Abstract
Purpose:
Patients with rare cancers (incidence less than 6 cases per 100,000 persons per year) commonly have less treatment opportunities and are understudied at the level of genomic targets. We hypothesized that patients with rare cancer benefit from approved anticancer drugs outside their label similar to common cancers.
Experimental Design:
In the Drug Rediscovery Protocol (DRUP), patients with therapy-refractory metastatic cancers harboring an actionable molecular profile are matched to FDA/European Medicines Agency–approved targeted therapy or immunotherapy. Patients are enrolled in parallel cohorts based on the histologic tumor type, molecular profile and study drug. Primary endpoint is clinical benefit (complete response, partial response, stable disease ≥ 16 weeks).
Results:
Of 1,145 submitted cases, 500 patients, including 164 patients with rare cancers, started one of the 25 available drugs and were evaluable for treatment outcome. The overall clinical benefit rate was 33% in both the rare cancer and nonrare cancer subgroup. Inactivating alterations of CDKN2A and activating BRAF aberrations were overrepresented in patients with rare cancer compared with nonrare cancers, resulting in more matches to CDK4/6 inhibitors (14% vs. 4%; P ≤ 0.001) or BRAF inhibitors (9% vs. 1%; P ≤ 0.001). Patients with rare cancer treated with small-molecule inhibitors targeting BRAF experienced higher rates of clinical benefit (75%) than the nonrare cancer subgroup.
Conclusions:
Comprehensive molecular testing in patients with rare cancers may identify treatment opportunities and clinical benefit similar to patients with common cancers. Our findings highlight the importance of access to broad molecular diagnostics to ensure equal treatment opportunities for all patients with cancer.
Translational Relevance.
Patients with rare cancers (incidence less than 6 cases per 100,000 persons per year) account for as much as 24% of all cancer diagnoses. However, this significant subgroup of patients with cancer have significantly less access to new treatment opportunities and are understudied at the level of genomic targets compared with patients with common cancers. In the Drug Rediscovery Protocol (DRUP), patients with therapy-refractory metastatic cancers, harboring an actionable molecular profile, are matched to an available FDA/European Medicines Agency–approved targeted therapy or immunotherapy. Our study shows that there is a significant overlap in genomic targets between common and rare cancers. Also, progression-free survival and overall survival for common cancers and rare cancers treated in DRUP were similar. Furthermore, exceptional responders have been observed in the rare cancer group highlighting the importance that rare cancers deserve similar access to diagnostics and novel treatment approaches to avoid inequality in care.
Introduction
Patients with rare cancers, commonly defined as cancers with an incidence rate of less than 6 cases per 100,000 persons per year (1), account for as much as 24% of all cancer diagnoses and thus represent a significant group of patients with specific diagnostic and treatment challenges (2). These patients have significantly less access to new treatment opportunities as patients with nonrare cancers. This is partly due to the limited number of tumor-specific clinical trials for patients with a defined rare cancer that contributes to a lack of comprehensive molecular profiling of these tumors. In addition, these studies have historically shown low accrual rates. As a consequence, the drug label generally does not include rare cancer types. This is a vicious circle that creates a serious and increasing problem for patients with rare cancer resulting in generally a significantly lower relative survival (2).
The European Society for Medical Oncology (ESMO) has been addressing the issue of rare cancers since the launch of “Rare Cancers Europe” in 2008 (3). Currently, the definition of rare cancers is solely based on histologic diagnoses, whereas precision oncology aims to identify patients with specific molecular targets. For example, NTRK fusions, microsatellite instability (MSI), and BRCA mutations are identified in 0.31%, 1.9%, and 4.9%, respectively, of all tumors (4–6). Whereas the definition of rare cancers based on histology is very valuable to make sure patients with “rare cancer” get similar treatment opportunities as patients with common cancer, in a broader context, testing for rare molecular targets will benefit all patients (7). This will further emphasize the importance of comprehensive molecular profiling of all patients. It is also underscored by the observation that even in common cancers, specific but rare molecular targets are not always part of the diagnostic workup resulting in a significant inequality of care (8–10). It is, therefore, inevitable that cancer care needs an infrastructure that provides access to adequate diagnostic tests to create treatment opportunities, not only for patients with rare cancers, but for all patients with tumors harboring rare molecular profiles.
We recently showed that the use of whole-genome sequencing (WGS; standard of care) in diagnostics leads to the identification of actionable genomic alterations in more than 60% of patients with metastatic cancer (11). In as much as 13% of these patients, commercially available FDA/European Medicines Agency (EMA)–approved drugs could be used to target these actionable alterations outside their label (10), which has been referred to as off-label use. We, therefore, initiated the Drug Rediscovery Protocol (DRUP) to facilitate the use of approved drugs outside their label. DRUP is an ongoing and open-end, multidrug, pan-cancer precision oncology trial designed to facilitate access for patients with treatment-refractory advanced or metastatic cancer to FDA/EMA-approved targeted therapies and immunotherapies based on the molecular profile of the tumor. The main objective of the DRUP study is to identify signals of efficacy in small cohorts of patients, defined by a molecular target, a tumor type, and a specific drug or combination of drugs (12). This approach using existing anticancer drugs beyond the approved indications has also successfully been tested in other precision oncology trials, such as the MyPathway study and I-PREDICT study (13, 14).
Expanding the use of approved drugs has already shown to be successful (15, 16). The best known example is the development of imatinib for the treatment of Philadelphia-positive chronic myeloid leukemia (CML) and c-KIT–positive gastrointestinal stromal tumor (GIST) that has changed our perspective on molecular-guided therapy in oncology (17). To gain insight in the distribution of actionable genomic targets in patients with rare cancers and to evaluate the outcome of treatment with targeted off-label use of approved anticancer drugs, we compared this with the same assessments in patients with nonrare cancers in the first 500 patients treated in DRUP.
