Abstract
Despite unprecedented amount of information now available in medical records, health data remain underexploited due to their heterogeneity and complexity. Simple charts and hypothesis-driven statistics can no longer apprehend the content of information-rich clinical data. There is, therefore, a clear need for powerful interactive visualization tools enabling medical practitioners to perceive the patterns and insights gained by state-of-the-art machine learning algorithms. Here, we report an interactive graphical interface for use as the front end of a machine learning causal inference server (MIIC), to facilitate the visualization and comprehension by clinicians of relationships between clinically relevant variables. The widespread use of such tools, facilitating the interactive exploration of datasets, is crucial both for data visualization and for the generation of research hypotheses. We demonstrate the utility of the MIIC interactive interface, by exploring the clinical network of a large cohort of breast cancer patients treated with neoadjuvant chemotherapy (NAC). This example highlights, in particular, the direct and indirect links between post-NAC clinical responses and patient survival. The MIIC interactive graphical interface has the potential to help clinicians identify actionable nodes and edges in clinical networks, thereby ultimately improving the patient care pathway.
Subject terms: Outcomes research, Risk factors, Breast cancer
Introduction
The availability of health data from patient medical records is increasing, and these data constitute, in theory, a rich resource for research purposes. However, despite the unprecedented amount of information now available, health data remain underexploited due to their heterogeneity and complexity. There is, therefore, an urgent need for innovative tools, based on intuitive and interactive graphical interfaces, specifically designed for the exploration of health data by medical practitioners. Data visualization is gradually emerging as a new field of research, and graphical representations are used for two main purposes: (i) explanatory illustration, to highlight novel scientific insights graphically and to ensure efficient communication between scientists1–4; and (ii) exploratory analysis, searching for relationships previously overlooked and leading to new discoveries, thereby maximizing the potential of information-rich databases. We present here an exploratory analysis of a global clinical network from a large breast cancer cohort, with a novel interactive graphical interface for the exploration of health data.
We previously developed an advanced computational method for graphical analyses, including causal relationships, from multivariate data5. The underlying MIIC (Multivariate Information-based Inductive Causation) algorithm, which was released as an online server6, uses a machine learning method combining constraint-based and information theory approaches to reconstruct causal, non-causal or mixed networks from large datasets. The MIIC algorithm was first developed to analyze categorical genomic data5,6 and has recently been extended to the analysis of more challenging heterogeneous datasets, such as medical records, combining both categorical and continuous variables, in which interdependence is notoriously difficult to assess7.
Breast cancer (BC) clinical datasets are particularly suitable for the type of exploratory analysis presented here, as BC is a complex heterogeneous disease highly variable in its aggressiveness and prognosis. BC remains one of the leading causes of cancer-related death among women. The BC patients included in the cohort analyzed here were treated with neoadjuvant (or preoperative) chemotherapy (NAC). NAC was originally restricted to patients with inflammatory or locally advanced BC, but is now the standard care for aggressive early-stage breast cancers, i.e., triple-negative (TNBC) and HER2-positive BCs8,9. From the patient’s viewpoint, the benefits of the neoadjuvant strategy include a greater feasibility of breast-conserving surgery and the prognostic stratification of risk obtained after analyses of the residual tumor burden at surgery. From the research and development standpoint, the neoadjuvant setting makes it possible to monitor the chemosensitivity of the tumor in vivo, and provides an opportunity for the rapid validation of research hypotheses and the acceleration of drug approval.
Results
The global network displayed in Fig. 1 is accessible at https://miic.curie.fr/job_results_showcase.php?id=NEOREP. We discuss below some of the links inferred in the NEOREP network after grouping according to several clinically relevant concepts identified from published studies on BC.
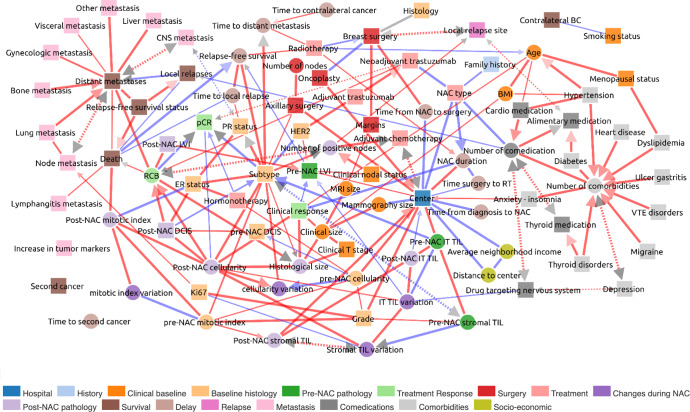
Fig. 1. MIIC global network for the NEOREP breast cancer cohort.
Each node corresponds to a variable of the dataset, with circles indicating continuous variables and squares indicating categorical variables. The colors define a category of variables, as detailed under the figure. Each edge corresponds to a “direct” association between two variables with different types of orientation described in Methods. BC breast cancer, BMI body mass index, DCIS ductal carcinoma in situ, ER estrogen receptor status, LVI lymphovascular invasion, NAC neoadjuvant chemotherapy, CNS central nervous system, pCR pathological complete response, PR progesterone receptor status, RCB residual cancer burden, TILs tumor-infiltrating lymphocytes. Blue edges indicate negative partial correlations, red edges indicate positive partial correlations.
MIIC performs quality control
MIIC first identifies relationships between a disease and the corresponding treatment. ER positivity—which is predictive of efficacy for anti-hormonal treatment10—is associated with the use of endocrine therapy (Supplementary Fig. 3A), and a similar association is observed for HER2-positivity and trastuzumab use (Supplementary Fig. 3B)11. Beyond cancer, significant associations are also found between depression and the use of psycholeptics (Supplementary Fig. 3C), between thyroid disorders and thyroid hormone use (Supplementary Fig. 3D), and between hypertension and drugs for the treatment of cardiovascular diseases (Supplementary Fig. 3E). More generally, comedication use is associated with the type of NAC (Supplementary Fig. 3F), reflecting the greater likelihood of less toxic regimens being prescribed to fragile patients (patients on other types of medication) than to patients without comedication12–14.
