Abstract
Gene expression specificity of homeobox transcription factors has remained paradoxical. WUSCHEL activates and represses CLAVATA3 transcription at lower and higher concentrations, respectively. We use computational modeling and experimental analysis to investigate the properties of the cis-regulatory module. We find that intrinsically each cis-element can only activate CLAVATA3 at a higher WUSCHEL concentration. However, together, they repress CLAVATA3 at higher WUSCHEL and activate only at lower WUSCHEL, showing that the concentration-dependent interactions among cis-elements regulate both activation and repression. Biochemical experiments show that two adjacent functional cis-elements bind WUSCHEL with higher affinity and dimerize at relatively lower levels. Moreover, increasing the distance between cis-elements prolongs WUSCHEL monomer binding window, resulting in higher CLAVATA3 activation. Our work showing a constellation of optimally spaced cis-elements of defined affinities determining activation and repression thresholds in regulating CLAVATA3 transcription provides a previously unknown mechanism of cofactor-independent regulation of transcription factor binding in mediating gene expression specificity.
Optimally spaced cis-elements of defined affinities regulate concentration-dependent transcriptional switching in stem cells.
INTRODUCTION
Spatiotemporal regulation of gene expression is critical for specifying different cell types during development (1–3). Eukaryotic gene regulation involves interactions among DNA sequences and proteins, many of which are transcription factors (TFs). Enhancers, the DNA sequences that bind a given TF or multiple TFs, can regulate transcription irrespective of their location in the gene (1, 3, 4). Since a given class of TFs binds similar DNA sequences, how they achieve gene expression specificity has been the subject of intense investigation. One of the possible mechanisms to achieve specificity is the binding of cofactors that may unmask latent binding specificity of TFs as shown in the case of homeobox-mediated regulation in anterior-posterior body patterning in Drosophila melanogaster (5). Another mechanism involves the utilization of the cis-regulatory modules (CRMs), a subset of enhancers that contain cis-elements for one or more TFs, which have been shown to determine the expression of neighboring genes in a variety of organisms (4, 6–9). In general, the CRMs can be classified into homotypic, where they bind a given type of TF, or heterotypic, where they bind different TFs (10, 11). The heterotypic CRMs largely have been thought to mediate spatiotemporal regulation of gene expression through their ability to recruit different collections of TFs in space and time (10, 12, 13).
Both the homotypic and heterotypic CRMs have been shown to regulate spatiotemporal gene expression patterns in response to TF gradients. The earliest examples of homotypic CRMs have been described in the promoters of genes activated by the TFs that accumulate in a graded manner during early embryonic development in Drosophila (14–17). Classically, the French flag model proposed by Wolpert has been applied to explain the expression of genes by TF gradients. According to this model, the target gene expression is highest in places of the highest concentration of the TF (18). Analysis of multiple CRMs has identified three recurring properties: cis-element number, affinity, and cooperativity, which determine gene expression (16). Essentially, decreasing any of the three CRM properties reduces the mean expression while increasing any of the properties leads to overexpression (15, 16, 19, 20).
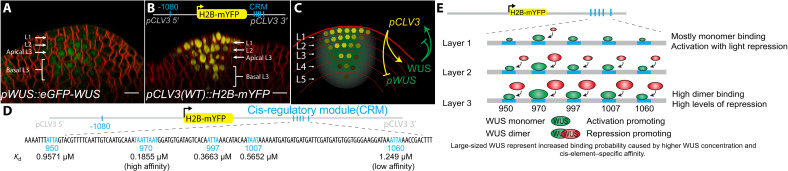
In Arabidopsis shoot apical meristems (SAMs), WUSCHEL (WUS) is a homeodomain TF expressed in the rib meristem (RM) (21, 22). WUS protein migrates into the overlying central zone (CZ), where it promotes stem cell fate by repressing differentiation and also activates its own negative regulator—CLAVATA3 (CLV3) (23, 24) (Fig. 1, A to C). CLV3 encodes a secreted peptide that activates a receptor kinase pathway to restrict WUS expression (25, 26). WUS has also been shown to bind to the promoters of key differentiation-promoting TFs to repress transcription (27). How the same TF activates some genes, such as CLV3, and represses other genes in the same cells is largely unknown. However, a recent study has provided some clues to this regulation. Perales et al. (24) showed that WUS binds a CRM, a collection of five closely spaced cis-elements, in the CLV3 enhancer region (Fig. 1D). The incremental deletion of cis-elements led to down-regulation of CLV3 in the outer layers of the CZ and misexpression in the inner layers of the RM, suggesting that same cis-elements mediate activation and repression of CLV3 at lower and higher WUS, respectively. Biochemical analysis revealed that WUS binds cis-elements as monomers at lower WUS concentrations and binds as dimers/multimers with increasing WUS concentrations, suggesting that dimerization/multimerization of WUS at higher levels may repress CLV3 (Fig. 1E). The biochemical analysis also revealed that DNA promotes homodimerization (28, 29). Furthermore, increasing the affinity of one of the cis-elements decreased the dimerization threshold and led to the repression of CLV3 in the CZ, supporting the hypothesis of affinity-based concentration-dependent activation-repression of transcription in maintaining CLV3 expression over a window of WUS levels. This concentration-dependent switching of CLV3 transcription is unique among the homotypic CRMs studied and forms an exception to the French flag model.
Fig. 1. CRM required for CLV3 activation and repression.
Side views of wild-type (WT) meristems with the WUS protein reporter pWUS::eGFP-WUS (A) and CLV3 transcriptional reporter containing all five WT cis-elements within the 3′ CRM pCLV3(WT)::H2B-mYFP (B). Scale bars, 10 μm. (C) Side view of a SAM cartoon showing WUS protein distribution (green) and CLV3 (yellow), which form a regulatory feedback loop across cell layers. The CLV3 CRM, a cluster of WUS binding cis-elements, interacts with the WUS concentration to repress and activate CLV3. CLV3 signals to WUS at both the posttranslational level, enriching the WUS protein, and the transcriptional level, repressing WUS expression. (D) Schematic of the CLV3 gene including the location and Kd of WUS binding TAAT cis-elements (cyan) of the CLV3 CRM. (E) Schematic of WUS monomer and dimer binding to the CLV3 cis-elements depending on the WUS concentration gradient (across SAM cell layers) and the relative affinities of cis-elements.
Understanding how the concentration-dependent transcriptional switch is established requires understanding how the CLV3 CRM functions as a unit. The complexity of the CLV3 CRM regulation involving five cis-elements and the bidirectional relationship between CLV3 and WUS can be challenging to untangle experimentally. The current experimental limitations cannot provide a direct real-time view of the actual WUS/CLV3 molecular dynamics under precisely defined conditions. A multiscale computational model capable of simulating the binding and unbinding dynamics of WUS to all five cis-elements and CLV3 transcription at the tissue level can be helpful in providing mechanistic insights into the WUS concentration–dependent functioning of the CLV3 CRM.
Different approaches have been developed for studying TF binding dynamics. The thermodynamic models are usually based on the occupancy of the promoter by the TFs, the statistical weights of possible configurations, and the free energy (30–34). However, when multiple cis-elements with different affinities and their interactions are involved as observed in CLV3 CRM, the number of possible configurations becomes large and it is not practical to use the thermodynamic approach. Instead, the stochastic simulation algorithm, i.e., Gillespie algorithm where the dynamics of WUS binding and unbinding to the cis-elements can be modeled as a series of probabilistic events occurring at random time steps determined, is ideal to explicitly model CLV3 transcription.
We developed a stochastic model to simulate the WUS binding to the CRM in a single cell. The single-cell model was applied to simulate WUS binding cis-elements under different concentrations and compared the simulation output with the experimental data on tissue-level expression patterns of different cis-element mutants of CLV3 to investigate the roles of WUS binding affinity, distance-dependent cooperativity among cis-elements, and RNA polymerase II (Pol II) recruitment in the transcription process. Subsequently the single-cell stochastic model was applied to multiple cells represented by unit spheres to develop a cell-based three-dimensional (3D) model representing the SAM. The 3D model was applied to further test the mechanisms identified in the single-cell model in generating the spatial patterns of CLV3 expression.
Using a WUS gradient consistent with the experimental data, both computational models suggested a role for residence time limit (see Results for details) of WUS monomer binding to the individual cis-elements of different affinities, which have been shown to activate CLV3 largely to a similar extent when acting alone. Beyond residence time limit, the aged WUS monomers fail to activate transcription and they are replaced with newly synthesized WUS monomers to sustain CLV3 activation. Our experimental observations showing a correlation between higher WUS turnover and increased CLV3 activation support such a mechanism. When multiple cis-elements are involved, we found that the cooperative binding of WUS monomers and dimers is required to achieve correct CLV3 activation patterns. The model simulations also suggested a nonhomogeneous cooperativity among cis-elements that depends on the intervening distance between cis-elements. The model prediction on distance-dependent cooperativity was tested in experiments by increasing the intervening distance between cis-elements, which revealed an increase in CLV3 activation. The corresponding biochemical experiments revealed that an increase in intervening distance between cis-elements increased their affinity to WUS monomers but did not alter the concentration at which WUS monomers switch to form stable dimers/higher molecular weight complexes. These results show the importance of optimal spacing between cis-elements in determining the concentration range over which an appropriate number of WUS monomers and dimers populate on cis-elements in setting up the activation-repression thresholds. The 3D model that incorporates multiple cis-elements of different affinities that are spaced optimally allowed independent manipulation of the monomer and dimer cooperativity. Our simulations revealed that monomer cooperativity was critical for expression of CLV3 at the lower WUS concentration, while the dimer cooperativity was critical for repression at the higher WUS concentration. Moreover, a balance of the monomer and dimer cooperativity levels was critical to achieve the wild-type CLV3 expression at a WUS concentration range observed in experiments.
