Abstract
A genomic library derived from the deep-sea bacterium Photobacterium profundum SS9 was conjugally delivered into a previously isolated pressure-sensitive SS9 mutant, designated EC1002 (E. Chi and D. H. Bartlett, J. Bacteriol. 175:7533–7540, 1993), and exconjugants were screened for the ability to grow at 280-atm hydrostatic pressure. Several clones were identified that had restored high-pressure growth. The complementing DNA was localized and in all cases found to possess strong homology to recD, a DNA recombination and repair gene. EC1002 was found to be deficient in plasmid stability, a phenotype also seen in Escherichia coli recD mutants. The defect in EC1002 was localized to a point mutation that created a stop codon within the recD gene. Two additional recD mutants were constructed by gene disruption and were both found to possess a pressure-sensitive growth phenotype, although the magnitude of the defect depended on the extent of 3′ truncation of the recD coding sequence. Surprisingly, the introduction of the SS9 recD gene into an E. coli recD mutant had two dramatic effects. At high pressure, SS9 recD enabled growth in the E. coli mutant strain under conditions of plasmid antibiotic resistance selection and prevented cell filamentation. Both of these effects were recessive to wild-type E. coli recD. These results suggest that the SS9 recD gene plays an essential role in SS9 growth at high pressure and that it may be possible to identify additional aspects of RecD function through the characterization of this activity.
Increasing hydrostatic pressure results in a distinctive physical perturbation to biological systems which can be used as a tool both to uncover pressure-sensitive cellular processes and to discern aspects of pressure acclimation in barotolerant organisms capable of growth over a broad pressure range. The former application is useful for the isolation and characterization of conditional mutants in a manner analogous to that associated with temperature sensitivity or osmosensitivity. The latter approach is employed to study biochemical adaptation in the largest portion of the biosphere, the deep sea, where pressures can exceed 1,000 times that present on the planet’s surface.
Studies with Escherichia coli indicate that elevated pressure can inhibit many cellular functions, including substrate transport (22); biosynthesis of DNA, RNA, and protein (44); and protein quaternary structure and function (27). Perhaps the most apparent effect of elevated pressure on E. coli and other mesophiles is the inhibition of cell division and the formation of highly filamentous cells at sublethal pressures (47). Furthermore, DNA synthesis, the most pressure sensitive of the macromolecular synthetic processes, is prevented at pressures above 500 atm (44). Lower pressures can enhance the synchronization of DNA synthesis and cell division in cell cultures, perhaps owing to a tendency of pressure to prolong the duration of a particular part of the cell cycle (44).
The strategies used by marine bacteria inhabiting the deep sea to cope with the high pressures associated with their environments remain poorly understood, particularly at the molecular level. Photobacterium sp. strain SS9 (recently designated Photobacterium profundum SS9 [31]) is the only barophilic or, more correctly, piezophilic (pressure-loving [41]) species for which genetic experiments have been undertaken. SS9 mutants impaired in pressure sensing or high-pressure growth have been isolated and used to uncover genes associated with these processes (9, 39).
This study describes the complementation of EC1002, a previously isolated pressure-sensitive mutant of SS9 (8). We demonstrate that the source of the mutation in EC1002 is in the recD gene. RecD, along with RecB and RecC (exonuclease V), plays a major role in the homologous recombination pathway of E. coli (reviewed in reference 29). The RecBCD complex possesses several different ATP-dependent functions including single- and double-stranded exonuclease activity, single-stranded endonuclease activity, and helicase activity. RecBCD has been proposed to function as a destructive nuclease of linear DNA until it encounters the octameric sequence Chi (χ; 5′ GCTGGTGG 3′) (38). At this sequence, RecD is hypothesized to dissociate from the complex, while the RecBC enzyme is proposed to convert to a χ-independent recombinogenic helicase, to produce single-stranded DNA that serves as a substrate for RecA-catalyzed recombination (12). E. coli recD mutants are hyperrecombinogenic, decreased in plasmid stability, and increased in plasmid multimer formation (5). In this study, experiments with both SS9 and E. coli strains revealed that SS9 RecD is required for normal growth and cell morphology at high pressure.
MATERIALS AND METHODS
Bacterial strains and growth media.
