Abstract
Background
Thyroid carcinoma (TC) is an increasingly common malignancy of endocrine organs, and its most frequently encountered histotype is papillary thyroid cancer (PTC). Identifying new potential gene alterations is important for completely elucidating the mechanism of PTC initiation and progression. Thus, we performed whole transcriptome sequence analysis (RNA-seq) on 79 PTC tissue samples and paired adjacent nontumor tissue samples to study the molecular mechanism of TC tumorigenesis and progression further. The results of RNA-seq analysis showed that spectrin beta, nonerythrocytic 2 (SPTBN2), was markedly overexpressed in PTC tissues relative to that in the paired nontumor tissues. Additionally, the analysis results for 502 PTC samples and 58 nontumor thyroid samples from The Cancer Genome Atlas dataset were consistent with our RNA-seq results. However, the molecular mechanisms and function of SPTBN2 in TC progression remain unknown.
Methods
We examined SPTBN2 gene expression in 48 papillary thyroid tumor tissues and paired adjacent normal thyroid tissues by using qRT-PCR. SPTBN2 expression in the TC cell lines was silenced by small interfering RNA. Then, the transfected TC cells were used to investigate the in vitro function of SPTBN2.
Result
The expression of SPTBN2 was significantly upregulated in our RNA-seq cohort, our local validated cohort, and TCGA RNA-seq cohort. The results of the in vitro experiment revealed that in TC cell lines, SPTBN2 downregulation considerably suppressed tumor cell proliferation, the cell cycle, migration, colony formation, and invasion and induced cell apoptosis. Furthermore, the protein levels of CCNE2, CDK2, CDK4, and Bcl-2 were downregulated, and those of P21, Bax, cleaved caspase-8, and cleaved caspase-3 had increased in transfected TC cells relative to in control TC cells.
Conclusion
The downregulation of SPTBN2 caused apoptosis and retarded G1/S cell cycle transition in TC cells. Thus, SPTBN2 may be a good candidate gene for TC diagnosis and therapy.
1. Introduction
Thyroid carcinoma (TC) is a highly common malignancy of endocrine organs; this disease accounted for approximately 2.9% of all new cases of malignancies in the United States of America in 2019 [1]. In the United States of America, the morbidity of thyroid cancer is now 2 times greater than that 30 years ago, and thyroid cancer is now the sixth most prevalent cancer in women [2, 3]. In the majority population, papillary thyroid cancer (PTC) is the most common subtype of TC and accounts for 85% of newly diagnosed TC cases [4]. Although the morbidity of thyroid cancer is high, PTC has a good prognosis with a >90% 10-year survival rate [5, 6]. However, a large number of patients with PTC experience metastasis and recurrence after conventional therapeutic strategies [7]. Therefore, further studies on the probable molecular mechanism of thyroid tumors are needed to provide early accurate diagnosis and appropriate treatment to patients with TC.
With the advent of whole transcriptome sequencing (RNA-seq) technology, next-generation sequencing has become widely utilized to identify changes in genes between tumors and normal tissues. Although numerous studies have been carried out to explore the probable mechanisms of thyroid tumors, the precise biomechanisms underlying the progression and occurrence of thyroid tumors remain to be elucidated fully. A growing number of reports have shown that genetic alterations, such as gene mutations, epigenetic modifications, and copy number variations, play an important role in the initiation of TC [8, 9]. For example, the point mutation of B-type Raf kinase can promote PTC tumorigenesis and progression via the initiation of the mitogen-activated protein kinase pathway [10, 11]. Furthermore, PAX8/PPARγ rearrangements, RET/PTC rearrangements, and RAS mutations are important for the initiation and progression of differentiated TC [12–14]. Several biomarkers have been identified as appropriate diagnostic markers and potential therapeutic targets for TC. However, the identification of new potential gene alterations remains important for completely elucidating the mechanism of TC initiation and progression.
We performed RNA-seq on 79 PTC tissue samples and paired adjacent nontumor tissue samples to further study the underlying molecular mechanisms of TC. Through bioinformatics analysis, we discovered that spectrin beta, nonerythrocytic 2 (SPTBN2), a gene located on chromosome 11q13 in humans and encoding the membrane scaffold protein beta-III spectrin, is a potential powerful candidate gene that is associated with TC initiation and progression. Beta-III spectrin maintains dendritic architecture and stabilizes several membrane proteins [15]. A mutation in SPTBN2 causes spinocerebellar ataxias type-5, a human neurodegenerative disorder that is characterized by progressive locomotor incoordination, slurred speech, and uncoordinated eye movements [16, 17]. Another study reported that the mutation of SPTBN2 can disrupt Hippo signaling activity in follicle cells [18]. However, the previous study mainly focused on the effect of SPTBN2 mutation on spinocerebellar ataxias type-5. The effect of the copy number variation of SPTBN2 on the tumorigenesis and metastasis of malignancies has never been studied. Here, we were the first to prove that SPTBN2 acts as a potential oncogene in the tumorigenesis and metastasis of thyroid malignancies by inhibiting apoptosis and promoting the G1/S cell cycle transition in malignant thyroid cells.
2. Materials and Methods
2.1. Tissues
The fresh malignant thyroid samples and paired nontumor thyroid samples that were used in this study were obtained from patients with PTC who underwent thyroid surgery at the First Affiliated Hospital of Wenzhou Medical University (China) from May 2017 to April 2019. The patients did not undergo chemo- or radiotherapy for pretreatment. All of the samples were snap-frozen in liquid nitrogen and stored at −80°C before RNA extraction. All of the tumor tissues used in the present study were examined blindly by three independent senior thyroid pathologists. Written consent/assent for the usage of the thyroid samples was given by the patients and their relatives. The procedures that were performed in our study were approved by and conducted in accordance with the ethical standards of the Institutional Review Board of the First Affiliated Hospital of Wenzhou Medical University (approval no. 2012-57).
2.2. The Cancer Genome Atlas Data
Data, including the expression profiles of the SPTBN2 gene (502 PTC samples and 58 nontumor samples) with complete clinicopathological characteristics, were collected from The Cancer Genome Atlas (TCGA) databank for further analysis. All data related to the specimens were downloaded from TCGA website (https://tcgadata.nci.nih.gov/tcga).
2.3. Cell Culture
HTORI-3 (normal thyroid epithelial cells; RRID, CVCL 4W02) and KTC-1 (poorly differentiated thyroid cancer; RRID, CVCL 6300) were obtained from the Stem Cell Bank of the Chinese Academy of Sciences. TPC-1 (thyroid gland papillary carcinoma; RRID, CVCL 6298) and BCPAP (thyroid gland papillary carcinoma; RRID, CVCL 0153) were generously donated by Prof. Ming-Zhao Xing (School of Medicine, Johns Hopkins University, US). The cells were all incubated in Roswell Park Memorial Institute 1640 (RPMI-1640) medium (Invitrogen, Waltham, MA, US) containing 100 μg/mL penicillin-streptomycin (Sigma Aldrich, US) and 10% fetal bovine serum (FBS; Invitrogen, Waltham, MA, US). The cells were kept in a humidified incubator (Thermo Fisher Scientific, Waltham, MA, US) at 37°C and 5% CO2.