RESULTS
Affinity and collective activity of multiple cis-elements determine CLV3 expression
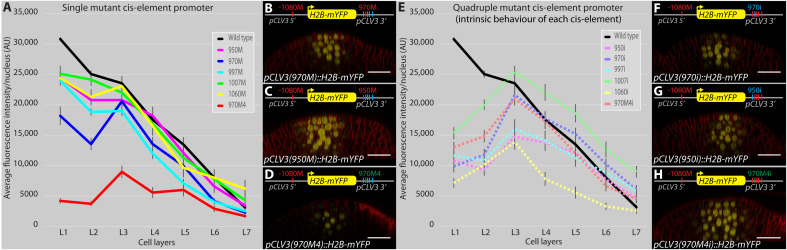
Incremental mutations of cis-elements within the CRM result in incremental down-regulation of CLV3 expression in outer cell layers of the CZ and up-regulation in the inner cell layers of the RM, suggesting interaction among cis-elements (24). To understand the collective behavior of cis-elements, we first deduced the contribution of each cis-element within the CRM to the regulation of CLV3 by analyzing the loss of binding mutations in each of the five cis-elements. Single loss of binding mutations in high-affinity cis-element 970 (Fig. 1D and table S1) led to drastic down-regulation in the outer layers of CZ (Fig. 2, A and B, and fig. S1). On the other hand, independent loss of binding mutations in the four lower-affinity cis-elements led to a minor down-regulation in the L1 layer (Fig. 2, A and C). These results show that all five elements contribute to the CLV3 expression, with the highest-affinity cis-element contributing maximally over the lower-affinity cis-elements CLV3. Moreover, our previous work shows that increasing the affinity of 970 cis-element alone down-regulated CLV3 expression, revealing the critical role of affinity of cis-elements in regulating the CLV3 expression (Fig. 2, A and D). These results suggest that each cis-element contributes to CLV3 expression and their affinities are critical to achieving proper spatial regulation.
Fig. 2. The number of cis-elements and affinity influences the collective behavior of the CRM in regulating CLV3 activation and repression.
Average fluorescence levels (mean ± SE) of H2B-mYFP in different cell layers of various pCLV3::H2b-mYFP promoter variants carrying a mutation in single cis-elements of the 3′ CRM (A). AU, arbitrary unit. (B to D) Side views of WT meristems showing various mutant pCLV3::H2B-mYFP reporter expression patterns. Single cis-element mutants 970M (B) and 950M (C) and a higher-affinity mutant 970M4 (D). Average fluorescence levels (mean ± SE) of H2B-mYFP in different cell layers of various pCLV3::H2b-mYFP promoter variants carrying mutations in four of the five cis-element mutants (quadruple mutants) (E). Side views of WT meristems showing various mutant pCLV3::H2B-mYFP reporter expression patterns. Quadruple mutant (mutants 950M, 997M, 1007M, and 1060M) referred to as 970 intrinsic (970i) (F), (mutants 970M, 997M, 1007M, and 1060M) referred to as 950 intrinsic (950i) (G), and (mutants 950M, 997M, 1007M, and 1060M) referred to as 970M4 intrinsic (970M4i) (H). All cis-element mutations within the CRM in the 3′ region were generated in the mutant-1080 cis-element background. In all images, scale bars = 20 μm. (A and E) The error bars represent the SE (in all cases, n = 4 represents independent transformants).
Single cis-elements can only activate CLV3 at a higher WUS level
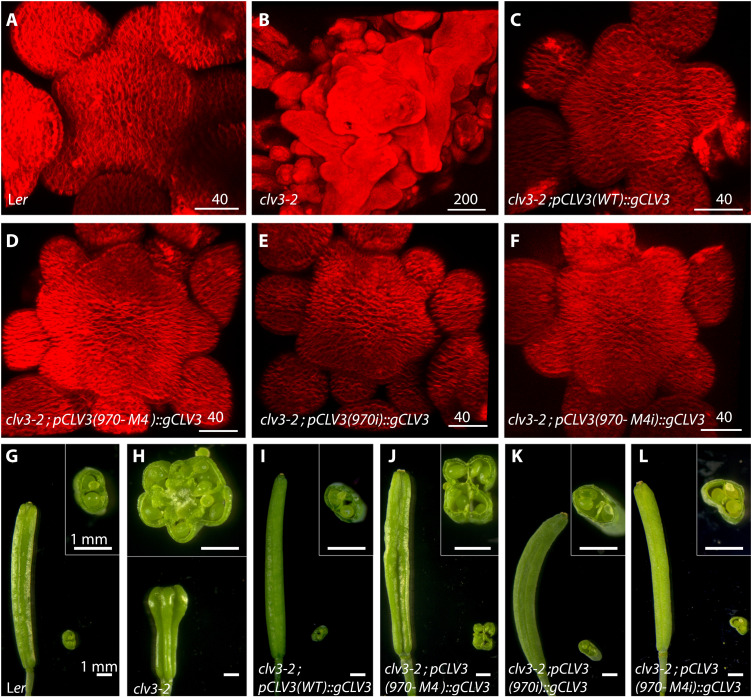
The subtle changes observed upon mutating individual lower-affinity cis-elements ruled out a simple additive interaction in regulating CLV3 expression. Therefore, to further understand the nature of interactions among cis-elements, we first determined the contribution of each one of the five cis-elements to CLV3 expression, referred to as the intrinsic (i) behavior. We generated a library of five mutant CLV3 reporters; each contained only one functional cis-element referred to as 970i, 997i, 1007i, 950i, and 1060i. The reporter expression analysis revealed a marked down-regulation of CLV3 expression in outer cell layers of CZ, including the higher-affinity cis-element 970i (Fig. 2, E and F, and fig. S1). To test further the importance of affinities in influencing intrinsic behavior, we analyzed the expression of 970M4i (Fig. 2, E and H). The 970M4 cis-element is a mutation in 970 that binds WUS with three times higher affinity, and it has been shown to repress CLV3 expression even at lower WUS in outer cell layers of CZ (24). The 970M4i reporter (Fig. 2, E and H, and fig. S1) was expressed at a notably higher level than 970M4 (Fig. 2, A and D). To further test whether the reactivation of CLV3 associated with 970M4i is functionally relevant, we examined its ability to complement clv3-2 null mutants by expressing CLV3 genomic version. The 970M4 mutants partially complement the SAM and the floral meristem (FM) phenotypes when compared to the wild-type CLV3 promoter (Fig. 3 and fig. S2). However, 970M4i was able to significantly better complement both the SAM and FM phenotypes, showing the reactivation of 970M4i (Fig. 3, F and L). Furthermore, both 970i (Fig. 3, E and K) and 970M4i (Fig. 3, F and L) complemented clv3-2 to a similar extent despite binding WUS with different affinities. Consistent with this conclusion, all single cis-elements irrespective of large differences in their WUS binding affinities largely activated CLV3 only in the inner layers of RM, where WUS accumulates at a higher level (fig. S3). However, cis-element affinity is important in the context of other functioning cis-elements in the CRM, as exemplified by the repression of 970M4. In summary, the affinity-dependent collective WUS binding to all five cis-elements is required for balancing activation and repression of transcription in regulating the spatial expression and levels of CLV3.
Fig. 3. Functional analysis reveals the importance of the collective behavior of the CLV3 CRM.
(A to F) Top views of 3D-reconstructed SAMs stained with plasma membrane dye FM4-64 (red). WT (A), clv3-2 (B), and clv3-2 complemented with WT genomic CLV3 (gCLV3) expressed from the WT CLV3 promoter [pCLV3(WT)::gCLV3;clv3-2] (C), CLV3 promoter carrying high-affinity 970M4 cis-element [pCLV3(970M4)::gCLV3;clv3-2] (D), CLV3 promoter carrying loss of binding mutation in 950, 997,1007, and 1060 [pCLV3(970i)::gCLV3;clv3-2] (E), and CLV3 promoter carrying high-affinity mutation 970M4 and loss of binding mutations in 950, 997, 1007, and 1060 [pCLV3(970M4i)::gCLV3;clv3-2] (F). (G to L) Side views of intact siliques and cross section of sliced siliques. Insets show a higher-magnification view of the cross section of the sliced siliques. WT (G), clv3-2 (H), and [pCLV3(WT)::gCLV3;clv3-2] (I), [pCLV3(970M4)::gCLV3;clv3-2] (J), [pCLV3(970i)::gCLV3;clv3-2] (K), and [pCLV3(970M4i)::gCLV3;clv3-2] (L). Scale bars (in micrometers) are given on individual panels in (A) to (F), and the scale bars in (G) to (L) are 1 mm.
Description of a stochastic single-cell model of CLV3 transcription
To investigate the mechanisms of interaction among five cis-elements, we developed a stochastic modeling framework to simulate the WUS binding to the CLV3 CRM in a single cell, together with the RNA Pol II recruitment and CLV3 mRNA synthesis (Fig. 4A). The model was applied to understand the mechanisms underlying the CLV3 activation by the individual cis-elements that bind WUS with different affinities and the interactions among multiple cis-elements in regulating the CLV3 expression together. The stochasticity was introduced by implementing the Gillespie algorithm (35) to simulate all possible WUS binding and unbinding events to form a monomer or dimer and recruitment of Pol II for activating CLV3 transcription. A sufficiently long time was allowed for all the simulations to reach the steady state. The CLV3 reporter analysis performed in the wild-type background uses a steady-state WUS gradient to quantify the effects of the number, affinity, and intervening distance between cis-elements on CLV3 expression. Since the focus of this study is to analyze concentration-dependent binding of WUS to the CLV3 CRM, the feedback regulation of CLV3 on WUS was disabled to maintain a constant WUS concentration gradient throughout simulations to match the reporter analysis. It was also assumed that WUS binding the CLV3 CRM alone would not change the overall WUS concentration. The stochastic time step and index for the next occurring event were generated by following the original Gillespie algorithm based on the assumption that binding to one cis-element was independent of the other cis-elements unless cooperativity among cis-elements exists. The average amount of CLV3 mRNA synthesized, at a fixed WUS concentration, from multiple simulations was calculated. The model was then applied to measure the total amount of CLV3 mRNA synthesized at different WUS concentrations (see the Supplementary Materials for details).
