRNA transcript but no protein (9). As such, Nfkbid qPCR primers were designed to amplify only the wild-type transcript by localization of the primers over the mutation site sequence (Nfkbid-F: TCC ATG CTA CCT ATA CAG AGG G and Nfkbid-R: AGA GAG TCC TCC ATC ACC AGG)(9). Il10, Bcl3, Ifit1 and Ifit3 primer pairs were: Il10-F: TAG AGC TGC GGA CTG CCT T and Il10-R: GGC AAC CCA AGT AAC CCT TAA A; Bcl3-F: CCG AAT ACT CAG CCT TTT CAA GC and Bcl3-R: TGG TAA TGT GGT GAT GAC AGC C; Ifit1-F: GCA AAC ATA GGC CAT CTC AA and Ifit1-R: ACT GAG GAC ATC CCG AAA CA (30); Ifit3-F: TCA GCC CAC ACC CAG CTT TT and Ifit3-R: TTG CAC ACC CTG TCT TCC ATA. Pooled cDNA was used to generate standard curves for each gene of interest and their expression was normalized to that of Gapdh expression and compared to control samples.
Western blotting
Spleen, thymus and liver were dissected and passed through nytex membranes to generate single cell suspensions. RBCs were lysed from spleen samples using Gey’s buffer as above and all tissue homogenates were suspended in 1x RIPA buffer (Cell Signaling Technologies, Danvers, MA) with 1x HALT Protease Inhibitor Cocktail (Thermo Fisher Scientific). Protein lysates were quantified by Bradford Assay (Thermo Fisher Scientific) and a Simple Wes automated western blotting system (ProteinSimple, San Jose, CA) was used to examine Nfkbid protein abundance (9). Briefly, 1mg/ml lysates were mixed with 5x Fluorescent Sample buffer and denatured at 95°C. Then biotinylated molecular weight standard, protein samples, blocking solution, Nfkbid rabbit polyclonal antibody (Ab) (1:100) (31), biotin labeling reagents, wash buffers, HRP-conjugated anti-rabbit secondary Ab and chemiluminescent substrate were loaded into a plate prefilled with capillary separation and stacking matrices. Protein samples were run in duplicate with one for Nfkbid detection (60-minute primary incubation) and one for Total Protein Detection (ProteinSimple). Chemiluminescent signal was captured, and images were analyzed with the Compass for Simple Western software package (ProteinSimple).
T1D incidence and insulitis
Beginning at 10 weeks of age, female mice were monitored for onset of hyperglycemia by weekly testing using Diastix urine glucose test strips (Bayer, Leverkusen, Germany). Mice with a positive glucosuria test result (corresponding to >300mg/dl in blood) were retested three days later and were deemed diabetic if still positive. Nondiabetic mice at 30 weeks of age were euthanized and pancreata were fixed in Bouin’s solution (Rowley Biochemical, Danvers, MA), embedded, sectioned and stained by aldehyde fuchsin and H&E. Insulitis scores were quantified by a blinded observer as previously described (32). Briefly, islets were scored using the following system: no lesions = 0; peri-insular aggregates = 1; <25% islet destruction = 2; 25–75% islet destruction = 3; 75–100% islet destruction = 4. Final mean insulitis score (MIS) was determined by dividing the total score for each pancreas by the number of islets examined (at least 20 per mouse). In one analysis, insulitis levels were determined in a cohort of mice at 10–12 weeks of age.
RNAseq analyses
Thymii from NOD-AI4, NOD-AI4-Nfkbid−/− and NOD-AI4-60A mice were dissected and passed through nytex to create single cell suspensions. Cells stained (as described above) as CD4+CD8+ double positive (DP) cells were sorted directly into FBS. DP thymocytes were resuspended in TRIzol and RNA was purified according to manufacturer’s protocol with RNeasy MiniElute Cleanup (Qiagen, Germantown, MD) for an RNA Integrity Number >9. KAPA stranded mRNA libraries were constructed and sequenced on a NextSeq 550 system (Illumina, San Diego, CA) yielding ~30 million single end 75bp reads per sample. Expected read counts were generated using the conda package emase 0.10.16 (EMASE: Expectation-Maximization algorithm for Allele Specific Expression). EMASE model 4 was run with bowtie 1.0.0 and samtools 0.1.18 using a transcriptome based on the GRCm38/mm10 reference genome and NOD/ShiLtJ-specific variants. The Bioconductor R package edgeR 3.24.3 was used to analyze differential expression (33). Differentially expressed genes with a fold-change (FC) >2 and false discovery rate (FDR) ≤0.05 (Supplemental Tables 1 and 2) were annotated using the ENRICHR online tool (34) to identify genes with murine NF-kB transcription factor binding sites within promoter regions (Supplemental Table 3). The lists of differentially expressed genes for each contrast (Nfkbid deficient or overexpressing vs control) were entered into the website tool (https://maayanlab.cloud/Enrichr/enrich#). Genes with mouse Nfkb1, Rel, and RelA binding sites were identified using the results from the position weight matrices from TRANSFAC and JASPAR library.
IL-10 ELISA
CD4+Foxp3+ splenic Tregs from NOD.Foxp3-GFP, NOD-Nfkbid−/−.Foxp3-GFP and NOD-60A.Foxp3-GFP mice were isolated by negative enrichment of CD4 T-cells as described above followed by sorting for Foxp3-GFP+ cells. Cells were plated in complete RPMI supplemented as above and additionally with 0–4μg/ml anti-CD3. After 3 days, cells were collected, pelleted and the supernatant was analyzed for IL-10 secretion using the ELISA MAX Standard Set Mouse IL-10 according to manufacturer’s protocol (Biolegend).
Treg suppression assay
Populations of CD4+CD25+ Tregs and CD4+CD25− responder T-cells were isolated from NOD, NOD-Nfkbid−/− and NOD-60A mice by negative enrichment for CD4 T-cells (as above) followed by cell sorting into FBS. Responder T-cells were labeled with cell proliferation dye eFluor670 (eBioscience, Thermo Fisher Scientific) and plated with Tregs at the indicated ratios. Splenocytes from NOD.scid mice were used as APC and cultures were incubated in RPMI 1640 supplemented as above along with 20μg/ml anti-CD3 (145–2C11, BD Biosciences). After 72 hours, cells were harvested, and responder T-cell proliferation assessed by flow cytometric determination of marker dye dilution. Percent suppression was calculated using the Proliferation Modeling tool within FlowJo.
Adoptive transfer studies
For transfer studies utilizing diabetogenic AI4 T-cells, NOD and NOD-60A mice were sub-lethally irradiated (600 cGy) and injected i.v. with ~1×107 NOD.Rag1null. AI4 splenocytes (normalized to 8×105 AI4 CD8 T-cells). A subset of NOD-60A mice were then i.p. injected with 62.5μg of the CD25 specific PC61 antibody (BioXCell, Lebanon, NH) to reduce but not completely eliminate Tregs. These groups of AI4 T-cell re-populated NOD and NOD-60A mice were then compared for T1D development daily over a 14-day period. Levels of Foxp3+CD25+ Tregs in Panc LN were measured by flow cytometry as described above along with anti-CD25 specific 7D4 antibody (BD Biosciences) upon T1D occurrence or at 14 days post-transfer.
