Abstract
The nirA gene encoding the blue dissimilatory nitrite reductase from Alcaligenes xylosoxidans has been cloned and sequenced. To our knowledge, this is the first report of the characterization of a gene encoding a blue copper-containing nitrite reductase. The deduced amino acid sequence exhibits a high degree of similarity to other copper-containing nitrite reductases from various bacterial sources. The full-length protein included a 24-amino-acid leader peptide. The nirA gene was overexpressed in Escherichia coli and was shown to be exported to the periplasm. Purification was achieved in a single step, and analysis of the recombinant Nir enzyme revealed that cleavage of the signal peptide occurred at a position identical to that for the native enzyme isolated from A. xylosoxidans. The recombinant Nir isolated directly was blue and trimeric and, on the basis of electron paramagnetic resonance spectroscopy and metal analysis, possessed only type 1 copper centers. This type 2-depleted enzyme preparation also had a low nitrite reductase enzyme activity. Incubation of the periplasmic fraction with copper sulfate prior to purification resulted in the isolation of an enzyme with a full complement of type 1 and type 2 copper centers and a high specific activity. The kinetic properties of the recombinant enzyme were indistinguishable from those of the native nitrite reductase isolated from A. xylosoxidans. This rapid isolation procedure will greatly facilitate genetic and biochemical characterization of both wild-type and mutant derivatives of this protein.
Dissimilatory nitrite reductase is a key enzyme in the denitrification process, in which nitrate undergoes stepwise reduction to the gaseous products nitrous oxide and dinitrogen (42). There are two distinct classes of periplasmic nitrite reductase: one containing cd1 heme as the prosthetic group and the other containing copper (42). The copper centers in nitrite reductases comprise type 1 centers, giving rise to the blue or green color of these enzymes, and type 2 centers, which do not contribute significantly to the visible spectrum. Copper-containing nitrite reductases can be distinguished as either green enzymes or blue enzymes depending on the electronic absorbance of their type 1 copper centers. Thus, the enzymes that have been isolated from Achromobacter cycloclastes, Alcaligenes faecalis, Pseudomonas sp., and Rhodobacter sphaeroides are green enzymes that have absorbance maxima at ∼460, 595, and 700 to 750 nm, while Alcaligenes xylosoxidans and Pseudomonas aureofaciens (42) have blue enzymes that show little absorbance in the 460-nm range. The X-ray structures of the Nir enzymes from A. cycloclastes (3, 11), A. faecalis (15), and A. xylosoxidans (6, 7, 14) have been determined at 2- to 2.1-Å resolution. This information, together with the deduced amino acid sequences of a number of nitrite reductases from various sources derived from the gene sequences, has been important in elucidating the type 1 and type 2 copper ligands (5, 9, 10, 22, 35, 36, 41). The availability of the high-resolution structures of the blue Nir enzyme from A. xylosoxidans has allowed comparison of the structures of the type 1 centers of the blue and green enzymes. The structures of the type 1 sites show the same ligands to the copper ions and little difference in the Cu ligand distances, including that of the Cu-Sγ (Met) bond, which had been suggested to be responsible for the differences in color (3). The major difference is in the His-Cu-Met angle of 17° between the two classes of type 1 Cu sites (7), a finding consistent with a theoretical analysis of the factors likely to affect the electronic structure of such Cu sites (16). In addition, it has been proposed that the difference in color may result from a shift in the polypeptide backbone around the Met caused by a Tyr residue in the neighborhood of the type 1 copper center of the green enzymes being replaced with a Thr in the blue enzymes (14).
Work originally done by Libby and Averill (19) with the A. cycloclastes enzyme and later studies with the enzyme from A. xylosoxidans (1, 2) indicated depletion of the type 2 copper center (type 2-depleted [T2D] enzyme) was associated with a dramatic reduction in enzyme activity. Indeed, both native and recombinant enzymes appear to lose the type 2 Cu during isolation (42). Reconstitution of the center by incubation with exogenous copper restores enzyme activity and type 2 Cu electron paramagnetic resonance (EPR) signals (1, 2, 15). These results are commensurate with the type 2 copper center being involved directly in catalysis (13, 23). Evidence has been presented that indicates that the function of the type 1 Cu center is to receive electrons from the physiological electron donor pseudoazurin (15) and to transfer them to the type 2 center (8, 31, 32, 38).
In order to dissect the molecular events involved in electron transfer between the copper centers and the reduction of nitrite at the type 2 copper center, it would be a significant advantage to have an expression system that permits ready manipulation of the gene. This would facilitate the introduction of specific mutations and thus provide a rapid means by which the enzyme could be recovered in large amounts for biochemical analysis. We describe here the cloning of the nirA gene from A. xylosoxidans, the purification of the enzyme from the periplasmic fraction of Escherichia coli, and the characterization of the recombinant enzyme. Among the many advantages presented by this heterologous expression system is that the recombinant enzyme on a biochemical basis is essentially indistinguishable from the native enzyme. Moreover, recombinant Nir can be readily isolated in a form that is wholly deficient in type 2 copper centers. Significantly, the enzyme can also be isolated with a full complement of type 1 and type 2 centers simply by incubation with copper sulfate prior to purification.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains used in this study were A. xylosoxidans subsp. xylosoxidans (NCIMB 11015) and E. coli JM109 (40) and BL21(DE3) (30). The E. coli strains were grown in Luria-Bertani medium. A. xylosoxidans was grown in a medium containing the following (per liter): nutrient broth, 8 g; NaCl, 0.5 g; yeast extract, 1 g; Na2CH3O2, 5 g; NaNO3, 5 g; and 1 μM concentrations of CuSO4, Na2MoO4, MnSO4, and FeCl3. Growth was performed in standing cultures at 30°C, while E. coli strains were grown aerobically at 37°C in vigorously shaking conical flasks filled to a maximum of 10% of their volumes with medium. When used, glucose was added to a final concentration of 10 mM, and isopropyl-β-d-thiogalactopyranoside (IPTG) was used at a final concentration of 0.25 mM. Antibiotics were used at the following final concentrations: ampicillin, 50 mg · liter−1; kanamycin, 50 mg · liter−1.