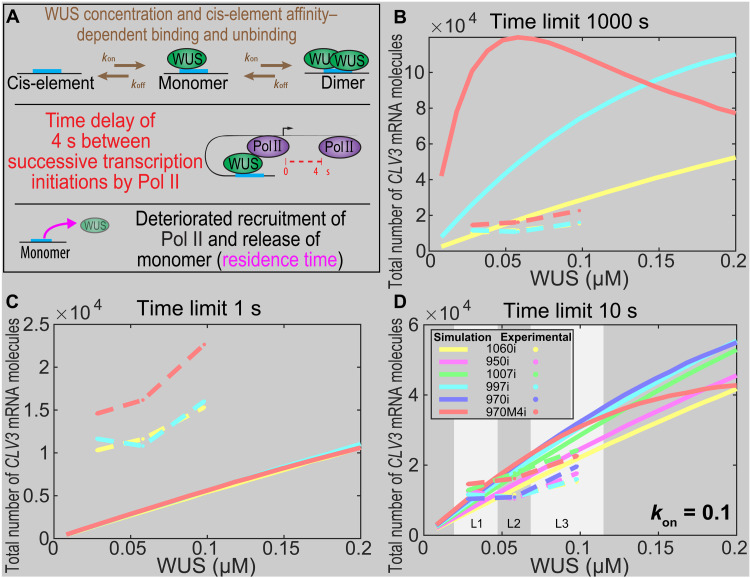
Fig. 4. WUS protein time limit on cis-elements determines the CLV3 levels and domain of expression.
(A) WUS concentration–dependent binding [konWUS], kon is the association rate, and cis-element affinity–dependent unbinding [koff = Kdkon] determine three possible WUS occupancy states: unbound (zero WUS), monomer bound (one WUS), and dimer bound (two WUS). We assume that only the monomer bound is able to recruit RNA Pol II. A 4-s gap between recruitment of successive Pol II molecules was estimated from the Pol II elongation rate and the size of Pol II footprint on the DNA. In addition, multiple rounds of Pol II recruitment by WUS monomer deteriorate the ability of WUS to recruit additional Pol II (residence time limit). (B to D) Single-cell model of WUS-mediated activation of CLV3 from single cis-element promoters (four mutated and only one functional cis-element). (B to D) Scaled simulation results of highest-affinity (970M4i), intermediate-affinity (997i), and lowest-affinity (1060i) cis-elements with the residence time limit of 1000 s (similar expression pattern as without the time limit since the time limit is extremely large) (B), 1 s (C), or 10 s (D). (D) All five cis-elements in addition to 970M4i and approximate WUS concentration range to reflect the corresponding WUS fold changes from L1, L2, and L3 layers.
Modeling WUS binding to the CRM
Our previous analysis revealed that each cis-element binds WUS at different concentrations as monomers first and then switches to forming dimers at increasing concentrations (24). Therefore, we first aimed to determine the binding and unbinding probabilities associated with each cis-element by reproducing the ratio of monomer- and dimer-bound cis-elements observed in electrophoretic mobility shift assay (EMSA) experiments (24). Since increasing the TF concentration decreases the search time of its binding to cis-elements (36), it was assumed that the probability of WUS binding to cis-elements increases with the increase in WUS concentration. In particular, the propensity of WUS binding to an empty cis-element or with a monomer is assumed to depend linearly on WUS concentration, i.e.,, where is the binding rate. Then, the unbinding propensity of WUS associated with each cis-element is calculated as = , where , the dissociation constant, was quantified in our previous work (table S1) (24). To test this assumption, we considered a wide range of WUS concentration that encompasses WUS monomer and dimer binding to each one of the five cis-elements observed in EMSA experiments (24). We first simulated WUS monomer binding to a single cis-element to determine , a free parameter, such that proportions of bound monomers obtained in the model were similar to those observed in the EMSA experiments with WUS that lacked the C-terminal homodimerization domain (HOD) (fig. S4A). Since dimerization occurs through sequential recruitment of WUS to the WUS monomer-DNA complex, we next modeled the dimer formation by recruiting the second WUS molecule to a monomer. In the absence of the experimental values on binding affinity associated with the WUS dimerization, we chose associated with the binding of the second WUS molecule to be the same as the one used to simulate monomer We chose for dimer binding such that proportions of monomer and dimer bound to the cis-elements matched the EMSA experiments with full-length WUS (fig. S4B) (24).
Modeling CLV3 transcription
We considered the recruitment of Pol II as another stochastic event in the model. It has been shown that the transition from monomer binding to dimer binding could be correlated to the transcriptional switch from activation to repression of CLV3. Therefore, we assumed that monomer binding recruits Pol II to activate CLV3 transcription, while the WUS dimers fail to recruit Pol II and activate CLV3 transcription. We introduced a time delay between two successive Pol II recruitment events due to the size of the Pol II complex occupying the transcription start site. The time delay calculated based on an 80–base pair (bp) footprint of RNA polymerase and mRNA elongation rate, which is estimated to be 1.2 kb/min (37), was approximated as . It is also assumed that, after transcription initiation, the WUS monomer can unbind or bind another WUS molecule to form a dimer. Moreover, we considered the Pol II recruitment rate as an uncalibrated parameter and carried out perturbations to examine its effect on the transcriptional output. The model was calibrated over a wide range of WUS concentrations. We then applied the model to simulate WUS binding/unbinding to a single cis-element and Pol II recruitment to generate the intrinsic expression of CLV3 at different WUS concentrations. By comparing the CLV3 mRNA production with the experimental quantification of the CLV3 expression in Fig. 2 (E to H), an optimal scale of WUS concentrations was obtained to capture the WUS gradient in different cell layers of the SAM. This optimal WUS concentration scale was used in all single-cell simulations to investigate possible mechanisms controlling the intrinsic behavior of each cis-element in regulating the CLV3 expression.
Mechanisms of the intrinsic behaviors of cis-elements in regulating CLV3
It has been observed that the transcriptional output depends on the affinity of cis-elements and the TF concentration (30, 32, 38–45). In general, a higher-affinity cis-element results in a longer TF occupancy than the lower-affinity cis-element at a given WUS concentration. Consequently, a longer TF occupancy leads to a higher mRNA production (46). Experiments reveal that WUS binds to 970M4i with approximately 21.4 times higher affinity than to the lowest-affinity cis-element 1060i. Therefore, a longer residence time of WUS on 970M4i was expected to produce much higher levels of CLV3 than 1060i. However, our experiments revealed that, although five cis-elements bound WUS with different affinities, intrinsically (950i, 970i, 997i, 1007i, and 1060i mutants), they were able to similarly activate CLV3 only in inner cell layers of RM where the WUS concentration is higher (Fig. 2, E to H). The initial attempt in modeling by assuming WUS occupancy based on affinities produced distinct CLV3 expression patterns for the highest 970M4i cis-element and the lowest 1060i cis-element (Fig. 4B). 970M4i produced a much sharper increase in CLV3 expression at the lower WUS concentration than did 1060i. With the increase in WUS concentration, 970M4i produced a lower amount of CLV3 mRNA, which is expected because of the WUS dimerization, while 1060i continued to yield higher CLV3 mRNA (Fig. 4B). Such markedly different CLV3 expression patterns produced by 970M4i and 1060i were not consistent with experimental observations, suggesting that additional mechanisms may regulate the intrinsic activation behavior of cis-elements in addition to their affinities.
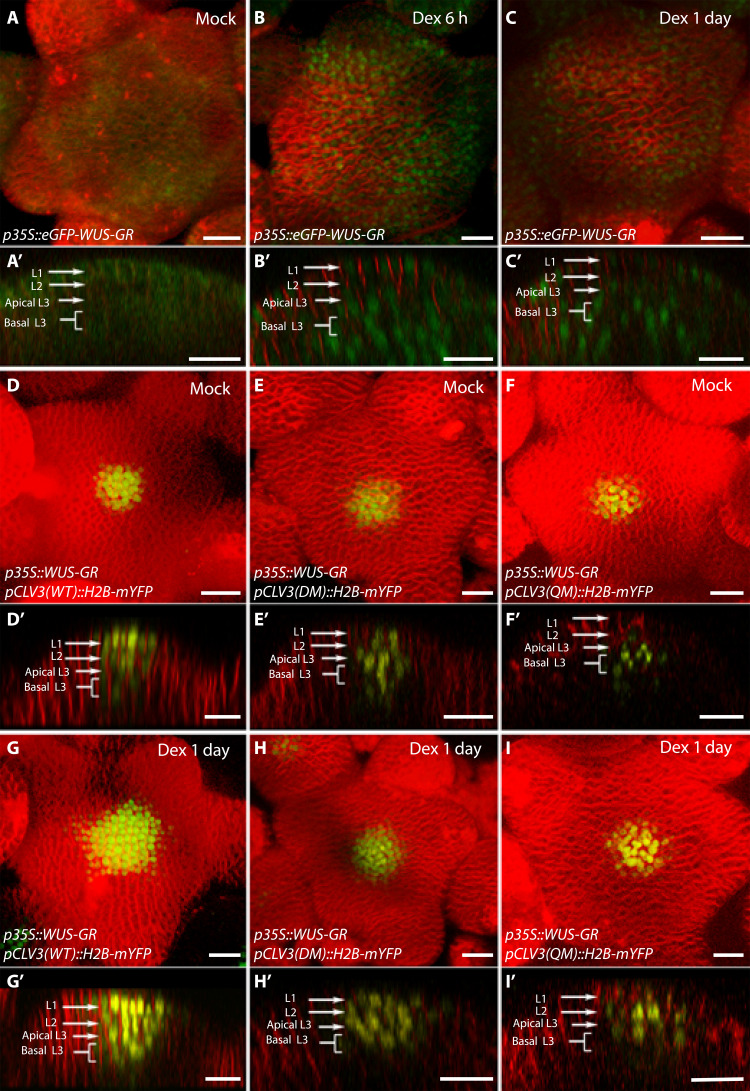
It has been noticed that for different types of TFs, including general control TF (GCN4) in yeast (47) and transcriptional coactivator NPR1 involved in systemic acquired resistance (SAR) in Arabidopsis (48), a higher turnover of TFs leads to a higher transcriptional activation. Furthermore, the transcriptional activation domains of GCN4 and other TFs have been shown to overlap with degradation domains, suggesting a possible correlation between transcriptional activation and TF turnover (47, 49). Moreover, transcription-dependent degradation has been shown for sterol regulatory element–binding protein (SREBP) family of TFs (50). These observations suggest that TFs when actively transcribing may get progressively modified (for example, phosphorylated) and become transcriptionally ineffective and marked for their degradation (47, 49). Although deep mechanistic links between WUS, protein phosphorylation, and protein destabilization machinery are still unknown, our earlier work suggests similarities between WUS and TFs described above. (i) The transcriptional regulatory domains [WUS-box and EAR-like (ethylene-responsive element binding factor associated amphiphilic repression) domains] function as degrons (51, 52). (ii) CLV3 activated at the lower WUS concentration in the CZ can be repressed by enriching and stabilizing the WUS protein in the nucleus (24, 51). (iii) The dexamethasone (Dex)–mediated nuclear translocation of WUS by using the 35S::eGFP-WUS-GR system led to an immediate destabilization of the protein in the CZ within 6 hours (53). By 24 hours of Dex application, the protein was only detected in the nuclei of cells in the edge of the peripheral zone (PZ) and deeper cell layers of the RM. The CLV3 activation and expansion into the PZ followed the centripetal pattern of rapid destabilization of the WUS protein [Fig. 5; (53)].