Results
T1D resistance in Nfkbid-transgenic NOD Mice
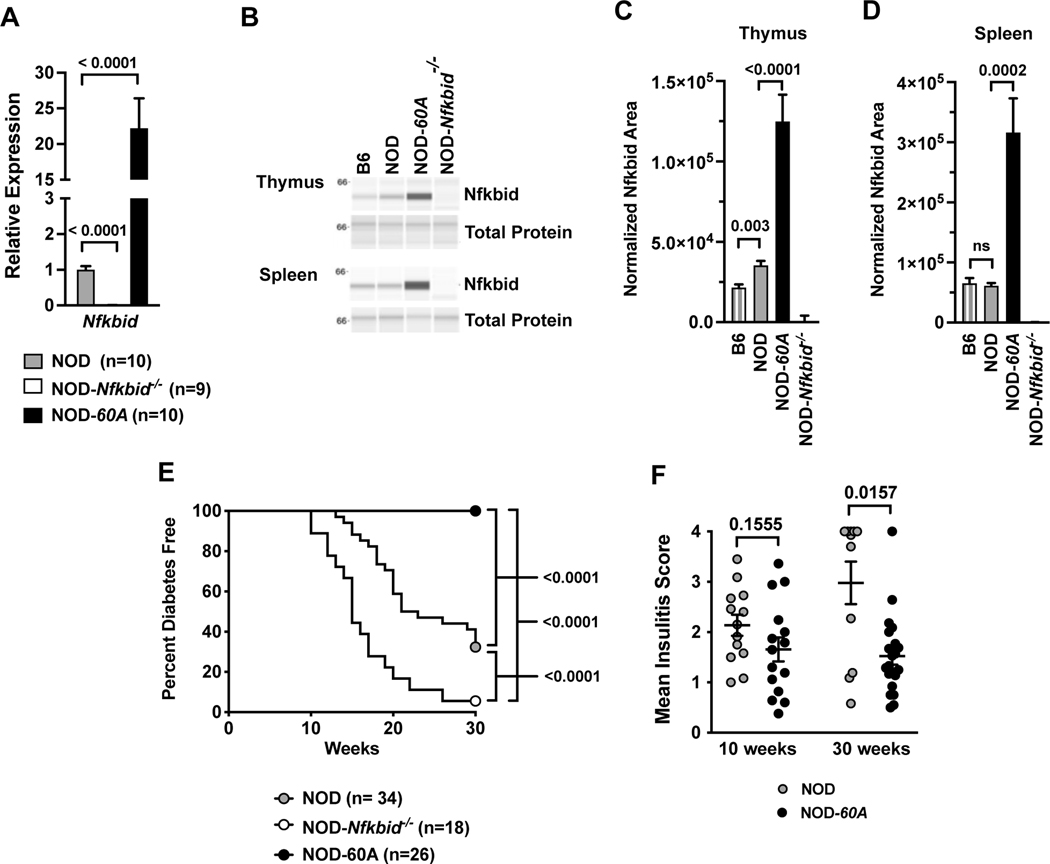
The protein coding regions of the NOD and B6 Nfkbid alleles are identical (35). Thus, polymorphisms in non-coding regulatory regions likely contribute to the enhanced expression of the NOD Nfkbid allele. The locations of SNPs distinguishing the NOD and B6 Nfkbid alleles are depicted in Table 1. To examine whether differences in Nfkbid expression could alter T1D progression by influencing autoreactive CD8 T-cell and Treg populations, we transgenically inserted a B6-derived Nfkbid genomic transgene into our previously developed NOD-Nfkbid−/− mice (9). Likely due to the insertion of multiple copies, a resultant transgenic line, designated NOD-60A, was found to be characterized by an approximately 20-fold elevated Nfkbid mRNA transcript expression in whole thymic extracts compared to standard NOD mice (Figure 1A). Nfkbid protein is readily detected as a 55kD band in B6 and NOD thymus and spleen (Figure 1B-D). As previously reported (9), Nfkbid protein levels are increased in NOD thymus compared to the B6 reference strain (Figure 1B, C). However, baseline splenic Nfkbid protein levels did not differ in NOD and B6 mice (Figure 1B, D). This may be due to the more heterogeneous cellular makeup of the spleen than thymus. NOD-60A transgene carriers show 3.53-fold and 5.12-fold increased expression of Nfkbid in thymus and spleen compared to NOD controls (Figure 1B-D). As expected, NOD.Nfkbid−/− mice lack both full length Nfkbid transcript and protein (Figure 1A-D). Nfkbid protein was absent in liver indicating the transgene is expressed in a normal range of tissues (data not shown). NOD-60A transgene carriers breed normally, though their spleen size is increased and enlarged follicles are noted (data not shown).
Table I:
Genetic variation between C57BL/6 Reference and NOD/ShiLtJ strains within and +/− 1kb of Nfkbid (Chr 7: 30,120,157–30,129,171 (GRCm39))
| Variant ID | Variant Class | Predicted Consequence | B6 | NOD |
|---|---|---|---|---|
| rs3147809 | SNP | upstream gene variant | T | C |
| rs3142468 | SNP | upstream gene variant | A | G |
| rs8261832 | SNP | intron variant | A | G |
| rs3147847 | SNP | intron variant | T | G |
| rs3142508 | SNP | intron variant | C | T |
| rs3147846 | SNP | synonymous variant | T | C |
| rs3147845 | SNP | intron variant | G | A |
| rs3142506 | SNP | intron variant | G | A |
| rs3147844 | SNP | synonymous variant | T | A |
| rs3142505 | SNP | intron variant | A | G |
| rs3142504 | SNP | intron variant | G | A |
| rs3147843 | SNP | intron variant | C | T |
| rs3147842 | SNP | intron variant | C | T |
| rs8261873 | insertion | intron variant | - | T |
| rs3142503 | SNP | intron variant | G | C |
| rs586927013 | SNP | intron variant | G | A |
| rs258934926 | SNP | intron variant | T | C |
| rs3142502 | SNP | intron variant | G | A |
| rs3142501 | SNP | intron variant | A | T |
| rs258679767 | deletion | splice region variant | C | - |
| rs3147840 | SNP | synonymous variant | C | T |
| rs3147839 | SNP | synonymous variant | G | A |
| rs3147838 | SNP | intron variant | C | T |
| rs3142500 | SNP | non coding transcript exon variant | G | T |
| rs585933734 | SNP | non coding transcript exon variant | G | A |
| rs3147837 | SNP | intron variant | C | T |
| rs3142498 | SNP | intron variant | A | G |
| rs3147836 | SNP | intron variant | C | T |
| rs3147835 | SNP | intron variant | C | G |
| rs3147834 | SNP | intron variant | T | C |
| rs3142497 | SNP | intron variant | G | A |
| rs3142496 | SNP | intron variant | C | T |
| rs3142495 | SNP | intron variant | T | G |
| rs3142494 | SNP | synonymous variant | A | G |
| rs3142493 | SNP | intron variant | A | G |
| rs252747946 | SNP | intron variant | G | C |
| rs225186782 | SNP | intron variant | G | A |
| rs46738579 | SNP | intron variant | T | G |
| rs8261888 | SNP | intron variant | C | T |
| rs8261887 | SNP | intron variant | C | G |
| rs8261886 | SNP | intron variant | T | C |
| rs8261885 | SNP | intron variant | G | A |
| rs32265778 | SNP | intron variant | A | T |
| rs263326630 | deletion | intron variant | TGATCTTAAATG | - |
| rs3142492 | SNP | splice region variant | C | T |
| rs3147832 | SNP | 3' UTR variant | C | T |
| rs3142491 | SNP | 3' UTR variant | G | A |
Figure 1. Enhanced expression of Nfkbid prevents T1D and reduces insulitis.
(A) Nfkbid mRNA levels were measured in whole thymus extracts taken from 8-week-old standard NOD, NOD-Nfkbid−/−, and NOD-60A female mice (n=9–10 per genotype). Data are normalized to Gapdh expression levels and standard NOD controls. (B) Nfkbid protein (55kDa) was detected by Simple Wes western blotting in lysates from spleen and thymus of B6, NOD and NOD-60A mice, but absent in all NOD-Nfkbid−/− extracts. A representative sample from each strain is shown run in parallel for Nfkbid detection (upper panel) and for total protein assay (lower panel). (C) Quantitation of thymic Nfkbid levels normalized to total protein expression for n=8 each for B6 and NOD, n=9 NOD-60A and n=6 NOD-Nfkbid−/−mice. (D) Quantitation of splenic Nfkbid levels normalized to total protein expression for n=8 each for B6, NOD, NOD-60A and NOD-Nfkbid−/−mice. (E) T1D development was monitored by weekly urine glucose testing of female NOD-60A mice, their Nfkbid-deficient nontransgenic littermates (NOD-Nfkbid−/−), and standard NOD mice. Survival curve comparisons were analyzed by log-rank (Mantel-Cox) test. (F) Insulitis level was assessed by a blinded trained observer in 10-week-old mice or those surviving to 30 weeks. For A, C, D and F, mean ± SEM is represented and statistical significance was determined by Mann-Whitney nonparametric comparison.
We monitored T1D development in standard NOD controls compared to both NOD-60A mice and littermates lacking the Nfkbid transgene (equivalent to the NOD-Nfkbid−/− strain). Strikingly, NOD-60A mice show complete resistance to T1D development out to 30 weeks of age (Figure 1E), and a separate small cohort survived beyond one year (data not shown). As previously observed nearly all non-transgenic Nfkbid deficient littermates developed accelerated disease (Figure 1E) (9). Analysis of pancreatic sections at 10 weeks of age revealed an equivalent moderate level of insulitis in NOD-60A and NOD control mice (Figure 1F) mice. However, when examined at 30 weeks of age the extent of insulitis in NOD-60A transgenic mice failed to progress to the much higher levels characterizing surviving NOD controls [30-week MIS: NOD=2.98, 60A=1.57, p=0.0157] (Figure 1F). It should be noted these described Nfkbid transgenic effects are manifest in a hemizygous state.
Nfkbid overexpression improves negative selection of autoreactive diabetogenic CD8 T-cells
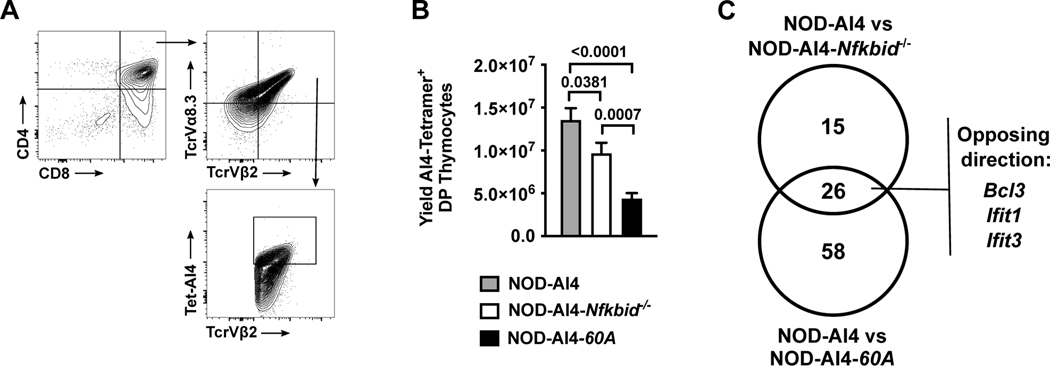
Nfkbid ablated NOD mice have improved thymic negative selection of autoreactive diabetogenic AI4 and NY8.3 CD8 T-cells at the DP stage of development (9). Therefore, we examined whether increasing Nfkbid expression might elicit effects on thymic selection of diabetogenic AI4 CD8 T-cells. We crossed the 60A transgene into NOD-Nfkbid−/− mice also carrying the transgenic TCR from the diabetogenic AI4 CD8 T-cell clone. The promiscuous AI4 receptor recognizes multiple antigens, including insulin (27). Surprisingly, the resultant NOD-AI4-60A mice also showed enhanced thymic deletion of diabetogenic AI4 T-cells compared to NOD-AI4 controls. Indeed, the extent of thymic AI4 T-cell deletion in the NOD-AI4-60A stock was even greater than in NOD-AI4-Nfkbid−/− mice (Figure 2A, B).
