Abstract
Purpose:
Although chemotherapies kill most cancer cells, stem cell–enriched survivors seed metastasis, particularly in triple-negative breast cancers (TNBC). TNBCs arise from and are enriched for tumor stem cells. Here, we tested if inhibition of DOT1L, an epigenetic regulator of normal tissue stem/progenitor populations, would target TNBC stem cells.
Experimental Design:
Effects of DOT1L inhibition by EPZ-5676 on stem cell properties were tested in three TNBC lines and four patient-derived xenograft (PDX) models and in isolated cancer stem cell (CSC)-enriched ALDH1+ and ALDH1− populations. RNA sequencing compared DOT1L regulated pathways in ALDH1+ and ALDH1− cells. To test if EPZ-5676 decreases CSC in vivo, limiting dilution assays of EPZ-5676/vehicle pretreated ALDH1+ and ALDH1− cells were performed. Tumor latency, growth, and metastasis were evaluated. Antitumor activity was also tested in TNBC PDX and PDX-derived organoids.
Results:
ALDH1+ TNBC cells exhibit higher DOT1L and H3K79me2 than ALDH1−. DOT1L maintains MYC expression and self-renewal in ALDH1+ cells. Global profiling revealed that DOT1L governs oxidative phosphorylation, cMyc targets, DNA damage response, and WNT activation in ALDH1+ but not in ALDH1− cells. EPZ-5676 reduced tumorspheres and ALDH1+ cells in vitro and decreased tumor-initiating stem cells and metastasis in xenografts generated from ALDH1+ but not ALDH1− populations in vivo. EPZ-5676 significantly reduced growth in vivo of one of two TNBC PDX tested and decreased clonogenic 3D growth of two other PDX-derived organoid cultures.
Conclusions:
DOT1L emerges as a key CSC regulator in TNBC. Present data support further clinical investigation of DOT1L inhibitors to target stem cell–enriched TNBC.
Translational Relevance.
Inherent or acquired resistance to conventional anticancer treatment results from persistence or emergence of tumor cell subsets with high tumor-initiating and metastatic capabilities. Eliminating this tumor-initiating stem cell (TISC)–like population has been a longstanding challenge in treatment of high-grade, TISC-rich, aggressive tumors such as triple-negative breast cancer. Here, we show that high DOT1L expression associates with poor survival and DOT1L inhibition targets the TISC population in TNBC. TISC-enriched ALDH1+ cells in TNBC express higher levels of DOT1L and H3K79me2, and DOT1L regulates cMyc expression, WNT activation, and DNA damage responses uniquely in ALDH1+ cells. DOT1L inhibition decreased TISC abundance in vitro and in vivo and repressed tumor growth and metastasis from ALDH1+ xenografts. The DOT1L inhibitor, EPZ-5676, also shows antitumor activity in TNBC PDX in vivo and PDX-derived organoid cultures. These findings support further clinical investigation of DOT1L inhibitors as a TISC-targeting strategy for TNBC with potential to prevent or limit the emergence of chemotherapy resistance.
Introduction
Conventional treatments with chemotherapeutic drugs and radiation efficiently kill highly proliferative tumor cells, reducing tumor burden. Effective cytoreduction by common chemotherapeutic agents can be as high as 98% (1). However, expansion of minor, intrinsically resistant subpopulations and emergence of acquired resistance lead to disease recurrence, metastasis, and ultimately patient mortality and represent the central limitation of current cancer therapy. Increasing evidence has emerged that cancer cells persisting after conventional therapeutic regimens show enrichment for stem cell–like populations with enhanced tumorigenic potential, drug efflux (2) and DNA repair properties (3). Enrichment of tumor-initiating stem cells (TISC) in chemotherapy- and or radiotherapy-resistant populations has been reported in human leukemia (4), melanoma (5), colorectal (6), brain (7), and breast cancers (8, 9). High ALDH1 expression and activity in primary breast cancers are associated with chemo- (9, 10) and radio-resistance (11). ALDH1 activity is a marker of breast cancer stem cells (CSC), and high intratumor ALDH1A1/A3 expression correlates with advanced-stage, early metastasis, and poor breast cancer patient outcome (12, 13).
Despite improvements in therapy over the last 30 years, breast cancer remains the leading cancer and second highest cause of cancer death in women (14). Triple-negative breast cancer (TNBC; lacking expression of estrogen and progesterone receptors, and HER2 amplification) disproportionately affects younger women, accounts for nearly 15% of all invasive cancers and comprises one of the most aggressive forms of the disease. TNBC exhibit high rates of both intrinsic and acquired therapy resistance, leading to early metastasis and patient demise (15). Gene profiling and marker studies suggest that TNBC are enriched for cells with a more primitive stem/progenitor profile that could account for their high chemoresistance (16). Despite the promise of therapies targeting EGFR (17), VEGF (18), PARP (19), mTOR/PI3K (20), and immune-checkpoint inhibitors (21), single pathway targeting strategies have shown variable responses and limited antitumor efficacy, often due to the emergence of bypass mechanisms. Because current cancer therapies fail to abolish aggressive tumor-initiating stem cells, new strategies to target stem cell drivers of malignancy would be beneficial.
Epigenetic regulators play functional roles to modulate gene transcription programs governing cell fate and identity (22). Epigenetic deregulation is frequently observed and critical to tumor initiation, progression, and therapy resistance in both hematologic and solid malignancies (23, 24). Hence, growing drug-discovery efforts have yielded a number of promising new drugs targeting the cancer epigenome, many of which are in clinical trials or approved for patient care. These drugs target DNA methyltransferases (DNMT), histone deacetylases (HDAC), and histone methyltransferases (HMT) and include EZH2 inhibitors and bromodomain and extraterminal motif (BET) inhibitors (23, 25, 26). Because epigenetic regulators govern stem cell fates in normal tissue development, there has been considerable interest in the possibility that epigenetic drugs might target malignant stem cells. Furthermore, the favorable therapeutic index observed for several epigenetic drugs would permit the development of novel drug combination strategies.
DOT1L (disruptor of telomeric silencing 1-like) is an evolutionarily conserved histone methyltransferase that catalyzes methylation on lysine 79 of histone H3 to regulate transcription activation and elongation. There is also evidence that DOT1L regulates cell-cycle progression and DNA repair (27). DOT1L plays essential roles during embryogenesis and normal development, maintaining normal tissue stem/progenitor population homeostasis. Germline deletion of DOT1L is embryonic lethal (28), and DOT1L loss led to the growth arrest of ES cells (29). DOT1L-mediated H3K79 methylation critically regulates gene-expression programs required for pluripotency in ES cells, and for postnatal cardiac cell fate and maturation in adult mice (30). DOT1L also maintains embryonic and adult hematopoietic stem and progenitor populations (31–33) and intestinal crypt stem cells (34).
DOT1L was first shown to be oncogenically activated in aggressive mixed lineage leukemia (MLL). DOT1L is constitutively recruited by MLL fusion oncoproteins to promote transcriptional activation of drivers of malignant transformation in MLLr leukemia (27, 35). The observation that DOT1L was an essential oncogene in MLL-rearranged leukemia stimulated the development of DOT1L inhibitors (36, 37). DOT1L has also been implicated in the maintenance of solid malignancies such as colorectal (38), neuroblastoma (39), ovarian (40), and breast cancers (41, 42). EPZ-5676 is a potent and highly selective inhibitor of DOT1L catalytic activity developed by Epizyme. Antitumor efficacy of EPZ-5676 has been demonstrated in vivo in models of MLL-rearranged leukemia (37, 43), ovarian cancer (40), and glioblastoma (44). The safety and tolerability of EPZ-5676 has been established in humans (45), and this drug is currently under investigation in clinical trials for leukemia together with standard chemotherapy. Although DOT1L is known to govern hematopoietic stem/progenitor homeostasis and to be required for cardiomyogenesis, its role in maintenance of stem-like or tumor-initiating cells in solid tumors has not been fully explored.
Here, we investigated whether DOT1L maintains TNBC stem cells. We found that the stem cell–enriched ALDH1+ population exhibits higher DOT1L and H3K79 dimethylation than the ALDH1− population. Gene-expression profiling revealed that DOT1L regulates oxidative phosphorylation, DNA damage responses, and the WNT pathway uniquely in the ALDH1+ population. EPZ-5676 decreased ALDH1+ cells, sphere formation, and MYC expression in vitro, and TISC frequency, tumor growth, and metastasis in xenografts generated from the ALDH1+ but not from the ALDH1− population, revealing its potential for targeting CSC in human cancer.
Materials and Methods
Cell lines
MDA-MB-231 (231) and MDA-MB-468 (468) cell lines were obtained from American Type Culture Collection in 2011 and were grown in Dulbecco's Modified Eagle's Medium supplemented with 10% FBS and 1% penicillin/streptomycin for in vitro work. SUM149 was originally sourced from Dr. Stephen Ethier's Lab, Medical University, South Carolina and cultured in Ham's F12, supplemented with 10% FBS, 1% penicillin/streptomycin, 1 μg/mL hydrocortisone, and 5μg/mL insulin. MDA-MB-468 cells were used for sorting distinct ALDH1+ and ALDH1− cell populations for the study. For in vivo work, MDA-MB-468-luciferease tagged cells were treated with the DOT1L inhibitor or DMSO for 10 days and sorted into ALDH1+ and ALDH1− populations. Cell lines were authenticated at ICBR Gene Expression and Genotyping at University of Florida, routinely verified for mycoplasma contamination and all experiments were performed on cells passaged less than 15 times.
Reagents and antibodies
EPZ-5676 was provided by Epizyme, and DMSO was purchased from Sigma (cat. no. D2438). Cells were dissociated using TrypLE Express Enzyme (Thermo Fisher Scientific, cat. no. 12604021). Primary antibodies used for histone blots were as follows: anti-H3K79me2 (Abcam; ab3594), anti-Histone H3 (Abcam; ab1791). Fluorophore-conjugated secondary antibody IRDye 680RD Donkey anti-Rabbit IgG (Licor, 92568073) was used for histone blots. Primary antibodies used for Western blots were as follows: anti-DOT1L (Cell Signaling; 77087S), anti-cMyc (Cell Signaling; 5605S), anti-SOX2 (Cell Signaling; 3579S), anti-H3K79me2 (Abcam; ab3594), anti-GAPDH (Cell Signaling; 2118S), and anti–β-actin (Cell Signaling; 4970S). HRP-conjugated anti-rabbit IgG (W4011) and anti-mouse IgG (W4021) were purchased from Promega.
Histone extraction and immunoblotting
Cells were treated with dimethyl sulfoxide (DMSO) as control and the DOT1L inhibitor EPZ-5676 in 1:2 titration ranging from 10 nmol/L to 10 μmol/L for 7 days. TrypLE-dissociated cells were washed in phosphate-buffered saline (PBS) and lysed in histone lysis buffer (10 mmol/L MgCl2, 25 mmol/L KCl, 1% TritonX-100, 8.6% sucrose in 10 mmol/L Tris-HCl with protease inhibitor, pH 6.5). Histones were acid extracted using ice-cold 0.4N H2SO4 and pelleted with acetone and resolved by 15% SDS-PAGE and then transferred onto a nitrocellulose membrane (Bio-Rad; 162-0112) at 70 V for 50 minutes. The membrane was blocked in 5% BSA for 1 hour and incubated in primary antibodies overnight followed by secondary antibody for 1 hour at room temperature. Western bands were imaged and quantified using LICOR imaging system. Levels of H3K79me2 were normalized against the total H3 levels for each blot.
Cell proliferation assay
Cells were plated in 6-well plates for 231 and 468 at day = 0 and treated with or without EPZ-5676 or DMSO. Cells were harvested and counted at days 2, 4, 6, 8, 10, and 12. Three different biological repeat assays were done. Viable cell counts were calculated using Trypan Blue solution (Sigma; T8154) at each time point and plotted as mean ± standard error of the mean (SEM).
Viability assay
The 231 and 468 cells were seeded in 24-well culture plates and allowed to grow for 7 and 10 days with or without the presence of the DOT1L inhibitor EPZ-5676. Cells were evaluated for viability using 120 μL of CellTiter 96 AQueous One Solution (MTS reagent, Promega; G3580) and incubated for 4 hours. Absorbance of live cells was measured at 490 nm on a BioTek plate reader.
Cell-cycle assay
Flow cytometry for cell-cycle distribution was performed as in (46). In brief, cells were pulse labeled with 10 μg/mL BrdUrd (Life Technologies, B23151) for 2 hours, dissociated in TrypLE express, and fixed by dropwise addition of ice-cold 70% ethanol. Fixed cells were washed twice with 2N HCl and 0.1M sodium borate before staining with anti-BrdUrd antibody (eBioscience Thermo Fisher, 11-5071-42) and propidium iodide solution (Miltenyi Biotec, 130-093-233). Flow cytometry analysis of DNA content and BrdUrd uptake indicating the percentage of cells in G1/S/G2–M phases was carried out using LSRII analyzer (BD Biosciences).
