Abstract
Early life stress (ELS) induces long-term phenotypic adaptations that contribute to increased vulnerability to a host of neuropsychiatric disorders. Epigenetic mechanisms, including DNA methylation, histone modifications and non-coding RNA, are a proposed link between environmental stressors, alterations in gene expression, and phenotypes. Epigenetic modifications play a primary role in shaping functional differences between cell types and can be modified by environmental perturbations, especially in early development. Together with contributions from genetic variation, epigenetic mechanisms orchestrate patterns of gene expression within specific cell types that contribute to phenotypic variation between individuals. To date, many studies have provided insights into epigenetic changes resulting from ELS. However, most of these studies have examined heterogenous brain tissue, despite evidence of cell-type-specific epigenetic modifications in phenotypes associated with ELS. In this review, we focus on rodent and human studies that have examined epigenetic modifications induced by ELS in select cell types isolated from the brain or associated with genes that have cell-type-restricted expression in neurons, microglia, astrocytes, and oligodendrocytes. Although significant challenges remain, future studies using these approaches can enable important mechanistic insight into the role of epigenetic variation in the effects of ELS on brain function.
Subject terms: Epigenetics in the nervous system, Genetics
Epigenetics and cellular programming
At the beginning of life, the unicellular fertilized egg, or zygote, gives rise to all the cells of an organism, possessing the property of “totipotency”. As the zygote becomes an embryo, and the embryo a fetus, there is a gradual decline in totipotency of the newly divided cells, narrowing the range of cell types they can become, rendering them “pluripotent”, a function of the plasticity of epigenetic marks. As the embryo develops and cells differentiate, pluripotency declines and epigenetic marks become more stable, determining and maintaining gene expression programs that underlie cell fate. In this way, cellular programming can be defined as the epigenetic process that contributes to stem cell differentiation into mature cell types [1, 2]. As the genetic sequence is virtually identical in all cells within each individual, the epigenome of each cell orchestrates the pattern of gene expression to confer cellular identity through histone modifications, DNA methylation (DNAm), and non-coding RNA (ncRNA).
Octamers of histone proteins coil DNA to form the nucleosomes, which are themselves wound to form chromatin. Modifications of histone N-terminal tails by acetylation, methylation, phosphorylation and ubiquitination play a role in determining DNA accessibility by modifying the positively charged N-terminal tails that tightly interact with the negatively charged DNA. For example, histone acetylation and deacetylation are understood to render chromatin more or less accessible to transcription factors (TFs), leading to enhanced or reduced transcriptional activity, respectively [3]. Histone methylation has also been associated with gene silencing or activation, depending on the amino acid modified [4, 5].
Methylation of DNA is a class of DNA modification that largely occurs at cytosine bases that are followed by guanine bases (CpG sites) in the mammalian genome. Non-CpG methylation, although less frequent, has been found in embryonic stem cells [6], neurons [7] and mature oocytes [8]. At gene promoters, first exons, and first introns, DNA methylation (DNAm) can suppress gene expression by inhibiting TF binding to regulatory elements [9]. DNAm within gene bodies and internal exons involves complex interactions with TF binding sites and conformational chromatin structure [10–12], the regulation of alternative splicing [13, 14], and is positively associated with transcription, especially for ubiquitously expressed genes [12, 15]. DNAm can also suppress gene expression through other mechanisms including histone deacetylase complex recruitment, which introduces histone modifications that result in chromatin silencing [16]. Conversely, TFs themselves can regulate DNAm by binding to specific DNA sequences to protect de novo methylation or recruit DNA methyltransferases to maintain, suppress, or initiate de novo DNAm [17].
Several types of ncRNAs, including micro-RNAs (miRs) and long non-coding RNA (lncRNAs) are sometimes considered an epigenetic mechanism due to their prominent roles in epigenetic regulation [18]. miRs are short sequences of nucleotides (~22) that largely repress gene expression post-transcriptionally through complementary binding to their target mRNAs, of which there can be hundreds, prompting the degradation of the corresponding mRNA and ultimately reducing its protein level [19]. The regulation of other genes at the RNA level by miRs is a property shared with other common post-transcriptional regulators of gene expression that are not typically considered part of epigenetics. Other ncRNAs are involved in epigenetic regulation at the nucleus, as is the case for lncRNA [20]. lncRNAs are >100 nucleotides in length, involved in processes including chromatin remodeling, modulation of histone and DNA methylation and acetylation, pre and post-transcription and translation, and have been functionally characterized mainly in the context of cancer [21, 22]. Generally, lncRNAs are processed and operate in the cytoplasm (see review [20]), however, lncRNAs involved in X-chromosome inactivation play a prominent role in the nucleus [22, 23].
The epigenetic program arises in response to fetal environmental signals that include extrinsic and intrinsic signaling molecules and growth factors (see review [24]), genomic imprinting through DNAm and histone modifications, and the DNA sequence itself. Genotypic differences can introduce DNAm sites (e.g. the presence of cytosines) and affect the binding of TFs, which in turn influences epigenetic modifications [17]. Cycles of epigenetic processes involved in genetic imprinting and sex-chromosome dosage compensation also occur in the zygote as discussed [25] and reviewed elsewhere [2, 26]. For example, soon after fertilization, in the pre-implanted embryo, or blastocyst, there is genome-wide demethylation in somatic cells, followed by a global wave of remethylation at implantation with locus-specific changes to methylation that continue into late gestation [27].
