Abstract
Purpose:
Uterine leiomyosarcoma is among the most aggressive gynecological malignancies. No effective treatment strategies have been established. This study aimed to identify novel therapeutic targets for uterine leiomyosarcoma based on transcriptome analysis and assess the preclinical efficacy of novel drug candidates.
Experimental Design:
Transcriptome analysis was performed using fresh-frozen samples of six uterine leiomyosarcomas and three myomas. The Ingenuity Pathway Analysis (IPA) was used to identify potential therapeutic target genes for uterine leiomyosarcoma. Afterward, our results were validated using three independent datasets, including 40 uterine leiomyosarcomas. Then, the inhibitory effects of several selective inhibitors for the candidate genes were examined using SK-UT-1, SK-LMS-1, and SKN cell lines.
Results:
We identified 512 considerably dysregulated genes in uterine leiomyosarcoma compared with myoma. The IPA revealed that the function of several genes, including CHEK1 and PLK1, were predicted to be activated in uterine leiomyosarcoma. Through an in vitro drug screening, PLK1 or CHEK1 inhibitors (BI-2536 or prexasertib) were found to exert a superior anticancer effect against cell lines at low nanomolar concentrations and induce cell-cycle arrest. In SK-UT-1 tumor-bearing mice, BI-2536 monotherapy remarkably suppressed tumorigenicity. Moreover, the prexasertib and cisplatin combination therapy inhibited tumor proliferation and prolonged the time to tumor progression.
Conclusions:
We identified upregulated expressions of PLK1 and CHEK1; their kinase activity was activated in uterine leiomyosarcoma. BI-2536 and prexasertib demonstrated a significant anticancer effect. Therefore, cell-cycle–related kinases may present a promising therapeutic strategy for the treatment of uterine leiomyosarcoma.
Translational Relevance.
The development of next-generation sequencing has contributed greatly to cancer research. However, the biological features of uterine leiomyosarcoma are not fully understood. Hence, there is a total absence of effective treatment strategies that are established on the basis of the molecular background of uterine leiomyosarcoma. In this study, we assessed the transcriptional profiles of 46 patients with uterine leiomyosarcoma using three independent datasets and through the assistance of our cohort. The integrative transcriptional analysis showed that the upregulation and activation of cell-cycle–related genes were the dominant features of uterine leiomyosarcoma. Afterward, we demonstrated that PLK1 or CHEK1 inhibition induced cell-cycle arrest and caused DNA damage, which resulted in cell death of leiomyosarcoma-derived cells. Moreover, these drugs portrayed a more significant anticancer effect in the mice model. These findings suggest that cell-cycle–dependent kinases are novel therapeutic targets, and their regulation could potentially improve the prognosis for patients with uterine leiomyosarcoma.
Introduction
Uterine sarcomas are a rare subset of gynecologic malignancies with extremely aggressive behavior. Uterine sarcomas are further classified into one of the following groups: leiomyosarcoma (LMS), endometrial stromal sarcoma, and adenosarcoma (1, 2). Among these sarcomas, LMS is the most common subtype. The annual incidence of uterine LMS (ULMS) is approximately 0.86 per 100,000 women and the majority of the patients are postmenopausal (3, 4). To achieve complete resection, total hysterectomy offers the best chance of cure for localized ULMS. However, the risk of recurrence after complete resection reaches within the range of 50%–70% (1, 2). It is pertinent to note that there is no proven benefit for adjuvant chemotherapy or radiotherapy (1, 2). Furthermore, for recurrent and metastatic ULMS, the docetaxel and gemcitabine combination therapy has been widely used and seems to be partially effective (2). However, in the past few decades, the median overall survival (OS) of patients with metastatic ULMS is found to be one or two years (4, 5). Recently, novel agents, such as trabectedin, pazopanib, and eribulin, were approved for soft-tissue sarcomas. Despite the high therapeutic expectations regarding these agents, the prognosis of patients with ULMS did not considerably improve (6–8). For instance, it has been observed from a subgroup analysis that the median progression-free survival (PFS) and OS for patients with ULMS who were treated with trabectedin were 4.0 and 13.4 months, respectively (6). Similarly, in other clinical trials, the use of pazopanib and eribulin for the treatment of ULMS showed a median OS of 17.5 and 12.7 months, respectively (7, 8). Therefore, the clinical outcome of ULMS remains unsatisfactory.
Recently, the development of next-generation sequencing has enabled the genomic landscape to provide more insight into the pathogenesis of several types of cancer. Several studies have demonstrated that alterations affecting TP53, RB1, ATRX, and PTEN frequently occur in ULMS (9–11). Moreover, in some cases, fusion genes, such as TNS1-ALK, ACTG2-ALK, and KAT6B-KANSL1, have been identified (11, 12). Therefore, the genomic features of ULMS may be indicative of its aggressiveness. In addition, gene expression profiles provide valuable information for understanding the biology of cancers. According to previous studies, LMS can be classed into several subtypes based on the gene expression profile (13–15). For instance, Hemming and colleagues (14) proposed the three subtypes of LMS: conventional, inflammatory, and uterogenic. Moreover, other findings also indicated that the uterogenic subtype was exclusively composed of gynecological LMS; however, ULMS is distributed over all subtypes (13–15). The gynecological subtype of LMS is characterized by the high expressions of ESR1 and PGR, and the protein expressions of ER or PGR were associated with a favorable outcome in ULMS (14, 16). Therefore, the biological characteristics of ULMS are currently being elucidated and have a possibility of being partly different from those of nonuterine LMS (17). Regarding the drug discovery research for ULMS, a previous study identified Aurora A kinase as potential therapeutic target; however, a subsequent phase II study was unable to demonstrate the single-agent activity of the Aurora kinase inhibitor in patients with ULMS (18, 19). Therefore, further studies are of great importance in the development of novel therapeutic agents against ULMS.
In this study, we found that cell-cycle–related genes were upregulated and that their kinase activity was predictively activated in ULMS compared with myoma and normal myometrium. Moreover, subsequent analyses indicate that PLK1 and CHEK1 inhibition strongly induced cell-cycle arrest and exerted superior anticancer effects.
Materials and Methods
Patients
Archival fresh-frozen tumor samples stored at the National Cancer Center Biobank (Tokyo, Japan) were used. Since 2011, six patients with ULMS had undergone surgery without neoadjuvant therapy. The sarcoma and adjacent myometrium tissues of the six patients were collected. Moreover, three patients with benign leiomyoma were included as controls. The study protocol was approved by the ethics committee at our institution (approval No. 2020–160). Written informed consent was obtained from all patients. This study was conducted in accordance with the Declaration of Helsinki.
RNA extraction and transcriptome analysis
Total RNA was extracted from six ULMS and three myoma samples using the miRNeasy Mini Kit (Qiagen). Pair-end sequencing was performed using a DNBSEQ-G400 (MGI Tech) by Azenta. Expression levels for each gene were quantified from the sequencing data by Kallisto (20). Afterward, the data were summarized using the tximport package (ver. 1.18.0) of R software (ver. 4.0.3) and RStudio (RStudio), and scaledTPM counts were used for further analysis. Excluding genes with low-read coverage (maximum read count: <100 reads), 3,070 differentially expressed genes (DEGs, |log2FC| > 1) between the ULMS and myoma samples were used for a heatmap analysis. The heatmap.2 function of the gplots package (ver. 3.1.0) was used after the data were converted to base 10 logarithms and z-scores. For volcano plots, the adjusted P values for each gene were calculated using the Wald test in DESeq2 (ver. 1.30.0) using the data for 23,353 annotated genes. Subsequently, the Ingenuity Pathway Analysis (IPA, Qiagen) was conducted using the significant DEGs identified on the volcano plot.
NCBI GEO and TCGA datasets
The three datasets, including GSE36610, GSE64763, and GSE68295, were downloaded from the NCBI GEO database. The three datasets were microarray-based transcriptional profiles and 10,641 common gene symbols were extracted. The expression data were converted to z-scores, and 1,683 DEGs (the difference between the mean z-scores of ULMS and myometrium is greater than 1) were used to generate the heatmap and principal component analysis (PCA). The PCA was visualized using the prcomp and plot3d functions of the rgl package (ver. 0.100.54). For volcano plot, log2FC and adjusted P values for each gene were calculated for each of the datasets. Afterward, pathway analysis was performed by IPA using the common DEGs identified on the volcano plots.
The TCGA data of LMS were downloaded from the Firehose (https://gdac.broadinstitute.org/) and differences in the gene expression between ULMS and nonuterine LMS were compared. Furthermore, the correlation of the gene expression was evaluated. In addition, the GEPIA (http://gepia.cancer-pku.cn/) was used to investigate gene expression in various cancers (21).
