Abstract
Purpose:
To determine whether metastatic castration–resistant prostate cancers (mCRPC) partition into molecular phenotypes corresponding to intrinsic differentiation states and ascertain whether these subtypes exhibit specific druggable features and associate with treatment outcomes.
Experimental Design:
We used RNAseq, digital spatial profiling, and histological assessments from metastatic biopsies and patient-derived xenografts to segregate mCRPCs into subtypes defined by the PAM50 breast cancer classification algorithm. Subtype associations with treatment responses in preclinical models and patients were determined.
Results:
Using the PAM50 algorithm, we partitioned 270 mCRPC tumors into LumA (42%), LumB (24%), and Basal (34%) subtypes with classification largely driven by proliferation rates and androgen receptor (AR) activity. Most neuroendocrine tumors classified as Basal. Pathways enriched in the LumA subtype include TGFß and NOTCH signaling. LumB subtype tumors were notable for elevated MYC activity. Basal subtype tumors exhibited elevated IL6-STAT3 signaling and features of adult stem cell states. In patients where multiple tumors were evaluated, the majority had concordant PAM50 subtype determination, though a subset exhibited marked inter- and intratumor heterogeneity, including divergent classifications between primary and metastatic sites. In preclinical models, LumA subtype tumors were highly responsive to androgen deprivation and docetaxel chemotherapy whereas Basal tumors were largely resistant. In clinical cohorts patients with Basal subtype tumors demonstrated a shorter time on treatment with AR signaling inhibitors and docetaxel relative to patients with luminal subtypes.
Conclusions:
Subtyping of mCRPC based on cell differentiation states has potential clinical utility for identifying patients with divergent expression of treatment targets and responses to systemic therapy.
Translational Relevance.
Metastatic prostate cancers are recognized to exhibit subtypes categorized by underlying genomic alterations and phenotypes largely partitioned by androgen receptor (AR) signaling and neuroendocrine activity. In the present study, we evaluated a phenotypic classification approach originally developed for subtyping breast carcinomas using the PAM50 gene signature. PAM50 subtypes associated with specific genotypes such as RB1 loss and phenotypes such as small-cell/neuroendocrine carcinoma as well as tumor histology, including cribriform morphology. In the context of clinical translation, PAM50 classification segregated tumors into groups with distinct druggable targets such as cell surface proteins amenable to antibody–drug conjugates. Classification into Luminal A, Luminal B, and Basal tumors is associated with time on AR signaling inhibitors, and responses to taxane chemotherapy. These findings support further clinical investigation of PAM50-based classification for prostate cancer patient stratification in therapeutic studies.
Introduction
Epithelial malignancies such as carcinoma of the prostate, bladder, and breast are well recognized to comprise subtypes that can be distinguished on basis of genomic and/or phenotypic characteristics. In primary tumors, the composite expression of gene sets can serve to reproducibly define “intrinsic” cancer subtypes that may indicate a cell of origin. Importantly, signatures of cancer subtypes serve to group tumors into categories that associate with prognosis and predict outcomes to specific therapeutics. Notable among primary tumor classification systems is the predictive analysis of microarrays 50 (PAM50) gene panel developed from large-scale transcriptome assessments of breast carcinoma (1–3). The PAM50 classifier partitions breast carcinomas into 5 categories: Normal-like (Norm-L), Luminal A (LumA), Luminal B (LumB), Basal-like (Basal) and HER2. These intrinsic subtypes have been shown to add prognostic information to breast cancer staging concerning the risk of relapse after primary treatment, and also predict the efficacy of treatment such as endocrine therapy and chemotherapy (4–7). Furthermore, breast tumors of Basal intrinsic subtype are enriched for specific immune cell signatures, including CD8 T cells, indicating that tumor intrinsic mechanisms may influence tumor microenvironments (8).
Though developed on basis of analyses of breast carcinomas, the PAM50 RNA expression classifier has also demonstrated the capability of partitioning other solid tumors, including Basal and Luminal urothelial cancers, and LumA, LumB and Basal prostate cancers (9–11). As with breast cancers, Basal-like bladder carcinomas exhibit aggressive features with a high frequency of advanced-stage or metastatic disease at presentation (11, 12). When applied to localized prostate cancers, PAM50-based segregation determined that LumB cancers exhibited poor clinical outcomes following primary therapy and the outcome of LumB tumors was favorably influenced by androgen deprivation therapy (ADT; ref. 9). Localized prostate cancers classified by PAM50 as Basal-like are highly enriched for tumors with very low androgen receptor (AR) signaling, a subtype associated with high recurrence rates following primary treatment (13).
In this study, we sought to determine whether PAM50 classification can partition metastatic castration–resistant prostate cancers (mCRPC) into subtypes that potentially reflect cell of origin and whether these subtypes correlate with specific genomic alterations or with previously characterized phenotypes such as tumors with neuroendocrine features. We also sought to determine whether PAM50 tumor classification exhibited heterogeneity across tumors within an individual, and whether particular PAM50 subtypes associated with clinical outcomes after a diagnosis of metastatic disease.
Materials and Methods
Human and animal study approval
All animal procedures were approved by the Fred Hutch Institutional Animal Care and Use Committee (IACUC) and performed in accordance with NIH guidelines. All procedures involving human subjects in the rapid autopsy program were approved by the Institutional Review Board of the University of Washington and of the Fred Hutchinson Cancer Research Center. Samples were obtained from patients who died of metastatic CRPC and who signed written informed consent for a rapid autopsy performed within 6 hours of death, under the aegis of the Prostate Cancer Donor Program at the University of Washington.
Transcriptome analysis
RNAseq data of bulk flash-frozen needle biopsies from the previously published SU2C/PCF cohort were sequenced and aligned as described previously in Abida and colleagues (14). RNAseq data of bulk tumors and digital spatial profiling (DSP) data from the previously published UW mCRPC cohort were sequenced and aligned as described previously in Brady and colleagues (15). Microarray data of laser-capture microdissected tumors from the previously published UW mCRPC cohort were profiled as described previously in Kumar and colleagues (16). LuCaP PDX (patient-derived xenograft) RNAseq data were sequenced and aligned as described previously in Labrecque and colleagues (17). In brief, sequencing reads were mapped to the hg38 human genome using STAR v2.7.3a (18). PDX data were also aligned to the mm10 mouse genome. All subsequent analyses were performed in R. PDX-sequencing reads deriving from mouse were subtracted using XenofilteR. Gene level abundance was quantitated using the GenomicAlignments Bioconductor package (19). Differential expression was assessed using transcript abundances as inputs to the edgeR (19) and limma (20) Bioconductor packages in R, filtered for a minimum expression level using the filterByExpr function with default parameters before testing, and using the Benjamin–Hochberg FDR adjustment. Gene expression results were ranked by their limma statistics and used to conduct Gene Set Enrichment Analysis (21) to determine patterns of pathway activity using the curated pathways from within the MSigDBv7.4 and custom gene sets. Single sample enrichment scores were calculated using GSVA (22) with default parameters using genome-wide log2 FPKM values as input and “AR,” “NE,” CCP,” and “FGF”-MEK” gene signatures, listed in Supplementary Table S13.
Tumors were assigned to PAM50 categories using the classification method described previously by Parker and colleagues (23). We restricted the classification to LumA, LumB and Basal, removing Her2 and Normal samples from the training set and centroid scores before classification. SU2C/PCF cohort polyA and capture RNAseq library data were analyzed separately. The PAM50 categories were merged for comparison with phenotypes and genomic groups by using the highest confidence PAM50 call for tumors on both platforms. For UW rapid autopsy cohort tumors, microarray and RNA-seq data were analyzed separately. PAM50 classification was assigned by patient for the time on docetaxel analysis based on the most frequent PAM50 across multiple tumors per patient or in the case of a tie, the PAM50 with the highest confidence. Microarray data were used for patients where no RNAseq data were available. For analysis of responses to treatment in PDX lines, PAM50 classification was assigned per line based on most frequent or highest confidence call for multiple tumors per line assessed.