Strains and plasmids used in this study are listed in Table 1. P. profundum SS9 strains were aerobically cultured in 2216 medium (28 g/liter; Difco Laboratories) at 15°C. Anaerobic growth of SS9 cultures was performed at 9°C in 2216 medium supplemented with 0.4 g of glucose per liter and 0.1 M HEPES, pH 7.5. E. coli strains were grown aerobically at 37°C in Luria-Bertani (LB) medium (26). Anaerobic growth of E. coli was performed with LB medium supplemented with 0.1 M Tris (pH 7.5) and 20 mM glucose. Antibiotics were used in the following concentrations: chloramphenicol, 25 μg/ml; ampicillin, 100 μg/ml; tetracycline, 20 μg/ml; rifampin, 100 μg/ml; and kanamycin, 50 μg/ml (E. coli) or 200 μg/ml (P. profundum). X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was added to solid medium at 40 μg/ml in N,N-dimethylformamide.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference(s) or source |
|---|---|---|
| Strains | ||
| P. profundum SS9 | ||
| EC10 | Rifr SS9 derivative | 8 |
| EC1002 | Pressure-sensitive EC10 derivative | 8 |
| KB3 | Complemented strain of EC1002, Kanr | This study |
| KB60 | EC10 recD disruption mutant, Kanr | This study |
| KB63 | EC10 recD disruption mutant, Kanr | This study |
| E. coli | ||
| DH5α | recA strain used for plasmid main-tenance | 19 |
| MC1061 | Host for pRK24, pRL528, and genomic library conjugal donor strain | 6, 7 |
| ED8654 | Used for pRK2073 maintenance | 28 |
| MG1655 | λ− | 18 |
| CAG12135 | recD::Tn10 | 37; CGSC7429 |
| Plasmids | ||
| pRK24 | Conjugal plasmid; derivative of RK2 | 25 |
| pRL528 | Helper plasmid, carrying mob | 13 |
| pRK2073 | Carries tra genes for conjugal transfer | 4 |
| pMUT100 | pBR322-derived suicide vector; Kanr | 6 |
| pCR2.1 | Vector for cloning PCR products, Kanr | Invitrogen |
| pKT231 | Broad-host-range vector, Kanr | 3 |
| pUC18 | High-copy-number vector, Ampr | 32 |
| pGL10 | Broad-host-range vector, Kanr | 1a |
| pB3 | Plasmid rescue from KB3, Kanr | This study |
| pKB26 | recBD fragment from pB3 in pGL10 | This study |
| pKB27 | recBD fragment from pB3 in pUC18 | This study |
| pKB60 | 470-bp recD fragment in pMUT100 | This study |
| pKB62 | recD from MG1655 in pUC18 | This study |
| pKB63 | 440-bp recD fragment in pMUT100 | This study |
| pKB65 | recD from MG1655 in pGL10 | This study |
| pCM1 | recD from EC1002 in pCR2.1 | This study |
High-pressure growth studies.
High-pressure growth of SS9 was performed as previously described (8). Briefly, late-exponential-phase cultures were diluted into fresh medium to an optical density at 600 nm (OD600) of 0.001 and used to fill 4.5-ml polyethylene transfer pipettes (Samco). Pipettes were then heat sealed with a handheld heat sealing clamp (Nalgene) and incubated at either 1 or 280 atm in stainless steel pressure vessels equipped with quick-connect fittings for rapid sampling and repressurization (45). High-pressure growth studies of E. coli were performed at 310 atm.
Construction of a genomic SS9 library.
All DNA preparations, enzymatic digestions, cloning, bacterial transformations, and gel electrophoresis were performed as previously described (2). Genomic DNA from EC10 was partially digested with Sau3A and size fractionated by sucrose density centrifugation (23). Fragments of DNA were size selected (∼5 to 7 kb) and cloned into the BamHI site of pMUT100, disrupting the tetracycline resistance cassette. The library was transformed into MC1061(pRK24, pRL528), and transformants were pooled and stored in LB medium-glycerol (85:15 [vol/vol]) at −80°C for future use.
Bacterial conjugations.
Plasmids from E. coli were transferred into SS9 via conjugations. Biparental matings were performed with E. coli MC1061(pRK24, pRL528), and triparental matings were performed with E. coli DH5α and ED8654(pRK2073). Stationary-phase cultures of E. coli and SS9 were harvested by centrifugation, cell pellets were resuspended in 2216 medium, and 100 μl of each strain was spotted onto 0.4-μm-pore-size polycarbonate membrane filters (Poretics) on dry 2216 plates. Matings were performed at room temperature for 12 to 16 h. After incubation, the cells were washed from the filters with 2216 medium and plated onto selective medium. Plates were incubated at 15°C for 3 to 5 days before the appearance of exconjugants. Exconjugants arising from pMUT100-based plasmids were the result of the entire plasmid integrating into the genome in a single-crossover event, conferring Kanr on the strain.
Plasmid rescues and secondary complementation analysis.
Genomic DNA from the complemented strain KB3 was prepared and digested with either BglII, HpaI, KpnI, SacI, or XbaI (Stratagene, La Jolla, Calif.). Digested genomic DNA was then circularized with T4 ligase (Gibco) and transformed into DH5α with selection for Kanr. Rescued plasmids, which averaged ∼8 to 15 kb in size, were then transformed into MC1061(pRK24, pRL528) and reconjugated into EC1002. Exconjugants were tested for the ability to grow at high pressure to verify that the complementing DNA had been isolated.
DNA sequencing.
DNA sequences were determined by double-stranded thermal cycle dideoxy sequencing with fluorescently labeled terminators (Perkin-Elmer) with an ABI 373A automated sequencer. Initial global similarity searches were performed with the BLAST network service (1). Individual alignments of amino acids were performed with the GCG University of Wisconsin Computer Group package version 7 BESTFIT program (11).
Construction of gene disruption mutants.
An internal portion of the recD gene was amplified from EC10 by PCR with the following primers: recBam, 5′ GTCAGGGATCCAGCCCACA 3′, and recEco, 5′ GGTTCGAATTCGTACCAGT 3′, or recD3, 5′ ACTGTGGCACCAAATCATCA 3′, and recDMUT2, 5′ CCAGAGCCTAAACATTGG 3′. Cycles of PCR were 92°C for 1 min, 48°C for 1 min, and 72°C for 1 min for 25 cycles. The PCR products were cloned into pCR2.1 (Invitrogen) and then subcloned into pMUT100. The resulting plasmids, designated pKB60 and pKB63, were conjugated into EC10 as described above.
Isolation of the EC1002 and E. coli recD genes.
The EC1002 recD gene was amplified with primers recDf, 5′ GTCGAAGCCATGCGTGAT 3′, and recDr, 5′ AGCCTATCCTAATAAACG 3′. The primers ECrecDf, 5′ CTTACACCCAACAGGCTA 3′, and ECrecDr, 5′ TAACACTCGTACGTCGCA 3′, were used to amplify the recD gene from MG1655. Cycles of PCR were 92°C for 1 min, 50°C for 1 min, and 72°C for 1.5 min for 25 cycles. Both recD genes were cloned into pCR2.1, and the E. coli recD gene was subsequently cloned into pUC18 and pGL10.