Cell lines
Three ULMS-derived cell lines were used for this study. SKN was purchased from the Japanese Cancer Research Resources Bank, and SK-UT-1 and SK-LMS-1 were purchased from the ATCC. According to the depmap portal (https://depmap.org/portal/), all cell lines contain different missense mutations in TP53. Moreover, SK-UT-1 contains frameshift mutations in RB1 and PTEN, whereas SKN contains a missense mutation in PTEN. SKN cells were maintained in Ham's F12 medium (Sigma-Aldrich) containing 10% FBS (Thermo Fisher Scientific) and antibiotics. SK-UT-1 and SK-LMS-1 cells were maintained in MEM (Nacalai Tesque) containing 10% FBS, 1 mmol/L sodium pyruvate (Thermo Fisher Scientific), and antibiotics. The cell lines tested negative to Mycoplasma contamination and were used between five and 40 passages for the experiments.
Chemicals
All selective kinase inhibitors were purchased from Selleck; their putative targets are shown in Supplementary Fig. S1A. Briefly, BI-2536 and volasertib are PLK1 inhibitors. Prexasertib and PF-477736 are CHEK1/2 inhibitors. Dinaciclib and flavopiridol are pan-CDK inhibitors. AT9283 and tozasertib are pan-Aurora inhibitors. JNJ-7706621 is a pan-CDK and potent Aurora A/B inhibitor, and BAY 1217389 is a TTK inhibitor. A majority of the inhibitors are being investigated in clinical trials in other malignancies. Pazopanib is an approved multi-target inhibitor, whereas olaparib is a selective PARP1/2 inhibitor. In addition, trabectedin (Taiho Pharmaceutical) and cisplatin (Nichi-Iko Pharmaceuticals) were used. Excluding cisplatin, the drugs were dissolved in DMSO as stock solutions and further diluted in the culture medium for experiments.
siRNAs
Silencer Select Pre-designed siRNAs for each gene and Negative Control No.2 siRNA (Thermo Fisher Scientific) were used. The following are the assay IDs: s448 (siPLK1 No. 1), s449 (siPLK1 No. 2), s503 (siCHEK1 No. 1), s504 (siCHEK1 No. 2), s22119 (siCHEK2 No. 1), and s22121 (siCHEK2 No. 2). Cells were transfected with 3 nmol/L siRNA using Lipofectamine RNAi Max (Thermo Fisher Scientific).
Cell viability assay
Cells were seeded into 96-well plates. After attachment, the cells were immediately treated with the inhibitors and incubated for 72 hours. For siRNAs, cells were transfected with the siRNAs and incubated for 24, 48, and 72 hours. Cell viability was assessed using the CellTiter-Glo 2.0 Cell Viability Assay (Promega). Luminescence measurements were taken 10 minutes after adding the reagent using a microplate reader (Molecular Devices). Viability was calculated with the percentage of untreated cells, and experiments were performed in triplicate and repeated three times. Synergy was determined using CompuSyn software (version 1.0; http://www.combosyn.com/index.html). In addition, caspase 3/7 activity was assessed using the Caspase-Glo 3/7 Assay System (Promega) after 48 hours treatment with inhibitors or siRNAs. Luminescence was measured 60 minutes after adding the reagent using the microplate reader.
Cell-cycle assay
Cells were seeded in 6-well plates and grown to approximately 80% confluence. Afterward, cells were treated with selective inhibitors or siRNAs for 24 hours. The cells were harvested and fixed in cold 70% ethanol. The cells were then resuspended in 3% FBS/PBS and stained with ReadiDrop Propidium Iodide (Bio-Rad Laboratories). A cell analyzer EC800 (Sony) was used, and the resulting data were analyzed with FlowJo software (BD Biosciences).
qRT-PCR
Paired ULMS and adjacent normal tissues were used, and cDNA was synthesized using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific). The QuantiTect SYBR Green PCR Kit (Qiagen) or THUNDERBIRD SYBR qPCR Mix (Toyobo) were used. Specific primers were synthesized by Fasmac. The primer sequences and amplification program are shown in Supplementary Fig. S1B.
Western blot analysis
Cells were treated with prexasertib for 16 hours; afterward, total protein extracts were prepared using the M-PER Mammalian Protein Extraction Reagent containing the Halt Protease and Phosphatase Inhibitor Single-Use Cocktail (Thermo Fisher Scientific). After quantification, 10 μg of total protein was separated on Mini-PROTEAN TGX gels (4%–20%, Bio-Rad Laboratories) and transferred to polyvinylidene difluoride membranes. The primary antibodies Chk1 (2G1D5) Mouse mAb #2360, Phospho-Chk1 (Ser296) antibody #2349 (Cell Signaling Technology), and β-actin (C4; MAB1501) were used. Horseradish peroxidase (HRP)–conjugated anti-mouse IgG (NA931) and HRP-conjugated anti-rabbit IgG (NA934) were purchased from GE Healthcare. Protein bands were visualized on ImmunoStar LD (Fujifilm Wako Pure Chemical) and ImageQuant LAS-4000 (Fujifilm).
Immunofluorescence
After 24 hours treatment, cells were fixed with 4% paraformaldehyde and treated with 0.3% Triton-X/Blocking One solution (Nacalai Tesque). Anti-Ki67 antibody (SP6) ab16667 (Abcam), Phospho-Histone H2A.X (Ser139) Antibody #2577 (Cell Signaling Technology), Alexa-Fluor 488 Goat anti-Rabbit IgG (H + L) cross-absorbed secondary antibody (Thermo Fisher Scientific), and Hoechst 33342, trihydrochloride, trihydrate (Thermo Fisher Scientific) were used for the detection and staining. Images were captured using a BZ-X700 fluorescence microscope (Keyence).
Animal studies
All mouse experiments were approved by the National Cancer Center Research Institute, Institute of Laboratory Animal Research (Number: T18-009). Four-week-old female BALB/C nude mice were used for the experiments and 3.0 × 106 SK-UT-1 cells were injected into the right flank of the mice. BI-2536 was dissolved in hydrochloric acid (0.1N) and diluted with 0.9% NaCl. Prexasertib was dissolved in a vehicle (5% DMSO + 40% PEG 300 + 5% Tween80 + ddH2O), whereas cisplatin was diluted with 0.9% NaCl. The drugs were intraperitoneally administered twice a week. The tumor volume was calculated using the modified ellipsoid formula (length × width2 × 0.5). The mice were sacrificed when the tumors reached a volume of 2,000 mm3.
Statistical analysis
Statistical analysis was performed with RStudio and R software (ver. 4.0.3). The Welch's t test was used to determine the significant difference between the means of two sets of data. The paired t test was used to determine the significant difference between the paired LMS and myometrium samples. The Dunnett's test was used for multiple comparisons with the control group using the multicomp package (ver. 1.4-17). To evaluate gene expression correlation, the Pearson correlation coefficient was calculated. The Kaplan–Meier curves and log-rank test were used for the survival analysis. The P value of less than 0.05 was considered statistically significant.
Data availability statement
The data generated in this study are available in Gene Expression Omnibus (GEO) at GSE185543.