2.4. Cell Transfection
Small interfering RNA (siRNA) for SPTBN2 and the negative control siRNA for cell interference experiments were synthesized by Gene Pharma Co., Ltd (Gene Pharma, Shanghai, China) (Table 1). KTC-1 (8 × 104 cells per well) and BCPAP (7 × 104 cells per well) were plated on six well-plates and incubated for 24 h before transfection. siRNA was transfected into the TC cell lines by using Lipofectamine RNAiMAX Reagent (13778, Invitrogen, Carlsbad, CA, US). The transfected TC cells were harvested from the six-well plates at 48 h posttransfection, and knockdown efficacy was determined via qRT-PCR.
Table 1.
siRNA targeting human SPTBN2.
| si-RNA name | Sequence (5′ to 3′) | |
|---|---|---|
| Sense | Antisense | |
| SPTBN2 Si-RNA1 | GCCAGUACAGUGACAUCAATT | UUGAUGUCACUGUACUGGCTT |
| SPTBN2 Si-RNA2 | GCUGCUGAUGCUGCCAUUATT | UAAUGGCAGCAUCAGCAGCTT |
2.5. RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from thyroid tissues or cell lines with TRIzol reagent (15596026, Invitrogen, US). Afterward, the A260/A280 ratio was used to evaluate RNA quality. By using a ReverTra Ace qPCR RT Kit (TOYOBO Life Science, Cat. No: FSQ-101, Japan), RNA was converted into cDNA. RNA expression levels were detected via qRT-PCR with an AB Prism 7500 fast system (Applied Biosystems, US) and SYBR qPCR Mix (QPS-201; TOYOBO Life Science, Japan). The primer pair details are shown in Table 2.
Table 2.
Real-time PCR primers.
| Primer name | Sequence (5′ to 3′) |
|---|---|
| SPTBN2-F | GGGCATCCTTGGCTGACT |
| SPTBN2-R | GACGGAAACCACCGACTG |
| GADPH-F | GGTCGGAGTCAACGGATTTG |
| GADPH-R | ATGAGCCCCAGCCTTCTCCAT |
2.6. Cell Counting Kit-8 Proliferation Assay
The Cell Counting Kit-8 (CCK-8, C0038; Beyotime Institute of Biotechnology, China) assay was performed to evaluate cell proliferation capability. KTC-1 and BCPAP-1 (1.5 × 103 cells/well in 96-well plates) transfected with Si-NC or SPTBN2 siRNA were cultured in RPMI-1640 medium+10% FBS for 1–4 days. Afterward, CCK-8 solution (10 μL) and RPMI-1640 medium (100 μL) were added into each well at 24, 48, 72, and 96 h, and the samples were incubated at 37°C in 5% CO2 for 3 h. Then, the absorbance at 450 nm was determined.
2.7. Colony Formation Assay
The treated TC cells (1.5 × 104 cells per well) were incubated in 10% FBS/RPMI-1640 medium in six-well plates for 7 days. Subsequently, the cells were washed three times by using phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde solution for 30 min. Lastly, all of the cells were rinsed with PBS three times and stained by using crystal violet (0.01%).
2.8. Migration and Invasion Assays
Migration capacity was detected via a Transwell migration assay. A Transwell cell culture chamber (8 μm pore size, 3422, Corning, NY, US) was used for the in vitro Transwell migration assay. A total of 3.5 × 104 transfected TC cells were suspended in 300 μL of medium (RPMI-1640, free serum) and placed in the upper insert. The lower outer well contained 650 μL of 10% serum RPMI-1640 medium. After 20 h of incubation, the nonmigrated cells on the upper side of each membrane were removed by using sterile cotton swabs, and the migrated cells on the lower surface were fixed in 4% paraformaldehyde solution and stained with crystal violet (0.01%). Cell invasion capacity was evaluated by using an invasion chamber (354480, Corning, NY, US). All of the steps in this procedure were the same as those followed in the Transwell migration assay.
2.9. Apoptosis Analysis
Cell apoptosis was measured by applying an Annexin V-fluorescein isothiocyanate/propidium iodide apoptosis detection kit (556547, BD, US). TC cells (5–6 × 10 cells per well) were cultured in six-well plates for 1 day and transfected for 2 days. The cells were collected and centrifuged at 1100 rpm for 5 min. Then, the supernatant was decanted, and the cells were washed with PBS (1 mL). These processes were repeated three times. Afterward, each tube was added with 4 μL of Annexin V-FITC and 4 μL of PI and then placed for 20 min in the dark. Apoptosis was studied with a BD FACSCalibur flow cytometer (Becton-Dickinson, CA, US).
2.10. Cell Cycle Assay
7-Amino-actinomycin D (7-AAD; 559925, BD Pharmingen, CA, US) was used for cell cycle analysis. Transfected TC cells (1 × 106 cells per well) were collected from six-well plates and washed with PBS. Then, the TC cells were immersed in 70% ethanol overnight (4°C). Finally, the cells were rewashed with PBS and stained with 7-AAD for 10 min. The cell cycle assay was performed with a BD FACSCalibur flow cytometer (Becton-Dickinson, CA, US).
2.11. Gene Set Enrichment Analysis
Gene set enrichment analysis (GSEA) 4.0.2 software (http://www.broadinstitute.org/gsea) was used to evaluate the pathways that were associated with SPTBN2 expression in TCGA dataset. For GSEA, PTC tissues (502) were divided equally into the high and low SPTBN2 groups. The C2: curated gene sets (CP: KEGG gene set) used in GSEA were downloaded from the MSigDB database (version 7.1, https://www.gsea-msigdb.org/gsea/msigdb) [15, 16].
2.12. Western Blot Analysis
The protein was prepared from the cells by using RIPA lysis buffer. Protein concentrations were quantified by using a bicinchoninic acid assay kit (23227, Thermo Fisher Scientific). The total cell lysate proteins were separated by using SDS/PAGE gel (10%, 80–120 V) and electrotransferred onto a 0.22 μm polyvinylidene fluoride membrane (PVDF; 300 mA for 70–100 min). Then, the PVDF membranes were incubated with 5% skimmed milk (232100, BD, MD, US) for 90 min at normal room temperature. Subsequently, the membranes were incubated with the primary antibody at 4°C overnight and incubated with the corresponding secondary antibody at room temperature for 1 h. The primary antibodies were as follows: SPTBN2 (1 : 500 dilution; human; 55107-1-AP, ProteinTech Group, Inc., China), Bax (1 : 2000 dilution; human; 50599-2-Ig, ProteinTech Group, Inc., China), caspase-8 (1 : 500 dilution; human; 13423-1-AP, ProteinTech Group, Inc., China), caspase-3 (1 : 500 dilution; human; 19677-1-AP, ProteinTech Group, Inc., China), Bcl-2 (1 : 500 dilution; human; ab182858, Abcam, Shanghai, China), CCNE2 (1 : 500 dilution; human; 507032, ZENBIO, Chengdu, China), CDK2 (1 : 500 dilution; human; 10122-1-AP, ProteinTech Group, Inc., China), CDK4 (1 : 500 dilution; human; 11026-1-AP, ProteinTech Group, Inc., China), and P21 (1 : 500 dilution; human; 10355-1-AP, ProteinTech Group, Inc., China).
