Abstract
Objectives
Observational analyses suggest that high bone mineral density (BMD) is a risk factor for osteoarthritis (OA); it is unclear whether this represents a causal effect or shared aetiology and whether these relationships are body mass index (BMI)-independent. We performed bidirectional Mendelian randomization (MR) to uncover the causal pathways between BMD, BMI and OA.
Methods
One-sample (1S)MR estimates were generated by two-stage least-squares regression. Unweighted allele scores instrumented each exposure. Two-sample (2S)MR estimates were generated using inverse-variance weighted random-effects meta-analysis. Multivariable MR (MVMR), including BMD and BMI instruments in the same model, determined the BMI-independent causal pathway from BMD to OA. Latent causal variable (LCV) analysis, using weight-adjusted femoral neck (FN)–BMD and hip/knee OA summary statistics, determined whether genetic correlation explained the causal effect of BMD on OA.
Results
1SMR provided strong evidence for a causal effect of BMD estimated from heel ultrasound (eBMD) on hip and knee OA {odds ratio [OR]hip = 1.28 [95% confidence interval (CI) = 1.05, 1.57], p = 0.02, ORknee = 1.40 [95% CI = 1.20, 1.63], p = 3 × 10–5, OR per standard deviation [SD] increase}. 2SMR effect sizes were consistent in direction. Results suggested that the causal pathways between eBMD and OA were bidirectional (βhip = 1.10 [95% CI = 0.36, 1.84], p = 0.003, βknee = 4.16 [95% CI = 2.74, 5.57], p = 8 × 10–9, β = SD increase per doubling in risk). MVMR identified a BMI-independent causal pathway between eBMD and hip/knee OA. LCV suggested that genetic correlation (i.e. shared genetic aetiology) did not fully explain the causal effects of BMD on hip/knee OA.
Conclusions
These results provide evidence for a BMI-independent causal effect of eBMD on OA. Despite evidence of bidirectional effects, the effect of BMD on OA did not appear to be fully explained by shared genetic aetiology, suggesting a direct action of bone on joint deterioration.
Keywords: Osteoarthritis, bone mineral density, Mendelian randomization, body mass index, UK Biobank
Key Messages.
Mendelian randomization (MR) analyses suggest that bone mineral density (BMD), assessed from heel ultrasound scans, is a risk factor for osteoarthritis, independently of adiposity.
Evidence for reverse causality (i.e. a causal effect of osteoarthritis on BMD) may reflect the shared biological pathways contributing to bone and joint development.
Latent causal variable (LCV) analysis provides evidence for a direct causal effect of BMD on osteoarthritis, which is not fully explained by genetic correlation between these two traits.
This paper illustrates the utility of methods such as LCV analysis and multivariable MR when examining causal pathways in situations in which complex relationships exist, such as those between BMD, body mass index and osteoarthritis.
Introduction
Although osteoarthritis (OA) is a major cause of morbidity worldwide, effective pharmacological treatment remains elusive. It may be possible to develop novel therapeutic approaches based on understanding of risk factors. Several large population-based studies have identified positive relationships between bone mineral density (BMD) and hip and knee OA. 1 Mendelian randomization (MR), which is commonly used to explore causal relationships,2–4 has recently obtained evidence for a causal role of BMD on hip and knee OA risk.5 Body mass index (BMI), a risk factor for OA6–8 and positively associated with BMD,9 may bias MR estimates for the relationship between BMD and OA. Funck-Brentano et al. addressed this by excluding instrument(s) associated with BMI.5 An alternative approach, yet to be applied in this context, is the use of multivariable MR (MVMR) to estimate the direct causal effect of the exposure on the outcome when the instrument(s) are associated with multiple risk factors.10 Alternatively, rather than a causal effect of BMD on OA, shared biological pathways may contribute to both traits. Consistently with this possibility, a genetic correlation between lumbar spine (LS)–BMD and OA has been observed.11 Genetic correlation may give rise to bidirectional causal relationships in MR analysis.
As well as the relationship between BMD and OA, relationships with BMI could be characterized by bidirectional relationships. A causal effect of BMI on BMD is well established; the skeleton adapts to the increased load placed upon it by increasing BMD. Alternatively, a causal pathway between BMD and BMI is plausible via the metabolic effects of bone turnover. Murine osteocalcin knockouts have increased fat mass and are insulin-resistant;12 in humans, higher BMD is associated with lower circulating osteocalcin, which may mediate the positive association between BMD and fat mass. However, an MR analysis found no evidence of a causal pathway between femoral neck (FN) or LS–BMD and BMI in children.9
To provide a more complete understanding of the relationship between BMD and OA, we tested bidirectional relationships between BMD, OA and BMI (Figure 1) using one-sample (1S) and two-sample (2S) MR, and aimed to determine the direct (i.e. unconfounded) causal pathways between these variables using MVMR.
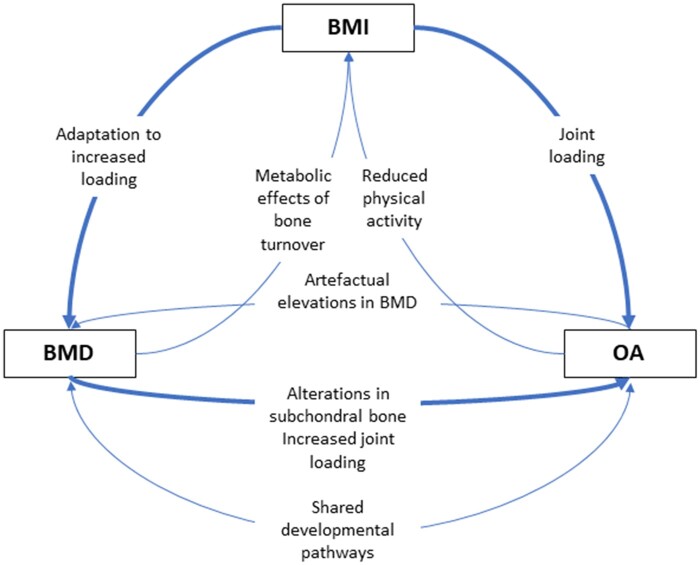
Figure 1.
Diagram summarizing hypothesized relationships between bone mineral density, body mass index and osteoarthritis
Thicker arrows represent stronger hypothesized relationships. Diagram does not take account of temporality of relationships due to the uncertainty in the temporal sequence, e.g. OA may first cause an increase in BMI due to reduced PA, leading to further OA through greater joint loading; however, it is equally possible that BMI leading to an increase in joint loading is the initiating event. BMD, bone mineral density; BMI, body mass index; OA, osteoarthritis.
Methods
Individual-level analyses
Individual-level analyses were performed in the UK Biobank population. UK Biobank is a UK-wide population-based health research resource consisting of ∼500 000 people, aged 38–73 years, who were recruited in 2006–2010.13 Participants provided a range of information [e.g. demographics, health status, lifestyle/physical activity (PA) measures] via questionnaires and interviews; anthropometric measures and blood samples were taken (data available at www.ukbiobank.ac.uk). A full description of the study design, participants and quality-control methods has been published.13 Methods for assessing BMD and ascertaining hospital-diagnosed OA status are described in the Supplementary Information (available as Supplementary data at IJE online). UK Biobank received ethical approval from the Research Ethics Committee (REC reference: 11/NW/0382).