Results
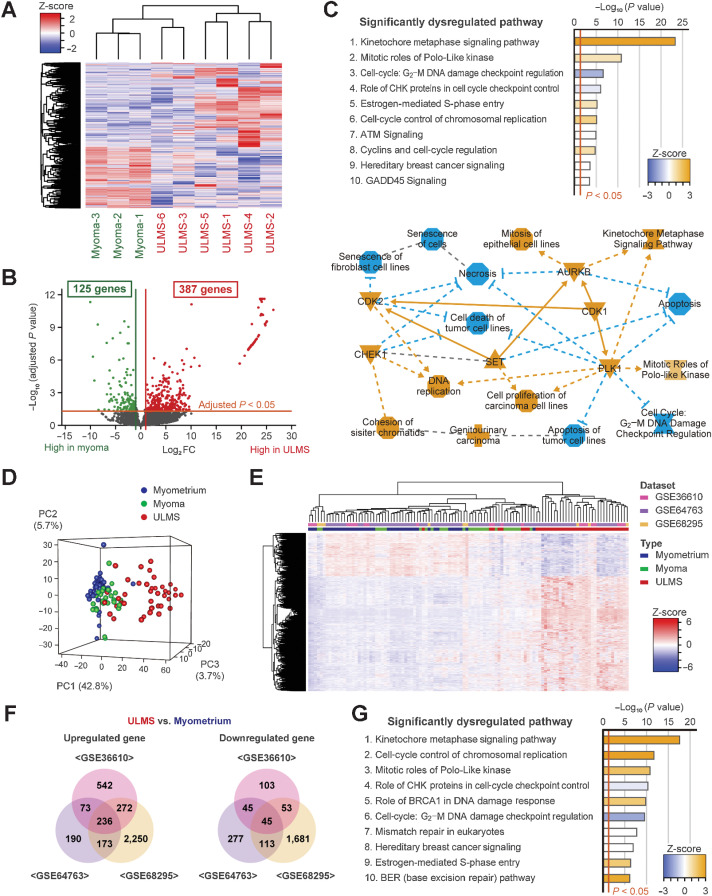
Transcriptome analysis of clinical samples
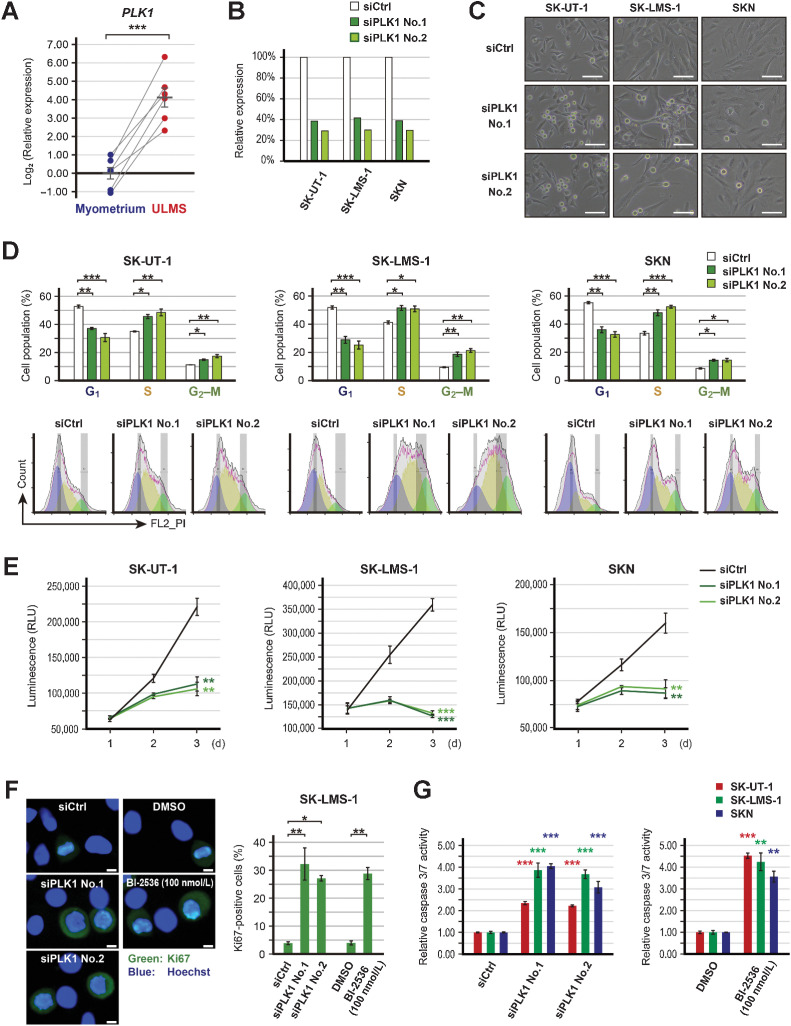
Transcriptome analysis was performed using six ULMS and three myoma samples. Clinical information of the patients is shown in Supplementary Fig. S2A. The heatmap showed that the gene expression profile of ULMS was quite different from that of myoma (Fig. 1A). However, according to hierarchical clustering, the gene expression profile of ULMS-3 and ULMS-6 was similar to that of myoma. We identified that the mitotic rate of ULMS-3 and ULMS-6 was relatively low, and the expression of ESR1 and PGR was higher in ULMS-3 and ULMS-6 than in other patients with ULMS (Supplementary Fig. S2A and S2B). Subsequently, the expression of 23,353 genes was compared by multivariate analysis. There were 387 and 125 significantly upregulated and downregulated genes, respectively, in ULMS based on a cutoff value of |log2FC| > 1 and an adjusted P value of <0.05 (Fig. 1B; Supplementary Table S1). To assess the putative function of the 512 DEGs, pathway analysis was performed using the IPA software, which revealed that several pathways associated with cell-cycle and DNA damage checkpoint were significantly dysregulated. For instance, these pathways included the Kinetochore Metaphase Signaling Pathway (P = 5.01E-24), Mitotic Roles of Polo-Like Kinase (P = 1.58E-11), and cell cycle: G2–M DNA Damage Checkpoint Regulation (P = 2.51E-7; Fig.1C). In addition, from the upstream regulator analysis using IPA, it was observed that the function of CDK1, AURKB, PLK1, CHEK2, CHEK1, CDK2, and PRKDC was significantly activated in ULMS (Fig.1C; Table 1).
Figure 1.
Transcriptome analysis of ULMS using the GEO datasets and our cohort. A, The hierarchical clustering and heatmap showing 3,070 DEGs between ULMS and myoma. The DEGs were defined as an absolute log2 fold change exceeding 1. B, The volcano plot showing significant DEGs between ULMS and myoma. The adjusted P values for each gene were calculated by the Wald test in DESeq2. C, The top 10 significantly dysregulated pathways and the graphical summary based on IPA for the significant DEGs. The orange and blue nodes represent the activated and inhibited genes or pathways, respectively. The orange arrows and blue inhibitory arrows indicate activation and suppression, respectively. D, The principal component analysis. E, The hierarchical clustering and heatmap for GSE36610, GSE64763, and GSE68295 datasets. Each data were converted to z-scores and merged. A total of 1,683 DEGs were used for analyses. F, The Venn diagrams showing the significantly upregulated and downregulated genes between ULMS and myometrium in each dataset. G, The top 10 significantly dysregulated pathways for the three datasets. IPA was performed using the 282 significant DEGs in common.
Table 1.
The list of upstream regulators and their target molecules.
| Upstream regulator (kinase) | Predicted activation state | Target molecules in dataset | P value of overlap |
|---|---|---|---|
| CDK1 | Activated | AURKB, BIRC5, BUB1, BUB1B, CDC20, CDC25A, CDC25C, CDC6, CDT1, FEN1, FOXM1, GAS2L3, H2AX, MCM4, PBK, PLK1, PTTG1, SGO1, STMN1, TTK | 7.63E−10 |
| AURKB | Activated | AURKB, BIRC5, CDCA8, CENPA, DTL, KIF2C, SGO2, SKA1, SKA3, TOP2A | 6.24E−09 |
| PLK1 | Activated | BIRC5, BUB1, BUB1B, CCNB1, CDC20, CDC25C, CEP55, FBXO5, GTSE1, H2AX, HAUS8, KIF2C, KIFC1, PTEN | 2.48E−08 |
| CHEK2 | Activated | BIRC5, CDC25A, CDC25C, CDK1, FOXM1, TTK | 4.84E−05 |
| CHEK1 | Activated | CDC25A, CDC25C, CHEK1, CLSPN, E2F7, E2F8, H2AX | 1.46E−04 |
| TTK | BUB1, BUB1B, KNL1, RMI2 | 1.53E−04 | |
| CDK2 | Activated | CCNA2, CDC25A, CDC6, CDK1, CDT1, CHEK1, KIF11, MCM4, MYBL2, PTTG1 | 5.66E−04 |
| CDKN1A | CDC6, CDK1, CDT1, CHEK1, H2AX | 8.10E−04 | |
| WEE1 | CDK1, H2AX, STMN1 | 0.002 | |
| PRKDC | Activated | CBX5, CHEK1, H2AX, PLK1, TPX2 | 0.002 |
| PLK3 | CDC25C, H2AX, PTEN | 0.003 | |
| MAPK9 | CBX7, CCND3, CDC25A, CDC25C, CHEK1, H2AX | 0.004 | |
| CCNB1 | BIRC5, BUB1, CDC25A, PBK | 0.006 | |
| RPS6KA3 | CHEK1, H2AX, NEK2, SHANK3, STMN1 | 0.009 | |
| BUB1B | BUB1B, CDC20 | 0.013 | |
| RPS6KA2 | CHEK1, FBXO5, NEK2 | 0.016 |
Validation analysis using datasets
To validate our results, we used three GEO datasets. The datasets comprised the expression of 10,641 genes in 40 ULMS, 28 myoma, and 42 normal myometrium samples (Supplementary Fig. S2C). The PCA and heatmap analysis revealed that the transcriptional profile of ULMS is different from that of myoma and normal myometrium (Fig. 1D and E). By comparing ULMS and myometrium samples in each dataset, we discovered that 236 and 45 genes were commonly upregulated and downregulated in ULMS, respectively (Fig. 1F; Supplementary Fig. S2D). The IPA analysis of 281 DEGs validated the activation of the Kinetochore Metaphase Signaling Pathway (P = 2.58E-18) and inhibition of Cell Cycle: G2–M DNA Damage Checkpoint Regulation (P = 2.82E-10; Fig. 1G). In addition, by comparing the ULMS and myoma samples in GSE64763 and GSE68295 datasets, similar results were also obtained (Supplementary Fig. S2D–S1F). Therefore, aberrant activation of the cell-cycle–related pathways in ULMS was a dominant feature. Furthermore, in all comparisons, the overexpression of AURKB, CHEK1, and PLK1 in ULMS was commonly observed (Supplementary Fig. S2G).
