Abstract
Purpose:
Vismodegib is approved for the treatment of locally advanced basal cell carcinoma (laBCC), but some cases demonstrate intrinsic resistance (IR) to the drug. We sought to assess the frequency of IR to vismodegib in laBCC and its underlying genomic mechanisms.
Experimental Design:
Response to vismodegib was evaluated in a cohort of 148 laBCC patients. Comprehensive genomic and transcriptomic profiling was performed in a subset of five intrinsically resistant BCC (IR-BCC).
Results:
We identified that IR-BCC represents 6.1% of laBCC in the studied cohort. Prior treatment with chemotherapy was associated with IR. Genetic events that were previously associated with acquired resistance (AR) in BCC or medulloblastoma were observed in three out of five IR-BCC. However, IR-BCCs were distinct by highly rearranged polyploid genomes. Functional analyses identified hyperactivation of the HIPPO-YAP and WNT pathways at RNA and protein levels in IR-BCC. In vitro assay on the BCC cell line further confirmed that YAP1 overexpression increases the cell proliferation rate.
Conclusions:
IR to vismodegib is a rare event in laBCC. IR-BCCs frequently harbor resistance mutations in the Hh pathway, but also are characterized by hyperactivation of the HIPPO-YAP and WNT pathways.
Translational Relevance.
We show in a cohort of 148 locally advanced basal cell carcinoma (laBCC) treated with vismodegib that 6% demonstrate intrinsic resistance (IR) and 9% acquired resistance (AR). Prior treatment with chemotherapy was associated with 8-times higher incidence of IR. Out of five IR-BCC subjected to whole-exome sequencing, two had vismodegib-resistance genetic events in the Hh pathway (SMO mutation p.W535L and GLI2 amplification), which were previously described in AR-BCC. All five IR-BCC demonstrated a particular hyperactivation of the HIPPO-YAP and WNT pathways as compared with other BCCs, suggesting that the inhibition of these pathways could be a treatment option in the future.
Introduction
Basal cell carcinoma (BCC) of the skin is the most common form of human cancer and is known to be driven by genetic alterations that result in hyperactivation of the Hedgehog (Hh) pathway. Mutations typically affect either the tumor suppressor Patched 1 (PTCH1) or the oncoprotein Smoothened (SMO; ref. 1). Most BCCs can be surgically resected; however, a subset of BCCs progress to a more advanced, life-threating stage, hereafter referred to as either locally advanced (laBCC) or metastatic BCC (mBCC; refs. 2–4). The Hh pathway inhibitors (HPI) vismodegib (v) and sonidegib (s) have been approved for the treatment of laBCC (v + s) and mBCC (v only) since 2012 and 2015, respectively (5, 6). Although most patients experience clinical benefit from such HPIs, some tumors never respond (intrinsic resistance, IR), whereas others initially regress but eventually become resistant and regrow while still exposed to the drug (refs. 7–9; acquired resistance, AR).
In BCC, the most common mechanism of AR to HPIs is mutations of the drug-binding pocket of SMO (8, 9). Other less frequent mechanisms of AR are genetic alterations of the effectors of the Hedgehog pathway, SUFU and GLI2 (8). At the same time, molecular mechanisms underlying IR to vismodegib have not been elucidated.
The frequency of IR and AR cases has not been systematically examined for laBCC. We evaluated the response to vismodegib of 148 laBCC patients included in the STEVIE, a phase II, international, multicenter study. Using uniform definitions between centers, we report frequency of BCC cases with IR and AR, as well as cases with complete response (CR), partial (PR) response, and stable disease (SD). To unravel genetic mechanisms of IR to HPI in BCC (IR-BCC), we performed in-depth genomic characterization of five IR-BCC cases and identified putative pathways associated with their progression and drug resistance.
Materials and Methods
Study population and treatment regimen
The signed informed consent was obtained from all patients prior to study start (clinicaltrials.gov: NCT01367665). Institutional review boards and independent ethics committees of participating study centers approved the study protocol, which was executed in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines.
This retrospective study is based on 148 laBCC patients recruited to the STEVIE study between 2011 and 2013 for whom the response was evaluated (Supplementary Table S1). laBCCs included tumors that recurred at the same location after two or more surgical procedures and for which curative resection was deemed unlikely, as well as tumors where substantial morbidity or deformity from surgery was anticipated. Both patients with measurable and nonmeasurable disease (RECIST; version1.1, https://recist.eortc.org/recist-1-1-2/) were included. Patients were given 150 mg vismodegib orally once a day and were seen every month during the treatment period and followed every 3 months at least one year after treatment discontinuation. MRI or CT were performed every 4 to 8 weeks during treatment. We established a consensus definition of resistance between participants in this study and according to the STEVIE protocol response criteria.
Clinical criteria of disease status
Tumors were defined as IR when target lesions exhibited at least a 20% increase in total diameter compared with baseline either shortly after treatment initiation in cases of hyperprogression or after at least 6 months of treatment. This delay was chosen based on the initial phase I study where most responses were observed within 6 months of treatment (10). These IR cases correspond to patients with disease progression in the STEVIE study. AR cases required 30% shrinkage of tumors with subsequent regrowth at the same site during treatment. For patients discontinuing vismodegib without resistance, response was assessed at time of treatment discontinuation. SD refers to tumors that had <30% change in during treatment. Complete response (CR) was defined as broad tumor remission proven clinically, radiologically, and/or histologically, whereas partial response (PR) cases experienced at least a 30% decrease in the total diameter of target lesions compared with baseline (Supplementary Table S2).
Statistical analysis of the patient cohort
Patients' characteristics were described as median and interquartile range for quantitative data and counts and percentage for qualitative data. The cumulative incidence of discontinuation following IR and AR was assessed within a competing risk framework, with vismodegib discontinuation for other reasons than IR or AR considered as a competing event. Association between baseline covariates and the occurrence of discontinuation for IR or AR were estimated using the direct model for the cumulative incidence model proposed by Fine and Gray and quantified by subdistribution hazard ratio (11). If no event was observed in a subgroup, Gray test for comparing cumulative incidences curves was used instead of the Fine and Gray model (12). All analyses were performed using R version 3.1.2 (RRID:SCR_001905; ref. 13).
Diagnostic hematoxylin and eosin (H&E) and IHC
All samples were analyzed by H&E before extraction to confirm the presence of BCC tumor and appreciate tumor density. Histologic diagnosis was conducted by two pathologists, who evaluated the samples independently of each other, using formalin-fixed and paraffin-embedded (FFPE) sections. Enzymatic IHC was performed for routine diagnosis using the following antibodies: antiepithelial antigen antibody (clone Ber-EP4, DAKO-Agilent, catalog number GA637 dilution 1:100) and anti-EMA (clone E29, Dako-Agilent, catalog number GA629, dilution 1:200) as markers for BCC and squamous cell carcinoma (SCC), respectively. The tumors exhibited histologic profiles without extra cutaneous progression in accordance with the diagnosis of BCC. Clinical and histologic characteristics of these IR patients are shown in Supplementary Table S3. Four of five IR-BCCs (NB020_T01, NB022_T01, NB023_T01, and SG005_T01) were classified as basosquamous (BSC) subtype, and one BCC (NB021_T01) as morpheaform (Supplementary Table S3). Immune environment was studied by CD4, CD8, CD20, and PD1 staining.