2.13. Statistical Analysis
The differential expression of SPTBN2 between TC and paired nontumor samples was analyzed through a t-test (2-tailed; paired t-test or independent t-test). Medcalc was used to perform receiver operating characteristic (ROC) analysis. The χ2 test and Fisher's exact test were used to evaluate the associations between clinicopathological characteristics and SPTBN2 expression. Univariate and multivariate Cox regression analyses were conducted to evaluate the association between SPTBN2 and tumor size. The analyses were performed with the statistical software SPSS (IBM SPSS, v23.0; SPSS Inc.) and Medcalc (v12.7.0; MedCalc Software, Ltd). P < 0.05 (∗) was considered statistically significant.
3. Results
3.1. SPTBN2 Was Highly Overexpressed in PTC
RNA-seq was performed on 79 PTC tissue samples and paired adjacent nontumor tissues to examine the potential mechanism of PTC. We found that the expression of SPTBN2 in the PTC samples was highly upregulated compared with that in the adjacent nontumor thyroid tissues (Figure 1(a), Supplementary Table 1). Tissues from patients with PTC and normal thyroid tissues in TCGA dataset were also investigated (Figure 1(c)). The mRNA expression level of SPTBN2 was also upregulated in our local validated cohort (48 PTC patients) as revealed by qRT-PCR (Figure 1(e)). Both results indicated that SPTBN2 was highly upregulated in the PTC samples relative to that in the nontumor thyroid samples. The diagnostic potential of SPTBN2 was evaluated on the basis of a ROC curve. ROC curve analysis was performed on all of the above cohorts (our RNA-seq cohort, our local validated cohort, and TCGA RNA-seq cohort) (Figures 1(b), 1(d), and 1(f)). The results indicated that SPTBN2 is a potential gene for the diagnosis of PTC.
Figure 1.
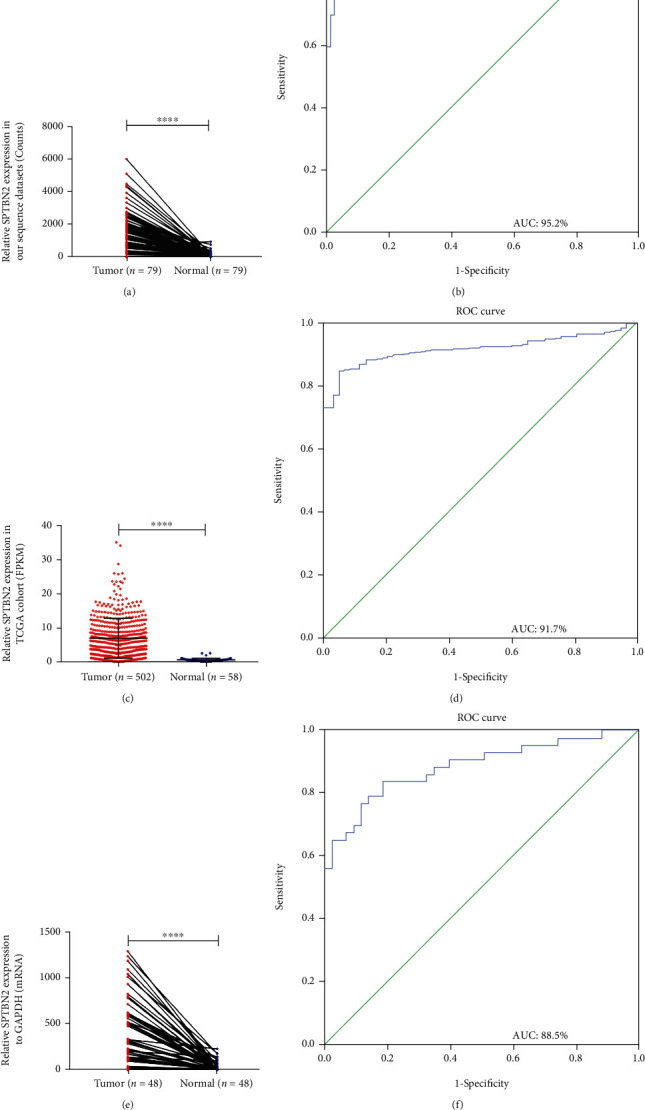
SPTBN2 in PTC samples was highly upregulated compared with that in nontumor tissues. (a) RNA-seq dataset containing 79 PTC tissue samples and paired adjacent nontumor tissue samples (P < 0.0001). (b) ROC for the expression of SPTBN2 in the RNA-seq cohort (AUC = 95.2%, sensitivity = 89.9%, and specificity = 2.4%) (P < 0.001). (c) TCGA dataset containing 502 PTC tissues and 58 normal thyroid tissues (P < 0.0001). (d) ROC for the expression of SPTBN2 in TCGA cohort (AUC = 91.7%, sensitivity = 85.1%, and specificity = 94.8%) (P < 0.001). (e) Local validated cohort containing 48 PTC tissue samples and paired adjacent nontumor tissue samples (P < 0.0001). (f) ROC for the expression of SPTBN2 in the local validated cohort (AUC = 88.5%, sensitivity = 83.7%, and specificity = 81.4%) (P < 0.001). The optimal cutoff value of SPTBN2 expression was determined by maximizing the Youden index value. Youden index = (sensitivity + specificity) − 1. The data are presented as the mean ± standard deviation (three independent experiments). ∗∗∗∗P < 0.0001.
3.2. SPTBN2 Expression Was Closely Related to Clinicopathological Characteristics in PTC
We examined the relationship between clinicopathological features and SPTBN2 expression in our local validated cohort and TCGA cohort to gain insights into the mechanism of SPTBN2 in the progression of PTC. The PTC tissues in TCGA cohort (502 patients with PTC) and our local validated cohort (48 patients with PTC) were classified individually into the high SPTBN2 expression group or the low SPTBN2 expression group in accordance with their median expression level value of SPTBN2. The results of the analyses showed that in TCGA cohort, histological type, LNM, tumor size, and disease stage were related to SPTBN2 upregulation (Table 3). Similar results were found for our validated cohort. Specifically, the upregulation of SPTBN2 was correlated with tumor size (P = 0.004), disease stage (P < 0.001), and LNM (P = 0.002) (Table 4). We omit the nonstatistically significant clinicopathologic features (Table 3) in Table 4. All analytical results provided support that SPTBN2 may act as an oncogene in PTC.
Table 3.