ALDEFLUOR assay and sorting ALDH1+ and ALDH1− populations
The ALDEFLUOR kit (Stem Cell Technologies, 1700) was used to identify and sort cell populations with a high ALDH1 enzymatic activity per the manufacturer's instructions. In brief, cells were treated with or without EPZ-5676 for the indicated time and harvested with TrypLE. Cells (1 × 106) were suspended in ALDEFLUOR assay buffer with ALDH substrate (BODIPY-aminoacetaldehyde-diethyl acetate or BAAA, 1 mmol/L). The ALDH inhibitor diethylaminobenzaldehyde (DEAB) was used to control background fluorescence as negative control. The samples were incubated for 45 minutes at 37°C. ALDEFLUOR was excited at 488 nm, and fluorescence emission was detected using standard fluorescein isothiocyanate (FITC) 530/30-nm band-pass filter using the CantoII flow cytometer (BD Biosciences). To sort ALDH1+ and ALDH1− cell populations, 10–20 × 106 MDA-MB-468 cells were subjected to the ALDEFLUOR assay as above. Cells were aliquoted as 1 × 106 cells/tube in 1 mL of ALDEFLUOR assay buffer. After incubation, samples were resuspended in assay buffer/tube on ice and then filtered through the cell strainer cap into 5 mL polystyrene flow cytometry tubes. Samples were then sorted using FACSAria IIu flow cytometer (BD Biosciences) and collected in ice-cold PBS in 5 mL polypropylene tubes, centrifuged, and plated into overnight culture to facilitate removal of dead cells and debris.
Mammosphere assay
Sphere assays were performed as in (47). Cells were treated with the inhibitor EPZ-5676 or DMSO at the indicated concentrations for 10 days and then harvested and seeded as 5,000 cells/well for MDA-MB-231 and 20,000 cells/well for MDA-MB-468 for in vitro sphere assay. Sorted ALDH1− and ALDH1+ cells were seeded as 10,000 cells/well for sphere assay. Mammosphere media were prepared fresh, and cells were plated in ultra-low attachment 6-well plates (Corning), with EPZ-5676/DMSO added at the time of seeding. Spheres were allowed to grow for 12–14 days, and spheres ≥ 75 μm in diameter were quantitated using a GelCount (Oxford Optronix). For analysis, background and isolated single cells were corrected for, using appropriate edge and center detection settings on GelCount optimizer.
Quantitative RT-PCR (qPCR)
RNA was extracted using TRIzol reagent (Ambion, 15596018) as per the manufacture's protocol. cDNA was synthesized from 1 μg RNA using iSCRIPT cDNA synthesis kit (Bio-Rad, 1706891). Quantitative real-time PCRs were set up using iQ SYBR green supermix (Bio-Rad, 1708882) and run on LightCycler 480 Instrument II (Roche). GAPDH or 18S were used as internal controls. All samples were performed in three technical replicates in each of three biological repeats and average Ct values were normalized with GAPDH or 18S RNA to derive relative expression level quantification using the 2–ΔΔCt method. Oligonucleotide sequences of primers used were as follows: MYC Forward 5′-GAGTCTGGATCACCTTCTGCTG-3′ and MYC Reverse 5′-AGGATAGTCCTTCCGAGTGGAG-3′; SOX2 Forward 5′-CGAGTAGGACATGCTGTAGGT-3′ and SOX2 Reverse 5′-TGGACAGTTACGCGCACAT-3′; POU5F1 Forward 5′-GGGAGATTGATAACTGGTGTGTT-3′ and POU5F1 Reverse 5′-GTGTATATCCCAGGGTGATCCTC-3′; SOX11 Forward 5′-AGGATTTGGATTCGTTCAGCG-3′ and SOX11 Reverse 5′-AGGTCGGAGAAGTTCGCCT-3′; ALDH1A1 Forward 5′-GCACGCCAGACTTACCTGTC-3′ and ALDH1A1 Reverse 5′-CCTCCTCAGTTGCAGGATTAAAG-3′; RARRES1 Forward 5′-AAACCCCTTGGAAATAGTCAGC-3′ and RARRES1 Reverse 5′-GGAAAGCCAAATCCCAGATGAG-3′; GAPDH Forward 5′-ATCAAGTGGGGCGATGCTG-3′ and GAPDH Reverse 5′-ACCCATGACGAACATGGGG-3′; 18S rRNA Forward 5′-AGGAATTGACGGAAGGGCAC-3′ and 18S rRNA Reverse 5′-GGACATCTAAGGGCATCACA-3′.
Western blotting
FACS-sorted ALDH1+ and ALDH1− cell lysates were prepared in RIPA buffer (Cell Signaling, 9806S) supplemented with 1× protease and phosphatase inhibitors (G-Biosciences), and lysates (20–30 μg/lane) were loaded onto 4% to 20% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad), and samples were resolved 100 V for 1 to 2 hours and then transferred onto PVDF membrane (Bio-Rad, 162-0177) overnight. Membranes were blocked for 1 hour at room temperature in 5% BSA, incubated with primary antibodies at suggested dilutions overnight at 4°C and subsequently reacted with appropriate secondary antibodies for 1 hour at room temperature. Protein bands were detected by chemiluminescent imaging using Pierce ECL (Thermo Fisher, 32106). All Western blots were quantified using densitometric analysis and relative quantification of test compared with control are mentioned in the Results section.
RNA sequencing
MDA-MB-468 cells were treated with 0.5 μmol/L EPZ-5676 or DMSO as control for 10 days, and then ALDH1+ and ALDH1− population were isolated by flow cytometry. Total RNA was extracted from the sorted populations using TRIzol reagent, and RNA quality was assayed by Nanodrop and Bioanalyzer at the UM Oncogenomics core facility. Three biological replicates with good quality RNA with RNA integrity number (RIN) >9 were used to make cDNA libraries. Libraries were constructed from 2 μg total RNA using NEBNext Ultra II Directional RNA Library Prep Kit (New England Biolabs, E7765S) following the manufacturer's protocol. Quality of the libraries was verified using fragment analyzer, and pair-end sequencing was performed on an Illumina' NextSeq platform by the Sylvester Comprehensive Cancer Center Oncogenomics Core facility. The run was passed through quality control before performing differential gene-expression analysis. The GEO submission ID number for this study is GSE194434.
Bioinformatic analysis of RNA-seq data
Raw FastQ files were put through two different software tools to complete an initial quality control screen. Reads were put through the FastQC and FastQ_screen programs to determine read quality. After sufficient QC was established, the raw reads were then put through an adapter trimming script TrimGalore. Next, alignment was carried out by the STAR RNA-seq aligner (v2.5.2) and gene counts were quantified simultaneously by using the GENCODE v19 gene features reference in the hg19 reference genome. Custom scripts then took the raw reads from the alignment and quantification, which was then put through the edgeR differential expression R software package. Significance was cutoff at false discovery rate (FDR) <0.05. CPM values obtained from the RNA-seq alignment were put into a matrix to be fed into the Broad Institute's gene set enrichment analysis (GSEA) software package. Filtering was set at 1 CPM to remove genes that were not expressed at a significant level to be ranked. GSEA was carried out with Hallmark references as well as C2 gene set references to obtain a list of significant gene sets. Biological processes and enrichment categories were searched for FDR significance on DAVID portal. Pathway analysis was also performed through the WIKI pathways, DAVID resource to look for significant enrichment with KEGG as well as Reactome reference databases.
In vivo tumor-initiating stem cell assays
Female NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice used for the pilot and main experiment were purchased from Jackson Labs. A pilot experiment was conducted to identify the lowest number of cells that could give rise to tumors from sorted ALDH1+ and ALDH1− populations isolated from a luciferase tagged MDA-MB-468 using a limiting dilution assay. The ALDEFLUOR assay was optimized for luciferase-tagged MDA-MB-468. Limiting dilutions of 100,000, 10,000, 1,000, and 100 cells from the flow-sorted ALDH1+ and ALDH1− populations (n = 4/group) were mixed 1:1 with matrigel (Corning) in a final 100 μL volume. Mice were anesthetized with isoflurane and a small left abdominal flank incision was made for orthotopic injection of cells into the fourth mammary fat pad of 6-week-old NSG mice. The abdominal wound was closed with metallic wound clips (Fine Science Tools). Mice were injected with a single dose of buprenorphine SR 1 mg/kg body weight via subcutaneous route as an analgesic after surgery. Tumor growth and volume were monitored by In Vivo Imaging System (IVIS, Xenogen Corporation). As all mice injected with 100 ALDH1+ cells developed tumors within 10 weeks, our experiment to quantitate drug effects on tumor-initiating stem cells in vivo, smaller cell numbers (50–10 cells as limiting dilution) was used. Assays of drug effects on tumor growth and metastasis used 5,000 cells/injection. To assay drug effects on tumor-initiating cell populations in vivo, 5-week-old female NSG mice were used. MDA-MB-468-luc cells were treated with DMSO or 1 μmol/L EPZ-5676 for 10 days and on day 10, cells were FACS sorted for the ALDH1+ and ALDH1− population as described earlier. Sorted ALDH1+ and ALDH1− populations were allowed to recover in culture overnight prior to injection into mice. FACS-sorted ALDH1+ and ALDH1− populations in limiting dilution of 5,000, 500, 50, and 10 cells with 1:1 matrigel in 100 μL were injected directly into the fourth mammary fat pad. For continuous drug treatment in vivo, mice were treated with six doses of either vehicle alone (DMSO control group) or EPZ-5676 (EPZ group) using 50 mg/kg EPZ-5676 suspended in 2% DMSO + 30% PEG 300 + 5% Tween80 + 63% PBS administered by intraperitoneal injection on alternate days. Animals were weighed weekly. All xenograft animal work was performed as per Institutional Animal Care and Use Committee (IACUC)-approved protocol (University of Miami).
IVIS imaging and data quantification
Animals were imaged weekly through the IVIS imaging system. Prior to imaging, animals were injected with Xenolight D-luciferin K+ salt (Perkinelmer) and anesthetized with 2.5% isoflurane. After 8 minutes of incubation with luciferin, bioluminescence was quantified for each animal. Tumor volumes were monitored as bioluminescence (photon flux/second) by IVIS using Living Image software. At sacrifice, the final tumor was measured using Vernier calipers, and the volume was calculated using the formula (long-side × short-side2)/2. For in vivo metastasis, primary tumor sites were covered, and bioluminescence was quantified as photon flux by IVIS.
Drug treatment on patient-derived xenograft (PDX) model in vivo
TNBC PDXs HCI-001 and HCI-010 (48) were obtained from Alana Welm. The PDX was fragmented into five pieces, and each piece was implanted into the fourth mammary fat pad of NSG mice (n = 5 mice/per group). Mice were treated with either 40 mg/kg of EPZ-5676 or vehicle (2% DMSO + 30% PEG 300 + 5% Tween80 + 63% PBS) daily via intraperitoneal injection for 5 weeks. Tumor growth was monitored by measuring palpable tumors using caliper every week. All PDX animal work was performed as per IACUC-approved protocol (Georgetown University).
PDX colony formation assays
Mammary tumor organoids were generated from two TNBC PDX lines, TM00089 and TM00096, from The Jackson Laboratory as previously described (49). In brief, PDX tumors were isolated from NSG mice, mechanically minced, then enzymatically digested in a collagenase and trypsin solution. A series of differential centrifugations were then used to separate the epithelial tumor organoids from contaminating stromal cells. The epithelial organoids were then further digested into epithelial cell clusters (2–10 cells in size) and embedded at a concentration of 200 clusters/μL into a Geltrex basement membrane matrix (Thermo Fisher; #A1413202). After treatment with DOT1L-targeting compounds, resulting colonies were either isolated and reseeded into matrix or fixed for analysis. For reseeding experiments, Geltrex matrix was dissolved in Cell Recovery Solution (Corning; #354253), and recovered colonies were digested to clusters and reseeded at a concentration of 200 clusters/μL into Geltrex. All reseeded colonies were split into two groups and received either further treatment with DOT1L inhibitors or vehicle control. Following treatment, colonies in matrix were fixed for 10 minutes in 1.0% paraformaldehyde supplemented with 0.25% glutaraldehyde. To facilitate high-content imaging, colonies were stained with Alexa Fluor Phalloidin (Thermo Fisher #A12379) and then imaged on a Molecular Devices ImageXpress Micro. Resulting colonies were identified using ImageJ, counted, and analyzed using custom Python 3.0 scripts. The percentage of colony formation in response to DOT1L inhibitor treatment was calculated by normalizing the number of colonies in the experimental condition to that of the vehicle control (DMSO).
Survival curve analysis
The comparison of DOT1L expression in breast tumor to normal breast tissue in Fig. 1A was analyzed on UALCAN webportal (50), whereas the comparison among breast cancer subtypes was analyzed on tumorsurvival.org portal (51). Breast cancer data sets from METABRIC and The Cancer Genome Atlas (TCGA) were downloaded from cbioportal.org. All breast cancer patients and the subgroups of TNBC patients who had disease recurrence or who died within 8 years were analyzed for association of DOT1L gene expression with relapse-free survival (RFS) or overall survival (OS), respectively, using the median DOT1L expression as cutoff for high and low. The number of patients with survival data in TGCA exceeds that with disease-free survival (DFS) data among “all breast cancers” and for TNBC patients.