Figure 2. Increased negative selection of developing autoreactive CD8 T-cells with both ablation and overexpression of Nfkbid.
(A) Flow cytometric gating strategy to quantify CD4+CD8+ DP thymocytes expressing the AI4 TCR using antibodies for TCRVα8.3 and TCRVβ2 in conjunction with AI4 tetramer binding. (B) Quantification of AI4 DP thymocytes from NOD-AI4, NOD-AI4-Nfkbid−/−, and NOD-AI4-60A mice (n= 9–13 mice per group). Statistical significance was determined by Mann-Whitney nonparametric comparison. (C) RNAseq was performed on DP thymocytes from NOD-AI4, NOD-AI4-Nfkbid−/− and NOD-AI4-60A mice. Among the differentially expressed genes, those harboring NF-κB transcription factor binding sites within promoter regions were identified.
Ablation or elevation of Nfkbid levels differentially alters NF-κB target gene expression in NOD DP thymocytes
We hypothesized the enhanced thymic deletion of diabetogenic CD8 T-cells elicited by either ablating or elevating Nfkbid expression in NOD mice likely occurs through differing mechanisms. To initially address this possibility, we performed RNA-seq analysis on sorted AI4 DP thymocytes from NOD-AI4, NOD-AI4-Nfkbid−/−, and NOD-AI4-60A mice in order to identify potential differential NF-κB target gene expression patterns (data submitted to GenBank Accession # GSE176566 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=). Compared to NOD-AI4 controls 237 and 526 total genes were differentially expressed (FC>2, FDR≤0.05) in Nfkbid deficient and overexpressing DP thymocytes respectively (Supplemental Tables 1 and 2). As expected, the level of Nfkbid mRNA levels were unchanged between NOD-AI4 and NOD-AI4-Nfkbid−/− samples due to non-productive transcripts emanating from the mutated locus. However, Nfkbid expression was 6.21-fold higher in transgenic samples compared to NOD-AI4. Using ENRICHR analysis (34), we annotated murine NF-κB target genes (Supplemental Table 3) based on Rel, RelA, or NFKB1 consensus binding sites within promoter regions of differentially expressed genes. Forty-one NF-κB target genes were differentially expressed between NOD-AI4-Nfkbid−/− and NOD-AI4 mice, and 84 NF-κB target genes were differentially expressed between NOD-AI4-60A and NOD-AI4 mice (Figure 2C and Supplemental Table 3). Interestingly, ablation or elevation of Nfkbid did not significantly enrich the frequency of differentially expressed genes harboring NF-κB binding sites (analysis not shown). However, the constitution of NF-κB targets within the gene sets shifted (Table 2).
Table II:
Geneset comparison of NF-κB target genes exhibiting differential expression in Nfkbid-ablated or -transgenic DP thymocytes compared to NOD-AI4 controls
| Shared between NOD-AI4-60A or NOD-AI4-Nfkbid−/− vs. NOD-AI4 | Unique to NOD-AI4-60A vs. NOD-AI4 | Unique to NOD-AI4-Nfkbid−/− vs. NOD-AI4 | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||
| Fold Change | ||||||||
| Gene | Transgenic vs NOD-AI4 | Ablated vs NOD-AI4 | Gene | Fold Change | Gene | Fold Change | Gene | Fold Change |
|
|
|
|
|
|||||
| Tgfb1 | 15.771 | 8.257 | Csf2rb | 10.951 | Fscn1 | 2.665 | Dnase1l3 | 4.190 |
| Wdfy4 | 9.071 | 7.193 | Pax5 | 9.950 | Apol7e | 2.650 | Phldb1 | 3.696 |
| Cd74 | 7.741 | 5.108 | Myadm | 9.702 | Ncf1 | 2.632 | Hunk | 3.610 |
| Ccl22 | 4.457 | 2.693 | Casp1 | 7.592 | Rarg | 2.601 | Acan | 2.786 |
| Pde2a | 3.991 | 3.570 | Serpina3g | 7.176 | S1pr1 | 2.456 | Ptpn13 | 2.742 |
| Miuc20 | 3.811 | 3.121 | Spib | 6.527 | B2m | 2.382 | Slc45a3 | 2.598 |
| Fads2 | 3.295 | 2.556 | C3 | 6.523 | Gbp2 | 2.376 | Heyl | 2.441 |
| Mettl1 | 2.986 | 2.585 | Nfkbid | 6.208 | Cfb | 2.351 | Gsto1 | 2.226 |
| Adam19 | 2.668 | 2.204 | Csf1r | 5.738 | Cd72 | 2.341 | Oaf | 2.103 |
| Gpr25 | 2.557 | 2.017 | Ccl5 | 5.703 | Il15ra | 2.301 | Tshz3 | 2.081 |
| Il7r | 2.534 | 3.438 | Cd180 | 5.616 | Psmb9 | 2.179 | Jdp2 | 2.057 |
| Hk2 | 2.190 | 2.256 | Tyrobp | 5.374 | Acsbg1 | 2.144 | Pde8a | 2.018 |
|
|
|
|||||||
| Ifit3 | 4.334 | −3.076 | Pltp | 5.017 | Ccnd2 | 2.121 | Cyp2t4 | −3.050 |
| Bcl3 | 4.219 | −2.430 | Napsa | 4.885 | Chst3 | 2.074 | Avil | −2.326 |
| Ifit1 | 3.370 | −2.717 | Nuak2 | 4.282 | Asph | 2.058 | Stac3 | −2.051 |
|
|
||||||||
| Spry1 | −2.084 | −2.051 | Btg3 | 3.754 | Oas1b | 2.053 | ||
| Serpinf2 | −2.139 | −2.338 | Ddx60 | 3.666 | Tmem176b | 2.019 | ||
| Zfp651 | −2.305 | −2.525 | Rab44 | 3.628 | H2-Q9 | 2.013 | ||
| Gpd1 | −2.400 | −2.107 | Coro2a | 3.553 | Enpp1 | 2.012 | ||
|
|
||||||||
| Dusp1 | −2.576 | −2.274 | Zeb2 | 3.096 | Rsph1 | −2.510 | ||
| Junb | −2.889 | −2.908 | Lilrb3 | 2.934 | Fzd3 | −2.343 | ||
| Klf4 | −3.592 | −3.500 | Mctp2 | 2.921 | Smoc1 | −2.196 | ||
| Ppp1r15a | −4.826 | −4.069 | Dhx58 | 2.899 | Rimkla | −2.170 | ||
| FosB | −9.715 | −4.218 | Ppfia4 | 2.881 | Kl | −2.170 | ||
| Tulp2 | −10.933 | −9.098 | Rassf4 | 2.860 | Rtkn | −2.142 | ||
| Fos | −13.609 | −8.465 | Epsti1 | 2.854 | Camk2n1 | −2.104 | ||
| Bst2 | 2.797 | Odf3b | −2.072 | |||||
| Xaf1 | 2.683 | Shank3 | −2.068 | |||||
| Gm6034 | 2.682 | Tmem88 | −2.006 | |||||
Gene set comparisons identified only 26 NF-κB target genes that were differentially expressed in DP thymocytes from both NOD-AI4-60A and NOD-AI4-Nfkbid−/− mice compared to NOD-AI4 controls (Fig. 2C, and Table 2). Twelve of these genes were upregulated in both strains compared to NOD-AI4 (Table 2). Eleven of these genes were downregulated in both strains compared to NOD-AI4 DP thymocytes (Table 2). Only three genes were expressed in opposing directions resulting from ablating or elevating Nfkbid levels (Ifit3, Bcl3, and Ifit1) (Table 2). Fifty-eight NF-κB target genes were unique to the NOD-AI4–60A versus NOD-AI4 comparison. Fifteen NF-κB target genes were unique to the NOD-AI4-Nfkbid−/− versus NOD-AI4 comparison (Fig. 2C and Table 2). Taken together these data reveal that Nfkbid does not solely function as a positive or negative regulator of NF-κB activity in DP thymocytes undergoing varying negative selection pressure, but rather depending on the quantitative level of this molecule tailors the makeup of NF-κB target gene expression.