MIIC then identifies clinical factors known to be epidemiologically related (Supplementary Fig. 4A). Menopause, a process occurring in older women, is directly linked to age (Supplementary Fig. 4B) (median age: 43 years for premenopausal, versus 58 years for postmenopausal women). Postmenopausal status is associated with dyslipidemia (Supplementary Fig. 4C)15. Consistent with these associations, body mass index (BMI) increases with age (Supplementary Fig. 4A, D) and both factors, which have been reported to increase cardiovascular risks, are linked to hypertension (Supplementary Fig. 4A, E). The number of drugs taken by a patient (comedication) increases with the number of comorbidities (Supplementary Fig. 4A, F).
MIIC identifies inherent associations between variables
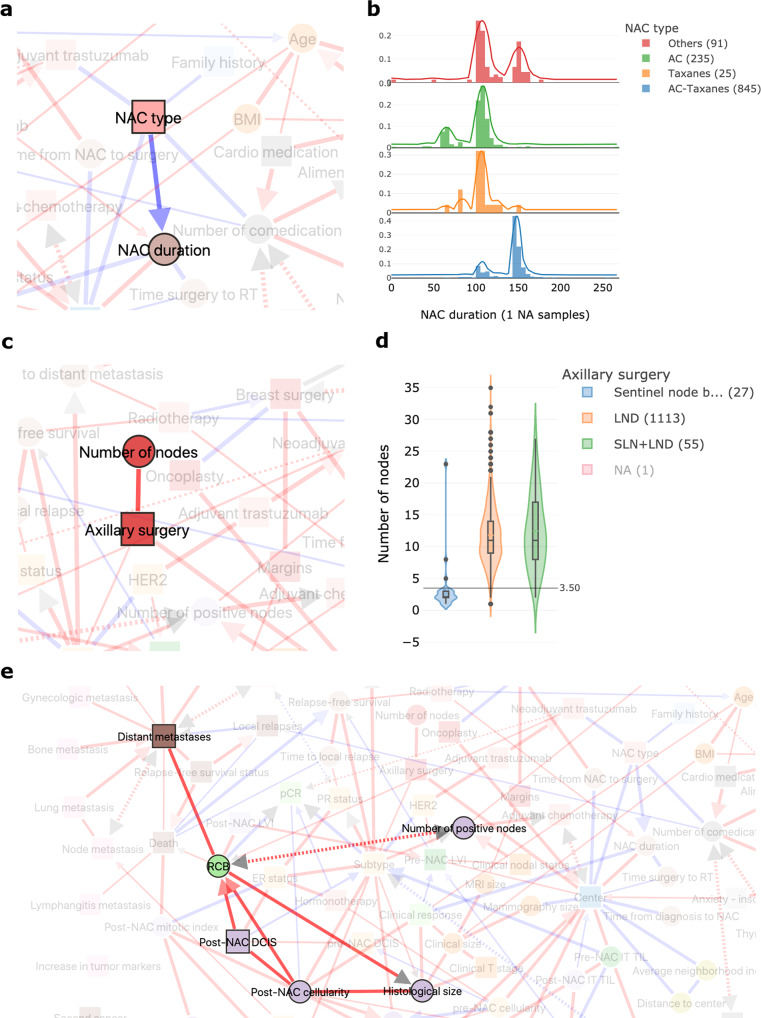
The duration of neoadjuvant treatment is directly linked to the type of NAC regimen delivered (Fig. 2a) reflecting the fact that anthracycline-based (AC) regimens usually include four cycles (median of 106 days, Fig. 2b), whereas sequential regimens in which anthracyclines are followed by taxanes are generally administered over six or eight cycles (median of 147 days, Fig. 2b). The number of nodes retrieved is associated with the type of axillary surgery (Fig. 2c), consistent with the fact that sentinel node (SLN) biopsy procedures were developed to reduce the number of lymph nodes removed during dissection (LND) (Fig. 2d)16. MIIC correctly represents the direct links between residual cancer burden (RCB) (Fig. 2e) and the patterns making up this score, derived from measurements on the primary tumor bed (size, fraction of invasive cancer, cellularity) and the regional lymph nodes (number of positive lymph nodes).
Fig. 2. The MIIC interactive online interface identifies inherent associations between variables.
a NAC type is directly correlated with NAC duration. NAC = neoadjuvant chemotherapy. b Distribution of neoadjuvant chemotherapy (NAC) duration (in days) according to the NAC regimen administered: anthracyclines (AC), taxanes or sequential AC-taxanes. c The number of axillary nodes in the histological specimen depends on the type of axillary surgery performed. d Boxplot showing the number of axillary nodes removed according to the type of surgery performed: lymph node dissection (LND), sentinel lymph node biopsy (SLN) or both. The boxes represent the IQR, the horizontal lines correspond to the median values and lines ends mark the upper and lower fence. The number of cases considered for the analysis are reported in the legend. e Network interactions of the RCB node with the five patterns making up the RCB score.
MIIC identifies intra- and inter-modality associations
For the variables derived from pathology records, MIIC found associations between tumor grade, Ki67, and mitotic index (Supplementary Fig. 5A–C), all of which are markers of tumor proliferation17. MIIC can also visualize links between patterns assessed in different ways. Measurements of pre-NAC tumor size evaluated clinically, by mammography and by MRI, were found to be closely related (Supplementary Fig. 5C–E) as previously reported18,19. Similarly, the response to treatment assessed clinically at NAC completion was found to be associated with histological size based on the surgical specimen (Supplementary Fig. 5F).
MIIC provides insight into tumor biology and response to treatment
The presence of lymphovascular invasion (LVI) in the post-NAC specimen is associated with a higher RCB index, consistent with the strong resistance to chemotherapy of these tumors20 (Supplementary Fig. 6A). TNBCs and HER2-positive tumors have a higher pre-NAC mitotic index and more stromal TIL infiltration (Supplementary Fig. 6B, C) than luminal BCs21,22. Consistently, high TIL levels are significantly associated with histological grade 3 tumors (Supplementary Fig. 6D).