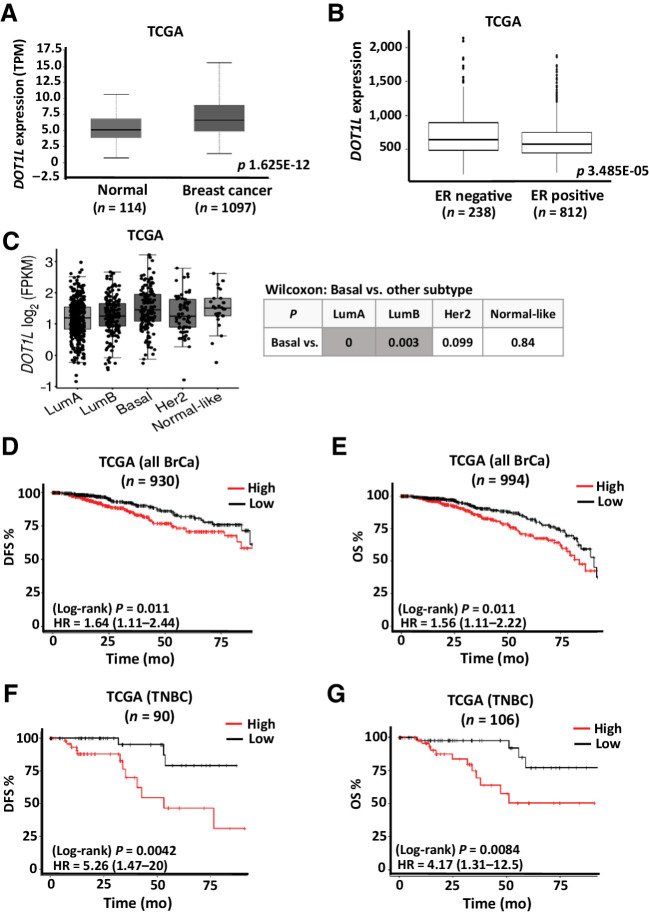
Figure 1.
Prognostic value of DOT1L expression in breast cancer. A, Box plot comparing DOT1L gene expression in breast cancer (n = 1,097) and normal breast tissue (n = 114) using TCGA data set. (The data figure was generated using UALCAN cancer OMICS web portal) (50). B, Box plot comparing DOT1L gene expression in estrogen receptor positive (n = 812) and estrogen receptor negative (n = 238) in TCGA data set downloaded from cBioPortal. C, Gene-expression analysis of DOT1L expression (FPKM) in breast cancer subtypes using TCGA data set. Nonparametric Wilcoxon test P values for comparison of basal vs. each subtype is tabulated. (The data figure was generated using tumorsurvival.org portal.) D and E, Breast cancer data from TCGA was downloaded from cbioportal.org. Kaplan–Meier analysis shows association of DOT1L gene expression above or below median expression with DFS or OS, for patients who had disease recurrence (D) or who died within 8 years (E). F and G, TNBC data from TCGA were downloaded from cbioportal.org. Kaplan–Meier analysis shows association of DOT1L gene expression above or below median expression with RFS (F) or OS (G), for TNBC patients who had disease recurrence or who died within 8 years, respectively. HA = hazard ratio; log-rank P values are shown. See also Supplementary Fig. S1.
Statistical analysis
All graphed data are presented as mean ± SEM from three independent biological replicates for each assay. Comparisons between two groups were done using a paired Student t test, and P values ≤ 0.05 were considered statistically significant. For RNA-seq analysis, FDR q ≤ 0.05 was taken as statistically significant. The TISC frequency was calculated by L-Calc limiting dilution software (http://www.stemcell.com/en/Products/All-Products/LCalc-Software.aspx) from STEMCELL Technologies Inc. The statistical differences between growth curves were calculated using “compareGrowthCurves” function of the statmod software package using the website (http://bioinf.wehi.edu.au/software/compareCurves). This analysis was applied for cellular proliferation assays and orthotopic tumor growth curves.
Data availability statement
All primary data can be obtained from the corresponding author. All raw RNA-seq data are available at the GEO accession number GSE194434.
Results
DOT1L overexpression is prognostic of poor breast cancer outcome
Among breast cancers, aggressive TNBC show greater abundance of more primitive stem cell/progenitor cells (16). Their treatment resistance is attributed in part to escape mechanisms unique to CSCs (9, 52). Analysis of TCGA data revealed higher DOT1L expression in breast cancer than in normal breast tissue (Fig. 1A). DOT1L expression was also higher in ER− than ER+ breast cancer in two independent data sets, TCGA and METABRIC (Fig. 1B; Supplementary Fig. S1A). Basal breast cancers, which comprise most TNBC, have higher DOT1L than the less aggressive ER+ luminal A or luminal B breast cancers (Fig. 1C). Kaplan–Meier analysis of TCGA breast cancer patients with recurrence or death within 8 years showed DOT1L expression above the median (high DOT1L) is associated with poor DFS (HR = 1.64; 95% CI, 1.11–2.44, P = 0.011, n = 930), and OS, HR = 1.56; 95% CI, 1.11–2.22, P = 0.011, n = 994) in all breast cancers (Fig. 1D and E). Similar analysis of over 900 patients in the independent METABRIC data set validated the prognostic significance of DOT1L expression (see Supplementary Fig. S1B and S1C). In the smaller subset of TNBC patients in TCGA, high DOT1L showed an even greater association with poor outcome, correlating strongly with both reduced DFS (HR = 5.26; 95% CI, 1.47–20, P = 0.0042) and OS (HR = 4.17; 95% CI, 1.31–12.5, P = 0.0084; Fig. 1F and G).
The DOT1L inhibitor EPZ-5676 effectively reduces H3K79me2 in TNBC lines
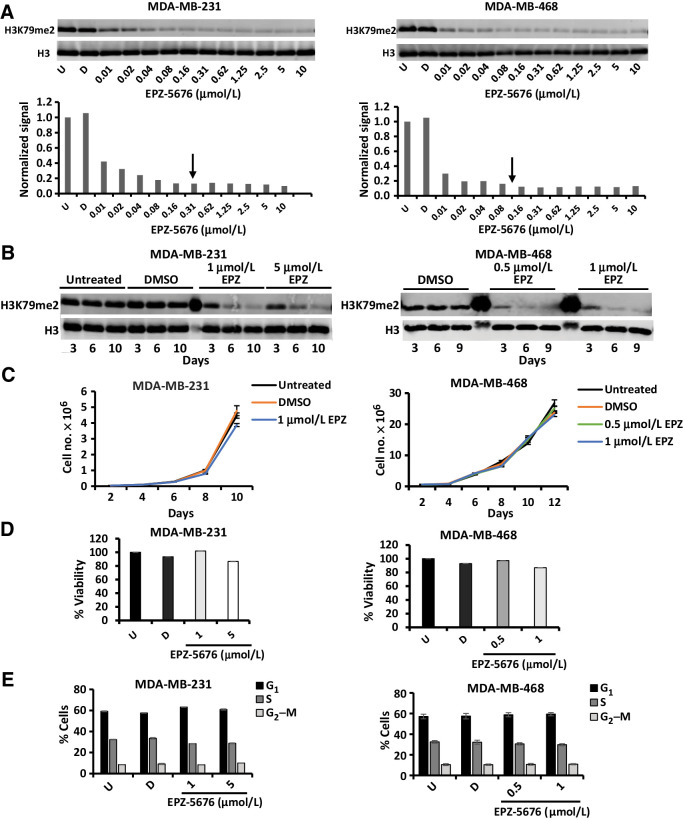
Previously, we identified two hierarchically linked populations in both TNBC lines and primary dissociated TNBC. The more primitive, TISC-enriched CD44+CD24low+ gave rise to CD44+CD24neg cells with lower TISC abundance and lacking metastatic potential (53). A drug screen comprising 60 epigenetic drugs suggested the stem cell–enriched population that contained cells with high ALDH1 activity was uniquely sensitive to DOT1L inhibition. EPZ-5676 is a potent DOT1L HMT inhibitor with high selectivity for DOT1L compared with other HMTs. It decreases H3K79 dimethylation with an inhibition constant (Ki) of 0.08 nmol/L, and exhibits low nanomolar IC50 values for DOT1L inhibition in both MLLr and non-MLLr human leukemic cell lines (37). To elucidate the role of DOT1L in TNBC stem cells, EPZ-5676 was evaluated further in two independent TNBC cell lines, MDA-MB-231 (hereafter 231) and MDA-MB-468 (hereafter 468). Cells were EPZ-5676–treated over a 10-fold concentration range for 7 days, and H3K79 dimethyl (H3K79me2) levels were quantified by Western blot, normalizing to total histone H3. H3K79me2 was maximally reduced with 300 nmol/L EPZ-5676 for 231 and 100 nmol/L for 468 (Fig. 2A), decreasing progressively over 10 days (Fig. 2B).
Figure 2.
DOT1L inhibition by EPZ-5676 does not significantly change proliferation, viability, or cell-cycle distribution. A, Cells were treated with indicated drug concentration or DMSO for 7 days. Histones were acid extracted and analyzed for H3K79me2 levels by Western blots as described in Materials and Methods. H3K79 me2 levels were normalized against total H3 and H3K79me2 quantification plotted. Arrows indicate IC90 concentration at which maximal inhibition was achieved. B, Cells were treated with indicated drug concentration or DMSO and analyzed for H3K79me2 and total H3 levels after 3, 6, 9–10 days. C, Cells were treated with indicated EPZ-5676 concentrations, and viable cells were counted after 2, 4, 6, 8, 10, and 12 days using Trypan Blue exclusion dye. Untreated and DMSO-treated cells served as controls. D, Cells were treated with or without indicated EPZ-5676 concentrations for 10 days followed by viability assay using MTS reagent. Absorbance was measured at 490 nm. Untreated (U) and DMSO (D)- treated cells served as controls. None of the experimental values differed significantly from U or D controls. E, Cells were treated with EPZ-5676 or DMSO (D) as vehicle or left untreated (U) for 10 days and were analyzed for cell-cycle distribution by flow cytometry following BrdUrd pulse labeling and propidium iodide staining. All graphed data represent the means ± SEM of triplicated repeats from at least three independent biological assays.
DOT1L inhibition has little effect on population growth, viability, or cell-cycle distribution
The tumor sphere assay, in which a sphere forms from a single cell in suspension, is an in vitro surrogate for stem cell self-renewal and relies on stem cell proliferation (47). Before testing effects of DOT1L inhibition on CSCs, we first determined if EPZ-5676 could decrease H3K279me2 without inhibiting global cell proliferation or viability, because dead or growth arrested cells cannot form spheres. Effects on cell viability, cell-cycle distribution, and population growth were assayed at drug doses 3× and 5× higher than the IC90 that maximally decreased H3K79me2. 231 cells received 1 μmol/L (3× IC90), and 468 cells were treated with 0.5 μmol/L (5× IC90) and 1 μmol/L (10× IC90) EPZ-5676 for 10 days, with DMSO as control. Cells were passaged to permit normal population doubling and counted every 2 days for 12 days. EPZ-5676 showed little or no effect on population growth (Fig. 2C) or viability by MTS assay (Fig. 2D). Over 10 days of treatment, EPZ-5676 had little effect on cell-cycle distribution in 231, with only a minor decrease in the percentage of S phase cells from 33% to 28% and no effect in treated 468 (Fig. 2E). Thus, prolonged DOT1L inhibition by EPZ-5676 does not cause global cell death or cell-cycle arrest in these lines.
DOT1L inhibition decreases ALDH1 activity, sphere formation, and stem cell transcription factor expression
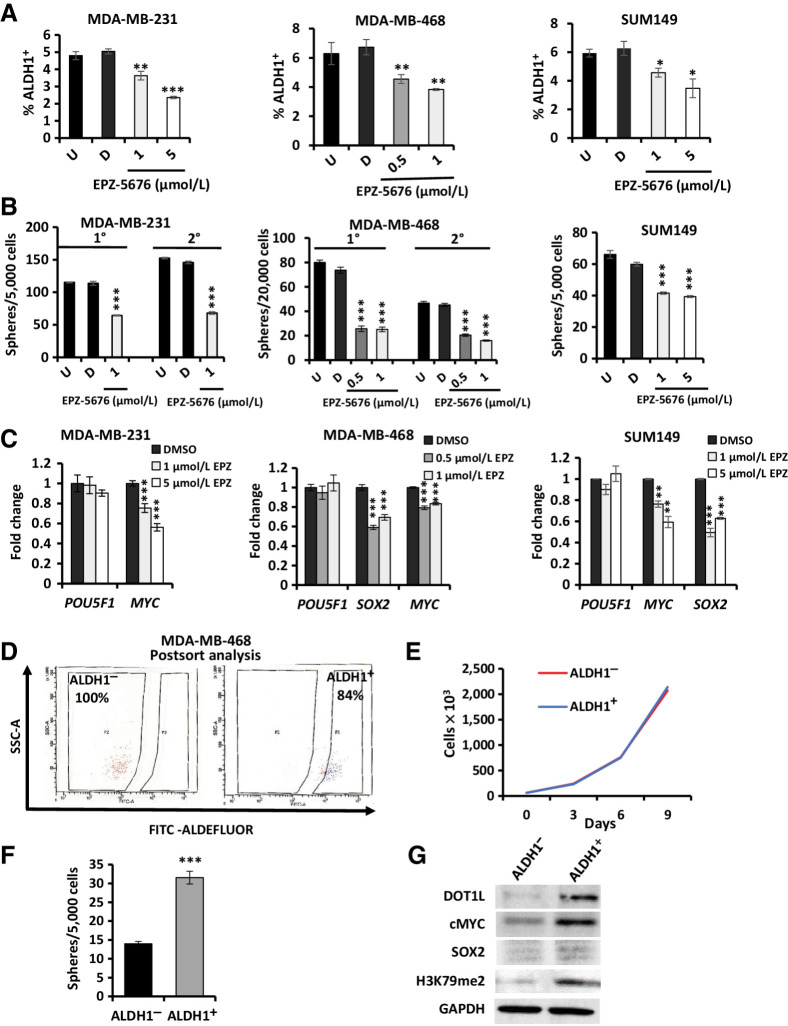
To test if DOT1L maintains stem cells in TNBC models, three independent TNBC lines, 231, 468, and SUM149, were treated with EPZ-5676 for 10 days and recovered to assay stem cell properties in vitro. The proportion of CSC-enriched ALDH1+ decreased significantly with 1 μmol/L EPZ-5676 in 231, and at all doses tested in the 468 and SUM149 cells (Fig. 3A) and CD44+CD24–/low+ cells were reduced in 231; Supplementary Fig. S2A). Addition of EZP5676 at plating decreased sphere formation by 40% in 231 and SUM149 cells, by 50% to 65% in 468 cells (Fig. 3B). Secondary sphere formation was also decreased by EPZ-5676 treatment in both 231 and 468 cells (Fig. 3B). Expression of pluripotency transcription factor genes MYC and SOX2 was downregulated by the DOT1L inhibitor, whereas POU5F1 remained unchanged (Fig. 3C). In addition, treatment with a second DOT1L inhibitor, EPZ4777, also significantly decreased the percentage of ALDH1+ cells, the abundance of sphere-forming cells and MYC and SOX2 expression levels in 231 and 468 cell lines (Supplementary Fig. S2B–S2D). Transient DOT1L knockdown using three independent siRNAs also reduced MYC and SOX2 levels in both lines (Supplementary Fig. S2E). Hence, EPZ-5676 concentrations that maximally inhibit DOT1L-mediated H3K79 dimethylation do not affect global proliferation but appear to reduce TNBC cells with stem cell properties in these models in vitro.