Nfkbid overexpression enhances Treg levels
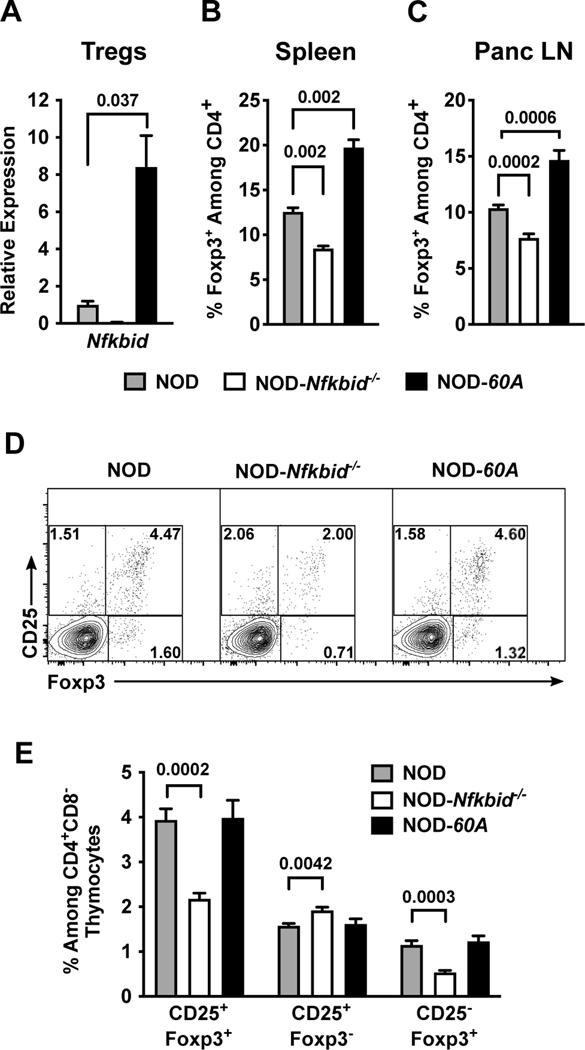
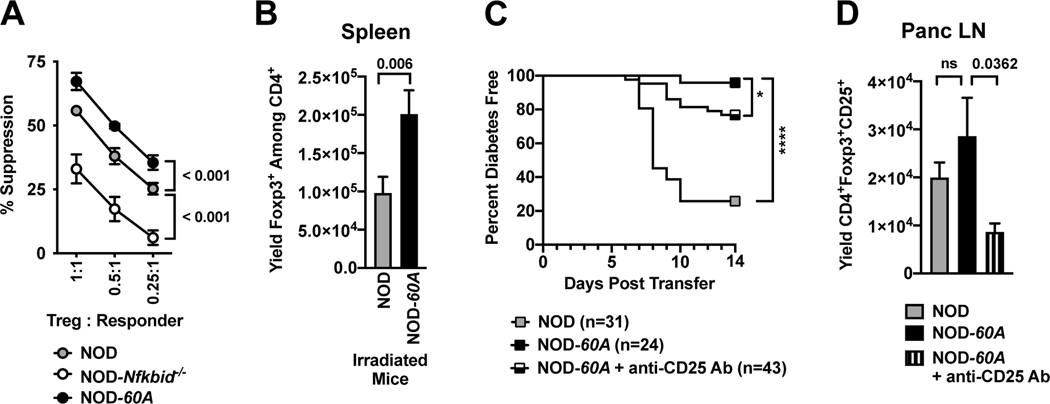
We previously found while Nfkbid deficient NOD mice were characterized by improved thymic deletion of diabetogenic CD8 T-cells, they also unexpectedly developed disease at an accelerated rate (9). Thus, we next evaluated how if both ablation and elevation of Nfkbid expression enhances the thymic deletion of diabetogenic CD8 T-cells, why does only the latter manipulation elicit a disease protective effect. The unexpected T1D acceleration in NOD-Nfkbid−/− mice was previously found to be associated with both a numerical decrease in Tregs and a decrease in the proportion of such cells producing IL-10 (9). Therefore, we examined the Treg compartment in NOD-60A mice for alterations that might additionally explain their striking protection from T1D development beyond the observed negative selection phenotype. Nfkbid mRNA is readily detected in Tregs isolated from NOD spleen and its expression is increased 8.4-fold by the presence of the 60A transgene (Figure 3A). The frequencies of CD4+Foxp3+ Tregs in spleen and Panc LN across the various analyzed NOD mouse models correlates with Nfkbid expression levels. Specifically, lost or elevated expression of Nfkbid respectively resulted in reduced and increased frequencies of Tregs in both the spleen and Panc LN compared to NOD controls (Figure 3B-C). Due to the enlarged spleen size in Nfkbid transgene carriers, the total yield of Tregs per mouse is also increased compared to NOD controls (data not shown).
Figure 3. Nfkbid overexpression increases the peripheral Treg compartment.
(A) Nfkbid mRNA expression was measured by qPCR and normalized to Gapdh expression in CD4+CD25+ Tregs sorted from the spleens of 6–8-week-old female NOD, NOD-Nfkbid−/− and NOD-60A mice (n=3–5 per strain). (B, C) Frequency of Foxp3+ Tregs among CD4 T-cells as measured by flow cytometry in spleens (B) and Panc LN (C) from 6–8-week-old female NOD, NOD-Nfkbid−/− and NOD-60A mice (n=8 per strain). (D, E) Frequency of mature CD25+Foxp3+ Tregs among CD4+ single positive thymocytes from NOD, NOD-Nfkbid−/− and NOD-60A mice and frequency of CD25+Foxp3− and CD25−Foxp3+ developing Tregs as measured by flow cytometry in 6–8-week-old female mice (n=8 per strain). Statistical significance in all panels was determined by Mann-Whitney nonparametric comparison.
In thymus, the frequency of mature CD4+ CD25+ Foxp3+ Tregs was diminished in NOD-Nfkbid−/− mice but were unchanged in the NOD-60A stock (Figure 3D-E). Immature CD25+Foxp3− and CD25−Foxp3+ thymocyte populations may represent two separate pathways for Treg development (36, 37). Ablation of Nfkbid significantly reduces the CD25−Foxp3+ population (Figure 3D-E) consistent with the known contribution of Nfkbid to Foxp3 transcription and Treg development (10, 38). Thus, the increased frequency of peripheral Tregs in NOD-60A mice may culminate from a quicker thymic exit, increased survival and/or conversion from another CD4 T-cell population.
Activated profile of Nfkbid transgenic Tregs
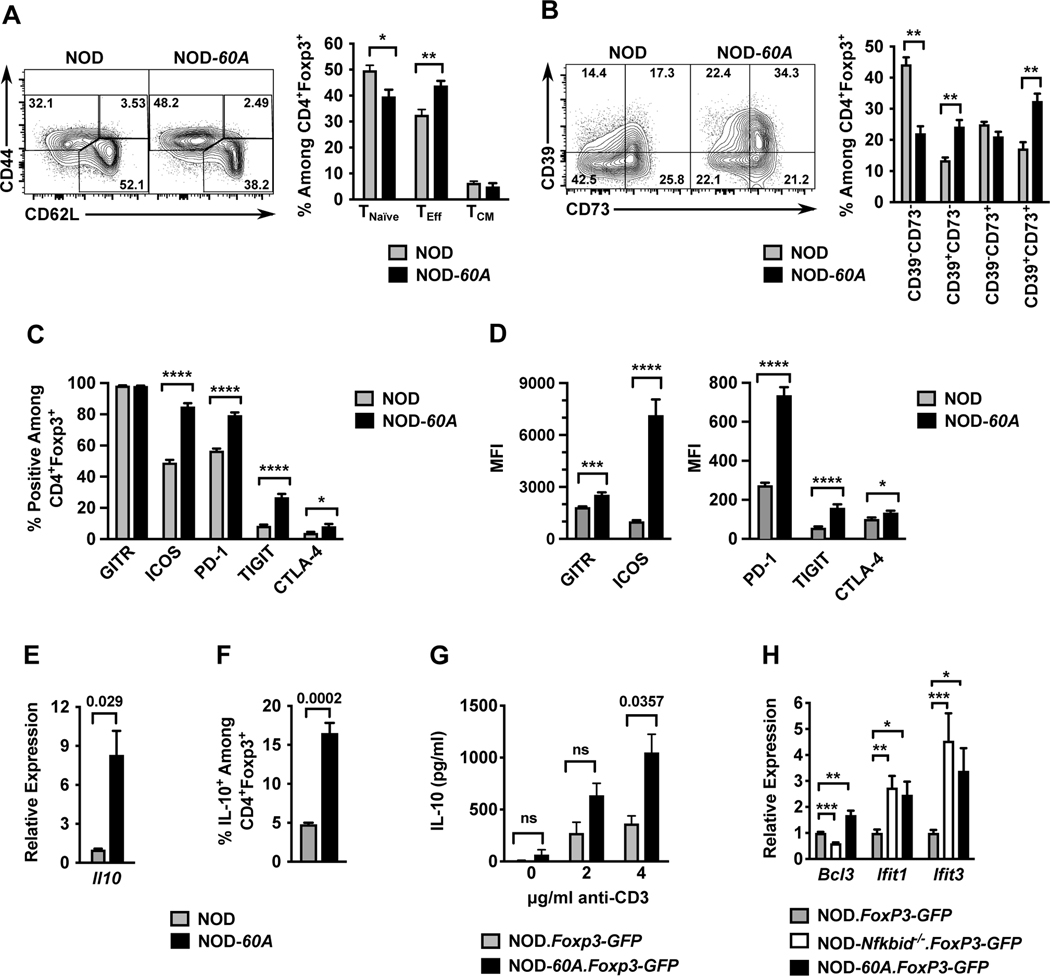
Peripheral Tregs are important for both the suppression of normal T-cell activity following pathogen clearance and the inhibition of those capable of mediating autoimmune responses (39, 40). Tregs use several strategies to manifest their immunosuppressive activity. Activated Tregs express functional markers, such as CD44, to aid migration and survival. Splenic CD4+Foxp3+ Tregs in NOD-60A mice have an increased frequency of CD44hiCD62L− effectors (TEff), and a lower level of naïve CD44loCD62L+ T-cells (Figure 4A). The CD44hiCD62L+ central memory (TCM) fraction was unchanged compared to NOD Tregs (Figure 4A). Upon activation, Tregs boost their ability to metabolize extracellular ATP released by cellular conversion to immunosuppressive adenosine (41). Therefore, we examined the expression of the ATP metabolizing transmembrane receptors CD39 (ATP→AMP) and CD73 (AMP→adenosine) in NOD control and NOD-60A transgenic Tregs. A higher proportion of Tregs from NOD-60A mice co-express CD39 and CD73 than in standard NOD controls (Figure 4B). Costimulatory and inhibitory accessory receptor molecules (e.g., GITR, ICOS PD-1, TIGIT, CTLA-4) modify TCR signal strength to summarily determine Treg reaction to MHC class II engagement on APCs. (39, 40, 42). We evaluated a panel of such receptors on Tregs from NOD control and NOD-60A mice to determine if Nfkbid overexpression may influence Treg signaling through altered coreceptor expression. As shown in Figure 4C-D, nearly all Tregs from NOD and NOD-60A mice express GITR. However, the median fluorescent intensity (MFI) of GITR antibody staining was higher on Tregs from NOD-60A mice. Further, NOD-60A Tregs showed a greatly increased percentage of ICOS+ cells and a 7-fold increase in the MFI of ICOS antibody staining (Figure 4C-D). Inhibitory molecules are upregulated after TCR signaling and can indicate the activation state of the cell. The percentage of PD-1+ Tregs and their expression level of this molecule were also increased in NOD-60A compared to standard NOD mice (Figure 4C-D). NOD-60A mice were also characterized by an approximately three times higher frequency of Tregs that were TIGIT+, with the expression level of this molecule also increased (Figure 4C-D). Increases in CTLA-4 expression were also noted. (Figure 4C-D). Taken together, these combined data indicates that NOD-60A Tregs have a more activated phenotype than their NOD counterparts.