Fig. 5. The number of cis-elements determines the sensitivity of CLV3 promoter to the dynamic changes in the WUS protein accumulation.
(A to C) SAMs showing WUS protein accumulation patterns (p35S::eGFP-WUS-GR) upon its Dex-induced nuclear translocation at 6 hours (B), at 24 hours (C), and upon mock treatment (A). (D to I) p35S::WUS-GR–expressing SAMs showing pCLV3::H2B-mYFP reporter expression of WT CLV3 promoter (D), the double mutant promoter (970 and 997 mutants) (E), and the quadruple mutant promoter (970, 997, 950, and 1060 mutants) (F) upon mock treatment. The pCLV3 reporter expression of the corresponding genotypes after 24-hour Dex treatment is shown in (G) to (I). (A to I) Three-dimensional reconstructed top views of SAMs and corresponding side views shown in (A′) to (I′). Plasma membrane stain–FM4-64 (red), eGFP-WUS-GR (green), and H2B-mYFP (yellow). Scale bars, 20 μm.
Perhaps degradation of WUS decreases the dimer concentration or creates a dynamic WUS that works favorably with the Pol II binding limit to increase CLV3 activation. Therefore, we considered an upper limit on the residence time of WUS beyond which WUS becomes inactive and fails to recruit Pol II, referred to as residence time limit in the model (Fig. 4A). The older/inactive WUS species need to be replaced with newly synthesized WUS monomers to maintain transcription. Therefore, we imposed the same WUS monomer residence time limit for all cis-elements. A markedly lower WUS monomer residence time limit substantially decreased CLV3 expression for all cis-elements (Fig. 4C and fig. S5). Simulations with a balanced residence time limit were able to generate a similar intrinsic expression pattern of CLV3 for all cis-elements. In particular, to generate similar expression patterns of 970M4i (highest affinity) and 1060i (lowest affinity) cis-elements, we chose the residence time limit to be 10 for all simulations involving multiple cis-elements discussed in the following sections (Fig. 4, B to D, and fig. S5).
The CLV3 CRM composition determines sensitivity to dynamic changes in WUS protein levels
The number of cis-elements may also determine the sensitivity of the CLV3 promoter to WUS levels to regulate spatial expression of CLV3. To test this, we analyzed the response of the mutant promoters lacking several WUS binding cis-elements to 35S::eGFP-WUS-GR system, upon 24 hours of Dex application, described in the previous section. The wild-type CLV3 promoter with five functional cis-elements expressed at high levels and the promoter activity expanded into the PZ (Fig. 5, D and G). The mutant promoter lacking the two functional WUS binding cis-elements (970M and 997M)-pCLV3(DM)::H2b-mYFP is initially expressed in the deeper cell layers, and the expression levels are below that of the wild-type promoter (Fig. 5E). The 24-hour Dex application was able to activate pCLV3(DM) in the CZ weakly but failed to expand into the PZ (Fig. 5H) (n = 8) when compared to the wild-type promoter, which revealed strong activation and radial expansion (Fig. 5G). The mutant promoter lacking four cis-elements (950M, 970M, 997M, and 1060M)-pCLV3(QM)::H2b-mYFP was expressed only in the deeper layers (Fig. 5F). After 24 hours of Dex application, the mutant promoter was mildly up-regulated in deeper layers; however, it failed to activate in the CZ and expand radially into the PZ (Fig. 5I). Together, rapid destabilization of WUS can lead to higher CLV3 activation, which is maintained even at undetectable WUS protein levels, showing that all five cis-elements working together increase the sensitivity of CLV3.
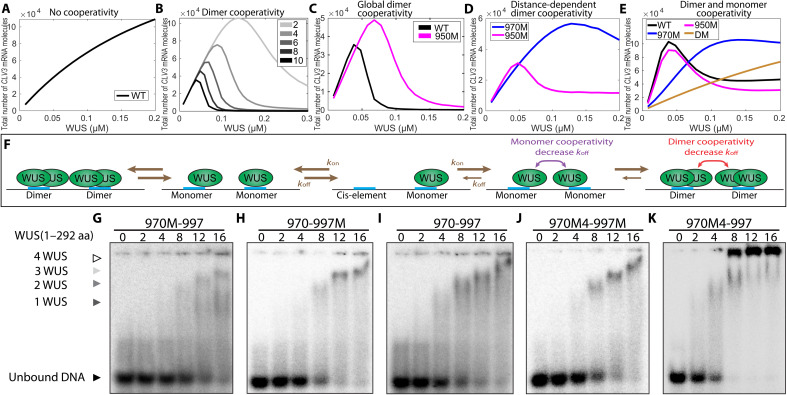
Cooperativity among cis-elements regulates CLV3 expression
Our experimental analysis showing different expression patterns of CLV3 for single cis-elements and multiple cis-elements suggested an interaction among cis-elements within the CRM (24). The same study also showed that an increase in cis-element affinity (970M4) resulted in a decrease in dimerization threshold and repressed CLV3 in outer cell layers of CZ where WUS accumulates at a lower level. These observations suggested that cis-element affinity is important in the context of the multiple cis-elements, possibly in inducing cooperative interactions among WUS dimers bound to multiple cis-elements within the CRM. To understand the multiple cis-element behaviors, we used the calibrated single-cell WUS binding model by extending it to include multiple cis-elements. Without any cooperative interactions among them, an increase in WUS concentration led to an increase in CLV3 expression, which can be interpreted as a linear combination of intrinsic behaviors of individual cis-elements, which is not consistent with the experimental analysis (Fig. 6A). Therefore, we introduced cooperativity among cis-elements into the model. First, we considered equal cooperativity among all cis-elements irrespective of the intervening distance. As the cooperativity increased, the CLV3 expression decreased at the higher WUS concentration, which could be due to increased dimerization (Fig. 6B). Then, we chose appropriate values for parameters involved in the dimer cooperativity to obtain the highest activation of CLV3 at a lower WUS concentration as observed in experiments. Next, we used the calibrated model with the chosen cooperativity parameters to simulate mutant CLV3 consisting of different number of cis-elements. In particular, our experimental analysis showed a weaker down-regulation of CLV3 upon mutating any one of the four lower-affinity cis-elements (950M, 997M, 1007M, and 1060M) for the low WUS concentration, compared to the highest affinity, i.e., 970 cis-element (970M) (Fig. 2, A to C). However, in the simulations with the calibrated equal dimer cooperativity, 950M was expressed at a much higher level than the wild type at the high WUS concentration (Fig. 6C), which was not consistent with the experimental observation, suggesting unequal cooperativity among those cis-elements in the CRM.
Fig. 6. Cooperativity among cis-elements regulates CLV3 expression.
Average CLV3 mRNA levels from single-cell simulations in response to WUS concentration without cooperativity (A), at different strengths of dimer cooperativity (B), when dimer cooperativity between every cis-element is considered (C), when the dimer cooperativity depends on the intervening distance between cis-elements (D), and when both WUS monomer and dimer cooperativity were considered (E). (F) Binding and unbinding dynamics of WUS monomer and dimer on cis-elements. (G to K) Gel shift assay of increasing concentrations of full-length WUS 1 to 292 amino acids (aa)] to probes of similar length that cover the 970 and 997 cis-elements. Probes with loss of binding mutations to the TAAT elements in the 970 cis-element (G) and the 997 cis-element (H). (I) Probe with WT copies of the 970 and 997 cis-elements. Probes that contain higher-affinity mutant 970M4 along with the mutant 997 (J) or WT 997 cis-element (K). (G to K) Arrowheads denote higher-order WUS complexes: monomer (dark gray), dimer (light gray), and higher complexes (white). The unbound probe (black).