Data collection, genotyping, and imputation and observational analyses in UK Biobank are described in the Supplementary Material (available as Supplementary data at IJE online).
MR
A summary of all MR analyses performed, along with the source of each of the instruments, is presented in Table 1 and of the assumptions of MR and how we tested these in Figure 2. We examined causal relationships with hip and knee OA separately, given the availability of separate genome-wide association studies (GWAS) for these outcomes, which have no overlap in terms of the most strongly associated single-nucleotide polymorphisms (SNPs).
Table 1.
Summary of all one-sample and two-sample Mendelian randomization analyses performed
| Exposure |
Outcome |
|||
|---|---|---|---|---|
| Source | Source | |||
| 1S | eBMD |
|
Knee OA | Individual-level HD knee OA status in UK Biobank |
| 2S | eBMD |
|
Knee OA |
|
| 1S | eBMD |
|
Hip OA | Individual-level HD hip OA status in UK Biobank |
| 2S | eBMD |
|
Hip OA |
|
| 1S | eBMD |
|
BMI | Individual-level BMI data in UK Biobank |
| 2S | eBMD |
|
BMI |
|
| 1S | BMI |
|
Knee OA | Individual-level HD knee OA status in UK Biobank |
| 2S | BMI |
|
Knee OA |
|
| 1S | BMI |
|
Hip OA | Individual-level HD hip OA status in UK Biobank |
| 2S | BMI |
|
Hip OA |
|
| 1S | BMI |
|
eBMD | Individual-level eBMD data in UK Biobank |
| 2S | BMI |
|
eBMD |
|
| 1S | Knee OA |
|
eBMD | Individual-level eBMD data in UK Biobank |
| 2S | Knee OA |
|
eBMD |
|
| 1S | Knee OA |
|
BMI | Individual-level BMI data in UK Biobank |
| 2S | Knee OA |
|
BMI |
|
| 1S | Hip OA |
|
eBMD | Individual-level eBMD data in UK Biobank |
| 2S | Hip OA |
|
eBMD |
|
| 1S | Hip OA |
|
BMI | Individual-level BMI data in UK Biobank |
| 2S | Hip OA |
|
BMI |
|
1S, one-sample MR; 2S, two-sample MR; I, instrumented by; ca, cases; co, controls; eBMD, estimated bone mineral density; GEFOS, Genetic Factors for Osteoporosis; GO, Genetics of Osteoarthritis; GWAS, genome-wide association study; BMI, body mass index; HD, hospital-diagnosed; FN–BMD, femoral neck bone mineral density; OA, osteoarthritis; GIANT, Genetic Investigation of Anthropometric Traits; arcOGEN, Arthritis Research UK Osteoarthritis Genetics.
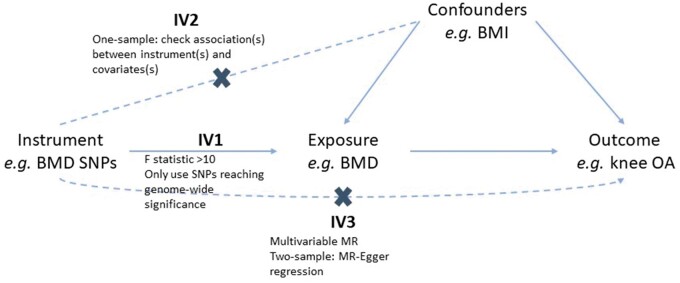
Figure 2.
Assumptions of Mendelian randomization and how we tested these assumptions
For a Mendelian randomization (MR) effect estimate to be valid, the instrument(s) must satisfy three key assumptions:3 IV1 [the instrument(s) must be robustly associated with the exposure], IV2 [the instrument(s) must not be associated with any confounders of the exposure–outcome relationship] and IV3 [the instruments(s) can only be associated with the outcome via the exposure and not via a different biological pathway independent of the exposure (i.e. horizontal pleiotropy)]. In one-sample analyses, IV1 was tested by calculating the F-statistic, which is a measure of instrument strength. A >10 threshold is used to determine sufficient instrument strength.2 IV2 was tested by determining the association between the instruments and potential confounders of the exposure–outcome relationship. In two-sample analyses, to satisfy IV1, we ensured that all instruments were robustly associated with the exposure by only including SNPs associated with the exposure at genome-wide significance. To address IV3, MR–Egger regression was performed to generate an estimate of horizontal pleiotropy (intercept) and a pleiotropy-robust estimate of the causal effect (slope). Weighted median regression was performed to determine the robustness of IVW estimates as weighted median estimates are valid even if ≤50% of the SNPs are not valid instruments.20 BMD, bone mineral density; BMI, body mass index; OA, osteoarthritis; SNP, single-nucleotide polymorphism.
One-sample MR
1SMR analyses were performed in the UK Biobank population using the instrumental-variable regression (‘ivreg’) function of the Applied Econometrics with R package.14 Exposures were instrumented by an unweighted genetic risk score (GRS), generated as the sum of the dosage for exposure-increasing alleles (data sources provided in Table 1). Analyses were adjusted for age at BMD/BMI assessment, sex, genotyping chip and 40 principal components. Continuous exposures (eBMD/BMI) were standardized before analysis. Effect estimates for binary outcomes (hip/knee OA) were generated from a linear two-stage least-squares regression and represent the increased probability of having OA per unit increase in the exposure. We generated an estimate of the odds ratio (OR) per standard deviation (SD) increase in the exposure, for comparison with 2SMR results, by first regressing the instruments on the exposure, generating predicted values of the exposure, and then regressing the predicted values of the exposure on the binary outcomes using a logistic-regression model. The standard errors for this estimate are likely to be underestimated.15
Two-sample MR
To maximize the sample size, and thus statistical power, we performed 2SMR using summary-level data from published GWAS. 2SMR analyses were performed using the TwoSampleMR R package, version 0.4.22.16 SNP–exposure estimates were extracted for all SNPs associated with the exposure at genome-wide significance. Details of the Genetic Factors for Osteoporosis (GEFOS), Genetic Investigation of Anthropometric Traits (GIANT) and the Genetics of Osteoarthritis (GO) consortium providing the summary statistics for eBMD, BMI and OA, respectively, are provided in the Supplementary Information (available as Supplementary data at IJE online). Summary statistics for the eBMD, BMI, hip and knee OA instruments are provided in Supplementary Tables S2–S7 (available as Supplementary data at IJE online). Clumping was performed to exclude non-independent SNPs based on a pairwise r2 > 0.001. SNP–outcome effect estimates were then extracted for independent SNPs. SNP–outcome effect estimates for each analysis are presented in Supplementary Tables S2–S7 (available as Supplementary data at IJE online). SNP–exposure and SNP–outcome data were harmonized to ensure that the effect estimates corresponded to the same allele. Palindromic SNPs with indeterminate allele frequencies [minor allele frequency (MAF) > 0.42] were excluded. Steiger filtering excluded SNPs that explained a greater proportion of the variance in the outcome than the exposure.17 The proportion of variance explained by each SNP was calculated using the p-value and sample size and the ‘get_r_from_pn’ function of the ‘TwoSampleMR’ package for continuous variables and the ‘get_r_from_lor’ function for dichotomous variables. The ‘get_r_from_lor’ function requires the case prevalence in the study population to be specified, which was calculated as the number of cases divided by the total sample size (15% for knee OA and 8% for hip OA). Seven, four and two eBMD SNPs were excluded for analyses with hip OA, knee OA and BMI outcomes, respectively. Two BMI SNPs explained a greater proportion of variance in hip OA risk, 1 for knee OA risk and 15 for eBMD. One knee OA SNP was excluded due to a greater r2 for eBMD. All Steiger-filtered SNPs are listed in Supplementary Tables S2–S7 (available as Supplementary data at IJE online). Estimates were generated using inverse-variance weighted (IVW) random-effects meta-analysis of the Wald ratios for each SNP.