Here we show that the DRUP fulfils an unmet need for patients with rare cancers and that these patients have similar treatment outcomes when treated with targeted drugs as patients with nonrare cancer. We also show that there is a significant overlap in drug targets present in rare cancers and nonrare cancers. Our study emphasizes that patients with rare cancers deserve similar access to diagnostics and novel treatment approaches to avoid inequality in care for this vulnerable patient group.
Materials and Methods
Study population
Patients submitted to DRUP (NCT02925234) had to have progressive advanced or metastatic cancer and exhausted all standard-of-care treatments. Patients were eligible for treatment if they had a somatic actionable target on either the primary tumor or metastasis for which an approved drug was available and if there were no contraindications for the proposed targeted treatment (12, 18). Actionable targets were identified by molecular profiling as part of regular diagnostics in the participating hospitals, or as part of a clinical trial such as the CPCT-02 trial (NCT01855477) in which WGS was performed on biopsies taken prior to regular systemic treatments. All patients underwent a mandatory pretreatment biopsy on which WGS was performed, which was used, among other things, for validation of the actionable target.
Study design
This trial was not randomized and investigators were not blinded to treatment allocation. The study was approved by the Medical Ethical Committee of the Netherlands Cancer Institute in Amsterdam, the Netherlands and was conducted in accordance with Good Clinical Practice guidelines and the Declaration of Helsinki's ethical principles for medical research. All patients provided written informed consent upon enrollment.
Drug selection was performed by the central study team, where needed, after consultation of the DRUP central molecular tumor board. If more than one potentially actionable variant was present in the tumor tissue or if there was more than one drug available in the trial that could target the genomic variant, the agent with the highest level of evidence was selected, as described previously (12, 18). On the basis of molecular target, patients were matched to one of the study drugs and enrolled in parallel cohorts, defined by tumor type, molecular profile, and study drug. Study drugs could not be combined with anticancer drugs outside of the study. The primary endpoint of the trial is clinical benefit, which is defined as a confirmed objective response or stable disease for more than 16 weeks. This is analyzed per cohort using Simon two-stage design (19). Secondary endpoints include progression-free and overall survival and duration of treatment. Cohorts considered to be successful after completion in stage 2 can, depending on discussions with payers and pharmaceutical companies, continue enrollment of patients in an extension stage (stage 3). This model allows for extended data collection aiming to increase the level of evidence for a certain drug – tumor type – molecular profile indication and provides prolonged access to these drugs, after completion of stage 2 (20). Safety and accrual data were regularly reviewed by the Independent Data Monitoring Committee, who could subsequently provide advice on the conduct of the trial.
For all analyses that were performed in this study, comparisons have been made between patients with rare cancer types (defined as tumors with an incidence of 6 per 100,000 cases) and patients with nonrare cancer types treated within the trial.
Statistical analysis
All statistical analyses were performed using R version 4.0.2 (http://www.R-project.org/). Patient characteristics, adverse events (AE), and tumor responses were summarized using descriptive statistics and comparisons have been made between patients with rare cancer types and nonrare cancer types. Baseline characteristics (including number of lines of previous treatment) were compared using the Wilcoxon rank-sum test for continuous variables, Fisher test for categorical variables, and a trend test for ordinal variables (WHO performance status). In addition, Fisher test was used to compare the percentages of patients for whom a matching target was found in the groups with rare and nonrare cancers. The number of patients with AEs was calculated counting the highest grade according to the CTC criteria (version 4.03), as a percentage of the total number of patients who received at least one dose of study medication.
Data availability
The data described in this study are available for academic use upon request. WGS in combination with clinical data can be obtained through the Netherlands Cancer Institute and the Hartwig Medical Foundation through standardized procedures. Request forms can be found at https://www.hartwigmedicalfoundation.nl/en. An independent data-access board will evaluate whether the intended use of the data is compatible with the consent given by the patients, and whether there would be any applicable legal or ethical constraints. Clinical data, treatment outcome, and safety data can be obtained at a per-patient level by emailing the Institutional Review Board of the Netherlands Cancer Institute (IRB@nki.nl).
Results
From September 1, 2016 until November 15, 2019, 500 patients (of 1,145 submitted and reviewed cases; 44%) fulfilled the inclusion criteria for treatment and were enrolled in DRUP. Data presented reflect follow-up until July 12, 2021.
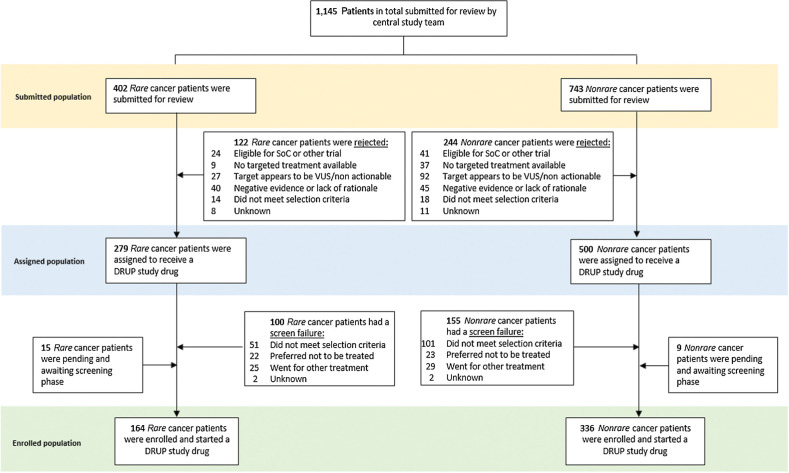
In total, 645 reviewed cases (56%) did not start treatment for several reasons, with harboring a nonactionable target (n = 119) or not meeting the inclusion criteria (n = 184) as the most frequent. The reasons for nonenrollment were comparable between patients with a rare cancer and a nonrare cancer type (Fig. 1).
Figure 1.
Flowchart of submitted and enrolled cases. SoC, standard-of-care treatments; VUS, variant of unknown significance.
In all enrolled patients, treatment with one of 25 available study drugs was initiated. Out of these, 164 patients (33%) were diagnosed with a rare cancer type according to the ESMO definition (1). The remainder were patients with common cancers.