In addition, we compared 31 ULMS and 234 nonuterine LMS using the TCGA data. Statistically, the expression of CDK1, CDK2, CHEK1, and TTK was significantly upregulated in ULMS than in nonuterine LMS (Supplementary Fig. S3A). According to previous reports, a subtype with the higher expression of ESR1 and PGR potentially showed a favorable clinical outcome (14, 16). Therefore, we evaluated the difference in the gene expression according to subtypes. The expression of ESR1 and PGR was moderately and positively correlated in ULMS (R2 = 0.663, P < 0.001; Supplementary Fig. S3B). Moreover, the expression of AURKB and PLK1 was negatively correlated with the expression of ESR1 in ULMS (AURKB, R2 = −0.418, P = 0.019; PLK1, R2 = −0.392, P = 0.029).
In vitro screening of selective inhibitors for the activated upstream regulators
We considered the activated key regulators as potential therapeutic targets for ULMS. On the basis of the upstream analysis of our cohort, we considered CDK1, AURKB, PLK1, CHEK2, CHEK1, TTK, and CDK2 as potential therapeutic targets (Table 1). Considering the clinical trials in other malignancies, we selected the inhibitors for the target genes. The anti-cancer effects were assessed using three cell lines derived from ULMS. First, we evaluated the efficacy of pazopanib, an approved drug used in the treatment of malignant soft-tissue tumors. Cells were treated with pazopanib for 72 hours and the IC50 value for SK-UT-1, SK-LMS-1, and SKN cells was 28.3, 56.4, and 4.7 µmol/L, respectively (Supplementary Fig. S4A). Afterward, we assessed the efficacy of selective inhibitors for the target genes and found that most of them were highly effective compared with pazopanib (Fig. 2A and B; Supplementary Fig. S4B). We selected inhibitors with high sensitivity based on the IC50 values and found that PLK1 inhibitors showed cytotoxicity at a lower nanomolar concentration in SK-UT-1 and SK-LMS-1 cells (Fig. 2A). Moreover, CHEK1/2 inhibitors also showed a higher sensitivity in SK-UT-1 cells, with an IC50 value below 10 nmol/L (Fig. 2B). However, both BI-2536 and prexasertib were less effective in SKN cells compared with SK-UT-1 cells, although their effect was at least 10 times greater than that of pazopanib. In addition, dinaciclib, a CDK1/2 inhibitor, was effective, regardless of the type of cell line (Supplementary Fig. S4B). However, dinaciclib was excluded from the subsequent analysis because it failed to prolong the survival of SK-LMS-1 tumor-bearing mice in a previous study (22).
Figure 2.
Inhibitory effects of PLK1 or CHEK1/2 inhibitors. A, The effect of PLK1 inhibitors: BI-2536 and volasertib. B, The effect of CHEK1/2 inhibitors: prexasertib and PF-477736. Cells were treated with each inhibitor for 72 hours. The red, green, and blue colors represent SK-UT-1, SK-LMS-1, and SKN, respectively. The experiments were performed in triplicate and repeated three times. The IC50 value was calculated using the following equation: IC50 = 10[log(A/B) × (50 − C)/(D − C) + log(B)], where A and B represent the highest and the lowest concentrations to cover an estimated IC50 value, respectively, and where C and D represent the cell viability at concentrations B and D, respectively. C, Cell-cycle distribution of BI-2536–treated cells. D, Cell-cycle distribution of prexasertib-treated cells. Cells were treated with each inhibitor for 24 hours. Cell-cycle distribution was calculated by FlowJo. The experiments were performed in three independent replicates and the percentage of cells was compared using the Dunnett's test. Error bars represent standard errors of the mean.
Afterward, we evaluated the effect of BI-2536 and prexasertib on the cell cycle. In the 24-hour treatment, 10 nmol/L BI-2536 showed little effect; however, 100 nmol/L BI-2536 considerably decreased the cell population in the G1 phase but increased that in the S and G2–M phases (Fig. 2C). Similarly, after treatment with 100 nmol/L prexasertib, the cell population in the G1 phase was considerably decreased, whereas that in the S and G2–M phases was considerably increased in all cell lines (Fig. 2D). In particular, SK-UT-1 cells were highly sensitive to prexasertib, such that 10 nmol/L prexasertib was enough to induce cell-cycle arrest.
Effect of PLK1 inhibition
According to the results of the integrative transcriptome analysis and drug screening, PLK1 was found to be the most yielding therapeutic target. In our cohort, the expression of PLK1 was significantly increased in ULMS compared with adjacent normal myometrium (P < 0.001; Fig. 3A). Thus, we performed gene silencing experiments using siRNAs. Two siRNAs for PLK1 reduced the expression of PLK1 to about 30%–40% and increased the number of round-shaped cells (Fig. 3B and C). The cell-cycle analysis revealed that siRNAs for PLK1 considerably decreased the cell population in the G1 phase and increased that in the S and G2–M phases in all cell lines (Fig. 3D). Therefore, PLK1 knockdown significantly hindered the cell proliferation (in SK-UT-1, SK-LMS-1, and SKN; P < 0.01, P < 0.001, and P < 0.01, respectively; Fig. 3E). Immunocytochemistry for Ki67 revealed that BI-2536 and the siRNAs for PLK1 significantly increased the percentage of Ki67-positive cells in SK-LMS-1 cells (Fig. 3F). Moreover, caspase 3/7 activity was significantly increased by BI-2536 and the siRNAs for PLK1 (Fig. 3G).
Figure 3.
Effects of PLK1 silencing. A, The relative expression of PLK1 in paired ULMS and myometrium. ACTB was used as a reference gene to normalize expression, and the relative expression was compared using the paired t test. B, Validation of PLK1 suppression following transfection with 3 nmol/L of siRNA for PLK1 (siPLK1). C, The representative images of siPLK1-transfected cells; scale bars, 100 µm. D, Cell-cycle distribution of siPLK1-transfected cells. Cell-cycle distribution was calculated by FlowJo. The experiments were performed in three independent replicates and the percentage of cells was compared using the Dunnett's test. E, The proliferation of siPLK1-transfected cells. Cell viability was measured at 24, 48, and 72 hours. Experiments were performed in triplicate and repeated three times. The luminescence was compared using the Dunnett's test. F, Immunofluorescent of Ki67 in SK-LMS-1 cells treated with PLK1 inhibition. Cells were treated with PLK1 inhibition for 24 hours. The experiments were performed in three independent replicates. The percentage of Ki67-positive cells was compared using Welch's t or Dunnett's tests. The green and blue colors indicate Ki67 and Hoechst, respectively; scale bars, 10 µm. G, The relative caspase 3/7 activity in cells treated with PLK1 inhibition. Caspase 3/7 activity was measured after 48 hours treatment. Experiments were performed in triplicate and repeated three times. The luminescence was compared using Welch's t or Dunnett's tests. Error bars represent standard errors of the mean, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
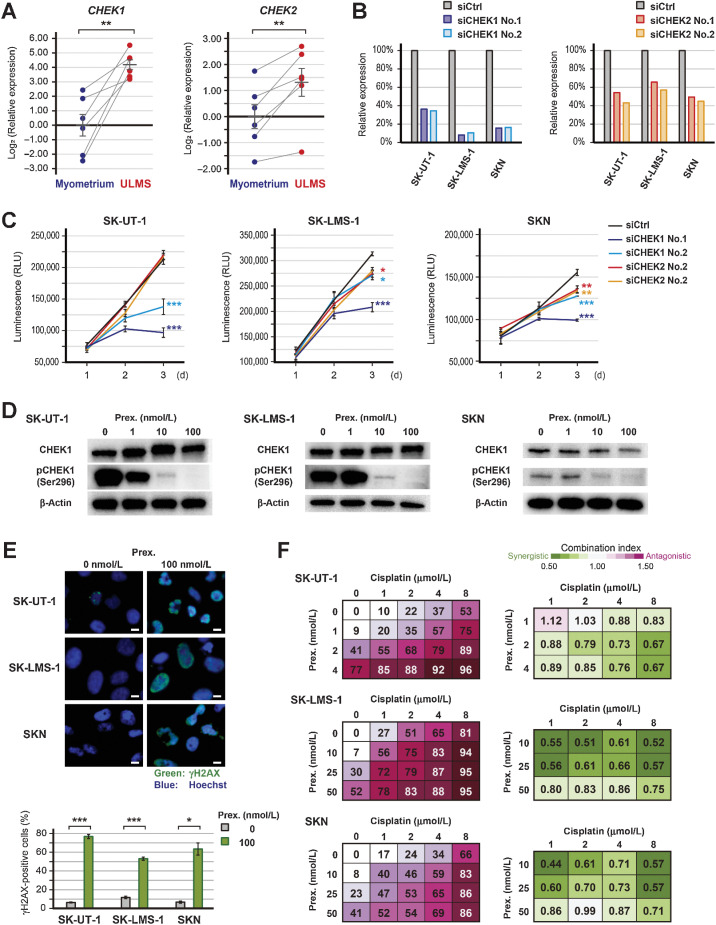
Effect of CHEK1 inhibition
In addition to PLK1 inhibition, CHEK1/2 inhibition is also a promising therapeutic strategy for impaired DNA damage response. Both CHEK1 and CHEK2 were upregulated in ULMS compared with adjacent normal myometrium (P < 0.01 and P < 0.01; Fig. 4A). The siRNA-mediated downregulation of CHEK1 and CHEK2 was confirmed by qRT-PCR, but the effect of the siRNAs was different depending on the cell types (Fig. 4B). In SK-UT-1 cells, the siRNAs for CHEK1 significantly inhibited cell proliferation (P < 0.001), whereas the siRNAs for CHEK2 showed no effect on proliferation (Fig. 4C). In SK-LMS-1 and SKN cells, the siRNAs for both CHEK1 and CHEK2 slightly inhibited the proliferation. Moreover, prexasertib decreased the expression of pCHEK1(Ser296) in a dose-dependent manner (Fig. 4D). Therefore, CHEK1 was more responsible for the prexasertib-induced growth arrest.