Next, we introduced unequal dimer cooperativity wherein the interaction between neighboring cis-elements was higher, and cooperativity decreased with increasing intervening distance (referred to as distance-dependent cooperativity). Similar kind of cooperativity was studied in thermodynamic models earlier (54). For simplicity, we simulated 970M and 950M expressions representing mutations in high- and low-affinity cis-elements, respectively. The model with distance-dependent dimer cooperativity was able to generate wild-type expression patterns. However, a similar expression behavior was observed for both 950M and 970M at lower WUS concentrations, which is inconsistent with the experimental data (Fig. 6D). This suggested that the cis-element affinity influences interactions among cis-elements and the higher-affinity cis-element 970 interacts differently than the lower-affinity cis-elements in activating CLV3 at lower WUS. Therefore, distance-dependent monomer cooperativity between cis-elements was considered. The monomer and dimer cooperativity were considered separately since one is responsible for activation and the other one is responsible for repression. Considering that the affinity plays a role when multiple cis-elements interact, the residence time limit associated with single cis-element was disabled. The additional WUS monomer cooperativity along with the dimer cooperativity between all cis-elements was able to generate expected wild-type and mutant (970M and 950M) cis-element behaviors at all WUS concentrations (Fig. 6E), showing the importance of both in regulating CLV3 transcription.
The neighboring cis-elements influence WUS DNA-protein complex formation
To test predictions of model simulations on the possible cooperative behavior of cis-elements, we performed EMSA with increasing concentration of WUS on probes that contain two adjacent cis-elements. We considered the two adjacent cis-elements 970 and 997 because mutating these two cis-elements has been shown to down-regulate CLV3 expression in outer cell layers of CZ and up-regulate expression in the inner layers of RM (24). Full-length WUS at lower concentration has been shown to bind as a monomer to single cis-elements, which shifts to a dimeric complex at the higher WUS concentration (24). We found that WUS bound the oligo that contains 970 and 997 cis-elements (Fig. 6I) at much lower concentrations than observed with the oligos of the same length that only contains one functional cis-element that is either the 970 (Fig. 6H) or 997 cis-element (Fig. 6G). In addition, the WUS shifted to form higher molecular weight complexes at much lower concentrations with the two functional cis-elements than one functional cis-element (Fig. 6I). To further test the nature of the protein and complex formation across multiple cis-elements, we tested the binding patterns of two WUS protein variants: WUS1-134 that only contained the DNA binding domain and lacked the centrally located HOD, and WUS1-208 that contains the centrally located HOD domain. Our earlier work has shown that these fragments bind cis-elements with comparable affinities to the full-length protein (24). With increasing concentration of WUS1-134, a gradual switch from monomeric to the higher molecular complex was observed, which is expected as previous work has shown that the DNA binding domain also participates in dimerization (24, 29). With WUS1-208, at the same protein concentration range, we observed a faster shift from the monomer form into the higher molecular weight complex. Testing these two protein versions on a probe containing only one functional 970 cis-element revealed higher molecular complex formation at a much higher concentration (24). These results suggest that the second dimerization domain may facilitate interaction between WUS molecules bound to the adjacent cis-elements in promoting higher molecular WUS complex formation.
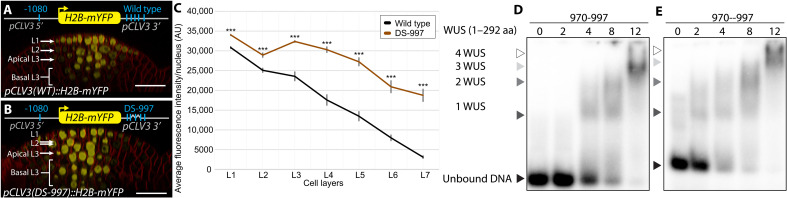
The distance between cis-elements is critical for CLV3 expression
The cooperativity observed in gel shift assays suggests that the neighboring cis-elements increase WUS binding, possibly through protein-protein interaction facilitated by the second HOD (HOD2). To test the influence of spacing between cis-elements without reducing the number or affinity, we duplicated the sequence between neighboring cis-elements. The increased distance might reduce the interaction of WUS bound to neighboring cis-elements without affecting the intrinsic binding to each independent cis-elements. Therefore, we duplicated the intervening sequence between 970-997 and 997-1007 (double space around 997) pCLV3(DS-997)::H2B-mYFP. Increasing the distance between neighboring cis-elements led to increased CLV3 expression in all cell layers, and an increase in the deeper layers was much higher than in the outer cell layers of CZ (Fig. 7, A to C). These results suggest that the distance between cis-elements is more critical for the repression of CLV3, likely through the formation of large WUS complexes across neighboring cis-elements. To test whether the increased distance between 970 and 997 cis-elements alters the binding dynamics, we analyzed WUS binding to the oligo with duplicated sequences that doubled the distance between 970 and 997 (970--997). The full-length WUS protein could bind the oligo (970--997) at lower WUS (Fig. 7, D and E). However, the transition from lower molecular weight complexes to higher molecular weight complexes occurred over a much wider WUS concentration range. Therefore, the increase in CLV3 expression in all cell layers seen in DS-997 could be explained by the larger WUS concentration range over which it remains as a lower molecular weight complex, showing that, in addition to the affinity of the cis-elements, the intervening distance is important in regulating the CLV3 expression.
Fig. 7. Spacing between cis-elements is critical for CLV3 repression.
Side view of SAM showing the WT pCLV3::H2B-mYFP expression (A) and mutant CLV3 reporter containing duplicated sequence to the left (5′) and right (3′) of the 997 cis-element (double sequence around 997—DS-997) (B). Scale bars, 20 μm. (C) Average H2B-mYFP fluorescence intensity (mean ± SE) in 10 centrally located nuclei/cell layers quantified from four independent transformants of WT and DS-997 (n = 4). ***P < 0.001. EMSAs showing increasing concentrations of full-length WUS (1 to 292 amino acids) bound to the probe containing the 970 and 997 cis-elements with WT intervening sequence (D) or a duplicated intervening sequence (E).
Description of a 3D cell–based model of CLV3 transcription
The single-cell model provided insights into the WUS binding dynamics with individual cis-elements, Pol II recruitment, and minimum cooperativity mechanisms required for CLV3 expression (fig. S6). However, the single-cell model can only provide average expression behavior at given WUS concentrations, without considering the tissue spatial organization and the stochasticity associated with individual cells within layers of the SAM under a broader range of WUS concentrations. Therefore, we expanded our scope of study by developing a 3D multicellular model to capture the tissue-level spatial dynamics.
The 3D model could help quantify the establishment of the CLV3 expression pattern throughout the tissue by simulating the stochastic single-cell model in individual cells simultaneously at different WUS concentrations. The 3D model was constructed on the basis of the framework used in our previous work (27) combined with new biological data and mechanisms identified by using the stochastic single-cell model. The computational domain consisted of a 3D matrix of unit spheres organized in a half-dome shape, corresponding to cells within the SAM from the L1 to L7 layers. At the tissue level, a spatial gradient of WUS proteins across different layers, which captured a similar fold change from deeper layers to outer layers observed in experiments (Fig. 8C and fig. S3), was introduced and maintained at this fixed concentration throughout each simulation (Figs. 1A and 8C). In individual cells, the single-cell stochastic model was applied to simulate WUS binding with cis-elements by using the local WUS concentrations to regulate CLV3 transcription. The same mechanisms identified by the single-cell stochastic model were implemented under wild-type and multiple cis-element mutant conditions. Each simulation was allowed to run long enough to achieve the steady-state behavior, and the parameters used in the simulations are listed in table S3.
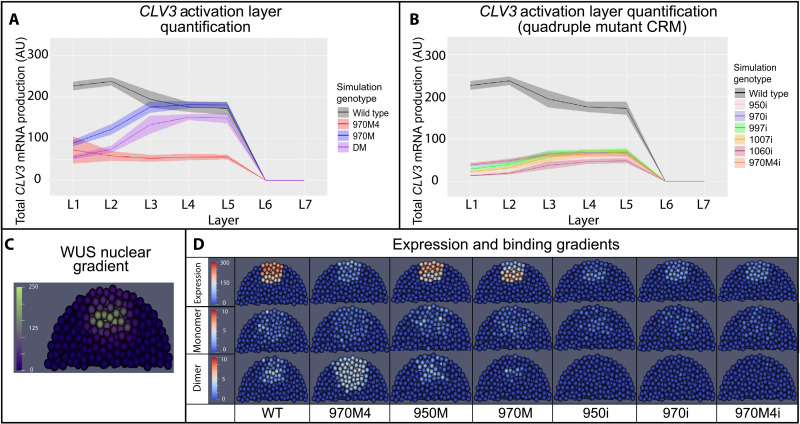
Fig. 8. Simulated CLV3 dynamics and WUS protein complexes in the 3D SAM model.
(A) Levels of CLV3 (mean = line, SD = shaded area) activation in WT system and system with selected mutated cis-elements. (B) Levels of CLV3 gene activation (mean = line, SD = shaded area) in WT system and system carrying all possible combinations of quadruple mutants (reflects intrinsic behavior of each functional cis-element). Line indicates mean CLV3 in different cell layers. Shaded area indicates SD of activation among the cells in a given cell layer. L1 to L7 indicate the layers of the SAM from outermost CZ to inner layers of the RM. (C) Spatial distribution of nuclear-localized WUS. (D) Median longitudinal sections of simulated SAMs showing WUS monomer (middle), WUS dimer (bottom), and CLV3 expression (top) in WT and system carrying various mutant cis-elements 970M4, 950M, 970M, 950i, 970i, and 970M4i.