Enrolled patients were treated in 33 centers throughout the Netherlands, including academic, teaching, and community hospitals. Patients with rare cancer types were primarily treated in the Netherlands Cancer Institute or in academic hospitals (81% of the patients), whereas patients with nonrare cancer were more evenly distributed across all participating hospitals. The median follow-up duration was 21.2 months for all patients.
Baseline characteristics
Baseline characteristics of all patients treated are depicted in Table 1, in which the subgroup of patients with rare cancer is outlined separately. The most frequent nonrare cancers were non–small cell lung cancer (n = 90) and colorectal cancer (n = 73). In the rare cancer subgroup, tumors originating primarily from the bile duct (n = 20) and central nervous system (n = 23) were most frequently included (Supplementary Table S1). Also, patients with ultrarare solid tumors (with an incidence of lower than 0.2/100,000/year; ref. 21) were enrolled, such as metastasized hidradenocarcinoma (n = 2), neuroendocrine carcinomas (n = 4), and salivary duct carcinomas (n = 10). Importantly, the median number of previous therapy lines was significantly lower in the subgroup of rare cancers compared with nonrare cancers [2 (IQR 1–3) vs. 3 (IQR 2–5); P < 0.001].
Table 1.
Baseline characteristics.
| Nonrare (N = 336) | Rare (N = 164) | Total (N = 500) | P | |
|---|---|---|---|---|
| Age at consent (IQR) | 64 (55–71) | 61 (49–68) | 63 (53–70) | <0.001a |
| Gender (%) | ||||
| Male | 166 (49%) | 103 (63%) | 269 (54%) | 0.006b |
| Female | 170 (51%) | 61 (37%) | 231 (46%) | |
| WHO PS (%) | ||||
| WHO 0 | 90 (30%) | 51 (35%) | 143 (32%) | 0.229c |
| WHO 1 | 186 (61%) | 89 (60%) | 275 (61%) | |
| WHO 2 | 25 (8%) | 8 (5%) | 33 (7%) | |
| Previous systemic therapy lines (IQR) | 3 (2–5) | 2 (1–3) | 3 (2–5) | <0.001a |
| Previous chemotherapy lines (IQR) | 2 (1–3) | 1 (0–3) | 2 (1–3) | <0.001a |
| Previous hormonal therapy lines (IQR) | 0 (0–1) | 0 (0–0) | 0 (0–0) | NA |
| Previous immunotherapy lines (IQR) | 0 (0–1) | 0 (0–0) | 0 (0–1) | NA |
| Previous targeted therapy lines (IQR) | 0 (0–0) | 0 (0–0) | 0 (0–0) | NA |
Note: Baseline characteristics of enrolled patients in the DRUP trial: rare versus nonrare cancer types.
Abbreviations: IQR, interquartile range; NA, nonapplicable; WHO PS, World Health Organization Performance Status.
aKruskal–Wallis ranks sum test.
bFisher exact test for count data.
cTrend test for ordinal variable.
Genomic targets and profiling techniques used for target identification and opened cohorts
In 52% of the enrolled patients, actionable targets matched to a study drug were identified by WGS. In the remaining cases (48%), targets were revealed by other molecular profiling techniques (e.g., smaller gene panels, IHC, FISH). The percentage of patients with genomic targets identified by WGS was comparable for rare and nonrare cancers (n = 82; 50% and n = 179; 53%, respectively, P = 0.40).
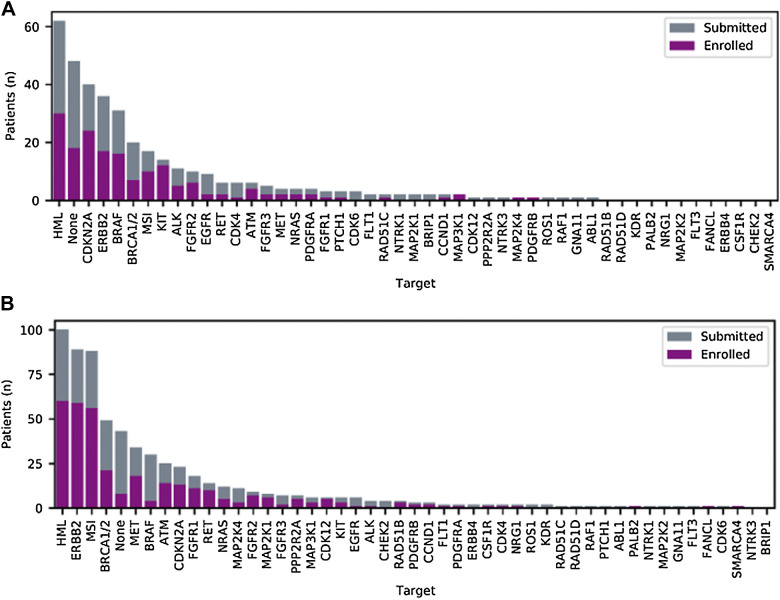
For both patients with rare cancer and nonrare cancer, high mutational load (HML) and alterations of ERBB2 (rare cancers n = 17; 10%, nonrare cancers n = 59; 18%) were the most frequently reported targets that could be matched to study treatment (Fig. 2). When breaking down the ERBB2 alterations, relatively more amplifications than mutations were observed in the rare cancer group (amplification n = 15; 88%, mutation n = 2; 12%), whereas this distribution was more evenly distributed in the nonrare cancer group (amplification n = 27; 46%, mutation n = 32; 54%). However, inactivating mutations or homozygous deletions of CDKN2A and activating BRAF mutations were observed more often in rare cancer cases compared with nonrare cancer cases (Fig. 2A), resulting in more matches to CDK4/6 inhibitors (rare cancers n = 24; 15%, nonrare cancers n = 13; 4%, P ≤ 0.001) or BRAF inhibitors (rare cancers n = 16; 10%, nonrare cancers n = 4; 1%, P ≤ 0.001) in the rare cancer subgroup We observed that patients with rare cancers were more frequently matched to anti-EGFR treatment based on the RAF/RAS wild-type status of the tumor (referred to as “none” in Fig. 2).