Figure 4.
Effects of CHEK1 silencing and prexasertib. A, The relative expression of CHEK1 and CHEK2 in paired ULMS and myometrium. ACTB was used as a reference gene to normalize expression, and the relative expression was compared using the paired t test. B, Validation of CHEK1 or CHEK2 suppression following transfection with 3 nmol/L of siRNA for CHEK1 (siCHEK1) or CHEK2 (siCHEK2). C, The proliferation of siCHEK1-transfected cells. Cell viability was measured at 24, 48, and 72 hours, and luminescence was compared using the Dunnett's test. Experiments were performed in triplicate and repeated three times. D, The expression of CHEK1 and pCHEK1(Ser296) protein in prexasertib-treated cells. Cells were treated with each concentration of prexasertib for 16 hours. E, Immunofluorescent of γH2AX in prexasertib-treated cells. Cells were treated with 0 or 100 nmol/L prexasertib for 24 hours. The experiments were performed in three independent replicates and the percentage of γH2AX-positive cells was compared using the Welch's t test. The green and blue colors indicate γH2AX and Hoechst, respectively; scale bars, 10 µm. F, The combination effect of prexasertib and cisplatin. Cells were treated with each drug concentration for 72 hours, and the percentage of growth inhibition is shown relative to untreated controls. Experiments were performed in triplicate and repeated three times. Drug synergy was analyzed using CompuSyn software. Error bars represent standard errors of the mean, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To confirm the prexasertib-induced DNA damage, immunocytochemistry for phospho-H2AX was performed. In SK-UT-1 and SKN cells, a 24-hour exposure to 100 nmol/L prexasertib caused structural abnormalities in the nucleus (Fig. 4E). Moreover, in all cell lines, the percentage of γH2AX-positive cells was significantly increased by the prexasertib treatment (in SK-UT-1, SK-LMS-1, and SKN; P < 0.001, P < 0.001, and P < 0.05, respectively). Similarly, the siRNAs for CHEK1 also increased the percentage of γH2AX-positive cells in SK-UT-1 cells (Supplementary Fig. S5A). Caspase 3/7 activity was significantly increased by prexasertib and the siRNAs for CHEK1 (Supplementary Fig. S5B).
Afterward, we assessed the effect of the prexasertib and cisplatin combination therapy because increasing DNA damage is expected to enhance the effect of prexasertib. Cells were treated with a combination of various concentrations of prexasertib and cisplatin, and the synergy was determined. As a result, cisplatin synergistically enhanced the effect of prexasertib (Fig. 4F). Similarly, trabectedin synergistically enhanced the effect of prexasertib in SKN cells; however, the effect was addictive in SK-UT-1 and SK-LMS-1 (Supplementary Fig. S5C). Furthermore, the combination index of prexasertib and olaparib was also synergistic (Supplementary Fig. S5D).
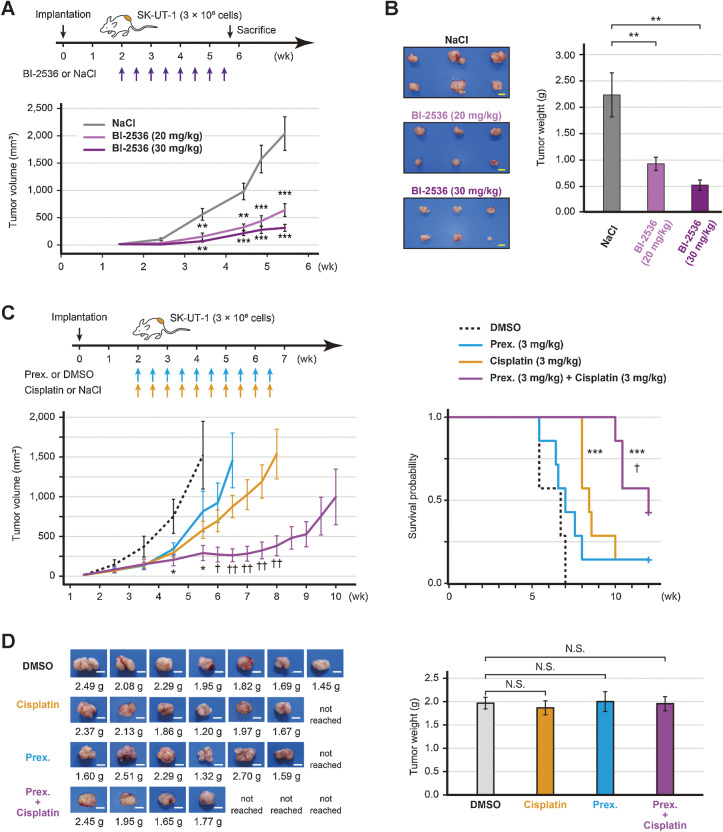
In vivo efficacy of PLK1 and CHEK1 inhibition
Finally, we investigated the in vivo efficacy of the inhibitors. After tumor implantation, SK-UT-1 tumor-bearing mice were treated with either BI-2536 (20 or 30 mg/kg) or saline. It was found that the mice treated with BI-2536 monotherapy significantly decreased the tumor volume (P < 0.001; Fig. 5A; Supplementary Fig. S6A). When the tumor volume of the control mice reached 2,000 mm3, all mice were sacrificed. The mean tumor weight of the high-dose BI-2536, low-dose BI-2536, and control groups was 0.53, 0.93, and 2.24 g, respectively. Therefore, BI-2536 monotherapy significantly decreased the tumor weight in a dose-dependent manner (low-dose and high-dose; P < 0.01 and P < 0.01, respectively; Fig. 5B).
Figure 5.
In vivo efficacy of BI-2536 or prexasertib. A, Estimated tumor volume of SK-UT-1 tumor-bearing mice treated with either BI-2536 monotherapy or saline (n = 6 per group). High-dose (30 mg/kg), low-dose (20 mg/kg) BI-2536, or saline was intraperitoneally administered twice a week for four weeks. B, The representative images of tumors and the mean tumor volume of SK-UT-1 tumor-bearing mice treated with either BI-2536 or saline. The mice were sacrificed when the tumors of the control mice reached a volume of 2,000 mm3. C, Estimated tumor volume and the Kaplan–Meier plot of SK-UT-1 tumor-bearing mice treated with the prexasertib and cisplatin combination therapy (n = 7 per group). Prexasertib monotherapy (3 mg/kg), cisplatin monotherapy (3 mg/kg), the prexasertib (3 mg/kg) and cisplatin (3 mg/kg) combination therapy, or vehicle (DMSO) was intraperitoneally administered twice a week for four weeks. The mice were sacrificed when the tumors reached a volume of 2,000 mm3. The tumor volume and weight were compared using Welch's t test, and survival was compared by a log-rank test. D, The representative images of tumors and the mean tumor volume of SK-UT-1 tumor-bearing mice treated with the prexasertib and cisplatin combination therapy. The tumor weight was compared using the Dunnett's test; scale bars, 1 cm. Error bars represent the standard errors of the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with control mice). †, P < 0.05; ††, P < 0.01 (compared with cisplatin-treated mice); N.S., not significant.