Cloning of the A. xylosoxidans nirA gene.
Two degenerate primers based on the amino acid sequence of the A. xylosoxidans NirA protein (36a) were designed. The forward primer corresponded to amino acid positions 33 to 38 of the protein and had the sequence 5′-AAGGARTTCACNATGAC-3′, and the reverse primer, which corresponded to amino acid positions 254 to 259, had the sequence 5′-CCANACCCARTCNCCRTG-3′. N represents any nucleotide, while R represents an A or a G. A PCR was performed with 20 pmol of each of the above primers and 1 ng of chromosomal DNA isolated from A. xylosoxidans. The resulting ∼680-bp DNA fragment was subcloned into SmaI-digested pUC19, yielding plasmid pUAX-1, and the authenticity of the DNA sequence of the insert was confirmed (27). In order to clone the wild-type nirA gene, 10-μg aliquots of chromosomal DNA from A. xylosoxidans were initially digested to completion with either BamHI or SalI and, after separation in a 0.8% (wt/vol) agarose gel, the DNA fragments were blotted onto a nitrocellulose membrane and hybridized with the 32P-labelled DNA insert from pUAX-1 (26). Two signals of 3.0 and 2.1 kb were detected after BamHI digestion, and fragments of 1.4 and 1.2 kb were detected after SalI digestion. The 2.1-kb BamHI DNA fragment was successfully cloned into pUC19 digested with BamHI, yielding pUB1.
Due to problems of instability the 3′ portion of the nirA gene could only be cloned by performing inverse PCR (25) on the 1.2-kb SalI DNA fragment. Subsequent to isolation of the DNA fragment, plasmid minicircles were generated by ligating the DNA fragments in a large volume (0.1 ml) to promote intramolecular ligation events. A 1-ng aliquot of the ligation mixture was used as the template in a PCR with two oligonucleotide primers (Nit-11, 5′-CCTGCTCGCCAGGGTTGA-3′; Nit-15, 5′-CCGCACCTGATCGGCGGC-3′) that were designed based on the nucleotide sequence of the nirA gene determined from pUB1. The PCR generated a 900-bp DNA fragment that was cloned into the SmaI site of pUC19, yielding plasmid pUS6. The nirA gene was sequenced completely on both strands (27).
The complete nirA gene was amplified by using Pfu DNA polymerase (Stratagene) from the chromosome of A. xylosoxidans with oligonucleotides NF-1 (5′-GGGAGCTCACATGAACGCATTACGGC-3′) and NF-2 (5′-GGAAGCTTCCAGTGCCAATCTGATTGC-3′). These oligonucleotides introduced a SacI restriction site at the 5′ end of the nirA gene and a HindIII site at the 3′ end. After digestion of the amplified product with SacI and HindIII, it was cloned into SacI-HindIII-digested pET28a, yielding pEnirsp-1. The nirA gene in pEnirsp-1 possesses the artificial ribosome-binding site GGAG, which was derived from the SacI restriction site and a G residue of the plasmid polylinker. The pEnirsp-2 derivative was created by digesting pEnirsp-1 with NdeI, filling in the protruding 5′ ends with the Klenow fragment of DNA polymerase and deoxynucleoside triphosphates according to the method of Sambrook et al. (26), and then ligating the product.
Overproduction and subcellular localization of NirA.
Overexpression of the nirA gene was achieved by introducing pEnirsp-1 into BL21(DE3) (30). Four 0.5-liter cultures of the transformed strain were grown at 30°C until an optical density at 600 nm of 0.5 was attained. Subsequently, IPTG was added to a final concentration of 0.25 mM, and the culture was incubated for a further 90 min with vigorous agitation. The cells were harvested by centrifugation at 8,000 × g for 20 min at 4°C. The cell pellet was resuspended in 15 ml of 50 mM potassium phosphate buffer (pH 7.0) and centrifuged again at 8,000 × g for 20 min. Spheroplasts and the periplasmic fraction were prepared according to the method of Osborn et al. (24). Briefly, the cell pellet was resuspended in 10 mM potassium phosphate buffer (pH 7.0) at a concentration of 1 ml/g (wet weight) of cells. To each milliliter of cell suspension 5 ml of 1 M sucrose, 0.4 ml of 1 M Tris-HCl (pH 8.0), 0.4 ml of 0.1 M EDTA (pH 8.0), and 1.2 ml of a freshly prepared 5-mg/ml solution of lysozyme were added. The mixture was stirred gently for 2 min, and then 1 ml of 0.1 M MgCl2 and 10 ml of H2O were slowly added. The mixture was gently stirred at 30°C for 30 min. Spheroplasts were separated from the periplasmic fraction by centrifugation at 12,000 × g for 30 min. The spheroplasts were resuspended in 20 ml of 10 mM potassium phosphate buffer and disrupted by two passages through a French press at 16,000 lb/in2 (1.03 × 102 MPa). The crude extract was prepared by centrifugation at 10,000 × g for 30 min.