Figure 4. Nfkbid overexpression imparts an activated cellular profile to Tregs.
Splenocytes from 6–8-week-old female standard NOD and NOD-60A mice were analyzed by multicolor flow cytometry for expression of markers of activation among CD4+Foxp3+ Tregs. (A) CD44 and CD62L staining was performed on CD4+ Foxp3+ Tregs to characterize activation profile by delineating CD44loCD62L+ naïve T-cells (TNaïve), CD44hiCD62L+ (TCM) and CD44hiCD62L− (TEff) populations. n=6–7 per group. Gating (left) and quantification (right). (B) Splenocytes were stained for CD39 and CD73 expression. Gating (left) and frequency (right) of CD39 CD73 double negative, single positive and DP populations among CD4+Foxp3+ cells are shown. n=6–7 per group. (C, D) Splenocytes (n=10 per group) were stained for GITR, ICOS, PD-1, TIGIT and CTLA-4. Frequencies of CD4+ Foxp3+ Tregs showing positive staining are shown (C) with median fluorescence intensities (MFI) (D) for these stains. (E) Il10 mRNA levels were measured by qPCR of RNA isolated from sorted CD4+CD25+ splenic Tregs and normalized to Gapdh (n=4 biological replicates per group). (F). Frequency of IL-10+ cells among CD4+Foxp3+ Tregs measured from splenocyte cultures stimulated for 5 hours with cell stimulation cocktail and monensin. (G). IL-10 secretion was measured by ELISA in culture supernatants of sorted CD4+Foxp3+ Tregs stimulated with varied amounts of anti-CD3 for 3 days (n=3–5 per strain). (H) Relative expression of Bcl3, Ifit1, and Ifit3 RNA transcripts in splenic CD4+ Foxp3-GFP+ Tregs from NOD-Nfkbid−/−.Foxp3-GFP and NOD-60A.Foxp3-GFP mice compared to those from NOD.Foxp3-GFP controls (n= 6–8 per strain). All P-values were determined by Mann-Whitney nonparametric comparison (*=p< 0.05; **=p< 0.01; ***=p< 001; ****=p< 0001).
One mechanism by which Tregs mediate immunosuppression is through the secretion of IL-10 (39). Nfkbid has been shown to bind the Il10 locus in Th17 cells (31). Therefore, we examined the effects of elevated Nfkbid expression on Il10 mRNA levels and protein abundance within and secretion from Tregs. Compared to those from NOD controls, Tregs isolated from Nfkbid overexpressing mice have greatly elevated Il10 mRNA expression, an increase in the proportion expressing IL-10, and upon activation with anti-CD3, increased secretion of this cytokine (Figure 4E-G). As noted earlier, only the Bcl3, Ifit1, and Ifit3 NF-κB target genes exhibited opposite direction expression changes in NOD DP thymocytes as a result of elevating or ablating Nfkbid levels. Of potential interest only Bcl3 exhibited a similar opposite direction expression pattern in NOD Tregs as a result of elevating or ablating Nfkbid levels (Fig. 4H).
Elevated Nfkbid expression increases suppressive capacity of Tregs that are at least partially responsible for T1D resistance in NOD-60A mice
Previous reports have shown on a per cell basis Tregs from Nfkbid deficient and intact B6 mice have an equivalent suppressive capacity (10). However, the increased activation profile of Tregs from NOD-60A mice suggested they had potential capacity to exert an enhanced per cell suppressive activity. CD4+CD25+ Tregs were purified from the spleens of NOD, NOD-Nfkbid−/− and NOD-60A mice and plated in varying ratios with CD4+CD25− responder T-cells purified from NOD mice labeled with cell proliferation dye. On a per cell basis Tregs from Nfkbid knockout NOD mice were significantly less able to inhibit responder T-cell proliferation (Figure 5A). Conversely, Tregs overexpressing Nfkbid demonstrated superior suppressive capacity (Figure 5A). These collective data indicated T1D protection in NOD-60A mice could be due, at least in part, to their increased numbers and activity of Tregs.
Figure 5. Elevated Nfkbid expression increases suppressive capacity of Tregs that are at least partially responsible for T1D resistance in NOD-60A mice.
(A) The capacity of CD4+CD25+ Tregs expressing varied levels of Nfkbid (standard NOD, NOD-Nfkbid−/− and NOD-60A, n=4 for each strain) to suppress T-cell proliferation was measured in cultures containing 5×104 CD4+CD25− NOD responder T-cells labeled with cell proliferation dye, 2.5×105 APC from NOD.scid mice and 20μg/ml CD-3 antibody. (B) At four days after sublethal irradiation (600 cGy) splenic Treg levels were 99% decreased in both NOD and NOD-60A mice compared to non-irradiated strain-matched controls (data not shown) but remained higher in the latter strain (n=13 for both strains). (C) Rag1null.AI4 CD8 T-cells transfer T1D at a significantly higher level to NOD control than 60A mice, and Treg depletion by i.p. injection with 62.5μg of the CD25 specific PC61 Ab partially abrogates disease protection in the latter strain. T1D developmental comparisons were analyzed by log-rank (Mantel-Cox) test. (D) Treg levels in Panc LNs at 14 days following sub-lethal irradiation in AI4 T-cell infused NOD (n=12) or NOD-60A mice that did (n=15) or did not (n=11) also receive a single anti-CD25 antibody injection. For A, B and D, statistical comparisons determined by Mann-Whitney nonparametric comparison.
We tested the capacity of Nfkbid overexpressing Tregs to limit T1D development by comparing the ability of engrafted TCR transgenic NOD.Rag1null. AI4 CD8 T-cells to induce disease in sub-lethally irradiated (600 cGy) NOD and NOD-60A recipients. It was previously found the ability of AI4 T-cells to induce T1D in sub-lethally irradiated NOD recipients was complete by 14 days post-transfer (43). Albeit lower than in untreated controls, at four days following sub-lethal irradiation, splenic Treg numbers remained greater in NOD-60A than NOD mice (Figure 5B). Thus, a subset of NOD-60A mice infused with AI4 T-cells were also treated with a single 62.5μg injection of the CD25 specific PC61 antibody to further diminish their Treg levels. Over a 14-day follow-up period transferred AI4 T-cells induced T1D in 74% (23/31) of NOD, but only 4% (1/24) of NOD-60A recipients (Figure 5C). Such protection against AI4 T-cell transferred T1D was significantly, but not completely abrogated in NOD-60A recipients treated with anti-CD25 (Figure 5C). An evaluation at the end of the study revealed anti-CD25 treatment did result in lower Treg levels in Panc LNs from NOD-60A recipients of AI4 T-cells (Figure 5D). Thus, T1D resistance in NOD-60A mice can be attributed at least in part to their elevated numbers and/or enhanced suppressive activity of Tregs. In addition, these AI4 transfer studies reveal that elevated Nfkbid expression may also contribute to complete T1D protection in NOD-60A mice through effects in cell types beyond Tregs and the previously described improved negative selection of pathogenic CD8 T-cells.