Multivariable MR
As we hypothesized that BMI is a confounder of the BMD–OA relationship (i.e. a common causes of both phenotypes), we determined the independent effect of BMD on OA outcomes by performing 1S MVMR including GRS for both BMI and BMD as instruments. Both instruments were regressed on each exposure to generate a predicted value for each exposure. The predicted values for each exposure were then included in a multivariable regression to generate the effect of one exposure on OA when conditioning on the other exposure. Analyses were adjusted for sex, genotyping chip and 40 principal components (PCs). Sanderson–Windmeijer conditional F-statistics were calculated as measures of instrument strength in MVMR analyses.18
Sensitivity analyses
MR–Egger regression was performed to generate an estimate of horizontal pleiotropy in the two-sample analyses.19 Weighted median regression determined the robustness of IVW estimates as weighted median estimates are valid as long as 50% of the information is derived from valid instruments.20 We repeated the 2SMR analyses restricted to eBMD SNPs also associated with FN–BMD (p < 5 × 10–8) in the GEFOS FN–BMD meta-analysis,21 to determine whether FN–BMD has a stronger effect than eBMD on hip or knee OA risk. We also performed a latent causal variable (LCV) model, as described by O’Connor and Price,22 to determine whether there is a true causal effect of BMD on OA, independently of the genetic correlation. Full methods are described in the Supplementary Information (available as Supplementary data at IJE online).
Results
Confirming observational relationships between BMD, OA and BMI in UK Biobank
A total of 334 061 individuals in UK Biobank with genotype data also had measurements of eBMD, BMI, covariates and hospital-diagnosed hip OA; 341 920 had data for knee OA. The mean (SD) age of those with hip OA was 61.7 (6.0), of those with knee OA was 60.2 (6.9) and of controls was 56.2 (8.1) years (Supplementary Table S8, available as Supplementary data at IJE online). Fifty-seven per cent of people with hip OA were female compared with 50% with knee OA and 54% of the controls. Both hip and knee OA cases were heavier than controls, with mean BMI 28.9 (5.0), 30.3 (5.4) and 27.1 (4.6) kg/m2, respectively. Descriptive statistics were virtually the same when restricting to individuals with complete data for eBMD, BMI and OA who were included in the multivariable MR analyses (Supplementary Table S8, available as Supplementary data at IJE online).
MR analyses provide evidence for bidirectional causal pathways between BMD and OA
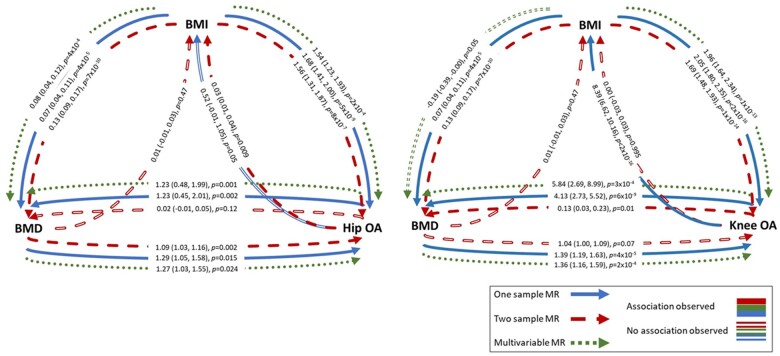
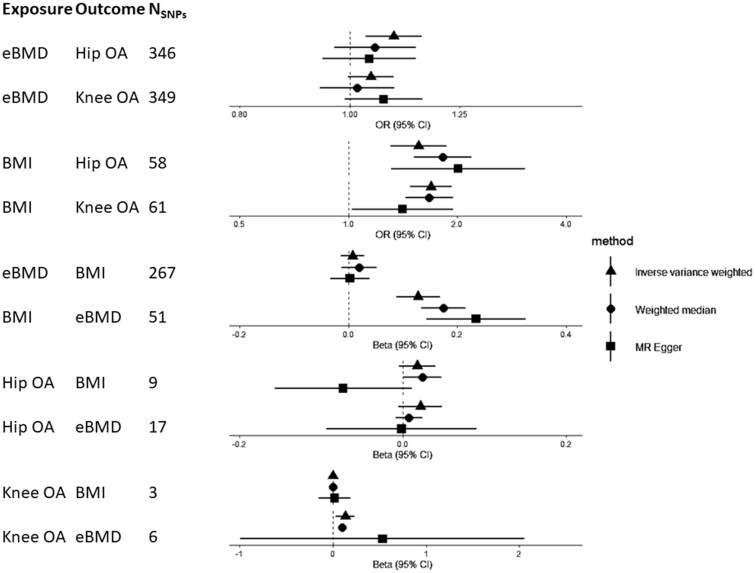
A summary of MR results is presented in Figure 3. In 1SMR, eBMD was causally related to both hip and knee OA, with an SD increase in eBMD related to a 29% [95% confidence interval (CI) = 5, 58] increased odds of having hip OA and 39% (95% CI = 19, 63) increased odds of having knee OA (Table 2). The F-statistic confirmed sufficient instrument strength (F > 3000). Univariable 1SMR results were unaltered by using individual SNPs rather than PRS as instruments, but F-statistics were lower, as was the effect estimate for the causal effect of eBMD on knee OA (Supplementary Table S9, available as Supplementary data at IJE online). The BMD risk score was related to BMI but was not related to PA or hormone-replacement-therapy use (Supplementary Table S10, available as Supplementary data at IJE online). In 2SMR analyses, IVW provided evidence for a causal effect of eBMD on hip OA [OR per SD increase = 1.09 (95% CI = 1.03, 1.16)], which was relatively consistent (in magnitude) across the three MR methods (Figure 4 and Supplementary Figure S1, available as Supplementary data at IJE online). Evidence for a causal effect on knee OA was weaker [OR = 1.04 (95% CI = 1.00, 1.09), Supplementary Figure S2, available as Supplementary data at IJE online]. Excluding two SNPs more strongly related to BMI than eBMD did not alter results (Supplementary Table S11, available as Supplementary data at IJE online). When restricting to 10 SNPs also associated with FN–BMD (p < 5 × 10–8) in GEFOS, the magnitude of the effect was stronger for hip OA [OR = 1.40 (95% CI = 1.12, 1.74)], but this effect estimate was less consistent with the MR–Egger and weighted median estimates (Supplementary Table S11, available as Supplementary data at IJE online). However, evidence for a causal effect on knee OA was more consistent (in magnitude) across the three methods [ORIVW = 1.21 (95% CI = 1.01, 1.44)].
Figure 3.
Summary of results of one-sample, two-sample and multivariable Mendelian randomization analyses
Effect estimates represent the SD increase in outcome per SD increase in exposure for BMD–BMI and BMI–BMD analyses, the odds ratio per SD increase in exposure for BMI–OA and BMD–OA analyses, and the SD increase in BMD or BMI per 1-unit increase in the log odds of OA. eBMD, estimated bone mineral density; BMI, body mass index; OA, osteoarthritis; MR, Mendelian randomization; SNPs, single-nucleotide polymorphisms.
