Abstract
Purpose:
PARP inhibitors (PARPi) have demonstrated efficacy in tumors with germline breast cancer susceptibility genes (gBRCA) 1 and 2 mutations, but further factors influencing response to PARPi are poorly understood.
Experimental Design:
Breast cancer tumor tissue from patients with gBRCA1/2 mutations from the phase III EMBRACA trial of the PARPi talazoparib versus chemotherapy was sequenced using FoundationOne CDx.
Results:
In the evaluable intent-to-treat population, 96.1% (296/308) had ≥1 tumor BRCA (tBRCA) mutation and there was strong concordance (95.3%) between tBRCA and gBRCA mutational status. Genetic/genomic characteristics including BRCA loss of heterozygosity (LOH; identified in 82.6% of evaluable patients), DNA damage response (DDR) gene mutational burden, and tumor homologous recombination deficiency [assessed by genomic LOH (gLOH)] demonstrated no association with talazoparib efficacy.
Conclusions:
Overall, BRCA LOH status, DDR gene mutational burden, and gLOH were not associated with talazoparib efficacy; however, these conclusions are qualified by population heterogeneity and low patient numbers in some subgroups. Further investigation in larger patient populations is warranted.
Translational Relevance.
While the efficacy of the PARP inhibitor talazoparib has been demonstrated in patients with germline breast cancer susceptibility gene (gBRCA1/2)-mutated HER2-negative locally advanced or metastatic breast cancer, less is known about tumor-related factors that might influence response to talazoparib. This retrospective analysis evaluated tumor tissue samples from patients enrolled in the EMBRACA study to explore potential tumor mutational and genomic factors that may influence response to talazoparib. The results from this analysis show that genetic/genomic characteristics, including BRCA loss-of-heterozygosity status, DNA damage response gene mutational burden, and tumor homologous recombination deficiency, demonstrate no association with talazoparib efficacy among patients with germline BRCA1/2 mutations. Further research is warranted and will be important in identifying patients in the clinic who may maximally benefit from talazoparib treatment.
Introduction
DNA double-strand break repair is a key process in maintaining genomic stability (1). Breast cancer susceptibility genes (BRCA) 1 and 2 are tumor-suppressing genes that play a critical role in this process via homologous recombination repair (2). Germline BRCA1/2 mutations (gBRCA1/2mut) are associated with an increased susceptibility to breast, ovarian, prostate, and pancreatic cancer (3). Cancer cells with BRCA1/2mut have homologous recombination deficiency (HRD) and become more reliant on PARP 1 and 2, which mediate base excision repair of single-strand DNA (ssDNA) breaks (2, 4–6).
PARP inhibitors act via direct catalytic inhibition and there is evidence that they trap PARP on sites of DNA damage, hindering transcription and inducing replication fork collapse (2, 7, 8). Inhibition of PARP results in persistent ssDNA breaks, culminating in accumulation of double-stranded DNA breaks and therefore, in cells with HRD, ultimately inducing synthetic lethality stemming from irreparable DNA damage (2).
The PARP inhibitor talazoparib has demonstrated efficacy in cancers with gBRCAmut (9–11), and has also shown higher PARP-trapping activity in vitro compared with other PARP inhibitors (2, 8, 12). In the phase III EMBRACA trial (NCT01945775), talazoparib significantly improved median progression-free survival (PFS) compared with chemotherapy in patients with HER2-negative locally advanced/metastatic breast cancer (LA/mBC) and a gBRCAmut [8.6 months; 95% confidence interval (CI), 7.2–9.3 vs. 5.6 months; 95% CI, 4.2–6.7; HR for disease progression or death 0.54; 95% CI, 0.41–0.71; P < 0.001); ref. 9]. On the basis of the EMBRACA results, talazoparib (oral; 1 mg, once daily) is approved in the United States, European Union, and other countries for patients with HER2-negative LA/mBC with gBRCA1/2mut (13, 14). Similarly, multiple guidelines recommend the use of PARP inhibitors in patients with gBRCA1/2mut and advanced HER2-negative breast cancer, as an alternative to chemotherapy, with the National Comprehensive Cancer Network (NCCN) guidelines including treatment with talazoparib or olaparib, as a category 1, preferred option for patients with gBRCA1/2mut (15, 16).
While the efficacy of PARP inhibitors in patients with gBRCA1/2mut HER2-negative advanced breast cancer has been demonstrated in clinical trials (9–11), little is known about tumor-related factors that might influence response to PARP inhibitors in such patients. The purpose of these analyses was to evaluate samples of tumor tissue from patients enrolled in EMBRACA to explore the prevalence of tumor BRCA1/2mut (tBRCAmut) in patients with a gBRCA1/2mut, the concordance of BRCA mutation between germline and tumor samples, and tBRCA zygosity. Non-BRCA DNA damage response (DDR) mutations; other common, mechanistically pertinent, non-BRCA mutations; and HRD as assessed by genomic loss of heterozygosity (gLOH) were also evaluated. In addition, potential associations of these factors with patient outcomes were assessed.
Materials and Methods
Study design and patients
EMBRACA was an open-label, randomized, international, phase III trial comparing the efficacy and safety of talazoparib with chemotherapy (capecitabine, eribulin, gemcitabine, or vinorelbine), assigned in a 2:1 ratio, in patients with HER2-negative and gBRCA-mutated LA/mBC. Details of the study have been previously published (9). Briefly, eligible patients were 18 years of age or older and had received ≤3 previous cytotoxic regimens for advanced breast cancer, and had been previously treated with a taxane, an anthracycline, or both, unless these treatments were contraindicated. The primary endpoint was radiographic PFS by blinded independent review facility (IRF) using RECIST version 1.1.
The trial protocol was approved by an independent ethics committee at each site before initiation of the trial, and the study was conducted in accordance with the principles of the Declaration of Helsinki, International Council for Harmonisation Guideline for Good Clinical Practice, the U.S. Code of Federal Regulations, and/or other national and local regulations. Written informed consent was obtained from each patient before entering the patient into the study.