Purification of mature, recombinant nitrite reductase.
Mature, recombinant NirA was purified to apparent homogeneity from the periplasmic fraction of BL21(DE3) in a single chromatographic step with a carboxymethyl cellulose CM52 matrix (Whatman). A 110-ml (14 by 2.5 cm) column of CM52 initially equilibrated with 20 mM Tris-HCl (pH 7.0) was equilibrated with H2O and 95 ml (∼40 mg of protein) of periplasmic fraction was applied. Unbound proteins were removed by washing the column with H2O. NirA appeared as a tight, dark blue band at the top of the column. Pure NirA was eluted with 20 mM Tris-HCl (pH 7.0) containing 50 mM NaCl. Subsequent to elution, NirA was dialyzed against 20 mM MES (morpholineethanesulfonic acid; pH 6.0) and concentrated to approximately 2 mg ml−1 (total volume, ∼0.5 ml) by using a Centricon-10 concentrator (Amicon).
Reconstitution of type II copper sites in recombinant nitrite reductase.
The periplasmic fraction was dialyzed at 4°C for 16 h against 1,000 volumes of 20 mM MES buffer (pH 6.0) containing 0.1 mM CuSO4. The periplasmic fraction was then dialyzed exhaustively against three changes of 20 mM MES buffer (pH 6.0) with a minimum equilibration period of 5 h between buffer changes.
Analysis of nitrite reductase enzyme activity.
Nitrite reductase enzyme activity was determined by using the discontinuous assay described previously (1). The thermostability of native and recombinant nitrite reductase was determined by measuring enzyme activity after incubation of 1 ml of a solution of the enzyme (40 μg of protein in 20 mM MES [pH 6.0]) in a water bath at 80°C and after 5-μl aliquots were withdrawn at various time intervals.
Spectroscopic methods.
UV-visible spectra were recorded at room temperature on a Hewlett-Packard 8452A diode array spectrophotometer. EPR spectra were recorded at 60 K, at a microwave power of 20 mW, a microwave frequency of 9.3 GHz, and a modulation amplitude of 4.637 G on a Bruker ER ER200 D-SRC spectrometer fitted with an ER042 MRH microwave bridge and by using an ER033C field frequency lock and an Oxford Instruments Ite503 temperature controller. Integrated intensities were obtained by comparison with aqueous Cu-EDTA with a g value correction applied as described previously (1). Simulations were performed by using a computer program based on that of Lowe (20).
Other methods.
The protein concentration was determined by the method of Lowry et al. (21). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of proteins was performed as described previously (17). Specific radioactive labelling of polypeptides with [35S]methionine was performed with strain BL21(DE3) as described previously (18, 33). Aliquots were removed different time points, and labelled polypeptides were analyzed by autoradiography after separation by SDS-PAGE. The copper content of purified nitrite reductase was determined on wet-ash samples by using induced coupled plasma emission spectroscopy exactly as described previously (1). Amino acid alignments were performed with the CLUSTAL W program (34).
RESULTS AND DISCUSSION
Cloning and sequence analysis of the nirA gene.
The amino acid sequence of A. xylosoxidans nitrite reductase has recently been determined by chemical analysis and mass spectrometry (37). To facilitate cloning of the complete nirA gene for a blue Nir, a DNA probe of the nirA gene was generated by PCR by using degenerate oligonucleotides, which were designed based on the Nir amino acid sequence, as described in Materials and Methods. Southern blot analysis of A. xylosoxidans chromosomal DNA failed to identify a suitably sized restriction fragment that carried the complete nirA gene (data not shown). Portions of the nirA gene could, however, be shown to reside on 3.0- and 2.1-kb BamHI fragments or on 1.4- and 1.2-kb SalI DNA fragments. Both the 2.1-kb BamHI fragment and the 1.4-kb SalI fragment could be readily cloned in pUC19. DNA sequence analysis of the BamHI fragment in pUB1 revealed that only the initial 548 bp of the nirA gene were present on the plasmid. It was not possible to clone either the 3-kb BamHI fragment or the 1.2-kb SalI DNA fragment in either a high- or a low-copy-number vector due to problems with plasmid instability. Instead, an inverse PCR approach with minicircles generated from the 1.2-kb SalI DNA fragment proved successful in amplifying the 3′ portion of the nirA gene. The complete nirA gene was subsequently cloned on a single DNA fragment, and determination of the nucleotide sequence on both strands failed to reveal a discrepancy in the sequence of several clones. Moreover, all plasmids constructed that contained the complete nirA gene were completely stable. This strongly suggests that the inability to clone the 3-kb BamHI fragment or the 1.2-kb SalI DNA fragment must be due to DNA sequences on these fragments that lie outside the nirA coding sequence.
Just prior to publication of this sequence, the DNA sequence of the A. xylosoxidans nirA sequence appeared in the database under accession number AF051831. The nucleotide sequence we determined was identical to that published in the GenBank database. Furthermore, our deduced amino acid sequence of Nir was identical to the amino acid sequence of the Nir protein recently determined by protein-sequencing methods (37).
The nirA gene encodes a 360-amino-acid protein, the first 24 amino acids of which constitute the signal peptide (reference 4 and see below). The signal sequence includes a single Arg residue near the N terminus and otherwise shares no significant similarity with the signal sequences of other copper-containing nitrite reductases (Fig. 1). The deduced amino acid sequence is identical to that determined by Edman degradation (37), except that the gene sequence predicts the first codon of the mature enzyme to be a glutaminyl residue, whereas the N-terminal amino acid of the native Nir enzyme isolated from A. xylosoxidans has been shown to be pyroglutamate (12a, 37).