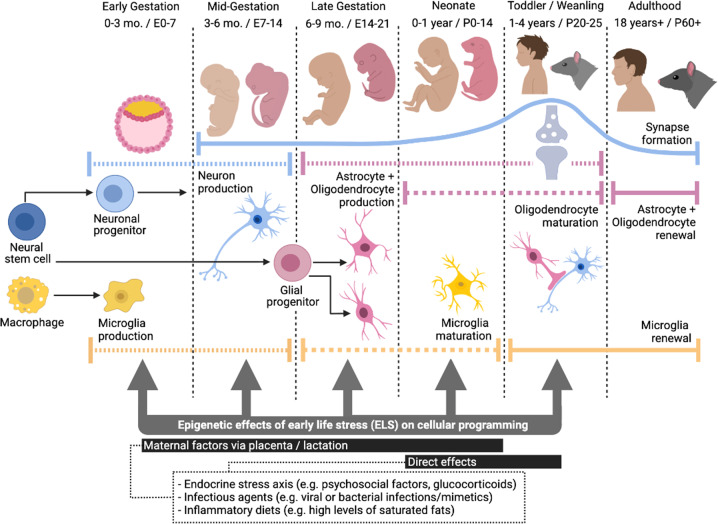
In the embryo, one of the first major phases of cellular programming is the differentiation of the embryonic stem cells into the three germ layers: the endoderm, mesoderm, and ectoderm, the latter of which gives rise to the neuroectoderm, the precursor tissue of the central nervous system (CNS) [28]. The neuroectodermal stem cells, also known as neural stem cells (NSCs), are “multipotent” and produce distinct cell types of the CNS (Fig. 1) [29]. During early gestation, NSCs self-renew and symmetrically divide into two identical daughter cells to increase the pool of multipotent cells. Early to mid-gestation, NSCs divide asymmetrically to produce a NSC and a neuronal progenitor, precursors for neurons. In early and mid-gestation in humans [30–32] and rats [33], myeloid-derived macrophages in the yolk sac, formed from the endoderm [34] invade the embryonic nervous system to become the resident CNS macrophages, or microglia [35, 36], which mature into the neonatal period [37]. In late gestation to toddler age in humans [38, 39] and weanling age in mice [40], NSCs give rise to glial progenitors, precursors for astrocytes and oligodendrocytes [1], with myelination occurring from birth to toddler age in humans [39] and weanling age in rodents [41].
Fig. 1. Timeline of cellular programming in the brain and windows of susceptibility for epigenetic effects by early life stress (ELS).
Early to mid-gestation, neural stem cells produce neuronal progenitors, precursors for neurons, which migrate and form synapses from mid-gestation to toddler/weanling age. Synaptogenesis starts mid-gestation, peaks in toddler/weanling age, and continues throughout life. In late gestation to toddler/weanling age, neural stem cells give rise to glial progenitors, precursors for astrocytes and oligodendrocytes, which mature from birth to toddler/weanling age. In early and mid-gestation, myeloid-derived macrophages invade the embryonic nervous system to become microglia, which mature into the neonatal period. Microglia and glia continue to have local self-renewal and proliferative capacity into adulthood. Overall, during these periods of cellular production and maturation, ELS factors (e.g. stress hormones, immune stimulants) can alter cell type proportions as well as induce epigenetic reprogramming, leading to long-term changes in the brain. E = embryonic day, P = post-natal day.
Neurons are largely post-mitotic cells; most production and migration takes place prenatally and continues to a limited degree in the postnatal period [42], with a few neurogenic zones remaining active in adulthood [43]. The formation of synapses starts mid-gestation, peaking at toddler age in humans [44], and weanling age in rodents [42], and continues throughout life. Microglia continue to have local self-renewal and proliferative capacity [35, 36], and glial progenitors continue to proliferate, migrate and mature into adulthood [45]. Neurodevelopment also involves substantial apoptosis and synaptic pruning. Most neuronal cell death occurs early to late gestation, while glial cell death, synaptic pruning and experience-dependent modification occurs mostly at the post-natal stage [29].
Epigenetic reprogramming by early life stress
Developmental programming is the process by which cellular programming is fine-tuned by environmental factors in fetal and early postnatal stages, referred to as “early life”. This fine-tuning of cellular programs involves epigenetic reprogramming, whereby the epigenetic plan is altered in a persistent manner within a given cell type, can occur in multiple cell types, does not change cell type, and is typically stable across mitotic cell divisions [46, 47]. Epigenetic reprogramming can alter transcript abundance through long-term modifications that persist even in the absence of the initial environmental trigger. Although epigenetic reprogramming can occur later in life, the early life period is more sensitive to stress, or disruptions to homeostasis, since cell fates are established during this time.
Early life stress (ELS) is an acute or chronic factor that takes place at the prenatal, perinatal and/or pre-pubertal postnatal stages and elicits or affects stress responses. The most studied forms of ELS are inflammation-based (e.g. infection, high-fat diets, xenobiotics) and psychosocial (e.g. emotional and physical abuse or neglect, and sexual abuse) [48]. ELS causes elevations in inflammatory cytokines and/or stress hormones that either impact offspring directly or indirectly via the placenta or breastmilk as maternal factors, inducing epigenetic modifications that impact neurodevelopmental trajectories [49]. In humans, inflammation-based and psychosocial ELS factors are most closely associated with neurodevelopmental conditions including autism spectrum disorder and psychiatric disorders related to anxiety, mood and psychosis (schizophrenia) [50–58]. In parallel, evidence from animal studies show that exposure to maternal high-fat diets, immunostimulants such as the bacterial mimic lipopolysaccharide (LPS) and psychosocial stress in early life lead to long-term changes in corresponding behavioral features of these disorders [59–65]. These findings are accompanied by evidence of long-lasting changes in microglial densities, cytokine expression and phagocytic actions involved in synaptic pruning [59, 66, 67], neurotransmission, neural connectivity, and neuron-glia interactions [68–70]. In this context, accumulating evidence indicates that lasting phenotypic changes induced by ELS involve modifications to cellular programs in the brain [57, 71].
Animal models of ELS permit investigation of in and ex vivo changes taking place in the brain that are not easily feasible in humans. However, the correspondence to human conditions of ELS may only be approximate. For example, laboratory rodents are immunologically naïve, unlike humans. Additional complications concern the assessment of different forms of psychosocial ELS (e.g. verbal versus physical abuse) and factors known to influence resiliency in humans (e.g. family income or educational attainment) [72, 73] that are difficult to model in animals. Nevertheless, given the complexities of studying the human brain in the context of ELS, rodent models appear necessary to discover causal pathways and enable the development of molecular targets for therapeutics.