MIIC reflects clinical practice
Several associations highlighted in the network reflect clinical practice decisions applied throughout BC centers. For example, the likelihood of performing conservative breast surgery depends on tumor histology (higher rates of mastectomy have been reported for patients with lobular or other histological types of tumor less likely to respond to NAC)23,24 (Supplementary Fig. 7A) and is positively associated with the practice of oncoplastic surgery25 (Supplementary Fig. 7B). Similarly, lumpectomy is more frequently associated with radiation therapy than with mastectomy (Supplementary Fig. 7C)26–29. After surgery, the addition of a second line of treatment by adjuvant chemotherapy, to decrease the risk of relapse, is driven by the identification of factors associated with a poor prognosis30, such as high levels of lymph node involvement (Supplementary Fig. 7D).
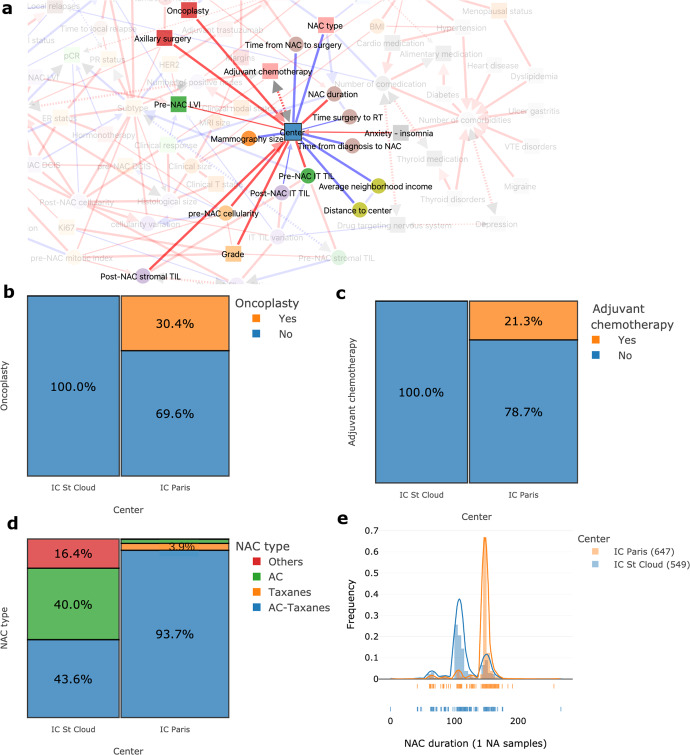
Beyond these well-established practices, MIIC also identified differences in clinical practices between the two centers of the cohort (Fig. 3a). For example, oncoplastic surgery and adjuvant chemotherapy were performed at only one of the two centers (Fig. 3b, c); the NAC regimen also differed between centers, with the Curie St Cloud center using more AC regimens than AC-taxane combinations, resulting in a shorter duration of NAC treatment (Fig. 3d, e).
Fig. 3. MIIC identifies differences in clinical practices between the two centers of the cohort.
a Network interactions around the node “center” of treatment. b Proportion of patients undergoing oncoplastic surgery, according to treatment center: Paris or St Cloud. c Proportion of patients receiving adjuvant chemotherapy according to treatment center: Paris or St Cloud. d Proportion of the various NAC regimens according to treatment center. e Distribution plot for NAC duration in days, according to treatment center.
MIIC traces the natural course of the disease
The natural course of BC may include local relapse, possibly followed by distant metastases, the trigger events leading to death31–35 (Fig. 4a–c). Contralateral BC is often used in composite survival endpoints, such as distant relapse-free survival36, but MIIC clearly identifies contralateral BC as an event being independent of other oncologic events and almost totally isolated from the rest of the network (Fig. 1). Luminal BC is known to recur and develop metastases later than HER2-positive BC and TNBC (Fig. 4d)21,22,37,38. The link between has also been found between PR negativity and a higher risk of brain metastasis39–43 (Fig. 4e).
Fig. 4. MIIC traces the natural course of the disease.
a Network interactions showing links between relapses, metastases and death in breast cancer. b Proportion of distant metastases according to the occurrence or absence of local relapses. c Proportion of deaths according to distant metastasis status. d Distribution plot for relapse-free survival (in months) according to breast cancer subtype. e Proportion plot displaying the relationship between central nervous system (CNS) metastasis and progesterone receptor (PR) status.
MIIC identifies unexpected associations, leading to new discoveries
With more than 15 associations involving treatment center (Fig. 3a), MIIC unmasked an unexpected “batch” effect relating to the site of BC treatment in this cohort. The observed differences reflect not only differences in therapeutic practice, but also in the characteristics of the population (differences in the proportion of women with psychological disorders, difference in incomes), in tumor presentation (tumor size), in pathological variable scoring (grade, presence of pre-NAC LVI, tumor cellularity, TILs), and in time to treatment within the care pathway.
MIIC identifies factors likely to improve prediction or prognosis
MIIC also favors new insights, e.g., comedication appears to protect against local relapse (Fig. 5a). Several retrospective studies have reported this association, with the use of statins44, NSAIDs45, or beta-blockers46 found to have indirect anticarcinogenic effects. It has recently been suggested that these non-oncological treatments may have immunomodulatory and chemosensitizing effects47.
Fig. 5. MIIC identifies factors likely to improve prediction or prognosis.
a Network interaction displaying the link between local relapse occurrence and the number of drugs taken (comedication). b Proportion plot showing the percentage of different clinical responses according to the presence or absence of pre-NAC lymphovascular invasion. c Boxplot of relapse-free survival according to the presence or absence of pre-NAC lymphovascular invasion. d Network interaction displaying the link between death, RCB and post-NAC mitotic index. e Boxplot of RCB values according to vital status. f Boxplot of post-NAC mitotic index according to vital status. The boxes represent the IQR, the horizontal lines correspond to the median values and lines ends mark the upper and lower fence. The number of cases considered for the analysis are reported in the legend.