FIG. 1.
Alignment of the deduced amino acid sequences of copper-containing nitrite reductases from different bacterial sources. The amino acid alignment was generated with the CLUSTAL W package (34). Asterisks represent amino acid identity, and dots indicate similar amino acids. Similar amino acids include aromatic amino acids, basic amino acids, acidic amino acids, and hydrophobic amino acids. The numerals 1 and 2 above the sequence alignment signify the type 1 and type 2 copper ligands, respectively. ALCXY, A. xylosoxidans accession number AF051831; ACHCY, A. cycloclastes accession number Z48635 (9); ALCFA, A. faecalis accession number D13155 (22); PSEsp, Pseudomonas sp. strain G-179 accession number M97294 (41); PSEAR, P. aureofaciens accession number Z21945 (10); RHIHE, R. hedysari accession number U65658 (35); RHOSPH, R. sphaeroides accession number U62291 (36).
An alignment of the deduced amino acid sequence of the nirA gene with those of other copper-containing nitrite reductases from various bacterial sources is shown in Fig. 1. Pairwise comparisons of Nir from A. xylosoxidans with other Nir sequences reveals a significant primary sequence identity ranging between 48 and 77%, with overall similarity ranging from 58 to 83%. The ligands to the type 1 and type 2 copper centers are highly conserved among all Nir enzymes. His-89, Cys-130, His-139, and Met-144 are the ligands to the type 1 copper site, while His-94, His-129, and His-300 are ligands to the type 2 copper site in the mature A. xylosoxidans Nir protein (Fig. 1).
Of the seven proteins shown in Fig. 1, only the Nir enzyme from A. xylosoxidans and that from P. aureofaciens (10) have short signal peptides with the characteristics typical of proteins secreted by the Sec-dependent export pathway (4). Notably, the mature Nir proteins from these two organisms also share the highest degree of amino acid similarity (Fig. 1). The remaining five nitrite reductases have long signal peptides of approximately 43 to 45 amino acids, with a double-arginine motif close to the N terminus. This type of signal sequence, including the double-arginine motif, has recently been shown to be recognized by a novel export pathway that is Sec independent and which has been proposed to secrete folded, redox-active proteins (4, 28, 29, 39). The possible physiological significance of this difference remains to be established.
Overproduction and subcellular localization of Nir in E. coli.
The nirA gene was subcloned from pUnirsp-1 into pET28a on a SacI-HindIII fragment (see Materials and Methods). The resulting plasmid (pEnirsp-1) was transformed into BL21(DE3), and the polypeptides derived from T7 RNA polymerase-directed transcription of the nirA gene were detected (Fig. 2A). After electrophoretic separation in a 12.5% acrylamide gel, two polypeptides with apparent molecular masses of 43 and 35 kDa were observed. The N-terminal amino acid sequence of the smaller polypeptide was determined to be Q-D-A-D-K-L, which is in perfect agreement with that predicted for the mature Nir protein lacking the signal sequence. Thus, mature recombinant Nir (35 kDa) migrated with a molecular mass that was in close agreement with the DNA-deduced molecular mass of 36.5 kDa. Clearly, E. coli, unlike A. xylosoxidans, is unable to modify the N-terminal glutamine residue of Nir to pyroglutamate.
FIG. 2.
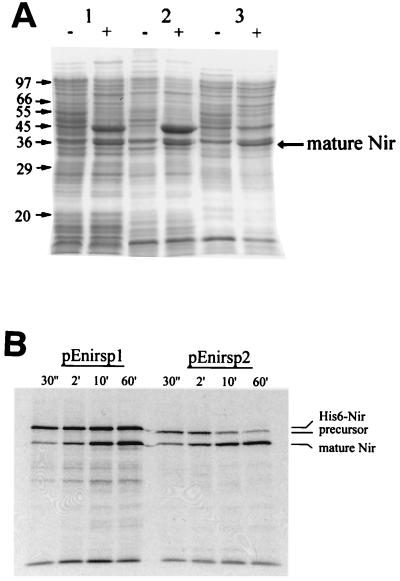
Nitrite reductase from A. xylosoxidans is exported to the periplasm in the heterologous host E. coli BL21(DE3). (A) Subcellular localization of recombinant Nir in E. coli BL21(DE3). Polypeptides (50 μg of protein per lane) were separated by SDS-PAGE in a gel containing 12.5% (wt/vol) acrylamide. Lanes: 1, whole-cell extracts; 2, insoluble material from the crude extract; 3, the periplasmic fraction. The minus sign indicates subcellular fractions derived from BL21(DE3)/pET28a, and the plus sign indicates subcellular fractions derived from BL21(DE3)/pEnirsp-1. The location of the mature Nir protein is indicated. The migration positions of the molecular mass markers are shown on the left of the diagram. (B) Pulse-chase experiment showing that mature Nir production is independent of the His-tagged Nir fusion protein. Samples were removed at the time points indicated, and the [35S]methionine-labelled polypeptides were separated by SDS-PAGE in 12.5% (wt/vol) acrylamide gels. After being dried, the gels were exposed to X-ray film. The migration positions of the insoluble His6-Nir fusion protein, the Nir precursor, and mature Nir are shown.
The N terminus of the larger polypeptide had the sequence G-S-S-H-H-H-H-H, which corresponds to the first 8 amino acids (minus the initiator Met residue) of the His tag peptide of pET28a. DNA sequence analysis of pEnirsp-1 confirmed that the nirA gene was fortuitously cloned in frame with the coding region of the polyhistidine tag, generating a hybrid His-tagged Nir protein.