Figure 3.
DOT1L inhibition decreases CSC properties and CSC-enriched ALDH1+ cells express high DOT1L and H3K79me2 levels. A, MDA-MB-231 (left), SUM149 (right), and MDA-MB-468 (center) cells were treated ± EPZ-5676 or DMSO for 10 days and then assayed for ALDH1 activity (%ALDH1+) by the ALDEFLUOR assay. B, MDA-MB-231 (left), SUM149 (right), and MDA-MB-468 (center) cells were pretreated ± EPZ-5676 or DMSO for 10 days and then seeded into sphere assays ± EPZ-5676 or DMSO added once to the media at seeding without further replenishment. Spheres ≥ 75 μm were counted after 12–14 days and graphed as mean ± SEM. C, Embryonic stem cell TFs POU5F1 (OCT4), MYC, and SOX2 expression was assayed by qPCR after 10-day exposure to EPZ-5676 or DMSO and mean values graphed ± SEM. D, ALDH1 activity was assayed in 468 cells by ALDEFLUOR assay, and ALDH1+ and ALDH1− cells were sorted by flow cytometry as described in Materials and Methods. Representative images show the purity of flow-sorted ALDH1+ and ALDH1− populations upon postsort analysis. E, Population growth curves show mean cell numbers of sorted ALDH1+ and ALDH1− 468 cells grown over 9 days in culture. F, FACS-sorted 468 ALDH1+ and ALDH1− cells were seeded into sphere assays. Spheres ≥ 75 μm were counted at 14 days, and mean sphere numbers are graphed ± SEM. G, Western blots show DOT1L, cMYC, SOX2, and global H3K79 dimethylation levels in sorted ALDH1+ and ALDH1− 468 populations. All assays with graphed data were performed as three technical replicates in each of three biological repeats, and mean numbers are graphed ± SEM. Student t test compares each drug condition with untreated control: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. See also Supplementary Fig. S2.
ALDH1+ cells have higher DOT1L, cMyc, and H3K79 dimethylation
The enrichment of stem cell–like properties in ALDH1+ cells has been demonstrated in different breast cancer lines in vitro and in xenografts (54). To test drug effects on the stem cell–enriched ALDH1+ population versus ALDH1− cells, these populations were isolated by flow cytometry (55). The proportion of ALDH1+ cells in 468 was higher (8%–10%) than in 231 (3%–4%); thus, it was technically more feasible to sort ALDH1+ and ALDH1− populations from 468, and this line was used for subsequent assays in ALDH1+ and ALDH1− cells. Isolated ALDH1+ were 82% to 85% pure and ALDH1− 99% to 100% pure after flow sorting (Fig. 3D). The sorted (untreated) ALDH1-positive and -negative populations showed similar growth rates in culture (Fig. 3E). The ALDH1+ subpopulation in 468 showed greater sphere formation than ALDH1− cells (Fig. 3F), as in other lines (54, 55). Notably, the ALDH1+ population exhibited higher DOT1L (6.1-fold) and H3K79me2 (3.6-fold) and cMyc (5.7-fold) and SOX2 (2.4-fold) levels than the ALDH1− population (Fig. 3G).
DOT1L inhibition decreases cMyc and impairs sphere formation in ALDH1+ but not in ALDH1− cells
ALDH1+ cells have been shown to undergo asymmetrical cell division generating ALDH1+ and ALDH1− progeny, whereas ALDH1− cells generate only ALDH1− cells (ref. 55; see model in Supplementary Fig. S2F). Single cells from flow-sorted ALDH1+ and ALDH1− populations were next plated into sphere assays with EPZ-5676 or DMSO control added once at seeding. Based on the known drug decay in vitro, seeded cells were thus exposed to active drug for only the initial 3 days. Interestingly, DOT1L inhibition decreased sphere formation from ALDH1+ cells, but not from ALDH1− (Fig. 4A).
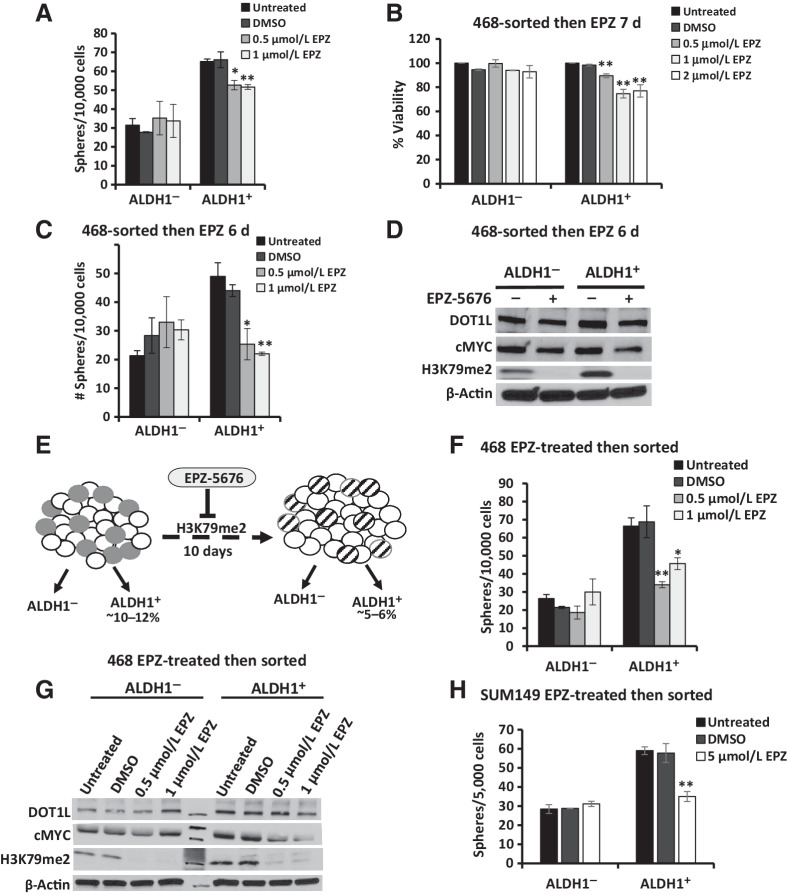
Figure 4.
Prolonged DOT1L inhibition attenuates sphere formation and cMyc expression in ALDH1+ cells. A, ALDH1+ and ALDH1− cells isolated from 468 were plated into sphere assays ± EPZ-5676 or DMSO added to sphere media at seeding. Mean sphere numbers ≥ 75 μm in diameter at 12–14 days with ± SEM are graphed. Total drug exposure was 3 days. B, Cell viability was assayed in sorted 468 ALDH1+ and ALDH1− cells treated ± EPZ-5676 or DMSO controls over 7 days. C, ALDH1+- and ALDH1−-sorted 468 cells were cultured ± EPZ-5676 or DMSO for 6 days, then plated into sphere assay conditions ± either DMSO or EPZ-5676 added once at seeding. Mean sphere numbers at 14 days are graphed ± SEM. Total drug exposure was approximately 9 days. D, Western blots show DOT1L, cMyc, and global H3K79me2 levels in ALDH1+ and ALDH1− populations treated with DMSO control or 0.5 μmol/L EPZ-5676 for 6 days. E, Model compares treatment-naïve ALDH1+ cells (gray, left image) with EPZ-5676–treated ALDH1+ cells that persist after 10 days EPZ-5676 (striped, right image). The decrease in ALDH1+ cells with treatment from 10% to 5% could result from ALDH1+ cell death or differentiation to generate more ALDH1− progeny. F, ALDH1+ and ALDH1− 468 cells were isolated after 468 treatment ± EPZ-5676 or DMSO for 10 days. Sorted cells were then plated into sphere assay with the drug or DMSO added once at seeding. Spheres ≥ 75 μm were counted at 12–14 days, and mean numbers ± SEM were plotted. Total drug exposure was 10 + 3 days or approximately 13 days. G, ALDH1+ and ALDH1− 468 cells were isolated after 468 treatment ± EPZ-5676 or DMSO for 10 days and levels of DOT1L, cMyc, and global H3K79me2 assayed by Western blots. H, SUM149 cells were treated with EPZ-5676 or DMSO for 10 days and FACS isolated into ALDH1+ and ALDH1− cells. Sorted cells were plated into sphere assay with drug/DMSO added at seeding, and spheres were counted at 12–14 days. Mean number of spheres is plotted with ± SEM. All assays were performed in three biological repeat sorts with three technical replicates within each assay and mean is graphed with ± SEM. Student t test compares each drug condition to DMSO controls: *, P ≤ 0.05; **, P ≤ 0.01.
The effect of longer drug exposure was next tested. When sorted cells were plated into drug and treated over 7 days with drug replenished at day 3, ALDH1+ but not ALDH1− cells showed a modest 22% to 25% decrease in viability only after 7 days (Fig. 4B), indicating a sensitivity of the ALDH1+ population to DOT1L catalytic inhibition.
Sorted ALDH1+ and ALDH1− cells were next treated with EPZ-5676 or DMSO control for 6 days in 2D culture before seeding into sphere assays, with further drug or DMSO added at the time of seeding. This provided a 9-day drug exposure (6 days in 2D and about 3 days in sphere culture). This longer EPZ-5676 exposure caused a greater decrease in sphere formation of >50% by ALDH1+ cells (Fig. 4C, compared with the 20% decrease after only 3 days of drug, Fig. 4A), whereas sphere formation by ALDH1− cells was not significantly different from DMSO controls. Notably, although both ALDH1+ and ALDH1− populations showed loss of H3K79me2, with methylation reduced by >90% in both populations after 6 days in 2D culture in 0.5 μmol/L EPZ-5676, cMyc levels were reduced by 50% in drug-treated ALDH1+ cells, but were unchanged ALDH1− cells, as assayed by densitometry of WB shown in Fig. 4D. Thus, DOT1L inhibition impedes ALDH1+ cell self-renewal in vitro and the effect of EPZ-5676 is enhanced by longer exposure, as would be expected from an epigenetic drug.
Prolonged inhibition of DOT1L activity alters stem cell properties of ALDH1+ cells
As DOT1L, cMyc, and H3K79me2 levels were higher in the ALDH1+ population and DOT1L inhibition reduced cMyc and sphere formation in ALDH1+ cells but not in ALDH1− cells, we next tested how stem cell properties of the ALDH1+ cells that persist after 10-day treatment differ from those of drug-naïve ALDH1+ cells in two independent TNBC lines. Unsorted 468 and SUM149 cells were first treated with EPZ-5676 or DMSO for 10 days. Drug-exposed ALDH1+ and ALDH1− cells were then isolated and compared with their drug-naïve counterparts (see model, Fig. 4E). ALDH1+ and ALDH1− groups were sorted by flow cytometry and seeded for sphere formation with either DMSO or fresh EPZ-5676 added once at seeding. As EPZ-5676 remains in culture media 2 to 3 days, total drug exposure was 12 to 13 days. This prolonged DOT1L inhibition reproducibly decreased sphere formation by 50% to 60% in the ALDH1+ population from the 468 cell line (Fig. 4F), whereas sphere formation from the ALDH1− populations from each of these lines was not significantly altered. As in Fig. 4D, untreated ALDH1+ cells showed higher DOT1L, cMyc, and global H3K79me2 levels than ALDH1− cells. Despite drug-mediated loss of H3K79me2 in both populations, DOT1L inhibition decreased cMyc only in the ALDH1+ population (by 60% with 0.5 μmol/L EPZ-5676, and by 80% with 1 μmol/L drug compared with untreated controls) and not in ALDH1− cells (Fig. 4G). DOT1L inhibition also decreased sphere formation by 40% in the ALDH1+ population from the SUM149 cell line (Fig. 4H). Taken together, these data suggest DOT1L-mediated H3K79 dimethylation maintains cMyc, viability, and self-renewal of the stem cell–enriched ALDH1+ cells in the 468 TNBC model in vitro.