Nucleotide sequence accession number.
The sequence of SS9 recD, along with the partial sequence of recB, is deposited in GenBank under accession no. AF064802.
RESULTS
Complementation analysis.
DNA complementing the pressure-sensitive phenotype of mutant EC1002 was identified following the introduction of an SS9 genomic library derived from EC10, the parental strain of EC1002. Since EC1002 grows slowly even at 1 atm, exconjugants arising on plates at atmospheric pressure were visually inspected for large colonies and individual clones were screened for high-pressure-adapted growth in liquid culture at 280 atm. According to this scheme, 4 piezophilic exconjugants were identified among the 15,000 exconjugants examined. Subsequent analysis of these clones revealed that all four of the exconjugants were complemented by overlapping segments of DNA (data not shown). One of the four clones was chosen for further study and was designated KB3.
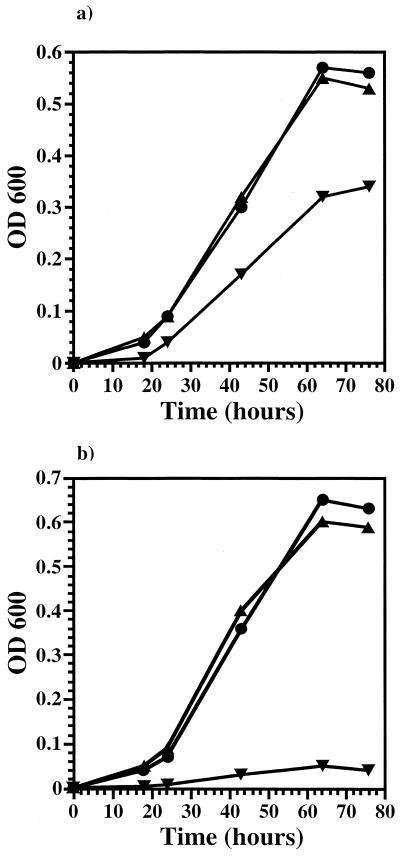
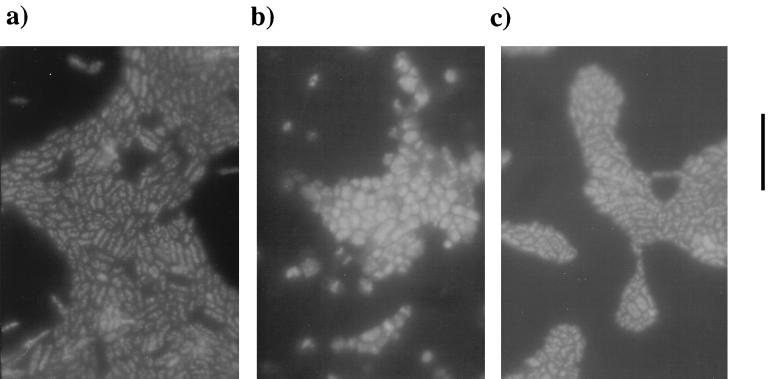
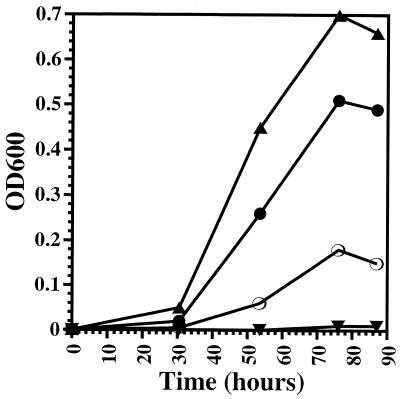
Comparison of the growth characteristics of KB3 at 1 and 280 atm with those of EC10 and EC1002 revealed that KB3 behaved similarly to EC10, displaying a restored high-pressure growth rate and final cell density (Fig. 1). Additionally, epifluorescence microscopy of the cells, stained with 4′,6′-diamidino-2-phenylindole (DAPI) and grown at high pressure, indicated that KB3 possessed a rod-shaped morphology and a size (∼2 by 4 μm) similar to that of EC10 (Fig. 2a and c). In contrast, EC1002 cells changed from normal SS9 rods at atmospheric pressure to large (>3- to 6-μm diameter [Fig. 2b]) irregularly shaped cells following high-pressure incubation. These data indicate that both pressure-altered phenotypes of EC1002, reduced growth rate and yield, as well as enlarged cell shape, are returned to those of wild-type cells in the complemented strain.
FIG. 1.
Growth profiles of three SS9 strains, EC10 (▴), EC1002 (▾), and KB3 (●), cultured at 9°C at 1 atm (a) or 280 atm (b).
FIG. 2.
Micrographs of DAPI-stained cells of P. profundum SS9. All cultures were grown at 280 atm and harvested at early stationary phase (OD600 = 0.51). (a) EC10; (b) EC1002; (c) KB3. Magnification, ×1,250. Bar, 10 μm.
Identification of the gene responsible for complementation in EC1002.