DNA and RNA extraction
RNA from frozen tissue sections was extracted using the All prep DNA/RNA mini kit (Qiagen, catalog number 80204) and TRIzol (Thermo Fisher Scientific, TRIzol Reagent-15596018). RNA quantity and quality were assessed using the NanoDrop-ND-1000 (Nanodrop Technologies). Constitutive DNA was extracted from blood.
Whole-exome sequencing and data processing
For the genomic analysis, we used fresh-frozen tumor biopsies taken before the treatment with HPI for four of five cases (NB020, NB021, NB022, and NB023), and sample SG005 was biopsied 6 months after vismodegib treatment initiation. FFPE samples for genomic analysis were taken 1 month, 1 year, and 8 years before the HPI treatment initiation for SG005, NB020, and NB021 patients, respectively. Cases NB023 and SG005 were treated with radiotherapy 18 and 27 years before the HPI treatment initiation. Chemotherapy was applied to cases NB020 and NB023 1 month and 1 year before the HPI treatment. We performed whole-exome sequencing of all 5 IR-BCC and compared them with 103 BCC naïve of treatment (N-BCC), 9 after treatment BCC with acquired resistance to vismodegib (AR-BCC) and 5 vismodegib-sensitive BCC (S-BCC) from previous studies (8, 14).
The exomes were sequenced using Illumina HiSeq 2500 or BGISEQ-500 (sample SG005_T01) sequencers according to the manufacturer's protocols to the mean coverage after deduplication equal to 42× for tumor and 54× for normal DNA (100 bp paired-end reads). Reads were mapped using BWA-MEM (v0.7.12; RRID:SCR_010910) software (15) to the GRCh37 human reference genome and then we used the standard GATK (RRID:SCR_001876) best practice pipeline (16) to process the samples and call somatic genetic variants. PCR duplicates were removed, and base quality score was recalibrated using GATK (17), MarkDuplicates, and BaseRecalibrator tools. Somatic SNVs and INDELs were called and filtered using GATK tools Mutect2, FilterMutectCalls, and FilterByOrientationBias and annotated with oncotator (RRID:SCR_005183; ref. 18). SCNAs calling was done with FACETS (19). Quality controls of FASTQ and mapping were done with FASTQC (RRID:SCR_014583; ref. 20), Samtools (v1.9; RRID:SCR_002105; ref. 21), GATK HSmetrics and multiqc (RRID:SCR_014982; ref. 22). All processing steps were combined in a pipeline built with snakemake (v5.4.0; RRID:SCR_003475; ref. 23).
Somatic mutations with PASS flag from GATK Mutect2 were additionally filtered to have at least one supporting read from each strand and three reads in total. Oncogenic mutations were annotated using the OncoKB database (RRID:SCR_014782) MAF annotator (24). Mutational profiles of the samples were inferred using SigProfilerMatrixGenerator software (25). Samples from the previous work (14) used for comparison were processed and analyzed in the similar way.
To identify temporal order of mutations for samples with available archival FFPE slides, we first identified somatic mutations in fresh-frozen tumors and then searched for reads supporting given alleles in the BAM files of archival FFPE samples using snp-pileup script from the FACETS package (19). This analysis was restricted to only single-nucleotide variants and regions with coverage higher than 30× in FFPE samples.
RNA-seq and data processing
We sequenced five transcriptomes of IR-BCC and compared them with the previously published N-BCC [n = 51; AR-BCC, n = 7; actinic keratosis/squamous cell carcinoma (AK/SCC), n = 19] and normal skin samples (n = 18). Samples of AK and SCC were retrieved from Hoang and colleagues (26).
IR-BCC samples were sequenced on the BGISEQ 500 sequencer (20 million reads). Reads were mapped using STAR (2.7.2b; RRID:SCR_004463; ref. 27) software to the GRCh37 human reference genome and then read count per transcript was done with HTSeq (v0.11.2; RRID:SCR_005514; ref. 28) on GENCODE annotation (v28lift37; ref. 29). Mapping quality controls were done with Samtools (21) and RSeQC (RRID:SCR_005275; ref. 30). All processing steps were combined in a pipeline built with snakemake (v5.4.0; RRID:SCR_003475; ref. 23).
RNA fusion analysis was performed with the nf-core rnafusion pipeline (v1; ref. 31), and fusions of interest were validated with PCR. The NF-CORE rnafusion pipeline runs five software for RNA fusion calling [STAR-Fusion v1.5.0 (32), Fusioncatcher v1.00 (33), Ericscript v0.5.5 (34), Pizzly v0.37.3 (35), and Squid v1.5 (36)] and summarizes results.
We focused our analysis on known oncogenic pathways using information relative to the activity of their core transcription factors (TF), which is inferred from the expression level of their direct targets (37, 38). To assess the activity of oncogenic TFs through expression level of their direct target genes we used Dorothea footprinting framework (37). First, we removed genes with fewer than 50 counts in average across all samples to reduce technical noise, then the read counts were converted into counts per million (CPM) with TMM approach of EdgeR (39) package (RRID:SCR_012802). Afterward, the CPM were fed into Dorothea package, and a matrix with CMP was transformed into the matrix of relative TF activity using Viper (38) approach and only high-quality TF–target pairs (A–D, at least 30 targets per TF).
Validation of the candidate RNA fusions
cDNA synthesis using the superscript III reverse transcriptase (Invitrogen; cat. no. 12574026) was performed according to the manufacturer's instructions. PCR fragments were amplified using gene-specific primers and 400 ng cDNA with Promega PCR master mix (Cat No./ID: A6001) according to the manufacturer's instructions. Initial PCR was programmed as follows: 94°C for 5 minutes, 35 amplification cycles, including a denaturation at 95°C for 30 seconds, annealing at 62°C for 30 seconds, extension at 72°C for 1 minute, and a final extension at 72°C for 10 minutes. Nested PCR using the Initial PCR product (4 μL) was set up as the following program: 95°C for 5 minutes, 20 amplification cycles, including a denaturation at 95°C for 30 seconds, annealing at 62°C for 30 seconds, extension at 72°C for 45 seconds, and a final extension at 72°C for 10 minutes. PCR products were separated on 1.8% agarose gel and analyzed by the Geldoc XR+ system (Bio-Rad). PCR products were sequenced by using the gene-specific primers. Applied primers for PCR were purchased from Invitrogen with the following sequences:
*C11orf70_YAP1_F
CCTCTTCTGTCTTTAAAGTTTCAGC
*C11orf70_YAP1_R
TGGTGGCTGTTTCACTGGAG
*TEAD2_SLC6A16_F
GCGATACATGATGAACAGCGT
*TEAD2_SLC6A16_R
CTGCAAGGGAGGGTTCATCT
*YAP1_CWF19L2_F
CAGATGAACGTCACAGCCCC
*YAP1_CWF19L2_R
TGAATGGACTCACGCTCTGG
Single epitope IHC for YAP1 and BCAT
To assess the level of activation of the HIPPO-YAP and WNT pathways, we assessed the cellular localization of YAP1 and BCAT in IR-BCC, N-BCC, and S-BCC using single epitope enzymatic IHC in FFPE slides and digital image analysis. Single epitope IHC was performed on tumors from 18 patients with a diagnosis of BCC included from three different centers (Saint-Louis Hospital, Paris, France; Cantonal Hospital St. Gallen, Switzerland; and Lausanne University Hospital, Switzerland) as follows: 8 patients with BCCs that responded to the treatment with vismodegib (all patients from Saint-Louis Hospital), 5 patients with vismodegib-resistant BCCs (4 from Saint-Louis Hospital and 1 from Cantonal Hospital St. Gallen), and 5 patients diagnosed with nodular BCC who did not receive vismodegib as a control group (Lausanne University Hospital). All the analyzed slides except SG005 were prepared from FFPE biopsies taken before vismodegib treatment initiation and represent baseline characteristics of the tumors relative to HPI.