Epigenetic reprogramming by ELS: the current landscape
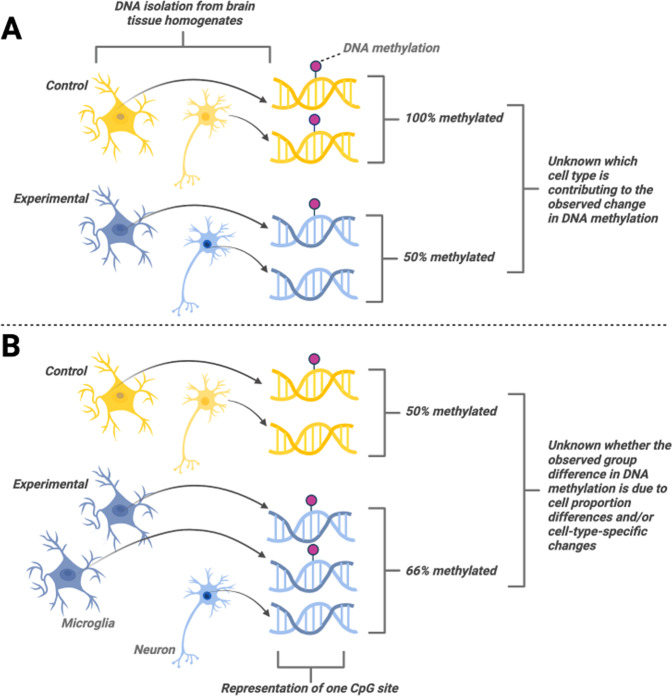
The conventional approach to studying epigenetic reprogramming by ELS involves quantifying epigenetic modifications to select genes, or whole genomes, in heterogenous brain tissue as reviewed elsewhere [48, 49, 74–77]. One classic finding is changes of levels of hippocampal DNAm in the promoter region of the glucocorticoid receptor (Nr3c1; GR), a gene linked to impaired negative feedback inhibition of the hypothalamic-pituitary-adrenal (HPA) axis [78], in response to ELS (e.g. childhood abuse, neglect). However, studies of heterogenous brain tissue pose limitations on defining the mechanistic roles of such epigenetic modifications (Fig. 2). First, differences in the epigenetic mark (and expression levels) of a particular gene that is expressed in multiple cell types may be difficult to interpret if the function of a given gene varies between cell types. Second, differences in the epigenetic modifications of a gene expressed in multiple cell types can be masked by ‘background noise’ resulting from constitutive epigenetic modifications related to cell type-specific functions. Returning to the previous example, GR is expressed in all cell types in the brain [79], thus levels of DNAm of the Nr3c1 promoter in heterogenous brain tissue are not indicative of which cell types are driving such changes. GR can exert distinct genomic and rapid non-genomic functions in different cell types, regulating inflammation in glia [80, 81] and excitability in neurons [82], for example. Furthermore, perturbations in early development can influence cell fate determination, leading to differences in cell type proportions within tissues between experimental groups [83]. For example, neonatal maternal separation has been found to deplete oligodendrocyte progenitors in adult male mice [84]. Epigenetic modifications (e.g., DNAm) measured in particular genes in heterogenous tissue could then be reflective of cell type proportion differences rather than epigenetic reprogramming of a particular cell type(s). To mitigate these potential confounds, some recent studies have examined cell-type-specific epigenetic modifications.
Fig. 2. Limitations of examining heterogenous brain tissue for measuring epigenetic modifications.
A Group differences in the epigenetic mark of a particular gene are not indicative of which cell type(s) are responsible for driving these changes. B Group differences in the epigenetic mark of a particular gene may be masked by differences in cell type proportions between groups.
Here we review studies that have examined the effects of ELS on epigenetic modifications in cell-type-enriched isolates, or in genes from heterogenous brain tissue where expression is known to be restricted to a single cell type in the brain (Table 1). We considered ELS studies that are inflammation-based or psychosocial that took place at the prenatal (maternal gestational), perinatal, or neonatal stages of development in female and male offspring aged post-natal day (PND)1 to adulthood. We considered both human and rodent studies, although only one human study was found within our scope. As our focus is on ELS, we did not review effects on offspring due to pre-conceptional parental or transgenerational stress as reviewed elsewhere in heterogeneous brain tissue samples [85–88]. In the brain, the primary categorization of cell types includes neurons, glia (microglia, astrocytes, oligodendrocytes), pericytes and brain epithelial cells [89, 90], however cells in these broad categories can be subdivided based on cellular functions. For example, neurons can be classified as being either glutamatergic or GABA-ergic, excitatory or inhibitory, as well as by cortical layer of origin [91–93]. For the purposes of this review, and consistent with available data in the literature, the primary categorization for cell type was used.
Table 1.
Summary of reviewed articles whereby epigenetic modifications of genes from cell-type-enriched isolates, or genes with cell-type-enriched expression from heterogenous brain tissue, were measured in offspring exposed to early life stress.