Genomic analysis
Bi-allelic loss of RB1 and TP53, and AR amplification and AR mutation in the SU2C/PCF cohort shown in Fig. 1 and Supplementary Tables S1–S4 were determined through custom curation of exome data, as described previously in Nyquist and colleagues (24). Bi-allelic loss of CDK12 and core homology directed DNA repair genes (HRG) as well as COSMIC mutational “Signature 3” (CSig3) status were determined as previously described (25). TMPRSS2-ERG (T2-ERG) fusion status was determined through custom curation by combining consortium calls available at cBioPortal (prad_su2c_2019) with additional calls detected using STAR-fusion v1.9.1. AR splice junctions were extracted from STAR-aligned BAM files using the sjFromSAMcollapseUandM.awk script in the STAR package. Spliced reads were quantified as spliced reads per million (SRPM). AR splice variants detected with SRPM greater than zero were assigned as positive.
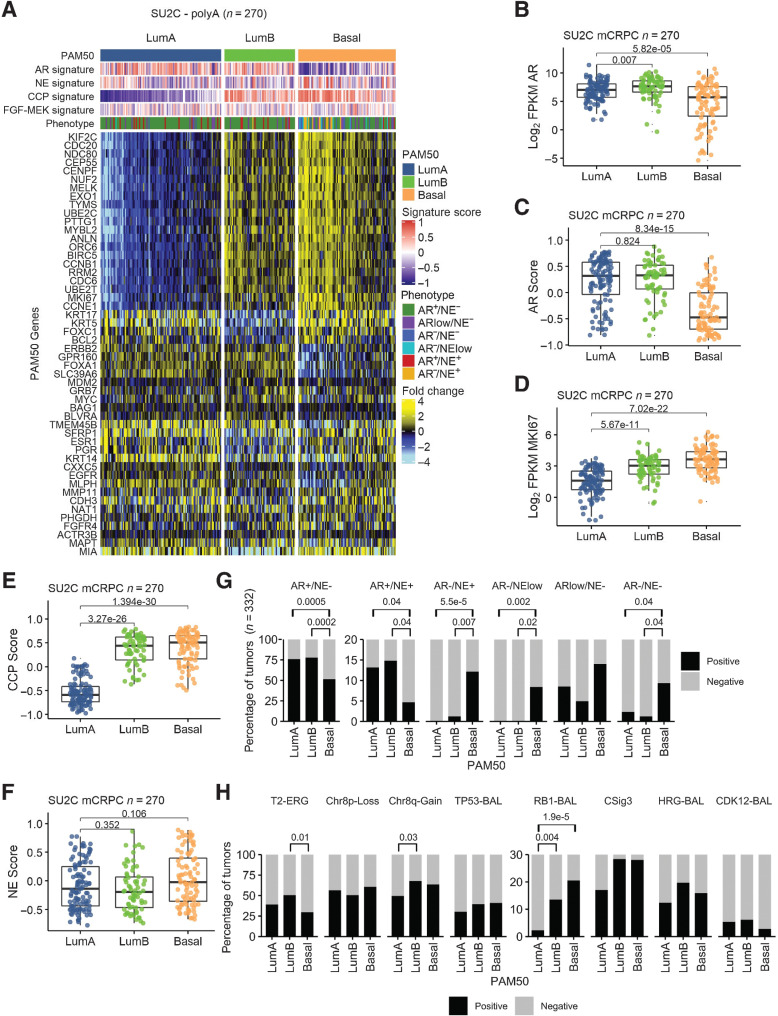
Figure 1.
PAM50 classification of metastatic prostate cancer associates with AR activity, proliferation, and genotype. A, PAM50 classification partitions mCRPC tumors from the SU2C cohort into LumA, LumB, and Basal subtypes. Fold difference scale reflects mean-centered log2 FPKM values from RNAseq. Molecular signature scores and phenotypes shown at the top of plot and colored according to legends at the right side. PAM50 subtypes associate with (B) AR expression and (C) activity and measures of proliferation determined by (D) MKI67 expression and (E) cell-cycle progression (CCP) score. F and G, mCRPCs with neuroendocrine (NE) characteristics are predominantly classified into the PAM50 Basal subtype whereas the common AR active/NE negative mCRPCs are subclassified into LumA, LumB, and Basal subtypes. H, PAM50 subtypes are not associated with common genomic alterations observed in mCRPC except for RB1 loss that is enriched in the PAM50 Basal subtype compared with LumA (P = 1.9e−5) and LumB (P = 0.004), TMPRSS2–ERG fusion event enrichment in LumB compared with Basal (P = 0.013), and Chr8q-gain in LumB compared with LumA tumors (P = 0.03). T2–ERG, TMPRSS2–ERG fusion; RB1-BAL, bi-allelic loss of RB1; TP53-BAL, bi-allelic loss of TP53; CSig3, COSMIC mutational “Signature 3”; HRG-BAL, bi-allelic loss of core homology directed DNA repair genes; CDK12-BAL, bi-allelic loss of CDK12. B–E, and F, Groups compared by Wilcoxon-rank tests with BH adjusted P values shown on plots. G and H, Proportions of groups compared by pairwise Fisher's exact tests with BH-adjusted P values shown on plots. Tumors with PAM50, phenotype, and TMPRSS2--ERG fusion assessment (n = 332). Tumors with PAM50 and genomic assessments (n = 317).
Statistical analysis
Statistical analyses pertaining to each figure are included within the figure legends. For categorical comparisons, we performed the Fisher's exact test or for pairwise comparisons using the fisher.multcomp function in R with Benjamini–Hochberg multiple testing correction. Gene expression (log2 FPKM) or GSVA signature scores versus PAM50 status were compared using Wilcoxon-rank tests with Benjamini–Hochberg multiple testing correction. Pearson's correlation coefficient was used to study the relationships between variables shown in scatterplots using the cor.test function in R. Univariate association tests between PAM50 group or docetaxel treatment and survival/time on treatment were generated from the Cox proportional hazards model. Kaplan–Meier curves were estimated using the survival survfit function in R and plotted using survminer overall survival (OS) from initiation of first line ARSI, time on ARSI therapy, and time on docetaxel. Tumors were stratified by PAM50 group or docetaxel treatment. The log-rank test was used to test for differences between survival curves. Multivariate analyses were performed for the association of PAM50 and clinical variables or AR aberrations as shown in Supplementary Tables S9, S10, and S12 with survival from or time on treatment by Cox proportional hazards model implemented by the coxph function in R.
Histomorphologic assessment
Hematoxylin and eosin–stained formalin-fixed paraffine-embedded tissue sections of 142 tumors for which corresponding RNAseq data were available were evaluated independently by two pathologists (M.P. Roudier and M.C. Haffner). All slides were digitized using a Ventana DP200 scanner (Roche) and tumor morphologies were assessed and categorized into adenocarcinoma, adenocarcinoma with cribriform morphology, high-grade carcinoma not otherwise specified (NOS), high-grade neuroendocrine carcinoma, carcinoid like tumor, hybrid tumor with neuroendocrine and adenocarcinoma features, high-grade carcinoma with pleomorphic giant cells and high-grade carcinoma with squamoid features. Morphology calls were integrated and in cases of discrepancies, slides were re-reviewed to achieve a consensus.
Data availability
The whole-exome sequencing and RNAseq data analyzed in the study are available in the cBioPortal (prad_su2c_2019) and Github under the accession: https://github.com/cBioPortal/datahub/tree/master/public/prad_su2c_2019. The UW mCRPC RNAseq data used in this study are available in the Gene Expression Omnibus repository (GEO) under accession number GSE147250. The DSP transcript data are available from Brady and colleagues (15) in Supplementary Data File S3. LuCaP PDX RNAseq data are available in GEO under accession number GSE199596.
Results
PAM50 gene expression signatures subclassify metastatic prostate cancers
To determine whether mCRPCs can be categorized into intrinsic subtypes with luminal-like and basal-like features, we analyzed RNAseq data from metastatic tumor biopsies from 270 patients with mCRPC (SU2C PolyA RNAseq dataset; refs. 14, 26). We used RNAseq reads to quantitate the expression levels of genes comprising the PAM50 gene set. Using the PAM50 algorithm, 34% (n = 91) of mCRPCs classified as Basal, 42% (n = 113) classified as LumA, and 24% (n = 66) classified as LumB (Fig. 1A). LumA, LumB, and Basal subtypes were also identified using a RNAseq probe-capture dataset of 212 mCRPC tumors (Supplementary Fig. S1A). The representation of mCRPCs broadly recapitulated results from PAM50 classification of localized prostate cancer in TCGA-PRAD: Basal 23%, LumA 35%, LumB 42% and an independent analysis of localized prostate cancer reported by Zhao et al: Basal 34%, LumA 33%, LumB 33% (9).