Tissue samples taken for diagnostic histologic examination were FFPE in the pathology departments of the respective hospitals using the standard processing protocols. Four-micron-thick serial sections were cut using a Leica HistoCore AUTOCUT rotary microtome (Leica Microsystems). Manual single epitope enzymatic IHC was performed on serial sections from the FFPE blocks in the Laboratory for Experimental Dermatology, Institute for Immunobiology, Cantonal Hospital St. Gallen, to assess YAP1 and BCAT expression within the tumor cells.
Tissue sections were placed on poly-L-lysine–coated slides. The slides were dewaxed in xylene, rehydrated, subjected to HIER using sodium citrate (pH 6) target retrieval solution for YAP1 and Tris-EDTA (pH 9) target retrieval solution for BCAT for 20 minutes in a microwave oven, and allowed to cool to room temperature. Endogenous horseradish peroxidase activity was blocked by a 10-minute incubation at room temperature with 3% hydrogen peroxide solution (catalog number 160-0-029, Laboratorium Dr. G. Bichsel AG, CH), followed by an avidin/biotin blocking step (catalog number SP-2001, Vector Laboratories) for 30 minutes at room temperature, a 30-minute incubation with 3% skim milk at room temperature, and an overnight incubation at 4°C with a rabbit anti-human YAP1 monoclonal primary antibody (catalog number 14074S, Cell Signaling, dilution 1:100, RRID:AB_2650491) and a mouse anti-human BCAT primary antibody (NCL-L-B-CAT, Leica Biosystems, CH, dilution 1:100, RRID:AB_563467). The next day, slides were incubated for 30 minutes at room temperature with an anti-rabbit biotinylated secondary antibody for YAP1 (catalog number D30-1, GBI Labs; dilution 1:200), and an anti-mouse biotinylated secondary antibody for BCAT (catalog number D30-0, GBI Labs; dilution 1:200), followed by another 30-minute incubation at room temperature with HRP-conjugated streptavidin (catalog number D30-1, GBI Labs; dilution 1:200). The signal was visualized using a DAB Substrate Kit High Contrast (catalog number DAB500plus, Zytomed Systems). The slides were dehydrated with alcohol, cleared with xylene, and mounted with Pertex (catalog number 41-4011-00, Medite) after methyl green counterstaining (catalog number H-3402, Vector Laboratories).
Image analysis
Quantitative morphometry was performed on all stained slides using the QuPath v0.2.3 software platform for whole-slide image analysis (RRID:SCR_018257; ref. 40). Whole-slide scans acquired with a Panoramic 250 Flash III digital slide scanner (3D Histech) were individually loaded into the software. For each staining vector (i.e., color), estimates of the background staining were applied to maximize differences in expression in QuPath by selecting a region of interest containing an area of background along with examples of strong nuclear counterstaining and chromogen staining and applying QuPath's “Estimate stain vectors” command to identify stain vectors within this region. The “Positive Cell Detection” command was then used to identify cells across the entire tissue section based upon nuclear identification. The full extent of each cell (area occupied by the cell) was estimated based upon measurements of intensity and morphology, including nuclear area, circularity, staining intensity for counterstaining and chromogen staining, and nucleus/cell area ratio. A two-way boosted decision-tree classifier was then interactively trained to distinguish tumor cells from all other detected artifacts (comprising nontumor epithelial cells, immune cells, stromal cells, necrosis, and any artifacts misidentified as cells). Cells were classified as positive or negative based upon a single-intensity threshold applied to the maximum optical density of the detected chromogen within the nucleus, cytoplasm, or membrane of the cell depending on the expression pattern (the threshold was adjusted for a random region of interest containing both negative and highly positive tumor cells until all the cells defined as positive by the software matched those considered positive by the evaluator). Summary scores were generated as the percentage of cells classified as positive after “other” detections were removed. The quantitative morphometric analysis was performed on the entire tumor tissue present on each section stained. We profiled 1361-134651 cells in each FFPE slide to receive an unbiased estimation of the nuclear or membranous staining.
YAP1 lentiviral transduction
To investigate the role of YAP1 in BCC, we transduced the ASZ001 cell line (RRID:CVCL_6251) with the mouse Yap1 expression lentivirus vector and assessed proliferation rate between ASZ001 and ASZ001 (YAP1+) cells at 5 and 24 hours. We used the mouse Yap1 expression lentivirus vector (mCherry:T2A:Puro) from VectorBuilder (pLV[Exp]-mCherry:T2A:Puro-EFS>mYap1[NM_001171147.1]; vector ID VB900000-0080jmw). Plasmids were isolated by incubating the bacterial stock for up to 24 hours in 50 mL of LB media supplemented with ampicillin (MCE, HY-B0522; 100 μg/mL). The isolation was performed with Machrey-Nagel NucleoBond Xtra Midi kit for transfection-grade plasmid DNA (Machrey-Nagel, ref.: 740410.50) according to the manufacturer's protocol. The virus was prepared using CaCl2 transfection (41). The day before transfection, Hek 293T LentiX (TakaraBio, 632180) were seeded on 5-cm dishes 2.8 mil/5 mL in DMEM. On the day of transfection, 25 μmol/L chloroquine was added to the media. Five μg of plasmid, 3.75 μg of psPax (addgene 12260), 2.5 μg pMD2.G (addgene 12259), and 31.25 μL 2M CaCl2 (Sigma-Aldrich, C4901) were added to 250 μL final volume of ddH2O. The mix was added dropwise into the 250 μL of 2 × HBS precipitation buffer while blowing air through, incubated for 10 minutes, and dropwise distributed on the HEK 293T lenti-X cells. The medium was changed after 6 hours for DMEM supplemented with 10% FCS, penicillin/streptomycin, and HEPES 25 mmol/L. After 72 hours, the medium containing virus was harvested and filtered through a 0.45-μm filter, incubated with PEG-it Virus Precipitation Solution (5×; SBI, # LV810A-1) overnight, and centrifuged (1,500 × g/30 min/4°C). The medium was removed followed by resuspension in 100× OPTIMEM media and stored at −80°C. For the lentiviral infection, 3 mil ASZ001 mouse BCC cells were harvested and placed in 12-well plates in 1 mL of medium, and polybrene was added (16 μg/mL, followed by 10 μL of virus). The cells were spinocculated for 2 hours at 1,000 × g/33°C. The medium was removed and cells were resuspended in fresh media and plated in T75 flasks. Seventy-two hours after seeding, 2 μg/mL puromycin (MCE, HY-B1743A) was applied, and cells were selected for at 1.5 weeks.
Cell culture
Both ASZ001 and ASZ001 (YAP1+) cell lines were cultured in complete BCC medium: 500 mL 154-CF medium (Gibco, catalog number M154CF500), 5 mL 100× penicillin/streptomycin (Corning, ref: 30-002-CI), 10 mL heat-inactivated chelex-treated FBS, and 125 μL CaCl2 0.2 mol/L. Cells were split at a 1:3 ratio once 70% to 80% confluency was reached.