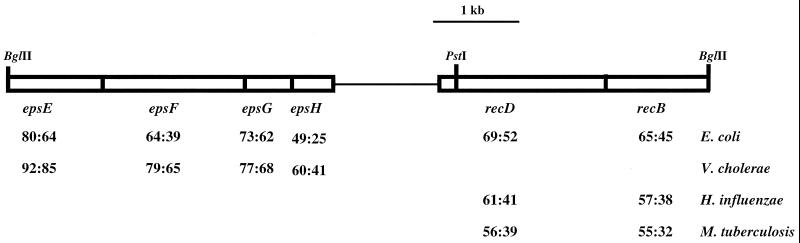
The plasmid integrant in KB3, along with flanking chromosomal DNA, was recovered following restriction and ligation of chromosomal DNA and transformation into E. coli with antibiotic resistance selection for the pMUT100 plasmid vector. Rescued plasmids, averaging 8 to 15 kb in size, were reconjugated into EC1002 to verify the recovery of complementing DNA. All exconjugants arising from introduction of the rescued plasmids into EC1002 were observed to restore high-pressure growth. Localization of the complementing DNA was further accomplished by generating subclones in the broad-host-range plasmid pGL10 (1a) with DNA derived from the rescued plasmid, pB3. After introduction of several different subclones into the mutant and examination of the exconjugants for high-pressure growth, it was established that one ∼2.8-kb PstI/BglII subclone, designated pKB26, complemented EC1002.
DNA sequence analysis revealed that pKB26 harbored open reading frames possessing a high degree of identity and similarity to RecB and RecD from several different bacteria. A substantial portion (∼1.95 of 2.1 kb) of the presumed recD coding sequence is present on pKB26, including its 5′ end and putative promoter region. Only a small portion of the 3′ end was missing from the gene (∼150 bp) compared to the sequence found in pB3, which contains the recD sequence in its entirety. The remainder of the pKB26 insert (∼900 bp) contained sequences upstream of recD which appear to encode the 3′ region of recB. Since less than one-third of the recB gene is present on this plasmid, it was concluded that the introduction of wild-type recD from EC10 was responsible for EC1002 complementation. The gene organization of the SS9 recD locus and a comparison of the predicted amino acid sequence of RecD, as well as additional open reading frames found on pB3, to homologous proteins present in other organisms are shown in Fig. 3. It is of note that the SS9 recD gene possesses ∼192 bp of unique sequence (located between nucleotides 469 and 660) that exhibits no homology with any of the seven reported recD genes, including those from E. coli and Mycobacterium tuberculosis (14, 34). The significance of this is unknown.
FIG. 3.
Schematic of the SS9 genomic DNA insert in pB3. The percents similarity and identity of SS9 RecBD to homologues from E. coli (14, 15), Haemophilus influenzae (16), and M. tuberculosis (34) are shown. Several open reading frames were found on pB3 that were homologous to extracellular protein secretion genes found in E. coli (17) and V. cholerae (33, 36), among others. The BglII and PstI sites delineate the region of DNA that is cloned into pKB26 and pKB27. The thin line that separates epsH and recD represents pMUT100 (not drawn to scale).
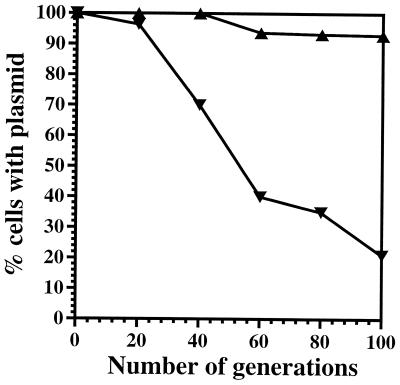
Plasmid stability tests in SS9.
E. coli recD mutants are defective in plasmid maintenance (5), apparently as a result of increased plasmid multimerization. Plasmid stability tests (24) were therefore performed with parental strain EC10 and the putative recD mutant EC1002 to determine if EC1002 has a similar defect. Plasmid pKT231 (3) was introduced into both strains, and cells were cultured in the absence of plasmid selection for 100 generations (doubling time, ∼2.5 h). Every 20 generations, cultures were diluted 1,000-fold and plated onto nonselective plates. Fresh colonies were patched from nonselective plates onto plates containing appropriate antibiotics, and the resulting fractions of antibiotic-resistant cells were scored. As shown in Fig. 4, EC1002 cells began losing the plasmid after 20 generations of growth while the plasmid was steadily maintained (>90%) in EC10 throughout the course of the experiment.
FIG. 4.
Stability of plasmid pKT231 in EC10 (▴) and EC1002 (▾).
Identification of the mutation in EC1002 and construction of a recD gene disruption mutant.
The above results indicated that the pressure sensitivity of strain EC1002 is due to a recD mutation. To verify this, the recD gene from EC1002 was isolated via PCR amplification and the sequences of three independently obtained clones were determined. In all cases, the EC1002 recD genes possessed a G-to-A transition at nucleotide position 258 in the recD coding sequence, resulting in the creation of an opal stop codon (TGA) at amino acid position 86. Ethyl methanesulfonate, which was employed in the mutagenesis of EC10 to generate EC1002, is known to generate transition mutations (26).
Two independent recD mutations were introduced into EC10 by gene disruption mutagenesis with the suicide vector pMUT100. One mutant, designated KB60, created an ∼420-bp deletion at the 3′ end of recD, eliminating ∼20% of the gene. The second mutant, designated KB63, created an ∼1.4-kb deletion at the 3′ end of recD, eliminating ∼67% of the gene. Growth curves were generated at 280 atm for both KB60 and KB63 and compared to those of EC10 and EC1002. KB60 was found to possess a high-pressure growth phenotype intermediate between that of EC10 and that of EC1002, while KB63, containing a substantial deletion in recD, was almost as pressure sensitive as EC1002 (Fig. 5). KB63 barely grew above an OD600 of 0.1 at 280 atm, and its growth rate, as reflected by changes in optical density over time, was reduced by ∼75% compared to wild-type EC10. The changes in optical density observed for cultures of KB63 may not be a true reflection of cell division since KB63 cells at high pressure developed large, irregular morphologies like those of EC1002 cells (Fig. 2b). For this reason, the 75% reduction in growth rate that is observed may actually be an underestimate.