Next-generation sequencing and mutational analysis
Molecular eligibility for enrollment in EMBRACA was supported by germline testing using the BRACAnalysis CDx test (Myriad Genetic Laboratories, Inc.; ref. 9). In this analysis, mandated tumor tissue samples from primary or metastatic sites from patients enrolled in EMBRACA were tested using FoundationOne CDx (Foundation Medicine Inc.). Mutations were defined as known/likely pathogenic single-nucleotide variants (SNV), insertions, deletions, or rearrangements. Copy-number alterations (CNA) were analyzed separately as these alterations often reflect larger genomic changes that are not necessarily associated with a specific gene (17, 18). Furthermore, non-BRCA DDR genes (ARID1A, ATR, ATM, BARD1, BRD4, BRIP1, CDK12, CHEK2, FANCA, FANCC, FANCG, NBN, PALB2, RAD51B, and STAG2) present in tumor tissue were included in a subset of correlative analyses based on their role in homologous recombination–mediated DNA response and/or demonstrated potential for these mutations to sensitize to PARP inhibitors in various nonclinical models (19–22), and based on the presence of mutations in these genes in the FoundationOne CDx dataset. gLOH and somatic-germline-zygosity (SGZ) assessments were performed by Foundation Medicine Inc. gLOH was used to evaluate HRD at the genome level (23, 24). SGZ was used to predict the homozygous versus heterozygous state of the BRCA mutations (25). Finally, germline versus tumor sequence comparisons were used to determine whether tumor BRCA mutations were of germline or somatic origin.
Patient populations included in biomarker analysis
The evaluable intent-to-treat (ITT) population included all patients in the ITT population with tumor samples suitable for analysis by FoundationOne CDx testing. Subsets of the ITT population were used for different analyses (Supplementary Fig. S1) and included the tBRCA-mutated ITT population: patients with ≥1 tBRCAmut (i.e., alterations consisting of known or likely pathogenic SNVs, insertions, deletions, or rearrangements, but excluding CNAs) identified by FoundationOne CDx; ITT population evaluable for BRCA zygosity: a subset of the tBRCA-mutated ITT population evaluable for zygosity by SGZ; and ITT population evaluable for gLOH: a subset of the evaluable ITT population in which gLOH could be assessed.
Clinical efficacy endpoints assessed in correlative analyses
Clinical efficacy endpoints assessed in correlative analyses included the clinical benefit rate [defined as a complete response (CR), partial response (PR), or stable disease (SD) lasting for ≥24 weeks (CBR24) per RECIST 1.1 by investigator assessment] and PFS per RECIST 1.1 by IRF assessment. Best percent change of sum of longest diameters of target lesions from baseline over time by investigator assessment, with best overall response assessed as CR, PR, SD, or progressive disease, was also evaluated. Patients with samples that were not evaluable due to tissue availability, sample quality, or tumor-cell fraction were excluded from these analyses.
Data availability statement
Pfizer will provide access to individual deidentified participant data and related study documents [e.g., protocol, statistical analysis plan (SAP), clinical study report (CSR)] upon request from qualified researchers, and subject to certain criteria, conditions, and exceptions.
Results
Patients
A total of 431 patients were included in the overall ITT population. This comprised 287 patients in the talazoparib arm and 144 patients in the chemotherapy arm. Tumor tissue was evaluable from 308 patients (71.5%) forming the evaluable ITT population for this analysis: 201 patients (70.0%) receiving talazoparib and 107 patients (74.3%) receiving chemotherapy. A summary of baseline characteristics in the overall ITT population and evaluable ITT population is presented in Supplementary Table S1, which demonstrates similar overall characteristics between these populations. Whether tumor tissue was taken from primary or metastatic sites was not recorded in the case report form.
Tumor molecular profiling
Among the 308 patients in the evaluable ITT population, 296 (96.1%) had ≥1 tBRCAmut: 135 (43.8%) had ≥1 tBRCA1mut with no tBRCA2mut, 157 (51.0%) had ≥1 tBRCA2mut with no tBRCA1mut, 4 (1.3%) had both tBRCA1mut and tBRCA2mut, and 12 (3.9%) did not have either tBRCA1mut or tBRCA2mut in tumor samples (Supplementary Fig. S2A). Known or likely pathogenic CNAs in BRCA1 and BRCA2 (not counted as BRCA mutations) were observed in 6 of 308 (1.9%) and 3 of 308 (1.0%) of the evaluable ITT population tumors, respectively.
Of the 12 patients recorded as lacking a tBRCA1 or tBRCA2 mutation (Supplementary Table S2), 3 were found to have tBRCA2 SNVs classified by FoundationOne CDx as “pathogenicity unknown”; for 2 of these 3, the same variants were originally identified in germline testing and used to support molecular eligibility for enrollment of these patients. Seven patients had tBRCA CNAs that were classified as pathogenic by FoundationOne CDx. Further analysis showed that these samples harbored partial deletions of BRCA genes, and gBRCA enrollment testing (BRACAnalysis CDx test) showed deletions in the corresponding gBRCA genes for these patients. Three patients lacked any tBRCA variants, including 2 with central gBRCA1/2mut used to support molecular eligibility for enrollment and one with no gBRCA1/2mut detected by Central testing, who was enrolled on the basis of local gBRCA test results. Clinical outcomes for patients who were recorded as lacking tBRCAmut are shown in Supplementary Table S2.
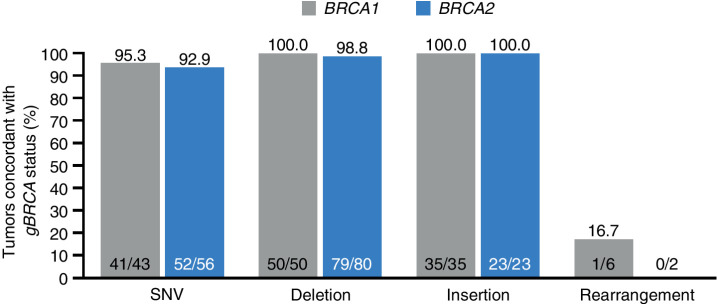
Concordance between gBRCA and tBRCA mutational status was assessed in 295 patients. A total of 281 of 295 (95.3%) patients exhibited concordance (Fig. 1). However, a majority (9) of the 14 patients described as nonconcordant were found to have the same/similar underlying variants in both the germline and tumor tests, and were only classified as nonconcordant due to differences in variant classification/nomenclature between tests (Fig. 1). Hence, the concordance rate would be 290 of 295 (98.3%) if these variant classification/nomenclature differences were factored into this concordance assessment. In addition to patients listed as lacking a tBRCA1/2mut, 3 patients had different gBRCA and tBRCA mutations (2 patients had a gBRCA2 but a tBRCA1 mutation and 1 patient had a gBRCA1 but a tBRCA2 mutation).