Figure 2.
Molecular targets used for submission/enrollment. Representation of all genes harboring somatic alterations that were reviewed by the study team and enrolled in the trial of rare cancer (A) and nonrare cancer (B) patients. For enrolled patients, the target depicted was matched for treatment. None indicates that only the RAF/RAS wild-type status could confer anti-EGFR treatment (panitumumab) or that there was no actionable target present.
The opposite holds true for the nonrare cancer subgroup where MSI was more frequently identified (n = 56; 17%) as target for immunotherapy compared with the rare cancer group (n = 10; 6%; Fig. 2B). In November 2019, DRUP had 134 parallel cohorts open for enrollment of patients, of which the majority (89 cohorts; 66%) for rare cancers (Supplementary Table S1).
AEs
Of all patients with rare cancer, 18% (n = 30) experienced a grade ≥3 AE related to the treatment; none of these were fatal. In the nonrare cancer group, 24% (n = 80) of the patients had a grade 3 or higher AE that could be linked to the treatment, of which 3 were fatal (Supplementary Table S2). The difference between the rare and nonrare cancers was nonsignificant (P = 0.11). In the nonrare cancer group, the most common reported related AEs were hypertension (n = 10) and abnormal liver biochemical tests (n = 16). For the rare cancer group, there was no clear predominance of specific AEs, related to the DRUP treatment.
Clinical benefit
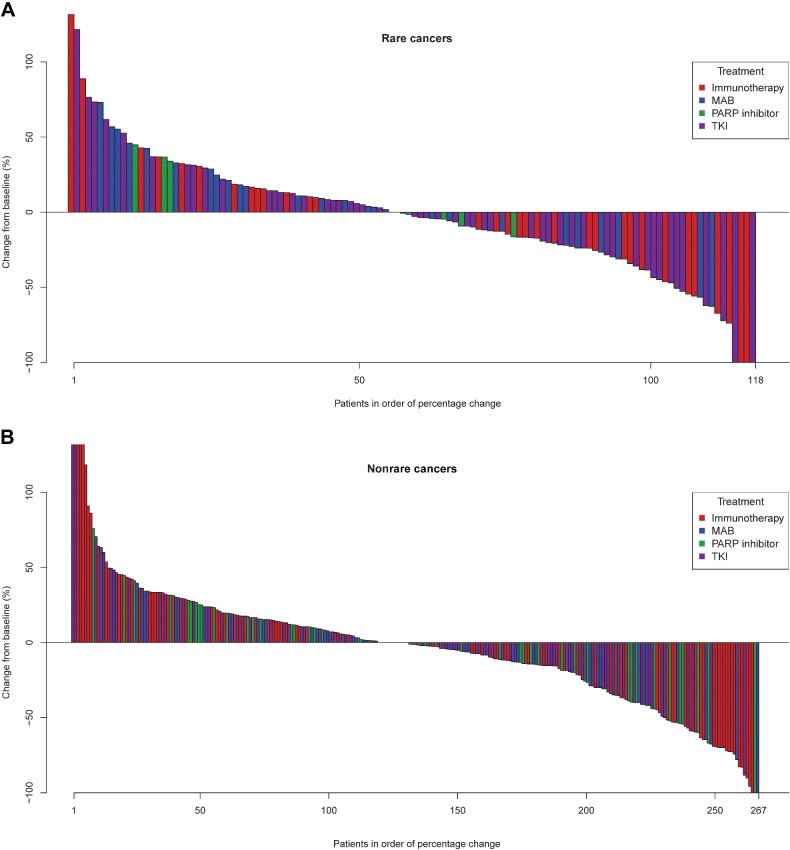
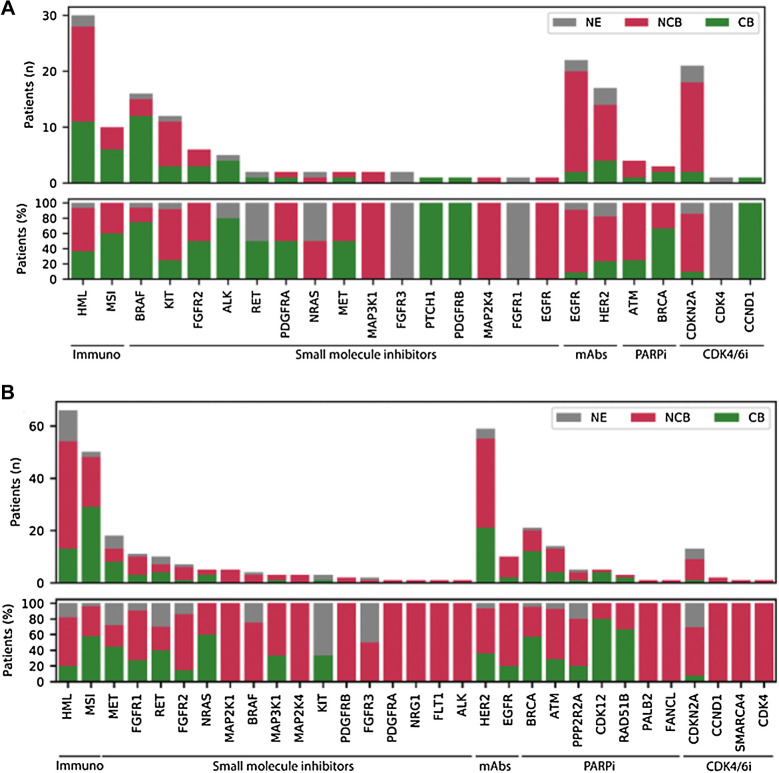
The first 500 patients treated in DRUP had an overall clinical benefit rate of 33%. This was observed across all treatment classes (Fig. 3) and similar between the rare cancer and the nonrare cancer subgroups (33% in both groups). When studying in more detail at the different treatment types targeting specific pathways for the rare cancer group, we observed that patients treated with small-molecule inhibitors targeting BRAF, experienced higher rates of clinical benefit (75%) compared with the nonrare cancer subgroup (0%) expressing the same mutated BRAF oncoprotein (Fig. 4A). These patients (n = 16) were enrolled in five parallel cohorts with various rare cancers and were treated with BRAF inhibitors alone or in combination with MEK inhibitors. Other cohorts, in which patients with rare cancer were treated with small-molecule inhibitors targeting ALK, FGFR2, RET, and PDGFRA, were also successful across different tumor types, although the total number of treated patients in these cohorts were small. Notable successful cohorts in the nonrare cancer subgroup were, for example, the NRAS-mutated tumors treated with MEK inhibitors or cohorts with PARP inhibitors for tumors with somatic inactivating alterations in homologous recombination genes (e.g., BRCA, CDK12, and RAD51B). It should be noted that the numbers of patients in these cohorts were small. In both patients with rare cancer and nonrare cancer, very limited benefit was seen in patients with tumors harboring inactivating alterations in CDKN2A treated with single-agent CDK4/6 inhibitors. (Fig. 4A and B).