FIG. 5.
Growth profiles of two different recD disruption mutants created in SS9, KB60 (●) and KB63 (○), compared with EC10 (▴) and EC1002 (▾), at 280 atm and 9°C.
Effects of the SS9 recD gene on high-pressure growth in E. coli.
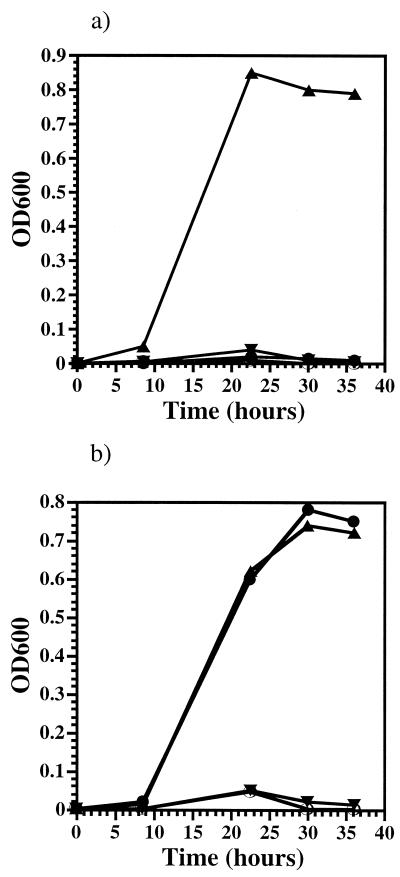
The increased pressure sensitivity of the P. profundum recD mutant EC1002 could reflect a particular evolutionary adaptation of the SS9 recD gene for piezophily. Alternatively, it could result from the perturbation of a physiological process whose alteration would lead to increased pressure sensitivity in multiple organisms, including those not found in high-pressure habitats. To discriminate among these possibilities, the effect of a recD mutation on the growth and morphology of E. coli at elevated pressure was investigated. Although the pressure optimum for E. coli is 1 atm under most culture conditions, it is piezotolerant, with growth, albeit filamentous, being reported at pressures up to 500 atm (44, 47). We examined the growth of two E. coli strains, a recD mutant, CAG12135 (37), and its isogenic parental strain, MG1655 (18). Both strains displayed piezotolerant growth at 310 atm when cultured in the absence of a plasmid (Fig. 6). In contrast, no growth was observed when either strain was transformed with pUC18 (32) and cultured at high pressure in the presence of antibiotic selection (Fig. 6). When antibiotic selection was removed from pUC18-bearing strains incubated at high pressure, growth was once again apparent (data not shown). These results suggest that elevated pressure leads to the impairment of plasmid maintenance in these strains.
FIG. 6.
High-pressure growth profiles of E. coli strains. (a) MG1655 (▴), MG1655(pUC18) (▾), MG1655(pKB27) (●), and MG1655(pKB62) (○) cultured at 310 atm and 37°C. (b) CAG12135 (▴), CAG12135(pUC18) (▾), CAG12135(pKB27) (●), and CAG12135(pKB62) (○) cultured at 310 atm and 37°C.
The SS9 recD gene was then tested for the ability to heterologously complement the high-pressure plasmid maintenance defect in both CAG12135 and MG1655. The SS9 recD gene was cloned onto plasmid pUC18, and the resulting plasmid, designated pKB27, was transformed into both E. coli strains. Under conditions of plasmid selection, pKB27 enabled piezotolerant growth in the case of the recD mutant CAG12135, but not of MG1655 containing its own functional recD gene (Fig. 6). The effect of the SS9 recD gene was apparently recessive to its homologue in E. coli. For comparison, the recD gene from MG1655 was cloned into pUC18, creating pKB62, which was introduced into both MG1655 and CAG12135, and the resulting strains were also tested for growth at high pressure. In both cases, the strains containing pKB62 were unable to grow at 310 atm as long as antibiotic selection was maintained (Fig. 6). These results suggest that SS9 recD has an activity at high pressure which is not provided by E. coli recD that influences plasmid maintenance.
The E. coli recD gene was also introduced into EC1002 on plasmid pKB65 to determine if it could complement the high-pressure growth defect of this mutant in a manner similar to that seen with SS9 recD. As expected, the E. coli recD gene was unable to restore growth at 280 atm to EC1002 (data not shown).
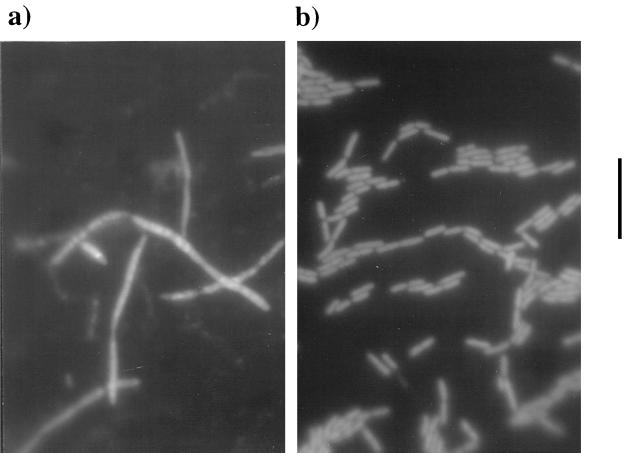
Numerous studies have documented that E. coli cells grown at pressures in excess of ∼250 atm are filamentous (21, 40, 46–49). Indeed, when we examined MG1655 and CAG12135 cells grown at 310 atm, filament formation was observed in both cases (data not shown). To determine if the SS9 recD gene had an effect on E. coli cell morphology at high pressure, epifluorescence microscopy was performed with DAPI-stained CAG12135 cells harboring either pUC18 or pKB27 grown at 310 atm. The cells containing pUC18 displayed a filamentous morphology (>20 by ∼0.5 μm) with an uneven distribution of DAPI-stained DNA in which nucleoid material appeared highly amplified in some filament sections and lacking in others (Fig. 7a). In contrast, the cells containing pKB27 possessed a typical E. coli rod-shaped morphology (∼0.5 by 2 to 3 μm) with chromosomal material evenly distributed throughout the cell (Fig. 7b). Microscopy was also performed on DAPI-stained preparations of CAG12135 cells transformed with pKB62 and MG1655 cells transformed with either pUC18, pKB27, or pKB62 that were grown under the conditions described above. In all cases, these cells were filamentous and displayed a DAPI staining pattern similar to that of the recD strain with pUC18 (Fig. 7a and data not shown).