Figure 1.
Tumor sequencing and concordance with germline BRCA status—evaluable ITT population. The proportion of patients with a known gBRCAmut by Central lab who have a BRCA1/2mut (defined as known or likely pathogenic variant, CNAs excluded) detected in a tumor using FoundationOne CDx. All patients showing concordant BRCA1 or BRCA2 mutational status exhibited the same mutation in tumor as originally detected in germline, as evidenced by mapping to a common Variation ID in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar) or other comparative means, with one exception (1 patient who exhibited a distinct BRCA2mut in germline and tumor). A total of 14 patients were discordant: 2 patients—same germline variants used to support enrollment were classified by the FoundationOne CDx test as pathogenicity unknown; 7 patients exhibited gBRCA deletion impacting one or multiple exons and were hence mapped to Rearrangement category. These patients exhibited alterations classified as pathogenic BRCA CNAs by FoundationOne CDx in the corresponding BRCA genes; 2 patients—gBRCA2mut, tBRCA1mut; 1 patient—gBRCA1mut, tBRCA2mut; 2 patients—no tBRCA variant detected.
Of the 296 patients in the tBRCA-mutated ITT population, 236 (79.7%) were evaluable for BRCA zygosity and 236 of 308 (76.6%) of the evaluable ITT population were evaluable for gLOH. BRCA LOH, with retention of a mutant BRCA allele, was predicted in 195 of 236 (82.6%) patients including 1 patient with mutations in both BRCA1 and BRCA2 with LOH predicted for both genes. Forty-one patients had a tBRCA1/2mut with no LOH (Supplementary Fig. S2B). Baseline characteristics of these patients are presented in Supplementary Table S3.
In the evaluable ITT population, non-BRCA mutations within the tumor were found in ARID1A (2.3%); CHEK2 and FANCA (each 1.6%); NBN (1.3%); ATM and BRIP1 (each 1.0%); ATR, BRD4, FANCC, PALB2, and RAD51B (each 0.6%); and BARD1, CDK12, FANCG, and STAG2 (each 0.3%). The most common non-BRCA mutations (≥10%) were TP53 and PIK3CA in both the evaluable ITT population (51.6% and 10.4%) and the tBRCA-mutated population (52.0% and 10.8%), respectively. TP53 mutations were more commonly observed with tBRCA1mut while PIK3CA mutations were more frequently observed in BRCA2-mutated tumors (Table 1). Known/likely pathogenic CNAs with a prevalence of ≥10% in the evaluable ITT population were MYC (21.4%) and RAD21 (21.1%) with similar values observed in the tBRCA-mutated ITT population (20.9% and 21.3.%, respectively; Table 1).
Table 1.
Most commonly mutated non-BRCA1/2 tumor tissue genes in patients with tBRCAmut (evaluable ITT population).
| Talazoparib n/N (%) | Chemotherapy n/N (%) | Combined n/N (%) | ||
|---|---|---|---|---|
| Mutations | ||||
| TP53 | ||||
| tBRCA1mut only | 76/90 (84.4) | 39/45 (86.7) | 115/135 (85.2) | |
| tBRCA2mut only | 23/102 (22.5) | 16/55 (29.1) | 39/157 (24.8) | |
| tBRCA1mut and tBRCA2mut | 0/1 (0.0) | 0/3 (0.0) | 0/4 (0.0) | |
| tBRCA1mut or tBRCA2mut | 99/193 (51.3) | 55/103 (53.4) | 154/296 (52.0) | |
| PIK3CA | ||||
| tBRCA1mut only | 6/90 (6.7) | 1/45 (2.2) | 7/135 (5.2) | |
| tBRCA2mut only | 16/102 (15.7) | 9/55 (16.4) | 25/157 (15.9) | |
| tBRCA1mut and tBRCA2mut | 0/1 (0.0) | 0/3 (0.0) | 0/4 (0.0) | |
| tBRCA1mut or tBRCA2mut | 22/193 (11.4) | 10/103 (9.7) | 32/296 (10.8) | |
| Known/likely pathogenic CNAs | ||||
| MYC | ||||
| tBRCA1mut | 16/90 (17.8) | 12/45 (26.7) | 28/135 (20.7) | |
| tBRCA2mut | 21/102 (20.6) | 11/55 (20.0) | 32/157 (20.4) | |
| tBRCA1mut and tBRCA2mut | 0/1 (0.0) | 2/3 (66.7) | 2/4 (50.0) | |
| tBRCA1mut or tBRCA2mut | 37/193 (19.2) | 25/103 (24.3) | 62/296 (20.9) | |
| RAD21 | ||||
| tBRCA1mut | 10/90 (11.1) | 13/45 (28.9) | 23/135 (17.0) | |
| tBRCA2mut | 23/102 (22.5) | 16/55 (29.1) | 39/157 (24.8) | |
| tBRCA1mut and tBRCA2mut | 0/1 (0.0) | 1/3 (33.3) | 1/4 (25.0) | |
| tBRCA1mut or tBRCA2mut | 33/193 (17.1) | 30/103 (29.1) | 63/296 (21.3) | |
Note: tBRCA-mutated ITT population includes all patients with tumor samples suitable for the genomic evaluation and analyzed using FoundationOne CDx who have BRCA mutations (variants with known or likely pathogenic impact, excluding CNAs). Genes shown exhibit mutations (known/likely pathogenic variant, CNAs excluded) or known/likely pathogenic CNAs in ≥10% of patients in combined arms.
In EMBRACA, median (range) gLOH scores were 21.8% (0.0–52.7) and 20.5% (0.2–40.5) for the talazoparib and chemotherapy arms, respectively. An analysis from the Foundation Medicine Insights database found similar gLOH scores in patients with HER2-negative BRCA1/2-mutated breast cancer (median 22.6%, based on N = 1,471 tumors), but gLOH was lower in the overall breast cancer population (median 12.5%, based on N = 17,261 tumors). The relationship between BRCA LOH and gLOH was also explored. In both treatment arms, gLOH was significantly higher in tumors exhibiting BRCA LOH than in tumors not exhibiting BRCA LOH: median (minimum, maximum): 22.8% (0.1, 52.7) and 12.7% (0.0, 21.2), P < 0.0001, for talazoparib; median (minimum, maximum): 21.8% (0.2, 40.5) and 15.8% (1.6, 23.6), P = 0.0028, for chemotherapy (Supplementary Fig. S3).