Table 2.
Results of one-sample univariable and multivariable Mendelian randomization
| Exposure | Outcome | NSNPs in GRS | F-statistic | N | Two-stage least-squares regression |
Two-stage linear and logistic regression |
|||
|---|---|---|---|---|---|---|---|---|---|
| Estimate (95% CI) | p-value | OR (95% CI) | p-value | ||||||
| UV MR | eBMD | Hip OA | 44 | 3439 | 190 181 | 0.008 (0.001, 0.014)rd | 0.018 | 1.29 (1.05, 1.58) | 0.015 |
| eBMD | Knee OA | 44 | 3534 | 194 410 | 0.016 (0.008, 0.024)rd | 4 × 10–5 | 1.39 (1.19, 1.63) | 4 × 10–5 | |
| BMI | Hip OA | 63 | 4201 | 333 828 | 0.016 (0.011, 0.022)rd | 8 × 10–9 | 1.68 (1.41, 2.00) | 5 × 10–9 | |
| BMI | Knee OA | 63 | 4446 | 341 686 | 0.036 (0.030, 0.043)rd | <2 × 10–16 | 2.05 (1.80, 2.35) | <2 × 10–16 | |
| BMI | eBMD | 63 | 2953 | 218 700 | 0.073 (0.038, 0.108)sd | 4 × 10–5 | |||
| Hip OA | eBMD | 10 | 104 | 190 181 | 1.23 (0.48, 1.99)sd | 0.001 | |||
| Hip OA | BMI | 10 | 201 | 333 828 | 0.52 (-0.01, 1.05)sd | 0.054 | |||
| Knee OA | eBMD | 4 | 50 | 194 410 | 4.13 (2.73, 5.52)sd | 6 × 10–9 | |||
| Knee OA | BMI | 4 | 89 | 341 686 | 8.39 (6.62, 10.16)sd | <2 × 10–16 | |||
| MV MR | eBMD | Hip OA | 44 | 1795 | 190 175 | 0.007 (0.001, 0.013)rd | 0.028 | 1.27 (1.03, 1.55) | 0.024 |
| BMI | Hip OA | 63 | 1487 | 190 175 | 0.014 (0.006, 0.021)rd | 3 × 10–4 | 1.54 (1.23, 1.93) | 2 × 10–4 | |
| eBMD | Knee OA | 44 | 1795 | 194 404 | 0.015 (0.007, 0.023)rd | 2 × 10–4 | 1.36 (1.16, 1.59) | 2 × 10–4 | |
| BMI | Knee OA | 63 | 1487 | 194 404 | 0.034 (0.025, 0.043)rd | 2 × 10–13 | 1.96 (1.64, 2.34) | 1 × 10–13 | |
| Hip OA | eBMD | 10 | 66 | 190 175 | 1.23 (0.45, 2.01)sd | 0.002 | |||
| BMI | eBMD | 63 | 1080 | 190 175 | 0.080 (0.035, 0.124)sd | 4 × 10–4 | |||
| Knee OA | eBMD | 4 | 53 | 194 404 | 5.84 (2.69, 8.99)sd | 3 × 10–4 | |||
| BMI | eBMD | 63 | 865 | 194 404 | –0.193 (–0.387, 3 × 10–4)sd | 0.050 | |||
Estimates represent the SD increase in outcome per SD increase in exposure. Estimates for binary exposures represent the SD increase in outcome per doubling in risk of exposure.
Estimates for binary outcomes represent the risk difference per SD increase in exposure. Due to the difficult interpretation of these results, odds ratios were also calculated by two-stage regression. Odds ratios are per SD increase in exposure.
MR, Mendelian randomization; BMD, bone mineral density; OA, osteoarthritis; UV, univariable; MV, multivariable; eBMD, estimated BMD.
Figure 4.
Results of two-sample Mendelian randomization analyses
eBMD, estimated bone mineral density; OA, osteoarthritis; BMI, body mass index; CI, confidence interval.
There was evidence for a causal pathway between hip OA and eBMD in 1SMR [SD increase per doubling in odds of hip OA = 1.23 (95% CI = 0.48, 1.99)] (Table 2), but not 2SMR analysis (Supplementary Figure S3, available as Supplementary data at IJE online). Evidence for a causal effect of knee OA on eBMD was provided by 1SMR [β = 4.13 (95% CI = 2.73, 5.52)] and 2SMR [β = 0.13 (95% CI = 0.03, 0.23)], with a positive effect observed for all three 2SMR methods, albeit weaker, with wide CIs overlapping the null for MR–Egger regression (Figure 4 and Supplementary Figure S4, available as Supplementary data at IJE online). The knee, but not hip, OA GRS was related to BMI, potentially invalidating instrumental-variable assumption 2 (IV2) (Supplementary Table S10, available as Supplementary data at IJE online).
BMI is a strong causal risk factor for BMD and OA with weaker evidence for bidirectionality
1SMR provided evidence that BMI has a strong causal effect on hip and knee OA, with an SD increase in BMI associated with a 68% (95% CI = 41, 100) increased odds of hip OA and 105% (95% CI = 80, 135) increased risk of knee OA (Table 2). 2SMR suggested that BMI is causally related to OA, with an SD increase in BMI related to a 56% (95% CI = 31, 87) increased odds of hip OA and a 69% (95% CI = 48, 93) increased odds of knee OA. These results were consistent across the three 2SMR methods (Supplementary Figures S5 and S6, available as Supplementary data at IJE online), other than the causal effect of BMI on knee OA estimated by MR–Egger, which was ∼30% weaker, albeit in the same direction (Figure 4). There was strong evidence, from 1SMR, that the causal pathway between BMI and knee OA was bidirectional, with weaker evidence for hip OA (Table 2). Additional adjustment for total weekly PA (assessed using the International Physical Activity Questionnaire) did not attenuate these relationships. 2SMR, however, provided weak and inconsistent evidence (across the three methods) of a causal effect of hip OA on BMI only (Figure 4 and Supplementary Figures S7 and S8, available as Supplementary data at IJE online).
We could not perform bidirectional 1SMR for BMD–BMI as the FN–BMD SNPs were identified by weight-adjusted GWAS, meaning the instrument for FN–BMD may be inversely related to weight and thus BMI.23 2SMR using summary statistics from the eBMD GWAS, not adjusted for weight, did not identify a causal effect of eBMD on BMI (Figure 4 and Supplementary Figure S9, available as Supplementary data at IJE online). There was robust evidence for a causal effect of BMI on eBMD in 1SMR, with an SD increase in BMI causing a 0.07SD (95% CI = 0.04, 0.11) increase in heel BMD (Table 2). This estimate was like that from 2SMR and the effect size was consistent for IVW, weighted median and MR–Egger analyses, although the MR–Egger intercept did reveal some evidence of horizontal pleiotropy (Figure 4, Supplementary Table S11 and Supplementary Figure S10, available as Supplementary data at IJE online).