Analysis of CLV3 expression and WUS complexes captured by the 3D model
Using a biologically relevant WUS gradient (Fig. 8C), CLV3 simulations were generated under a variety of different conditions, including wild type, four cis-elements (970M), three cis-elements (DM), and single cis-element (e.g., 970i). The behaviors of several cis-element mutants are shown in Fig. 8 (A and B). CLV3 expression in wild type was generally higher than in other mutants, similar to the experimental data shown in Fig. 2A. In particular, wild-type CLV3 activation was highest in the L1 layer and lowest in the inner layers of RM. 970M showed a higher expression in the inner layers of RM than in the outer L1 layer. Of particular interest was 970M4, in which the affinity was strengthened over the default 970 affinity, expressed at a lower level in all cell layers. When simulating the mutants with a single functional cis-element in the CRM, e.g., 950i, 970i, 997i, 1007i, and 1060i, the CLV3 expression was detected in only the inner layers of RM. Other than the minor difference in the magnitude, all single cis-element mutants expressed only in the inner cell layers (Fig. 8B), similar to the experimental results. Simulations also showed an impairment in the spatial patterns of CLV3 expression as more cis-elements were deleted. For example, the deletion of a single lower-affinity cis-element 950 (950M) had a relatively minimal effect on CLV3 activation (Fig. 8D). In contrast, deletion of the higher-affinity cis-element 970 (970M) shifted CLV3 expression to the inner layers (Fig. 8D). The more drastic shift in CLV3 expression into deeper layers occurred when deleting four cis-elements (e.g., 950i or 970i) regardless of their WUS binding affinity (Fig. 8D). Therefore, the cooperativity mechanism identified by the single-cell stochastic model was able to generate the expected CLV3 expression behavior in the 3D model.
Bimolecular fluorescence complementation (BiFC) assays in plants expressing split enhanced green fluorescent protein (eGFP)–WUS constructs expressed from the native WUS promoter revealed very few fluorescent positive cells in the L3 and L2 layers of SAMs (fig. S7). These results show that WUS dimerizes in cells that accumulate higher levels of WUS, supporting the correlation observed in biochemical analysis. However, the observed dimerization in BiFC assays does not distinguish between DNA-bound WUS complexes and unbound complexes. Moreover, it likely represents WUS complexes with cis-elements of many target genes (27). Therefore, we used the 3D model to visualize the spatiotemporal distributions of WUS complexes including monomers and dimers on the CLV3 promoter across cell layers in SAMs (Fig. 8D). A higher concentration of WUS monomers in the outer layers of CZ and higher dimers in the inner layers of RM were observed for the wild-type and lower-affinity cis-element 950M. Deleting the 970 cis-element showed lower levels of WUS monomers in the outer layers of CZ and lower levels of dimers in the inner layers of RM (Fig. 8D). This suggested that the higher-affinity cis-element exerts a stronger influence on CLV3 transcription, but it was not sufficient to completely activate in the outer layers of the CZ or repress the inner layers of RM on its own, showing that cis-elements interact with each other in maintaining specific amounts of WUS monomer and dimer complexes in different layers in regulating CLV3 expression. The 970M4 results resolved the seemingly paradoxical expression patterns of this mutant. A massive amount of WUS dimers in all layers can explain a marked reduction of CLV3 expression. In contrast, both monomers and dimers accumulated at a lower level when only one cis-element was functional, showing that WUS failed to populate at higher levels on cis-elements likely due to the lack of cooperativity. Overall, the 3D model simulations showed the spatial distributions of WUS complex formation at a quantitative level in different cell layers of SAMs. The WUS complex formation could be correlated to WUS concentration in different cell layers and the affinity-dependent cooperative behavior of cis-elements in expressing CLV3 in the CZ.
Effect of cooperativity on the spatial patterns of CLV3 transcription
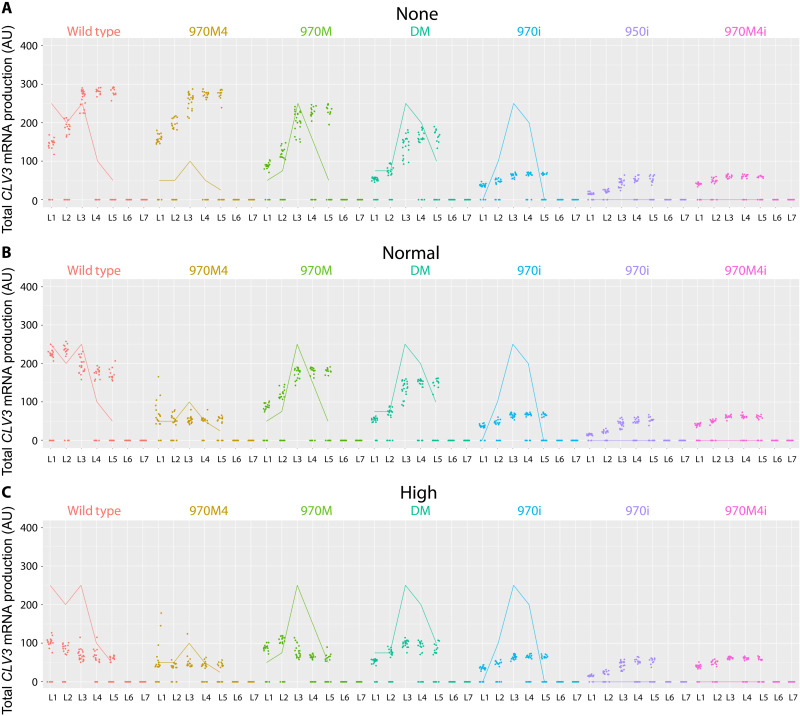
The experimental evidence suggested that the cooperativity among cis-elements is critical to achieving proper spatial patterns of CLV3 expression. To better understand the role of cooperativity in the robust regulation of CLV3 expression quantitatively, we imposed different levels of cooperativity between monomers or dimers for both wild-type and mutant conditions. A complete removal of cooperativity led to a higher CLV3 expression in the inner cell layers of RM and a lower expression in outer cell layers of CZ under all conditions (Fig. 9A). In contrast, increasing cooperativity led to CLV3 down-regulation (Fig. 9C), showing that strength of cooperativity influences CLV3 expression. Our experimental analysis shows that increasing the cis-element affinity (970M4) leads to down-regulation of CLV3 expression, which could be due to a higher cooperativity among cis-elements leading to the repression. To test this hypothesis, we removed cooperativity from 970M4, which led to an increase in CLV3 expression, and the pattern of expression resembled that of wild type (Fig. 9A). These results show the importance of cooperativity in modulating CLV3 expression, which in turn depends on the cis-element affinity.
Fig. 9. Cooperativity levels influence CLV3 activation.
CLV3 activation level of WT CLV3 CRM and mutant CLV3 CRMs under various cooperativity levels. (A) No cooperativity: simulations had no cooperativity between cis-elements. (B) Normal cooperativity: normal cooperativity values (0.01 monomer cooperativity, 0.2 dimer cooperativity) used in the default simulations. (C) High cooperativity: simulations with 10× the cooperativity of the default values in the simulation, i.e., 0.001 monomer cooperativity and 0.02 dimer cooperativity. Dots are simulation values for a cell. Lines are corresponding average expression values from experimental studies. Colors represent different mutants.
Our experimental analysis also showed that decreasing the number of cis-elements leads to a decrease in CLV3 expression in outer cell layers of CZ and an increase in inner cell layers of RM, suggesting that the number of cis-elements may also aid in inducing cooperativity. Consistent with the requirement of multiple cis-elements in mediating cooperativity, the effects of cooperativity levels on CLV3 expression diminished with the deletions of multiple cis-elements (Fig. 9).
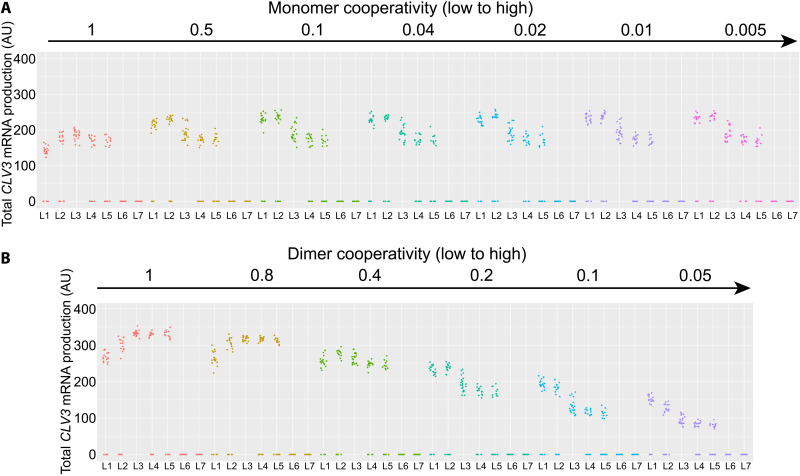
As shown above, removing the overall cooperativity that includes both the monomer and dimer cooperativity leads to the internalization of CLV3, which is not entirely consistent with the in vivo observed overall increase of CLV3 expression even in the outer cell layers of pCLV3(DS-997) (Fig. 7, B and C). Removing the overall cooperativity that also included the monomer cooperativity might have caused the down-regulation of CLV3 in outer cell layers of CZ. Therefore, we perturbed monomer and dimer cooperativity independently. At a constant dimer cooperativity, increasing monomer cooperativity alone led to a gradual increase in CLV3 expression in outer cell layers and expression maxima shifted to outer cell layers (Fig. 10A and figs. S8 and S9). In contrast, increasing the dimer cooperativity alone led to an overall decrease in CLV3 expression, which was more pronounced in the inner layers of RM and a shift in the expression maxima to the outer layers of CZ (Fig. 10B and figs. S8 and S9). This suggests that CLV3 expression is regulated through a balance between dimer and monomer cooperativity mediating the repression and activation, respectively. These simulation results could also help us to understand the experimental data, in which the increased expression of CLV3 in all cell layers observed upon doubling the distance (DS-997) could be attributed to lower dimer cooperativity leading to derepression. Together, these results show that cooperativity plays a critical role in regulating CLV3 expression when all five cis-elements are functional.
Fig. 10. Independent perturbations of the monomer or dimer cooperativity.