Figure 3.
Waterfall plot. Waterfall plots depicting best RECIST 1.1 response of rare cancer subgroup (A) and nonrare cancer subgroup (B). Colors denote the different treatment types, as defined in the legend.
Figure 4.
Clinical benefit. Boxplot showing clinical benefit (CB; yes or no) per drug type and involved pathway in absolute numbers (n; upper figure) and percentage (%; bottom figure) of rare cancers (A) and nonrare cancers (B). i, inhibitor; NE, not evaluable.
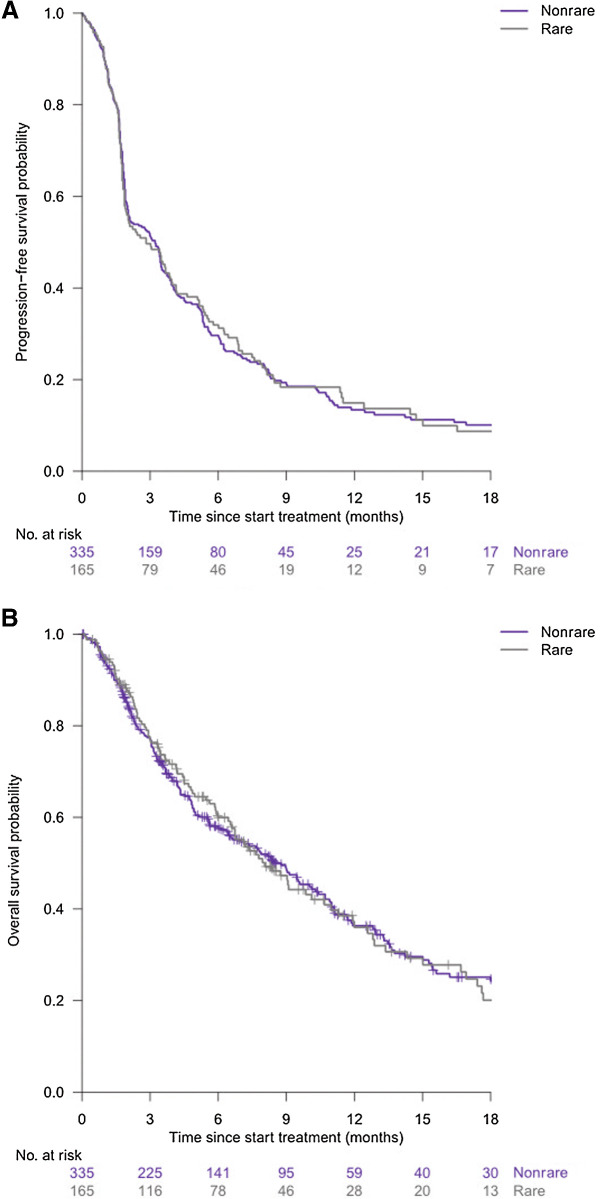
Progression-free survival and overall survival did not significantly differ between patients with a rare cancer type or a nonrare cancer type (Fig. 5A and B).
Figure 5.
Progression-free survival (A) and overall survival curves (B) (rare versus nonrare cancers).
To illustrate the potential value of our approach for individual patients, several patients with rare cancers had prolonged treatment benefit that could serve as a basis for further studies. For example, a 64-year-old female with BRAF p.V600E mutated poorly differentiated large cell carcinoma who was treated with BRAF/MEK inhibition. After only four cycles of treatment, no measureable lesions were detectable. This complete remission is still ongoing, > 2 years after treatment initiation (Supplementary Fig. S1). Other exceptional responders included a BRAF-mutated salivary duct carcinoma (treated with BRAF/MEK inhibition) experiencing an ongoing complete remission for 96 weeks and a patient with atypical fibroxanthoma with HML > 450 in a microsatellite-stable tumor with an ongoing partial remission for > 2.5 years.
Discussion
In this study, we show that patients with rare cancers have a similar benefit from off-label targeted agents as patients with nonrare cancers. This is illustrated by the analysis of the first 500 patients included in DRUP, in which rare cancers were relatively overrepresented (33% in DRUP compared with 24% of all patients with cancer as described in literature; ref. 2). The clinical benefit experienced by all treated patients is in line with the initial analysis of the first 215 patients treated in DRUP, and comparable to other precision oncology studies (12, 22). The high percentage of patients with rare cancer in DRUP is possibly explained by the fact that these patients often do not have many standard-of-care treatment options and are generally excluded from other clinical trials, whereas DRUP provided access to off-label treatment that was otherwise not available. In the past, only very few clinical trials have reported on outcomes of patients withrare cancer treated with genomics-based therapies. Most precision oncology trials enroll patients with rare cancer types, but have not yet shared focused outcomes of this specific subgroup or have analyzed their outcomes in a tumor agnostic manner (13, 14, 23). A recent publication by the German Cancer Consortium paved the way by showing that whole-genome/exome and RNA sequencing enables molecularly informed treatments that lead to clinical benefit when treated with molecularly matched agents (24).