The association between SPTBN2 expression and clinicopathologic features in TCGA cohort.
| Clinicopathologic features | High expression (n = 251) | Low expression (n = 251) | X 2 | P value |
|---|---|---|---|---|
| Gender | 0.253 | 0.615 | ||
| Female | 186 | 181 | ||
| Male | 65 | 70 | ||
| Age (years) | ||||
| Mean ± SD | 46.876 ± 15.236 | 47.809 ± 16.363 | 0.128 | 0.721 |
| ≤45 | 116 | 120 | ||
| >45 | 135 | 131 | ||
| Multi-nodularity | 0.805 | 0.37 | ||
| Yes | 118 | 108 | ||
| No | 133 | 143 | ||
| Histological type | 26.605 | <0.001a | ||
| Classical | 202 | 149 | ||
| Other types | 49 | 102 | ||
| Tumor size (mm) | 18.594 | <0.001a | ||
| ≥20 | 120 | 73 | ||
| <20 | 131 | 178 | ||
| Lymph-node metastasis | 27.167 | <0.001a | ||
| Yes | 140 | 82 | ||
| No | 111 | 169 | ||
| Distant metastasis | 0 | 1 | ||
| Yes | 5 | 4 | ||
| No | 246 | 247 | ||
| Disease stage (AJCC7) | 15.084 | <0.001a | ||
| I+II | 147 | 188 | ||
| III+IV | 104 | 63 |
Chi-square test. aP value < 0.05. AJCC: American Joint Committee on Cancer.
Table 4.
The association between SPTBN2 expression and clinicopathologic features in the local validated cohort.
| Clinicopathologic features | High expression (n = 24) | Low expression (n = 24) | X 2 | P value |
|---|---|---|---|---|
| Tumor size (mm) | 2.064 | 0.004a | ||
| ≥20 | 18 | 8 | ||
| <20 | 6 | 16 | ||
| Lymph-node metastasis | 12.343 | <0.001a | ||
| Yes | 20 | 8 | ||
| No | 4 | 16 | ||
| Disease stage (AJCC7) | 5.371 | 0.020a | ||
| I+II | 9 | 17 | ||
| III+IV | 15 | 7 |
Chi-square test. aP value < 0.05. AJCC: American Joint Committee on Cancer.
3.3. SPTBN2 Overexpression Was Correlated with Increased Tumor Size
Logistic regression studies were applied to evaluate the relationship between SPTBN2 and tumor size in PTC. In the univariate logistic regression analysis, age, gender, SPTBN2 expression, LNM, and disease stage were significant risk factors for increased tumor size (Table 5). Subsequently, multivariate logistic regression analysis was conducted on the variables that were found to be statistically significant in the univariate logistic regression analysis. The results indicated that age, gender, SPTBN2 expression, and disease stage were significant independent risk factors for increased tumor size (Table 6).
Table 5.
Univariate logistic regression analysis for the risk of increased tumor size.
| Factor | OR | 95% CI | P value |
|---|---|---|---|
| Age (≤45 vs. >45) | 1.969 | 1.362-2.845 | <0.001a |
| Gender (male vs. female) | 0.553 | 0.371-0.825 | 0.004a |
| SPTBN2 expression (low vs. high.) | 2.234 | 1.546-3.228 | <0.001a |
| Histological type (PTC vs. others.) | 0.963 | 0.651-1.424 | 0.85 |
| Lymph-node metastasis | 2.686 | 1.855-3.889 | <0.001a |
| Disease stage (AJCC7) | 11.117 | 7.196-17.199 | <0.001a |
Chi-square test. aP value < 0.05. AJCC: American Joint Committee on Cancer.
Table 6.
Multivariate logistic regression analysis for the risk of increased tumor size.
| Factor | OR | 95% CI | P value |
|---|---|---|---|
| Age (≤45 vs. >45) | 0.127 | 0.050-0.326 | <0.001a |
| Gender (male vs. female) | 0.611 | 0.376-0.993 | 0.047a |
| SPTBN2 expression (low vs. high) | 1.664 | 1.071-2.583 | 0.023a |
| Disease stage (AJCC7) | 51.751 | 19.838-135.004 | <0.001a |
Chi-square test. aP value < 0.05. AJCC: American Joint Committee on Cancer.
3.4. Effects of the Underexpression of SPTBN2 on the Proliferation Capacity of TC Cell Lines
We evaluated the RNA and protein expression levels of SPTBN2 in three malignant thyroid cell lines and HTORI-3. As shown in Figures 2(a) and 2(c), the mRNA expression level of SPTBN2 in the malignant cell lines was markedly greater than that in the normal cell lines (HTORI-3). Clearly, SPTBN2 was commonly overexpressed in thyroid malignancy tissues and cell lines. siRNA was used to knock down SPTBN2 in malignant thyroid cell lines (BCPAP and KTC-1) with high SPTBN2 expression to further study the function of SPTBN2 in thyroid malignancies. The transfection efficiency of the siRNA was investigated by using qRT-PCR and Western blot (WB) analyses. The results indicated that the siRNA transfection efficiency exceeded 50% (Figures 2(b) and 2(d)). Then, the proliferation capacity of the malignant thyroid cells was investigated by using CCK-8 and colony formation assays to evaluate the function of SPTBN2 in TC. The results of the CCK-8 and colony formation assays showed that the downregulation of SPTBN2 markedly suppressed the proliferation of the KTC-1 and BCPAP cell lines (Figures 2(e)–2(g)).
Figure 2.