Correlative analysis
CBR24 was generally comparable for tumors with BRCA1/2mut: in the talazoparib arm; CBR24 was 64% (n/N = 58/90; 95% CI, 54–74) and 76% (n/N = 78/102; 95% CI, 67–84) for tBRCA1mut and tBRCA2mut, respectively. In the chemotherapy arm, CBR24 was 36% (n/N = 16/45; 95% CI, 22–51) and 31% (n/N = 17/55; 95% CI, 19–45) for tBRCA1mut and tBRCA2mut, respectively. In patients with or without tBRCA1/2 LOH treated with talazoparib, CBR24 was 74.6% (n/N = 91/122) and 66.7% (n/N = 18/27), respectively.
In the talazoparib arm, no significant differences in median PFS were observed between the 122 patients with tBRCA LOH and the 27 patients without tBRCA LOH [9.0 months vs. 6.9 months; HR (95% CI) = 0.868 (0.512–1.470); P = 0.597; Table 2]. Similarly, no significant difference in median PFS was observed between patients with or without tBRCA LOH in the chemotherapy arm [5.8 vs. 5.6 months; HR (95% CI) = 1.797 (0.751–4.298); P = 0.179; Table 2]. Differences in PFS between patients with and without tBRCA LOH were also evaluated by lines of prior chemotherapy and tumor subtype where subgroup sample size was ≥10 patients. In the talazoparib arm, a trend favoring patients with tBRCA LOH was observed in the hormone receptor–positive subgroup [median PFS: 13.0 months vs. 8.5 months; HR (95% CI) = 0.542 (0.285–1.032); P = 0.058; Table 2]; however, the triple-negative breast cancer (TNBC) subgroup was not evaluable for this analysis as most evaluable patients (56/60, 93.3%) exhibited BRCA LOH.
Table 2.
PFS of patients with BRCA-mutant tumors: BRCA1 or BRCA2 LOH versus no BRCA1 or BRCA2 LOH (tBRCA-mutant ITT population evaluable for BRCA zygosity).
| Median PFS,a mo | |||||||
|---|---|---|---|---|---|---|---|
| Group | Evaluable for BRCA zygosity, n | BRCA1 or BRCA2 LOH, n | No BRCA1 or BRCA2 LOH, n | BRCA1 or BRCA2 LOH | No BRCA1 or BRCA2 LOH | HR (95% CI) | P value |
| Talazoparib arm | |||||||
| ITT | 149 | 122 | 27 | 9.0 | 6.9 | 0.868 (0.512–1.470) | 0.597 |
| 1 prior line of chemotherapy | 56 | 40 | 16 | 6.9 | 5.8 | 0.879 (0.407–1.899) | 0.740 |
| HR+ BC | 89 | 66 | 23 | 13.0 | 8.5 | 0.542 (0.285–1.032) | 0.058 |
| Chemotherapy arm | |||||||
| ITT | 87 | 73 | 14 | 5.8 | 5.6 | 1.797 (0.751–4.298) | 0.179 |
| 1 prior line of chemotherapy | 26 | 23 | 3 | 3.6 | 5.6 | 0.809 (0.177–3.702) | 0.783 |
| HR+ BC | 54 | 45 | 9 | 6.7 | 5.6 | 0.696 (0.238–2.030) | 0.500 |
Note: Cox proportional hazards model with no BRCA1/2 LOH as the reference group was used to calculate HR and 95% CI. HR < 1 indicates better PFS in the BRCA1 or BRCA2 LOH group, whereas HR > 1 indicates better PFS in the no BRCA1 or BRCA2 LOH group. Log-rank two-sided test was performed to compare between the two groups. Evaluable ITT population includes all patients with tumor samples suitable for the genomic evaluation and analyzed using FoundationOne CDx who have known or likely pathogenic BRCAmut (BRCA CNAs excluded) and who are evaluable for BRCA zygosity. Subgroups shown are of the evaluable ITT population defined by previous line of chemotherapy or cancer subtype. PFS is per RECIST 1.1 by IRF assessment.
Abbreviations: HR+ BC, hormone receptor–positive breast cancer; mo, months.
aBased on Kaplan–Meier estimates.
Additional analyses explored the relationship between number of DDR mutations with tumor response in the evaluable ITT population and the impact of alteration status on commonly altered non-DDR genes on PFS in patients with tBRCAmut. No association was observed between the total number of DDR mutations (1 vs. ≥2) and best overall response to talazoparib or chemotherapy [OR (95% CI): 0.76 (0.31–1.87), P = 0.55 for talazoparib; 0.98 (0.27–3.51), P = 0.97 for chemotherapy; Supplementary Fig. S4]. In the talazoparib group, PFS was significantly shorter in patients with TP53 mutations than in those without [HR (95% CI) = 1.693 (1.186–2.418); P = 0.0033]. In the chemotherapy group, PFS was shorter in patients with TP53 mutations than in those without; however, this did not reach statistical significance [HR (95% CI) = 1.439 (0.859–2.411); P = 0.1614]. PTEN, PIK3CA, RAD21, and MYC mutational and/or CNA status were not associated with PFS in either arm (Supplementary Table S4).