Effects of DOT1L inhibition on gene-expression profiles in ALDH1+ and ALDH1− cells
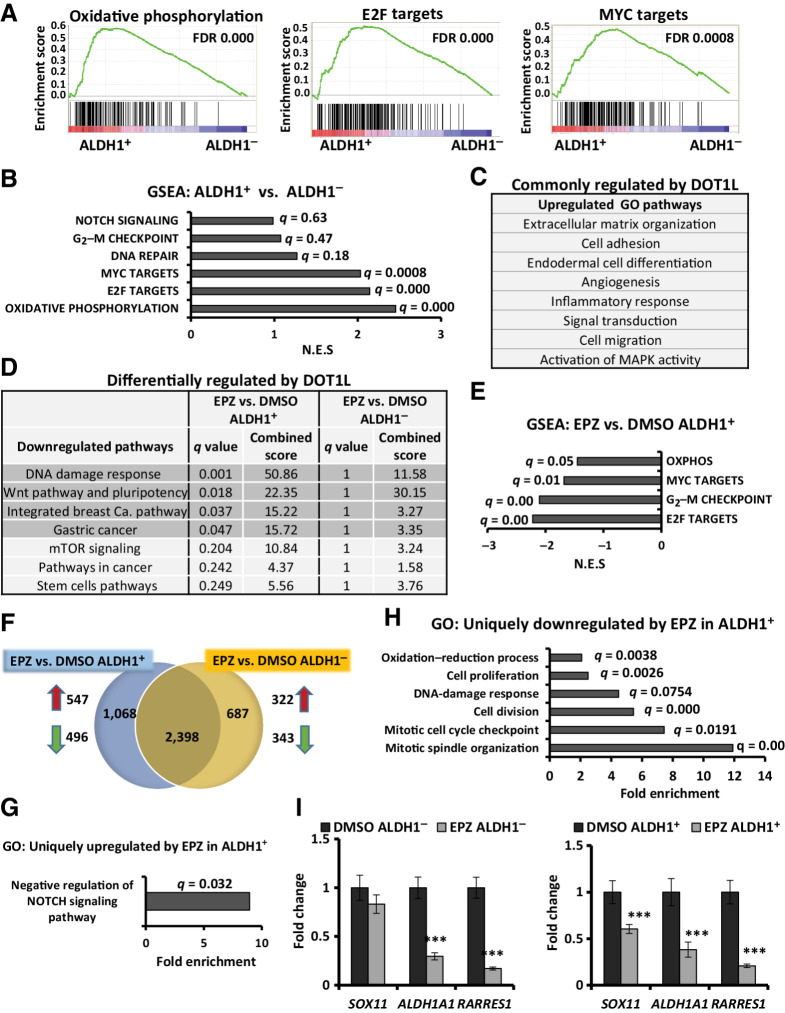
To further investigate how DOT1L maintains stem-like cells with high ALDH1 activity, we aimed to identify a gene-expression profile unique to this rare subset and how DOT1L inhibition changes it. Thus, 468 cells were first treated with either 0.5 μmol/L EPZ-5676 or DMSO over 10 days, renewing drug every three days, and then sorted based on ALDH1 activity. Global gene-expression patterns were compared and DOT1L-regulated pathways were identified. RNA-seq was carried out on cDNA libraries constructed from each of three biologically independent repeat assays (see Materials and Methods). Differences in patterns of gene expression between ALDH1+ and ALDH1− cells and after drug treatment are shown in Supplementary Table S1.
ALDH1+ cells express profiles of oxidative phosphorylation and cMyc and E2F target genes
Global gene expression in 468-derived ALDH1+ and ALDH1− cells was compared. Using an FDR of 0.05, only 71 genes were differentially expressed between ALDH1+ and ALDH1− populations. On GSEA, the three most strongly represented gene profiles in ALDH1+ cells versus ALDH1− were associated with oxidative phosphorylation (FDR, 0.000), E2F targets (FDR, 0.000), and cMyc targets (FDR, 0.00079; Fig. 5A and B). Gene sets involved in NOTCH signaling, DNA repair, and G2–M checkpoint showed a trend to upregulation in ALDH1+ cells compared with ALDH1− (Supplementary Fig. S3A). ALDH1+ cells showed significant enrichment of gene signatures identified in embryonic and mammary stem cells, breast cancer clusters, poorly differentiated cancers and metastasis (Supplementary Fig. S3B).
Figure 5.
DOT1L inhibition downregulates gene profiles of oxidative phosphorylation, cell division, and WNT pathway specifically in ALDH1+ cells. RNA-seq was performed in three independent biological ALDH1+ and ALDH1− cell populations collected after 10 days of treatment with DMSO or 0.5 μmol/L EPZ-5676. A, GSEA plots representing the three most significantly enriched clusters in 468 ALDH1+ cells vs. ALDH1− populations. B, Gene profiles overrepresented in ALDH1+ vs. ALDH1− cells, with respective FDR/q values and normalized enrichment scores of data graphed in A and Supplementary Fig. S3A. C, GO analysis for genes upregulated (fold change ≥ 2; q ≤ 0.05) in both EPZ-5676–treated ALDH1+ and ALDH1− populations compared with respective DMSO controls. D, WIKI/KEGG pathway analysis of the downregulated genes (fold change ≤ 0.7; q ≤ 0.05) in drug-treated vs. control cells. Gray highlighted pathways are significantly downregulated in EPZ-5676–treated ALDH1+ cells (q ≤ 0.05) but not in drug-treated ALDH1− cells (q = 1). E, GSEA comparing EPZ-5676–treated ALDH1+ vs. DMSO-treated ALDH1+ cells shows gene sets downregulated by treatment. F, Venn diagram displays genes whose expression is either commonly or differently altered by EPZ-5676 (EPZ) treatment in isolated ALDH1+ and ALDH1− populations compared with their respective DMSO controls: 547 genes were uniquely upregulated and 496 genes were uniquely downregulated in drug-treated ALDH1+ cells; 322 genes were uniquely upregulated and 343 genes were uniquely downregulated in response to drug in ALDH1− cells. G, GO analysis for genes significantly upregulated only in drug-treated ALDH1+ cells (547 genes). Biological processes shown with FDR q ≤ 0.05. H, GO analysis for genes significantly downregulated only in drug-treated ALDH1+ cells (496 genes). Biological processes shown with FDR q ≤ 0.05. I, qPCR gene expression of SOX11, ALDH1A1, and RARRES1 in drug- or DMSO-treated ALDH1− (left) and ALDH1+ populations (right). ***, P ≤ 0.001. See also Supplementary Fig. S3.
EPZ-5676 downregulates programs of oxidative phosphorylation, cell division, and proliferation in ALDH1+ cells
H3K79 dimethylation by DOT1L maintains active gene transcription (56–58). Hence, inhibition of this histone methyltransferase would impair DOT1L-activated target gene transcription. Comparison of transcript abundance in ALDH1+ and ALDH1− populations identified gene sets commonly or differentially modified after 10d EPZ-5676 treatment. Although DOT1L stimulates gene expression, unexpectedly, more genes were upregulated than downregulated after drug exposure. Gene ontology (GO) analysis showed that most biological processes/pathways upregulated by drug were similar in both drug-treated ALDH1− and ALDH1+ populations. DOT1L directly or indirectly governs gene programs associated with ECM organization, cell adhesion, angiogenesis, cell migration, inflammation, and MAPK activity in all cells (Fig. 5C). Intriguingly, DOT1L inhibition significantly downregulated gene programs associated with stem cell pluripotency, WNT signaling, DNA damage response, and breast cancer (q < 0.05) uniquely in ALDH1+ cells (Fig. 5D; see also Supplementary Table S2). Furthermore, programs of oxidative phosphorylation, E2F, and cMyc target gene that were overexpressed in ALDH1+ compared with ALDH1− were significantly downregulated by drug in ALDH1+ cells (Fig. 5E). In addition, GSEA showed DOT1L inhibition induces programs directing epithelial differentiation, while inhibiting stem cell, breast cancer, and metastatic gene signatures in ALDH1+ cells (Supplementary Fig. S3C).
A second analysis identified gene profiles whose regulation by DOT1L differed in the two populations. Here, we evaluated whether the significantly differentially expressed genes (DEG) in drug-treated ALDH1+ were also significantly altered in drug-treated ALDH1− populations and vice versa. Although most of the DEGs were commonly altered in both groups, 1,068 genes were uniquely and significantly altered in response to drug in the ALDH1+ cells, whereas 687 genes were uniquely changed (up or downregulated) by drug in ALDH1− cells (Fig. 5F, for uniquely up or downregulated genes in ALDH1+ cells, see also Supplementary Table S3). The DAVID pathway revealed a profile of genes associated with negative regulation of NOTCH signaling uniquely in EPZ-5676–treated ALDH1+ cells (Fig. 5G). This included mediators of NOTCH inhibition, BEND6, CHAC1, and MMP14, and DLL4, which can serve as an inhibitory NOTCH ligand, all of which were significantly increased in EPZ-5676–treated ALDH1+ cells only. Expression data also demonstrated downregulation of NOTCH target genes HES1, and HEY2 only in EPZ-5676 treated ALDH1+ cells. Thus, DOT1L appears to activate NOTCH signaling uniquely in ALDH1+ cells. Genes associated with mitotic cell-cycle checkpoint, mitotic spindle organization, cell division, and proliferation were significantly downregulated by drugs in ALDH1+ cells but not in ALDH1− cells (Fig. 5H; see also Supplementary Table S4). It is noteworthy that these processes are also regulated by E2F and cMyc (59, 60). This second analysis also showed that the oxidation–reduction pathways upregulated in ALDH1+ relative to ALDH1− cells (Fig. 5A) were also significantly and specifically downregulated by DOT1L catalytic inhibition in ALDH1+ cells (Fig. 5E and H). Thus, DOT1L-mediated H3K79 methylation appears to regulate gene drivers of DNA integrity, proliferation, and self-renewal in ALDH1+ CSC-enriched cells.
A subset of DEGs identified by expression profiling was validated by qPCR. SOX11 is upregulated in ER-negative breast cancers and promotes breast cancer invasion and progression (61). Notably, SOX11 was uniquely and significantly repressed in drug-treated ALDH1+ cells (Fig. 5I). ALDH1A1 activity is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome (12). It also plays pleiotropic roles in cell metabolism, many of which are unrelated to stem cells (62). Retinoic acid receptor responder protein 1 (RARRES1) is an RA-inducible gene and the protein product interacts with retinoic acid receptors upon retinoic acid activation by the ALDH1 pathway (63). ALDH1A1 and RARRES1 expression was confirmed to be significantly downregulated by DOT1L inhibition in both populations (Fig. 5I). Thus, DOT1L promotes ALDH1A1 induction in this TNBC model.
DOT1L inhibition decreases 468 xenograft growth in vivo
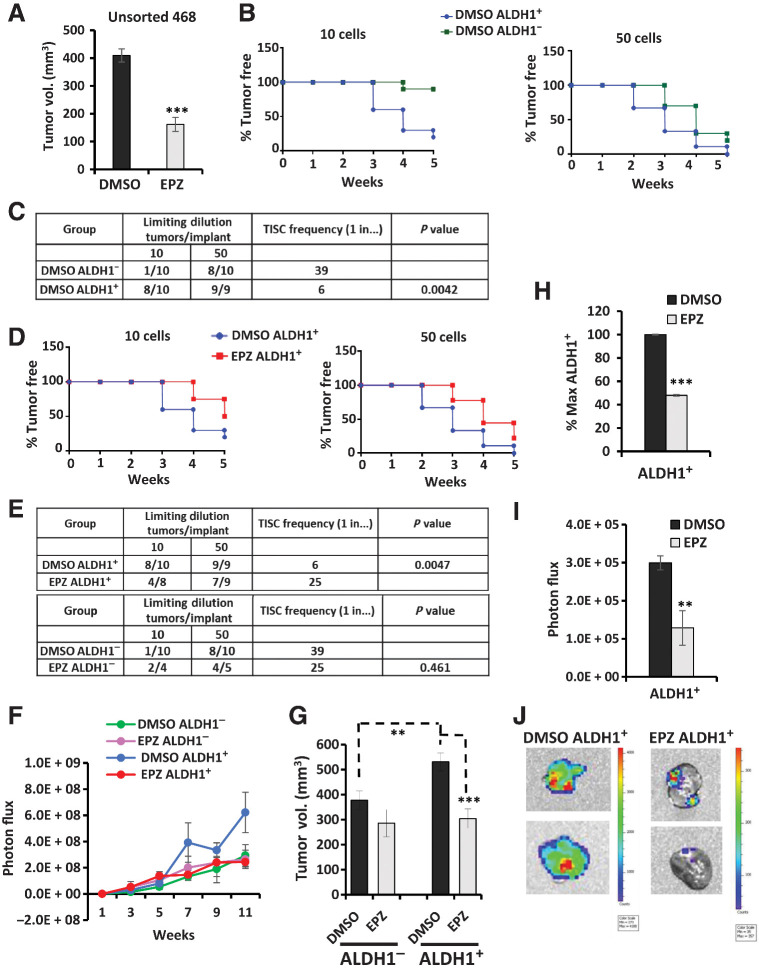
Although our ultimate goal was to test if DOT1L inhibition can target the stem cell–enriched ALDH1+ cells in vivo, in a first experiment, we first tested if EPZ-5676 could inhibit 468 growth in vivo. EPZ-5676 is not bioavailable orally and has a very short half-life after intraperitoneal (i.p.) injection (37, 43). Thus, luciferase-tagged 468 cells (468-luc) were pretreated in vitro for 10 days with either EPZ-5676 1 μmol/L or DMSO control (n = 6/group); then 5 × 105 cells were injected orthotopically into NODSCIDγ mice. In an effort to prolong drug exposure and prevent the recovery of H3K79 dimethylation during tumor formation, drug/vehicle treatment was extended after orthotopic injections by giving EPZ-5676 50 mg/kg i.p. on alternate days for six doses. Thus, total drug treatment extended for 21 days (10 days in 2D culture and 11 days in vivo). There was no evidence for systemic toxicity or weight loss in drug-treated mice compared with controls (Supplementary Fig. S4A). EPZ-5676 significantly decreased tumor volume by 10 weeks (Fig. 6A) and metastasis from primary tumors (P < 0.05; Supplementary Fig. S4B).