FIG. 7.
Micrographs of DAPI-stained cells of E. coli. Cultures were grown at 310 atm and 37°C, and cells were harvested at early stationary phase (OD600 = 0.73). (a) CAG12135(pUC18). (b) CAG12135(pKB27). Bar, 10 μm. Magnification, ×1,250.
DISCUSSION
Pressure provides a useful physiochemical parameter for the isolation and characterization of novel conditional mutants. The effects of elevated pressure on cell processes are a consequence of system volume changes associated with the equilibria and rates of biochemical reactions (42); therefore, an alteration in pressure exerts a fundamentally different influence from that of other changes in state including pH, temperature, or osmolarity. In this study, we have presented the results of genetic analyses with the pressure-sensitive mutant EC1002 derived from the marine piezophile P. profundum SS9. These results provide an important clue towards the elucidation of the molecular bases of pressure adaptation in an ecologically significant group of extremophilic microorganisms, as well as insight into additional aspects of RecD function.
The conclusion that recD gene expression is important for piezophily in SS9 is based on the following evidence. First, SS9 recD complements the poor growth and enlarged cell morphology of EC1002 cells at high pressure (Fig. 1 and 2). Second, the pressure-sensitive mutant EC1002 has a mutation within the recD gene. Third, two different gene replacement mutants possessing a 3′-truncated recD gene were found to be pressure sensitive and possessed aberrations in morphology similar to those of EC1002 at high pressure, albeit to various extents, depending on the amount of recD coding sequence removed (Fig. 5). Fourth, at high pressure the SS9 recD gene greatly enhanced the growth of E. coli recD mutants under conditions of plasmid selection and prevented cell filamentation (Fig. 6 and 7).
The SS9 recD gene contains 192 bp of sequence not found in any of the previously reported recD genes, resulting in an additional 64 amino acids after amino acid position 156 in the RecD protein. Curiously, we have found an open reading frame encoding a RecD homologue present in the genome of Vibrio cholerae (available through The Institute for Genomic Research) whose deduced protein sequence is similar in size and sequence to that encoded by the recD gene from SS9. Furthermore, we have found that V. cholerae cells, which do not normally encounter extremes in pressure in their natural environment, do not form filaments when incubated at elevated pressure (our unpublished results). Thus, the unique structural motif of RecD from SS9, V. cholerae, and perhaps other related bacteria could be important for some aspect of RecD function at high pressure. As to why V. cholerae may possess a RecD that may be adapted for function under high-pressure conditions, it is thus far unclear.
Unlike the unusual response of V. cholerae cells to elevated pressure, most mesophilic bacterial cells respond to a moderate pressure increase (200 to 300 atm) by developing highly filamentous cell structures (21, 40, 46–49). Indeed, filament formation is certainly one of the most dramatic effects of elevated pressure on bacteria. Cell division in piezophiles can also be highly sensitive to the effects of pressure. However, in these bacteria cell filamentation has been found to occur at pressures not only above their optima but below them as well (20, 43). Therefore, it is noteworthy that the SS9 recD gene can function in a heterologous system to restore a 1-atm cell division phenotype to E. coli cells lacking their own recD gene. This represents the first introduction of a piezophilic trait into an atmospheric pressure-adapted organism. To our knowledge, this is also the first case where any extremophilic attribute has been successfully conferred upon a mesophile.
In the course of this study, the SS9 recD gene was also found to be necessary for high-pressure plasmid maintenance in E. coli. For the SS9 recD gene to complement this defect, the absence of a functional E. coli recD gene was necessary, a requirement also needed to complement the defects in cell division. This is most likely explained by the fact that an E. coli strain that already possesses its own functional copy of RecD will preferentially incorporate it into the formation of the RecBCD (ExoV) complex to the exclusion of a foreign RecD. A previous study by Rinken et al. (35) demonstrated that active hybrid enzymes of ExoV could be formed from different RecBCD components taken from E. coli, Serratia marcescens, and/or Proteus mirabilis. In each combination examined, however, the most efficient enzyme was formed from the three components that originated from the same organism. It is possible that the SS9 RecD, while fully capable of forming an active ExoV complex along with E. coli RecB and RecC, is not preferred to E. coli RecD as it most likely does not possess the same level of intersubunit affinity.
Not all E. coli strains have plasmid maintenance problems at elevated pressure. Kato et al. (21) have reported small increases in plasmid copy number for an E. coli strain incubated at a 300-atm pressure. In that study, a variety of plasmids were examined, including a pUC-derived plasmid similar to that employed in this study. However, the genotype of the parent strain used by Kato and colleagues, JM109, was different from that of the isogenic E. coli strains used here. In particular, the recA mutation in JM109 would be expected to exert a substantial influence on plasmid stability, as RecA is essential for homologous recombination and plasmid instability is eliminated in E. coli recD recA double mutants (5).