To determine whether the mature Nir protein resulted from cleavage of the native precursor protein and not the larger polypeptide, a pulse-chase experiment with [35S]methionine was carried out (Fig. 2B). Two radioactive polypeptides with molecular masses of 43 and 35 kDa were detected with BL21(DE3) transformed with pEnirsp-1. Both polypeptides accumulated over the time period of the experiment and did not show a typical precursor-product relationship (Fig. 2B). Surprisingly, we were unable to detect the native 39-kDa precursor of Nir.
Filling in the NdeI restriction site in pEnirsp-1 created plasmid pEnirsp-2, which carries a frameshift in the coding region for the larger polypeptide carrying the polyhistidine tag (see Materials and Methods). Analysis of the products from this derivative revealed that, as anticipated, the large 43-kDa protein was not synthesized (Fig. 2B). However, the processed, mature 35-kDa Nir polypeptide was still produced at a level similar to that observed with pEnirsp-1. This result strongly suggests that the precursor of mature Nir was indeed the native Nir polypeptide whose translation initiated at the wild-type ribosome-binding site and was not the 43-kDa polypeptide. This finding was further supported by the appearance of a polypeptide of ∼41 kDa, which had a size slightly larger than that expected for the wild-type precursor (Fig. 2B). Nevertheless, this polypeptide exhibited a precursor-product relationship with the 35-kDa protein, suggesting that it is indeed the native precursor of mature Nir. It is currently unclear why the native precursor was not observed as a product from the pEnirsp-1 derivative.
In A. xylosoxidans Nir is a periplasmic enzyme (42). Separation of the subcellular fractions of E. coli BL21(DE3) containing pEnirsp-1 revealed that approximately 50% of mature Nir was in the periplasmic fraction, whereas the uncleaved, His-tagged Nir fusion polypeptide was exclusively found in inclusion bodies (Fig. 2A). Clearly, the native Nir signal peptide is recognized and efficiently cleaved by the E. coli export apparatus. This is not the case with the A. faecalis nitrite reductase, which was poorly exported into the periplasm of E. coli at a low temperature and not at all at higher growth temperatures (22). This is perhaps also indicative of the A. xylosoxidans enzyme being exported via the Sec export pathway and the A. faecalis enzyme via the double-arginine Sec-independent pathway.
The results shown in Fig. 2A also provide further support for the contention that mature Nir results from cleavage of the native precursor, since the hybrid His-tagged fusion protein is in inclusion bodies and therefore is inaccessible for export to the periplasm. Moreover, the extra 37 amino acids added to the signal peptide presumably prevent it from being recognized as a substrate by the Sec system (4).
Purification and spectroscopic analysis of mature recombinant Nir.
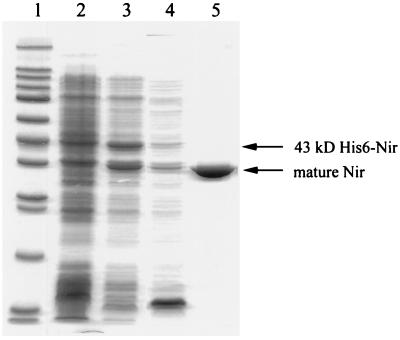
Nir was purified from the periplasmic fraction prepared from approximately 2 g (wet weight) of cells. The specific nitrite reductase activity in the periplasmic fraction was 1.35 μmol of nitrite reduced min−1 mg of protein−1. After activation with copper sulfate the specific activity increased to 5.48 U mg of protein−1 for the periplasmic fraction. Purification of nitrite reductase from the periplasmic fraction was achieved in a single step after chromatography on carboxymethyl cellulose (Fig. 3). Purified nitrite reductase was dark blue and had a specific activity of 167.7 U mg of protein−1. It was not possible to determine the yield of Nir recovered after purification because there was a discrepancy between the total activity of the periplasmic fraction (18.3 U) and the total activity of the purified enzyme (160 U). It appears that an unidentified activity present in the periplasmic fraction of E. coli BL21(DE3) interferes with the accurate determination of nitrite reductase enzyme activity as measured by the methyl viologen-linked assay.
FIG. 3.
Purification of recombinant nitrite reductase. The photograph shows polypeptides separated in a 12.5% (wt/vol) acrylamide gel and stained with Coomassie brilliant blue R-250. Lanes: 1, molecular mass markers (Sigma wide molecular-weight range); 2, crude extract from BL21(DE3)/pET28a (60 μg of protein); 3, crude extract from BL21(DE3)/pEnirsp-1 (50 μg of protein); 4, periplasmic fraction from BL21(DE3)/pET28a (20 μg of protein); 5, purified Nir after carboxymethyl cellulose chromatography (5.5 μg of protein).
Recombinant Nir was indistinguishable from the native enzyme when chromatographed on a Superdex-200 gel filtration column (data not shown). This indicates that purified, activated, recombinant Nir had a trimeric structure, a finding which is in agreement with previous observations (1, 12).
Nitrite reductase that had been purified without prior activation by copper sulfate had a specific activity of 10.8 U mg of protein−1. This indicates that an approximately 16-fold activation had occurred. The copper content of the purified, nonactivated enzyme was 1.97 mol of Cu/mol of enzyme, while that of the activated enzyme was 5.97 mol of Cu/mol of enzyme. This indicates that activation restored a full complement of type 2 Cu sites to the enzyme. Since the type 2 Cu centers are the sites of catalysis, these data are in accord with the nonactivated enzyme having lost the type 2 Cu center and accounts for the reduced enzyme activity observed (see also below).