| Cell type | Analysis | Stress timing | Animal | Early life stress type | Age point(s) | Sex | Brain region | Findings | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Neuron | Isolates | Postnatal | Rat | Maternal separation | PND56 | Male | DG | ↑ Retinoic acid receptor promoter DNAm of CpG island | [102] |
| Neonatal stress | PND90 | Both | mPFC | ↑ Bdnf IV DNAm across 12 CpG sites (females only) | [100] | ||||
| Enriched expression | Prenatal | Rat | Maternal stress | PND0 | Male | Whole brain | ↑ miR-323 | [108] | |
| Maternal stress | PND60 | Male | HPC & PFC |
↑ miR-133b in HPC ↓ miR-133b in PFC |
[103] | ||||
| Postnatal | Mouse | Maternal separation | PND42–56 | Both | AMY | ↑ Htr1a promoter DNAm (females) | [113] | ||
| Maternal separation | PND16 | Male | HPC | ↑ Arc promoter histone H4ac | [109] | ||||
| Rat | Maternal separation | PND60 | Both | PVN & CeA | ↓ Crh promoter CpG1 and CpG2 methylation in PVN | [119] | |||
| Maternal separation and/or CUS | Adult | Male | NAc & striatum |
↑ miR-326 in NAc ↓ miR-326 in striatum (for CUS & MS+CUS only) |
[117] | ||||
| Maternal separation | Adult | Undeclared | Ca1 |
↑ Crh promoter histone H3ac ↓ Crh promoter CpG methylation |
[64] | ||||
| Microglia | Isolates | Prenatal | Mouse | Maternal allergic asthma | PND35 | Female | Whole brain | WGBS, DMR, functional annotation, pathway analysis | [129] |
| Postnatal | Rat | Maternal separation+ handling | ~PND60 | Male | NAc | ↓ Il10 gene CpG island methylation | [131] | ||
| Neonatal alcohol | PND6 & 90 | Combined | Mediobasal hypoT |
↑ Tnfa promoter histone H3K9ac ↑ Il6 promoter histone H3K9ac |
[130] | ||||
| Enriched expression | Prenatal | Rat | Maternal LPS | PND1 | Undeclared | Whole brain | ↓ miR-146 & miR-126 | [132] | |
| Oligoden-drocyte | Isolates | Postnatal | Human | Childhood abuse |
Childhood abuse: 19–85 years Controls: 15–81 years |
Combined | ACC |
↓ LINGO3 gene CpG methylation ↓ POU3F1 gene CpG methylation |
[139] |
| Enriched expression | Prenatal | Rat | Maternal stress | PND0 | Male | Whole brain | ↑ miR-219 | [108] | |
| Astrocyte | Enriched expression | Postnatal | Rat | Maternal separation | PND35 | Male | Frontal cortex | ↓ Gfap promoter CpG methylation | [138] |
ac acetylation, ACC anterior cingulate cortex, AMY amygdala, Arc activity-regulated cytoskeleton-associated protein, Bdnf brain derived neurotrophic factor, CeA central amygdala, Crh corticotropin releasing hormone, CUS chronic unpredictable stress, DG dentate gyrus, DNAm DNA methylation, DMR differentially methylated regions, Gfap glial fibrillary acidic protein, HPC hippocampus, Htr1a serotonin receptor 1A, HypoT hypothalamus, Il10 interleukin 10, LINGO3 leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 3, LPS lipopolysaccharide, miR micro-RNA, mPFC medial prefrontal cortex, MS maternal separation, NAc nucleus accumbens, PND postnatal day, POU3F1 POU class 3 homeobox 1, PVN paraventricular nucleus of the hypothalamus, WGBS whole genome bisulfite sequencing.
Epigenetic modifications induced by ELS in neurons
In the adult human brain, there is approximately a 1:1 ratio of neurons to glia, although this varies across brain regions [94, 95], similar to rodents [96]. Epigenetic alterations to particular neurons as a consequence of ELS can alter phenotypes associated with neural circuits involved in the response to stress, fear memory, and cognition [97–99]. Dysregulation or impairments of related behaviors are associated with epigenomic alterations in the brain in response to ELS. For example, there are differences in DNAm modifications of genes regulating the HPA axis, monoamines, and neuropeptides in humans with childhood trauma and in animals exposed to ELS [77, 97, 99]. However, it remains unclear whether neurons are ultimately the cell types driving the observed epigenetic changes.
To date, only a few studies have isolated neurons from rodent models of ELS to assess epigenetic modifications. One study found that an adverse caregiving environment from PND1–7 led to increased brain-derived neurotrophic factor (Bdnf) exon IV DNAm across 12 CpG sites in neurons isolated from the medial prefrontal-cortex (PFC) of adult female but not in male rats [100], mirroring findings found in heterogenous PFC tissue [101]. BDNF is involved in learning and memory and is a vital factor in neurodevelopment. Retinoic acid receptor, also critical to neurodevelopment, specifically neural differentiation, was found in another study to have increased promoter CpG island methylation in adult neural precursor cells isolated from the dentate gyrus of adult male rats (females unexamined) that underwent neonatal maternal separation (MS) [102]. While additional findings are needed to characterize the impacts of ELS on neurons, these studies indicate that neonatal psychosocial stressors may lead to aberrant gene expression programs essential for neural development.
Some studies that have used whole brain tissue to measure ELS-induced epigenetic changes have measured levels of miRs or DNAm of genes that are only expressed in a single cell type. In adult male rats born to dams with gestational psychosocial stress, miR-133b was found to have increased levels in the hippocampus (HPC) and decreased levels in the PFC [103]. miR-133b shows neuron-enriched expression in the brain [104, 105] and is involved in promoting neurite outgrowth [106]. The same ELS in another study in adult male rats was found to increase whole brain levels of the neuron-specific miR-323 [107], which is involved in host-pathogen interactions with viruses [108]. Together, these findings provide evidence that gestational ELS induces neuron-specific epigenetic reprogramming associated with neuronal growth and immune regulation.
Effects of postnatal stress on the neuronal epigenome have mostly been examined through MS paradigms, whereby neonatal rodents are typically separated from their dam for 3–4 hours/day from PND1–10 or 16. In one study, at PND16 immediately after MS, male mice were found to have increased hippocampal histone H4 acetylation at the promoter of activity-regulated cytoskeleton-associated protein (Arc) [109], which is only expressed in neurons and plays a critical role in learning and experience-induced synaptic plasticity [110, 111]. In adulthood, in the amygdala of female but not male mice, MS led to increased DNAm at the promoter of the neuron-specific serotonin-1A receptor (Htr1a) [112], which modulates emotional behavior [113]. MiR-326, which has neuron-enriched expression in the neocortex [105, 114, 115] and targets the dopamine D2 receptor [116] was found to be increased in the nucleus accumbens (NAc) and reduced in the striatum of adult male rats exposed to MS alone, and MS combined with maternal chronic unpredictable stress [117]. Also in adult rats (sex undeclared) with neonatal MS, there was increased hippocampal CA1 histone H3 acetylation and decreased methylation of CpGs in the promoter of the neuron-specific [118] corticotropin releasing hormone (Crh) [64]. Similar to the latter study, which also found evidence indicative of increased CRH expression, another study in adult rats of both sexes exposed to MS showed reduced Crh promoter CpG1 and CpG2 methylation, but in the paraventricular nucleus (PVN) of the hypothalamus [119]. PVN secretion of CRH kickstarts the HPA axis stress response, while CRH binding in the HPC is primarily responsible for regulating glutamatergic transmission and memory function [120]. Altogether, these findings suggest that MS leads to long-term alterations to emotion and learning processes via epigenetic reprogramming of neurotransmitter receptors and neurohormones.