The observation that patients with rare cancers received generally less standard-of-care treatment lines reflects limited treatment opportunities and may have resulted in more sensitivity to targeted agents, as routinely untreated cancers seem less heterogenous in their genomic drivers (25). Furthermore, in rare cancers the natural course of the disease can be variable as it is less well understood as for common cancers.
We showed that the clinical benefit of genomically matched targeted therapies in patients with rare cancer was comparable with patients with nonrare cancer, as well as similar toxicity profiles. Anecdotally, patients with rare cancer treated with SMI even showed long lasting benefit. This underpins the need for comprehensive genomic testing in these patients to guarantee they have the same treatment opportunities as patients with nonrare cancers (22, 26). In addition, this also shows the importance of systematic, high-quality data collection in this patient group to be able to learn from these exceptional responders. The DRUP platform could serve as a drug development path for patients with rare cancer, where clinical data can be combined with systematic genomic data to expedite its progress.
This may also facilitate the use of combinatorial therapeutic strategies in the absence of single-agent activity sometimes found in clinical trials.
Several examples are reported in which single-agent therapy failed to elicit clinical responses, whereas these drugs proved successful in combination with (multiple) other agents targeting different signaling pathways (27). Because genomics can only provide part of the required biological insights to develop successful (combinatorial) treatment strategies, it is likely, in the (near) future, DRUP and similar initiatives will incorporate more extensive molecular profiles, such as RNA sequencing, proteomics, or functional assays. One important issue that needs to be addressed is the potential overlapping toxicities, as most drug combinations will not have been investigated in phase I clinical trials. Although new combinations should be given with caution to patients with cancer, the I-PREDICT study has shown the feasibility of this approach (28). Although our analyses provide a strong case of support for broad molecular testing for patients with rare cancers and subsequent access to targeted treatment as part of studies, similar to patients with more common cancers, this trial also has limitations. The DRUP was designed to identify signals of activity in very small but defined groups of patients. Even when the percentage of patients with clinical benefit was similar in both groups, lumping data-cross cohorts may lead to unjustified generalizations, and may distract from either clear signs or of activity or lack of activity of a specific treatment. Overlapping survival curves may also obscure results of subgroups.
Another limitation is the absence of a rare cancer control group: as all patients qualifying for DRUP have exhausted standard-of-care treatments and would have otherwise entered a phase I clinical trial or received best supportive care, it remains challenging to create comparable control groups of patients with similar tumor characteristics. Moreover, considering the molecular diversity of tumors, it is extremely challenging to generate a truly comparable control group. It is, therefore, essential that real-life data collections are created, in which molecular profiles, treatment and survival data are registered in a structured manner. This should preferably be done as part of an international data sharing network. At present, an international data sharing network for studies with a similar design as DRUP is being established including the TAPUR and CAPTUR studies and various other more recently initiated studies (e.g., NCT04341181, NCT04185831; refs. 29, 30). Such a network and systematic data collection will not only allow for generation of sufficient numbers to address specific questions, it will hopefully also lead to better treatment opportunities for our patients.
In summary, we show that patients with metastatic rare cancers benefit from comprehensive molecular diagnostics. Systematic testing, data collection and access to off-label drugs will contribute to alleviate the current inequality in care from which these patients suffer.
Supplementary Material
Acknowledgments
The DRUP trial is supported by the Barcode for Life Foundation; the Dutch Cancer Society (grant number 10014); and all participating pharmaceutical companies: Amgen, AstraZeneca, Bayer, Boehringer Ingelheim, Bristol-Myers Squibb, Clovis Oncology, Eisai, Ipsen, Merck Sharp & Dohme, Novartis, Pfizer, and Roche. We thank the Hartwig Medical Foundation for their in-kind support by performing sequencing and biomarker analyses on baseline biopsies; the Multidisciplinary Expert Board for supporting the central case-review process; the Independent Data Monitoring Committee for their advice on cohort decisions and monitoring of preliminary safety data; the Netherlands Cancer Institute's Biobank Facility, Scientific Department and Pharmacy for their facilitating services; and lastly, we thank the patients and the investigators and research staff from all participating hospitals.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