Discussion
T1D originates with the evasion of autoreactive T-cells from thymic negative selection during their initial development, in addition to a diminished ability to activate peripheral Treg mediated immunoregulatory mechanisms that normally limit the activation of such pathogenic effectors. Recent strategies to combat T1D include cellular Treg-based approaches to restrain the activity of autoreactive T-cells (44–46). Treg-based therapy for T1D faces many hurdles including the difficulty culturing sufficient numbers of such cells for reinfusion and the generation of appropriate antigen specific populations. Here we report that genetic elevation of Nfkbid levels completely inhibits T1D in NOD-60A mice through mechanisms at least partly entailing enhanced thymic deletion of pathogenic CD8 T-cells and numeric and functional increases in Tregs. Given that the protein coding sequence of Nfkbid is highly conserved between mice and humans (35), similar pathways may be relevant for T1D in humans and may advance the development of Treg-based therapies for T1D and other autoimmune disorders. Of potential significance, we mined data from a recent meta-analysis and found three single nucleotide polymorphisms (SNPs) residing in possible regulatory regions of the human NFKBID gene (rs181302331, rs75062784, and rs73590895) that at a significance level of p<0.005 may contribute to T1D susceptibility or resistance (47).
Using the combined approaches of congenic mapping, mRNA transcript profiling, and CRISPR/Cas9 gene editing in NOD-AI4 TCR transgenic mice we previously identified the NF-κB signaling modifier Nfkbid as an Idd7 localized allelically varying gene contributing to the level diabetogenic CD8 T-cells undergo thymic negative selection (9). The NOD Nfkbid allele was found to be expressed at higher levels in thymocytes than that characterizing T1D resistant B6 mice. Thus, it was unexpected that ablation of Nfkbid in NOD mice resulted in accelerated T1D development (9). Importantly however, Nfkbid ablation also produced deficiencies within the Treg compartment, presumably allowing surviving autoreactive effectors to act unimpeded (9). We will also note at this point that the NOD Idd7 locus effect was originally reported to contribute to T1D resistance rather than susceptibility (48). However, this was in the context of an outcross of NOD with the NON, rather than the B6 control strain. Our current study indicates Nfkbid alleles are likely responsible for an Idd7 locus effect with the NOD and B6 variants relatively contributing to T1D susceptibility and resistance. Here we report elevation of Nfkbid expression in NOD mice through a genomic transgene approach results in complete T1D protection characterized by a moderate level of insulitis present at 10 weeks of age that fails to subsequently progress to the level required for clinical disease onset.
Interestingly, Nfkbid overexpression elicited by the 60A transgene in NOD-AI4 mice results in a higher level of autoreactive CD8 T-cells succumbing to thymic negative selection compared to both NOD-AI4-Nfkbid−/− mice and NOD-AI4 controls. RNAseq analysis revealed three classes of NF-κB target genes affected by ablating or increasing Nfkbid expression levels in DP AI4 thymocytes (Table 2). The first of these was shared same direction expression changes compared to NOD-AI4. Another was gene expression changes uniquely resulting from ablation or elevation of Nfkbid levels. Differentially expressed NF-κB target genes that are shared between both NOD-AI4-60A and NOD-AI4-Nfkbid−/− mice (Table 2) may be responsible for the improved negative selection observed in both strains compared to NOD controls. Finally, and potentially most importantly, there were three genes, Bcl3, Ifit1, and Ifit3 respectively exhibiting increased or decreased expression in DP thymocytes as a result of elevating or ablating Nfkbid levels. Of these three genes only Bcl3 exhibited similar opposite direction expression effects in Tregs as a result of elevating or ablating Nfkbid levels. It has been previously reported that Bcl3 ablation accelerates T1D development in NOD mice (49). For this reason, their diminished Bcl3 expression could also be a contributor to accelerated T1D development in NOD-Nfkbid−/− mice. Conversely, overexpression of Bcl3 specifically within B6 background T-cells has been previously reported to decrease Treg levels and function (50). However, whether this Bcl3 overexpression effect on Tregs in B6 mice results from decreased levels and/or activity of Nfkbid is unknown. Thus, allelic variants of Bcl3 currently represent a prime candidate through which ablation or elevation of Nfkbid expression respectively accelerates and inhibits T1D development in NOD mice. We will address this possibility in future studies.
Our current study indicates increased numbers and/or activity of Tregs at least in part contributes to the halted insulitis progression and complete T1D resistance characterizing NOD-60A mice with elevated Nfkbid expression. We also found mice lacking Nfkbid show reduced Tregs in the thymus, as well as in secondary lymphoid organs such as spleen and Panc LN. Ablation of Nfkbid impairs Treg development as evidenced by increased levels of CD25+Foxp3− cells, decreased CD25−Foxp3+ cells and fewer mature CD25+Foxp3+ Tregs in the thymus. This is in accordance with the role of Nfkbid in supporting Foxp3 gene expression (10). Interestingly, complimentary changes are not seen in developing Tregs in NOD-60A mice. This indicates Nfkbid overexpression does not alter thymic Treg development. However, it remains possible Nfkbid overexpression may increase the speed by which Tregs traverse the thymus and emigrate to the periphery where their levels in NOD-60A mice are increased in both spleen and Panc LN compared to standard NOD controls. While it is still unknown if the higher frequency of Tregs in NOD-60A mice results from their enhanced survival and/or seeding the periphery, or increased conversion from other naïve T-cells, they exhibit numerous advantageous molecular changes resulting in increased functional capacity.
Peripheral Tregs from NOD-60A mice display altered proportions of naïve (CD44loCD62L+), activated TEff (CD44hiCD62L−) and TCM fractions (CD44hiCD62L+) compared to those from NOD controls. These findings indicate a shift towards increased active Tregs in T1D resistant NOD-60A mice. Intriguingly, increased frequencies of CD39+, CD73+, ICOS+, PD-1+, TIGIT+, CTLA-4+ and IL-10+ cells further support a more activated profile of Tregs in NOD-60A mice than those in NOD controls. Furthermore, Nfkbid overexpressing Tregs from NOD-60A mice make 8.3-fold higher levels of Il10 mRNA and secreted higher levels of this immunosuppressive cytokine in response to anti-CD3 treatment compared to those of NOD control origin, in line with previous data showing Il10 is a direct target of Nfkbid (31). The increased activated profile and elevated levels of NOD-60A Tregs likely contribute to the higher capacity of these cells to suppress the proliferation of CD4+CD25− effector T-cells on a per cell basis, and their at least partial contribution in eliciting T1D resistance in this strain.
While to date NOD-60A mice have proved to be completely T1D free, we have found that at best purified splenic T-cells from this strain transfer disease to NOD.scid recipients at only a marginally lower rate than those from NOD controls (Supplemental Figure 1). There is a known role of Nfkbid in supporting T-cell proliferation and secretion of IL-2 in response to TCR stimulation (13). Thus, within the lymphopenic environment of NOD.scid mice, the homeostatic expansion of autoreactive T-cells may outpace the ability of Tregs also present in the transfer inoculum to suppress such effectors potentially accounting for why T1D development was not efficiently inhibited in these recipients of purified NOD-60A T-cells. The ability of purified T-cells from NOD-60A mice to transfer T1D to at least a proportion of NOD.scid recipients also suggests complete disease protection in the donors may be due to elevated Nfkbid expression in other cell types. One possibility is complete T1D resistance in NOD-60A mice is at least in part due to elevated Nfkbid levels in myeloid cells enhancing their ability to serve as APC increasing Treg numbers and/or activity in this strain. In this scenario myeloid cells in NOD.scid recipients would express Nfkbid at wild type NOD levels and thus have a lesser capacity to serve as APC for Tregs present in the transferred NOD-60A T-cell inoculum. It is also possible elevated Nfkbid expression levels diminish the ability of myeloid APCs to activate diabetogenic T-cells. The above discussed possibilities will be assessed in deeper detail in future studies.
Transfer studies utilizing AI4-induced T1D indicate Tregs have at least a partial role in Nfkbid-mediated T1D protection. This was demonstrated by the finding that infusion of diabetogenic AI4 cells into sub-lethally irradiated NOD-60A recipients results in near complete T1D protection compared to NOD control recipients. This protection is reduced with anti-CD25 antibody mediated diminution of Tregs in NOD-60A recipients. However, the partial loss - as opposed to complete reversal - of T1D protection with Treg depletion also argues additional factors, such as other cell types expressing Nfkbid, likely contribute to disease resistance. It is also possible that anti-CD25 treatment decreased levels of highly activated diabetogenic effector T-cells thus limiting the extent disease resistance was broken in the NOD-60A recipients.
The NF-κB activity pathways govern a wide range of cellular functions across many cell types (18). However, Nfkbid expression is limited primarily within immune cells in both mice and humans (BioGPS, accessed March 22, 2022). This indicates Nfkbid may be a key regulator of NF-κB function within the immune system. For this reason, identifying Nfkbid-dependent transcriptional targets in Tregs and/or other immune cell populations may provide further understanding of the pathogenic basis for T1D development. Our studies also indicate elevating Nfkbid levels may inhibit T1D development in NOD mice through a mechanism involving elevating Bcl3 expression in developing effector T-cells and/or Tregs. Thus, finding a means to pharmacologically enhance Nfkbid and/or Bcl3 expression could ultimately provide a clinically effective T1D intervention approach.
Supplementary Material
Key points.