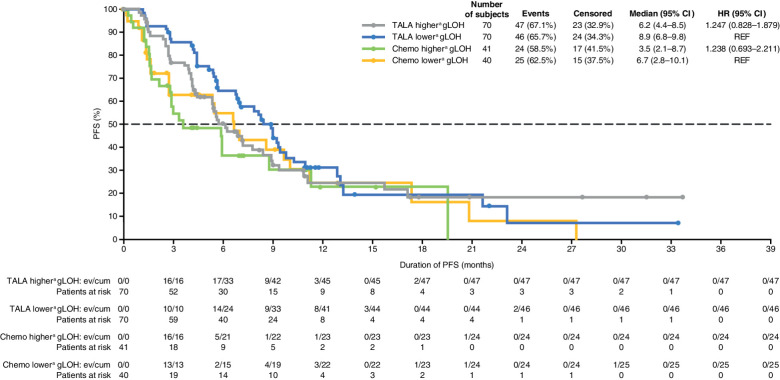
In either treatment arm, no differences in gLOH were observed between patients who did and did not achieve a clinical benefit, and no relationship was evident between gLOH and best overall response category based on the Jonckheere–Terpstra trend test (Supplementary Fig. S5 and S6; Table 3). The association between gLOH and clinical benefit was also investigated in subgroups of patients by lines of prior chemotherapy and tumor subtype (TNBC or hormone receptor–positive breast cancer) where subgroup sample size was ≥30 patients (Table 3). No association was observed between gLOH and clinical benefit with talazoparib by lines of prior chemotherapy. In patients with TNBC, gLOH was significantly higher in patients who had clinical benefit versus no clinical benefit in the talazoparib arm. No association between gLOH and clinical benefit was observed in patients with hormone receptor–positive breast cancer in either study arm. In both treatment arms, patients with gLOH more than or equal to the median exhibited PFS similar to that seen in patients with gLOH below the median: HR (95% CI) = 1.247 (0.828–1.879) for talazoparib; 1.238 (0.693–2.211) for chemotherapy (Fig. 2). In patients with TNBC, PFS was similar between patients with gLOH ≥ median and patients with gLOH less than median treated with talazoparib [HR (95% CI) = 0.913 (0.526–1.587)] (Supplementary Fig. S7). In patients treated with chemotherapy, a numerically longer PFS was observed in patients with gLOH less than median versus in patients with a gLOH ≥ median, although CIs overlapped with 1.0 [HR (95% CI) 2.036 (0.838–4.950)]. Finally, when gLOH was assessed in a stratified Cox regression model with treatment and gLOH as the covariates, no association between gLOH and PFS was evident (Supplementary Table S5).
Table 3.
Clinical benefit by gLOH for talazoparib and chemotherapy by line of therapy or breast cancer subtype—ITT population evaluable for clinical benefit and gLOH.
| Talazoparib | Chemotherapy | |||||
|---|---|---|---|---|---|---|
| gLOH (%) for clinical benefit—yes Median (range), n | gLOH (%) for clinical benefit—no Median (range), n | P value | gLOH (%) for clinical benefit—yes Median (range), n | gLOH (%) for clinical benefit—no Median (range), n | P value | |
| Evaluable ITT | 21.3 (0.0–52.7), 100 | 22.5 (0.0–45.1), 40 | 0.9762 | 20.1 (0.3–35.9), 26 | 20.9 (0.2–40.5), 55 | 0.4917 |
| No prior lines of chemotherapy | 22.0 (0.3–52.7), 44 | 26.4 (11.0–45.1), 10 | 0.1022 | 26.3 (0.3–35.9), 10 | 19.2 (0.2–40.5), 21 | 0.5771 |
| 1 prior line of chemotherapy | 20.4 (0.5–45.2), 33 | 18.7 (0.0–31.9), 16 | 0.1872 | Not shown (total n < 30) | ||
| 2 prior lines of chemotherapy | 23.1 (0.0–50.3), 21 | 22.7 (10.9–28.1), 11 | 0.5406 | Not shown (total n < 30) | ||
| HR+ BC | 18.1 (0.0–50.3), 62 | 18.91 (6.33–31.9), 15 | 0.8171 | 19.1 (1.6–35.9), 21 | 17.5 (0.2–39.0), 24 | 0.2633 |
| TNBC | 30.8 (0.3–52.7), 37 | 23.0 (0.0–45.1), 27 | 0.0456 | 23.6 (0.3–31.3), 5 | 26.7 (0.2–40.5), 32 | 0.3445 |
Note: Clinical benefit is based on target, nontarget, and new lesions per RECIST 1.1, and confirmation of CR, PR, and SD is not required. Clinical benefit is defined as best overall response of CR, PR, or SD lasting ≥24 weeks from randomization per RECIST 1.1 as determined by investigator. Subgroups shown are subgroups of the evaluable ITT population defined by previous lines of chemotherapy or cancer subtype. P value from two-tailed t test.
Abbreviation: HR+ BC, hormone receptor–positive breast cancer.
Figure 2.
Kaplan–Meier curves for duration of radiographic PFS by IRF assessment—ITT population evaluable for PFS and gLOH. Chemo, chemotherapy; cum, cumulative; ev, events; REF, reference; TALA, talazoparib. aHigher and lower indicate that gLOH is above or below the median, respectively. HR is based on unstratified Cox regression model and is relative to talazoparib gLOH < median or chemotherapy gLOH < median with <1 favoring higher gLOH.
Discussion
In the EMBRACA study, 96.1% of patients with evaluable tumor samples exhibited ≥1 tBRCA1mut or tBRCA2mut. Strong concordance (95.3%) between tBRCA mutational status based on FoundationOne CDx and gBRCA mutational status based on the Myriad BRACAnalysis CDx test was observed. When factoring in differences in classification/nomenclature between the two tests, the concordance rate was 98.3%. Strong concordance was also observed in the phase II ABRAZO trial in which 96.7% of patients had ≥1 BRCA1 or BRCA2 tumor mutation, and 96.4% of patients exhibited concordance between tBRCA and gBRCA mutational status (11, 26). This is unsurprising given the importance of BRCA mutations in breast cancer. However, it should be acknowledged that high positive concordance of tumor to germline BRCA mutational status might not translate in patients not preselected for gBRCAmut.
No differences in clinical benefit for tumors with BRCA1mut compared with BRCA2mut were noted in these analyses. The potential for clinical benefit observed in the small fraction of patients lacking tBRCAmut may have been due to the fact that the majority of patients classified as lacking tBRCAmut (n = 12) did in fact have a known/likely pathogenic tBRCA or gBRCA alteration: 7 patients who exhibited deleterious gBRCA deletions per BRACAnalysis CDx had known/likely pathogenic CNAs (losses) in the corresponding BRCA genes per Foundation Medicine nomenclature; 3 patients lacked any tBRCA variants, and 2 patients each had a tBRCA variant defined as being of unknown pathogenicity per Foundation Medicine with identical corresponding gBRCA variants defined as known/likely pathogenic per Myriad.