In addition to the expected expression of luminal lineage markers such as KRT18 and NKX3–1 in LumA and LumB cancers, and basal lineage markers such as ITGA6 in tumors classified as PAM50 Basal, the previous assessments of localized prostate cancer determined that Basal prostate cancers featured tumors with significantly lower levels of AR activity (9). mCRPCs classified as Basal also expressed significantly lower levels of AR and loss of AR activity as determined by a panel of AR-regulated genes (Fig. 1B and C). Notably, LumB and Basal cancers exhibited significantly higher proliferation indices as determined by increased expression of individual genes associated with proliferation such as MKI67 and a 31 gene cell-cycle progression score (Fig. 1D and E; ref. 27). Broadly, the partitioning of mCRPC tumors into LumA, LumB, and Basal subtypes reflects the relative activity of AR and proliferation with LumA categorized by ARHigh;ProliferationLow, LumB categorized by ARHigh;ProliferationHigh and Basal categorized by ARLow;ProliferationHigh.
Using transcriptional signatures that reflect cell differentiation states, we have previously partitioned mCRPCs into six subgroups based on AR and neuroendocrine activity and confirmed that these categories can be ascertained using immunohistochemical stains for cognate proteins (17). We next sought to determine how PAM50-based classification relates to mCRPC classification based on AR/NE program activity. Overall, the expression of NE-associated genes was marginally enriched in Basal tumors that broadly comprises tumors with and without neuroendocrine characteristics (P = 0.1; Fig. 1F). The majority of mCRPCs in this SU2C cohort (68%) classified as AR+/NE− with close to 50% classifying as LumA and the remaining split between LumB (28%) and Basal (25%; Fig. 1G; Supplementary Fig. S1). Three other phenotypic categories are also comprised of each PAM50 subtype with the amphicrine AR+/NE+ subtype represented by a greater proportion of LumA and LumB tumors, and the ARlow/NE− and AR−/NE− subtypes represented by greater proportion of Basal tumors. In contrast, tumors classified as AR−/NE+ and AR−/NElow were almost exclusively PAM50 Basal tumors (93% and 100%, respectively; Fig. 1G; Supplementary Fig. S1).
Deep genomic-profiling studies of mCRPCs have identified recurrent aberrations in cancer drivers and tumor suppressors of which a subset associate with outcomes and others associate with responses to specific therapeutics (14, 26, 28, 29). We next evaluated whether the PAM50 subtypes were enriched for any particular genomic aberration that could provide a mechanistic understanding for phenotype allocation. For most common aberrations such as loss of the TP53 tumor suppressor and mutations in homology-directed DNA repair genes, the representation across PAM50 subtypes was not significantly different (Fig. 1H; Supplementary Fig. S1). However, tumors with RB1 loss were significantly enriched for the Basal (P = 1.9e-05) and LumB (P = 0.004) subtypes. Tumors with gene rearrangements involving the TMPRSS2 and ERG genes (T2-ERG) and Chr8q gain were enriched for the LumB subtype (P = 0.01 and P = 0.03, respectively; Fig. 1H; Supplementary Fig. S1; Supplementary Table S1). Mutations in SPOP did not associate with a PAM50 subtype (Supplementary Tables S1 and S2).
Genomic alterations in the AR also commonly occur in mCRPC. These include AR genomic amplification, gain-of-function point mutations, and the expression of AR splice variants (AR-SV). The frequency of AR copy gains was significantly higher in LumB tumors compared with LumA or Basal subtypes (P < 0.003; Supplementary Tables S1 and S3). The frequency of AR mutations was not associated with any PAM50 group (Supplementary Tables S1 and S4). AR-SVs were detected commonly in all phenotypes ranging from 98% of LumA tumors to 86% of basal tumors (Supplementary Tables S1 and S5). LumA and LumB tumors were enriched for the presence of the AR-V7 splice variant compared with Basal tumors (P < 0.002; Supplementary Tables S1 and S6). In addition, comparison of AR variant expression abundance by PAM50 subtype identified higher levels in the Luminal B tumors (Supplementary Fig. S2A–S2D). The four most frequently detected AR variants in the SU2C cohort polyA transcriptome data were AR-V7 (exon 3-CE3), AR-V3 (exon 2-CE4), AR-V9 (exon3-CE5), and AR exon 2-exon 4. For each of these variants, LumB tumors had significantly higher levels of variant compared with LumA, despite similar detection levels (P = 0.002–0.03). Basal tumors displayed the lowest median expression levels of the four variants.
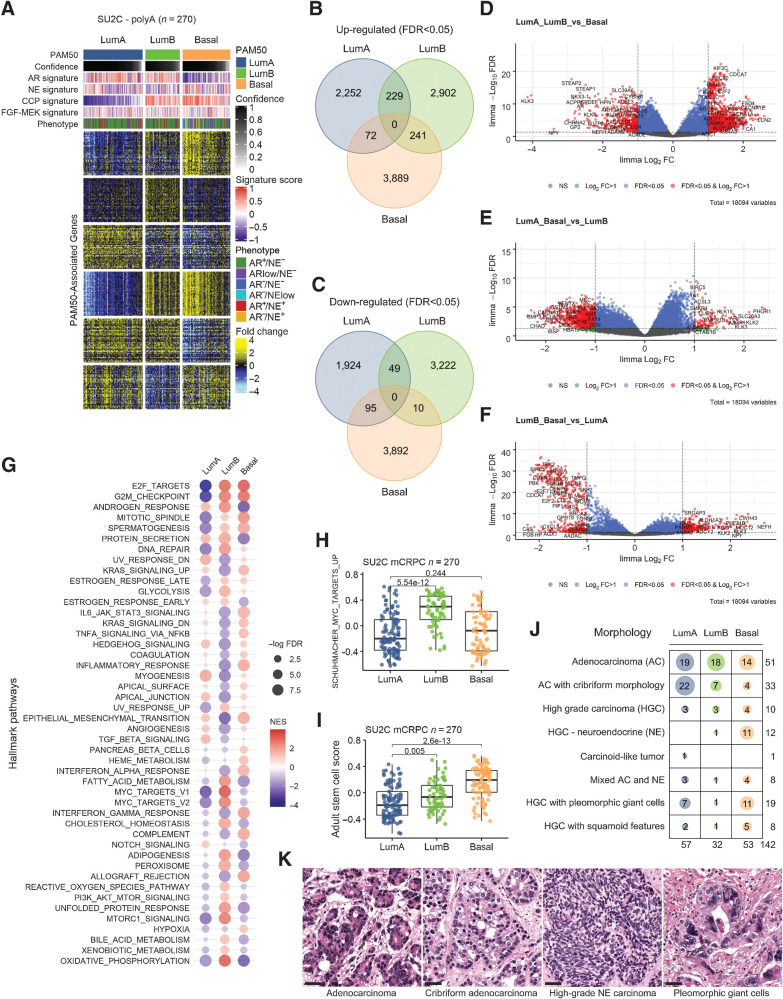
mCRPCs classified by PAM50 exhibit distinctive features
We next sought to identify additional features of the tumor subtypes partitioned on basis of PAM50 classification. Using global RNAseq data to quantitate gene expression, we identified differentially expressed genes that were unique to each PAM50 subtype after excluding the original 50 genes comprising the PAM50 classifier (Fig. 2A; Supplementary Table S7). These comparisons identified 3,889 upregulated and 3,892 downregulated genes in the Basal subtype, 2,252 genes upregulated and 1,924 genes downregulated in the LumA subtype and 2,902 genes upregulated and 3,222 genes downregulated in the LumB subtype (FDR < 0.05; Fig. 2B and C). In agreement with studies of localized prostate cancer, the AR-regulated genes were differentially reduced in the Basal subtype whereas proliferation-associated genes were differentially reduced in the LumA subtype (Fig. 2D–F). Notably, subtype-associated genes were also enriched in other pathways that provide insights into tumor biology and potentially to therapeutic strategies. Pathways enriched in the LumA subtype include TGFß signaling, angiogenesis, and NOTCH signaling with reduced activity of DNA repair and glycolysis pathways (Fig. 2G). LumB subtype tumors are notable for the increase in MYC activity, fatty acid metabolism, cholesterol homeostasis, oxidative phosphorylation, glycolysis and PI3K/MTOR signaling (Fig. 2G and H). Basal subtype tumors exhibit elevated IL6-JAK-STAT3 signaling, TNFA pathway activity, inflammatory response and an increase in gene expression associated with adult stem cell states (Fig. 2G and I; ref. 30).