L.R. Hoes reports grants and personal fees from Dutch Cancer Society; grants from Barcode for Life; non-financial support from Bayer; and grants and non-financial support from AstraZeneca, Amgen, Boehringer Ingelheim, Bristol Myers Squibb, Clovis Oncology, Eisai, Ipsen, Merck Sharp & Dohme, Novartis, Pfizer, and Roche during the conduct of the study. J.M. van Berge Henegouwen reports grants and non-financial support from Amgen, AstraZeneca, Bayer, Boehringer Ingelheim, Bristol Myers Squibb, Clovis Oncology, Eisai, Ipsen, Merck Sharp &Dohme, Novartis, Pfizer, and Roche during the conduct of the study. H. van der Wijngaart reports other support from Amgen, AstraZeneca, Bayer, Boehringer Ingelheim, Bristol Myers Squibb, Clovis Oncology, Eisai, Ipsen, Merck Sharp & Dohme, Novartis, Pfizer, Roche, Barcode for Life Foundation, and Dutch Cancer Society during the conduct of the study. L.J. Zeverijn reports grants and non-financial support from Amgen, AstraZeneca, Bayer, Bristol Myers Squibb, Clovis Oncology, Eisai, Ipsen, Merck Sharp & Dohme, Novartis, Pfizer, and Roche, as well as grants from Barcode for Life Foundation and Dutch Cancer Society during the conduct of the study. W.J. de Leng reports non-financial support from Amgen, AstraZeneca, Bayer, Boehringer Ingelheim, Bristol Myers Squibb, Clovis Oncology, Eisai, Ipsen, Merck Sharp & Dohme, Novartis, Pfizer, and Roche, as well as grants from Roche, Bristol Myers Squibb, Pfizer, Barcode for Life Foundation, and Dutch Cancer Society during the conduct of the study; in addition, W.J. de Leng reports personal fees from Janssen, Novartis, and AbbVie outside the submitted work. A.D.R. Huitema reports grants from Amgen, AstraZeneca, Bayer, Boehringer Ingelheim, Bristol Myers Squibb, Clovis Oncology, Eisai, Ipsen, Merck Sharp & Dohme, Novartis, Pfizer, Roche, Barcode for Life Foundation, and Dutch Cancer Society during the conduct of the study. E.H. Gort reports other support from Dutch Cancer Society, Barcode for Life Foundation, Amgen, AstraZeneca, Bayer, Boehringer Ingelheim, Bristol Myers Squibb, Clovis Oncology, Eisai, Ipsen, Merck Sharp & Dohme, Novartis, Pfizer, and Roche during the conduct of the study. J.W.B. de Groot reports personal fees from BMS, Servier, and Pierre Fabre outside the submitted work. D.J. de Groot reports grants from Ipsen during the conduct of the study. M.P. Hendriks reports grants and non-financial support from Amgen, AstraZeneca, Bayer, Boehringer Ingelheim, Bristol Myers Squibb, Clovis Oncology, Eisai, Ipsen, Merck Sharp & Dohme, Novartis, Pfizer, and Roche, as well as grants from Barcode for Life Foundation and Dutch Cancer Society during the conduct of the study. E.F. Smit reports grants from AstraZeneca, Bristol Myers Squibb, Boehringer Ingelheim, Daichii Sankyo, Eli Lilly, Merck Serono, MSD, Takeda, Roche Genentech, Novartis, Pfizer, Pharmamar, and Amgen outside the submitted work. W.T.A. van der Graaf reports grants from Amgen, AstraZeneca, Bayer, Boehringer Ingelheim, Bristol Meyers Squibb, Clovis Oncology, Eisai, Ipsen, Merck Sharp & Dohme, Novartis, Pfizer, Roche, Barcode for Life Foundation, and Dutch Cancer Society during the conduct of the study, as well as grants from GSK, Lilly, and Springworks outside the submitted work. M. Labots reports personal fees from Bristol Myers Squibb and MSD outside the submitted work. M.P. Lolkema reports grants and personal fees from Astellas, J&J, MSD, and Sanofi, as well as personal fees from Incyte, Amgen, Bayer, Servier, Roche, INCa, Pfizer, AstraZeneca, Novartis, Julius Clinical, and Elipsis Pharma during the conduct of the study. E. Cuppen reports other support from Illumina and personal fees from InteRNA Technologies outside the submitted work. E.E. Voest reports grants from Pfizer, Roche, Novartis, Clovis, Amgen, AstraZeneca, GSK, BMS, MSD, Seagen, Bayer, and BI during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
L.R. Hoes: Conceptualization, resources, data curation, formal analysis, investigation, writing–original draft, project administration. J.M. van Berge Henegouwen: Resources, data curation, investigation, project administration, writing–review and editing. H. van der Wijngaart: Resources, data curation, investigation, project administration, writing–review and editing. L.J. Zeverijn: Resources, data curation, investigation, project administration, writing–review and editing. D.L. van der Velden: Conceptualization, project administration, writing–review and editing. J. van de Haar: Formal analysis, writing–review and editing. P. Roepman: Investigation, molecular tumor board. W.J. de Leng: Investigation, molecular tumor board. A.M.L. Jansen: Investigation, writing–review and editing, molecular tumor board. E. van Werkhoven: Formal analysis, writing–review and editing. V. van der Noort: Formal analysis, writing–review and editing. A.D.R. Huitema: Writing–review and editing, pharmacist. E.H. Gort: Resources, writing–review and editing. J.W.B. de Groot: Resources, writing–review and editing. E.D. Kerver: Resources, writing–review and editing. D.J. de Groot: Resources, writing–review and editing. F. Erdkamp: Resources, writing–review and editing. L.V. Beerepoot: Resources, writing–review and editing. M.P. Hendriks: Resources, writing–review and editing. E.F. Smit: Resources, writing–review and editing. W.T.A. van der Graaf: Resources, writing–review and editing. C.M.L. van Herpen: Resources, writing–review and editing. M. Labots: Resources, writing–review and editing. A. Hoeben: Resources, writing–review and editing. H. Morreau: Methodology, writing–review and editing. M.P. Lolkema: Resources, writing–review and editing. E. Cuppen: Conceptualization, formal analysis, writing–review and editing. H. Gelderblom: Conceptualization, resources, supervision, funding acquisition, investigation, writing–review and editing, principle investigator. H.M.W. Verheul: Conceptualization, resources, supervision, funding acquisition, investigation, writing–review and editing, principle investigator. E.E. Voest: Conceptualization, resources, supervision, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing, principle investigator.