Proliferation assay of the ASZ001 cell lines
For the proliferation assay, 5,000 cells per well from each cell line (ASZ001 cells as well as YAP1-overexpressing ASZ001 cells) were plated in a 96-well plate (biological quadruplicates) and incubated overnight, followed by assessment at 5 and 24 hours. Cell proliferation was assessed using Cell Counting Kit-8 (MedChemExpress; catalog number HY-K0301), which utilizes a highly water-soluble tetrazolium salt. This produces a water-soluble formazan dye (yellow) upon reduction in the presence of an electron mediator, allowing for sensitive colorimetric assays (absorbance at 450 nm) for the determination of the number of viable cells.
Results
Frequency of the IR and AR to vismodegib in laBCC
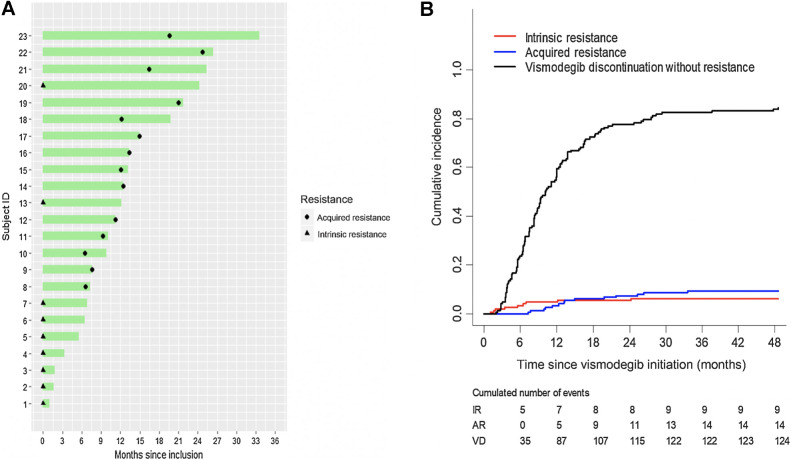
We evaluated the responses to vismodegib of 148 laBCC patients recruited in the STEVIE study between 2011 and 2013 (10). Characteristics of the patients are shown in Supplementary Table S1. The median time to treatment discontinuation was 9.1 months with a minimum of 1 month and a maximum of 47.8 months (Fig. 1A). The reason for discontinuation was IR to vismodegib for 9 patients (6.1%) and AR for 14 patients (9.5%). Clinical criteria of IR and AR are reported in the Methods and Supplementary Table S2.
Figure 1.
Incidence of the IR and AR to vismodegib in the studied cohort of 148 locally advanced BCC. A, Vismodegib treatment duration and diagnostic of resistance. B, Cumulative incidence of vismodegib discontinuation for IR, AR, and vismodegib discontinuation (VD) without resistance.
The remaining patients were in either CR (61%), PR (12%), or SD (11%) when discontinuing vismodegib. The cumulative incidence of IR was 4.8% (7 cases) at 12 months (Fig. 1B). For AR, the cumulative incidence was 3.4% (5 cases) and 7.5% (11 cases) at 12 and 24 months, respectively (Fig. 1). Seven percent of laBCCs from the study had received prior chemotherapy and 16% radiotherapy. Treatment with chemotherapy was associated with higher incidence of IR [SHR = 8.85 (2.25–34.8), P = 0.002], but not with higher incidence of AR [SHR = 1.01 (0.15–7.06), P = 0.99; Supplementary Table S4].
IR-BCC are characterized by high level of genomic instability
In order to characterize the mechanisms associated with the IR in BCC, we performed a comprehensive analysis of five IR-BCC samples that represented relapses of the surgically removed untreated with HPI tumors (Supplementary Table S3).
In line with previous observations, two BCCs (NB021_T01 and SG005_T01) displayed a clear BerEp4 staining, an IHC BCC marker, whereas three other BCCs (NB020_T01, NB022_T01, and NB023_T01) showed patchy stains or absence of Ber-Ep4 (42). Tumors were either negative for SCC (cSCC) IHC marker EMA or showed a patchy positivity (NB023_T01 and SG005_T01; Supplementary Fig. S1).
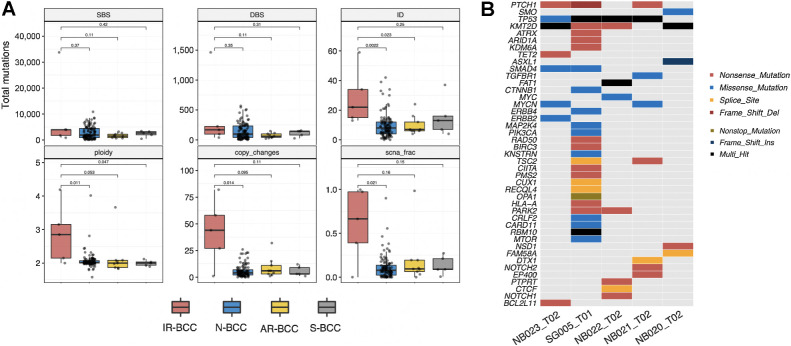
Analysis of the mutational load revealed a significantly increased number of short deletions and insertions in IR-BCC in comparison with N-BCC (2.98, P = 0.002, Mann–Whitney U test, two-sided) and AR-BCC (2.92, P = 0.023, Mann–Whitney U test, two-sided), whereas the number of single-base substitutions (SBS) and double-base substitutions was not different between groups (Fig. 2A). IR-BCCs demonstrated high level of genomic instability, which is represented by numerous SCNAs, increased ploidy (median = 2.85n), and high fraction of genome with copy-number changes (Fig. 2A).
Figure 2.
Genomic analysis of the five IR-BCC cases. A, Total number of different classes of mutations (SBS, single-base substitutions; DBS, double-base substitutions; ID, indels) and copy-number alterations (ploidy, average genomic ploidy; copy changes, number of copy-number alteration events longer than 5 Mb; scna_frac, fraction of autosomal regions which is different from diploid state) compared between IR-BCC and N-BCC, AR-BCC, or S-BCC. Significance of comparisons measured by two-sided Mann–Whitney U test is indicated. B, Oncoplot for oncogenic and likely oncogenic mutations identified in IR-BCC samples according to the OncoKB database.
Among five IR-BCC samples, four had typical BCC oncogenic mutations in the Hh pathway and its target genes (three inactivating PTCH1 mutations, one activating SMO mutation, and two activating MYCN mutations; Fig. 2B). Ultramutated sample SG005_T01 harbored 28 oncogenic mutations including gain-of-function mutations in CTNNB1, PIK3CA, and MTOR genes, which are not frequently mutated in BCC.
We specifically searched for events that can explain resistance to vismodegib in our samples and identified mutation p.W535 L in SMO in the NB020_T02 sample, which is a known cause of AR to vismodegib in BCC (9). The sample NB021_T02 had focal amplification on chromosome 2 with up to 17 copies of GLI2—another described mechanism of AR to HPI in BCC (8) and in medulloblastoma (43).