Figure 2.
PAM50 classification of metastatic prostate cancers associates with distinct molecular pathways and tumor histology. A, Heatmap of gene expression differences between PAM50 subtypes of mCRPC tumors determined by RNAseq. Fold difference scale reflects mean-centered log2 FPKM values from RNAseq. PAM50 output confidence score for classification is shown in black/gray. Molecular signature scores and phenotypes shown at the top of plot and colored according to legends at the right side. B, Venn diagram of transcripts differentially increased in each PAM50 subtype relative to others. Shown are transcripts with FDR < 0.05. C, Venn diagram of transcripts differentially decreased in each PAM50 subtype relative to others. Shown are transcripts with FDR < 0.05. D–F, Volcano plots of differentially expressed genes in pairwise PAM50 comparisons. G, Differentially enriched Hallmark Pathways across PAM50 subtypes of mCRPC. H, Enrichment of MYC-target genes in the PAM50 LumB subtype of mCRPC. I, Enrichment of genes comprising an Adult Stem Cell signature in the PAM50 Basal subtype of mCRPC. H and I, Groups compared by Wilcoxon-rank tests with BH-adjusted P values shown on plots. J, PAM50 subtype associations with histological classification of mCRPC tumors determined by hematoxylin and eosin staining and morphology. Number of tumors exhibiting each morphology displayed on plot. K, Micrographs show mCRPC tumors of the rapid autopsy cohort with adenocarcinoma, cribriform adenocarcinoma, high-grade neuroendocrine (NE) and high-grade carcinoma with pleomorphic giant cells morphology; scale bar, 20 μm. AC, adenocarcinoma; HGC, high-grade carcinoma; NE, neuroendocrine carcinoma.
To determine whether the mCRPC phenotypes partitioned by PAM50 associate with any distinctive morphological features, we evaluated a set of 142 mCRPC tumors acquired from 76 men at the time of rapid autopsy where PAM50 classifications were determined by RNAseq analysis. Consistent with the PAM50 subtype proportions from SU2C RNAseq subtyping, 40% classified as LumA, 23% classified as LumB and 37% classified as Basal. The largest fraction, comprising 36% of these mCRPC tumors, exhibited morphologic features of adenocarcinoma and the PAM50 distribution within these tumors was 37% LumA, 35% LumB and 28% Basal (P = 2.95e−05 for comparisons; Fisher's exact test). We observed that ACs with cribriform features (n = 33) were strongly enriched in the PAM50 LumA subtype (P = 0.0004; Fisher's exact test; Fig. 2J and K). Of tumors with high-grade neuroendocrine carcinoma morphology (including classical small-cell carcinoma), 75% (n = 15) were classified as Basal. Several other rarer variant histologies were identified, including tumors with features of high-grade carcinoma with pleomorphic giant cells and tumors with squamous differentiation: Collectively, 60% of tumors (35 of 58) with variant morphologies classified predominantly into the PAM50 Basal subtype (P = 1.33e−05).
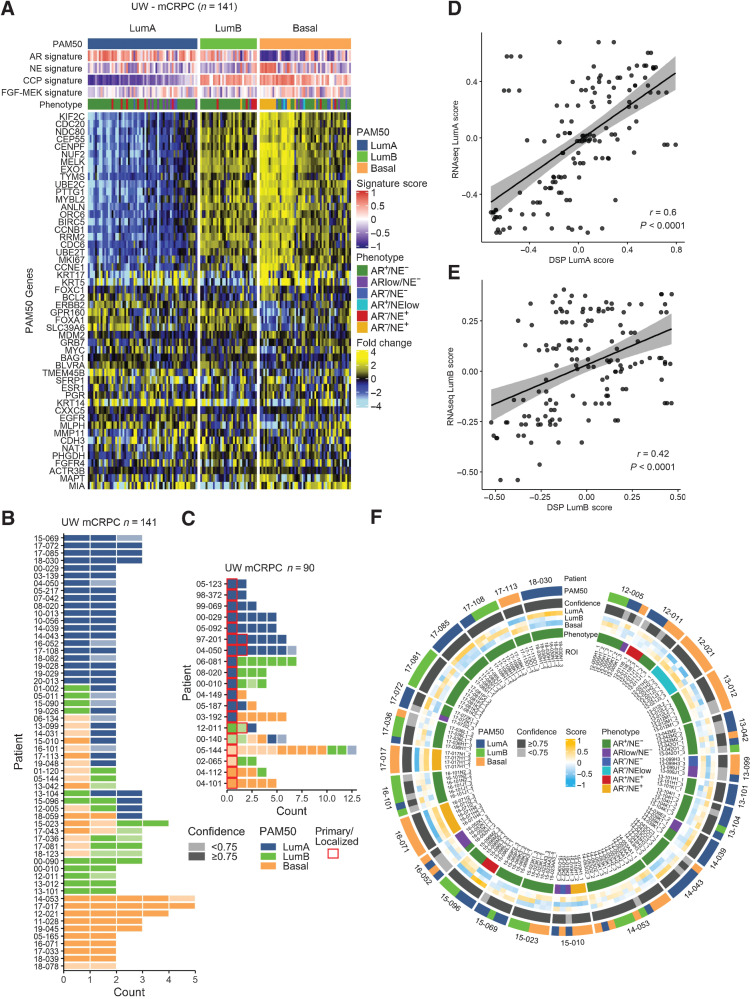
Intraindividual and intratumoral concordance and discordance of PAM50 classification
A notable feature of mCRPC is the induction of cell plasticity and the acquisition of different phenotypes following resistance to AR pathway repression (31–35). To determine whether PAM50 classification varied within an individual patient across different metastatic sites, we evaluated tumors (n = 141) from 58 men with mCRPC who underwent a rapid autopsy and where multiple metastatic tumors were acquired (range, 2–5). This dataset largely overlapped with the cohort described above (Fig. 2J), but excluded 25 men (25 tumors) where only a single tumor was analyzed and included 7 additional men (24 tumors) where detailed morphology was not available. The spectrum of PAM50 classifications also recapitulated the SU2C classifications with 43% LumA, 22% LumB and 35% Basal (Fig. 3A). For 60% of patients (n = 35) all tumors received the same PAM50 classification, supporting previous studies that reported a low overall degree of intra-individual tumor heterogeneity and a monoclonal origin of multiple metastases (Fig. 3B; refs. 16, 36, 37). However, for 40% of men (n = 23) at least one tumor received a discordant classification from other tumors. In addition to assigning a tumor to a particular PAM50 subtype, the PAM50 classification algorithm provides a confidence score for the assignment. For 16 of these discordant cases (70%), the confidence of the PAM50 annotation was below 0.75, indicating that a particular tumor may exhibit intratumor heterogeneity or harbor characteristics that challenge high confidence allocation into a single subtype (Fig. 3B). Consequently, when excluding tumors with lower confidence PAM50 classification (<0.75), concordance across tumors within an individual was 78%.
Figure 3.
Inter- and intratumor heterogeneity of PAM50 classification in mCRPC. A, PAM50 classification partitions mCRPC tumors from the University of Washington rapid autopsy cohort into LumA, LumB, and Basal subtypes. Fold difference scale reflects mean-centered log2 FPKM values from RNAseq. Molecular signature scores and phenotypes shown at the top of plot and colored according to legends at the right side. B and C, RNAseq-based PAM50 classifications of multiple tumors from patients with mCRPC. Tumor counts are the number of tumors evaluated per individual patient. Colors reflect PAM50 classification and shading reflects the confidence of the PAM50 allocation based on the output of the PAM50 algorithm. D and E, Scatterplot of RNAseq-based PAM50 classification and digital spatial profiling (DSP) PAM50 classification from the same metastatic sample. Two-sided test for association using Pearson's correlation coefficient, r; P value shown on plots. F, Overall concordance and discordance of PAM50 tumor classifications shown by individual ROIs. Outer circle shows PAM50 classification in each ROI within and between individual ROIs, second circle shows confidence level of PAM50 classification, individual tumors and individual patients. Inner circle shows tumor phenotype classification by AR/NE gene expression status.