Afterward, we assessed the anticancer effect of the prexasertib (3 mg/kg) and cisplatin (3 mg/kg) combination therapy. We found that prexasertib monotherapy showed no considerable tumor regression effect, whereas cisplatin monotherapy significantly prolonged the time to tumor progression compared with DMSO treatment (P < 0.001; Fig. 5C). In the combination of prexasertib with cisplatin, the anticancer effect was enhanced and marked growth inhibition was observed. Therefore, compared with DMSO treatment, the combination therapy significantly reduced the tumor volume (P < 0.05) and prolonged the time-to-tumor progression (P < 0.001; Fig. 5C; Supplementary Fig. S6B). Following the administration of the combination therapy group, the tumors of three mice did not reach a volume of 2,000 mm3 within 12 weeks. There was no significant difference in the tumor weights of sacrificed mice between the groups. This indicates that the experiment was conducted fairly (Fig. 5D). On the other hand, the survival period in the combination group was significantly longer than that in the cisplatin monotherapy group (P < 0.05; Fig. 5C and D).
During the treatment periods, there was no difference in the body weight of mice treated with low- or high-dose BI-2536 (Supplementary Fig. S6C). However, mice treated with cisplatin monotherapy and the prexasertib and cisplatin combination therapy showed mild body weight loss (Supplementary Fig. S6C). None of the mice were sacrificed due to toxicity. Therefore, BI-2536 monotherapy and the prexasertib and cisplatin combination therapy exerted growth inhibitory effects in the ULMS mouse model.
Discussion
In the present study, we identified the key regulators involved in the cell-cycle and DNA damage response activated in ULMS. Both in vitro and in vivo, the regulators PLK1 and CHEK1 are potential therapeutic targets and their selective inhibitors suppressed the progression of ULMS cells. According to the GEPIA database, PLK1 and CHEK1 are universally upregulated in various cancer tissues compared with normal tissues (Supplementary Fig. S7A and S7B). However, the importance of these genes in ULMS is yet to be discussed in detail.
ULMS is among the most aggressive gynecological malignancies. Therefore, the activation of cell-cycle–related genes in ULMS is consistent with this phenotype. Previous reports have also indicated the alterations in cell-cycle–related signaling pathways in ULMS (9, 19). Thus, the activation of these pathways is a dominant feature of ULMS, which may represent novel therapeutic targets. For instance, Aurora kinase A–targeted therapy hindered the growth of ULMS in preclinical models, which prompted a clinical trial of alisertib, an Aurora kinase inhibitor (18, 19). However, in a phase II trial, including 23 patients with recurrent/persistent ULMS, the median PFS was 1.7 months, which indicates that alisertib did not demonstrate a clinically significant single-agent activity (18). Therefore, it is important to investigate other cell-cycle–related target molecules and drugs.
PLK1 is a highly conserved serine/threonine–protein kinase and is involved in the regulation of cell division (23, 24). It has been reported that PLK1 is overexpressed in various kinds of cancers and that cancer cells often show the elevated activity of PLK1 (23, 24). Therefore, PLK1 is considered a promising therapeutic target. On the basis of this fact, BI-2536, a prototype PLK1 inhibitor, was developed (25). Studies have demonstrated that BI-2536 induces cell-cycle arrest and apoptosis and shows a greater anticancer effect in mice models (25–27). Therefore, several clinical trials have been conducted. In the phase II trial for advanced non–small cell lung cancer (NSCLC), BI-2536 monotherapy showed a modest efficacy and 4.2% of the patients experienced a partial response (28). However, in the other phase II trials, BI-2536 monotherapy showed limited efficacy in various solid tumors (29–31). Afterward, volasertib was developed, and phase II trials demonstrated an insufficient single-agent activity in metastatic urothelial cancer and NSCLC (32, 33). Thus, to maximize the therapeutic effect of PLK inhibitors, several studies have investigated the efficacy of combination therapies. The PLK1- and mTOR-targeting therapy showed a synergistic effect in squamous cell carcinoma; in addition, histone deacetylase inhibitors also enhanced the effect of PLK inhibitors synergistically in prostate cancer cells (34, 35). Moreover, an alternative approach is to explore predictive biomarkers for PLK1 inhibition. In NSCLC, more mesenchymal-like cancer cells were more sensitive to PLK1 inhibitors, whereas PIM1-overexpressing prostate cancer cells were highly sensitive to BI-2536 (36, 37). PIM kinases were reported to play a certain role in the development of sarcoma in an experimental model (38). Therefore, it is expected that ULMS would be highly sensitive to PLK1 inhibition.
CHEK1 and CHEK2 constitute the central regulators of the DNA damage response signaling. In brief, ATR and ATM act as sensors of single-strand and double-strand breaks, respectively, which activate CHEK1 and CHEK2 by phosphorylation. CHEK1 and CHEK2 prevent the removal of phosphates on CDK1 and CDK2 by suppressing CDC25A and CDC25C phosphatases (39). Therefore, the activation of CHEK1 and CHEK2 provides the cell time to repair the DNA damage. In this study, we demonstrated that CHEK1 was more responsible for sarcoma cell proliferation than CHEK2. Moreover, previous reports have indicated that CHEK1 inhibition results in the inappropriate activation of the CDC25A–CDK2 axis, as well as various abnormalities, such as increased double-stranded DNA breaks, accumulation of aberrant replication fork structures, and permission to enter the G2–M phase with damaged DNA (40, 41). Furthermore, a homologous recombination deficiency was observed in 25% (12 of 48) of the patients with ULMS and in LMS cell lines (11, 15). Hence, CHEK1 inhibition may serve as a novel therapeutic candidate for ULMS. Prexasertib (LY2606368) is an ATP-competitive protein kinase inhibitor with a Ki value of 0.9 nmol/L against purified CHEK1. Studies have shown its excellent antitumor effects in a variety of cancer cells (41–45). Consequently, consistent with our results, a synergistic effect of the combination of prexasertib and cytotoxic drugs or PARP inhibitors has been reported (42–45). In a clinical trial, prexasertib monotherapy showed single-agent activity in heavily pretreated squamous cell carcinoma (46). Moreover, another phase II study also showed the efficacy of prexasertib in BRCA wild-type, recurrent high-grade serous ovarian carcinoma (HGSOC), with 8 out of 24 patients showing partial responses (47). However, the clinical efficacy of prexasertib was modest in advanced BRCA wild-type triple-negative breast cancer, despite having similar molecular features with HGSOC (48). The post hoc analysis of HGSOC indicated that prexasertib activity might be associated with the amplification and overexpression of CCNE1 (47). This result is interesting because CCNE1 is amplified and overexpressed in ULMS (9). Thus, we highly anticipated the clinical benefit of prexasertib in patients with ULMS.
According to clinical trials, the toxicity of BI-2536, volasertib, and prexasertib was well tolerated. In patients administered BI-2536 or volasertib, neutropenia was the most frequently observed adverse event and about 30%–40% of the patients experienced grade III or IV neutropenia (28–30, 32, 33). Similarly, hematological adverse events were frequently seen in patients treated with prexasertib and almost all patients experienced grade III or IV neutropenia (46–48). Therefore, the hematological toxicity of these drugs should be kept in mind. However, it is important to note that all clinical trials have concluded the safety profile.
Our study had several limitations. First, we did not assess the regulatory mechanisms underlying the activation of the cell-cycle in ULMS. This would be interesting because it may lead to the identification of novel therapeutic targets. Second, kinase inhibitors potentially have off-target effects. In this study, we showed the importance of PLK1 or CHEK1 using siRNAs in ULMS cells but it was difficult to rule out potential off-target effects. Third, the IC50 values of the inhibitors differed according to the cell lines. Generally, cell lines are unable to completely recapitulate their primary tumor. Therefore, we considered that the genetic background and the resulting cell-cycle speed may contribute to the response to the kinase inhibitors. However, the associating factors implicated in the response to the inhibitors remain largely unknown. Finally, the clinical impact of BI-2536 and prexasertib in ULMS remains uncertain. According to the result from Phase I trials, the peripheral blood concentration of BI-2536 and prexasertib could reach more than 100 nmol/L after 24 hours of drug administration (49, 50). These results indicated that the drugs may attain a sufficient concentration to induce cell-cycle arrest of ULMS cells in humans. Therefore, the clinical effect of the drugs is highly expected and should be evaluated in clinical trials. Moreover, other cell-cycle genes are promising targets for cancer therapy (40). In particular, CDK inhibition is an attractive treatment strategy. However, according to a study, 20 mg/kg dinaciclib could not prolong the SK-LMS-1 tumor-bearing mice (22). Therefore, further optimization of CDK inhibitors would be required for clinical application.