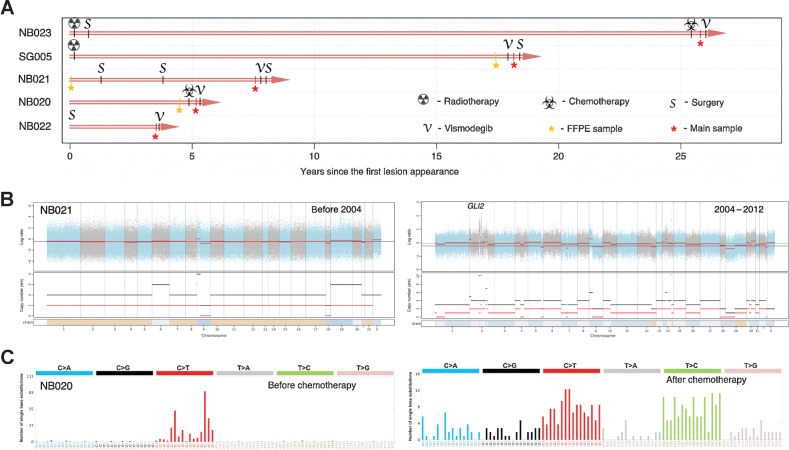
Evolution of the IR-BCC tumors
The studied IR-BCCs were characterized by a long history of their evolution ranging from 4 to 27 years (average, 10.8 years) and prior radiotherapy and/or chemotherapy in three cases (Fig. 3A; Supplementary Table S3). To better understand the evolution of the malignant aggressive phenotype in IR-BCC, we additionally sequenced three exomes of available archival FFPE blocks for patients SG005, NB020, and NB021, which were taken 1 month, 5 months, and 8 years before the biopsy of IR-BCC sample and before treatment with vismodegib.
Figure 3.
IR-BCC tumor evolution and associated genomic changes. A, Time course of the studied IR-BCC samples since the first diagnosis or lesion appearance. B, Copy-number profile of the two tumor samples of patient NB021: from 2004 (initial BCC; left) and 2012 (IR-BCC; right). Log ratio indicates difference in normalized coverage between normal and tumor samples. Copy number depicted for the total number of DNA copies (black) and minor allele (red). C, Trinucleotide-context mutational profiles of the two tumor samples of patient NB020: before (left) and after (right) chemotherapeutic treatment with cisplatin and adriamycin.
Evolution of BCC in patient NB021 was characterized by dramatic changes in the genome copy number and ploidy between samples taken 8 years before the biopsy of IR-BCC. A stable nearly diploid tumor genome was found in 2004 sample, but it became highly rearranged and polyploid 8 years later during tumor progression (Fig. 3B). Genomic instability resulted in a focal amplification of the TF GLI2, a major transducer of the Hh signal, which was associated with resistance to HPI (8, 43). Another cancer-related gene that was focally amplified (15 copies) after chemotherapy is MAP3K2, but its role in BCC is unknown. Sequencing of an archival FFPE sample of patient NB020, which was taken before chemotherapeutic treatment with cisplatin and adriamycin, revealed four of the five oncogenic mutations identified in IR-BCC. Only a frameshift indel in the ASXL1 gene was acquired after chemotherapy. The most interesting observation was a drastic change of the mutational spectrum between the two biopsies: while in the early BCC sample, the mutational spectrum was typical for skin cancer and dominated by UV-induced mutations, the IR-BCC sample had an 85% increase in mutations (198 SNVs per exome) that did not display any signature of UV-induced mutagenesis (Fig. 3C). Sample SG005_T01 had an ultramutated phenotype with 28 mutations annotated as oncogenic or likely oncogenic in the OncoKb database. We had sufficient coverage for 26 of these mutations, and all of them were present in a corresponding archival FFPE sample taken just before vismodegib treatment was initiated.
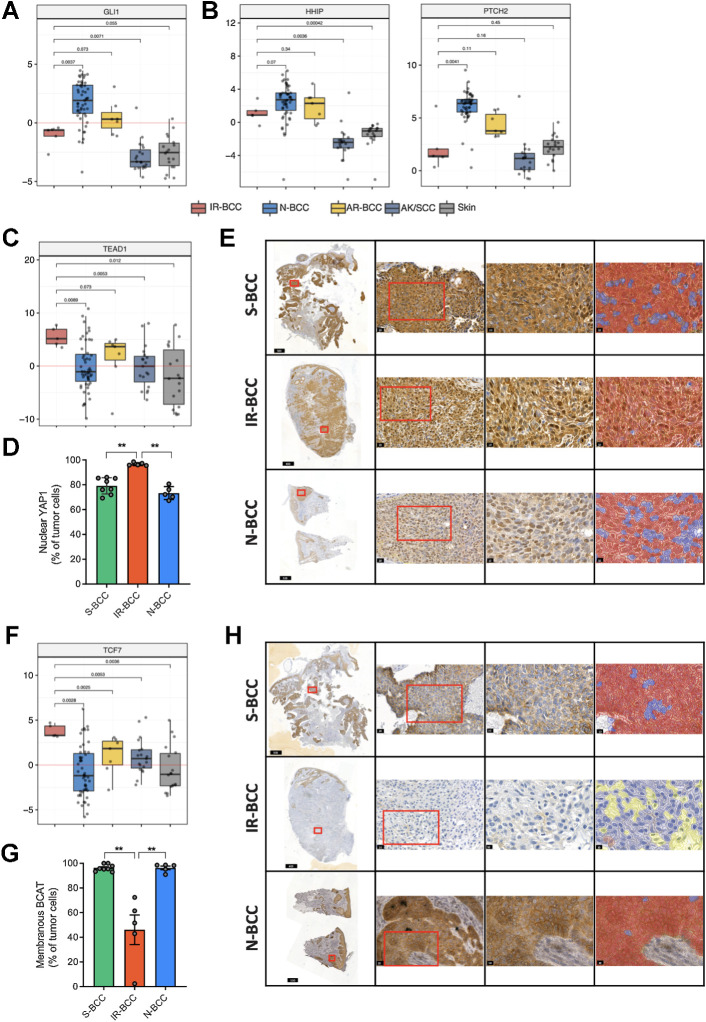
Upregulated activity of HIPPO-YAP and WNT but not Hh pathway in IR-BCC
The activity of the Hh pathway was measured as relative expression of direct target genes of one of its main TFs, GLI1. This analysis revealed activation of the Hh pathway in IR-BCC as compared with skin; however, it was significantly less activated when compared with N-BCC (Fig. 4A). In line with this, we detected significantly lower expression of Hh pathway target genes such as HHIP and PTCH2 in IR-BCC as compared with N-BCC (Fig. 4B).
Figure 4.
Transcriptional activity and immunohistochemical analysis of the Hedgehog, Hippo-YAP, and WNT markers in BCC. A, Relative activity of the central Hedgehog TF GLI1 based on the expression of its transcription targets in different groups of skin tumors and normal skin. B, RNA expression (logTPM values) of the HHIP and PTCH2 genes which are usually activated in BCC. C, Relative activity of the central HIPPO-YAP TF TEAD1 based on the expression of its transcription targets in different groups of skin tumors and normal skin. D, Quantification of nuclear YAP1 protein expression in tumor cells (% of total tumor cells) among the three groups of samples assessed by YAP1 enzymatic IHC. E, Representative micrographs for YAP1 expression in BCC groups at various magnifications: tumor overview on the whole-slide scan (first column), details of the regions marked with a red square in the first column (20× magnification, second column), and details of regions marked with red square in the second column (40× magnification, third column) with overlapping color mask (fourth column) marking nuclear YAP1-positive cells (red mask). Nuclear YAP1-negative tumor cells are marked in blue. The parts of the image are intentionally duplicated in Supplementary Figs. S2–S4 for S-BCC, IR-BCC, and N-BCC samples, respectively, as the same slides represented. F, Relative activity of the central WNT TF TCF7 based on the expression of its transcription targets in different groups of samples. G, Quantification of membranous BCAT expression in the tumor cells (% of total tumor cells) in the three groups of samples assessed by BCAT enzymatic IHC. H, Representative micrographs for BCAT expression in BCC groups at various magnifications: tumor overview on the whole-slide scan (first column), details of the regions marked with a red square in the first column (20× magnification, second column), and details of regions marked with red square in the second column (40× magnification, third column) with overlapping color mask (fourth column) marking membranous BCAT-positive cells (red mask). BCAT-negative tumor cells are marked in blue and nontumor cells are marked in yellow. The parts of the image are intentionally duplicated in Supplementary Figs. S6–S8 for S-BCC, IR-BCC, and N-BCC samples, respectively, as the same slides represented.