To evaluate the relationships between the phenotypes of primary tumors and corresponding metastases, we identified 19 patients in the UW rapid autopsy cohort where transcript profiles were ascertained by gene expression microarray analysis from both primary tumors and metastases resected at the time of autopsy. These primary tumors were exposed to the same systemic treatments as the metastases. Overall, 68% of the primary prostate cancers classified as LumA, 11% classified as LumB and 21% classified as Basal (Fig. 3C). For 8 patients (42%), the PAM50 classification for the primary tumor and all metastases were concordant. For three patients, two primary tumor samples were evaluated and in each case PAM50 classifications were concordant. Notably, for 6 of the 13 primary prostate cancers classified as LumA, the metastases exhibited a different phenotype: 3 patients with LumB metastases and 3 with Basal metastases, indicative of phenotypic plasticity (Fig. 3C).
To further evaluate the extent of heterogeneity with respect to PAM50 classification, we analyzed a gene expression dataset we acquired by DSP of multiple inter- and intratumoral regions of interest (ROI) from metastatic CRPC tumors (15). This DSP dataset quantitated the expression of 2,106 genes across 141 ROIs (range, 1–3 ROIs/tumor) from 53 tumors from 26 patients. Of the 50 genes comprising the PAM50 classifier, 46 passed a filter for expression in at least one sample above background levels. The 4 genes not detectable were ESR1, PGR, FGFR4 and MIA. We previously demonstrated a high concordance between tumor phenotype classification based on AR− and NE-associated genes using RNAseq and DSP-based transcript quantitation (15). We confirmed that RNAseq and DSP-based PAM50 classification was also largely concordant (r = 0.6 and r = 0.42; P < 0.0001 for LumA and LumB, respectively; Fig. 3D and E), though several tumors exhibited discordant calls, potentially reflecting intratumoral heterogeneity that may be masked by bulk RNAseq. By DSP analysis, all ROIs from all tumors from 11 men (range, 3–6 ROIs; 42%) received the same PAM50 classification (Fig. 3F). Of the remaining 15 men, tumors from 3 individuals were discordant for PAM50 calls, reflecting intra-individual heterogeneity, but the ROIs within each tumor were concordant, reflecting intratumor homogeneity. Of the remaining 12 men, ROIs within a given tumor were discordant for PAM50 classification, reflecting intratumoral heterogeneity. For 9 out of 12 of these tumors (75%) at least one ROI had a PAM50 confidence below 0.75 (Fig. 3F).
To further evaluate the potential mechanisms influencing PAM50 heterogeneity, we curated RB1, TP53, and AR genomic alterations for the matched tumor specimens evaluated by DSP. For the analysis of intra-individual heterogeneity, we scored whether PAM50 or genomic calls were heterogeneous for multiple tumors within an individual and found a significant overlap of heterogeneity in PAM50 subtype and heterogeneity in genomic calls across tumors within an individual (Fisher's exact P = 0.035; Supplementary Table S8A). To assess intratumor relationships, we scored whether PAM50 calls were heterogenous across multiple ROIs within a tumor and compared this assessment with genomic alterations that were clonal versus subclonal as determined by bulk whole-exome sequence analysis. The association between heterogeneous intratumoral PAM50 phenotypes with subclonal genomic alterations in AR, RB1 or TP53 was not significant (Fisher's Exact P = 0.72; Supplementary Table S8B).
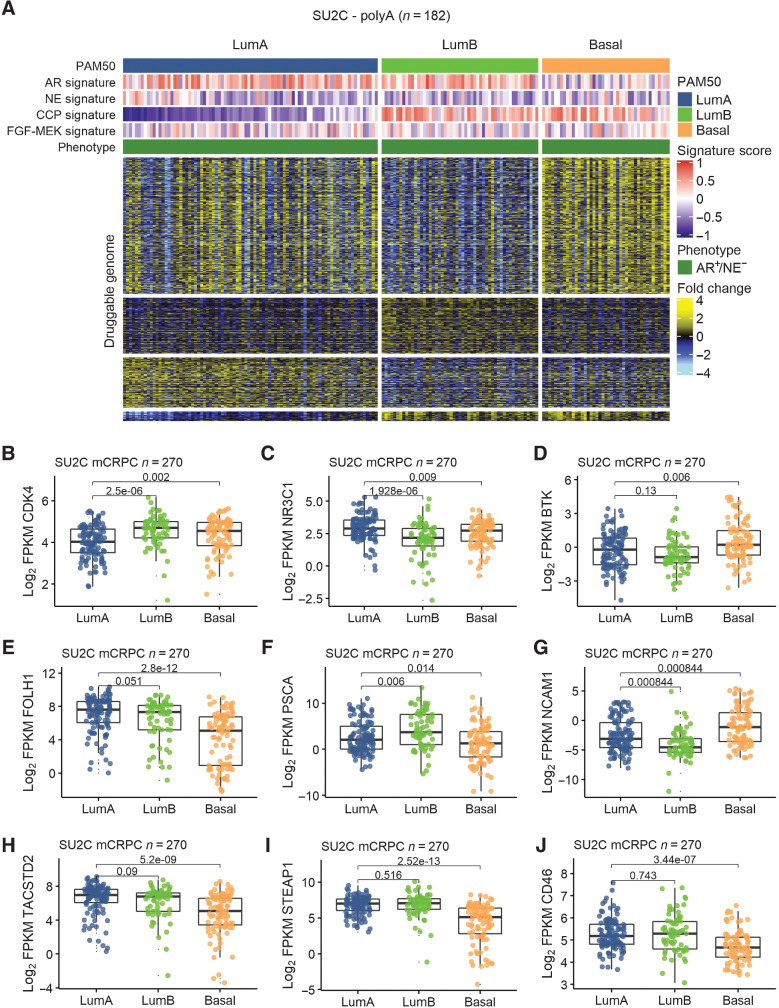
PAM50 classification associates with druggable targets and treatment outcomes
Previous studies have determined that PAM50 classification of localized prostate cancer is associated with differential responses to treatment, with LumB tumors exhibiting a benefit from ADT following radical prostatectomy and docetaxel in the setting of metastatic disease (9, 38). Because of the marked gene expression differences between mCRPC subtypes classified by PAM50 (Fig. 2A), we next sought to determine whether there were additional subtype-associated druggable pathways or potential treatment targets. We profiled the expression of genes comprising the druggable genome based on PAM50 tumor segregation (39). Of 3,687 druggable targets measured, 2,469 were differentially expressed between PAM50 subtypes (FDR < 0.05). For the Basal subtype, a substantial number of the putative druggable targets associated with neuroendocrine differentiation that is well-known to exhibit unique therapeutic targets relative to ARPC such as DLL3, BCL2, AURKA and others (40). We removed all non-AR+/NE− tumors from the dataset leaving 182 tumors, and repeated the analysis. Substantial differences in druggable pathways and genes were confirmed between PAM50 subtypes with 1,134 genes differentially expressed (FDR < 0.05; Fig. 4A). As noted previously, the MYC pathway was significantly increased in LumB tumors (Fig. 2H) as was expression of CDK4 (Fig. 4B), a well-established target in ER-positive HER2-negative breast carcinoma (41). The expression of the glucocorticoid receptor (GR; NR3C1) was differentially upregulated in LumA tumors (Fig. 4C). Previous studies have demonstrated GR activation to promote AR pathway resistance and regulate canonical AR program genes, thus serving as a potential treatment target in mCRPC (42). The expression of Bruton's tyrosine kinase (BTK), a critical regulator of B-cell development (43), was differentially increased in Basal tumors (Fig. 4D). Several BTK inhibitors are clinically approved for the treatment of lymphoma or CLL with clinical trials underway in other malignant and autoimmune diseases (44).
Figure 4.
PAM50-associated druggable targets in mCRPC. A, Heatmap of transcripts encoding putative druggable targets partitioned by PAM50 subtypes in AR+/NE− mCRPC colored according to mean-centered log2 FPKM values. Molecular signature scores and phenotypes shown at the top of plot and colored according to legends at the right side. B–D, PAM50-associated differential expression of small-molecule drug targets CDK4, NR3C1/GR, and BTK. E–J, PAM50-associated differential expression of cell surface proteins as targets for immune-based therapeutics, including chimeric antigen receptor (CAR) T cells and antibody–drug conjugates (ADC). B–J, Groups compared by Wilcoxon-rank tests with BH-adjusted P values shown on plots.