MIIC suggests relevant combinations of predictive of prognostic biomarkers
MIIC may provide clues to combinations of new prognostic biomarkers likely to improve the prediction of response to chemotherapy, or post-NAC prognosis. Pre-NAC lymphovascular invasion (LVI) was found to be associated with both lower rates of clinical response (Fig. 5b) and shorter relapse-free survival (Fig. 5c). Both RCB (Fig. 5d, e) and post-NAC mitotic index (Fig. 5d–f), a parameter rarely used in practice but nevertheless reported to be a predictor of BC recurrence48,49, appear to be strongly associated with the risk of death. MIIC may, therefore, be an efficient tool for identifying features likely to improve prognosis, by combining gold standard indicators with other parameters, such as post-NAC mitotic index, and post-NAC LVI, for example. Finally, MIIC also makes it possible to optimize the binning of residual cancer burden (RCB). RCB is a post-NAC histological score calculated as an increasing continuous index, and then subdivided into four classes (0, I, II, and III)50. Our analysis based on information maximization principles suggested a new unsupervised classification of RCB scores into three categories (Fig. 5e), with RCB = 0 with low RCB values merged, in particular, into a single class associated with a good prognosis.
Discussion
When applied to a large cohort of BC patients, the MIIC algorithm successfully (i) performed quality controls; (ii) identified intra- and inter-modality correlations; (iii) highlighted differences in clinical practice, including center specificities; (iv) traced the natural course of the disease; (v) highlighted unsuspected and hidden associations, leading to new discoveries. The interactive visualization and causal analyses provided by this algorithm make it a promising tool for fast and effective explorations of the increasing amount of available health data.
The amount of exploitable health data is increasing exponentially. The best known health data resource for cancer studies remains the SEER (Surveillance, Epidemiology, and End Results) database, which collects data from population-based cancer registries covering approximately 34.6% of the US population51,52. By 2016, the National Cancer Database (NCDB) had amassed more than 34 million hospital records from cancer patients (almost four times the size of the SEER database), to become the largest clinical cancer registry in the world53. In France, the French administrative healthcare database, the SNDS (Système National des Données de Santé), is one of the largest administrative databases in the domain of medicine, providing many opportunities for medical research54,55, as it covers 99% of the French population (about 66 million people). The French government is planning to ease access to this almost exhaustive population research resource, through release as part of the “Health data hub” project. Finally, beyond these structured databases, the largest mine of untapped data worldwide remains the content of electronic health records (EHRs), encompassing a full range of data (clinical notes, laboratory results, imaging, genetic data, etc.) relating to patient care. Recent advances in information technology have made it easier for both hospitals and healthcare institutions to collect large amounts of healthcare data.
Biomedical scientists are now facing new challenges in the management and analysis of massive, heterogeneous datasets56. These challenges include the development of tools for exploration and visualization, analytical methods, integration into a comprehensive overview, and translation of the findings into public health impact. The visualization of information makes it possible for users to find profound patterns in clinical data, through visual recognition. Simple charts cannot represent the complexity of big data analyses and fail to support multifaceted tasks effectively3,4. There is, therefore, a need for sophisticated visualization tools dealing with many elements simultaneously and enabling users to perceive the patterns and insight generated by the algorithm57. Supplementary Table 1 shows the main data visualization tools used to present medical data. Many of the visual methods have been adopted directly from the field of data mining, but others, specific to the healthcare domain, have also been designed (Supplementary Table 2). For example, Happe and Drezen built the ePEPs toolbox, which displays relevant patterns extracted by eye from patient reimbursement data in the SNDS database, and supporting interactive exploration by researchers58. CARRE provides web-based components for interactive health data (fitness and biomarkers) visualization and risk analysis for the management of cardiorenal diseases59. The MITRE Corporation has also developed a web-based solution that provides an overview of an individual’s health through graphical representations of EHR data, highlighting abnormal values60. None of these visualization programs has yet managed to bridge the gap between of the large amounts of clinical data available and the discovery of clinical knowledge or paths for scientific research. By processing large heterogeneous sets of variables inherent to clinical records, MIIC provides physicians with a full picture of BC disease. It will be interesting to see how extending the present cohort of BC patients to larger BC cohorts treated with similar NAC therapy will allow us to refine the visual clinical network presented here, Fig. 1.
In addition to this use for visualization, the MIIC algorithm presents several other advantages for analyses, including its unsupervised nature, overcoming the need for training or human involvement. This feature makes it possible to obtain new knowledge through the automatic identification of patterns and dependences in the data, highlighting new interactions, and it may be of use for feature selection in machine learning models.
In conclusion, MIIC, an open-access, interactive, multitask tool, is designed to visualize datasets to help clinicians and researchers to understand the relationships between the variables within them. It opens up promising perspectives for guiding the generation of new hypotheses, helping clinicians identify actionable nodes and edges in clinical networks, and revealing new clues to relationships of interest for research purposes. Its widespread use in the field of health data could increase the accuracy of prediction for treatment responses and prognosis. This tool has the potential to improve the care pathway and, ultimately, the survival of patients.
Methods
Patients and treatment
We analyzed a cohort of 1197 patients with non-metastatic BC treated by NAC, with or without trastuzumab, followed by surgery, at either of the two Institut Curie sites (Paris and Saint Cloud) between 2002 and 2012 (NEOREP Cohort, CNIL declaration number 1547270. The methods were performed in accordance with relevant guidelines and regulations and approved by CNIL and the Breast Cancer Group of Institut Curie on September 11, 2020. Owing to the retrospective nature of this study, the ethics committee granted a waiver of informed consent for the included participants.). We included unilateral, non-recurrent, non-inflammatory, non-metastatic tumors, and excluded T4 tumors. This retrospective study was conducted in accordance with institutional and ethical rules regarding research on tissue specimens and patients. Information on family history, clinical characteristics (age; menopausal status; body mass index) and tumor characteristics (clinical tumor stage and grade; histology; clinical nodal status; ER, PR and HER2 status; BC subtype; mitotic index; Ki67; lymphovascular invasion) was retrieved from electronic medical records. All the patients of the cohort received NAC, and additional treatments were decided in accordance with national guidelines.