Overall, 82.6% of patients with evaluable tBRCAmut exhibited BRCA LOH, which is consistent with previous reports that also showed a high prevalence of LOH for BRCA1/2mut (27, 28). In a study that analyzed whole-genome sequences of 560 breast cancer samples, 88.9% of samples with a germline or somatic BRCA1/2mut exhibited LOH (27). In another analysis, BRCA LOH was seen in 90.2% of BRCA1 carriers compared with 54.3% of BRCA2 carriers (28). A high rate of BRCA LOH (85.1%) was also observed in the evaluable tBRCA-mutated population in ABRAZO (11, 26). The high frequency of LOH in the above studies demonstrates the strong selective drive in breast cancer tumors to retain mutated gBRCA alleles and lose the wildtype allele, although mechanisms such as BRCA1 promoter methylation may also contribute to silencing the wildtype BRCA allele (27–29). Therefore, the absence of BRCA LOH is not necessarily a robust indicator of BRCA functionality in tumors. However, in both treatment arms of EMBRACA, gLOH was significantly higher in tumors exhibiting BRCA LOH than in those not exhibiting BRCA LOH. This relationship between BRCA LOH status and HRD is consistent with similar associations documented by others (28). Nonetheless, although there were some differences in PFS within the gLOH high versus low groups, particularly notable in the chemotherapy-treated group (median PFS 3.5 vs. 6.7 months), numbers of patients at risk were small, with wide CIs on median PFS, and no statistically significant association was found between PFS and BRCA LOH status. In addition, a trend in PFS appeared to favor patients with tBRCA LOH in the hormone receptor–positive subgroup, although this association did not reach statistical significance (P = 0.058). Overall, this may suggest a lack of predictive utility of BRCA LOH status in patients with HER2-negative LA/mBC and a gBRCAmut.
Mutations in non-BRCA genes implicated in homologous recombination, DDR, and/or sensitization to PARP inhibitor detected in this analysis included, but were not limited to, ARID1A, ATR, BARD1, BRD4, BRIP1, CHEK2, FANCC, and STAG2. The presence of these non-BRCA DDR mutations did not appear to be associated with differential sensitivity to talazoparib in patients with tBRCAmut; however, due to small patient numbers in this subgroup, confirmation in a larger study is warranted.
Mutational profiles of non-BRCA genes in EMBRACA differed between tBRCA1- and tBRCA2-related cancers. TP53 mutations were very frequent in BRCA1-mutated tumors (85.2%), but less frequent in BRCA2-mutated tumors (24.8%), similar to the mutational profiles observed in ABRAZO (75.9% for BRCA1- and 14.3% for BRCA2-mutated tumors; ref. 26). This difference may be due in part to the high prevalence of both TP53 and BRCA1 mutations in TNBC (30, 31). Conversely, PIK3CA mutations were more commonly seen in BRCA2-mutated tumors than in BRCA1-mutated tumors, which may be attributed to the relatively high prevalence of both PIK3CA mutations (32) and BRCA2 mutations in hormone receptor–positive mBC (33, 34).
No associations were evident between the alteration status of non-BRCA genes and PFS, with the exception of TP53 where the presence of TP53 mutations appeared to be associated with shorter PFS in the talazoparib arm, with a similar, albeit nonsignificant, trend evident in the chemotherapy arm; the significance of this finding could have been limited by the lower number of patients in the chemotherapy arm. While the prognostic significance of TP53 mutations is variable according to tumor type (35), TP53 mutations are associated with worse outcomes in metastatic breast cancer (36), which may reflect the close association between TP53 and BRCA1 mutations, and in turn reflect a common association with TNBC (30, 31).
In this analysis, gLOH was assessed as a genomic-level marker of HRD. gLOH scores in EMBRACA patients were similar to those found in HER2-negative BRCA1/2-mutant breast cancer from the Foundation Medicine Insights database and were much higher than those seen for the overall breast cancer population, reflecting HRD associated with BRCAmut. In both treatment arms, patients with gLOH greater than or equal to median exhibited PFS similar to that seen in patients with gLOH less than median. However, in the talazoparib arm, gLOH was higher in patients with TNBC who exhibited clinical benefit than in those who did not. Caution is needed when drawing a direct relationship between biological variation and clinical significance, as other, still unknown, parameters could be involved in the final response. Therefore, this observation would need to be confirmed in additional studies because of the relatively small numbers of patients in the EMBRACA subgroup being compared and the wide interpatient variability in gLOH scores. Overall, based on these retrospective, exploratory analyses from EMBRACA, there is no clear evidence that gLOH is associated with clinical benefit in this patient population, with the potential exception of the subgroup of patients with TNBC.
The limitations of the EMBRACA study have been previously reported (9). Regarding this analysis, the DNA sequencing used cannot detect sequence-independent functional deficiencies in DDR genes (e.g., promoter methylation). It was also not possible to analyze data based on whether tumor samples were collected from primary or metastatic sites as this information was not recorded in the study and, therefore, may have compromised our ability to resolve potential contributions of BRCA LOH status or gLOH to efficacy since these might change during tumor progression. Similarly, all patients in the EMBRACA study had to have a germline mutation in BRCA1 or BRCA2 to enroll; therefore, efficacy based on somatic-only mutations could not be assessed. Finally, small patient numbers in some mutational subpopulations warrant further investigation in larger datasets.
In summary, 96% of tumors exhibited ≥1 BRCA1/2mut and there was 95% concordance between known gBRCA and tBRCA mutational status. Genetic and genomic characteristics, including BRCA LOH status (with BRCA LOH evident in 83% of evaluable patients) and DDR gene mutational burden, demonstrated no apparent differential association with talazoparib efficacy. There was also no clear evidence that tumor HRD (as assessed by gLOH) was associated with clinical benefit, with the potential exception of the subgroup of patients with TNBC, although this subgroup was small and there was high interpatient variability. Taken together, these results showing high concordance between gBRCA and tBRCA mutations, high prevalence of BRCA LOH, and overall lack of association of BRCA LOH or HRD with outcomes in patients with gBRCA mutations are consistent with recent findings from the OlympiAD trial (37).
Supplementary Material
Acknowledgments
The authors would like to thank Cindy Cheng and her Clinical Programming Team at Pfizer Inc. for analyses generated in support of this manuscript.