Epigenetic modifications induced by ELS in microglia
Microglia comprise about 5–12% of the total glia population in the CNS in adult humans [121] and rodents [122] depending on the brain region. In resting states, microglia are involved in synaptic remodeling, maintenance and monitoring of the CNS environment with cell surface receptors that bind to antigens, antibodies, cytokines, and hormones. When potential insults are recognized, such as infection, inflammation, and neurodegeneration, microglia become more ‘activated’, altering their morphology to become phagocytic and releasing inflammatory cytokines that alert neighboring cells and influence their functioning [123]. Towards the end of the insult, microglia release anti-inflammatory cytokines and phagocytose cellular debris. When the insult diminishes, microglia return to their ‘resting’ state. Potentiated and dysregulated states of microglial inflammation are harmful to the tissue environment and can kill healthy neurons. Aberrant microglial activation has been associated with epigenetic dysregulation in anxiety, mood and autism spectrum disorders [124, 125] as well as neurodegenerative diseases [126, 127]. Notably, a similar phenotype indicative of exaggerated inflammation has been found in studies examining epigenetic effects of ELS in animal models [49, 108]. However, these studies utilized heterogenous brain tissue to form their epigenetic links. In addition to microglia, inflammation-related genes including cytokines are expressed by neurons, astrocytes and pericytes [128], making it difficult to determine the extent to which microglia drive neuroinflammatory changes in response to ELS.
Few studies have documented microglia-specific epigenetic changes in response to ELS. In microglia isolated from the whole brain of adolescent female mice subject to maternal allergic asthma exposure, whole-genome-bisulfite sequencing of differentially-methylated regions showed enrichment of gene sets associated with cytokine signaling pathways [129]. In microglia isolated from the medial hypothalamus of PND6 and adult rats (sexes combined), neonatal alcohol exposure was found to increase acetylation of histone H3K9 at the promoter regions of the pro-inflammatory cytokines tumor necrosis factor alpha (Tnfa) and interleukin (Il)6 in baseline conditions and 2 h post-LPS challenge [130]. Similar to these findings that suggest there is an increased pro-inflammatory phenotype in offspring exposed to ELS, the anti-inflammatory cytokine Il10 showed reduced levels of CpG island methylation in microglia isolated from the NAc of adult male rats that underwent MS as well as handling, a change that was absent in whole tissue from the NAc [131]. In the whole brain of PND1 rats (sex undeclared) subject to maternal LPS exposure 48 h earlier, there was reduced miR-126 and miR-146 [132], which regulate microglial inflammatory processes [107]. Since changes to both pro- and anti-inflammatory cytokines have been observed in these cell-type-specific studies, future studies can delineate how inflammatory pathways in microglia are epigenetically reprogrammed in response to ELS.
Epigenetic modifications induced by ELS in astrocytes and oligodendrocytes
In the human cerebral cortex, the glial population includes approximately 20% astrocytes and 75% oligodendrocytes [133], densities of which have also been examined in various brain regions in mice [134]. Astrocytes play critical roles in maintenance of homeostasis through ion buffering, immune signaling, blood-brain-barrier maintenance, regulation of neuronal synaptogenesis and removal [135]. Epigenetic dysregulation of astrocytes and reduced astrocyte cell proportions have been linked to psychiatric disorders associated with ELS, including major depressive disorder [136]. One study found that maternal separation led to reduced promoter CpG methylation of glial fibrillary acidic protein (Gfap), an intermediate filament protein expressed only in astrocytes [137], in the frontal cortex of adolescent male rats [138]. There is also some evidence of ELS-induced epigenetic reprogramming of oligodendrocytes, the myelinating cells of the CNS that enable saltatory nerve conduction and axon integrity. Isolated oligodendrocytes from the anterior cingulate cortex of human suicide completers with a history of childhood abuse exhibited decreased CpG methylation of myelination-regulating genes, specifically, leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor- interacting protein 3 (LINGO3) and POU class 3 homeobox 1 (POU3F1), compared to controls who died of non-suicide causes [139]. An opposite effect was observed in male rats exposed to maternal gestational psychosocial stress, where there were increased levels of whole-brain miR-219 [108], which shows oligodendrocyte-enriched expression [107] and is necessary for enabling and promoting the maturation of oligodendrocyte precursor cells into myelinating oligodendrocytes [140, 141].
Challenges and outlook
Converging evidence supports the role of epigenetic modifications as mediators of the impact of ELS on long-term neurobiological alterations. Unlike genetic factors, the epigenome is potentially dynamic throughout life. This malleable quality of the epigenome may enable the identification of cell-specific epigenetic biomarkers of disease and aid in the development of targeted therapeutic approaches for neuropsychiatric or neurodevelopmental disorders, as is the case for certain cancers [142]. Examples of these developments include epigenetic reprogramming of rodent neuronal stem cells to ameliorate neurodegeneration [143] and de-methylation of the fragile X mental retardation (FMR) gene in human neurons derived from Fragile X syndrome patients to normalize neuronal activity [144]. Possible epigenetic treatments for neuropsychiatric conditions have also been reviewed elsewhere [145–147].
The study of cell-type-specific epigenetic modifications requires sophisticated technical approaches. Isolating a purified cell population with intact DNA/RNA/protein from the brain can necessitate large sample sizes and volumes of fresh tissue, requiring tools such as primary cell culture, density-based centrifugation, magnetic bead separation, flow cytometry, and single-cell sequencing. These methods require advanced expertise and may be difficult to access. While computational deconvolution tools exists for obtaining cell-type-specific information from bulk sequence reads from heterogenous tissue, they nonetheless provide a less accurate estimation [148].