Tumor samples and pathological review
In accordance with the guidelines used in France (Group for Evaluation of Prognostic Factors using Immunohistochemistry in Breast Cancer61), cases were considered estrogen receptor (ER)-positive or progesterone receptor (PR)-positive if at least 10% of the tumor cells expressed estrogen and/or progesterone receptors (ER/PR). Endocrine therapy was prescribed when this threshold was exceeded. HER2-negative status was defined as a score of 0 or 1+ for the tissue section stained by immunohistochemistry (IHC). Tissue sections with scores of IHC 2+ or IHC 3+ were then analyzed by fluorescence in situ hybridization (FISH) to confirm HER2 positivity. BC tumors were classified into subtypes (TNBC, HER2-positive, and luminal HER2-negative [referred to hereafter as “luminal”]). BC subtypes were defined as follows: luminal, ER+ or PR+/HER2−; TNBC, ER−/PR−/HER2−; HER2-positive BC, HER2+. Pretreatment core needle biopsy specimens and/or the corresponding post-NAC surgical specimens were reviewed independently by breast disease experts for research purposes, to assess residual cancer burden index, and the levels of tumor-infiltrating lymphocytes. The pathological reviews of these specimens are described in detail elsewhere20,62,63. Pathological complete response (pCR) was defined as the absence of residual invasive cancer cells in the breast and axillary lymph nodes (ypT0/is þ/ypN0).
Survival endpoints
Relapse-free survival (RFS) was defined as the time from surgery to death, loco-regional recurrence or distant recurrence, whichever occurred first. Overall survival (OS) was defined as the time from surgery to death. The date of last known contact was retained for patients for whom none of these events were recorded. The cutoff date for survival analysis was March, 13th, 2019.
Variables of interest
The care pathway of BC patients eligible for neoadjuvant chemotherapy can be summarized as follows: (i) pretreatment biopsy for BC diagnosis; (ii) administration of chemotherapy as the first-line treatment; (iii) removal of the tumor by surgery; (iv) histological analysis of the specimens obtained; (v) prescription of adjuvant treatments, if indicated (radiotherapy, hormonotherapy, chemotherapy); (vi) patients follow-up to monitor for relapse or death. We identified 94 clinically relevant variables from clinical, radiological, pathological and outcome data, which we grouped into 14 categories (hospital, history, comedication, comorbidities, clinical baseline, baseline histology, pre-NAC pathology, treatment response, surgery, treatment, changes during NAC, post-NAC pathology, delayed relapse/survival, metastasis). For composite variables derived from raw variables (e.g., BC subtype, constructed from a combination of ER status, PR status, HER2 status), both derived and raw variables were represented on the network.
MIIC algorithm
The functioning of the algorithm has been described in detail elsewhere5,7. Briefly, starting from a fully connected network, the MIIC algorithm first removes dispensable edges by iteratively subtracting the most significant information contributions from indirect paths between each pair of variables. The remaining edges, the underlying effect of which cannot be explained by indirect paths, are then oriented based on the causality signature in the data, corresponding to the simultaneous head-to-head orientations of so-called “v-structures”, X→Z←Y. In principle, propagation of v-structure orientations to downstream edges can also be implemented to fulfill underlying model class assumptions64,65 but are not applied on the NEOREP clinical network to ensure that MIIC algorithmic decisions are only based on information actually contained in the data.
Each edge corresponds to a “direct” association between two variables, that is, a statistical association that cannot be entirely explained by indirect effects involving other variables. Red and blue edges correspond to positive and negative (i.e., anti-correlated) associations, respectively. Four types of edge orientations are distinguished by the MIIC online server: (i) directed edges with a gray arrowhead represent inferred causal relationships; (ii) bidirected edges (drawn with dashed lines) reflect the presence of a latent common cause (L) unobserved in the available dataset, i.e., X←(L)→Y; (iii) directed edges with a colored (red or blue) arrowhead are consistentent with either a causal or a latent common cause relationship; and (iv) undirected edges, whose orientation if it exists, cannot be inferred from non-perturbative data. The original algorithm was restricted to categorical variables5, but MIIC has recently been extended to include continuous variables, the values of which are partitioned into optimal bins, maximizing mutual information with another (continuous or categorical) variable of interest, while preventing the overfitting of datasets of finite size due to the use of too many bins7. In particular, each continuous variable may have different information-maximizing partitions depending on the associated variable of interest. For instance, MIIC finds three maximally informative bins for the residual cancer burden (RCB) score in association with patient survival status (Supplementary Fig. 1A), whereas eight RCB bins are required to estimate its mutual information with post-NAC cellularity correctly (Supplementary Fig. 1B).
MIIC online server
The MIIC online server is freely accessible at https://miic.curie.fr and can be used with the Google Chrome, Mozilla Firefox, Edge, and Safari browsers. The user guide summarizing the main steps for running the MIIC algorithm is accessible at https://miic.curie.fr/user_guide.php, and an online video tutorial is available at: https://miic.curie.fr/tutorial.php. The workbench is available from https://miic.curie.fr/workbench.php. As input data, the user can upload a dataset formatted as a table with commas, semicolons, tabs, pipes or colons, as separators, without row names. Each variable can be either categorical or quantitative (discrete or continuous). Variables can be grouped into families, identified with different colors on the network. Missing values are allowed in the dataset and their possible statistical biases are taken into account by MIIC7. They should be indicated as “NA” in the dataset table. Once the dataset has been prepared, the user runs the algorithm, and an e-mail is sent when the job is completed.
MIIC output
The MIIC online server generates a visualization of the global network of the dataset. An example based on the NEOREP dataset is displayed in Fig. 1, and is accessible as an interactive network at https://miic.curie.fr/job_results_showcase.php?id=NEOREP.
Interactive exploration of the network
The distributions and neighborhoods of each node and edge of the inferred network can be explored through an interactive interface, through the mouse-over right- or left-click buttons on the browser page, as detailed in the online tutorials. Briefly, any variable can be highlighted by clicking on the network or through the “Search” toolbox (Supplementary Fig. 2A). The corresponding plots can be downloaded as.png or.svg images. Each node can be explored individually in terms of counts (categorical variables, Supplementary Fig. 2B, C) or distribution (continuous variables Supplementary Fig. 2D, E). Each edge can be explored by a right click and the choice of “plot join distribution” or “plot discretization”. The resulting plots are (i) proportion plots, with the edge representing the total association between two categorical variables (Supplementary Fig. 2F); (ii) distribution histograms (Supplementary Fig. 2G) or boxplots (Supplementary Fig. 2H), in which the edge represents the total association between a categorical and a continuous variable or (iii) scatter plots (Supplementary Fig. 2I), in which the edge represents the total association between two continuous variables. Additional options include inverting the x and y axes, the choice of frequency or absolute counts, or NA removal (proportion plots), and faceting or superimposing the variables (distribution histograms). All the figures presented here were generated with the MIIC online interactive visualization tool.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
N.S. acknowledges support from Sorbonne University (ATER), V.C. from ARC foundation and HI from ITMO Cancer, Institut Curie and CNRS.