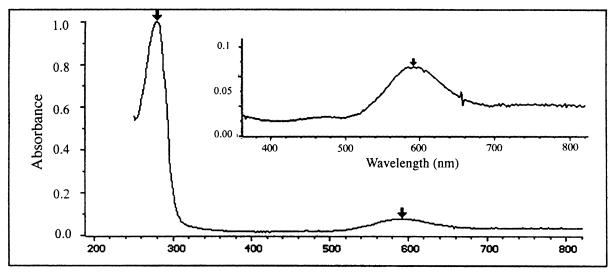
The absorbance spectrum of purified, reconstituted, recombinant Nir is shown in Fig. 4 and is similar to that obtained for the native enzyme (1). The spectrum shows an absorption maximum at ∼595 nm, which is characteristic of type 1 copper centers. The 280 nm/595 nm ratio is 11.6 for the recombinant enzyme and is in good agreement with a similar ratio of 12 observed for the native enzyme (7a). This ratio is indicative of a full occupancy of type 1 copper sites, a conclusion which is in accord both with the metal analysis and the EPR data (see below).
FIG. 4.
UV and visible absorption spectrum of recombinant nitrite reductase. Spectra were recorded in 20 mM MES buffer (pH 6.0) at room temperature. The spectrum was recorded at a 0.75-mg/ml protein concentration.
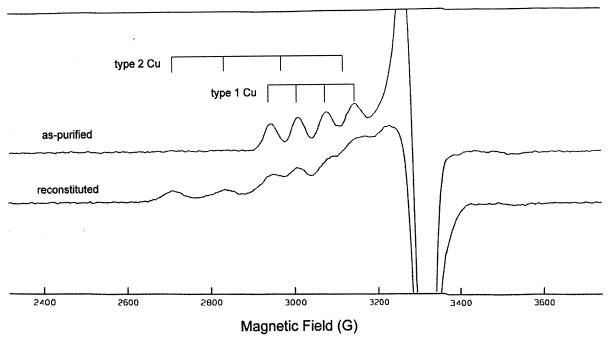
The EPR spectra of recombinant Nir that was purified without prior activation with CuSO4 and of reconstituted Nir are shown in Fig. 5. The nonactivated preparation shows only type 1 copper signals with no detectable type 2 copper. This indicates that the enzyme is T2D. Reconstitution with CuSO4 restored the type 2 Cu signals and delivered a spectrum with features very similar to those of the spectrum of the native enzyme (1). The total amount of EPR-detectable copper gave results comparable to the results determined by metal analysis and indicates that the nonactivated enzyme lacks type 2 copper. Thus, the low nitrite reductase enzyme activity associated with the nonactivated enzyme is due to the absence of the catalytic type 2 sites.
FIG. 5.
EPR spectra of type 2-deficient and reconstituted recombinant nitrite reductase. The experimentally obtained spectra are shown and were recorded at 60 K and at a 10-mW microwave frequency of 9.312 GHz. The simulation (not shown) determined the type 1 Cu to have g" ≈ 1.24, g⊥ ≈ 2.05, and A" ≈ 6.3 mT and the type 2 Cu to have g" ≈ 2.38, g⊥ ≈ 2.05, and A" ≈ 12.7 mT. Approximately equal amounts of type 1 and type 2 copper were determined to be present in the reconstituted sample. The protein concentration of the sample was 9.4 mg/ml for the “as-purified” enzyme (255 μM monomer) and 4.8 mg/ml for the reconstituted enzyme (135 μM monomer). The location of the features corresponding to the type 1 and type 2 Cu centers is shown by the stick diagram.
Thermostability of recombinant and native nitrite reductases.
Nir when purified from A. xylosoxidans is heat stable and when heated at 80°C retains 50% of the initial activity after a 20-min exposure to these potentially denaturing conditions. This resistance to denaturation may arise from the relatively large area of the monomer-monomer interface, which the crystal structure shows to be 4,500 Å2, and is stabilized by extensive interactions (7). The purified recombinant enzyme showed a similar temperature stability (data not shown). This, together with the similarity of the apparent Km for nitrite (35 μM at pH 7.5 [1]) and activity pH profile, indicates that the properties of the catalytic sites and the global features responsible for the thermostability of the enzyme are retained in the recombinant enzyme.
Conclusions.
We were able to isolate approximately 1 mg of pure recombinant Nir from the periplasmic fraction of E. coli BL21(DE3) cells derived from a 2-liter culture. Kinetic and thermostability analyses demonstrated that the recombinant enzyme was indistinguishable from the enzyme isolated directly from A. xylosoxidans. Based on EPR spectroscopy and metal analysis the enzyme that had not been activated with CuSO4 prior to purification was found to possess only type 1 copper. As a consequence, this enzyme had low nitrite reductase enzyme activity. Reconstitution of the type 2 copper sites was achieved by incubation of the periplasmic fraction with CuSO4 prior to enzyme purification. This enzyme was trimeric, had six copper atoms per trimer, exhibited an EPR spectrum with characteristics of type 1 and type 2 copper centers and had a high nitrite reductase specific activity. Taken together with the fact that processing of the signal peptide in the heterologous host occurred at the same site as in A. xylosoxidans, these data indicate that the recombinant enzyme is indistinguishable from the native protein (1). This newly developed isolation procedure with E. coli will greatly facilitate the analysis of the ligands to, and the pathway of electron transfer between, the copper centers. Moreover, the ability to isolate “clean” T2D enzyme will permit analyses of the electron transfer reactions to the type 1 center.