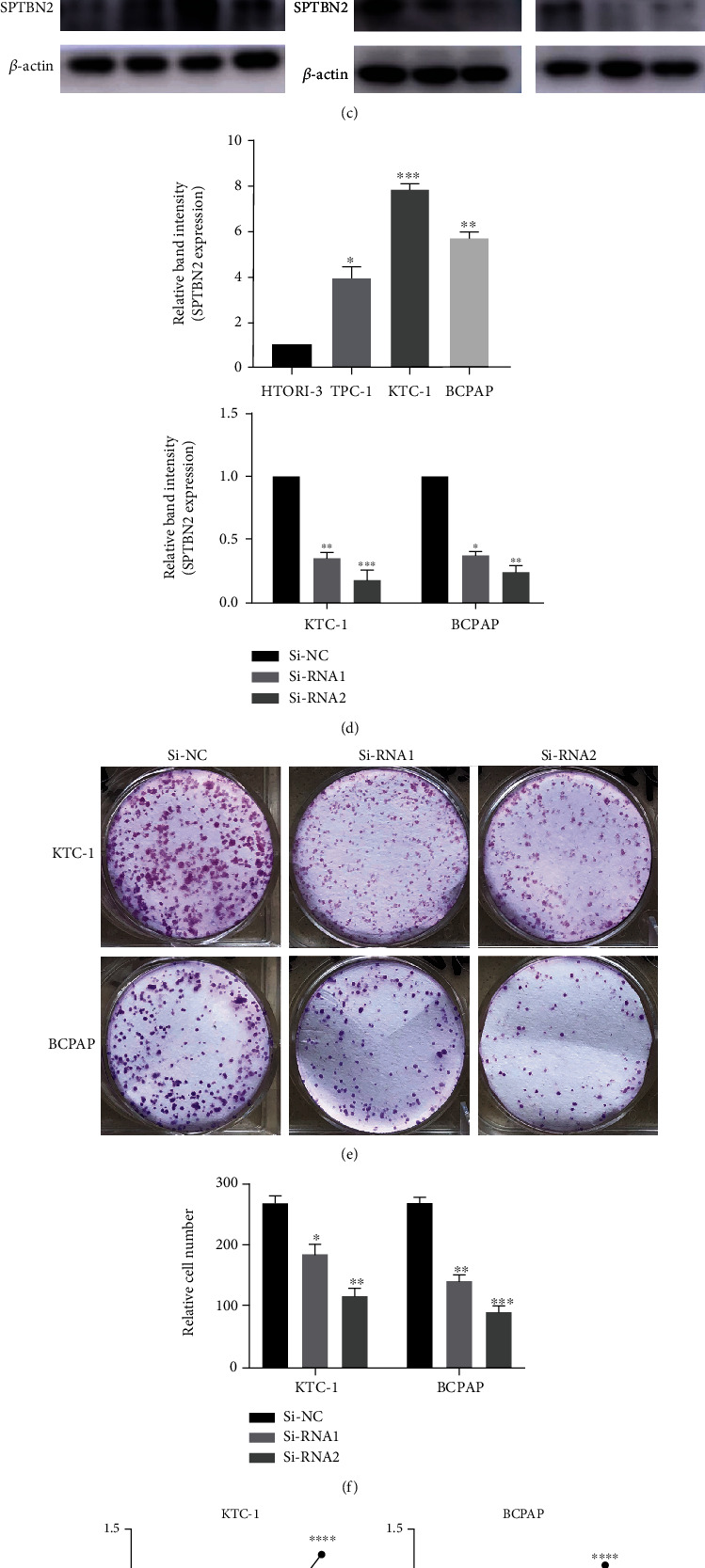
SPTBN2 expression in thyroid cells and siRNA transfection efficiency in KTC-1 and BCPAP. SPTBN2 knockdown inhibited TC cell line proliferation. (a) Relative expression of SPTBN2 (normalized to GADPH) in the three malignant thyroid cell lines and HTORI-3 (normal thyroid follicular epithelial cell line). (b) Relative expression of SPTBN2 (normalized to GADPH) in the TC cell lines (KTC-1 and the BCPAP) after siRNA treatment as detected via qRT-PCR. (c) SPTBN2 protein level in the thyroid cell lines HTORI-3, TPC-1, KTC-1, and BCPAP and the siRNA-transfected KTC-1 and BCPAP cell lines as determined via WB analysis. (d) Quantitative results of the (C) WB analysis (normalized to β-actin). (e) Colony formation assay. TC cells treated with siRNA were plated onto six-well plates and incubated for 7 days. Compared with that of the Si-NC group, the proliferation capacity of the malignant thyroid cell lines in the Si-RNA1 and Si-RNA2 groups was exceedingly high. (f) Relative quantification of the (E) colony formation assay. (g) CCK-8 proliferation assay was conducted to quantify the proliferation of transfected cells. The data are presented as the mean ± standard deviation (three independent experiments). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
3.5. Knockdown of SPTBN2 Inhibited TC Cell Migration and Invasion In Vitro
By subjecting TCGA cohort and our validated cohort to the χ2 test, we found that LNM was significantly correlated with SPTBN2 expression (Tables 3 and 4). Therefore, we studied the function of SPTBN2 in the migration/invasion capacities of TC. Migration capacity was assessed by using migration assays. The results demonstrated that in malignant thyroid cell lines, migration capacity was significantly suppressed by the knockdown of SPTBN2 (Figures 3(a) and 3(b)). The invasion capacity of the transfected TC cell lines was detected via invasion assays. The results showed that the invasion capability of the TC cell lines was also inhibited by the knockdown of SPTBN2 (Figures 3(c) and 3(d)).
Figure 3.
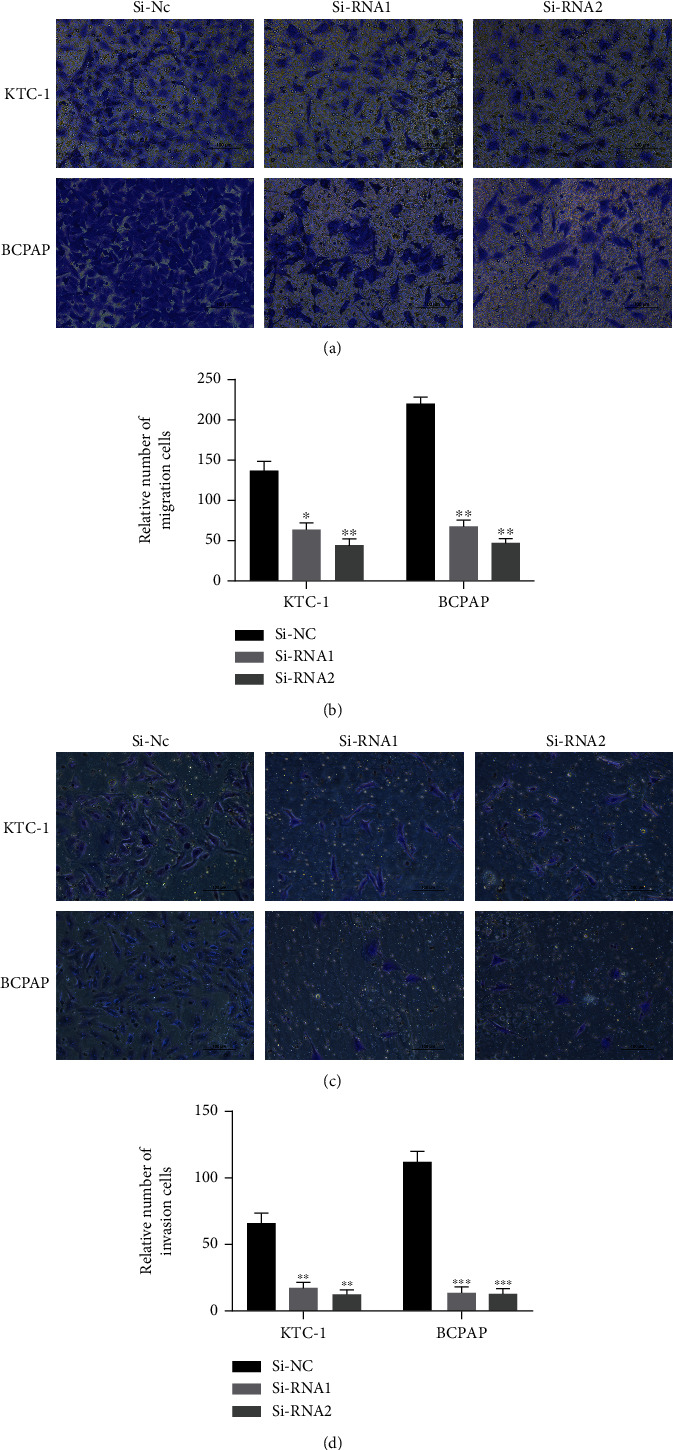
Underexpression of SPTBN2 suppressed cell migration and invasion capability in malignant thyroid cell lines (KTC-1 and BCPAP). (a) Cell migration assays. The migration capability of TC cells was suppressed by the downregulation of SPTBN2 (compared with Si-NC). (b) Quantitative results of the cell migration assay. (c) Cell invasion assays. The invasion capability of TC cells was inhibited by the knockdown of SPTBN2 (compared with Si-NC). (d) Quantitative results of the cell invasion assay. The data are presented the mean ± standard deviation (three independent experiments). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
3.6. Knockdown of SPTBN2 Induced Apoptosis and Retarded G1/S Cell Cycle Transition in TC Cell Lines In Vitro
The GSEA results revealed that in TCGA cohort, the cell cycle/apoptosis pathway was closely associated with SPTBN2 expression in thyroid cancer (Figure 4). Given that cell cycle regulation is closely associated with cell proliferation, we analyzed the function of SPTBN2 in the TC cell cycle. The proportion of G1-phase cells increased with the knockdown of SPTBN2, whereas that of S-phase KTC-1 and BCPAP cells decreased (Figures 5(a) and 5(b)). Hence, the G1/S transition was significantly suppressed with the knockdown of SPTBN2. Cell apoptosis is another cause of cell growth inhibition. Therefore, the percentage of apoptotic cells after siRNA transfection was assessed. The ratio of apoptotic cells after transfection with SPTBN2-siRNA significantly increased relative to that after transfection with the negative control (Figures 5(c) and 5(d)). These results indicated that SPTBN2 markedly promoted cell cycle transition and suppressed TC cell apoptosis.