Elevating expression of the NF-κB regulatory Nfkbid gene inhibits T1D in NOD mice.
Elevated Nfkbid levels enhances thymic deletion of diabetogenic CD8 T-cells.
Elevating Nfkbid increases Treg numbers and activity partly inhibiting T1D.
Acknowledgments
We would like to thank the staff at The Jackson Laboratory’s Flow Cytometry service, Research Animal Facility, Genetic Engineering Technologies and Genome Technology groups for technical support. We thank the NIH Tetramer Facility for providing AI4 tetramer. We are also grateful for a generous donation from Carl Stewie and his wife Maike Rohde toward T1D research at The Jackson Laboratory.
4. Abbreviations:
- B6
C57BL/6J
- Chr.
chromosome
- TCM
central memory T-cell
- CRISPR
clustered regularly interspaced short palindromic repeats
- Cas9
CRISPR associated protein 9
- DP
double positive
- TEff
effector T-cell
- MIS
Mean Insulitis Score
- MFI
median fluorescent intensity
- TNaïve
naïve T-cell
- NOD
NOD/ShiLtDvs
- NOD.Foxp3-GFP
NOD.129X1(Cg)-Foxp3tm2Tch/DvsJ
- NOD.Nfkbid−/−
NOD/ShiLtDvs-Nfkbidem3Dvs/Dvs
- NOD-60A
NOD/ShiLtDvs-Nfkbidem3Dvs Tg(Nfkbid)60–2Dvs/Dvs
- NOD.Rag1null. AI4
NOD.Cg-Rag1tmMom Tg(TcraAI4)1Dvs Tg(TcrbAI4)1Dvs/Dvs
- NOD-AI4
NOD/ShiLtDvs Tg(TcraAI4)1Dvs Tg(TcrbAI4)1Dvs/Dvs
- NOD-AI4-Nfkbid−/−
NOD/ShiLtDvs-Nfkbidem3Dvs Tg(TcraAI4)1Dvs Tg(TcrbAI4)1Dvs/Dvs
- NOD-AI4–60A
NOD/ShiLtDvs-Nfkbidem3Dvs Tg(Nfkbid)60–2Dvs Tg(TcraAI4)1Dvs Tg(TcrbAI4)1Dvs/Dvs
- NOD-Nfkbid−/−.Foxp3-GFP
NOD.Cg-Nfkbidem3Dvs Foxp3tm2Tch/Dvs
- NOD-60A.Foxp3-GFP
NOD.Cg-Nfkbidem3Dvs Foxp3tm2Tch Tg(Nfkbid)60–2Dvs/Dvs
- Panc LN
pancreatic lymph node
- Treg
regulatory T-cell
- T1D
type 1 diabetes
Footnotes
JRD was supported by NIDDK Supplemental Funding to Promote Re-Entry into Biomedical and Behavioral Research Supplement to DK-46266. JJR was supported for parts of this work by JDRF Fellowship 3-PDF-2017–372-A-N. IS was supported by grants of the German Research Council (project A23 within CRC854 and SCHM1586/6–2). DVS was supported by NIH grants DK-46266, DK-95735, and OD-5U4OD020351, Juvenile Diabetes Research Foundation grant 2018–568 as well as by Mark Foundation Grant 21–010-PPM. This work was also partly supported by Cancer Center Support Grant CA34196.
JRD designed and conducted experimentation, analyzed and interpreted data, and wrote the manuscript. JJR designed and conducted experimentation, analyzed and interpreted data, and contributed to the writing of the manuscript. HDC designed and conducted experimentation. AQ conducted experimentation. MP designed and conducted experimentation, analyzed and interpreted data. GS analyzed differential gene expression and performed NF-κB target gene analysis. IS contributed to study conception. DVS contributed to study conception and supervised experimental efforts and the writing of the manuscript.
References
- 1.Onengut-Gumuscu S, Chen WM, Burren O, Cooper NJ, Quinlan AR, Mychaleckyj JC, Farber E, Bonnie JK, Szpak M, Schofield E, Achuthan P, Guo H, Fortune MD, Stevens H, Walker NM, Ward LD, Kundaje A, Kellis M, Daly MJ, Barrett JC, Cooper JD, Deloukas P, C. Type 1 Diabetes Genetics, Todd JA, Wallace C, Concannon P, and Rich SS. 2015. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet 47: 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pociot F, Akolkar B, Concannon P, Erlich HA, Julier C, Morahan G, Nierras CR, Todd JA, Rich SS, and Nerup J. 2010. Genetics of type 1 diabetes: what’s next? Diabetes 59: 1561–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark M, Kroger CJ, Ke Q, and Tisch RM. 2020. The Role of T Cell Receptor Signaling in the Development of Type 1 Diabetes. Front Immunol 11: 615371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gomez-Tourino I, Arif S, Eichmann M, and Peakman M. 2016. T cells in type 1 diabetes: Instructors, regulators and effectors: A comprehensive review. J Autoimmun 66: 7–16. [DOI] [PubMed] [Google Scholar]
- 5.Varanasi V, Avanesyan L, Schumann DM, and Chervonsky AV. 2012. Cytotoxic mechanisms employed by mouse T cells to destroy pancreatic beta-cells. Diabetes 61: 2862–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azoury ME, Tarayrah M, Afonso G, Pais A, Colli ML, Maillard C, Lavaud C, Alexandre-Heymann L, Gonzalez-Duque S, Verdier Y, Vinh J, Pinto S, Buus S, Dubois-Laforgue D, Larger E, Beressi JP, Bruno G, Eizirik DL, You S, and Mallone R. 2020. Peptides Derived From Insulin Granule Proteins Are Targeted by CD8(+) T Cells Across MHC Class I Restrictions in Humans and NOD Mice. Diabetes 69: 2678–2690. [DOI] [PubMed] [Google Scholar]
- 7.Knight RR, Kronenberg D, Zhao M, Huang GC, Eichmann M, Bulek A, Wooldridge L, Cole DK, Sewell AK, Peakman M, and Skowera A. 2013. Human beta-cell killing by autoreactive preproinsulin-specific CD8 T cells is predominantly granule-mediated with the potency dependent upon T-cell receptor avidity. Diabetes 62: 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atkinson MA, Eisenbarth GS, and Michels AW. 2014. Type 1 diabetes. Lancet 383: 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Presa M, Racine JJ, Dwyer JR, Lamont DJ, Ratiu JJ, Sarsani VK, Chen YG, Geurts A, Schmitz I, Stearns T, Allocco J, Chapman HD, and Serreze DV. 2018. A Hypermorphic Nfkbid Allele Contributes to Impaired Thymic Deletion of Autoreactive Diabetogenic CD8(+) T Cells in NOD Mice. J Immunol 201: 1907–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuster M, Glauben R, Plaza-Sirvent C, Schreiber L, Annemann M, Floess S, Kuhl AA, Clayton LK, Sparwasser T, Schulze-Osthoff K, Pfeffer K, Huehn J, Siegmund B, and Schmitz I. 2012. IkappaB(NS) protein mediates regulatory T cell development via induction of the Foxp3 transcription factor. Immunity 37: 998–1008. [DOI] [PubMed] [Google Scholar]
- 11.Annemann M, Plaza-Sirvent C, Schuster M, Katsoulis-Dimitriou K, Kliche S, Schraven B, and Schmitz I. 2016. Atypical IkappaB proteins in immune cell differentiation and function. Immunol Lett 171: 26–35. [DOI] [PubMed] [Google Scholar]
- 12.Frentzel S, Katsoulis-Dimitriou K, Jeron A, Schmitz I, and Bruder D. 2019. Essential role of IkappaBNS for in vivo CD4(+) T-cell activation, proliferation, and Th1-cell differentiation during Listeria monocytogenes infection in mice. Eur J Immunol 49: 1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Touma M, Antonini V, Kumar M, Osborn SL, Bobenchik AM, Keskin DB, Connolly JE, Grusby MJ, Reinherz EL, and Clayton LK. 2007. Functional role for I kappa BNS in T cell cytokine regulation as revealed by targeted gene disruption. J Immunol 179: 1681–1692. [DOI] [PubMed] [Google Scholar]
- 14.Schuster M, Annemann M, Plaza-Sirvent C, and Schmitz I. 2013. Atypical IkappaB proteins - nuclear modulators of NF-kappaB signaling. Cell Commun Signal 11: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirotani T, Lee PY, Kuwata H, Yamamoto M, Matsumoto M, Kawase I, Akira S, and Takeda K. 2005. The nuclear IkappaB protein IkappaBNS selectively inhibits lipopolysaccharide-induced IL-6 production in macrophages of the colonic lamina propria. J Immunol 174: 3650–3657. [DOI] [PubMed] [Google Scholar]
- 16.Kuwata H, Matsumoto M, Atarashi K, Morishita H, Hirotani T, Koga R, and Takeda K. 2006. IkappaBNS inhibits induction of a subset of Toll-like receptor-dependent genes and limits inflammation. Immunity 24: 41–51. [DOI] [PubMed] [Google Scholar]
- 17.Miura M, Hasegawa N, Noguchi M, Sugimoto K, and Touma M. 2016. The atypical IkappaB protein IkappaB(NS) is important for Toll-like receptor-induced interleukin-10 production in B cells. Immunology 147: 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerondakis S, Fulford TS, Messina NL, and Grumont RJ. 2014. NF-kappaB control of T cell development. Nat Immunol 15: 15–25. [DOI] [PubMed] [Google Scholar]