Author contributions
N.S., V.C., and H.I. designed and implemented the machine learning and interactive exploration tools; A.S.H., L.D., M.L. performed research; A.S.H. and F.R. verified the data; N.S., V.C., H.I., L.D., F.R., and A.S.H. contributed to data analysis; A.S.H. and F.R. contributed to expert review. All authors contributed to data interpretation. L.D., A.S.H., N.S., and H.I. wrote the paper. N.S. worked on the paper while being affiliated at 2 and 3. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Data availability
All images and the associated network are publicly available at: https://miic.curie.fr/job_results_showcase.php?id=NEOREP. Data corresponding to the NEOREP cohort study will be available upon reasonable request.
Code availability
The code corresponding to the MIIC algorithm can be found at the following address: https://github.com/miicTeam/miic_R_package.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Nadir Sella, Anne-Sophie Hamy, Vincent Cabeli.
Contributor Information
Fabien Reyal, Email: fabien.reyal@curie.fr.
Hervé Isambert, Email: herve.isambert@curie.fr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41746-022-00647-0.
References
- 1.Bärtschi, M. Health Data Visualization-A review * Seminar Collaborative Data Visualization in 2015 (2015).
- 2.Luo J, Wu M, Gopukumar D, Zhao Y. Big data application in biomedical research and health care: a literature review. Biomed. Inf. Insights. 2016;8:1–10. doi: 10.4137/BII.S31559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ola O, Sedig K. Beyond simple charts: Design of visualizations for big health data [Internet] Online J Public Health Inform. 2016;8:e195. doi: 10.5210/ojphi.v8i3.7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shneiderman B, Plaisant C, Hesse BW. Improving healthcare with interactive visualization. Computer. 2013;46:58–66. doi: 10.1109/MC.2013.38. [DOI] [Google Scholar]
- 5.Verny L, Sella N, Affeldt S, Singh PP, Isambert H. Learning causal networks with latent variables from multivariate information in genomic data. PLoS Comput. Biol. 2017;13:e1005662. doi: 10.1371/journal.pcbi.1005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sella N, Verny L, Uguzzoni G, Affeldt S, Isambert H. MIIC online: a web server to reconstruct causal or non-causal networks from non-perturbative data. Bioinformatics. 2018;34:2311–2313. doi: 10.1093/bioinformatics/btx844. [DOI] [PubMed] [Google Scholar]
- 7.Cabeli V, et al. Learning clinical networks from medical records based on information estimates in mixed-type data [Internet] PLoS Comput. Biol. 2020;16:e1007866. doi: 10.1371/journal.pcbi.1007866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandão M, Reyal F, Hamy A-S, Piccart-Gebhart M. Neoadjuvant treatment for intermediate/high-risk HER2-positive and triple-negative breast cancers: no longer an “option” but an ethical obligation. ESMO Open. 2019;4:e000515. doi: 10.1136/esmoopen-2019-000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyal F, Hamy AS, Piccart MJ. Neoadjuvant treatment: the future of patients with breast cancer. ESMO Open. 2018;3:e000371. doi: 10.1136/esmoopen-2018-000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burstein HJ, et al. Adjuvant endocrine therapy for women with hormone receptor–positive breast cancer: American Society of Clinical Oncology Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2014;32:2255–2269. doi: 10.1200/JCO.2013.54.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson FR, et al. Herceptin® (trastuzumab) in HER2-positive early breast cancer: protocol for a systematic review and cumulative network meta-analysis. Syst. Rev. 2017;6:196. doi: 10.1186/s13643-017-0588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aaldriks AA, et al. Prognostic factors for the feasibility of chemotherapy and the Geriatric Prognostic Index (GPI) as risk profile for mortality before chemotherapy in the elderly. Acta Oncol. 2016;55:15–23. doi: 10.3109/0284186X.2015.1068446. [DOI] [PubMed] [Google Scholar]
- 13.van Leeuwen RWF, et al. Potential drug interactions in cancer therapy: a prevalence study using an advanced screening method. Ann. Oncol. 2011;22:2334–2341. doi: 10.1093/annonc/mdq761. [DOI] [PubMed] [Google Scholar]
- 14.Popa MA, Wallace KJ, Brunello A, Extermann M, Balducci L. Potential drug interactions and chemotoxicity in older patients with cancer receiving chemotherapy. J. Geriatr. Oncol. 2014;5:307–314. doi: 10.1016/j.jgo.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang N, Qin MZ, Cui J. Lipid profile comparison between pre- and post-menopausal women. Zhonghua Xin Xue Guan Bing. Za Zhi. 2016;44:799–804. doi: 10.3760/cma.j.issn.0253-3758.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Veronesi U, et al. Sentinel-lymph-node biopsy as a staging procedure in breast cancer: update of a randomised controlled study. Lancet Oncol. 2006;7:983–990. doi: 10.1016/S1470-2045(06)70947-0. [DOI] [PubMed] [Google Scholar]
- 17.Weidner N, Moore DH, Vartanian R. Correlation of Ki-67 antigen expression with mitotic figure index and tumor grade in breast carcinomas using the novel “paraffin”-reactive MIB1 antibody. Hum. Pathol. 1994;25:337–342. doi: 10.1016/0046-8177(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 18.Cortadellas T, et al. Estimation of tumor size in breast cancer comparing clinical examination, mammography, ultrasound and MRI—correlation with the pathological analysis of the surgical specimen. Gland Surg. 2017;6:330–335. doi: 10.21037/gs.2017.03.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berg WA, et al. Diagnostic accuracy of mammography, clinical examination, US, and MR imaging in preoperative assessment of breast cancer. Radiology. 2004;233:830–849. doi: 10.1148/radiol.2333031484. [DOI] [PubMed] [Google Scholar]