This study was sponsored by Pfizer Inc. Medical writing support was provided by Daniela DiBiase, MS, MPH, and Maddie Higgins, MBiolSci, of CMC AFFINITY, McCann Health Medical Communications, and was funded by Pfizer Inc.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
J.L. Blum reports personal fees from Pfizer Inc. during the conduct of the study; personal fees from Amgen, AstraZeneca, Athenex, Biotheranostics, Daiichi Sankyo, Genomic Health, Immunomedics, Myriad Genetics, Novartis, Puma Biotechnology, and Research to Practice outside the submitted work. A.D. Laird reports other support from Pfizer Inc. during the conduct of the study; other support from Pfizer Inc. outside the submitted work. J.K. Litton reports grants from Medivation/Pfizer Inc. during the conduct of the study; grants and other support from Pfizer Inc.; grants from Novartis, Genentech, GlaxoSmithKline, EMD-Serono, AstraZeneca, Zenith, and Merck; and other support from Medlearning, Physicians Education Resource, Prime Oncology, Medscape, Clinical Care Options, Medpage, Uptodate, Society for Immunotherapy of Cancer, National Comprehensive Cancer Network, American Society of Clinical Oncology, NIH PDQ, and Ayala outside the submitted work. H.S. Rugo reports grants from Pfizer Inc. during the conduct of the study; grants from Eli Lilly and Company, Novartis, Roche, Odonate, Merck, Macrogenics, Sermonix, Daiichi Sankyo, AstraZeneca, Gilead, Ayala, and Seagen; and personal fees from Samsung, Napo, and Puma outside the submitted work. J. Ettl reports other support from Biomarin, Medivation, and Pfizer Inc. during the conduct of the study; and personal fees from Pfizer Inc., Eli Lilly and Company, Novartis, Daiichi Sankyo, AstraZeneca, Roche, Gilead, and Seagen outside the submitted work. S.A. Hurvitz reports contracted research paid to institution from Ambrx, Amgen, AstraZeneca, Arvinas, Bayer HealthCare, Cytomx, Daiichi Sankyo, Dignitana, Genentech/Roche, Gilead, GlaxoSmithKline, Immunomedics, Eli Lilly and Company, Macrogenics, Novartis, Pfizer Inc., OBI Pharma, Orinove, Pieris, PUMA, Radius, Sanofi-Aventis, Seattle Genetics/Seagen, Zymeworks, and Phoenix Molecular Designs, Ltd. S. Hurvitz also reports the following relationships: preclinical work (grant paid to UCLA) for Ambrx and Samumed; national/international Principal Investigator for Novartis, Daiichi Sankyo, Seagen, and Genentech/Roche; steering committee for Novartis, Eli Lilly and Company, Daiichi Sankyo/AstraZeneca, Genentech/Roche, and Sanofi-Aventis; travel expenses from Eli Lilly and Company (2019); and uncompensated consulting/ad boards for 4DPharma, Ambrx, Amgen, Artios, Arvinas, Daiichi Sankyo, Dantari, Genentech/Roche, Immunomedics, Macrogenics, Eli Lilly and Company, Novartis, NK Max, Pieris, Pyxis, Seagen, and Biotheranostics. M. Martin reports grants and personal fees from Roche; grants from PUMA; personal fees from Novartis, Pfizer Inc., AstraZeneca, Daiichi Sankyo, Seagen, Eli Lilly and Company, and Taiho outside the submitted work. K.-H. Lee reports personal fees from AstraZeneca, Bayer HealthCare, Eisai, Ono Pharmaceutical, Roche, and Surface Oncology outside the submitted work. S. Lanzalone reports other support from Pfizer Italia Srl. during the conduct of the study, and other support from Pfizer Inc. outside the submitted work. A. Czibere reports other support from Pfizer Inc. during the conduct of the study. J.F. Hopkins is an employee of Foundation Medicine, Inc. and a stockholder of Roche Holdings AG. L.A. Albacker is an employee of Foundation Medicine, Inc. and a stockholder of Roche Holdings AG. No disclosures were reported by the other authors.
Authors' Contributions
J.L. Blum: Conceptualization, data curation, formal analysis, writing–review and editing. A.D. Laird: Data curation, formal analysis, writing–review and editing. J.K. Litton: Data curation, formal analysis, writing–review and editing. H.S. Rugo: Conceptualization, data curation, formal analysis, writing–review and editing. J. Ettl: Conceptualization, formal analysis, writing–review and editing. S.A. Hurvitz: Data curation, formal analysis, writing–review and editing. M. Martin: Data curation, formal analysis, writing–review and editing. H.H. Roché: Data curation, formal analysis, writing–review and editing. K.-H. Lee: Data curation, formal analysis, writing–review and editing. A. Goodwin: Data curation, formal analysis, writing–review and editing. Y. Chen: Formal analysis, writing–review and editing. S. Lanzalone: Conceptualization, data curation, formal analysis, writing–review and editing. J. Chelliserry: Data curation, writing–review and editing. A. Czibere: Conceptualization, data curation, formal analysis, writing–review and editing. J.F. Hopkins: Formal analysis, writing–review and editing. L.A. Albacker: Formal analysis, writing–review and editing. L.A. Mina: Data curation, formal analysis, writing–review and editing.
References
- 1. Scully R, Panday A, Elango R, Willis NA. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol 2019;20:698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017;355:1152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol 2008;26:3785–90. [DOI] [PubMed] [Google Scholar]
- 4. Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol 2011;5:387–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Javle M, Curtin NJ. The potential for poly (ADP-ribose) polymerase inhibitors in cancer therapy. Ther Adv Med Oncol 2011;3:257–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morales J, Li L, Fattah FJ, Dong Y, Bey EA, Patel M, et al. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr 2014;24:15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murai J, Huang SN, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res 2012;72:5588–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murai J, Huang SY, Renaud A, Zhang Y, Ji J, Takeda S, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther 2014;13:433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Litton JK, Rugo HS, Ettl J, Hurvitz SA, Gonçalves A, Lee K-H, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med 2018;379:753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Bono J, Ramanathan RK, Mina L, Chugh R, Glaspy J, Rafii S, et al. Phase I, dose-escalation, two-part trial of the PARP inhibitor talazoparib in patients with advanced germline BRCA1/2 mutations and selected sporadic cancers. Cancer Discov 2017;7:620–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Turner NC, Telli ML, Rugo HS, Mailliez A, Ettl J, Grischke EM, et al. A Phase II study of talazoparib after platinum or cytotoxic nonplatinum regimens in patients with advanced breast cancer and germline BRCA1/2 mutations (ABRAZO). Clin Cancer Res 2019;25:2717–24. [DOI] [PubMed] [Google Scholar]
- 12. Zandarashvili L, Langelier MF, Velagapudi UK, Hancock MA, Steffen JD, Billur R, et al. Structural basis for allosteric PARP-1 retention on DNA breaks. Science 2020;368:eaax6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. European Medicines Agency . TALZENNA® (talazoparib) summary of product characteristics. November 2020. Available from: https://www.ema.europa.eu/en/documents/product-information/talzenna-epar-product-information_en.pdf.