Multivariable MR identifies an independent causal effect of eBMD on OA
Overall, the 1S and 2S analyses provided consistent evidence that BMI is a confounder of the relationship between BMD and hip/knee OA (i.e. a common cause of both phenotypes, Figure 3). We therefore used 1SMVMR to examine the causal effect of eBMD on OA after accounting for BMI. Following adjustment for BMI, eBMD was found to be an independent causal risk factor for both hip and knee OA with a similar magnitude of effect to that observed in MR analyses not accounting for BMI. BMI had a stronger effect than eBMD for both hip and knee OA (Table 2). Sanderson–Windmeijer F-statistics were >1000 for both instruments. Results were generally consistent when using individual SNPs as instruments, although the evidence for a causal effect of eBMD on knee OA was weakened, as was the instrument strength estimated by F-statistics (Supplementary Table S9, available as Supplementary data at IJE online).
MVMR provided evidence for a BMI-independent causal effect of OA on eBMD [βhip = 1.23 (95% CI = 0.45, 2.01), βknee = 5.84 (95% CI = 2.69, 8.99), Table 2]. The causal effect of BMI on BMD was independent of hip OA [β = 0.08 (0.04, 0.12)]. When conditioning on knee OA, an inverse effect of BMI on BMD was observed [β = –0.19 (95% CI = –0.39, 0.00)]. This is unlikely to be bias due to conditioning on a common outcome (i.e. collider bias), as genetically predicted OA is not a common outcome.18 The BMI-independent causal effect of knee OA on eBMD was not observed when using individual SNPs as instruments (Supplementary Table S9, available as Supplementary data at IJE online).
LCV analyses provide evidence for a non-pleiotropic causal effect of BMD on OA
To determine whether shared underlying genetic aetiology fully explained the observed causal effect of BMD on OA, we performed LCV modelling using weight-adjusted summary statistics for both FN/LS–BMD and hip/knee OA. The LCV analysis identified evidence for genetic correlations between BMD (measured at both the FN and LS) and OA at both the hip and knee (rho = 0.16–0.23, Supplementary Table S12, available as Supplementary data at IJE online). There was evidence for a partial causal effect of BMD at both sites on OA at both sites, independently of genetic correlation and weight, with the largest magnitude of causal effect observed for FN–BMD and knee OA, with a genetic causality proportion of 0.64.
Discussion
We have found strong evidence for a causal effect of BMD on hip and knee OA using 1SMR, which was relatively consistent with 2SMR. MVMR confirmed that the effect of BMD on OA is independent of BMI. Our results also suggest that there is a bidirectional causal effect between OA and eBMD. We have confirmed strong causal effects of BMI on eBMD, hip and knee OA, with no causal effect of eBMD on BMI. Finally, we have found some evidence of a positive causal effect of knee and hip OA on BMI. The observed causal effect of BMI on eBMD in this adult population is consistent with a previous analysis of a paediatric population (mean age 10 years), in which a causal effect of BMI on FN–BMD was observed.9 As seen in this current analysis, Kemp et al. found no evidence for a causal effect of BMD on BMI.9 The strong causal effect of BMI on both hip and knee OA corroborates previous MR analyses.5,24
The causal effect of eBMD on hip and knee OA that we observed is consistent with previous MR analyses identifying causal effects of FN and LS–BMD on hip and knee OA5,24 and our recent analyses showing that generalized high bone mass (BMD Z-score >3.2 at the hip or L1) is related to greater worsening of osteophyte and joint space narrowing (JSN) severity at both the hip and knee.25,26 Taken together, these findings suggest that bone parameters in general have a causal effect on OA, regardless of the site or method of measurement. However, the magnitude of the effect of eBMD on OA was larger in 2S analyses restricted to SNPs associated with FN–BMD. There are two potential explanations for a stronger effect of BMD on OA when restricting to FN–BMD loci. First, FN–BMD measured by dual-energy X-ray absorptiometry may be a more accurate representation of the biological pathways between bone and cartilage, compared with eBMD, which represents a combination of speed of sound and broadband ultrasound attenuation. Alternatively, since the FN primarily comprises cortical bone, whereas heel BMD is predominantly trabecular,27,28 these findings may reflect the fact that cortical bone is more strongly related to OA pathogenesis compared with trabecular bone. For example, cortical BMD might be expected to correlate more strongly with subchondral plate sclerosis compared with trabecular BMD, which is implicated in the progression of OA.29 Inconsistently with the results of our analysis, some previous studies have provided evidence to suggest that high BMD is related to reduced progression of OA,30,31 although this could be explained by index-event bias, in which conditioning on OA leads to spurious associations between OA risk factors.32
We have found some evidence for reverse causality in the relationship between eBMD and OA. The positive direction of effect is as expected from artefactual elevation, rather than loss of bone mass due to reduced PA. However, as we do not expect BMD measurements at the heel to be artefactually elevated by features of OA, the observed causal effect of OA on eBMD in 1SMR may reflect biological pleiotropy (i.e. the same underlying biological pathways may be contributing to both phenotypes). Consistently with shared biological mechanisms contributing to both BMD and OA, Hackinger et al. identified a genetic correlation between LS–BMD (but not FN) and OA.11 By performing a cross-phenotype meta-analysis between OA and LS–BMD, the authors identified a number of known loci, as well as a novel locus in the SMAD3 gene.11 SMAD3 is part of the transforming growth factor β (TGFβ) signalling pathway, which regulates osteoblast differentiation. The first discovered OA loci, growth differentiation factor-5 (GDF5), is a ligand for this pathway.33 The canonical Wnt signalling pathway is involved in the regulation of osteoblasts and mutations in this pathway can lead to high or low BMD; e.g. activating mutations in low-density lipoprotein receptor-related protein 5 (LRP5, the receptor involved in Wnt signalling activation) cause high BMD.34 This signalling pathway has been implicated in OA pathogenesis;35 increased levels of a Wnt signalling inhibitor, DKK1, were associated with reduced progression of hip OA in a population of Caucasian women.36
However, we did find stronger, more consistent, evidence for an effect of eBMD on OA, as opposed to vice versa. This could reflect the stronger instrument for BMD, but our LCV analyses using the full set of summary statistics provided further evidence for a causal pathway between BMD and OA, not driven by genetic correlation (or confounding by weight as evidenced by the MVMR), suggesting that bone may still have a direct effect on OA, e.g. via increased joint loading or through related structural alterations in the subchondral bone, such as denser subchondral trabecular bone, which has been linked to the progression of JSN.37
Strengths and limitations
We have utilized the largest data sets possible to maximize the power to detect causal effects. We have ensured that there is no overlap between our exposure and outcome populations. We have examined individual-level data in UK Biobank to perform 1SMR to strengthen evidence.