Figure 6.
EPZ-5676 treatment decreases TISC abundance, tumor growth, and metastasis in ALDH1+ cell xenografts in vivo. A, Unsorted 468-luc cells pretreated with EPZ-5676 or DMSO for 10 days were orthotopically injected into mammary fat pad of NSG mice and then mice were treated with 50 mg/kg EPZ-5676 or vehicle via intraperitoneal injections every alternate day for six doses. Final tumor volumes at sacrifice are graphed. ***, P = 0.00053. B, Tumor formation from limiting dilutions of 10 ALDH1+ cells compared with 10 ALDH1− cells and 50 ALDH1+ cells compared with 50 ALDH1− cells is graphed as % tumor-free mice over time. C, Tumor formation in mice and TISC frequency was calculated using L-Calc limiting dilution software (STEMCELL Technologies) comparing control DMSO ALDH1+ with DMSO ALDH1− mice groups. D, Tumor formation in mice injected with 10 EPZ-5676–treated ALDH1+ cells compared with 10 DMSO-treated ALDH1+ cells and 50 EPZ-5676–treated ALDH1+ cells compared with 50 DMSO controls is graphed as % tumor-free mice over weeks. E, Tumor formation and TISC frequency are tabulated for the drug-treated ALDH1+ mice group and the vehicle control ALDH1+ group (top) and for drug-treated ALDH1− cell–injected mice and control ALDH1− cell–injected mice. F, Tumor growth is plotted as mean tumor bioluminescence over time in mice injected with 5,000 EPZ-5676–treated or vehicle-treated ALDH1+ and ALDH1− cells. G, Mean final tumor volumes from 5,000 cell injection groups at sacrifice are graphed ± SEM. Control ALDH1+ tumors vs. control ALDH1− **, P = 0.01; EPZ-5676 ALDH1+ tumors vs. DMSO control ALDH1+ ***, P = 0.0009. H, Tumors from mice injected with 5,000 EPZ-5676 (EPZ) or vehicle–treated ALDH1+ cells were excised at sacrifice, digested into single cells and analyzed for % ALDH1+ cells and graphed as % max ALDH1+ cells ± SEM. ***, P = 0.00012. I, Primary orthotopic tumor sites in mice injected with 5,000 EPZ- or vehicle-treated ALDH1+ cells were covered, and bioluminescence from metastasis outside the primary tumor bed was measured. **, P = 0.01. J, Representative images of lung metastasis imaged ex vivo at the time of sacrifice. The signal bioluminescence is indicated in the side bar.
EPZ-5676 increases tumor latency and decreases TISC frequency of ALDH1+ cells in vivo
Because prolonged EPZ-5676 exposure decreased ALDH1+ cells in vitro, we aimed to test if this loss of stem cell activity would be manifest in vivo by reduced tumor formation. As the TISC frequency in 468 cells was unknown, a pilot study tested TISC frequency in 100, 1,000, 10,000, and 100,000 sorted ALDH1+ or ALDH1− 468-luc cells (4 mice/group), as detailed in Materials and Methods. As few as 100 ALDH1+ cells generated tumors in all mice injected, whereas only 50% of mice injected with 100 ALDH1− cells formed tumors (Supplementary Fig. S4C). Hence, limiting dilutions of 50 and 10 sorted ALDH1− and ALDH1+ cells were chosen for subsequent TISC assays with and without drug treatment.
Given the limited bioavailability of EPZ-5676, we next tested if drug-mediated targeting of the ALDH1+ population ex vivo would be manifest as a loss of tumor-initiating cell abundance in vivo. To test if DOT1L inhibition reduces tumor initiation by stem cell–enriched ALDH1+ cells in vivo, drug effects on tumor-initiating frequency were compared in xenografts formed from either ALDH1+ or ALDH1− cells. After 10 days of 1 μmol/L EPZ-5676 or DMSO treatment in vitro, ALDH1+ and ALDH1− 468-luc cells were flow sorted, and limiting dilutions of 10 and 50 cells (n = 10 mice/group) were inoculated orthotopically, and TISC frequency was measured by serial IVIS over time. To monitor tumor growth and metastasis, 5,000 ALDH1+ or ALDH1− cells were also injected in groups of eight mice with or without DMSO or drug. These mice also received 6 i.p. injections of vehicle/EPZ-5676 as above.
In the 10 cell injection groups, 8/10 mice injected with DMSO-treated ALDH1+ cells developed palpable tumors within 5 weeks, whereas only 1/10 mice formed tumors from ALDH1− cells (% tumor-free plotted; Fig. 6B, left). In the 50 cell groups, all nine ALDH1+-injected mice and 8/10 ALDH1−-injected developed tumors by 5 weeks (Fig. 6B, right graph). TISC frequency was 1 in 6 cells in untreated ALDH1+ cells, significantly higher than that in control ALDH1− cells (1 in 39 cells, P = 0.0042; Fig. 6C). Thus, as in other breast cancer lines (54), ALDH1+ 468-luc cells form tumors with shorter latency and have higher TISC frequency than do ALDH1− cells.
DOT1L inhibition by EPZ-5676 prolonged tumor latency and significantly decreased tumor initiation only in the ALDH1+ group. Only 50% (n = 4/8) of ALDH1+ injected drug-treated mice in the 10-cell group formed tumors by 5 weeks compared with 80% (8/10) in ALDH1+ DMSO controls (Fig. 6D, left graph). Similarly, in the 50 cell-injected groups, EPZ-5676 treatment prolonged ALDH1+ tumor latency and decreased tumor initiation over 5 weeks with 78% (7/9) of drug-treated mice forming tumors compared with 100% (9/9) in untreated hosts (Fig. 6D, right). The TISC frequency declined significantly from 1/6 cells in controls to 1/25 in EPZ-5676–treated ALDH1+ cells (P = 0.0047; Fig. 6E). In contrast, tumor initiation cell frequencies did not differ significantly between drug- and DMSO-treated ALDH1− cells (P = 0.461; Fig. 6E). Thus, TISC frequency is higher and tumor latency reduced in the CSC-enriched ALDH1+ population compared with ALDH1− cells, and the DOT1L inhibitor EPZ-5676 selectively targets the stem cell–enriched ALDH1+ population and failed to affect tumor initiation by ALDH1− cells.
Tumors formed from 5,000 DMSO-treated ALDH1+ cells showed more rapid growth (serial IVIS assays; Fig. 6F) and greater final volume than the ALDH1− group (Fig. 6G). EPZ-5676 slowed growth of ALDH1+ cell–generated tumors, but not that of ALDH1− cell–derived tumors (Fig. 6F), again supporting the specific antitumor action of EPZ-5676 in the stem cell–enriched ALDH1+ population. EPZ-5676 significantly decreased final tumor volume only in ALDH1+ cell xenografts (Fig. 6G).
EPZ-5676 reduces the percentage of ALDH1+ tumor cells and metastasis in vivo
If DOT1L inhibition reduces the self-renewal and/or viability of ALDH1+ cells, drug-treated tumors formed from ALDH1+ cells should be enriched for ALDH1− cells compared with untreated controls. Tumors formed from 5,000 ALDH1+ cell injections were excised, dissociated, and the percent ALDH1+ cells evaluated. EPZ-5676 significantly reduced the percentage of ALDH1+ cells in ALDH1+ xenografts compared with untreated DMSO controls (Fig. 6H). Thus, DOT1L inhibition effectively targets ALDH1 high cells in tumors formed from ALDH1+-enriched populations, validating our in vitro findings.
To elucidate effects of DOT1L inhibition on tumor metastasis, mice injected with 5,000 cells were followed for tumor spread beyond the primary tumor site by weekly IVIS. All mice injected with ALDH1+ control cells developed metastasis by 11 weeks. EPZ-5676 significantly decreased metastasis from tumors arising from ALDH1+ cells (Fig. 6I), but not from ALDH1− tumors (Supplementary Fig. S4D). Representative images of lung metastasis from ALDH1+ xenografts are shown in Fig. 6J. Thus, DOT1L inhibition by EPZ-5676 decreased not only CSC abundance in the ALDH1+ population but also their growth and metastasis in vivo.
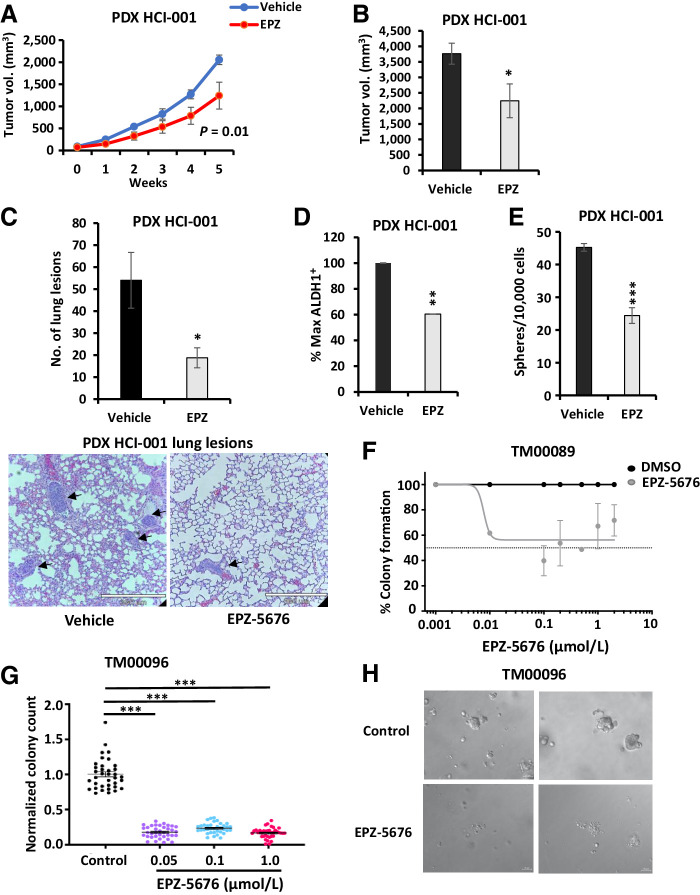
EPZ-5676 treatment decreases TNBC PDX tumor growth in vivo
To test the antitumor efficacy of EPZ-5676 in established TNBC tumors, effects on two well-characterized established TNBC PDX, HCI001 and HCI010 (48), were assayed. When implanted PDX mammary tumors reached 5-mm diameter, mice were treated with either 40 mg/kg EPZ-5676 or vehicle (n = 5 mice/group) i.p. daily for 5 weeks and tumor growth was measured weekly. DOT1L inhibition significantly slowed PDX HCI001 tumor growth and decreased formation of lung metastasis in vivo (Fig. 7A–C). To validate DOT1L action on stem cells in vivo, PDX tumors from vehicle- and drug-treated mice were dissociated and the percentage of ALDH1+ and sphere formation assayed. There was a significant decrease in the percentage of ALDH1+ cells (Fig. 7D), and the proportion of sphere-forming cells (Fig. 7E) in dissociated EPZ-5676–treated HCI001 PDX tumors compared with vehicle-treated controls. Notably, a second, more slowly growing HCI010 PDX model showed no significant decrease in tumor formation by EPZ-5676 (Supplementary Fig. S4E). There was no difference in host weight gain over time in treatment versus control groups for either of these PDX models.
Figure 7.
Treatment with EPZ-5676 decreases TNBC PDX tumor growth in vivo and organoid colony formation. A, TNBC PDX HCI-001 pieces were implanted into 5 NSG mice and mice received EPZ-5676 or vehicle treatment over 5 weeks via i.p. injections daily. Tumor growth is plotted as measured by caliper over time. Statistical analysis of growth curve with P value is indicated. B, Mean final tumor volume measured by caliper at sacrifice in mice injected with PDX HCI-001 is graphed ± SEM. EPZ-treated tumor vs. DMSO control P = 0.04. C, Number of micrometastatic lung lesions from PDX HCI-001 in vehicle- and EPZ-treated mice are graphed and representative images are shown below with arrows pointing to the lung lesion. D, Tumors from mice injected with PDX HCI-001 were excised at sacrifice, digested into single cells, and analyzed for % ALDH1+ cells. Normalized % max ALDH1+ cells are graphed ± SEM. P = 0.005. E, Tumors from mice injected with PDX HCI-001 were excised at sacrifice, digested into single cells, and plated into sphere assay. Number of spheres from EPZ-5676– or vehicle-treated tumors is graphed ± SEM. P = 0.00005. F, TNBC PDX-derived organoids were digested into 3–10 cell clusters and seeded with different concentrations of EPZ-5676 or DMSO for 14 days and allowed to grow colonies. The percentage of colony formation for EPZ-5676–treated compared with DMSO control is plotted. P = 0.0003. G, TNBC PDX-derived organoids were digested into 3–10 cell clusters and seeded to form colonies with EPZ-5676 or DMSO treatment for 12 days. Colonies were recovered, digested, and reseeded with DMSO for 12 days, and colonies were counted as described in the Materials and Methods section. Normalized colony counts in pretreatment with EPZ-5676 at different concentrations as compared with DMSO control are graphed. P < 0.0001. H, Representative images of colonies in DMSO control and EPZ-5676 treated samples derived from TNBC PDX organoids.