In what fashion might SS9 RecD influence plasmid stability, and cell growth and morphology, particularly at high pressure? Although additional studies will be needed to answer this question, we would like to offer the following working hypothesis. Since E. coli recD mutants are known to possess increased frequencies of homologous recombination which can lead to plasmid concatemerization and plasmid maintenance defects (5), we propose that SS9 recD mutants possess a similar phenotype. If a recD mutant in SS9 exhibits an increased frequency of interchromosomal recombination and genome concatemerization, then elevated pressure could compromise cell growth by exacerbating this condition. This could be accomplished by further increasing the rate of chromosome multimerization or by inhibiting the rate of resolution of entangled chromosomes. Many examples exist that connect mutations inhibiting chromosome partitioning with defects in cell division, including mukB (30) and xerC (10). Thus, impaired chromosome segregation could explain the poor growth and enlarged swollen cells of SS9 recD mutants grown at high pressure, as well as pressure-induced E. coli filamentation. Although this model is attractive since it explains all of the phenotypes observed here for the SS9 recD mutants and the E. coli cells at high pressure as stemming from a single activity, namely, homologous recombination, it must be stressed that no evidence linking RecD function to interchromosomal recombination in any microorganism is present at this time. Detailed studies of recombination in both SS9 and E. coli as a function of pressure will be necessary to validate or reject the model.
ACKNOWLEDGMENTS
We thank Christina McMann for help with cloning and sequencing the EC1002 recD gene and Kay Bidle for help with microscopy and photography. We also thank Christopher Francis and Kay Bidle for helpful comments and discussions regarding the manuscript.
This work was supported by NSF grant MCB96-30546 to D.H.B. and by NSF postdoctoral research fellowship BIR-9627134 to K.A.B.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers W E, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 1a.Andersen, K. Unpublished data.
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1993. [Google Scholar]
- 3.Bagdasarian M, Lurz R, Ruckert B, Franklin F C, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 4.Better M, Helinski D R. Isolation and characterization of the recA gene of Rhizobium meliloti. J Bacteriol. 1983;155:311–316. doi: 10.1128/jb.155.1.311-316.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biek D P, Cohen S N. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986;167:594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brahamsha B. A genetic manipulation system for oceanic cyanobacteria of the genus Synechococcus. Appl Environ Microbiol. 1996;62:1747–1751. doi: 10.1128/aem.62.5.1747-1751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 8.Chi E, Bartlett D H. Use of a reporter gene to follow high-pressure signal transduction in the deep-sea bacterium Photobacterium sp. strain SS9. J Bacteriol. 1993;175:7533–7540. doi: 10.1128/jb.175.23.7533-7540.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi E, Bartlett D H. An rpoE-like locus controls outer membrane protein synthesis and growth at cold temperatures and high pressures in the deep-sea bacterium Photobacterium SS9. Mol Microbiol. 1995;17:713–726. doi: 10.1111/j.1365-2958.1995.mmi_17040713.x. [DOI] [PubMed] [Google Scholar]
- 10.Colloms S D, Sykora P, Szatmari G, Sherratt D J. Recombination at ColE1 cer requires the Escherichia coli xerC gene product, a member of the lambda-integrase family of site-specific recombinases. J Bacteriol. 1990;172:6973–6980. doi: 10.1128/jb.172.12.6973-6980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon D A, Churchill J J, Kowalczykowski S C. Reversible inactivation of the Escherichia coli RecBCD enzyme by the recombination hotspot χ in vitro: evidence for functional inactivation or loss of the RecD subunit. Proc Natl Acad Sci USA. 1994;91:2980–2984. doi: 10.1073/pnas.91.8.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elhai J, Wolk C P. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 1988;167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 14.Finch P W, Storey A, Brown K, Hickson E D, Emmerson P T. Complete nucleotide sequence of recD, the structural gene for the alpha subunit of exonuclease V of Escherichia coli. Nucleic Acids Res. 1986;14:8583–8594. doi: 10.1093/nar/14.21.8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finch P W, Storey A, Chapman K E, Brown K, Hickson E D, Emmerson P T. Complete nucleotide sequence of the Escherichia coli recB gene. Nucleic Acids Res. 1986;14:8573–8582. doi: 10.1093/nar/14.21.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C A, Gocayne J D, Scott J D, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 17.Francetic O, Pugsley A P. The cryptic general secretory pathway (gsp) operon of Escherichia coli K-12 encodes functional proteins. J Bacteriol. 1996;178:3544–3549. doi: 10.1128/jb.178.12.3544-3549.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guyer M S, Reed R R, Steitz J A, Low K B. Identification of a sex-factor-affinity site in E. coli as γδ. Cold Spring Harbor Symp Quant Biol. 1981;45:135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 20.Jannasch H W. Effects of hydrostatic pressure on growth of marine bacteria. In: Jannasch H W, Marquis R E, Zimmerman A M, editors. Current perspectives in high pressure biology. Toronto, Ontario, Canada: Academic Press; 1987. pp. 1–15. [Google Scholar]