Differentially expressed cell surface proteins have emerged as strong candidates for immune-based therapeutics that include antibody–drug conjugates (ADC) and chimeric antigen receptor–engineered T cells. Several cell surface proteins were differentially expressed across PAM50 subtypes, including STEAP1, expressed highly on LumA and LumB tumors relative to Basal; FOLH1/PSMA expressed highly by LumA subtype tumors; and NCAM1 expressed highly by Basal tumors (Fig. 4E–J).
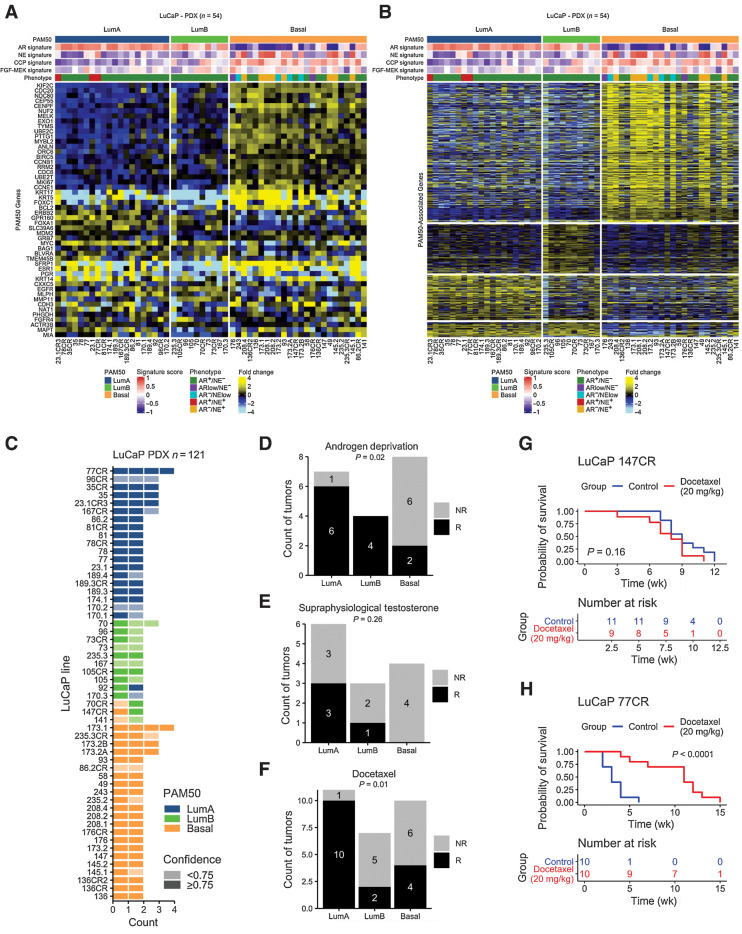
To determine whether prostate cancer subtypes associate with treatment responses, we applied the PAM50 classifier to a panel of prostate cancer PDX using RNAseq data. Of 54 PDX lines, 20 classified as LumA, 10 classified as LumB and 24 classified as Basal (Fig. 5A). All NEPC and AR-low tumors were designated Basal (Fig. 5A and B). We next assessed differential expression between PAM50 groups in PDX tumors and determined overlap with genes previously identified in patient tumors (Fig. 2A, Fig. 5B). There were 905 genes commonly upregulated in both PDX and patient LumA tumors, 1,055 in LumB and 2,502 in Basal (Fig. 5B). We included multiple passages for each line, and PAM50 calls were concordant for all passages for 49 lines (91%; Fig. 5C). For 5 lines, PAM50 subtype classification differed between passages. Notably, for 10/13 PDX pairs with a castration-sensitive and castration-resistant line, the PAM50 calls did not change. The majority of these lines have previously been evaluated for tumor responses to therapeutics that include castration/ADT, supraphysiological testosterone (SPT), and docetaxel chemotherapy (45, 46). PDXs classified as LumA and LumB had significantly greater response rates to ADT compared with Basal subtype tumors (Fig. 5D). Responses to SPT trended toward significance with no PAM50 Basal PDX responding (Fig. 5E). For docetaxel, LumA tumors exhibited high response rates (90%) compared with LumB (43%) or Basal (36%) tumors (P = 0.01; Fig. 5F–H).
Figure 5.
PAM50 subtypes associate with prostate cancer treatment outcomes in preclinical models. A, PAM50 classification and (B) PAM50-associated gene expression differences in common to patient tumors in 54 prostate cancer patient-derived xenograft (PDX) lines (121 tumors). Heatmap of PAM50 signature transcripts colored according to mean-centered log2 FPKM values. Molecular signature scores and phenotypes shown at the top of plot and colored according to legends at the bottom. C, Maintenance of PAM50 subtypes over different PDX passages. D–F, Responses of PDX lines by PAM50 classification to (D) androgen deprivation/castration (P = 0.02); E, Supraphysiological testosterone (P = 0.26). F, Docetaxel chemotherapy (P = 0.01). LuCap147CR is classified as Basal and LuCaP77CR is classified as LumB. Proportions of groups compared by the Fisher's exact test. Number of tumors shown in plots. G and H, Representative plots of response to docetaxel in PDX lines in (G) a nonresponsive basal line, LuCaP 147CR (P = 0.16 by log-rank test), and (H) a responsive LumA line, 77CR (P < 0.0001 by log-rank test). G–H, Number of samples are indicated on plots.
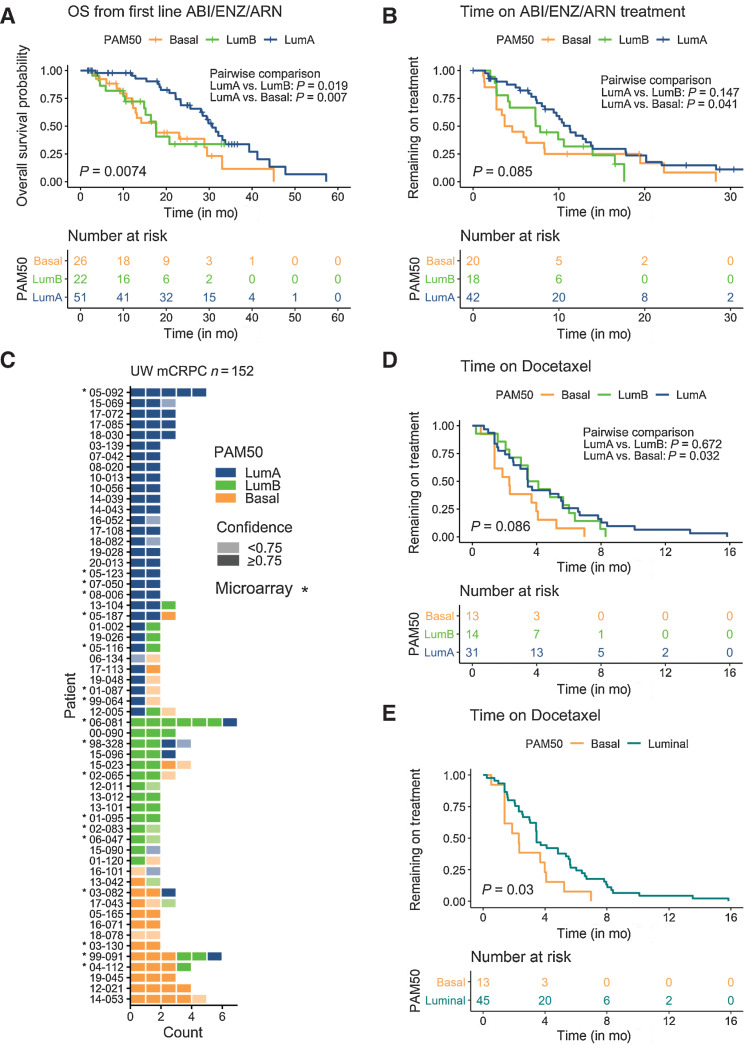
Of the patients in the SU2C mCRPC cohort, a subset (n = 99) received an AR pathway signaling inhibitor such as abiraterone or enzalutamide (14). Patients with tumors classified as LumA had a significantly greater OS of 30.7 months after initiating treatment with one of these agents compared with 17.6 months for patients with LumB tumors (P = 0.019) and 17.7 months for patients with Basal tumors (P = 0.007; Fig. 6A; Supplementary Table S9A). On multivariate analysis with clinical variables, tumors of LumB and Basal subtype and previous chemotherapy associated with shorter OS after initiating abiraterone or enzalutamide (n = 93; Supplementary Table S9B). The median time on treatment was also longer for patients with LumA subtype tumors (11.3 months) compared with Basal (4.1 months) tumors (P = 0.041; Fig. 6B; Supplementary Table S9C). In multivariate analyses with AR alterations, the Basal subtype, AR amplification and AR mutation associated with shorter time on abiraterone and enzalutamide treatment (n = 80; Supplementary Table S10B). Of patients in the UW rapid autopsy cohort, 108 men received docetaxel. Gene expression profiling was available for 89 patients and of these, 58 had more than one tumor evaluated (Fig. 6C; Supplementary Table S11). Time on docetaxel treatment was greater for patients with LumA tumors relative to the Basal subtype (P = 0.032; Fig. 6D; Supplementary Table S12A). When combining LumA and LumB tumors together, time on docetaxel treatment was significantly shorter for patients with Basal compared with Luminal tumors (P = 0.033; Fig. 6E). However, on multivariate analysis no other clinical features associated significantly with shorter time on docetaxel treatment (n = 53; Supplementary Table S12B and S12C).