However, we were unable to use eBMD instruments for 1SMR as they were identified in the same population used for analysis; reassuringly, F-statistics suggested that our FN–BMD instrument was of reasonable strength. We did not use the largest available meta-analysis as the source of the BMI instruments due to significant sample overlap with UK Biobank.38 However, the Locke et al. European-only meta-analysis, which we used for our instrument source for both 1S and 2SMR, still included >300 000 individuals and identified 77 loci; the PRS generated from these SNPs had a strong F-statistic suggesting that the magnitude of effects identified in 1SMR analyses are unlikely to be explained by bias due to weak instruments. Our OA outcomes for 1SMR were based on hospital diagnosis; it is unclear how this phenotype relates to radiographic features of OA, such as JSN, which are commonly used as clinical trial outcomes. Using a severe phenotype as the outcome means reduced power in GWAS and leads to fewer genome-wide significant loci and a greater chance of weak instrument bias (as highlighted by the much smaller F-statistics for the OA instruments). The OA outcomes from the GO consortium included a range of definitions of hip and knee OA, including hospital diagnosis, radiographic evidence and self-reported OA definitions. Heterogeneity in phenotype also reduces the power to detect loci in GWAS. The ORs from 1SMR are estimates and standard errors (SEs) are likely underestimated,15 so caution should be taken when interpreting these effect sizes. There may be additional risk factors related to the genetic variants that we did not include in our MVMR models. The UK Biobank population is limited by a latent population structure even after restricting to White Europeans and adjusting for PCs,39 which may confound estimates generated by 1SMR. The UK Biobank population examined was White British and all instruments were derived from predominantly White European populations, meaning that we were unable to examine causal effects in non-European populations, limiting generalizability to other ethnicities for whom the prevalence of, and therefore risk factors for, osteoarthritis may differ.40–43 The prevalence of OA is higher in men from UK Biobank compared with women, despite evidence in the general population suggesting a higher prevalence of knee OA in women.44 This could be explained by selection bias, as women and healthy individuals (i.e. free of OA) are more likely to participate in UK Biobank. Although we adjusted for sex in our analyses, it is possible that there are other variables related to participation in UK Biobank that we could not account for in our analyses. Individuals with OA have a higher risk of premature mortality than the general population,45 which could cause further selection bias if those with severe OA are less likely to survive to participate in UK Biobank. However, this selection bias is unlikely to explain the observed positive causal effect of OA on BMI, but may explain the positive causal effect estimate for OA on eBMD.
Conclusions
We have found evidence for a BMI-independent causal effect of BMD on hip and knee OA and some evidence for a bidirectional causal effect, which we hypothesize to reflect shared underlying genetic aetiology. We have confirmed strong causal effects of BMI on BMD and hip and knee OA, and have found novel evidence for a causal effect of knee OA on BMI, which did not appear to be mediated by pain-associated reductions in PA. Further analyses are required to determine the shared pathways contributing to both BMD and OA, and to determine the mechanisms by which higher BMD causes OA.
Notes
The Genetics of Osteoarthritis consortium: Lilja Stefánsdóttir,1 Yanfei Zhang,2 Rodrigo Coutinho de Almeida,3 Tian T. Wu,4 Jie Zheng,5 Maris Teder-Laving,6 Anne-Heidi Skogholt,7 Chikashi Terao,8 Eleni Zengini,9 George Alexiadis,10 Andrei Barysenka,11 Gyda Bjornsdottir,1 Maiken E. Gabrielsen,7 Arthur Gilly,11 Thorvaldur Ingvarsson,12,13 Marianne B. Johnsen,7,14,15 Helgi Jonsson,12,16 Margreet G. Kloppenburg,17 Almut Luetge,7 Reedik Mägi,6 Massimo Mangino,18 Rob R.G.H.H. Nelissen,19 Manu Shivakumar,20 Julia Steinberg,11,21,22,23 Hiroshi Takuwa,24,25 Laurent Thomas,26,7,27,28 Margo Tuerlings,1 George Babis,29 Jason Pui Yin Cheung,30 Dino Samartzis,30 Steve A. Lietman,31 P. Eline Slagboom,3 Kari Stefansson,3,12 André G. Uitterlinden,32 Bendik Winsvold,7,33,34 John-Anker Zwart,7,33 Pak Chung Sham,35 Gudmar Thorleifsson,1 Tom R. Gaunt,5 Andrew P. Morris,36 Ana M. Valdes,37 Aspasia Tsezou,38 Kathryn S.E. Cheah,39 Shiro Ikegawa,24 Kristian Hveem,7,40 Tõnu Esko,6 J. Mark Wilkinson,41 Ingrid Meulenbelt,3 Ming Ta Michael Lee2,42 and Unnur Styrkársdóttir1
1deCODE genetics/Amgen Inc., Reykjavik, Iceland; 2Genomic Medicine Institute, Geisinger Health System, Danville, PA 17822, USA; 3Department of Biomedical Data Sciences, Section Molecular Epidemiology, Leiden University Medical Center, Leiden, The Netherlands; 4Department of Psychiatry, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong; 5MRC Integrative Epidemiology Unit (IEU), Bristol Medical School, University of Bristol, Oakfield House, Oakfield Grove, Bristol, BS8 2BN, UK; 6Estonian Genome Center, Institute of Genomics, University of Tartu, Tartu, Estonia; 7K. G. Jebsen Center for Genetic Epidemiology, Department of Public Health and Nursing, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology, Trondheim, Norway; 8Laboratory for Statistical and Translational Genetics, RIKEN Center for Integrative Medical Sciences, Kanagawa, Japan; 95th Psychiatric Department, Dromokaiteio Psychiatric Hospital, Haidari, Athens, Greece; 101st Department of Orthopaedics, KAT General Hospital, Athens, Greece; 11Institute of Translational Genomics, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany; 12Faculty of Medicine, University of Iceland, Reykjavik, Iceland; 13Department of Orthopedic Surgery, Akureyri Hospital, Akureyri, Iceland; 14Research and Communication Unit for Musculoskeletal Health (FORMI), Department of Research, Innovation and Education, Division of Clinical Neuroscience, Oslo University Hospital, Oslo, Norway; 15Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, Oslo, Norway; 16Department of Medicine, Landspitali The National University Hospital of Iceland, Reykjavik, Iceland; 17Department of Rheumatology, Leiden University Medical Center, Leiden, The Netherlands; 18Department of Twin Research and Genetic Epidemiology, Kings College London, London, UK; 19Department of Orthopaedics, Leiden University Medical Center, Leiden, The Netherlands; 20Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA; 21Cancer Research Division, Cancer Council NSW, Sydney, NSW, Australia; 22School of Public Health, Faculty of Medicine and Health, The University of Sydney, Australia; 23Human Genetics, Wellcome Genome Campus, Wellcome Sanger Institute, Cambridge, UK; 24Laboratory for Bone and Joint Diseases, RIKEN Center for Integrative Medical Sciences, Tokyo, Japan; 25Department of Orthopedic Surgery, Shimane University, Izumo, Japan; 26Department of Clinical and Molecular Medicine, Norwegian University of Science and Technology, Trondheim, Norway; 27BioCore—Bioinformatics Core Facility, Norwegian University of Science and Technology, Trondheim, Norway; 28Clinic of Laboratory Medicine, St. Olavs Hospital, Trondheim University Hospital, Trondheim, Norway; 292nd Department of Orthopaedics, National and Kapodistrian University of Athens, Medical School, Nea Ionia General Hospital, ‘Konstantopouleio’, Athens, Greece; 30Department of Orthopaedics and Traumatology, The University of Hong Kong, Hong Kong; 31Musculoskeletal Institute, Geisinger, Danville, USA; 32Department of Internal Medicine, Erasmus MC, Medical Center, Rotterdam, The Netherlands; 33Department of Research, Innovation and Education, Division of Clinical Neuroscience, Oslo University Hospital, Oslo, Norway; 34Department of Neurology, Oslo University Hospital, Oslo, Norway; 35Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong; 36Centre for Genetics and Genomics Versus Arthritis, Centre for Musculoskeletal Research, University of Manchester, Manchester, UK; 37Faculty of Medicine & Health Sciences, School of Medicine, University of Nottingham, Nottingham, Nottinghamshire, UK; 38Laboratory of Cytogenetics and Molecular Genetics, Faculty of Medicine, University of Thessaly, Larissa, Greece; 39School of Biomedical Sciences, The University of Hong Kong, Hong Kong; 40HUNT Research Center, Department of Public Health and Nursing, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology, Trondheim, Norway; 41Department of Oncology & Metabolism and Healthy Lifespan Institute, University of Sheffield, Sheffield, UK; 42Institute of Biomedical Sciences, academia Sinica, Taipei, Taiwan.