DOT1L inhibitor abrogates clonogenic growth of PDX-derived organoid cultures
Assays of drug action were also carried out in two different PDX-derived organoid cultures. TNBC PDX TM00089 was digested into 3–10 cell clusters, seeded into 3D culture and grown with a range of EPZ-5676 concentrations or DMSO and colony formation evaluated after 14 days as in (49). As little as 10 nmol/L EPZ-5676 significantly decreased the percentage of colony formation from PDX TM00089-derived organoids by 40% (Fig. 7F). In a second experiment, the clonogenic ability of drug-exposed organoid colonies was assayed. Clonogenic organoid growth of PDX TM00096 was tested in the presence of drug or DMSO for 12 days, and then drug-treated organoids were digested and reseeded with either vehicle or drug exposure for a further 12 days. After the first 12 days in drug, cells reseeded from colonies that emerged in the presence of DOT1L inhibitor had completely lost clonogenic ability compared with DMSO controls (Fig. 7G; representative images of DMSO and EPZ-5676–treated colonies shown in Fig. 7H). Notably, after the first 12 days of EPZ-5676 treatment, there was no further loss of clonogenic growth induced by additional drug treatment between days 13 and 24 (Supplementary Fig. S4F). Assays with both PDX-derived organoid models were each carried out in triplicate biological repeats.
Discussion
Epigenetic mechanisms control chromatin packaging and direct patterns of gene transcription across the genome, thereby determining cell identity and fate as cell division occurs (64). Epigenomic regulators are central to both embryonic and adult tissue stem cell maintenance and govern the crucial balance between their self-renewal and progressive restriction to committed progenitor and mature populations (65). DOT1L has been shown to be an essential epigenetic modulator of stem and progenitor populations during normal development. This H3K79 HMT is essential for embryonic development, as murine embryos with homozygous DOT1L deletion do not survive beyond E10.5 (28). Similarly, cardiac-specific conditional DOT1L knockout causes cardiac dilation, and postnatal and adult lethality (66) as DOT1L-mediated H3K79me2 regulates essential, specific genes that define cardiac cell fate and maturation (30). Pharmacologic inhibition and knockdown of DOT1L impairs neural stem cell survival through altered transcriptional regulation of ER stress response programs (67). DOT1L also acts as a coactivator of TCF4/β-catenin to stimulate WNT target genes in intestinal crypt progenitor cells (34), suggesting a role in intestinal SC maintenance. Finally, conditional DOT1L deletion in embryonic, postnatal, and adult mice leads to depletion of hematopoietic stem cells, progenitors, and impaired bone marrow homeostasis (31–33).
CSCs hijack properties unique to normal tissue stem/progenitor cells to enable tumorigenesis and therapy resistance. To target CSC, we need to better understand mechanisms that sustain them. Oncogenic activation of DOT1L has been implicated in several cancers. DOT1L is a therapeutic target in MLL (MLL-rearranged AML; refs. 37, 68), wherein translocations of MLL1 with a number of gene partners, cause constitutive DOT1L recruitment to activate genes crucial to leukemogenesis (35). DOT1L overexpression has prognostic and therapeutic implications in ovarian (40), colorectal (38), and breast cancers (41, 42) and in neuroblastoma (39).
EPZ-5676 potently targets MLL-rearranged leukemic cells in vivo (37, 43), but these studies did not specifically address whether DOT1L is required for malignant stem cell maintenance or expansion. Here, we make the unprecedented finding that DOT1L is a potential therapeutic target in ALDH1+ TNBC stem cells. Elevated DOT1L, observed in TNBC from two independent data sets, is associated with poor patient outcome. This finding supports the biological and potential clinical relevance of DOT1L in this disease. DOT1L inhibition significantly decreases ALDH1+ cell abundance, sphere formation, and MYC and SOX2 expression—all properties of CSC—in three independent TNBC cell line models, with greater effects upon prolonged drug exposure. Although prior work has demonstrated DOT1L inhibitors have antitumor activity, here, the specific sensitivity of CSCs to DOT1L inhibition was revealed by sorting the CSC-enriched ALDH1+ from ALDH1− populations. ALDH1+ cells in MDA-MB-468 exhibit higher DOT1L, H3K79 dimethylation, and cMyc levels than ALDH1− cells. EPZ-5676 downregulates cMyc and impairs ALDH1+ cell self-renewal, but not that of ALDH1− cells in vitro. EPZ-5676 not only decreases ALDH1+ cell abundance in vitro, but also specifically decreases TISC frequency, to increase tumor latency and attenuate in vivo growth of tumors generated from ALDH1+, but not from ALDH1− populations. EPZ-5676 also reduces surviving ALDH1+ cells in posttreatment tumors, either through loss of ALDH1+ cell viability or by shifting cell fate toward generation of more ALDH1− progeny, and only decreases metastasis from xenografts generated from ALDH1+ cells. Moreover, EPZ-5676 treatment of established TNBC HCI001 PDX tumors in vivo not only decreased tumor growth, but also significantly depleted the abundance of ALDH1+ cells and sphere-forming cells in EPZ-5676–treated tumors. The failure of the HCI010 TNBC PDX model to respond to EPZ-5676 might reflect intrinsic heterogeneity of TNBC, but could also reflect the limited drug bioavailability and a requirement for higher dosing. In addition to effects in vivo in PDX, clonogenic growth of two independent TNBC PDX-derived organoid cultures was strongly inhibited following treatment with the DOT1L inhibitor. Although in vitro and in vivo work with sorted ALDH1+ cells indicates that DOT1L is a therapeutic target in CSC-enriched populations, drug responses in vivo in PDX and in PDX-derived organoids demonstrate the potential for DOT1L inhibitors to treat established tumors.
Our data strongly support earlier findings that implicate DOT1L in stem/progenitor regulation in solid tumors. In colorectal cancer, IL22 signaling upregulates DOT1L expression to promote activation of core stem cell genes and govern stem-like cell maintenance and tumorigenic potential (38). DOT1L overexpression transformed the nontumorigenic, immortal MCF10A breast epithelial line and led to acquisition of invasive properties (41). Notably, DOT1L was also shown to regulate TISC abundance in both ER+ and ER− breast cancer models (41). Genome-wide CRISPR-Cas9 screens in patient-derived glioblastoma recently identified DOT1L as a potential therapeutic target and EPZ-5676 pretreatment of primary GBM stem cell cultures yielded reduced tumor growth in vivo (44).
Our global expression profiling showed upregulation of oxidative phosphorylation programs and E2F and cMyc targets in the ALDH1+ compared with ALDH1−TNBC cells. ALDH1+ normal mammary epithelial cells also show upregulation of genes associated with oxidative phosphorylation compared with more differentiated ALDH1− cells (69). Notably, leukemic stem cells appear to rely on oxidative phosphorylation for survival and proliferation (70). MYC amplification in chemotherapy-resistant TNBC was shown to maintain the CSC population by enhancing mitochondrial oxidative phosphorylation and HIF-1α levels (71). The ALDH1+ 468 TNBC subpopulation had higher DOT1L, global H3K79 dimethylation and cMyc than the more differentiated ALDH1− population. DOT1L inhibition significantly depleted cMyc levels and downregulated cMyc targets and gene programs of oxidative phosphorylation, only in ALDH1+ cells. Thus, DOT1L-regulated MYC expression might be required to maintain oxidative phosphorylation in ALDH1+ breast CSCs.
Many E2F and cMyc targets, which are highly expressed in CSC-enriched ALDH1+ cells, regulate DNA repair, cell-cycle checkpoints, cell division, and proliferation (59). Notably, gene profiles associated with these biological processes were significantly downregulated upon DOT1L inhibition in ALDH1+ cells but not in ALDH1− cells. The role of cMyc in regulating ES cell pluripotency and CSC self-renewal is well established. cMyc-driven epigenetic reprogramming induces a stem-like state with metastatic capacity in breast cancer cells (72) and promotes progenitor cell reprogramming to preserve the stem cell–like phenotype in basal breast cancer models (73). DOT1L-mediated H3K79me2 is required for cMyc to recognize its target genes (74), and coregulation of gene expression by DOT1L and cMyc has been reported in cancer cells (41, 74, 75). Here, we show DOT1L maintains both cMyc levels and self-renewal in the minor ALDH1+ subpopulation but not the bulk population. Further investigations are needed to address how DOT1L and cMyc critically mediate ALDH1+ cell maintenance in TNBC.
DOT1L-mediated H3K79 dimethylation is linked to active transcription, and H3K79me2 annotates pluripotency genes in mouse ES cells (29, 30). NOTCH signaling, which is important for mammary stem cell self-renewal, appears to be regulated by DOT1L in ALDH1+ TNBC cells. Notably, negative regulators of NOTCH signaling, including BEND6, CHAC1, and MMP14, were upregulated in EPZ-5676–treated ALDH1+ cells, whereas critical NOTCH target genes HES1 and HEY2 were significantly downregulated only in drug-treated ALDH1+ cells. In 468 ALDH1+ but not in the ALDH1− TNBC population, DOT1L inhibition downregulates WNT signaling and pathways that regulate stem cell pluripotency. CSCs exhibit higher DNA damage response and repair mechanisms than bulk cancer cells and these have been implicated in their chemoresistance (3). Notably, DOT1L inhibition downregulates gene programs of DNA damage response, DNA repair, cell-cycle checkpoints, and mitotic spindle organization in ALDH1+ cells but not in ALDH1− cells. DOT1L inhibition was shown to attenuate DNA repair and sensitize MLLr AML cells to chemotherapeutic drugs, mitoxantrone and cytarabine (76). These findings raise the intriguing possibility that DOT1L inhibitors might be effectively combined with chemotherapeutic drugs to overcome CSC chemoresistance.
Global changes in epigenetic regulation occur during dedifferentiation and tumor progression (24, 77). The finding that epigenetic mechanisms are frequently aberrantly regulated in human malignancies has fueled the development of drugs targeting them (23). Inhibitors of histone deacetylases (HDAC), DNA methyltransferases (DNMT), and histone methyltransferases (HMT) have been most widely studied in cancer. EZH2 inhibitors showed high preclinical efficacy (78, 79), and clinical trials for lymphoma were recently completed. The DOT1L inhibitor EPZ-5676 is among the first HMT inhibitors to enter clinical trials. Notably, a phase I EPZ-5676 trial, completed in the course of this work, showed the drug is well tolerated, safe, and has therapeutic potential, despite only modest single-agent efficacy for refractory AML and myelodysplastic syndrome (45). A phase Ib/II clinical trial with EPZ-5676 and standard chemotherapy (daunorubicin and cytarabine) for AML is currently ongoing. TNBC appear to be enriched for and to possibly arise from stem-like cells or progenitors with high self-renewal (15, 16). These breast cancers also exhibit greater de novo and acquired therapy resistance, early metastasis, and mortality than most other forms of the disease (15). Specific and robust targeted treatments have remained elusive, in part due to incomplete mechanistic understanding of TNBC drivers. The present study identifies DOT1L as key epigenetic regulator of ALDH1+ cells in TNBC and demonstrates that the selective DOT1L inhibitor EPZ-5676 targets a high ALDH1 stem cell–enriched TNBC population to extend tumor-free survival and reduce metastasis using cell line, PDX, and organoid models. Our finding that DOT1L-mediated H3K79 methylation is elevated in a CSC-enriched TNBC subpopulation and that pharmacologic DOT1L inhibition can be exploited to target TNBC TISCs in vivo have important implications for therapy and warrant further clinical investigation in this aggressive, treatment-refractory form of breast cancer.
Authors' Disclosures
A.J. Ewald reports grants from the Breast Cancer Research Foundation during the conduct of the study; other support from Immunocore outside the submitted work; in addition, A.J. Ewald has a patent for US20140336282A1 issued and a patent for WO2016183183A1 pending. G. Pearson reports grants from the NIH during the conduct of the study. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
This work was supported by grant W81XWH-15-1-0581 from the US Department of Defense Breast Cancer Research Program to J. Slingerland and C. Wahlestedt and by a Breast Cancer Research Foundation grant to AJE (BCRF-21-048). EPZ-P5676 was provided for these studies by Epizyme. Epizyme did not provide any funding for this work. We thank Lluis Morey, Maria Figueroa, and Ralf Landgraf for useful discussions. We acknowledge the Imaging Core at Lombardi Comprehensive Cancer Center at Georgetown University, the Flow Cytometry Shared Resources at both Lombardi and Sylvester Comprehensive Cancer Centers and the Onco-Genomics Shared Resource at Sylvester Comprehensive Center Center, and the Research Informatics Core of the JP Hussman Institute for Human Genomics at University of Miami Miller School of Medicine.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
H. Kurani: Conceptualization, data curation, formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. Seyedeh Fatemeh Razavipour: Formal analysis, validation, investigation, writing–review and editing, co-second author in second position among authors. K.B. Harikumar: Conceptualization, formal analysis, validation, investigation, writing–review and editing, co-second author in third authorship position. M. Dunworth: Formal analysis, validation, investigation. A.J. Ewald: Conceptualization, funding acquisition, methodology, writing–review and editing. A. Nasir: Investigation. G. Pearson: Methodology, writing–review and editing. D. Van Booven: Formal analysis. Z. Zhou: Validation, investigation. D. Azzam: Conceptualization, investigation, methodology. C. Wahlestedt: Conceptualization, resources. J. Slingerland: Conceptualization, formal analysis, funding acquisition, methodology, writing–original draft, writing–review and editing.