Figure 4.
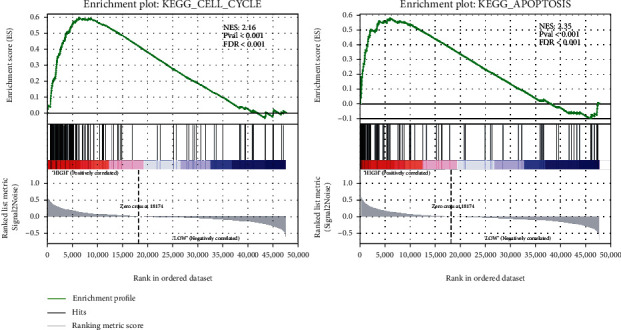
GSEA was used to evaluate the pathways that were associated with SPTBN2 expression in TCGA dataset.
Figure 5.
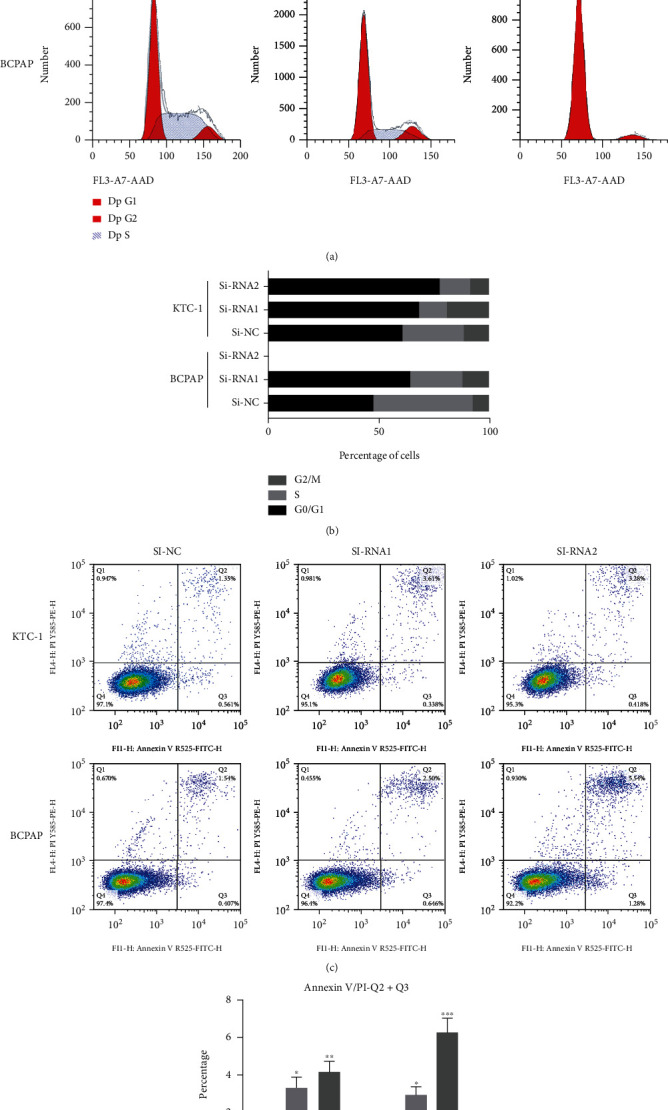
SPTBN2 gene knockdown induced apoptosis and retarded G1/S cell cycle transition in TC cells. (a) Flow cytometry assay of cell cycle distribution after SPTBN2 downregulation. (b) Distribution of cell cycle stages. (c) Flow cytometry analysis revealed the apoptosis rates of siRNA-transfected KTC-1 and BCPAP cells. (d) Quantitative analysis of apoptotic cell percentages. Percentage of cell apoptosis = Q2 (early apoptosis) + Q3 (late apoptosis). ∗P < 0.05, ∗∗P < 0.01, and∗∗∗P < 0.001. The data are presented as the mean ± standard deviation (three independent experiments).
3.7. SPTBN2 Knockdown Suppressed the Proliferation of PTC Cells by Arresting G1/S Transition and Promoting Cell Apoptosis
After transfecting TC cells with siRNA, we performed WB analysis to detect the expression levels of signaling biomarker proteins that were correlated with the cell cycle/apoptosis in TC cell lines. The results illustrated that in TC cells, the knockdown of SPTBN2 significantly inhibited the expression levels of cell cycle/anti-apoptotic proteins, including CCNE2, CDK2, CDK4, and Bcl-2 (Figure 6(a)). Furthermore, the downregulation of SPTBN2 significantly increased the expression levels of P21, cleaved caspase-8, cleaved caspase-3, and Bax in TC cells (Figures 6(a) and 6(b)). These results indicated that the knockdown of SPTBN2 inhibited the G1/S phase transition via the upregulation of P21 and the downregulation of CCNE2, CDK2, and CDK4. Moreover, it promoted TC cell apoptosis via the upregulation of Bax, cleaved-caspase-3, and cleaved-caspase-8 and the downregulation of Bcl-2.
Figure 6.