Figure 6.
PAM50 subtypes associate with prostate cancer treatment outcomes in men with mCRPC. A and B, PAM50 subtype associates with overall survival after the initiation of first-line AR signaling inhibitors and time on treatment by Kaplan–Meier analysis in the SU2C cohort. Number of samples, overall and pairwise log-rank test P values are indicated on plots. C, Gene expression microarray-based PAM50 classifications of multiple tumors from patients with mCRPC. Tumor counts are the number of tumors evaluated per individual patient. Colors reflect PAM50 classification and shading reflects the confidence of the PAM50 allocation based on the output of the PAM50 algorithm. D, Association of PAM50 subtype and time on docetaxel chemotherapy in the UW rapid autopsy cohort (overall and pairwise log-rank P values shown on plot). E, Association of PAM50 subtype and time on docetaxel chemotherapy in the UW rapid autopsy cohort in patients with concordant PAM50 tumor classifications and LumA and LumB tumors combined into a single luminal tumor category (P = 0.03 by log-rank test; A, B, D, and E). Number of samples are indicated on plots.
Discussion
Though developed initially as a classification system for primary breast cancer, subsequent work with the PAM50 classifier has demonstrated that localized carcinomas arising in diverse organ sites can be subclassified into luminal and basal subtypes using the same set of 50 genes (10). In the present work, we have extended this classification approach to metastatic disease, and determined that mCRPCs also retain subtypes with features associating with luminal and basal epithelium with a subdivision within luminal subtyping corresponding to Luminal A and Luminal B features. Other phenotype classification systems for mCRPC have been developed, primarily based on AR activity, and evidence of neuroendocrine gene expression (17). Although essentially all mCRPCs exhibiting neuroendocrine characteristics classify as Basal, it is notable that tumors retaining AR signaling and lacking neuroendocrine gene expression, termed AR+/NE− carcinomas, can be partitioned into all three PAM50 subtypes in near-equal proportions, indicating that AR is not the sole driver of mCRPC phenotype. In a small sample set, the majority of primary tumors and their derived metastasis shared the same classification, a finding that suggests a degree of fixed phenotype, perhaps dictated by distinct cells of origin and/or hardwired features dictated by genomic mutations or structural variants. Notably, comparing isogenic PDX models representing castration-sensitive and castration-resistant variants, the PAM50 classification generally remained consistent. Although it appears that phenotypes generally remain intact despite therapeutic pressures from ADT, docetaxel chemotherapy and AR signaling inhibitors, there were notable instances of discordance between the classification of primary tumors and metastases as well as diversity across metastases. These findings are consistent with previously described phenotypic plasticity involving neuroendocrine transdifferentiation, but also include luminal subtype divergence without the acquisition of neuroendocrine features.
A primary feature distinguishing the luminal subtypes that both exhibit high AR activity is the higher proliferation rate in the LumB subtype, which approximates that of Basal tumors. Both Basal and LumB mCRPCs are enriched for loss of the RB1 tumor suppressor, which may underlie the accelerated cell-cycle rates in a subset of these tumors. A feature distinguishing LumB from LumA tumors is the upregulation of MYC activity and gain of 8q24 which harbors the cMYC gene. PAM50 subtypes also exhibit differential therapeutic targets, particularly the expression of cell surface proteins that serve as biomarkers for radioligands, ADCs, and immune cell directed treatments.
Prior assessments of intra-individual tumor heterogeneity in men with mCRPC based on genomic parameters such as structural variants have reported a high degree of intertumor homogeneity (16, 36, 47). Notable exceptions involve disparate genomic alterations within patients that drive resistance to AR-directed therapeutics (37). We found that phenotypic classification of PAM50 subtypes was also largely concordant within individual patients, though a subset exhibited clear divergence in tumor phenotype. We also found that for a subset of tumors, the assignment to a particular phenotype lacked confidence, indicating that differentiation status may reflect a continuum or possibly other phenotypic features not readily captured by the PAM50 gene set. Further refinement based on additional genes/pathways could provide additional insights into tumor cell states.
A recent study described the subtyping of metastatic prostate cancer into luminal and basal subtypes (48). Although the PAM50 geneset was used, the PAM50 algorithm was not, and the methods used were not able to partition mCRPCs into LumA and LumB subtypes. In agreement with our analyses, patients with mCPRC classified with basal tumors had poorer outcomes on AR signaling inhibitors relative to those with luminal tumors. However, as with breast carcinoma, we find that distinguishing LumA and LumB tumors may be important as these subtypes exhibit different clinical outcomes and are also notable for significant differences in druggable genes and pathways. In contrast with the studies reporting worse outcomes in mCRPC patients with Basal type tumors, an assessment of PAM50 subtypes derived from primary tumors in men with metastatic castration-sensitive prostate cancer found overall worse outcomes to ADT in men with LumB versus Basal tumors, but found men with a LumB subtype benefitted from docetaxel therapy (38).
There are a number of limitations to this study. Associations between PAM50 classification and treatment outcomes were retrospective and not pre-specified. The survival analyses should be interpreted with caution in view of the limited sample sizes involved. Accepted clinical parameters of treatment outcomes such as radiographic assessments were not available and time on drug treatment was used as a surrogate for treatment response. The lack of available data for several known clinical prognostic variables is a limitation and predictive value of phenotype classification for specific therapeutics is not established. Though associations between PAM50 phenotypes and several molecular pathways were significant, the relationships are not yet understood at a mechanistic level. The study population consisted primarily of Caucasian men and the PAM50 subtype classification of mCRPC and treatment outcomes may not be broadly representative across race and ethnicity.
In conclusion, classification of mCRPCs using the PAM50 gene set and algorithm is feasible and reveals mCRPC phenotypes with distinctive features that provide a basis for subtype-specific allocation of therapy.
Supplementary Material
Acknowledgments
We are grateful to the patients who participated in these studies. We thank our colleagues who participated in the SU2C/PCF Dream Team for their contributions to generating datasets used in this study. We thank Lori Kollath and Agnes Gawne for organizing clinical annotations. We thank the IPCR and the patients, clinicians and staff that support the University of Washington autopsy program. This work was supported by the Prostate Cancer Foundation and by a Stand Up to Cancer—Prostate Cancer Foundation Prostate Cancer Dream Team Translational Cancer research grant. Stand Up to Cancer is a division of the Entertainment Industry Foundation. The indicated Stand Up To Cancer grant was administered by the American Association for Cancer Research Award. We also gratefully acknowledge research support from the NCI Cancer Center Support Grant P30CA015704, the Pacific Northwest Prostate Cancer SPORE P50CA97186, P01CA163227, R50CA221836, U54CA224079, W81XWH-18-1–0347 and W81XWH-18-1–0354. N. DeSarkar is a recipient of a CDMRP Prostate Cancer Research Program (PCRP) Early Investigator Research Award (W81XWH-17-1–0380) and PCF-VAL or YI award. Scientific Computing Infrastructure at Fred Hutch is funded by ORIP grant S10OD028685.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
N. De Sarkar reports grants from the Prostate Cancer Foundation and the Department of Defense during the conduct of the study; and grants from The Brotman Baty Institute outside the submitted work; and reports a patent for 21–195-WO-PCT pending. D. Li reports grants from National Institutes of Health, Department of Defense, and Pacific Prostate Cancer SPORE during the conduct of the study. E. Corey reports grants and other support from Bayer Pharmaceuticals, Forma Therapeutics, Kronos Bio, MacroGenics, Janssen Research, Foghorn Therapeutics, Arvinas, and AstraZeneca outside the submitted work. P.S. Nelson reports grants from the National Cancer Institute, Department of Defense, and the Prostate Cancer Foundation during the conduct of the study; and personal fees from Janssen, Bristol Myers Squibb, Pfizer, and Venable LLC outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
I.M. Coleman: Conceptualization, resources, data curation, validation, methodology, writing–original draft, writing–review and editing. N. DeSarkar: Data curation, formal analysis, writing–review and editing. C. Morrissey: Resources, writing–review and editing. L. Xin: Investigation, writing–review and editing. M.P. Roudier: Formal analysis, writing–review and editing. E. Sayar: Formal analysis, writing–review and editing. D. Li: Resources, investigation, writing–review and editing. E. Corey: Resources, writing–review and editing. M.C. Haffner: Formal analysis, writing–review and editing. P.S. Nelson: Conceptualization, resources, supervision, funding acquisition, validation, writing–original draft, project administration, writing–review and editing.