The effect of changes in monomer cooperativity (from 1 to 0.005) and dimer cooperativity (from 1 to 0.05) on CLV3 activation. The direction of the arrows indicates an increase in cooperativity. In row (A), dimer cooperativity was held constant at 0.2, while monomer cooperativity was varied (1 to 0.005). In row (B), monomer cooperativity was held at 0.01 and dimer cooperativity was varied (1 to 0.05). A complete table of changes in monomer and dimer cooperativity is presented in fig. S6. The individual graphs represent the CLV3 activation in different cell layers (L1 to L7) of simulated SAMs under the cooperativity levels noted for each simulation. The dots represent the values of the CLV3 signal for individual simulated cells.
DISCUSSION
A homotypic cluster of five cis-elements with different WUS binding affinities regulates levels and spatial expression of CLV3. WUS has been shown to activate and repress CLV3 at lower and higher levels, respectively. Our work reveals that the relative affinities of each element, the number of cis-elements, and intervening distance contribute to the collective effect. Moreover, the collective activity of the CRM arises not only because of the individual affinity but also because of cooperative binding of multiple neighboring cis-elements to WUS. WUS was previously shown to form a mixture of monomers, dimers, and oligomers in solution over a wide concentration range (24). Moreover, DNA/cis-elements have been shown to promote dimerization or multimerization of WUS over a small two- to fourfold increase in the WUS level.
Our biochemical analysis presented here reveals that two adjacent cis-elements can increase the binding sensitivity of WUS at lower levels than the single cis-elements, suggesting that the cis-elements cooperate in increasing the binding probability of WUS monomers, which could contribute to boost activation. Our biochemical work also shows that the two cis-elements working together allows the formation of higher-order WUS complexes at lower WUS levels, which depends on the second HOD (Fig. 6, J and K). This suggests that the second HOD may allow interaction of WUS species bound to the adjacent cis-elements in forming higher-order complexes. WUS has two dimerization domains, one of which is near the DNA binding domain and the other is found outside the DNA binding domain (24). The second dimerization domain may allow protein-protein interaction across neighboring cis-elements, which then allows cooperative binding across the cis-elements. Our analysis also reveals that cis-element affinity plays a critical role in inducing cooperativity across cis-elements. The increased affinity of 970M4 cis-element contributed to higher cooperativity, leading to the repression of CLV3. However, such repression requires other functional cis-elements in the CRM, showing that the collective behavior arises as a result of the number of cis-elements and the WUS binding affinities. The collective behavior of a low-affinity homotypic CRM has been shown to be critical in a recent study of the Drosophila SHAVENBABY locus. Increasing the binding affinity of one of the cis-elements resulted in a strong ectopic activation, suggesting that low-affinity homotypic CRMs may lead to higher specificity (9). Our work showing the importance of the number of cis-elements in regulating gene expression agrees with the fundamental concept of having multiple cis-elements organized in a constellation leading to gene expression specificity. However, the CLV3 CRM regulation differs from other homotypic CRMs such as SHAVENBABY locus where CLV3 expression is regulated through a concentration-dependent activation-repression switching mechanism. The C terminus of WUS has been shown to bind at least three proteins: HAIRYMERISTEM (55), SHOOT-MERISTEMLESS (56), and TOPLESS (57). Earlier analysis shows that the C terminus of WUS is not required for the regulation of DNA binding affinity and dimerization (24) and DNA binding specificity (29). Therefore, we suggest that WUS binding to the CLV3 CRM is a cofactor-independent mechanism that depends on the organization of cis-elements in the CRM. Besides CLV3, WUS has been shown to activate and repress several hundred genes (27). Our bioinformatics search for “TAAT” core-containing cis-element clusters (see the Supplementary Materials for details of the algorithm) identified multiple clusters in 152 of 154 WUS up-regulated genes and 298 of 303 WUS down-regulated genes (tables S6 and S7). This resource should guide future in vivo analysis to refine our understanding of the relationship between CRMs and gene expression specificity.
Our analysis also shows that the interaction between cis-elements in promoting higher molecular WUS complexes also depends on the distance between cis-elements. Increasing the distance between cis-elements unexpectedly decreased the WUS detection threshold, suggesting that distance may also play a role in sensing WUS concentration through an unknown mechanism. This might increase the probability of WUS monomer binding to adjacent cis-elements. However, the stabilization of WUS into a higher molecular weight complexes still occurred at the same WUS levels as observed with the wild-type distance. Thus, the increase in CLV3 expression observed upon increasing the distance could be due to increased activation and not entirely due to the reduced repression. Together, our results show that the cis-element affinity plays a dominant role in CLV3 repression, while it appears that the system can withstand an increase in intervening distance in forming higher WUS complexes.
The computational model developed in this study allows us to recreate and, in a sense, verify the plausibility of our mechanistic explanations of experimental results. It was possible to quantify properties that are very difficult to obtain through experimental means such as the residence time of WUS on cis-elements to calibrate the model and visualization of concentration-dependent ratios of WUS monomer and dimer/higher-order complexes on the CLV3 cis-elements. The upper limit on the residence time of WUS was critical to explain individual cis-element behaviors that differ in their binding affinities. Our experimental analysis shows that a higher WUS turnover leads to a higher CLV3 activation, suggesting that older WUS species may become ineffective and may unbind. The nuclear export of WUS has been shown to play a crucial role in regulating the WUS nuclear concentration (51). It has also been shown that a nuclear export signal is required for WUS degradation in the cytoplasm. Perhaps the older WUS molecules that unbind are exported and degraded in the cytoplasm, which may create space for newly synthesized WUS that moves into the outer layer of CZ to bind cis-elements to sustain CLV3 activation. CLV3 has been shown to offset nuclear export of WUS, which forms an additional feedback mechanism in regulating the nuclear concentration (51). Whether CLV3 levels also independently determine residence time of WUS by influencing its unbinding from cis-elements perhaps by regulating the WUS protein modifications remains to be explored. Nevertheless, a seamless connection involving WUS binding, unbinding, export, and degradation could lead to a robust maintenance of CLV3 transcription. However, the current model assumes a constant WUS gradient and is limited to exploring the mechanisms underlying the CLV3 expression without considering the feedback regulations of CLV3 signaling on WUS. Our recent study developed a model involving both transcriptional and posttranslational regulations of WUS by the CLV3 signaling (51). This model used a generic function of WUS concentration to represent the CLV3 transcription. The model perturbations revealed that the dual control of WUS transcription and nuclear levels by the CLV3 signaling when coupled to the WUS concentration–dependent transcriptional activation and repression of CLV3 leads to a robust maintenance of the WUS protein gradient. Our results show that the cis-element mutant reporter 970i was markedly reset into the outer layers of CZ in the clv3 null mutants complemented with the 970i genomic construct (fig. S10, A and B). Perhaps this is due to the effects of altered CLV3 signaling on the expression and nuclear accumulation of WUS establishing a new gradient. In the future, coupling the 3D stochastic model of CLV3 transcription developed here with the CLV3 signaling model of the regulation of WUS transcription and the WUS protein dynamics should allow assessment of the influence of different properties of the CLV3 CRM, including the number of cis-elements in regulating the robustness of the WUS gradient.
MATERIALS AND METHODS
Experimental design
Plants were grown under continuous light as described earlier in (24). Imaging was performed on the Zeiss 880 AIRYSCAN upright under a 40× objective. eGFP-WUS was excited at 488 nm and collected with filter 495 to 550 nm. Histone 2B modified yellow fluorescent protein (H2B-mYFP) was excited at 514-nm filtered with main beam splitter (MBS) 458/514/561/633 and collected with band-pass (BP) filter 495 to 550 nm. FM4-64 was excited at 561 nm and collected with BP 570 to 620 nm.
Stochastic single-cell model and the 3D cell–based model
Description of two computational models developed in this study is provided in detail in the Supplementary Materials. Parameters used in the stochastic single-cell model can be found in tables S1 and S2. Parameters used in the 3D cell-based model can be found in tables S1 and S3.
Statistical analysis
The source data associated with all experiments are presented in the additional data files. In addition, the means, N, and P values are included within each dataset.
Acknowledgments
We thank K. Hill, V. Ceja, D. Nejad, I. Gutierrez, V. Franco, and P. Rubiro for supporting experimental work and A. Plong for comments on the manuscript. The 3D computational model used the simulation framework developed by H. Jonsson (23) as its base framework. Additional helpful comments were provided by N. Korsbo.
Funding: This work was supported by NSF grant IOS-2055690 (to G.V.R., W.C., and M. Digman), RSAP-AES mission funding (to G.V.R.), and NSF grant DMS-1762063 through the joint NSF DMS/NIH NIGMS Initiative (to M.A., G.V.R., and W.C.).
Author contributions: Conceived research: K.R., G.V.R., and W.C. Experimental investigation: K.R., A.D., M.P., and G.V.R. Single-cell model and simulation: B.S.-A. and W.C. 3D model and simulation: A.D., B.S.-A., and W.C. Writing—original draft: K.R., A.D., B.S.-A., W.C., and G.V.R. Writing—review and editing: K.R., A.D., B.S.-A., M.P., M.A., W.C., and G.V.R.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Code availability: 1. An open-source MATLAB/C++ implementation of the computational model of transcription is available at GitHub (https://github.com/weitaoc/SAM.git and https://github.com/Ado012/MeristemBasic_p) or in Zenodo (DOI: 10.5281/zenodo.6632514 and DOI: 10.5281/zenodo.6629834) (58, 59). 2. Code for cis-element analysis is available at https://github.com/Ado012/BasicCisElementAnalyzer or in Zenodo (DOI: 105201/zenodo.6632279) (60).