ACKNOWLEDGMENTS
We thank S. A. Fairhurst for performing the EPR analyses.
M.P. was supported by a studentship (BD 5451/95) from Praxis XXI Fundação para Ciência e Tecnologia, Lisbon, Portugal. This work was supported by the BBSRC via a grant-in-aid to the John Innes Centre.
REFERENCES
- 1.Abraham Z H L, Lowe D J, Smith B E. Purification and characterization of the dissimilatory nitrite reductase from Alcaligenes xylosoxidans subsp. xylosoxidans (N.C.I.M.B.): evidence for the presence of both type 1 and type 2 copper centres. Biochem J. 1993;295:587–593. doi: 10.1042/bj2950587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham Z H L, Smith B E, Howes B D, Lowe D J, Eady R R. pH-dependence for binding a single nitrite ion to each type-2 copper centre in the copper-containing nitrite reductase of Alcaligenes xylosoxidans. Biochem J. 1997;324:511–516. doi: 10.1042/bj3240511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adman E T, Godden J W, Turley S. The structure of copper nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2− bound and type II copper depleted. J Biol Chem. 1995;270:27458–27474. doi: 10.1074/jbc.270.46.27458. [DOI] [PubMed] [Google Scholar]
- 4.Berks B C. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 5.Chen J-Y, Chang W-C, Chang T, Chang W-C, Liu M-Y, Payne W J, LeGall J. Cloning, characterization, and expression of the nitric oxide-generating nitrite reductase and of the blue copper protein genes of Achromobacter cycloclastes. Biochem Biophys Res Commun. 1996;219:423–428. doi: 10.1006/bbrc.1996.0249. [DOI] [PubMed] [Google Scholar]
- 6.Dodd F E, Hasnain S S, Abraham Z H L, Eady R R, Smith B E. Structures of a blue copper nitrite reductase and its substrate bound complex. Acta Crystallogr. 1997;D53:406–418. doi: 10.1107/S0907444997002667. [DOI] [PubMed] [Google Scholar]
- 7.Dodd F E, Van Beeumen J, Eady R R, Hasnain S S. X-ray structure of a blue nitrite reductase in two crystal forms. The nature of the copper sites, mode of substrate binding and recognition by redox partner. J Mol Biol. 1998;282:369–382. doi: 10.1006/jmbi.1998.2007. [DOI] [PubMed] [Google Scholar]
- 7a.Eady, R. Unpublished data.
- 8.Farver O, Eady R R, Abraham Z H L, Pecht I. The intramolecular electron transfer between copper sites of nitrite reductase: a comparison with ascorbate oxidase. FEBS Lett. 1998;436:239–242. doi: 10.1016/s0014-5793(98)01120-x. [DOI] [PubMed] [Google Scholar]
- 9.Fenderson F F, Kumar S, Adman E T, Liu M-Y, Payne W J, Legall J. Amino acid sequence of nitrite reductase: a copper protein from Achromobacter cycloclastes. Biochemistry. 1991;30:7180–7187. doi: 10.1021/bi00243a020. [DOI] [PubMed] [Google Scholar]
- 10.Glockner A B, Jüngst A, Zumft W G. Copper-containing nitrite reductase from Pseudomonas aureofaciens is functional in a mutationally cytochrome cd1-free background (NirS−) of Pseudomonas stutzeri. Arch Microbiol. 1993;160:18–26. doi: 10.1007/BF00258141. [DOI] [PubMed] [Google Scholar]
- 11.Godden J W, Turley S, Teller D C, Adman E T, Liu M Y, Payne W J, LeGall J. The 2.3 angstrom X-ray structure of nitrite reductase from Achromobacter cycloclastes. Science. 1991;253:438–442. doi: 10.1126/science.1862344. [DOI] [PubMed] [Google Scholar]
- 12.Grossmann J G, Abraham Z H L, Adman E T, Neu M, Eady R R, Smith B E, Hasnain S S. X-ray scattering using synchrotron radiation shows nitrite reductase from Achromobacter xylosoxidans to be a trimer in solution. Biochemistry. 1993;32:7360–7366. doi: 10.1021/bi00080a005. [DOI] [PubMed] [Google Scholar]
- 12a.Hasnain, S. S. Personal communication.