We then analyzed the RNA-seq data from the IR-BCC samples for activation of other cancer pathways. A strong increase in the signature of the HIPPO-YAP pathway TF TEAD1 (Fig. 4C) was observed. IHC demonstrated nuclear YAP1 positivity in a significantly higher fraction of IR-BCC tumor cells than in N-BCC (96.08% vs. 72.18%, P = 0.0079, Mann–Whitney U test) samples, and in S-BCC (79.61%, P = 0.0016, Mann–Whitney U test; Fig. 4D and E; Supplementary Figs. S2–S4; red, positive tumor cells; blue, negative tumor cells; yellow, nontumor cells). We did not detect any putative driver mutations in HIPPO-YAP pathway components in our set of IR-BCC samples. One of the mechanisms of HIPPO-YAP pathway activation involves YAP1 gene fusions (44). We searched for gene fusions from RNA-seq and found putative fusions in three samples, which can potentially explain hyperactivation of HIPPO-YAP pathway (TEAD2–SLC6A16 in two samples; YAP1–CWF19L2, C11orf70–YAP1). We validated the presence of these fusions by rtPCR (Supplementary Fig. S5). The fusion TEAD2–SLC6A16 was previously identified and validated in rhabdomyosarcoma (45), and C11orf70–YAP1 was identified in TCGA data set of lung adenocarcinoma (46).
Another oncogenic pathway with statistically significantly elevated activity in IR-BCC was WNT (represented by TCF7 TF; Fig. 4F). Immunostaining with anti-βcatenin (BCAT) revealed significant loss of membranous BCAT protein (average, 51.62%) in IR-BCC as compared with S-BCC (95.62%, P = 0.016, Mann–Whitney U test) and N-BCC (98.26%, P = 0.0079, Mann–Whitney U test; Fig. 4G and H; Supplementary Figs. S6–S8; red, positive tumor cells; blue, negative tumor cells; yellow, nontumor cells). Previously, such staining pattern was associated with increased aggressiveness in the other tumor types (47).
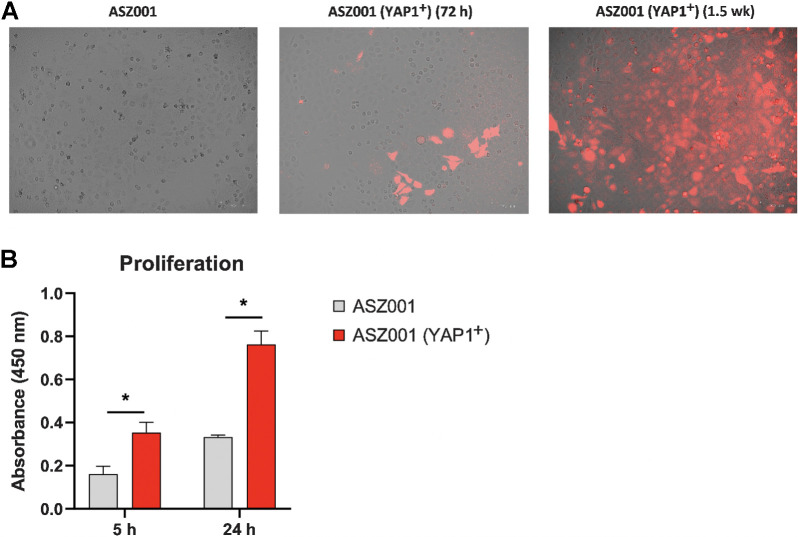
YAP1 overexpression leads to an increased proliferation in the murine BCC cell line
To better understand the role of the HIPPO-YAP pathway in IR-BCC pathogenesis, we transfected murine ASZ001 BCC cell line with a YAP1 overexpression vector. We then compared YAP1-overexpressing cells with untransfected ASZ001 cells used as a control by proliferation rate. Our analysis of the proliferation rate of murine BCC cell line transfected with YAP1 overexpression vector (Fig. 5A) demonstrated that YAP1+ cells had a 2.1-fold significant increase of proliferation rate at 5 hours in comparison with control cells (Fig. 5B; P < 0.006, two-tailed t test with alpha 0.05), and a 2.29-fold increase in proliferation rate at 24 hours (Fig. 5B; P < 0.001, two-tailed t test with alpha 0.05).
Figure 5.
Proliferation for ASZ001 Yap1-overexpressing and ASZ001 cell lines for two time intervals (5 and 24 hours). A, ASZ001 cell line in culture (control; left); YAP1-transfected ASZ001 72 hours after transfection (mCherry positivity indicates successful transfection; middle); YAP1-transfected ASZ001 1.5 weeks after transfection and after puromycin treatment for selection of transfected cells (mCherry positivity indicates successful transfection; right). B, Proliferation assay for ASZ001 and ASZ001 YAP1-overexpressing cell lines after 5 and 24 hours of culturing time.
Discussion
We present here the largest study addressing the frequency of intrinsic and acquired HPI resistance in laBCC and genomic features associated with the IR. Previous reports based on a small number of cases estimated resistance to affect anywhere from 21% to over 50% of patients treated with HPIs (7) and has been cited as a significant problem. We observed a 6.1% of IR, which is in accordance with previous reports showing that the majority of BCC patients experience clinical benefit from treatment with vismodegib (10, 48) and also consistent with the rate of progression in the previous studies (5, 10, 49).
A limitation of our study is that we considered the date of vismodegib discontinuation, which is a medical decision, rather than a prespecified regular assessment of resistance. Thus, some IR cases could be treated for long periods in this study (Fig. 1A).
Our results also indicate a remarkably low frequency of AR (9%) compared with other targeted agents, such as single-agent BRAF inhibitor therapy in melanoma where >80% of patients experience tumor regrowth during treatment (50).
The genomic mechanisms of IR in BCC were not assessed previously. We found that IR-BCC cases are characterized by increased levels of genomic instability and aneuploidy in comparison with N-BCC. In two of five samples, the IR phenotype can be explained by the same mechanism as in AR cases—mutation in SMO and GLI2 amplification. Another ultramutated sample SG005_T01 harbored in total 28 mutations that can be considered as likely oncogenic drivers. Among mutations in SG005_T01, we identified likely gain-of-function missense substitution S629F in PIK3CA (51). It was shown earlier on the Hh medulloblastoma model that mutations in the PI3K pathway affect the tumor phenotype and ability to respond to HPIs (52).