In conclusion, the overexpression of PLK1 and CHEK1 is among the hallmarks of ULMS. Both BI-2536 and prexasertib strongly induced cell-cycle arrest and inhibited the proliferation of ULMS cells. Therefore, PLK1 or CHEK1 inhibition is a promising therapeutic strategy that might improve clinical outcomes in patients with ULMS.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the Program for Promoting the Enhancement of Research Universities as young researcher units for the advancement of new and undeveloped fields at Nagoya University. We thank the National Cancer Center Biobank for providing biological resources. We received technical support from Yuko Fujiwara at the Laboratory of Molecular Carcinogenesis, National Cancer Center Research Institute. Moreover, we thank Enago (www.enago.jp) for the English language review. This study was supported by JSPS KAKENHI grant numbers 21H02721, 21H03075, and 21K16789. Moreover, YOKOYAMA Foundation for Clinical Pharmacology (YRY-2115), Japan Research Foundation for Clinical Pharmacology (2021A18), and Foundation for Promotion of Cancer Research in Japan supported as well. Furthermore, this work is partially supported by Nagoya University Research Fund.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
K. Yoshida: Conceptualization, data curation, formal analysis, funding acquisition, investigation, visualization, methodology, writing–original draft. A. Yokoi: Conceptualization, supervision, funding acquisition, writing–review and editing. T. Yamamoto: Investigation, methodology. Y. Hayashi: Investigation, methodology. J. Nakayama: Software, formal analysis, investigation, methodology. T. Yokoi: Writing–review and editing. H. Yoshida: Resources. T. Kato: Conceptualization, resources. H. Kajiyama: Conceptualization, supervision. Y. Yamamoto: Conceptualization, supervision, funding acquisition, visualization, writing–review and editing.
References
- 1. Roberts ME, Aynardi JT, Chu CS. Uterine leiomyosarcoma: a review of the literature and update on management options. Gynecol Oncol 2018;151:562–72. [DOI] [PubMed] [Google Scholar]
- 2. George S, Serrano C, Hensley ML, Ray-Coquard I. Soft tissue and uterine leiomyosarcoma. J Clin Oncol 2018;36:144–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Skorstad M, Kent A, Lieng M. Uterine leiomyosarcoma - incidence, treatment, and the impact of morcellation. A nationwide cohort study. Acta Obstet Gynecol Scand 2016;95:984–90. [DOI] [PubMed] [Google Scholar]
- 4. Seagle BL, Sobecki-Rausch J, Strohl AE, Shilpi A, Grace A, Shahabi S. Prognosis and treatment of uterine leiomyosarcoma: a National Cancer Database study. Gynecol Oncol 2017;145:61–70. [DOI] [PubMed] [Google Scholar]
- 5. Abeler VM, Røyne O, Thoresen S, Danielsen HE, Nesland JM, Kristensen GB. Uterine sarcomas in Norway. A histopathological and prognostic survey of a total population from 1970 to 2000 including 419 patients. Histopathology 2009;54:355–64. [DOI] [PubMed] [Google Scholar]
- 6. Hensley ML, Patel SR, von Mehren M, Ganjoo K, Jones RL, Staddon A, et al. Efficacy and safety of trabectedin or dacarbazine in patients with advanced uterine leiomyosarcoma after failure of anthracycline-based chemotherapy: subgroup analysis of a phase 3, randomized clinical trial. Gynecol Oncol 2017;146:531–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blay JY, Schöffski P, Bauer S, Krarup-Hansen A, Benson C, D'Adamo DR, et al. Eribulin versus dacarbazine in patients with leiomyosarcoma: subgroup analysis from a phase 3, open-label, randomised study. Br J Cancer 2019;120:1026–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Benson C, Ray-Coquard I, Sleijfer S, Litière S, Blay JY, Le Cesne A, et al. Outcome of uterine sarcoma patients treated with pazopanib: a retrospective analysis based on two European Organisation for Research and Treatment of Cancer (EORTC) Soft Tissue and Bone Sarcoma Group (STBSG) clinical trials 62043 and 62072. Gynecol Oncol 2016;142:89–94. [DOI] [PubMed] [Google Scholar]
- 9. Cuppens T, Moisse M, Depreeuw J, Annibali D, Colas E, Gil-Moreno A, et al. Integrated genome analysis of uterine leiomyosarcoma to identify novel driver genes and targetable pathways. Int J Cancer 2018;142:1230–43. [DOI] [PubMed] [Google Scholar]
- 10. Hensley ML, Chavan SS, Solit DB, Murali R, Soslow R, Chiang S, et al. Genomic landscape of uterine sarcomas defined through prospective clinical sequencing. Clin Cancer Res 2020;26:3881–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choi J, Manzano A, Dong W, Bellone S, Bonazzoli E, Zammataro L, et al. Integrated mutational landscape analysis of uterine leiomyosarcomas. Proc Natl Acad Sci U S A 2021;118:e2025182118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mas A, Alonso R, Garrido-Gómez T, Escorcia P, Montero B, Jiménez-Almazán J, et al. The differential diagnoses of uterine leiomyomas and leiomyosarcomas using DNA and RNA sequencing. Am J Obstet Gynecol 2019;221:320.e1–e23. [DOI] [PubMed] [Google Scholar]
- 13. Guo X, Jo VY, Mills AM, Zhu SX, Lee CH, Espinosa I, et al. Clinically relevant molecular subtypes in leiomyosarcoma. Clin Cancer Res 2015;21:3501–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hemming ML, Fan C, Raut CP, Demetri GD, Armstrong SA, Sicinska E, et al. Oncogenic gene-expression programs in leiomyosarcoma and characterization of conventional, inflammatory, and uterogenic subtypes. Mol Cancer Res 2020;18:1302–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anderson ND, Babichev Y, Fuligni F, Comitani F, Layeghifard M, Venier RE, et al. Lineage-defined leiomyosarcoma subtypes emerge years before diagnosis and determine patient survival. Nat Commun 2021;12:4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leitao MM Jr, Hensley ML, Barakat RR, Aghajanian C, Gardner GJ, Jewell EL, et al. Immunohistochemical expression of estrogen and progesterone receptors and outcomes in patients with newly diagnosed uterine leiomyosarcoma. Gynecol Oncol 2012;124:558–62. [DOI] [PubMed] [Google Scholar]
- 17. Cancer Genome Atlas Research Network. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell 2017;171:950–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hyman DM, Sill MW, Lankes HA, Piekarz R, Shahin MS, Ridgway MR, et al. A phase 2 study of alisertib (MLN8237) in recurrent or persistent uterine leiomyosarcoma: an NRG oncology/gynecologic oncology group study 0231D. Gynecol Oncol 2017;144:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shan W, Akinfenwa PY, Savannah KB, Kolomeyevskaya N, Laucirica R, Thomas DG, et al. A small-molecule inhibitor targeting the mitotic spindle checkpoint impairs the growth of uterine leiomyosarcoma. Clin Cancer Res 2012;18:3352–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34:525–7. [DOI] [PubMed] [Google Scholar]
- 21. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rello-Varona S, Fuentes-Guirado M, López-Alemany R, Contreras-Pérez A, Mulet-Margalef N, García-Monclús S, et al. Bcl-x(L) inhibition enhances dinaciclib-induced cell death in soft-tissue sarcomas. Sci Rep 2019;9:3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schöffski P. Polo-like kinase (PLK) inhibitors in preclinical and early clinical development in oncology. Oncologist 2009;14:559–70. [DOI] [PubMed] [Google Scholar]
- 24. Archambault V, Lépine G, Kachaner D. Understanding the Polo Kinase machine. Oncogene 2015;34:4799–807. [DOI] [PubMed] [Google Scholar]
- 25. Steegmaier M, Hoffmann M, Baum A, Lénárt P, Petronczki M, Krssák M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol 2007;17:316–22. [DOI] [PubMed] [Google Scholar]
- 26. Ding Y, Huang D, Zhang Z, Smith J, Petillo D, Looyenga BD, et al. Combined gene expression profiling and RNAi screening in clear cell renal cell carcinoma identify PLK1 and other therapeutic kinase targets. Cancer Res 2011;71:5225–34. [DOI] [PubMed] [Google Scholar]
- 27. Maire V, Némati F, Richardson M, Vincent-Salomon A, Tesson B, Rigaill G, et al. Polo-like kinase 1: a potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res 2013;73:813–23. [DOI] [PubMed] [Google Scholar]