Of course, epigenetic changes found in a certain cell type do not necessarily mean that similar changes are absent in other cell types. While measuring epigenetic changes in one cell type may provide information regarding mechanisms associated with its biological function, measuring them in multiple cell types would be needed to assess the specificity of such modifications. Absent the heroic effort of quantifying all major cell types within a brain region of interest, comparing epigenetic changes in a single cell type to those of heterogenous tissue is another potential strategy. In such a context, two types of information would be needed: 1) information pertaining to the epigenetic modification of the particular cell type (e.g., microglia) compared to whole tissue from the brain region of interest, and 2) quantification of the proportion of the cell type of interest relative to other cell types in the region of interest. This information would indicate both relative levels of the epigenetic modification in the cell type in the brain region of interest and cell proportion differences that may also contribute to the overall levels of the epigenetic modification (see Fig. 2). This could be particularly important in interpreting the observed epigenetic profiles, which may result from changes in the relative levels of the epigenetic modification in the cell type of interest (potentially indicating cellular reprogramming) and/or in the proportion of the cell type of interest within the region of interest. Notably, potential cell-proportion differences alone may constitute an epigenetically-determined phenotype [83, 84].
Based on the available literature, it is evident that cell-specific epigenetic changes in glia are presently very limited in the context of ELS studies, making this fertile ground for future discovery. As well, measurement of ncRNA has been limited to miRNA in the context of the analysis of cell-specific epigenetic changes due to ELS. Given the prominent role of lncRNA in cell type differentiation in the brain [149], and its association with ELS [150] and stress-related disease [22], cell-type-specific changes in lncRNA levels in the context of ELS is another important area for future inquiry.
Studying epigenetic modifications at a primary cell-specific resolution, while important, does not evade interpretational issues. As noted above, beyond the broad categorization of cell types in the brain, definitions of specific cell types are still contested [151]. Neurons can be further subdivided into categories that consider molecular, morphological, connectional, and functional properties, leading to conceptual difficulty in defining a cell type and a lack of a consensus on their taxonomy [152, 153]. Microglia are known to exist in a resting or an activation state, which can be divided into M1 and M2 states of activation that define the bounds of a spectrum of intermediate phenotypes that can vary according to where in the brain the microglia reside [123]. Thus, it is not guaranteed that similar epigenetic modifications will exist within defined cell types, even within a brain region. While single-cell sequencing can alleviate this problem by examining cells at an individual level, this does not seem to solve the problem entirely. To draw an example from cancer biology, histone methylation differences can exist among the same cell types at the center and periphery of the tumor [154]. As such, isolating particular cells involved in a ‘neural circuit’ or biological pathway may be more informative in delineating epigenetic changes contributing to particular phenotypes.
Analysis of the temporal dynamics of epigenetic modifications may also provide important information. Epigenetic modifications detected in temporal proximity to environmental factors are not always the same as the ones detected later. For example, one study found that in the PFC of female rats exposed to neonatal caregiver maltreatment, there was reduced Bdnf exon IV methylation at adolescence compared to normal care controls, but an increase at adulthood, and no differences at infancy [101]. Therefore, the temporal dynamics of epigenetic responses to environmental factors that lead to persistent effects on phenotypes is another important avenue for future research.
Another challenge concerns the interpretation of observed epigenetic modifications in relation to their role in gene expression and behavioral phenotypes. For example, such a relationship may not manifest as steady-state increases/reductions in transcript and may only be detected in specific conditions, such as after stress exposure. A study described earlier demonstrates this possibility; neonatal alcohol exposure was found to increase histone acetylation at Tnfa and Il6 promoters in hypothalamic microglia in adult rats at baseline and post-LPS challenge, however increased transcript abundance of microglial mRNA of these genes only occurred in the post-LPS condition and not at baseline [130]. Interpretational challenges also extend to determining whether epigenetic modifications at select loci are ‘causal’ mechanisms of behavior, which may only be possible by altering target epigenetic modifications in vivo [155–157]. Tools used for cell-type-specific epigenetic editing include zinc-finger proteins, which can be fused with histone modifiers and target specific DNA sequences, and transcriptional activator-like effectors (TALEs), DNA binding proteins from bacteria that can be targeted to regulate gene transcription (see review [156]). More recent tools to alter epigenetic modifications include the CRISPR (clustered regularly interspaced short palindromic repeat)/Cas9 system, whereby the prokaryotic RNA-guided endonuclease can be targeted to a specific genomic locus using designed single guide RNA, and a catalytically dead (dCas9) is used to avoid genetic double-strand breaks [155]. The cell-type-specific expression of epigenetic modifiers (e.g., histone acetylation or methylation proteins) fused to dCas9 are then inducible by Cre recombinase, which can be transgenically or virally co-expressed [155, 156]. A recent study in mice found that dopamine D2 receptor neuron-specific targeting of histone acetylation/methylation at the Fosb gene within the NAc led to a phenotype of stress susceptibility or resilience, respectively [158]. Epigenetic editing with CRISPR/dCas9 is not without its limitations, as it can involve off-target effects [155, 159]. However, the use of CRISPR, fusion and DNA binding proteins in combination with cell-specific analysis may help delineate the epigenetic mechanisms through which ELS leads to long-term perturbations on behavior, and improve therapeutic approaches for a variety of neuropsychiatric and neurodevelopmental disorders.
Acknowledgements
This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) to POM and an Ontario Graduate Scholarship to MFR.