The studied IR-BCCs were characterized by long history of disease and application of radiotherapy (patient SG005), chemotherapy (patient NB020), or both (patient NB023). We revealed dramatic changes in chromosomal copy number for biopsy (patient NB021) taken 8 years before the main sample where a stable genome became highly rearranged at relapse, and acquired a focal amplification of GLI2, which contributed to resistance to vismodegib therapy.
HIPPO-YAP pathway activity has been associated with highly aggressive tumor phenotypes and is mutated in 23% of sporadic BCCs (14). Recent work showed that YAP-TEAD signaling promotes BCC (53). Nuclear YAP1 localization was significantly increased in the studied IR-BCC. In addition, we found gene fusions involving the core HIPPO-YAP genes (YAP1, TEAD2) in three of five samples, which can be a genetic basis for observed activity of the pathway.
The WNT pathway is poorly characterized in the context of BCC, but we identified its aberrant activity in IR-BCC in comparison with N-BCC, AR-BCC, SCC, and skin (Fig. 4F and G). Loss of membranous staining of BCAT is a known biomarker of aggressiveness in cancer and was previously associated with metastatic state in melanoma (47), poor prognosis and metastasis in colorectal cancer (54–56), and aggressive forms of breast cancer (57). Here we have shown that loss of BCAT membranous staining can serve as a predictor of highly aggressive BCC phenotype.
We have demonstrated here that IR and AR to vismodegib are relatively rare events after treatment initiation in laBCC (15%) compared with other targeted therapies in various solid tumors. Our genomic analysis of five IR-BCCs identified that three of them had known mechanisms of resistance to vismodegib previously identified in AR-BCC and medulloblastoma. IR-BCC samples were characterized by increased genomic instability in two cases explained by application of chemo/radiotherapy, high nuclear YAP1 level, and loss of membranous BCAT staining. The mechanisms of IR to vismodegib appeared to be heterogeneous from results of our work, although these overlap with mechanisms of AR. The small number of samples analyzed in this study do not allow to identify all the aspects of resistance to vismodegib, which seem to be complex, and larger cohorts of IR-BCC cases would be needed to fully understand this phenomenon.
Supplementary Material
Acknowledgments
S.I. Nikolaev was supported by grant Foundation ARC 2017, Foundation Gustave Roussy and Swiss Cancer Research Foundation KFC-3985-08-2016. N. Basset-Seguin was supported by a Roche Genentech Grant. L. Flatz was supported by the Swiss National Science Foundation grant PP00P3_157448 and Research Fund of the Kantonsspital St. Gallen (11/19).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
M. Ighilahriz reports grants from Genentech during the conduct of the study. H.J. Sharpe reports personal fees from OMass Therapeutics outside the submitted work; in addition, H.J. Sharpe has a patent for US-2019083646-A1 pending, a patent for EP-3102197-B1 issued, and a patent for US-10330683-B2 issued. M. Battistella reports personal fees from Innate Pharma and Bristol Myers Squibb, as well as grants and personal fees from Takeda and Kyowa Kirin outside the submitted work. S. Mourah reports personal fees from Roche, Pierre Fabre, Biocartis, and Novartis outside the submitted work. F. Bouquet reports other support from F. Hoffmann-La Roche during the conduct of the study, as well as other support from F. Hoffmann-La Roche outside the submitted work. A. Savina reports nonfinancial support from AstraZeneca during the conduct of the study, as well as personal fees from AstraZeneca outside the submitted work. S. Monestier reports personal fees from BMS, MSD, Sanofi, Pierre Fabre Oncologie, and Roche outside the submitted work. L. Mortier reports personal fees from Sun Pharma during the conduct of the study, as well as personal fees from MSD, Novartis, Pierre Fabre, and BMS outside the submitted work. N. Meyer reports grants and personal fees from MSD and BMS, as well as personal fees from Novartis, Sun Pharma, Pierre Fabre, Sanofi, and Merck outside the submitted work. C. Robert reports personal fees from Roche, Novartis, BMS, MSD, Pierre Fabre, and Sanofi outside the submitted work. F. Herms reports personal fees from Sun Pharma and Sanofi outside the submitted work. F.J. de Sauvage reports other support from Genentech and Roche during the conduct of the study. L. Flatz reports grants from Swiss National Science Foundation and Swiss Cancer League during the conduct of the study, as well as grants from Hookipa Pharma and personal fees from Novartis, Bristol Myers Squibb, and Sanofi outside the submitted work. N. Basset-Seguin reports grants from Genentech during the conduct of the study. No disclosures were reported by the other authors.
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.
Authors' Contributions
A.A. Yurchenko: Conceptualization, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. O.T. Pop: Resources, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. M. Ighilahriz: Resources, data curation, formal analysis, investigation, writing–original draft. I. Padioleau: Resources, data curation, software. F. Rajabi: Resources, validation, investigation. H.J. Sharpe: Conceptualization, data curation. N. Poulalhon: Resources, data curation, investigation. B. Dreno: Resources, data curation, investigation. A. Khammari: Resources, data curation, investigation. M. Delord: Resources, data curation, investigation. A. Alberti: Resources, data curation, investigation. N. Soufir: Resources, data curation, investigation. M. Battistella: Resources, data curation, investigation. S. Mourah: Resources, data curation, investigation. F. Bouquet: Resources, data curation, investigation. A. Savina: Resources, data curation, investigation. A. Besse: Resources, data curation, investigation. M. Mendez-Lopez: Resources, data curation, investigation. F. Grange: Resources, data curation, investigation. S. Monestier: Resources, data curation, investigation. L. Mortier: Resources, data curation, investigation. N. Meyer: Resources, data curation, investigation. C. Dutriaux: Resources, data curation, investigation. C. Robert: Resources, data curation, investigation. P. Saiag: Resources, data curation, investigation. F. Herms: Resources, data curation, investigation. J. Lambert: Resources, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft. F.J. de Sauvage: Conceptualization, resources, supervision, writing–original draft, writing–review and editing. N. Dumaz: Conceptualization, resources, data curation, supervision, investigation, writing–original draft, project administration. L. Flatz: Conceptualization, supervision, investigation, writing–original draft, project administration. N. Basset-Seguin: Conceptualization, resources, data curation, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing. S.I. Nikolaev: Conceptualization, resources, data curation, supervision, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing.
References
- 1. Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat Rev Cancer 2008;8:743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peris K, Licitra L, Ascierto PA, Corvò R, Simonacci M, Picciotto F, et al. Identifying locally advanced basal cell carcinoma eligible for treatment with vismodegib: an expert panel consensus. Futur Oncol 2015;11:703–12. [DOI] [PubMed] [Google Scholar]
- 3. Amici JM, Battistella M, Beylot-Barry M, Chatellier A, Dalac-Ra S, Dreno B, et al. Defining and recognising locally advanced basal cell carcinoma. Eur J Dermatology 2015;25:586–94. [DOI] [PubMed] [Google Scholar]
- 4. Lear JT, Corner C, Dziewulski P, Fife K, Ross GL, Varma S, et al. Challenges and new horizons in the management of advanced basal cell carcinoma: a UK perspective. Br J Cancer 2014;111:1476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med 2012;366:2171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sekulic A, Von Hoff D. Hedgehog pathway inhibition. Cell 2016;164:831. [DOI] [PubMed] [Google Scholar]
- 7. Chang ALS, Oro AE. Initial assessment of tumor regrowth after vismodegib in advanced basal cell carcinoma. Arch Dermatol 2012;148:1324–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sharpe HJ, Pau G, Dijkgraaf GJ, Basset-Seguin N, Modrusan Z, Januario T, et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell 2015;27:327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Atwood SX, Sarin KY, Whitson RJ, Li JR, Kim G, Rezaee M, et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell 2015;27:342–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Basset-Seguin N, Hauschild A, Grob J-J, Kunstfeld R, Dréno B, Mortier L, et al. Vismodegib in patients with advanced basal cell carcinoma (STEVIE): A pre-planned interim analysis of an international, open-label trial. Lancet Oncol 2015;16:729–36. [DOI] [PubMed] [Google Scholar]
- 11. Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc 1999;94:496–509. [Google Scholar]
- 12. Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat 1988;19:1141–54. [Google Scholar]
- 13. R Core Development Team. R: A language and environment for statistical computing. Vienna, Austria 2019.