- 14. U.S. Food and Drug Administration. TALZENNA® (talazoparib) prescribing information. 2021. Available from:http://labeling.pfizer.com/ShowLabeling.aspx?id=11046.
- 15. Tung NM, Boughey JC, Pierce LJ, Robson ME, Bedrosian I, Dietz JR, et al. Management of hereditary breast cancer: American Society of Clinical Oncology, American Society for Radiation Oncology, and Society of Surgical Oncology guideline. J Clin Oncol 2020;38:2080–106. [DOI] [PubMed] [Google Scholar]
- 16. National Comprehensive Cancer Network (NCCN). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®): Breast cancer. Version 5. 2020. Available from: https://www.nccn.org/professionals/physician_gls/pdf/breast_blocks.pdf. [Google Scholar]
- 17. Ooi A, Inokuchi M, Horike SI, Kawashima H, Ishikawa S, Ikeda H, et al. Amplicons in breast cancers analyzed by multiplex ligation-dependent probe amplification and fluorescence in situ hybridization. Hum Pathol 2019;85:33–43. [DOI] [PubMed] [Google Scholar]
- 18. Kneissig M, Bernhard S, Storchova Z. Modelling chromosome structural and copy number changes to understand cancer genomes. Curr Opin Genet Dev 2019;54:25–32. [DOI] [PubMed] [Google Scholar]
- 19. Heeke AL, Pishvaian MJ, Lynce F, Xiu J, Brody JR, Chen WJ, et al. Prevalence of homologous recombination-related gene mutations across multiple cancer types. JCO Precis Oncol 2018;2018:PO.17.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun C, Yin J, Fang Y, Chen J, Jeong KJ, Chen X, et al. BRD4 inhibition is synthetic lethal with PARP inhibitors through the induction of homologous recombination deficiency. Cancer Cell 2018;33:401–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chung JH, Dewal N, Sokol E, Mathew P, Whitehead R, Millis SZ, et al. Prospective comprehensive genomic profiling of primary and metastatic prostate tumors. JCO Precis Oncol 2019;3:PO.18.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mondal G, Stevers M, Goode B, Ashworth A, Solomon DA. A requirement for STAG2 in replication fork progression creates a targetable synthetic lethality in cohesin-mutant cancers. Nat Commun 2019;10:1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sokol ES, Pavlick D, Khiabanian H, Frampton GM, Ross JS, Gregg JP, et al. Pan-cancer analysis of BRCA1 and BRCA2 genomic alterations and their association with genomic instability as measured by genome-wide loss of heterozygosity. JCO Precis Oncol 2020;4:442–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, Sun J, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol 2017;18:75–87. [DOI] [PubMed] [Google Scholar]
- 25. Sun JX, He Y, Sanford E, Montesion M, Frampton GM, Vignot S, et al. A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal. PLoS Comput Biol 2018;14:e1005965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Turner NC. Next-generation DNA sequencing (NGS) results for tumors from Phase 2 ABRAZO study of talazoparib after platinum or cytotoxic nonplatinum regimens in patients (pts) with advanced breast cancer (ABC) and germline BRCA1/2 (gBRCA) mutations. In: Proceedings of the European Society for Medical Oncology 2019 Congress; 2019Sept 27–Oct 1; Barcelona, Spain. Lugano, Switzerland: Annals of Oncology; 2019. Abstract nr 2575. [Google Scholar]
- 27. Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maxwell KN, Wubbenhorst B, Wenz BM, De Sloover D, Pluta J, Emery L, et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat Commun 2017;8:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Polak P, Kim J, Braunstein LZ, Karlic R, Haradhavala NJ, Tiao G, et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet 2017;49:1476–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holstege H, Joosse SA, van Oostrom CT, Nederlof PM, de Vries A, Jonkers J. High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Cancer Res 2009;69:3625–33. [DOI] [PubMed] [Google Scholar]
- 31. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mollon L, Aguilar A, Anderson E, Dean J, Davis L, Warholak T, et al. Abstract 1207: A systematic literature review of the prevalence of PIK3CA mutations and mutation hotspots in HR+/HER2- metastatic breast cancer. Cancer Res 2018;78:1207. [Google Scholar]
- 33. Atchley DP, Albarracin CT, Lopez A, Valero V, Amos CI, Gonzalez-Angulo AM, et al. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol 2008;26:4282–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Comen E, Davids M, Kirchhoff T, Hudis C, Offit K, Robson M. Relative contributions of BRCA1 and BRCA2 mutations to "triple-negative" breast cancer in Ashkenazi Women. Breast Cancer Res Treat 2011;129:185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2010;2:a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meric-Bernstam F, Zheng X, Shariati M, Damodaran S, Wathoo C, Brusco L, et al. Survival outcomes by TP53 mutation status in metastatic breast cancer. JCO Precis Oncol 2018;2018:PO.17.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hodgson D, Lai Z, Dearden S, Barrett JC, Harrington EA, Timms K, et al. Analysis of mutation status and homologous recombination deficiency in tumors of patients with germline BRCA1 or BRCA2 mutations and metastatic breast cancer: OlympiAD. Ann Oncol 2021;32:1582–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Pfizer will provide access to individual deidentified participant data and related study documents [e.g., protocol, statistical analysis plan (SAP), clinical study report (CSR)] upon request from qualified researchers, and subject to certain criteria, conditions, and exceptions.