- 19.Dhar A, Chawla M, Chattopadhyay S, Oswal N, Umar D, Gupta S, Bal V, Rath S, George A, Arimbasseri GA, and Basak S. 2019. Role of NF-kappaB2-p100 in regulatory T cell homeostasis and activation. Sci Rep 9: 13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grinberg-Bleyer Y, Caron R, Seeley JJ, De Silva NS, Schindler CW, Hayden MS, Klein U, and Ghosh S. 2018. The Alternative NF-kappaB Pathway in Regulatory T Cell Homeostasis and Suppressive Function. J Immunol 200: 2362–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh H, Grinberg-Bleyer Y, Liao W, Maloney D, Wang P, Wu Z, Wang J, Bhatt DM, Heise N, Schmid RM, Hayden MS, Klein U, Rabadan R, and Ghosh S. 2017. An NF-kappaB Transcription-Factor-Dependent Lineage-Specific Transcriptional Program Promotes Regulatory T Cell Identity and Function. Immunity 47: 450–465 e455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Messina N, Fulford T, O’Reilly L, Loh WX, Motyer JM, Ellis D, McLean C, Naeem H, Lin A, Gugasyan R, Slattery RM, Grumont RJ, and Gerondakis S. 2016. The NF-kappaB transcription factor RelA is required for the tolerogenic function of Foxp3(+) regulatory T cells. J Autoimmun 70: 52–62. [DOI] [PubMed] [Google Scholar]
- 23.Simecek P, Churchill GA, Yang H, Rowe LB, Herberg L, Serreze DV, and Leiter EH. 2015. Genetic Analysis of Substrain Divergence in Non-Obese Diabetic (NOD) Mice. G3 (Bethesda) 5: 771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serreze DV, Leiter EH, Hanson MS, Christianson SW, Shultz LD, Hesselton RM, and Greiner DL. 1995. Emv30null NOD-scid mice. An improved host for adoptive transfer of autoimmune diabetes and growth of human lymphohematopoietic cells. Diabetes 44: 1392–1398. [DOI] [PubMed] [Google Scholar]
- 25.Haribhai D, Lin W, Relland LM, Truong N, Williams CB, and Chatila TA. 2007. Regulatory T cells dynamically control the primary immune response to foreign antigen. J Immunol 178: 2961–2972. [DOI] [PubMed] [Google Scholar]
- 26.Presa M, Chen YG, Grier AE, Leiter EH, Brehm MA, Greiner DL, Shultz LD, and Serreze DV. 2015. The Presence and Preferential Activation of Regulatory T Cells Diminish Adoptive Transfer of Autoimmune Diabetes by Polyclonal Nonobese Diabetic (NOD) T Cell Effectors into NSG versus NOD-scid Mice. J Immunol 195: 3011–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graser RT, DiLorenzo TP, Wang F, Christianson GJ, Chapman HD, Roopenian DC, Nathenson SG, and Serreze DV. 2000. Identification of a CD8 T cell that can independently mediate autoimmune diabetes development in the complete absence of CD4 T cell helper functions. J Immunol 164: 3913–3918. [DOI] [PubMed] [Google Scholar]
- 28.DiLorenzo TP, Lieberman SM, Takaki T, Honda S, Chapman HD, Santamaria P, Serreze DV, and Nathenson SG. 2002. During the early prediabetic period in NOD mice, the pathogenic CD8(+) T-cell population comprises multiple antigenic specificities. Clinical immunology 105: 332–341. [DOI] [PubMed] [Google Scholar]
- 29.Julius MH, and Herzenberg LA. 1974. Isolation of antigen-binding cells from unprimed mice: demonstration of antibody-forming cell precursor activity and correlation between precursor and secreted antibody avidities. The Journal of experimental medicine 140: 904–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xing Y, Wang X, Jameson SC, and Hogquist KA. 2016. Late stages of T cell maturation in the thymus involve NF-kappaB and tonic type I interferon signaling. Nat Immunol 17: 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Annemann M, Wang Z, Plaza-Sirvent C, Glauben R, Schuster M, Ewald Sander F, Mamareli P, Kuhl AA, Siegmund B, Lochner M, and Schmitz I. 2015. IkappaBNS regulates murine Th17 differentiation during gut inflammation and infection. J Immunol 194: 2888–2898. [DOI] [PubMed] [Google Scholar]
- 32.Johnson EA, Silveira P, Chapman HD, Leiter EH, and Serreze DV. 2001. Inhibition of autoimmune diabetes in nonobese diabetic mice by transgenic restoration of H2-E MHC class II expression: additive, but unequal, involvement of multiple APC subtypes. J Immunol 167: 2404–2410. [DOI] [PubMed] [Google Scholar]
- 33.Raghupathy N, Choi K, Vincent MJ, Beane GL, Sheppard KS, Munger SC, Korstanje R, Pardo-Manual de Villena F, and Churchill GA. 2018. Hierarchical analysis of RNA-seq reads improves the accuracy of allele-specific expression. Bioinformatics 34: 2177–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, Jeon M, and Ma’ayan A. 2021. Gene Set Knowledge Discovery with Enrichr. Curr Protoc 1: e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, Fernandez Banet J, Billis K, Garcia Giron C, Hourlier T, Howe K, Kahari A, Kokocinski F, Martin FJ, Murphy DN, Nag R, Ruffier M, Schuster M, Tang YA, Vogel JH, White S, Zadissa A, Flicek P, and Searle SM. 2016. The Ensembl gene annotation system. Database (Oxford) 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owen DL, Mahmud SA, Sjaastad LE, Williams JB, Spanier JA, Simeonov DR, Ruscher R, Huang W, Proekt I, Miller CN, Hekim C, Jeschke JC, Aggarwal P, Broeckel U, LaRue RS, Henzler CM, Alegre ML, Anderson MS, August A, Marson A, Zheng Y, Williams CB, and Farrar MA. 2019. Thymic regulatory T cells arise via two distinct developmental programs. Nat Immunol 20: 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owen DL, Sjaastad LE, and Farrar MA. 2019. Regulatory T Cell Development in the Thymus. J Immunol 203: 2031–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuster M, Plaza-Sirvent C, Visekruna A, Huehn J, and Schmitz I. 2019. Generation of Foxp3(+)CD25(−) Regulatory T-Cell Precursors Requires c-Rel and IkappaBNS. Front Immunol 10: 1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vignali DA, Collison LW, and Workman CJ. 2008. How regulatory T cells work. Nat Rev Immunol 8: 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar P, Bhattacharya P, and Prabhakar BS. 2018. A comprehensive review on the role of co-signaling receptors and Treg homeostasis in autoimmunity and tumor immunity. J Autoimmun 95: 77–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antonioli L, Pacher P, Vizi ES, and Hasko G. 2013. CD39 and CD73 in immunity and inflammation. Trends Mol Med 19: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee DJ 2020. The relationship between TIGIT(+) regulatory T cells and autoimmune disease. Int Immunopharmacol 83: 106378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Driver JP, Racine JJ, Ye C, Lamont DJ, Newby BN, Leeth CM, Chapman HD, Brusko TM, Chen YG, Mathews CE, and Serreze DV. 2017. Interferon-gamma Limits Diabetogenic CD8(+) T-Cell Effector Responses in Type 1 Diabetes. Diabetes 66: 710–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bluestone JA, Buckner JH, Fitch M, Gitelman SE, Gupta S, Hellerstein MK, Herold KC, Lares A, Lee MR, Li K, Liu W, Long SA, Masiello LM, Nguyen V, Putnam AL, Rieck M, Sayre PH, and Tang Q. 2015. Type 1 diabetes immunotherapy using polyclonal regulatory T cells. Sci Transl Med 7: 315ra189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferreira LMR, Muller YD, Bluestone JA, and Tang Q. 2019. Next-generation regulatory T cell therapy. Nat Rev Drug Discov 18: 749–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perdigoto AL, Chatenoud L, Bluestone JA, and Herold KC. 2015. Inducing and Administering Tregs to Treat Human Disease. Front Immunol 6: 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiou J, Geusz RJ, Okino ML, Han JY, Miller M, Melton R, Beebe E, Benaglio P, Huang S, Korgaonkar K, Heller S, Kleger A, Preissl S, Gorkin DU, Sander M, and Gaulton KJ. 2021. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature 594: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McAleer MA, Reifsnyder P, Palmer SM, Prochazka M, Love JM, Copeman JB, Powell EE, Rodrigues NR, Prins JB, Serreze DV, and et al. 1995. Crosses of NOD mice with the related NON strain. A polygenic model for IDDM. Diabetes 44: 1186–1195. [DOI] [PubMed] [Google Scholar]
- 49.Ruan Q, Zheng SJ, Palmer S, Carmody RJ, and Chen YH. 2010. Roles of Bcl-3 in the pathogenesis of murine type 1 diabetes. Diabetes 59: 2549–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reissig S, Tang Y, Nikolaev A, Gerlach K, Wolf C, Davari K, Gallus C, Masri J, Mufazalov IA, Neurath MF, Wunderlich FT, Schattenberg JM, Galle PR, Weigmann B, Waisman A, Glasmacher E, and Hovelmeyer N. 2017. Elevated levels of Bcl-3 inhibits Treg development and function resulting in spontaneous colitis. Nat Commun 8: 15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.