- 21.Kato C, Sato T, Smorawinska M, Horikoshi K. High pressure conditions stimulate expression of chloramphenicol acetyltransferase regulated by the lac promoter in Escherichia coli. FEMS Microbiol Lett. 1994;122:91–96. doi: 10.1111/j.1574-6968.1994.tb07149.x. [DOI] [PubMed] [Google Scholar]
- 22.Landau J V. Induction, transcription, and translation in Escherichia coli: a hydrostatic pressure study. Biochim Biophys Acta. 1967;149:506–512. doi: 10.1016/0005-2787(67)90178-5. [DOI] [PubMed] [Google Scholar]
- 23.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 24.Meacock P A, Cohen S N. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980;20:529–545. doi: 10.1016/0092-8674(80)90639-x. [DOI] [PubMed] [Google Scholar]
- 25.Meyer R, Figurski D, Helinski D R. Physical and genetic studies with restriction endonucleases on the broad host range plasmid RK2. Mol Gen Genet. 1977;152:129–135. doi: 10.1007/BF00268809. [DOI] [PubMed] [Google Scholar]
- 26.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 27.Mozhaev V V, Heremans K, Frank J, Masson P, Balny C. High pressure effects on protein structure and function. Proteins. 1996;24:81–91. doi: 10.1002/(SICI)1097-0134(199601)24:1<81::AID-PROT6>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 28.Murray N E, Brammar W J, Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977;150:53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- 29.Myers R S, Stahl F W. Chi and the RecBCD enzyme of Escherichia coli. Annu Rev Genet. 1994;28:49–70. doi: 10.1146/annurev.ge.28.120194.000405. [DOI] [PubMed] [Google Scholar]
- 30.Niki H, Jaffe A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of Escherichia coli. EMBO J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nogi Y, Masui N, Kato C. Photobacterium profundum sp. nov., a new, moderately barophilic bacterial species isolated from a deep-sea sediment. Extremophiles. 1998;2:1–7. doi: 10.1007/s007920050036. [DOI] [PubMed] [Google Scholar]
- 32.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 33.Overbye L J, Sandkvist M, Bagdasarian M. Genes required for extracellular secretion of enterotoxin are clustered in Vibrio cholerae. Gene. 1993;132:101–106. doi: 10.1016/0378-1119(93)90520-d. [DOI] [PubMed] [Google Scholar]
- 34.Philipp W J, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom B R, Jacobs W R J, Cole S T. An integrated map of the genome of the tubercle bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rinken R, de Vries J, Weichenhan D, Wackernagel W. The recA-recBCD dependent recombination pathways of Serratia marcescens and Proteus mirabilis in Escherichia coli: functions of hybrid enzymes and hybrid pathways. Biochimie. 1991;73:375–384. doi: 10.1016/0300-9084(91)90104-9. [DOI] [PubMed] [Google Scholar]
- 36.Sandkvist M, Overbye-Michel L, Hough L P, Morales V M, Bagdasarian M, Koomey M, DiRita V J, Bagdasarian M. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J Bacteriol. 1997;179:6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove M, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith G R, Kunes S M, Schultz D W, Taylor A, Triman K L. Structure of chi hotspots of generalized recombination. Cell. 1981;24:429–436. doi: 10.1016/0092-8674(81)90333-0. [DOI] [PubMed] [Google Scholar]
- 39.Welch T J, Bartlett D H. Identification of a regulatory protein required for pressure-responsive gene expression in the deep-sea bacterium Photobacterium species strain SS9. Mol Microbiol. 1998;27:977–985. doi: 10.1046/j.1365-2958.1998.00742.x. [DOI] [PubMed] [Google Scholar]
- 40.Welch T J, Farewell A, Neidhardt F C, Bartlett D H. Stress response of Escherichia coli to elevated hydrostatic pressure. J Bacteriol. 1993;175:7170–7177. doi: 10.1128/jb.175.22.7170-7177.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yayanos A A. Microbiology to 10,500 meters in the deep sea. Annu Rev Microbiol. 1995;49:777–805. doi: 10.1146/annurev.mi.49.100195.004021. [DOI] [PubMed] [Google Scholar]
- 42.Yayanos A A. Empirical and theoretical aspects of life at high pressure in the deep sea. In: Horikoshi K, Grant W D, editors. Extremophiles. Microbial life in extreme environments. New York, N.Y: John Wiley and Sons, Inc.; 1998. pp. 47–92. [Google Scholar]
- 43.Yayanos A A, DeLong E F. Deep-sea bacterial fitness to environmental temperatures and pressures. In: Jannasch H W, Marquis R E, Zimmerman A M, editors. Current perspectives in high pressure biology. Toronto, Ontario, Canada: Academic Press; 1987. pp. 17–32. [Google Scholar]
- 44.Yayanos A A, Pollard E C. A study of the effects of hydrostatic pressure on macromolecular synthesis in Escherichia coli. Biophysics. 1969;9:1464–1482. doi: 10.1016/S0006-3495(69)86466-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yayanos A A, Van Boxtel R. Coupling device for quick high pressure connections to 1000 MPa. Rev Sci Instrum. 1982;53:704–705. [Google Scholar]
- 46.ZoBell C E. Pressure effects on morphology and life processes of bacteria. London, United Kingdom: Academic Press; 1970. [Google Scholar]
- 47.ZoBell C E, Cobet A B. Growth, reproduction, and death rates of Escherichia coli at increased hydrostatic pressures. J Bacteriol. 1962;84:1228–1236. doi: 10.1128/jb.84.6.1228-1236.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.ZoBell C E, Cobet A B. Filament formation by Escherichia coli at increased hydrostatic pressures. J Bacteriol. 1964;87:710–719. doi: 10.1128/jb.87.3.710-719.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.ZoBell C E, Oppenheimer C H. Some effects of hydrostatic pressure on the multiplication and morphology of marine bacteria. J Bacteriol. 1950;60:771–781. doi: 10.1128/jb.60.6.771-781.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]