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S11
Tables S1 to S5
References
Other Supplementary Material for this manuscript includes the following:
Tables S6 and S7
Data S1
REFERENCES AND NOTES
- 1.Levine M., Transcriptional enhancers in animal development and evolution. Curr. Biol. 20, R754–R763 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ong C.-T., Corces V. G., Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 12, 283–293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spitz F., Furlong E. E. M., Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Banerji J., Rusconi S., Schaffner W., Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299–308 (1981). [DOI] [PubMed] [Google Scholar]
- 5.Slattery M., Riley T., Liu P., Abe N., Gomez-Alcala P., Dror I., Zhou T., Rohs R., Honig B., Bussemaker H. J., Mann R. S., Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147, 1270–1282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Struhl G., Struhl K., Macdonald P. M., The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell 57, 1259–1273 (1989). [DOI] [PubMed] [Google Scholar]
- 7.Fakhouri W. D., Ay A., Sayal R., Dresch J., Dayringer E., Arnosti D. N., Deciphering a transcriptional regulatory code: Modeling short-range repression in the Drosophila embryo. Mol. Syst. Biol. 6, 341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joshi R., Passner J. M., Rohs R., Jain R., Sosinsky A., Crickmore M. A., Jacob V., Aggarwal A. K., Honig B., Mann R. S., Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell 131, 530–543 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crocker J., Abe N., Rinaldi L., McGregor A. P., Frankel N., Wang S., Alsawadi A., Valenti P., Plaza S., Payre F., Mann R. S., Stern D. L., Low affinity binding site clusters confer Hox specificity and regulatory robustness. Cell 160, 191–203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman B. P., Nibu Y., Pfeiffer B. D., Tomancak P., Celniker S. E., Levine M., Rubin G. M., Eisen M. B., Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. U.S.A. 99, 757–762 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lifanov A. P., Makeev V. J., Nazina A. G., Papatsenko D. A., Homotypic regulatory clusters in Drosophila. Genome Res. 13, 579–588 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Small S., Blair A., Levine M., Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 11, 4047–4057 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ip Y. T., Park R. E., Kosman D., Bier E., Levine M., The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 6, 1728–1739 (1992). [DOI] [PubMed] [Google Scholar]
- 14.Driever W., Nüsslein-Volhard C., The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature 337, 138–143 (1989). [DOI] [PubMed] [Google Scholar]
- 15.Gaudet J., Mango S. E., Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science 295, 821–825 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Jiang J., Levine M., Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell 72, 741–752 (1993). [DOI] [PubMed] [Google Scholar]
- 17.Rowan S., Siggers T., Lachke S. A., Yue Y., Bulyk M. L., Maas R. L., Precise temporal control of the eye regulatory gene Pax6 via enhancer-binding site affinity. Genes Dev. 24, 980–985 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolpert L., Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol. 25, 1–47 (1969). [DOI] [PubMed] [Google Scholar]
- 19.Jiang J., Kosman D., Ip Y. T., Levine M., The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes Dev. 5, 1881–1891 (1991). [DOI] [PubMed] [Google Scholar]
- 20.Parker D. S., White M. A., Ramos A. I., Cohen B. A., Barolo S., The cis-regulatory logic of Hedgehog gradient responses: Key roles for gli binding affinity, competition, and cooperativity. Sci. Signal. 4, ra38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer K. F., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T., Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Schoof H., Lenhard M., Haecker A., Mayer K. F. X., Jürgens G., Laux T., The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Yadav R. K., Perales M., Gruel J., Girke T., Jönsson H., Reddy G. V., WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 25, 2025–2030 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perales M., Rodriguez K., Snipes S., Yadav R. K., Diaz-Mendoza M., Reddy G. V., Threshold-dependent transcriptional discrimination underlies stem cell homeostasis. Proc. Natl. Acad. Sci. U.S.A. 113, E6298–E6306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark S. E., Williams R. W., Meyerowitz E. M., The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Brand U., Fletcher J. C., Hobe M., Meyerowitz E. M., Simon R., Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Yadav R. K., Perales M., Gruel J., Ohno C., Heisler M., Girke T., Jönsson H., Reddy G. V., Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol. Syst. Biol. 9, 654 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.C. Koppermann, “Crystal structure of the WUSCHEL homeodomain,” thesis, Universität zu Köln, Cologne, Germany (2017). [Google Scholar]
- 29.Sloan J., Hakenjos J. P., Gebert M., Ermakova O., Gumiero A., Stier G., Wild K., Sinning I., Lohmann J. U., Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL. Nat. Commun. 11, 2223 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinitz J., Hou S., Sharp D. H., Transcriptional control in Drosophila. Complexus 1, 54–64 (2003). [Google Scholar]
- 31.Shea M. A., Ackers G. K., The OR control system of bacteriophage lambda. A physical-chemical model for gene regulation. J. Mol. Biol. 181, 211–230 (1985). [DOI] [PubMed] [Google Scholar]
- 32.He X., Samee M. A. H., Blatti C., Sinha S., Thermodynamics-based models of transcriptional regulation by enhancers: The roles of synergistic activation, cooperative binding and short-range repression. PLOS Comput. Biol. 6, e1000935 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu D., Zabet N. R., Mitavskiy B., Models of transcription factor binding: Sensitivity of activation functions to model assumptions. J. Theor. Biol. 257, 419–429 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Sherman M. S., Cohen B. A., Thermodynamic state ensemble models of cis-regulation. PLOS Comput. Biol. 8, e1002407 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gillespie D. T., A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 22, 403–434 (1976). [Google Scholar]
- 36.Swift J., Coruzzi G. M., A matter of time—How transient transcription factor interactions create dynamic gene regulatory networks. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 75–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Brien T., Lis J. T., Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol. Cell. Biol. 13, 3456–3463 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ackers G. K., Johnson A. D., Shea M. A., Quantitative model for gene regulation by lambda phage repressor. Proc. Natl. Acad. Sci. U.S.A. 79, 1129–1133 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia H. G., Phillips R., Quantitative dissection of the simple repression input-output function. Proc. Natl. Acad. Sci. U.S.A. 108, 12173–12178 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gertz J., Siggia E. D., Cohen B. A., Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature 457, 215–218 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segal E., Raveh-Sadka T., Schroeder M., Unnerstall U., Gaul U., Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature 451, 535–540 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Zinzen R. P., Girardot C., Gagneur J., Braun M., Furlong E. E. M., Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462, 65–70 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Amit R., Garcia H. G., Phillips R., Fraser S. E., Building enhancers from the ground up: A synthetic biology approach. Cell 146, 105–118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.E. Davidson, The Regulatory Genome (Academic Press, 2006). [Google Scholar]
- 45.Garcia H. G., Sanchez A., Boedicker J. Q., Osborne M., Gelles J., Kondev J., Phillips R., Operator sequence alters gene expression independently of transcription factor occupancy in bacteria. Cell Rep. 2, 150–161 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lickwar C. R., Mueller F., Hanlon S. E., McNally J. G., Lieb J. D., Genome-wide protein–DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 484, 251–255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lipford J. R., Smith G. T., Chi Y., Deshaies R. J., A putative stimulatory role for activator turnover in gene expression. Nature 438, 113–116 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Spoel S. H., Mou Z., Tada Y., Spivey N. W., Genschik P., Dong X., Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137, 860–872 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geng F., Wenzel S., Tansey W. P., Ubiquitin and proteasomes in transcription. Annu. Rev. Biochem. 81, 177–201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sundqvist A., Ericsson J., Transcription-dependent degradation controls the stability of the SREBP family of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 100, 13833–13838 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plong A., Rodriguez K., Alber M., Chen W., Reddy G. V., CLAVATA3 mediated simultaneous control of transcriptional and post-translational processes provides robustness to the WUSCHEL gradient. Nat. Commun. 12, 6361 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Snipes S. A., Rodriguez K., DeVries A. E., Miyawaki K. N., Perales M., Xie M., Reddy G. V., Cytokinin stabilizes WUSCHEL by acting on the protein domains required for nuclear enrichment and transcription. PLOS Genet. 14, e1007351 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodriguez K., Perales M., Snipes S., Yadav R. K., Diaz-Mendoza M., Reddy G. V., DNA-dependent homodimerization, sub-cellular partitioning, and protein destabilization control WUSCHEL levels and spatial patterning. Proc. Natl. Acad. Sci. U.S.A. 113, E6307–E6315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sayal R., Dresch J. M., Pushel I., Taylor B. R., Arnosti D. N., Quantitative perturbation-based analysis of gene expression predicts enhancer activity in early Drosophila embryo. Elife 5, e08445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Y., Liu X., Engstrom E. M., Nimchuk Z. L., Pruneda-Paz J. L., Tarr P. T., Yan A., Kay S. A., Meyerowitz E. M., Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517, 377–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Su Y. H., Zhou C., Li Y. J., Yu Y., Tang L. P., Zhang W. J., Yao W. J., Huang R., Laux T., Zhang X. S., Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U.S.A. 117, 22561–22571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kieffer M., Stern Y., Cook H., Clerici E., Maulbetsch C., Laux T., Davies B., Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 18, 560–573 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.B. Senay-Aras, W. Chen. Stochastic cis-elements binding model (2022); https://zenodo.org/badge/latestdoi/349283086.
- 59.A. Do, MeristemBasic_p (2022); https://zenodo.org/badge/latestdoi/501822165.
- 60.A. Do, BasicCisElementAnalyzer (2022); https://zenodo.org/badge/latestdoi/498280124.
- 61.Roussel M. R., Zhu R., Validation of an algorithm for delay stochastic simulation of transcription and translation in prokaryotic gene expression. Phys. Biol. 3, 274–284 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Azpeitia E., Wagner A., Short residence times of DNA-bound transcription factors can reduce gene expression noise and increase the transmission of information in a gene regulation system. Front. Mol. Biosci. 7, 67 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gutierrez P. S., Monteoliva D., Diambra L., Cooperative binding of transcription factors promotes bimodal gene expression response. PLOS ONE 7, e44812 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brand U., Grünewald M., Hobe M., Simon R., Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129, 565–575 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S11
Tables S1 to S5
References
Tables S6 and S7
Data S1