Author contributions
MFR and POM contributed to the conception, design, drafting, revisions, and final approval of the article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Olynik BM, Rastegar M. The genetic and epigenetic journey of embryonic stem cells into mature neural cells. Front Genet. 2012; 3. 10.3389/fgene.2012.00081. [DOI] [PMC free article] [PubMed]
- 2.Reik W, Surani MA. Germline and pluripotent stem cells. Cold Spring Harbor Perspect Biol. 2015;7. 10.1101/CSHPERSPECT.A019422. [DOI] [PMC free article] [PubMed]
- 3.Perry M, Chalkley R. Histone acetylation increases the solubility of chromatin and occurs sequentially over most of the chromatin. A novel model for the biological role of histone acetylation. J Biol Chem. 1982;257:7336–47. doi: 10.1016/S0021-9258(18)34382-5. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Tsukada YI, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2005;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 6.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozlenkov A, Roussos P, Timashpolsky A, Barbu M, Rudchenko S, Bibikova M, et al. Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic Acids Res. 2014;42:109. doi: 10.1093/nar/gkt838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu B, Dong X, Gravina S, Kartal Ö, Schimmel T, Cohen J, et al. Genome-wide, single-cell DNA methylomics reveals increased non-CpG methylation during human oocyte maturation. Stem Cell Rep. 2017;9:397. doi: 10.1016/j.stemcr.2017.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anastasiadi D, Esteve-Codina A, Piferrer F. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenetics Chromatin. 2018;11:37. doi: 10.1186/s13072-018-0205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuang T-J, Chen F-C, Chen Y-Z. Position-dependent correlations between DNA methylation and the evolutionary rates of mammalian coding exons. Proc Natl Acad Sci USA. 2012;109:15841–6. doi: 10.1073/pnas.1208214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, Zhang J, Huang S, He X. Genome-wide analysis reveals that exon methylation facilitates its selective usage in the human transcriptome. Brief Bioinform. 2018;19:754–64. doi: 10.1093/bib/bbx019. [DOI] [PubMed] [Google Scholar]
- 12.Jjingo D, Conley A, Yi S, Lunyak V, Jordan I. On the presence and role of human gene-body DNA methylation. Oncotarget. 2012;3:462–74. doi: 10.18632/oncotarget.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23:1256–69. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shayevitch R, Askayo D, Keydar I, Ast G. The importance of DNA methylation of exons on alternative splicing. RNA. 2018;24:1351–62. doi: 10.1261/rna.064865.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ball MP, Li JB, Gao Y, Lee JH, Leproust EM, Park IH, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–8. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–9. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 17.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aristizabal MJ, Anreiter I, Halldorsdottir T, Odgers CL, McDade TW, Goldenberg A, et al. Biological embedding of experience: a primer on epigenetics. Proc Natl Acad Sci USA. 2020;117:23261–9. doi: 10.1073/pnas.1820838116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2020;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends Cancer. 2015;1:93–109. doi: 10.1016/j.trecan.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daskalakis NP, Provost AC, Hunter RG, Guffanti G. Noncoding RNAs: stress, glucocorticoids, and posttraumatic stress disorder. Biol Psychiatry. 2018;83:849–65. doi: 10.1016/j.biopsych.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leckman JF. Commentary: What does immunology have to do with brain development and psychopathology? - A commentary on O’Connor et al. (2014) J Child Psychol Psychiatry Allied Discip. 2014;55:632–4. doi: 10.1111/jcpp.12259. [DOI] [PubMed] [Google Scholar]
- 25.Rens W, Wallduck MS, Lovell FL, Ferguson-Smith MA, Ferguson-Smith AC. Epigenetic modifications on X chromosomes in marsupial and monotreme mammals and implications for evolution of dosage compensation. Proc Natl Acad Sci USA. 2010;107:17657–62. doi: 10.1073/pnas.0910322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang H, Disteche CM, Berletch JB. X Inactivation and escape: epigenetic and structural features. Front Cell Dev Biol. 2019;7:219. doi: 10.3389/fcell.2019.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 28.Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, et al. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med. 2000;6:88. doi: 10.1007/BF03401776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stiles J, Jernigan TL. The basics of brain development. Neuropsychol Rev. 2010;20:327. doi: 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andjelkovic AV, Nikolic B, Pachter JS, Zecevic N. Macrophages/microglial cells in human central nervous system during development: an immunohistochemical study. Brain Res. 1998;814:13–25. doi: 10.1016/S0006-8993(98)00830-0. [DOI] [PubMed] [Google Scholar]
- 31.Monier A, Adle-Biassette H, Delezoide AL, Evrard P, Gressens P, Verney C. Entry and distribution of microglial cells in human embryonic and fetal cerebral cortex. J Neuropathol Exp Neurol. 2007;66:372–82. doi: 10.1097/nen.0b013e3180517b46. [DOI] [PubMed] [Google Scholar]
- 32.Menassa DA, Gomez-Nicola D. Microglial dynamics during human brain development. Front Immunol. 2018;9. 10.3389/fimmu.2018.01014. [DOI] [PMC free article] [PubMed]
- 33.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–5. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross C, Boroviak TE. Origin and function of the yolk sac in primate embryogenesis. Nat Commun. 2020;11:1–14. doi: 10.1038/s41467-020-17575-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Utz SG, See P, Mildenberger W, Thion MS, Silvin A, Lutz M, et al. Early fate defines microglia and non-parenchymal brain macrophage development. Cell. 2020;181:557–573.e18. doi: 10.1016/j.cell.2020.03.021. [DOI] [PubMed] [Google Scholar]
- 36.Askew K, Li K, Olmos-Alonso A, Garcia-Moreno F, Liang Y, Richardson P, et al. Coupled proliferation and apoptosis maintain the rapid turnover of microglia in the adult brain. Cell Rep. 2017;18:391–405. doi: 10.1016/j.celrep.2016.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA. 2016;113:E1738–E1746. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi BH, Lapham LW. Radial glia in the human fetal cerebrum: a combined golgi, immunofluorescent and electron microscopic study. Brain Res. 1978;148:295–311. doi: 10.1016/0006-8993(78)90721-7. [DOI] [PubMed] [Google Scholar]