References
- 1. Tannock IF, Hill RP, Carey RW. The basic science of oncology. Plast Reconstr Surg 1989;83:920. [Google Scholar]
- 2. Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP–dependent transporters. Nat Rev Cancer 2002;2:48–58. [DOI] [PubMed] [Google Scholar]
- 3. Vitale I, Manic G, De Maria R, Kroemer G, Galluzzi L. DNA damage in stem cells. Mol Cell 2017;66:306–19. [DOI] [PubMed] [Google Scholar]
- 4. Wulf GG, Wang RY, Kuehnle I, Weidner D, Marini F, Brenner MK, et al. A leukemic stem cell with intrinsic drug efflux capacity in acute myeloid leukemia. Blood 2001;98:1166–73. [DOI] [PubMed] [Google Scholar]
- 5. Frank NY, Margaryan A, Huang Y, Schatton T, Waaga-Gasser AM, Gasser M, et al. ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma. Cancer Res 2005;65:4320–33. [DOI] [PubMed] [Google Scholar]
- 6. Dylla SJ, Beviglia L, Park I-K, Chartier C, Raval J, Ngan L, et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS One 2008;3:e2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006;444:756–60. [DOI] [PubMed] [Google Scholar]
- 8. Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A 2009;106:13820–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tanei T, Morimoto K, Shimazu K, Kim SJ, Tanji Y, Taguchi T, et al. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res 2009;15:4234–41. [DOI] [PubMed] [Google Scholar]
- 10. Raha D, Wilson TR, Peng J, Peterson D, Yue P, Evangelista M, et al. The cancer stem cell marker aldehyde dehydrogenase is required to maintain a drug-tolerant tumor cell subpopulation. Cancer Res 2014;74:3579–90. [DOI] [PubMed] [Google Scholar]
- 11. Duru N, Fan M, Candas D, Menaa C, Liu H-C, Nantajit D, et al. HER2-associated radioresistance of breast cancer stem cells isolated from HER2-negative breast cancer cells. Clin Cancer Res 2012;18:6634–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007;1:555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, et al. Aldehyde dehydrogenase 1–positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res 2010;16:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 15. Anders C, Carey LA. Understanding and treating triple-negative breast cancer. Oncology 2008;22:1233–43. [PMC free article] [PubMed] [Google Scholar]
- 16. Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 2009;15:907–13. [DOI] [PubMed] [Google Scholar]
- 17. Carey LA, Rugo HS, Marcom PK, Irvin W, Ferraro M, Burrows E, et al. TBCRC 001: EGFR inhibition with cetuximab added to carboplatin in metastatic triple-negative (basal-like) breast cancer. J Clin Oncol 2008;26:1009. [Google Scholar]
- 18. Miller K, Wang M, Gralow J, Dickler M, Cobleigh M, Perez EA, et al. Paclitaxel plus bevacizumab versus paclitaxel alone for metastatic breast cancer. N Engl J Med 2007;357:2666–76. [DOI] [PubMed] [Google Scholar]
- 19. Kopetz S, Mita MM, Mok I, Sankhala KK, Moseley J, Sherman BM, et al. First in human phase I study of BSI-201, a small molecule inhibitor of poly ADP-ribose polymerase (PARP) in subjects with advanced solid tumors. J Clin Oncol 2008;26:3577. [Google Scholar]
- 20. Wander SA, Hennessy BT, Slingerland JM. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J Clin Invest 2011;121:1231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med 2018;379:2108–21. [DOI] [PubMed] [Google Scholar]
- 22. Gates LA, Foulds CE, O'Malley BW. Histone marks in the ‘Driver's Seat’: functional roles in steering the transcription cycle. Trends Biochem Sci 2017;42:977–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Toh TB, Lim JJ, Chow EK-H. Epigenetics in cancer stem cells. Mol Cancer 2017;16:29–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Michalak EM, Visvader JE. Dysregulation of histone methyltransferases in breast cancer – opportunities for new targeted therapies? Mol Oncol 2016;10:1497–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pfister SX, Ashworth A. Marked for death: targeting epigenetic changes in cancer. Nat Rev Drug Discov 2017;16:241–63. [DOI] [PubMed] [Google Scholar]
- 26. Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov 2009;8:724–32. [DOI] [PubMed] [Google Scholar]
- 27. Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev 2011;25:1345–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet 2008;4:e1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barry ER, Krueger W, Jakuba CM, Veilleux E, Ambrosi DJ, Nelson CE, et al. ES cell cycle progression and differentiation require the action of the histone methyltransferase Dot1L. Stem Cells 2009;27:1538–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cattaneo P, Kunderfranco P, Greco C, Guffanti A, Stirparo GG, Rusconi F, et al. DOT1L-mediated H3K79me2 modification critically regulates gene expression during cardiomyocyte differentiation. Cell Death Differ 2016;23:555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feng Y, Yang Y, Ortega MM, Copeland JN, Zhang M, Jacob JB, et al. Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood 2010;116:4483–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood 2011;117:4759–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nguyen AT, He J, Taranova O, Zhang Y. Essential role of DOT1L in maintaining normal adult hematopoiesis. Cell Res 2011;21:1370–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mahmoudi T, Boj SF, Hatzis P, Li VSW, Taouatas N, Vries RGJ, et al. The leukemia-associated Mllt10/Af10-Dot1l are Tcf4/β-catenin coactivators essential for intestinal homeostasis. PLoS Biol 2010;8:e1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 2007;7:823–33. [DOI] [PubMed] [Google Scholar]
- 36. Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 2011;20:53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood 2013;122:1017–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kryczek I, Lin Y, Nagarsheth N, Peng D, Zhao L, Zhao E, et al. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity 2014;40:772–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wong M, Tee AEL, Milazzo G, Bell JL, Poulos RC, Atmadibrata B, et al. The histone methyltransferase DOT1L promotes neuroblastoma by regulating gene transcription. Cancer Res 2017;77:2522–33. [DOI] [PubMed] [Google Scholar]
- 40. Liu D, Zhang X-X, Li M-C, Cao C-H, Wan D-Y, Xi B-X, et al. C/EBPβ enhances platinum resistance of ovarian cancer cells by reprogramming H3K79 methylation. Nat Commun 2018;9:1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cho M-H, Park J-H, Choi H-J, Park M-K, Won H-Y, Park Y-J, et al. DOT1L cooperates with the c-Myc-p300 complex to epigenetically derepress CDH1 transcription factors in breast cancer progression. Nat Commun 2015;6:7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang L, Deng L, Chen F, Yao Y, Wu B, Wei L, et al. Inhibition of histone H3K79 methylation selectively inhibits proliferation, self-renewal and metastatic potential of breast cancer. Oncotarget 2014;5:10665–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang W-T, Han C, Sun Y-M, Chen Z-H, Fang K, Huang W, et al. Activation of the lysosome-associated membrane protein LAMP5 by DOT1L serves as a bodyguard for MLL fusion oncoproteins to evade degradation in leukemia. Clin Cancer Res 2019;25:2795–808. [DOI] [PubMed] [Google Scholar]
- 44. MacLeod G, Bozek DA, Rajakulendran N, Monteiro V, Ahmadi M, Steinhart Z, et al. Genome-wide CRISPR-Cas9 screens expose genetic vulnerabilities and mechanisms of temozolomide sensitivity in glioblastoma stem cells. Cell Rep 2019;27:971–86. [DOI] [PubMed] [Google Scholar]
- 45. Stein EM, Garcia-Manero G, Rizzieri DA, Tibes R, Berdeja JG, Savona MR, et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 2018;131:2661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sandhu C, Garbe J, Bhattacharya N, Daksis J, Pan CH, Yaswen P, et al. Transforming growth factor beta stabilizes p15INK4B protein, increases p15INK4B-cdk4 complexes, and inhibits cyclin D1-cdk4 association in human mammary epithelial cells. Mol Cell Biol 1997;17:2458–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 2003;17:1253–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. DeRose YS, Wang G, Lin Y-C, Bernard PS, Buys SS, Ebbert MTW, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med 2011;17:1514–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Padmanaban V, Grasset EM, Neumann NM, Fraser AK, Henriet E, Matsui W, et al. Organotypic culture assays for murine and human primary and metastatic-site tumors. Nat Protoc 2020;15:2413–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19:649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu S, Feng Y, Zhao S. Proteins with evolutionarily hypervariable domains are associated with immune response and better survival of basal-like breast cancer patients. Comput Struct Biotechnol J 2019;17:430–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Calcagno AM, Salcido CD, Gillet J-P, Wu C-P, Fostel JM, Mumau MD, et al. Prolonged drug selection of breast cancer cells and enrichment of cancer stem cell characteristics. J Natl Cancer Inst 2010;102:1637–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Azzam DJ, Zhao D, Sun J, Minn AJ, Ranganathan P, Drews‐Elger K, et al. Triple negative breast cancer initiating cell subsets differ in functional and molecular characteristics and in γ-secretase inhibitor drug responses. EMBO Mol Med 2013;5:1502–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res 2009;69:1302–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jang K, Kim M, Gilbert CA, Simpkins F, Ince TA, Slingerland JM. VEGFA activates an epigenetic pathway upregulating ovarian cancer-initiating cells. EMBO Mol Med 2017;9:304–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 2008;40:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol 2008;28:2825–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 2002;109:745–56. [DOI] [PubMed] [Google Scholar]
- 59. Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev 2002;16:245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol 1999;19:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shepherd JH, Uray IP, Mazumdar A, Tsimelzon A, Savage M, Hilsenbeck SG, et al. The SOX11 transcription factor is a critical regulator of basal-like breast cancer growth, invasion, and basal-like gene expression. Oncotarget 2016;7:13106–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vassalli G. Aldehyde dehydrogenases: not just markers, but functional regulators of stem cells. Stem Cells Int 2019;2019:3904645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tomita H, Tanaka K, Tanaka T, Hara A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget 2016;7:11018–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Barth TK, Imhof A. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem Sci 2010;35:618–26. [DOI] [PubMed] [Google Scholar]
- 65. Chen T, Dent SYR. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet 2014;15:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nguyen AT, Xiao B, Neppl RL, Kallin EM, Li J, Chen T, et al. DOT1L regulates dystrophin expression and is critical for cardiac function. Genes Dev 2011;25:263–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Roidl D, Hellbach N, Bovio PP, Villarreal A, Heidrich S, Nestel S, et al. DOT1L activity promotes proliferation and protects cortical neural stem cells from activation of ATF4-DDIT3-mediated ER stress in vitro. Stem Cells 2016;34:233–45. [DOI] [PubMed] [Google Scholar]
- 68. Klaus CR, Iwanowicz D, Johnston D, Campbell CA, Smith JJ, Moyer MP, et al. DOT1L inhibitor EPZ-5676 displays synergistic antiproliferative activity in combination with standard of care drugs and hypomethylating agents in MLL-rearranged leukemia cells. J Pharmacol Exp Ther 2014;350:646–56. [DOI] [PubMed] [Google Scholar]
- 69. Colacino JA, Azizi E, Brooks MD, Harouaka R, Fouladdel S, McDermott SP, et al. Heterogeneity of human breast stem and progenitor cells as revealed by transcriptional profiling. Stem Cell Reports 2018;10:1596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jones CL, Stevens BM, D'Alessandro A, Reisz JA, Culp-Hill R, Nemkov T, et al. Inhibition of amino acid metabolism selectively targets human leukemia stem cells. Cancer Cell 2018;34:724–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee K-M, Giltnane JM, Balko JM, Schwarz LJ, Guerrero-Zotano AL, Hutchinson KE, et al. MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab 2017;26:633–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Poli V, Fagnocchi L, Fasciani A, Cherubini A, Mazzoleni S, Ferrillo S, et al. MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state. Nat Commun 2018;9:1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Santoro A, Vlachou T, Luzi L, Melloni G, Mazzarella L, D'Elia E, et al. p53 loss in breast cancer leads to Myc activation, increased cell plasticity, and expression of a mitotic signature with prognostic value. Cell Rep 2019;26:624–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol 2006;8:764–70. [DOI] [PubMed] [Google Scholar]
- 75. Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS One 2008;3:e3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liu W, Deng L, Song Y, Redell M. DOT1L inhibition sensitizes MLL-rearranged AML to chemotherapy. PLoS One 2014;9:e98270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gupta PB, Pastushenko I, Skibinski A, Blanpain C, Kuperwasser C. Phenotypic plasticity: driver of cancer initiation, progression, and therapy resistance. Cell Stem Cell 2019;24:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Knutson SK, Kawano S, Minoshima Y, Warholic NM, Huang K-C, Xiao Y, et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther 2014;13:842–54. [DOI] [PubMed] [Google Scholar]
- 79. McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012;492:108–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All primary data can be obtained from the corresponding author. All raw RNA-seq data are available at the GEO accession number GSE194434.