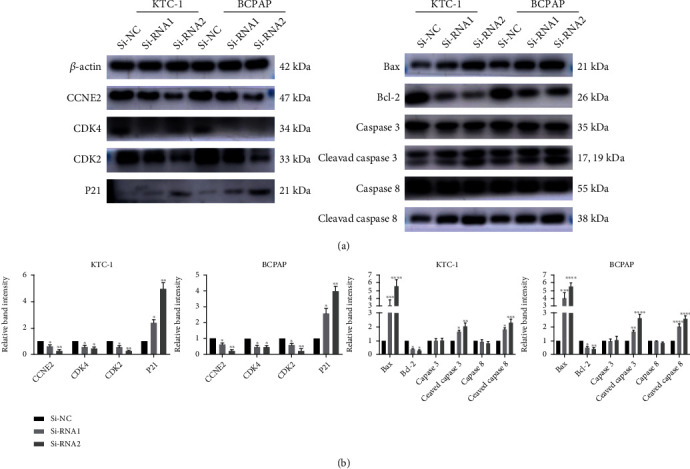
Downregulation of SPTBN2 inhibited the tumorigenesis and progression of TC cell lines by arresting G1/S transition and promoting cell apoptosis. (a) Levels of CCNE2, CDK2, CDK4, Bcl-2, P21, caspase-3, cleaved-caspase3, caspase-8, cleaved-caspase8, and Bax were evaluated via WB analysis with β-actin as the loading control. (b) Quantitative results of WB analysis (normalized to β-actin). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and∗∗∗∗P < 0.0001. The data are presented as the mean ± standard deviation (three independent experiments).
4. Discussion
Thyroid cancer is a heavy burden on society, and its morbidity has continuously increased on an annual basis [17]. PTC, the most frequent thyroid cancer type (>85%), has been attracting growing attention [18]. Approximately 10% of patients with PTC cannot benefit from conventional therapies due to metastasis and recurrence [19]. Thus, exploring the fundamental molecular mechanisms of TC is essential for the identification and management of this malignancy. A number of studies have reported that numerous genes play a vital role in the tumorigenesis of TC [20–22]. However, the current understanding of the molecular mechanism of TC is far from comprehensive.
The widely used next-generation sequencing technology provides an opportunity for elucidating the molecular mechanisms of TC. Previously, we subjected 79 paired PTC samples and nontumor thyroid tissues to whole transcriptome sequencing. Through RNA-seq data analysis, we discovered that SPTBN2 expression in PTC samples was highly upregulated compared with that in adjacent nontumor thyroid samples. Furthermore, in patients with PTC in TCGA cohort and our local validated cohort (48 PTC patients), clinicopathological characteristics (tumor size, LNM, and disease stage) showed a high positive correlation with the high expression of SPTBN2.
The SPTBN2 gene is located on chromosome 11q13.2 [23]. It codes for a protein called SPTBN2 or beta-III spectrin. Beta-III spectrin functions in diverse cellular functions, including the determination of cell form, the arrangement of transmembrane proteins, and the association of organelles. SPTBN2 mutation can result in spinocerebellar ataxias type-5. This disease is characterized by neural degeneration, uncoordinated eye movements, and locomotor incoordination. SPTBN1, which is an important paralog of SPTBN2, and other nonerythroid spectrins exert either oncogenic or tumor suppressor activities by affecting various cancer-related biological processes, including cell differentiation, DNA damage repair, and epithelial-to-mesenchymal transition [24]. A growing body of evidence has extensively proven that SPTBN2 plays a vital role in the diagnosis of cholangiocarcinoma. For example, by using a bioinformatics approach, Feng et al. demonstrated that SPTBN2 is highly associated with poor prognosis in ovarian cancer [25]. Zhang et al. and Yue et al. reported that SPTBN2 may play a vital role in the progression of colorectal cancer [26, 27]. Wu et al. reported that SPTBN2 is positively correlated with the poor prognosis of lung adenocarcinoma [28]. However, the probable mechanisms of SPTBN2 in this malignancy has never been investigated before [29–31].
Although bioinformatics studies have revealed that SPTBN2 may be correlated with the initiation and progression of malignancies, the molecular mechanism of SPTBN2 in thyroid cancer has never been investigated. In this study, we discovered that SPTBN2 was exceedingly overexpressed in PTC tissues in our RNA-seq and TCGA datasets. Then, we evaluated the expression of SPTBN2 in 48 PTC samples and paired adjacent nontumor tissues via qRT-PCR and explored the biological function of SPTBN2 in vitro. The results of qRT-PCR were consistent with those of previous RNA-seq and TCGA analyses.
Previous in vitro studies on TC cell lines (BCPAP and KTC-1) knocked down SPTBN2 expression by using siRNA. The results of the cell experiments indicated that the knockdown of SPTBN2 could inhibit proliferation and metastasis capacity and induce apoptosis in the TC cell line in vitro. Furthermore, the signaling pathways that might be associated with SPTBN2 were analyzed via the GSEA. Since the GSEA showed downregulation of SPTBN2 related to the apoptosis and cell cycle signaling pathways in TCGA dataset, we focused on the expression level of the apoptosis relative protein (Bax, Bcl-2, caspase-3, caspase-8, caspase-9, cleaved caspase-3, cleaved caspase-8, and cleaved caspase-9) and the cell-cycle relative protein (P21, CCNE2, CDK2, and CDK4) in the transfected TC cell line. Moreover, the downregulation of SPTBN2 led to a substantial increase in the protein levels of Bax, cleaved-caspase-8, and cleaved-caspase-3 and a reduction in CCNE2, CDK2, CDK4, and Bcl-2. This discovery suggested that SPTBN2 could promote the progression of TC cell lines by inhibiting G1/S transition and promoting apoptosis.
We showed that SPTBN2 was closely associated with tumor size, LNM, and disease stage. Furthermore, in TC cells, the knockdown of SPTBN2 inhibited G1/S transition and promoted apoptosis. These findings proved that SPTBN2 is a good candidate gene for TC diagnosis, treatment, and prevention.
Acknowledgments
This study was funded by the Wenzhou Science and Technology Planning Project (Y20180460 and Y20190204).
Data Availability
The datasets generated and/or analyzed during the current study are not publically available due to restrictions on data sharing imposed by the funding body but are available from the corresponding author upon reasonable request.
Ethical Approval
The present study was approved by the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University (approval no. 2012-57).
Consent
Written informed consent was obtained from all patients.
Conflicts of Interest
The authors declare no conflict of interest.
Authors' Contributions
XZ and LL performed the main experiments and wrote the manuscript. YQ and MX collected and analyzed the raw data. QX and YW performed partial experiments and revised the article. JQ designed the study and was responsible for confirming the authenticity of the data. All authors read and approved the final manuscript. Xiaofen Zhou and Lizhi Lin contributed equally to this work.
Supplementary Materials
Comparison of SPTBN2 mRNA expression between thyroid tumors and normal thyroid tissues in our local RNA-seq dataset.