- 28. Sebastian M, Reck M, Waller CF, Kortsik C, Frickhofen N, Schuler M, et al. The efficacy and safety of BI 2536, a novel Plk-1 inhibitor, in patients with stage IIIB/IV non–small cell lung cancer who had relapsed after, or failed, chemotherapy: results from an open-label, randomized phase II clinical trial. J Thorac Oncol 2010;5:1060–7. [DOI] [PubMed] [Google Scholar]
- 29. Mross K, Dittrich C, Aulitzky WE, Strumberg D, Schutte J, Schmid RM, et al. A randomised phase II trial of the Polo-like kinase inhibitor BI 2536 in chemo-naïve patients with unresectable exocrine adenocarcinoma of the pancreas—a study within the Central European Society Anticancer Drug Research (CESAR) collaborative network. Br J Cancer 2012;107:280–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Awad MM, Chu QS, Gandhi L, Stephenson JJ, Govindan R, Bradford DS, et al. An open-label, phase II study of the polo-like kinase-1 (Plk-1) inhibitor, BI 2536, in patients with relapsed small-cell lung cancer (SCLC). Lung Cancer 2017;104:126–30. [DOI] [PubMed] [Google Scholar]
- 31. Schöffski P, Blay JY, De Greve J, Brain E, Machiels JP, Soria JC, et al. Multicentric parallel phase II trial of the polo-like kinase 1 inhibitor BI 2536 in patients with advanced head and neck cancer, breast cancer, ovarian cancer, soft tissue sarcoma and melanoma. The first protocol of the European Organization for Research and Treatment of Cancer (EORTC) Network Of Core Institutes (NOCI). Eur J Cancer 2010;46:2206–15. [DOI] [PubMed] [Google Scholar]
- 32. Stadler WM, Vaughn DJ, Sonpavde G, Vogelzang NJ, Tagawa ST, Petrylak DP, et al. An open-label, single-arm, phase 2 trial of the Polo-like kinase inhibitor volasertib (BI 6727) in patients with locally advanced or metastatic urothelial cancer. Cancer 2014;120:976–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ellis PM, Leighl NB, Hirsh V, Reaume MN, Blais N, Wierzbicki R, et al. A randomized, open-label phase II trial of volasertib as monotherapy and in combination with standard-dose pemetrexed compared with pemetrexed monotherapy in second-line treatment for non–small cell lung cancer. Clin Lung Cancer 2015;16:457–65. [DOI] [PubMed] [Google Scholar]
- 34. Wissing MD, Mendonca J, Kortenhorst MS, Kaelber NS, Gonzalez M, Kim E, et al. Targeting prostate cancer cell lines with polo-like kinase 1 inhibitors as a single agent and in combination with histone deacetylase inhibitors. FASEB J 2013;27:4279–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu TT, Yang KX, Yu J, Cao YY, Ren JS, Hao JJ, et al. Co-targeting PLK1 and mTOR induces synergistic inhibitory effects against esophageal squamous cell carcinoma. J Mol Med 2018;96:807–17. [DOI] [PubMed] [Google Scholar]
- 36. Ferrarotto R, Goonatilake R, Yoo SY, Tong P, Giri U, Peng S, et al. Epithelial-mesenchymal transition predicts polo-like kinase 1 inhibitor-mediated apoptosis in non–small cell lung cancer. Clin Cancer Res 2016;22:1674–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van der Meer R, Song HY, Park SH, Abdulkadir SA, Roh M. RNAi screen identifies a synthetic lethal interaction between PIM1 overexpression and PLK1 inhibition. Clin Cancer Res 2014;20:3211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Narlik-Grassow M, Blanco-Aparicio C, Cecilia Y, Peregrina S, Garcia-Serelde B, Muñoz-Galvan S, et al. The essential role of PIM kinases in sarcoma growth and bone invasion. Carcinogenesis 2012;33:1479–86. [DOI] [PubMed] [Google Scholar]
- 39. Lin AB, McNeely SC, Beckmann RP. Achieving precision death with cell-cycle inhibitors that target DNA replication and repair. Clin Cancer Res 2017;23:3232–40. [DOI] [PubMed] [Google Scholar]
- 40. Otto T, Sicinski P. Cell-cycle proteins as promising targets in cancer therapy. Nat Rev Cancer 2017;17:93–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. King C, Diaz HB, McNeely S, Barnard D, Dempsey J, Blosser W, et al. LY2606368 causes replication catastrophe and antitumor effects through CHK1-dependent mechanisms. Mol Cancer Ther 2015;14:2004–13. [DOI] [PubMed] [Google Scholar]
- 42. Parmar K, Kochupurakkal BS, Lazaro JB, Wang ZC, Palakurthi S, Kirschmeier PT, et al. The CHK1 inhibitor prexasertib exhibits monotherapy activity in high-grade serous ovarian cancer models and sensitizes to PARP inhibition. Clin Cancer Res 2019;25:6127–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lowery CD, Dowless M, Renschler M, Blosser W, VanWye AB, Stephens JR, et al. Broad spectrum activity of the checkpoint kinase 1 inhibitor prexasertib as a single agent or chemopotentiator across a range of preclinical pediatric tumor models. Clin Cancer Res 2019;25:2278–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sen T, Tong P, Stewart CA, Cristea S, Valliani A, Shames DS, et al. CHK1 inhibition in small-cell lung cancer produces single-agent activity in biomarker-defined disease subsets and combination activity with cisplatin or olaparib. Cancer Res 2017;77:3870–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heidler CL, Roth EK, Thiemann M, Blattmann C, Perez RL, Huber PE, et al. Prexasertib (LY2606368) reduces clonogenic survival by inducing apoptosis in primary patient-derived osteosarcoma cells and synergizes with cisplatin and talazoparib. Int J Cancer 2020;147:1059–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hong DS, Moore K, Patel M, Grant SC, Burris HA III, William WN Jr, et al. Evaluation of prexasertib, a checkpoint kinase 1 inhibitor, in a phase Ib study of patients with squamous cell carcinoma. Clin Cancer Res 2018;24:3263–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee JM, Nair J, Zimmer A, Lipkowitz S, Annunziata CM, Merino MJ, et al. Prexasertib, a cell cycle checkpoint kinase 1 and 2 inhibitor, in BRCA wild-type recurrent high-grade serous ovarian cancer: a first-in-class proof-of-concept phase 2 study. Lancet Oncol 2018;19:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gatti-Mays ME, Karzai FH, Soltani SN, Zimmer A, Green JE, Lee MJ, et al. A phase II single arm pilot study of the CHK1 inhibitor prexasertib (LY2606368) in BRCA wild-type, advanced triple-negative breast cancer. Oncologist 2020;25:1013–e1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hong D, Infante J, Janku F, Jones S, Nguyen LM, Burris H, et al. Phase I study of LY2606368, a checkpoint kinase 1 inhibitor, in patients with advanced cancer. J Clin Oncol 2016;34:1764–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mross K, Frost A, Steinbild S, Hedbom S, Rentschler J, Kaiser R, et al. Phase I dose escalation and pharmacokinetic study of BI 2536, a novel Polo-like kinase 1 inhibitor, in patients with advanced solid tumors. J Clin Oncol 2008;26:5511–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available in Gene Expression Omnibus (GEO) at GSE185543.



![Figure 2. Inhibitory effects of PLK1 or CHEK1/2 inhibitors. A, The effect of PLK1 inhibitors: BI-2536 and volasertib. B, The effect of CHEK1/2 inhibitors: prexasertib and PF-477736. Cells were treated with each inhibitor for 72 hours. The red, green, and blue colors represent SK-UT-1, SK-LMS-1, and SKN, respectively. The experiments were performed in triplicate and repeated three times. The IC50 value was calculated using the following equation: IC50 = 10[log(A/B) × (50 − C)/(D − C) + log(B)], where A and B represent the highest and the lowest concentrations to cover an estimated IC50 value, respectively, where C and D represent the cell viability at concentrations B and D, respectively. C, Cell-cycle distribution of BI-2536-treated cells. D, Cell-cycle distribution of prexasertib-treated cells. Cells were treated with each inhibitor for 24 hours. Cell-cycle distribution was calculated by FlowJo. The experiments were performed in three independent replicates and the percentage of cells was compared using the Dunnett's test. Error bars represent standard errors of the mean.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/bdea/9365385/372a131e2204/2147fig2.jpg)