Supplementary data
Supplementary data are available at IJE online.
Ethics approval
UK Biobank received ethical approval from the Research Ethics Committee (REC reference : 11/NW/0382).
Funding
This work was supported by the Wellcome Trust [grant ref. 20378/Z/16/Z]. C.L.G. was funded by Versus Arthritis [grant ref. 20000]. J.Z. receives salary and start-up funding from the University of Bristol (Vice-Chancellor's fellowship). J.Z. is also supported by the Academy of Medical Sciences (AMS) Springboard Award, the Wellcome Trust, the Government Department of Business, Energy and Industrial Strategy (BEIS), the British Heart Foundation and Diabetes UK [SBF006\1117]. A.H., E.S., R.G., L.P., J.Z., G.D.S., C.L.G. and J.H.T. work in or are affiliated with a University of Bristol and MRC funded unit [MC_UU_00011/1, MC_UU_00011/4].
Supplementary Material
Acknowledgements
Individual-level analyses have been conducted using the UK Biobank resource under Application Number 17295. We are grateful to all individuals who provided data for these analyses.
Author contributions
A.H., R.G., L.P., C.L.G. and J.H.T. designed the study; E.S., J.Z. and G.D.S. provided code, analytical advice and critical interpretation of findings; L.S., K.H., C.G.B., J.V.M., E.Z. and the GO consortium provided summary statistics for MR analyses; A.H. conducted all analyses and drafted the manuscript; all authors edited and approved the manuscript.
Conflict of interest
None declared.
Contributor Information
April Hartley, MRC Integrative Epidemiology Unit, Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK; Musculoskeletal Research Unit, Translational Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK.
Eleanor Sanderson, MRC Integrative Epidemiology Unit, Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK.
Raquel Granell, MRC Integrative Epidemiology Unit, Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK.
Lavinia Paternoster, MRC Integrative Epidemiology Unit, Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK.
Jie Zheng, MRC Integrative Epidemiology Unit, Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK.
George Davey Smith, MRC Integrative Epidemiology Unit, Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK.
Lorraine Southam, Institute of Translational Genomics, Helmholtz Zentrum München, German Research Center for Environmental Health, 85764, Neuherberg, Germany.
Konstantinos Hatzikotoulas, Institute of Translational Genomics, Helmholtz Zentrum München, German Research Center for Environmental Health, 85764, Neuherberg, Germany.
Cindy G Boer, Department of Internal Medicine and Epidemiology, Erasmus MC, Rotterdam, The Netherlands.
Joyce van Meurs, Department of Internal Medicine and Epidemiology, Erasmus MC, Rotterdam, The Netherlands.
Eleftheria Zeggini, Institute of Translational Genomics, Helmholtz Zentrum München, German Research Center for Environmental Health, 85764, Neuherberg, Germany.
Celia L Gregson, Musculoskeletal Research Unit, Translational Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK.
Jon H Tobias, MRC Integrative Epidemiology Unit, Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK; Musculoskeletal Research Unit, Translational Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK.
The Genetics of Osteoarthritis Consortium:
Lilja Stefánsdóttir, Yanfei Zhang, Rodrigo Coutinho de Almeida, Tian T Wu, Jie Zheng, Maris Teder-Laving, Anne-Heidi Skogholt, Chikashi Terao, Eleni Zengini, George Alexiadis, Andrei Barysenka, Gyda Bjornsdottir, Maiken E Gabrielsen, Arthur Gilly, Thorvaldur Ingvarsson, Marianne B Johnsen, Helgi Jonsson, Margreet G Kloppenburg, Almut Luetge, Reedik Mägi, Massimo Mangino, Rob R G H H Nelissen, Manu Shivakumar, Julia Steinberg, Hiroshi Takuwa, Laurent Thomas, Margo Tuerlings, George Babis, Jason Pui Yin Cheung, Dino Samartzis, Steve A Lietman, P Eline Slagboom, Kari Stefansson, André G Uitterlinden, Bendik Winsvold, John-Anker Zwart, Pak Chung Sham, Gudmar Thorleifsson, Tom R Gaunt, Andrew P Morris, Ana M Valdes, Aspasia Tsezou, Kathryn S E Cheah, Shiro Ikegawa, Kristian Hveem, Tõnu Esko, J Mark Wilkinson, Ingrid Meulenbelt, Ming Ta Michael Lee, and Unnur Styrkársdóttir
Data Availability
UK Biobank data are available through a procedure described at https://www.ukbiobank.ac.uk/principles-of-access/. BMD GWAS summary statistics are available to download from the GEFOS website at http://www.gefos.org/?q=content/gefos-data-release. GIANT BMI GWAS summary statistics are available to download at https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files#GIANT_Consortium_2012-2015_GWAS_Summary_Statistics. Hip and knee OA GWAS summary statistics from UK Biobank and arcOGEN are publicly available at https://www.ebi.ac.uk/gwas/.
References
- 1. Hardcastle SA, Dieppe P, Gregson CL, Davey Smith G, Tobias JH.. Osteoarthritis and bone mineral density: are strong bones bad for joints? BoneKEy Rep 2015;4:624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith GD, Ebrahim S.. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003;32:1–22. [DOI] [PubMed] [Google Scholar]
- 3. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G.. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133–63. [DOI] [PubMed] [Google Scholar]
- 4. Richmond RC, Davey Smith G.. Mendelian randomization: concepts and scope. Cold Spring Harbor Perspectives in Medicine 2021; doi: 10.1101/cshperspect.a040501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Funck-Brentano T, Nethander M, Movérare-Skrtic S, Richette P, Ohlsson C.. Causal factors for knee, hip, and hand osteoarthritis: a Mendelian randomization study in the UK Biobank. Arthritis Rheumatol 2019;71:1634–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zheng H, Chen C.. Body mass index and risk of knee osteoarthritis: systematic review and meta-analysis of prospective studies. BMJ Open 2015;5:e007568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang L, Rong J, Wang Y. et al. The relationship between body mass index and hip osteoarthritis: a systematic review and meta-analysis. Joint Bone Spine 2011;78:150–55. [DOI] [PubMed] [Google Scholar]
- 8. Hardcastle SA, Dieppe P, Gregson CL. et al. Individuals with high bone mass have an increased prevalence of radiographic knee osteoarthritis. Bone 2015;71:171–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kemp JP, Sayers A, Smith GD, Tobias JH, Evans DM.. Using Mendelian randomization to investigate a possible causal relationship between adiposity and increased bone mineral density at different skeletal sites in children. Int J Epidemiol 2016;45:1560–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burgess S, Thompson SG.. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol 2015;181:251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hackinger S, Trajanoska K, Styrkarsdottir U, arcOGEN Consortium, GEFOS Consortium et al. Evaluation of shared genetic aetiology between osteoarthritis and bone mineral density identifies SMAD3 as a novel osteoarthritis risk locus. Hum Mol Genet 2017;26:3850–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee NK, Sowa H, Hinoi E. et al. Endocrine regulation of energy metabolism by the skeleton. Cell 2007;130:456–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Allen NE, Sudlow C, Peakman T, Collins R, UK Biobank UK biobank data: come and get it. Sci Transl Med 2014;6:224ed4. [DOI] [PubMed] [Google Scholar]
- 14. Kleiber C, Zeileis A, AER: Applied Econometrics with R. R package version 1.2–7. 2019.