- 13.Howes B D, Abraham Z H L, Lowe D J, Brüser T, Eady R R, Smith B E. EPR and electron nuclear double resonance (ENDOR) studies show nitrite binding to the type 2 copper centers of the dissimilatory nitrite reductase of Alcaligenes xylosoxidans (NCIMB 11015) Biochemistry. 1994;33:3171–3177. doi: 10.1021/bi00177a005. [DOI] [PubMed] [Google Scholar]
- 14.Inoue T, Gotowda M, Deeliger S, Kataoka K, Yamaguchi K, Suzuki S, Watanabe H, Gohow M, Kai Y. Type 1 Cu structure of blue nitrite reductase from Alcaligenes xylosoxidans GIFU 1051 at 2.05 Å resolution: comparison of blue and green nitrite reductase. J Biochem. 1998;124:876–879. doi: 10.1093/oxfordjournals.jbchem.a022201. [DOI] [PubMed] [Google Scholar]
- 15.Kukimoto M, Nishiyama M, Murphy M E P, Turley S, Adman E T, Horinouchi S, Beppu T. X-ray structure and site-directed mutagenesis of a nitrite reductase from Alcaligenes faecalis S-6: roles of two copper atoms in nitrite reduction. Biochemistry. 1994;33:5246–5252. doi: 10.1021/bi00183a030. [DOI] [PubMed] [Google Scholar]
- 16.LaCroix L B, Shadle S E, Wang Y, Averill B A, Hedman B, Hodgson K O, Solomon E I. Electronic structures of the perturbed blue copper site in nitrite reductase: spectroscopic properties, bonding, and implications for the entatic/rack state. J Am Chem Soc. 1996;188:7755–7768. [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Leinfelder W, Forchhammer K, Zinoni F, Sawers G, Mandrand-Berthelot M A, Böck A. Escherichia coli genes whose products are involved in selenium metabolism. J Bacteriol. 1988;170:540–546. doi: 10.1128/jb.170.2.540-546.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Libby E, Averill B A. Evidence that the type 2 copper centres are the site of nitrite reduction by Achromobacter cycloclastes nitrite reductase. Biochem Biophys Res Commun. 1992;187:1529–1535. doi: 10.1016/0006-291x(92)90476-2. [DOI] [PubMed] [Google Scholar]
- 20.Lowe D J. Electron paramagnetic resonance spectroscopy: computer simulation of spectra from frozen aqueous samples. Biochem J. 1978;171:649–651. doi: 10.1042/bj1710649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 22.Nishiyama M, Suzuki J, Kukimoto M, Ohnuki T, Horinouchi S, Beppu T. Cloning and characterization of a nitrite reductase gene from Alcaligenes faecalis and its expression in Escherichia coli. J Gen Microbiol. 1993;139:725–733. doi: 10.1099/00221287-139-4-725. [DOI] [PubMed] [Google Scholar]
- 23.Olesen K, Veselov A, Zhao Y, Wang Y, Danner B, Scoles C P, Shapleigh J P. Spectroscopic, kinetic, and electrochemical characterization of heterogeneously expressed wild-type and mutant forms of copper-containing nitrite reductase from Rhodobacter sphaeroides 2.4.3. Biochemistry. 1998;37:6086–6094. doi: 10.1021/bi971603z. [DOI] [PubMed] [Google Scholar]
- 24.Osborn M J, Gander J E, Parisi E. Mechanism of assembly of the outer membrane of Salmonella typhimurium. J Biol Chem. 1972;247:3973–3986. [PubMed] [Google Scholar]
- 25.Pang K M, Knecht D A. Partial inverse PCR: a technique for cloning flanking sequences. BioTechniques. 1997;22:1046–1048. doi: 10.2144/97226bm07. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santini C-L, Ize B, Chanal A, Müller M, Giordano G, Wu L-F. A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 1998;17:101–112. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sargent F, Bogsch E G, Stanley N R, Wexler M, Robinson C, Berks B C, Palmer T. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Studier F W, Moffat B A. Use of bacteriophage T7 RNA polymerase to direct high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki S, Deeliger S, Yamaguchi K, Kataoka K, Kobayashi K, Tagawa S, Kohzuma T, Shidara S, Iwasaki H. Spectroscopic characterization and intramolecular electron transfer processes of native and type 2 Cu-depleted nitrite reductases. J Biol Inorg Chem. 1997;2:265–274. [Google Scholar]
- 32.Suzuki S, Kohzuma T, Deeliger S, Yamaguchi K, Nakmura N, Shidara S, Kobayashi K, Tagawa S. Pulse radiolysis studies on nitrite reductase from Achromobacter cycloclastes IAM 1013: evidence for intramolecular electron transfer from type 1 Cu to type 2 Cu. J Am Chem Soc. 1994;116:11145–11146. [Google Scholar]
- 33.Tabor S, Richardson C C. A bacteriophage T7 polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toffanin A, Wu Q, Maskus M, Casella S, Abruña H D, Shapleigh J P. Characterization of the gene encoding nitrite reductase and the physiological consequences of its expression in the nondenitrifying Rhizobium “hedysari” strain HCNT1. Appl Environ Microbiol. 1996;62:4019–4025. doi: 10.1128/aem.62.11.4019-4025.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tosques I E, Kwiatkowski A V, Shi J, Shapleigh J P. Characterization and regulation of the gene encoding nitrite reductase in Rhodobacter sphaeroides 2.4.3. J Bacteriol. 1997;179:1090–1095. doi: 10.1128/jb.179.4.1090-1095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Van Beeumen, J. J. Personal communication.
- 37.Vandenberghe I H M, Meyer T E, Cusanovich M A, Van Beeumen J J. The covalent structure of the blue copper-containing nitrite reductase from Achromobacter xylosoxidans. Biochem Biophys Res Commun. 1998;247:734–740. doi: 10.1006/bbrc.1998.8847. [DOI] [PubMed] [Google Scholar]
- 38.Veselov A, Olesen K, Sienkiewicz A, Shapleigh J P, Scholes C P. Electronic structural information from Q-band ENDOR on the type 1 and type 2 copper liganding environment in wild-type and mutant forms of copper-containing nitrite reductase. Biochemistry. 1998;37:6095–6105. doi: 10.1021/bi971604r. [DOI] [PubMed] [Google Scholar]
- 39.Weiner J H, Bilous P T, Shaw G M, Lubitz S P, Frost L, Thomas G H, Cole J A, Turner R J. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 40.Yanish-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp19 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 41.Ye R W, Fries M R, Bezborodnikov S G, Averill B A, Tiedje J M. Characterization of the structural gene encoding a copper-containing nitrite reductase and homology of this gene to DNA of other denitrifiers. Appl Environ Microbiol. 1993;59:250–254. doi: 10.1128/aem.59.1.250-254.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zumft W G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]