- 20.Hamy A-S, et al. Lymphovascular invasion after neoadjuvant chemotherapy is strongly associated with poor prognosis in breast carcinoma. Breast Cancer Res. Treat. 2018;169:295–304. doi: 10.1007/s10549-017-4610-0. [DOI] [PubMed] [Google Scholar]
- 21.Meyers MO, et al. Impact of breast cancer molecular subtypes on locoregional recurrence in patients treated with neoadjuvant chemotherapy for locally advanced breast cancer. Ann. Surg. Oncol. 2011;18:2851–2857. doi: 10.1245/s10434-011-1665-8. [DOI] [PubMed] [Google Scholar]
- 22.Lowery AJ, Kell MR, Glynn RW, Kerin MJ, Sweeney KJ. Locoregional recurrence after breast cancer surgery: a systematic review by receptor phenotype. Breast Cancer Res. Treat. 2012;133:831–841. doi: 10.1007/s10549-011-1891-6. [DOI] [PubMed] [Google Scholar]
- 23.Waljee JF, Hu ES, Newman LA, Alderman AK. Predictors of re-excision among women undergoing breast-conserving surgery for cancer. Ann. Surg. Oncol. 2008;15:1297–1303. doi: 10.1245/s10434-007-9777-x. [DOI] [PubMed] [Google Scholar]
- 24.Truin W, et al. Differences in response and surgical management with neoadjuvant chemotherapy in invasive lobular versus ductal breast cancer. Ann. Surg. Oncol. 2016;23:51–57. doi: 10.1245/s10434-015-4603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munhoz AM, Montag E, Gemperli R. Oncoplastic breast surgery: indications, techniques and perspectives. Gland Surg. 2013;2:143–157. doi: 10.3978/j.issn.2227-684X.2013.08.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchholz TA. Radiation therapy for early-stage breast cancer after breast-conserving surgery. N. Engl. J. Med. 2009;360:63–70. doi: 10.1056/NEJMct0803525. [DOI] [PubMed] [Google Scholar]
- 27.Carlson RW, et al. Invasive breast cancer. J. Natl. Compr. Canc. Netw. 2011;9:136–222. doi: 10.6004/jnccn.2011.0016. [DOI] [PubMed] [Google Scholar]
- 28.Eifel P, et al. National Institutes of Health Consensus Development Conference Statement: adjuvant therapy for breast cancer, November 1-3, 2000. J. Natl. Cancer Inst. 2001;93:979–989. doi: 10.1093/jnci/93.13.979. [DOI] [PubMed] [Google Scholar]
- 29.Halberg FE, et al. Conservative surgery and radiation in the treatment of stage I and II carcinoma of the breast. American College of Radiology. ACR Appropriateness Criteria. Radiology. 2000;215(Suppl):1193–1205. [PubMed] [Google Scholar]
- 30.Masuda N, et al. Adjuvant capecitabine for breast cancer after preoperative chemotherapy. N. Engl. J. Med. 2017;376:2147–2159. doi: 10.1056/NEJMoa1612645. [DOI] [PubMed] [Google Scholar]
- 31.Dent R, et al. Factors associated with breast cancer mortality after local recurrence. Curr. Oncol. 2014;21:e418–e425. doi: 10.3747/co.21.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whelan T, Clark R, Roberts R, Levine M, Foster G. Ipsilateral breast tumor recurrence postlumpectomy is predictive of subsequent mortality: results from a randomized trial. Investigators of the Ontario Clinical Oncology Group. Int. J. Radiat. Oncol. Biol. Phys. 1994;30:11–16. doi: 10.1016/0360-3016(94)90513-4. [DOI] [PubMed] [Google Scholar]
- 33.Kurtz JM, et al. The prognostic significance of late local recurrence after breast-conserving therapy. Int. J. Radiat. Oncol. Biol. Phys. 1990;18:87–93. doi: 10.1016/0360-3016(90)90271-K. [DOI] [PubMed] [Google Scholar]
- 34.Sopik V, Nofech-Mozes S, Sun P, Narod SA. The relationship between local recurrence and death in early-stage breast cancer. Breast Cancer Res. Treat. 2016;155:175–185. doi: 10.1007/s10549-015-3666-y. [DOI] [PubMed] [Google Scholar]
- 35.Witteveen A, Kwast ABG, Sonke GS, IJzerman MJ, Siesling S. Survival after locoregional recurrence or second primary breast cancer: impact of the disease-free interval. PLoS ONE. 2015;10:e0120832. doi: 10.1371/journal.pone.0120832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hudis CA, et al. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J. Clin. Oncol. 2007;25:2127–2132. doi: 10.1200/JCO.2006.10.3523. [DOI] [PubMed] [Google Scholar]
- 37.Voduc KD, et al. Breast cancer subtypes and the risk of local and regional relapse. JCO. 2010;28:1684–1691. doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- 38.Wu X, et al. Pattern of Local Recurrence and Distant Metastasis in Breast Cancer By Molecular Subtype [Internet] Cureus. 2016;8:e924. doi: 10.7759/cureus.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snell CE, et al. Absent progesterone receptor expression in the lymph node metastases of ER-positive, HER2-negative breast cancer is associated with relapse on tamoxifen. J. Clin. Pathol. 2017;70:954–960. doi: 10.1136/jclinpath-2016-204304. [DOI] [PubMed] [Google Scholar]
- 40.Nishimura R, et al. Changes in the ER, PgR, HER2, p53 and Ki-67 biological markers between primary and recurrent breast cancer: discordance rates and prognosis. World J. Surg. Oncol. 2011;9:131. doi: 10.1186/1477-7819-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimura R, et al. Evaluation of factors related to late recurrence–later than 10 years after the initial treatment–in primary breast cancer. Oncology. 2013;85:100–110. doi: 10.1159/000353099. [DOI] [PubMed] [Google Scholar]
- 42.Darlix A, et al. Hormone receptors status: a strong determinant of the kinetics of brain metastases occurrence compared with HER2 status in breast cancer. J. Neurooncol. 2018;138:369–382. doi: 10.1007/s11060-018-2805-9. [DOI] [PubMed] [Google Scholar]