- 15. Burgess S, Small DS, Thompson SG.. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res 2017;26:2333–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hemani G, Zheng J, Elsworth B. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hemani G, Tilling K, Davey Smith G.. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet 2017;13:e1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sanderson E, Davey Smith G, Windmeijer F, Bowden J.. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int J Epidemiol 2019;48:713–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bowden J, Davey Smith G, Burgess S.. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bowden J, Davey Smith G, Haycock PC, Burgess S.. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol 2016;40:304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Estrada K, Styrkarsdottir U, Evangelou E. et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 2012;44:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Connor LJ, Price AL.. Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat Genet 2018;50:1728–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holmes MV, Davey Smith G.. Problems in interpreting and using GWAS of conditional phenotypes illustrated by ‘alcohol GWAS’. Mol Psychiatry 2019;24:167–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zengini E, Hatzikotoulas K, Tachmazidou I. et al. Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nat Genet 2018;50:549–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hartley A, Hardcastle SA, Paternoster L. et al. Individuals with high bone mass have increased progression of radiographic and clinical features of knee osteoarthritis. Osteoarthritis Cartilage 2020;28:1180–90. [DOI] [PubMed] [Google Scholar]
- 26. Hartley A, Hardcastle SA, Frysz M. et al. Increased development of radiographic hip osteoarthritis in individuals with high bone mass: a prospective cohort study. Arthritis Res Ther 2021;23:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wasnich R, Davis J, Ross P, Vogel J.. Effect of thiazide on rates of bone mineral loss: a longitudinal study. BMJ 1990;301:1303–05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crilly RG, Cox L.. A comparison of bone density and bone morphology between patients presenting with hip fractures, spinal fractures or a combination of the two. BMC Musculoskelet Disord 2013;14:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burr DB, Gallant MA.. Bone remodelling in osteoarthritis. Nat Rev Rheumatol 2012;8:665–73. [DOI] [PubMed] [Google Scholar]
- 30. Hart DJ, Cronin C, Daniels M, Worthy T, Doyle DV, Spector TD.. The relationship of bone density and fracture to incident and progressive radiographic osteoarthritis of the knee: the Chingford Study. Arthritis Rheum 2002;46:92–99. [DOI] [PubMed] [Google Scholar]
- 31. Zhang Y, Hannan MT, Chaisson CE. et al. Bone mineral density and risk of incident and progressive radiographic knee osteoarthritis in women: the Framingham Study. J Rheumatol 2000;27:1032–7. [PubMed] [Google Scholar]
- 32. Zhang YQ, Neogi T, Hunter D, Roemer F, Niu JB.. What effect is really being measured? An alternative explanation of paradoxical phenomena in studies of osteoarthritis progression. Arthritis Care Res (Hoboken) 2014;66:658–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cibrian Uhalte E, Wilkinson JM, Southam L, Zeggini E.. Pathways to understanding the genomic aetiology of osteoarthritis. Hum Mol Genet 2017;26:R193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gregson CL, , HardcastleSA, , CooperC, , Tobias JH.. Friend or foe: high bone mineral density on routine bone density scanning, a review of causes and management. Rheumatology (Oxford) 2013;52:968–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lories RJ, , CorrM, , Lane NE. To Wnt or not to Wnt: the bone and joint health dilemma. Nat Rev Rheumatol 2013;9:328–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lane NE, , NevittMC, , LuiL-Y, , De LeonP, , Corr M.. Wnt signaling antagonists are potential prognostic biomarkers for the progression of radiographic hip osteoarthritis in elderly Caucasian women. Arthritis Rheum 2007;56:3319–25. [DOI] [PubMed] [Google Scholar]
- 37. Lo GH, Schneider E, Driban JB. et al. Periarticular bone predicts knee osteoarthritis progression: data from the Osteoarthritis Initiative. 1532-866X (Electronic). [DOI] [PMC free article] [PubMed]
- 38. Yengo L, Sidorenko J, Kemper KE, the GIANT Consortium et al. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum Mol Genet 2018;27:3641–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haworth S, Mitchell R, Corbin L. et al. Apparent latent structure within the UK Biobank sample has implications for epidemiological analysis. Nat Commun 2019;10:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jordan JM, Helmick CG, Renner JB. et al. Prevalence of knee symptoms and radiographic and symptomatic knee osteoarthritis in African Americans and Caucasians: the Johnston County Osteoarthritis Project. J Rheumatol 2007;34:172–80. [PubMed] [Google Scholar]
- 41. Jordan JM, Helmick CG, Renner JB. et al. Prevalence of hip symptoms and radiographic and symptomatic hip osteoarthritis in African Americans and Caucasians: the Johnston County Osteoarthritis Project. J Rheumatol 2009;36:809–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang Y, Xu L, Nevitt MC. et al. Comparison of the prevalence of knee osteoarthritis between the elderly Chinese population in Beijing and whites in the United States: the Beijing Osteoarthritis Study. Arthritis Rheum 2001;44:2065–71. [DOI] [PubMed] [Google Scholar]
- 43. Nevitt MC, Xu L, Zhang Y. et al. Very low prevalence of hip osteoarthritis among Chinese elderly in Beijing, China, compared with whites in the United States: the Beijing Osteoarthritis Study. Arthritis Rheum 2002;46:1773–79. [DOI] [PubMed] [Google Scholar]
- 44. Hunter DJ, Bierma-Zeinstra S.. Osteoarthritis. Lancet 2019;393:1745–59. [DOI] [PubMed] [Google Scholar]
- 45.Osteoarthritis Research Society International. Osteoarthritis. A serious disease. Submitted to the US Food and Drug Administration, 1 December 2016. https://oarsi.org/sites/default/files/library/2018/pdf/oarsi_white_paper_oa_serious_disease121416_1.pdf (25 November 2021, date last accessed).
- 46. Morris JA, Kemp JP, Youlten SE, 23andMe Research Team et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat Genet 2019;51:258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boer CG, Hatzikotoulas K, Southam L, Regeneron Genetics Center et al. Deciphering osteoarthritis genetics across 826,690 individuals from 9 populations. Cell 2021;184:4784–818. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Locke AE, Kahali B, Berndt SI, The LifeLines Cohort Study et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tachmazidou I, Hatzikotoulas K, Southam L, arcOGEN Consortium et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat Genet 2019;51:230–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
UK Biobank data are available through a procedure described at https://www.ukbiobank.ac.uk/principles-of-access/. BMD GWAS summary statistics are available to download from the GEFOS website at http://www.gefos.org/?q=content/gefos-data-release. GIANT BMI GWAS summary statistics are available to download at https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files#GIANT_Consortium_2012-2015_GWAS_Summary_Statistics. Hip and knee OA GWAS summary statistics from UK Biobank and arcOGEN are publicly available at https://www.ebi.ac.uk/gwas/.