References
- 1.Torre L. A., Siegel R. L., Ward E. M., Jemal A. Global cancer incidence and mortality rates and trends--an update. Cancer Epidemiology, Biomarkers & Prevention . 2016;25(1):16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2019. CA: a Cancer Journal for Clinicians . 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Morris L. G., Sikora A. G., Tosteson T. D., Davies L. The increasing incidence of thyroid cancer: the influence of access to care. Thyroid . 2013;23(7):885–891. doi: 10.1089/thy.2013.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabanillas M. E., McFadden D. G., Durante C. Thyroid cancer. Lancet . 2016;388(10061):2783–2795. doi: 10.1016/S0140-6736(16)30172-6. [DOI] [PubMed] [Google Scholar]
- 5.Degroot L. J., Kaplan E. L., McCormick M., Straus F. H. Natural history, treatment, and course of papillary thyroid carcinoma. The Journal of Clinical Endocrinology and Metabolism . 1990;71(2):414–424. doi: 10.1210/jcem-71-2-414. [DOI] [PubMed] [Google Scholar]
- 6.Leboulleux S., Rubino C., Baudin E., et al. Prognostic factors for persistent or recurrent disease of papillary thyroid carcinoma with neck lymph node metastases and/or tumor extension beyond the thyroid capsule at initial diagnosis. The Journal of Clinical Endocrinology and Metabolism . 2005;90(10):5723–5729. doi: 10.1210/jc.2005-0285. [DOI] [PubMed] [Google Scholar]
- 7.Schneider D. F., Chen H. New developments in the diagnosis and treatment of thyroid cancer. CA: a Cancer Journal for Clinicians . 2013;63(6):373–394. doi: 10.3322/caac.21195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelstein B., Papadopoulos N., Velculescu V. E., Zhou S., Diaz L. A., Kinzler K. W. Cancer genome landscapes. Science . 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croce C. M. Oncogenes and cancer. The New England Journal of Medicine . 2008;358(5):502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 10.Xing M., Alzahrani A. S., Carson K. A., et al. Association between BRAF V600E mutation and recurrence of papillary thyroid cancer. Journal of Clinical Oncology . 2015;33(1):42–50. doi: 10.1200/JCO.2014.56.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wells S. A., Jr., Robinson B. G., Gagel R. F., et al. Vandetanib in patients with locally advanced or metastatic medullary thyroid cancer: a randomized, double-blind phase III trial. Journal of Clinical Oncology . 2012;30(2):134–141. doi: 10.1200/JCO.2011.35.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z., Hou P., Ji M., et al. Highly prevalent genetic alterations in receptor tyrosine kinases and phosphatidylinositol 3-kinase/akt and mitogen-activated protein kinase pathways in anaplastic and follicular thyroid cancers. The Journal of Clinical Endocrinology and Metabolism . 2008;93(8):3106–3116. doi: 10.1210/jc.2008-0273. [DOI] [PubMed] [Google Scholar]
- 13.Nikiforov Y. E. Molecular analysis of thyroid tumors. Modern Pathology . 2011;24(Supplement 2):S34–S43. doi: 10.1038/modpathol.2010.167. [DOI] [PubMed] [Google Scholar]
- 14.Nikiforov Y. E. RET/PTC rearrangement in thyroid tumors. Endocrine Pathology . 2002;13(1):3–16. doi: 10.1385/EP:13:1:03. [DOI] [PubMed] [Google Scholar]
- 15.Mootha V. K., Lindgren C. M., Eriksson K. F., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics . 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 16.Subramanian A., Tamayo P., Mootha V. K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America . 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies L., Welch H. G. Increasing incidence of thyroid cancer in the United States, 1973-2002. JAMA . 2006;295(18):2164–2167. doi: 10.1001/jama.295.18.2164. [DOI] [PubMed] [Google Scholar]
- 18.Kuo S. F., Lin S. F., Chao T. C., Hsueh C., Lin K. J., Lin J. D. Prognosis of multifocal papillary thyroid carcinoma. International Journal of Endocrinology . 2013;2013:6. doi: 10.1155/2013/809382.809382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu F. H., Kuo S. F., Hsueh C., Chao T. C., Lin J. D. Postoperative recurrence of papillary thyroid carcinoma with lymph node metastasis. Journal of Surgical Oncology . 2015;112(2):149–154. doi: 10.1002/jso.23967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhan A., Soleimani M., Mandal S. S. Long noncoding RNA and cancer: a new paradigm. Cancer Research . 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Younis E. Oncogenesis of thyroid cancer. Asian Pacific Journal of Cancer Prevention . 2017;18(5):1191–1199. doi: 10.22034/APJCP.2017.18.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lian E. Y., Maritan S. M., Cockburn J. G., et al. Differential roles of RET isoforms in medullary and papillary thyroid carcinomas. Endocrine-Related Cancer . 2017;24(1):53–69. doi: 10.1530/ERC-16-0393. [DOI] [PubMed] [Google Scholar]
- 23.Wen J. X., Li X. Q., Chang Y. Signature gene identification of cancer occurrence and pattern recognition. Journal of Computational Biology . 2018;25(8):907–916. doi: 10.1089/cmb.2017.0261. [DOI] [PubMed] [Google Scholar]
- 24.Chen S., Li J., Zhou P., Zhi X. SPTBN1 and cancer, which links? Journal of Cellular Physiology . 2020;235(1):17–25. doi: 10.1002/jcp.28975. [DOI] [PubMed] [Google Scholar]
- 25.Feng P., Ge Z., Guo Z., Lin L., Yu Q. A comprehensive analysis of the downregulation of miRNA-1827 and its prognostic significance by targeting SPTBN2 and BCL2L1 in ovarian cancer. Frontiers in Molecular Biosciences . 2021;8, article 687576 doi: 10.3389/fmolb.2021.687576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z., Wang Q., Zhang M., et al. Comprehensive analysis of the transcriptome-wide m6A methylome in colorectal cancer by MeRIP sequencing. Epigenetics . 2021;16(4):425–435. doi: 10.1080/15592294.2020.1805684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yue M., Liu T., Yan G., Luo X., Wang L. LINC01605, regulated by the EP300-SMYD2 complex, potentiates the binding between METTL3 and SPTBN2 in colorectal cancer. Cancer Cell International . 2021;21(1):p. 504. doi: 10.1186/s12935-021-02180-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu C., Dong B., Huang L., et al. SPTBN2, a new biomarker of lung adenocarcinoma. Frontiers in Oncology . 2021;11, article 754290 doi: 10.3389/fonc.2021.754290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu S., Jue D., Albanese J., Wang Y., Liu Q. Utilization of spectrins βI and βIII in diagnosis of hepatocellular carcinoma. Annals of Diagnostic Pathology . 2019;39:86–91. doi: 10.1016/j.anndiagpath.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Bian X., Wang S., Jin S., et al. Two novel missense variants in SPTBN2 likely associated with spinocerebellar ataxia type 5. Neurological Sciences . 2021;42(12):5195–5203. doi: 10.1007/s10072-021-05204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romaniello R., Citterio A., Panzeri E., et al. Novel SPTBN2 gene mutation and first intragenic deletion in early onset spinocerebellar ataxia type 5. Annals of Clinical Translational Neurology . 2021;8(4):956–963. doi: 10.1002/acn3.51345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of SPTBN2 mRNA expression between thyroid tumors and normal thyroid tissues in our local RNA-seq dataset.
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publically available due to restrictions on data sharing imposed by the funding body but are available from the corresponding author upon reasonable request.