- 43.Zhou L, et al. Progesterone suppresses triple-negative breast cancer growth and metastasis to the brain via membrane progesterone receptor α. Int. J. Mol. Med. 2017;40:755–761. doi: 10.3892/ijmm.2017.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahern TP, et al. Statin prescriptions and breast cancer recurrence risk: a Danish nationwide prospective cohort study. J. Natl. Cancer Inst. 2011;103:1461–1468. doi: 10.1093/jnci/djr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwan ML, Habel LA, Slattery ML, Caan B. NSAIDs and breast cancer recurrence in a prospective cohort study. Cancer Causes Control. 2007;18:613–620. doi: 10.1007/s10552-007-9003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Powe DG, et al. Beta-blocker drug therapy reduces secondary cancer formation in breast cancer and improves cancer specific survival. Oncotarget. 2010;1:628–638. doi: 10.18632/oncotarget.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamy A-S, et al. Comedications influence immune infiltration and pathological response to neoadjuvant chemotherapy in breast cancer. OncoImmunology. 2020;9:1677427. doi: 10.1080/2162402X.2019.1677427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farrugia DJ, et al. Mitotic index to predict breast cancer recurrence after neoadjuvant systemic therapy. JCO. 2016;34:e23265. doi: 10.1200/JCO.2016.34.15_suppl.e23265. [DOI] [Google Scholar]
- 49.Pattali S, et al. Value of mitotic index in residual tumors following neoadjuvant therapy for breast cancer: Single institution experience. JCO. 2016;34:548. doi: 10.1200/JCO.2016.34.15_suppl.548. [DOI] [Google Scholar]
- 50.Symmans WF, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J. Clin. Oncol. 2007;25:4414–4422. doi: 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 51.Duggan MA, Anderson WF, Altekruse S, Penberthy L, Sherman ME. The Surveillance, Epidemiology and End Results (SEER) program and pathology: towards strengthening the critical relationship. Am. J. Surg. Pathol. 2016;40:e94–e102. doi: 10.1097/PAS.0000000000000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu JB, Smith BD. NCI SEER public-use data: applications and limitations in oncology research [internet] Oncology. 2009;23:3. [PubMed] [Google Scholar]
- 53.Boffa DJ, et al. Using the National Cancer Database for Outcomes Research: a review. JAMA Oncol. 2017;3:1722–1728. doi: 10.1001/jamaoncol.2016.6905. [DOI] [PubMed] [Google Scholar]
- 54.Bezin J, et al. The national healthcare system claims databases in France, SNIIRAM and EGB: Powerful tools for pharmacoepidemiology. Pharmacoepidemiol. Drug Saf. 2017;26:954–962. doi: 10.1002/pds.4233. [DOI] [PubMed] [Google Scholar]
- 55.Tuppin, P. et al. Value of a national administrative database to guide public decisions: From the système national d’information interrégimes de l’Assurance Maladie (SNIIRAM) to the système national des données de santé (SNDS) in France [Internet]. Rev. Epidemiol. Sante Publique. 65 Suppl 4, S149–S167 (2017). [DOI] [PubMed]
- 56.Margolis R, et al. The National Institutes of Health’s Big Data to Knowledge (BD2K) initiative: capitalizing on biomedical big data. J. Am. Med. Inf. Assoc. 2014;21:957–958. doi: 10.1136/amiajnl-2014-002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keim, D. et al. Information Visualization (eds Kerren, A., Stasko, J. T., Fekete, J.-D., et al.) 154–175 (Springer, 2008).
- 58.Happe, A. & Drezen, E. A visual approach of care pathways from the French nationwide SNDS database—from population to individual records: the ePEPS toolbox [Internet]. Available from: https://hal-univ-rennes1.archives-ouvertes.fr/hal-01697626 (2018). [DOI] [PubMed]
- 59.Zhao, Y. et al. Visual analytics for health monitoring and risk management in CARRE. E-Learning and Games; 10th International Conference, Edutainment 2016. Hangzhou, China, April 14–16, 2016, Revised selected papers 9654, 380–391 (2016).
- 60.Ledesma A, Al-Musawi M, Nieminen H. Health figures: an open source JavaScript library for health data visualization [Internet] BMC Med. Inform. Decis. Mak. 2016;16:38. doi: 10.1186/s12911-016-0275-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.[Recommendations for the immunohistochemistry of the hormonal receptors on paraffin sections in breast cancer. Update 1999. Group for Evaluation of Prognostic Factors using Immunohistochemistry in Breast Cancer (GEFPICS-FNCLCC)]. Ann. Pathol. 19, 336–343 (1999). [PubMed]
- 62.Hamy A-S, et al. Stromal lymphocyte infiltration after neoadjuvant chemotherapy is associated with aggressive residual disease and lower disease-free survival in HER2-positive breast cancer. Ann. Oncol. 2017;28:2233–2240. doi: 10.1093/annonc/mdx309. [DOI] [PubMed] [Google Scholar]
- 63.Hamy-Petit A-S, et al. Pathological complete response and prognosis after neoadjuvant chemotherapy for HER2-positive breast cancers before and after trastuzumab era: results from a real-life cohort. Br. J. Cancer. 2016;114:44–52. doi: 10.1038/bjc.2015.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Affeldt, S. & Isambert, H. Robust reconstruction of causal graphical models based on conditional 2-point and 3-point information. In Proceedings of the 31th conference on Uncertainty in Artificial Intelligence (UAI) (Amsterdam, The Netherlands, 2015).
- 65.Affeldt S, Verny L, Isambert H. 3off2: a network reconstruction algorithm based on 2-point and 3-point information statistics. BMC Bioinforma. 2016;17(Suppl):12. doi: 10.1186/s12859-015-0856-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All images and the associated network are publicly available at: https://miic.curie.fr/job_results_showcase.php?id=NEOREP. Data corresponding to the NEOREP cohort study will be available upon reasonable request.
The code corresponding to the MIIC algorithm can be found at the following address: https://github.com/miicTeam/miic_R_package.