References
- 1. Casali PG, Trama A. Rationale of the rare cancer list: a consensus paper from the Joint Action on Rare Cancers (JARC) of the European Union (EU). ESMO Open 2020;5:e000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gatta G, Capocaccia R, Botta L, Mallone S, De Angelis R, Ardanaz E, et al. Burden and centralised treatment in Europe of rare tumours: results of RARECAREnet-a population-based study. Lancet Oncol 2017;18:1022–39. [DOI] [PubMed] [Google Scholar]
- 3. Gatta G, van der Zwan JM, Casali PG, Siesling S, Dei Tos AP, Kunkler I, et al. Rare cancers are not so rare: the rare cancer burden in Europe. Eur J Cancer 2011;47:2493–511. [DOI] [PubMed] [Google Scholar]
- 4. Okamura R, Boichard A, Kato S, Sicklick JK, Bazhenova L, Kurzrock R. Analysis of NTRK alterations in pan-cancer adult and pediatric malignancies: implications for NTRK-targeted therapeutics. JCO Precis Oncol 2018;2018:PO.18.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang RSP, Haberberger J, Severson E, Duncan DL, Hemmerich A, Edgerly C, et al. A pan-cancer analysis of PD-L1 immunohistochemistry and gene amplification, tumor mutation burden and microsatellite instability in 48,782 cases. Mod Pathol 2021;34:252–63. [DOI] [PubMed] [Google Scholar]
- 6. Jonsson P, Bandlamudi C, Cheng ML, Srinivasan P, Chavan SS, Friedman ND, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature 2019;571:576–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boyd N, Dancey JE, Gilks CB, Huntsman DG. Rare cancers: a sea of opportunity. Lancet Oncol 2016;17:e52–61. [DOI] [PubMed] [Google Scholar]
- 8. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Akcakanat A, Zheng X, Cruz Pico CX, Kim T, Chen K, Korkut A, et al. Genomic, transcriptomic and proteomic profiling of metastatic breast cancer. Clin Cancer Res 2021;27:3243–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Priestley P, Baber J, Lolkema MP, Steeghs N, de Bruijn E, Shale C, et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 2019;575:210–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. AACR Project GENIE Consortium. AACR project GENIE: Powering precision medicine through an international consortium. Cancer Discov 2017;7:818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van der Velden DL, Hoes LR, van der Wijngaart H, van Berge Henegouwen JM, van Werkhoven E, Roepman P, et al. The Drug Rediscovery protocol facilitates the expanded use of existing anticancer drugs. Nature 2019;574:127–31. [DOI] [PubMed] [Google Scholar]
- 13. Hainsworth JD, Meric-Bernstam F, Swanton C, Hurwitz H, Spigel DR, Sweeney C, et al. Targeted therapy for advanced solid tumors on the basis of molecular profiles: results from mypathway, an open-label, phase IIa multiple basket study. J Clin Oncol 2018;36:536–42. [DOI] [PubMed] [Google Scholar]
- 14. Sicklick JK, Kato S, Okamura R, Schwaederle M, Hahn ME, Williams CB, et al. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat Med 2019;25:744–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. D'Andrea AD. Susceptibility pathways in Fanconi's anemia and breast cancer. N Engl J Med 2010;362:1909–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feinberg AP, Williams BR. Wilms' tumor as a model for cancer biology. Methods Mol Biol 2003;222:239–48. [DOI] [PubMed] [Google Scholar]
- 17. Eck MJ, Manley PW. The interplay of structural information and functional studies in kinase drug design: insights from BCR-Abl. Curr Opin Cell Biol 2009;21:288–95. [DOI] [PubMed] [Google Scholar]
- 18. Meric-Bernstam F, Brusco L, Shaw K, Horombe C, Kopetz S, Davies MA, et al. Feasibility of large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials. J Clin Oncol 2015;33:2753–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simon R. Optimal two-stage designs for phase II clinical trials. Control Clin Trials 1989;10:1–10. [DOI] [PubMed] [Google Scholar]
- 20. van Waalwijk van Doorn-Khosrovani SB, Pisters-van Roy A, van Saase L, van der Graaff M, Gijzen J, Sleijfer S, et al. Personalised reimbursement: a risk-sharing model for biomarker-driven treatment of rare subgroups of cancer patients. Ann Oncol 2019;30:663–5. [DOI] [PubMed] [Google Scholar]
- 21. Regulation (EC) No 141/2000 of the European Parliament and of the council of 16 December 1999 on orphan medicinal products. [cited 2021 Dec 31]. Available from:http://data.europa.eu/eli/reg/2000/141/2019-07-26.
- 22. Cobain EF, Wu YM, Vats P, Chugh R, Worden F, Smith DC, et al. Assessment of clinical benefit of integrative genomic profiling in advanced solid tumors. JAMA Oncol 2021;7:525–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Colwell J. NCI-MATCH trial draws strong interest. Cancer Discov 2016;6:334. [DOI] [PubMed] [Google Scholar]
- 24. Horak P, Heining C, Kreutzfeldt S, Hutter B, Mock A, Hüllein J, et al. Comprehensive genomic and transcriptomic analysis for guiding therapeutic decisions in patients with rare cancers. Cancer Discov 2021;11:2780–95. [DOI] [PubMed] [Google Scholar]
- 25. Reiter JG, Makohon-Moore AP, Gerold JM, Heyde A, Attiyeh MA, Kohutek ZA, et al. Minimal functional driver gene heterogeneity among untreated metastases. Science 2018;361:1033–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blay JY, Coindre JM, Ducimetière F, Ray-Coquard I. The value of research collaborations and consortia in rare cancers. Lancet Oncol 2016;17:e62–e9. [DOI] [PubMed] [Google Scholar]
- 27. Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med 2019;381:1632–43. [DOI] [PubMed] [Google Scholar]
- 28. Settleman J, Neto JMF, Bernards R. Thinking differently about cancer treatment regimens. Cancer Discov 2021;11:1016–23. [DOI] [PubMed] [Google Scholar]
- 29. Mangat PK, Halabi S, Bruinooge SS, Garrett-Mayer E, Alva A, Janeway KA, et al. Rationale and design of the targeted agent and profiling utilization registry (TAPUR) study. JCO Precis Oncol 2018;2018:10.1200/PO.18.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Skamene T, Siu LL, Renouf DJ, Laskin JJ, Bedard PL, Jones SJM, et al. Canadian profiling and targeted agent utilization trial (CAPTUR/PM.1): a phase II basket precision medicine trial. J Clin Oncol 2018;36:TPS12127. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data described in this study are available for academic use upon request. WGS in combination with clinical data can be obtained through the Netherlands Cancer Institute and the Hartwig Medical Foundation through standardized procedures. Request forms can be found at https://www.hartwigmedicalfoundation.nl/en. An independent data-access board will evaluate whether the intended use of the data is compatible with the consent given by the patients, and whether there would be any applicable legal or ethical constraints. Clinical data, treatment outcome, and safety data can be obtained at a per-patient level by emailing the Institutional Review Board of the Netherlands Cancer Institute (IRB@nki.nl).