- 14. Bonilla X, Parmentier L, King B, Bezrukov F, Kaya G, Zoete V, et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet 2016;48:398–406. [DOI] [PubMed] [Google Scholar]
- 15. Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, et al. Oncotator: cancer variant annotation tool. Hum Mutat 2015;36:E2423–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andrews, S. FastQC: A quality control tool for high throughput sequence data. 2010. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 21. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016;32:3047–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Köster J, Rahmann S. Snakemake: a scalable bioinformatics workflow engine. Bioinformatics 2012;28:2520–2. [DOI] [PubMed] [Google Scholar]
- 24. Chakravarty D, Gao J, Phillips S, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017;2017:PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bergstrom EN, Huang MN, Mahto U, Barnes M, Stratton MR, Rozen SG, et al. SigProfilerMatrixGenerator: a tool for visualizing and exploring patterns of small mutational events. BMC Genomics 2019;20:685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoang VLT, Tom LN, Quek X-C, Tan J-M, Payne EJ, Lin LL, et al. RNA-seq reveals more consistent reference genes for gene expression studies in human non-melanoma skin cancers. PeerJ 2017;5:e3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anders S, Pyl PT, Huber W. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 2019;47:D766–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 2012;28:2184–5. [DOI] [PubMed] [Google Scholar]
- 31. Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 2020;38:276–8. [DOI] [PubMed] [Google Scholar]
- 32. Haas BJ, Dobin A, Stransky N, Li B, Yang X, Tickle T, et al. STAR-fusion: Fast and accurate fusion transcript detection from RNA-Seq. bioRxiv 2017. doi: 10.1101/120295. [DOI]
- 33. Nicorici D, Satalan M, Edgren H, Kangaspeska S, Murumägi A, Kallioniemi O, et al. FusionCatcher - a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv 2014. doi: 10.1101/011650. [DOI]
- 34. Benelli M, Pescucci C, Marseglia G, Severgnini M, Torricelli F, Magi A. Discovering chimeric transcripts in paired-end RNA-seq data by using EricScript. Bioinformatics 2012;28:3232–9. [DOI] [PubMed] [Google Scholar]
- 35. Melsted P, Hateley S, Joseph IC, Pimentel H, Bray N, Pachter L. Fusion detection and quantification by pseudoalignment. bioRxiv 2017. doi: 10.1101/166322 [DOI]
- 36. Ma C, Shao M, Kingsford C. SQUID: transcriptomic structural variation detection from RNA-seq. Genome Biol 2018;19:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Garcia-Alonso L, Holland CH, Ibrahim MM, Turei D, Saez-Rodriguez J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res 2019;29:1363–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alvarez MJ, Shen Y, Giorgi FM, Lachmann A, Ding BB, Ye BH, et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet 2016;48:838–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weber K, Thomaschewski M, Benten D, Fehse B. RGB marking with lentiviral vectors for multicolor clonal cell tracking. Nat Protoc 2012;7:839–49. [DOI] [PubMed] [Google Scholar]
- 42. Linskey KR, Gimbel DC, Zukerberg LR, Duncan LM, Sadow PM, Nazarian RM. BerEp4, cytokeratin 14, and cytokeratin 17 immunohistochemical staining aid in differentiation of basaloid squamous cell carcinoma from basal cell carcinoma with squamous metaplasia. Arch Pathol Lab Med 2013;137:1591–8. [DOI] [PubMed] [Google Scholar]
- 43. Dijkgraaf GJP, Alicke B, Weinmann L, Januario T, West K, Modrusan Z, et al. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res 2011;71:435–44. [DOI] [PubMed] [Google Scholar]
- 44. Szulzewsky F, Arora S, Hoellerbauer P, King C, Nathan E, Chan M, et al. Comparison of tumor-associated YAP1 fusions identifies a recurrent set of functions critical for oncogenesis. Genes Dev 2020;34:1051–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xie Z, Babiceanu M, Kumar S, Jia Y, Qin F, Barr FG, et al. Fusion transcriptome profiling provides insights into alveolar rhabdomyosarcoma. Proc Natl Acad Sci U S A 2016;113:13126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gao Q, Liang W-W, Foltz SM, Mutharasu G, Jayasinghe RG, Cao S, et al. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep 2018;23:227–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Demunter A, Libbrecht L, Degreef H, De Wolf-Peeters C, Van den Oord JJ. Loss of membranous expression β-catenin is associated with tumor progression in cutaneous melanoma and rarely caused by exon 3 mutations. Mod Pathol 2002;15:454–61. [DOI] [PubMed] [Google Scholar]
- 48. Dreno B, Basset-Seguin N, Caro I, Yue H, Schadendorf D. Clinical benefit assessment of vismodegib therapy in patients with advanced basal cell carcinoma. Oncologist 2014;19:790–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sekulic A, Migden MR, Basset-Seguin N, Garbe C, Gesierich A, Lao CD, et al. Long-term safety and efficacy of vismodegib in patients with advanced basal cell carcinoma: final update of the pivotal ERIVANCE BCC study. BMC Cancer 2017;17:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010;468:973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ng PK-S, Li J, Jeong KJ, Shao S, Chen H, Tsang YH, et al. Systematic functional annotation of somatic mutations in cancer. Cancer Cell 2018;33:450–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Metcalfe C, Alicke B, Crow A, Lamoureux M, Dijkgraaf GJP, Peale F, et al. PTEN loss mitigates the response of medulloblastoma to hedgehog pathway inhibition. Cancer Res 2013;73:7034–42. [DOI] [PubMed] [Google Scholar]
- 53. Maglic D, Schlegelmilch K, Dost AF, Panero R, Dill MT, Calogero RA, et al. YAP-TEAD signaling promotes basal cell carcinoma development via a c-JUN/AP1 axis. EMBO J 2018;37:e98642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bruun J, Kolberg M, Nesland JM, Svindland A, Nesbakken A, Lothe RA. Prognostic significance of β-catenin, E-cadherin, and SOX9 in colorectal cancer: results from a large population-representative series. Front Oncol 2014;4:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang S, Wang Z, Shan J, Yu X, Li L, Lei R, et al. Nuclear expression and/or reduced membranous expression of β-catenin correlate with poor prognosis in colorectal carcinoma: A meta-analysis. Med 2016;95:e5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li T, Guo H, Song Y, Zhao X, Shi Y, Lu Y, et al. Loss of vinculin and membrane-bound β-catenin promotes metastasis and predicts poor prognosis in colorectal cancer. Mol Cancer 2014;13:263. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57. Geyer FC, Lacroix-Triki M, Savage K, Arnedos M, Lambros MB, MacKay A, et al. Β-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol 2011;24:209–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.