References
- 1. Wallden B, Storhoff J, Nielsen T, Dowidar N, Schaper C, Ferree S, et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genet 2015;8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature 2000;406:747–52. [DOI] [PubMed] [Google Scholar]
- 3. Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001;98:10869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res 2010;16:5222–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chia SK, Bramwell VH, Tu D, Shepherd LE, Jiang S, Vickery T, et al. A 50-gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin Cancer Res 2012;18:4465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prat A, Chaudhury A, Solovieff N, Pare L, Martinez D, Chic N, et al. Correlative biomarker analysis of intrinsic subtypes and efficacy across the MONALEESA phase III studies. J Clin Oncol 2021;39:1458–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fitzal F, Filipits M, Fesl C, Rudas M, Greil R, Balic M, et al. PAM-50 predicts local recurrence after breast cancer surgery in postmenopausal patients with ER+/HER2− disease: results from 1204 patients in the randomized ABCSG-8 trial. Br J Surg 2021;108:308–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hammerl D, Massink MPG, Smid M, van Deurzen CHM, Meijers-Heijboer HEJ, Waisfisz Q, et al. Clonality, antigen recognition, and suppression of CD8(+) T cells differentially affect prognosis of breast cancer subtypes. Clin Cancer Res 2020;26:505–17. [DOI] [PubMed] [Google Scholar]
- 9. Zhao SG, Chang SL, Erho N, Yu M, Lehrer J, Alshalalfa M, et al. Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA Oncol 2017;3:1663–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao SG, Chen WS, Das R, Chang SL, Tomlins SA, Chou J, et al. Clinical and genomic implications of luminal and basal subtypes across carcinomas. Clin Cancer Res 2019;25:2450–7. [DOI] [PubMed] [Google Scholar]
- 11. Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014;25:152–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, Wobker SE, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A 2014;111:3110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spratt DE, Alshalalfa M, Fishbane N, Weiner AB, Mehra R, Mahal BA, et al. Transcriptomic heterogeneity of androgen receptor activity defines a de novo low AR-active subclass in treatment naive primary prostate cancer. Clin Cancer Res 2019;25:6721–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019;116:11428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brady L, Kriner M, Coleman I, Morrissey C, Roudier M, True LD, et al. Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling. Nat Commun 2021;12:1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 2016;22:369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Labrecque MP, Coleman IM, Brown LG, True LD, Kollath L, Lakely B, et al. Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J Clin Invest 2019;129:4492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 2013;9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nyquist MD, Corella A, Coleman I, De Sarkar N, Kaipainen A, Ha G, et al. Combined TP53 and RB1 loss promotes prostate cancer resistance to a spectrum of therapeutics and confers vulnerability to replication stress. Cell Rep 2020;31:107669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. De Sarkar N, Dasgupta S, Chatterjee P, Coleman I, Ha G, Ang LS, et al. Genomic attributes of homology-directed DNA repair deficiency in metastatic prostate cancer. JCI insight 2021;6:e152789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, et al. Prognostic value of an RNA expression signature derived from cell-cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol 2011;12:245–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rodrigues DN, Rescigno P, Liu D, Yuan W, Carreira S, Lambros MB, et al. Immunogenomic analyses associate immunological alterations with mismatch repair defects in prostate cancer. J Clin Invest 2018;128:5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med 2020;382:2091–102. [DOI] [PubMed] [Google Scholar]
- 30. Smith BA, Balanis NG, Nanjundiah A, Sheu KM, Tsai BL, Zhang Q, et al. A human adult stem cell signature marks aggressive variants across epithelial cancers. Cell Rep 2018;24:3353–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beltran H, Hruszkewycz A, Scher HI, Hildesheim J, Isaacs J, Yu EY, et al. The role of lineage plasticity in prostate cancer therapy resistance. Clin Cancer Res 2019;25:6916–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017;355:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 2017;355:84–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 2017;32:474–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bishop JL, Thaper D, Vahid S, Davies A, Ketola K, Kuruma H, et al. The master neural transcription factor BRN2 is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov 2017;7:54–71. [DOI] [PubMed] [Google Scholar]
- 36. Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med 2009;15:559–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, et al. The evolutionary history of lethal metastatic prostate cancer. Nature 2015;520:353–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hamid AA, Huang HC, Wang V, Chen YH, Feng F, Den R, et al. Transcriptional profiling of primary prostate tumor in metastatic hormone-sensitive prostate cancer and association with clinical outcomes: correlative analysis of the E3805 CHAARTED trial. Ann Oncol 2021;32:1157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Finan C, Gaulton A, Kruger FA, Lumbers RT, Shah T, Engmann J, et al. The druggable genome and support for target identification and validation in drug development. Sci Transl Med 2017;9:eaag1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Corella AN, Cabiliza Ordonio MVA, Coleman I, Lucas JM, Kaipainen A, Nguyen HM, et al. Identification of therapeutic vulnerabilities in small-cell neuroendocrine prostate cancer. Clin Cancer Res 2020;26:1667–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wolff AC. CDK4 and CDK6 inhibition in breast cancer—a new standard. N Engl J Med 2016;375:1993–4. [DOI] [PubMed] [Google Scholar]
- 42. Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 2013;155:1309–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zi F, Yu L, Shi Q, Tang A, Cheng J. Ibrutinib in CLL/SLL: from bench to bedside (Review). Oncol Rep 2019;42:2213–27. [DOI] [PubMed] [Google Scholar]
- 44. Zhang D, Gong H, Meng F. Recent advances in BTK inhibitors for the treatment of inflammatory and autoimmune diseases. Molecules 2021;26:4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nguyen HM, Vessella RL, Morrissey C, Brown LG, Coleman IM, Higano CS, et al. LuCaP prostate cancer patient-derived xenografts reflect the molecular heterogeneity of advanced disease and serve as models for evaluating cancer therapeutics. Prostate 2017;77:654–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lam HM, Nguyen HM, Labrecque MP, Brown LG, Coleman IM, Gulati R, et al. Durable response of enzalutamide-resistant prostate cancer to supraphysiological testosterone is associated with a multifaceted growth suppression and impaired DNA damage response transcriptomic program in patient-derived xenografts. Eur Urol 2020;77:144–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Aryee MJ, Liu W, Engelmann JC, Nuhn P, Gurel M, Haffner MC, et al. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci Transl Med 2013;5:169ra10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aggarwal R, Rydzewski NR, Zhang L, Foye A, Kim W, Helzer KT, et al. Prognosis associated with luminal and basal subtypes of metastatic prostate cancer. JAMA Oncol 2021;7:1644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole-exome sequencing and RNAseq data analyzed in the study are available in the cBioPortal (prad_su2c_2019) and Github under the accession: https://github.com/cBioPortal/datahub/tree/master/public/prad_su2c_2019. The UW mCRPC RNAseq data used in this study are available in the Gene Expression Omnibus repository (GEO) under accession number GSE147250. The DSP transcript data are available from Brady and colleagues (15) in Supplementary Data File S3. LuCaP PDX RNAseq data are available in GEO under accession number GSE199596.