- 39.Jakovcevski I, Filipovic R, Mo Z, Rakic S, Zecevic N. Oligodendrocyte development and the onset of myelination in the human fetal brain. Front Neuroanat. 2009;3:5. doi: 10.3389/neuro.05.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–9. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiggins RC. Myelination: a critical stage in development. Neurotoxicology. 1986;7:103–20. [PubMed] [Google Scholar]
- 42.Semple BD, Blomgren K, Gimlin K, Ferriero DM, Noble-Haeusslein LJ. Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog Neurobiol. 2013;0:1. doi: 10.1016/j.pneurobio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urbán N, Guillemot F. Neurogenesis in the embryonic and adult brain: same regulators, different roles. Front Cell Neurosci. 2014;8:396. doi: 10.3389/fncel.2014.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peter RH. Synaptic density in human frontal cortex - developmental changes and effects of aging. Brain Res. 1979;163:195–205. doi: 10.1016/0006-8993(79)90349-4. [DOI] [PubMed] [Google Scholar]
- 45.Domingues HS, Portugal CC, Socodato R, Relvas JB. Oligodendrocyte, astrocyte, and microglia crosstalk in myelin development, damage, and repair. Front Cell Dev Biol. 2016;4:71. doi: 10.3389/fcell.2016.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheray M, Joseph B. Epigenetics control microglia plasticity. Front Cell Neurosci. 2018;12:243. doi: 10.3389/fncel.2018.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neal M, Richardson JR. Epigenetic regulation of astrocyte function in neuroinflammation and neurodegeneration. Biochim Biophys Acta Mol Basis Dis. 2018;1864:432–43. doi: 10.1016/j.bbadis.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bale TL, Baram TZ, Brown AS, Goldstein JM, Insel TR, McCarthy MM, et al. Early life programming and neurodevelopmental disorders. Biol Psychiatry. 2010;68:314–9. doi: 10.1016/j.biopsych.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woods RM, Lorusso JM, Potter HG, Neill JC, Glazier JD, Hager R. Maternal immune activation in rodent models: a systematic review of neurodevelopmental changes in gene expression and epigenetic modulation in the offspring brain. Neurosci Biobehav Rev 2021. 10.1016/J.NEUBIOREV.2021.07.015. [DOI] [PubMed]
- 50.Han VX, Patel S, Jones HF, Dale RC. Maternal immune activation and neuroinflammation in human neurodevelopmental disorders. Nat Rev Neurol 2021;1–16. [DOI] [PubMed]
- 51.Contu L, Hawkes C. A review of the impact of maternal obesity on the cognitive function and mental health of the offspring. Int J Mol Sci. 2017;18:1093. doi: 10.3390/ijms18051093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore GS, Kneitel AW, Walker CK, Gilbert WM, Xing G. Autism risk in small- and large-for-gestational-age infants. Am J Obstet Gynecol. 2012;206:314.e1–314.e9. doi: 10.1016/j.ajog.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hagberg H, Gressens P, Mallard C. Inflammation during fetal and neonatal life: Implications for neurologic and neuropsychiatric disease in children and adults. Ann Neurol. 2012;71:444–57. doi: 10.1002/ana.22620. [DOI] [PubMed] [Google Scholar]
- 54.Khandaker GM, Dalman C, Kappelmann N, Stochl J, Dal H, Kosidou K, et al. Association of childhood infection with IQ and adult nonaffective psychosis in Swedish men a population-based longitudinal cohort and co-relative study. JAMA Psychiatry. 2018;75:356–62. doi: 10.1001/jamapsychiatry.2017.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown AS, Vinogradov S, Kremen WS, Poole JH, Deicken RF, Penner JD, et al. Prenatal exposure to maternal infection and executive dysfunction in adult schizophrenia. Am J Psychiatry. 2009;166:683–90. doi: 10.1176/appi.ajp.2008.08010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin Y, Xu J, Huang J, Jia Y, Zhang J, Yan C, et al. Effects of prenatal and postnatal maternal emotional stress on toddlers’ cognitive and temperamental development. J Affect Disord. 2017;207:9–17. doi: 10.1016/j.jad.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 57.Young ES, Farrell AK, Carlson EA, Englund MM, Miller GE, Gunnar MR, et al. The dual impact of early and concurrent life stress on adults’ diurnal cortisol patterns: a prospective study. Psychological Sci. 2019;30:739–47. doi: 10.1177/0956797619833664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khashan AS, Abel KM, McNamee R, Pedersen MG, Webb RT, Baker PN, et al. Higher risk of offspring schizophrenia following antenatal maternal exposure to severe adverse life events. Arch Gen Psychiatry. 2008;65:146–52. doi: 10.1001/archgenpsychiatry.2007.20. [DOI] [PubMed] [Google Scholar]
- 59.Bilbo SD, Tsang V. Enduring consequences of maternal obesity for brain inflammation and behavior of offspring. FASEB J. 2010;24:2104–15. doi: 10.1096/fj.09-144014. [DOI] [PubMed] [Google Scholar]
- 60.Sasaki A, de Vega WC, St-Cyr S, Pan P, McGowan PO. Perinatal high fat diet alters glucocorticoid signaling and anxiety behavior in adulthood. Neuroscience. 2013;240:1–12. doi: 10.1016/j.neuroscience.2013.02.044. [DOI] [PubMed] [Google Scholar]
- 61.Peleg-Raibstein D, Luca E, Wolfrum C. Maternal high-fat diet in mice programs emotional behavior in adulthood. Behav Brain Res. 2012;233:398–404. doi: 10.1016/j.bbr.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 62.Tishkina A, Stepanichev M, Kudryashova I, Freiman S, Onufriev M, Lazareva N, et al. Neonatal proinflammatory challenge in male Wistar rats: Effects on behavior, synaptic plasticity, and adrenocortical stress response. Behav Brain Res. 2016;304:1–10. doi: 10.1016/j.bbr.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 63.Walker FR, Knott B, Hodgson DM. Neonatal endotoxin exposure modifies the acoustic startle response and circulating levels of corticosterone in the adult rat but only following acute stress. J Psychiatr Res. 2008;42:1094–103. doi: 10.1016/j.jpsychires.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 64.Wang A, Nie W, Li H, Hou Y, Yu Z, Fan Q et al. Epigenetic upregulation of corticotrophin-releasing hormone mediates postnatal maternal separation-induced memory deficiency. PLoS ONE 2014;9. 10.1371/journal.pone.0094394. [DOI] [PMC free article] [PubMed]
- 65.Weinstock M. Prenatal stressors in rodents: effects on behavior. Neurobiol Stress. 2017;6:3–13. doi: 10.1016/j.ynstr.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Catale C, Gironda S, Iacono LL, Carola V. Microglial function in the effects of early-life stress on brain and behavioral development. J Clin Med 2020;9. 10.3390/JCM9020468. [DOI] [PMC free article] [PubMed]
- 67.Sominsky L, Walker AK, Ong LK, Tynan RJ, Walker FR, Hodgson DM. Increased microglial activation in the rat brain following neonatal exposure to a bacterial mimetic. Behavioural Brain Res. 2012;226:351–6. doi: 10.1016/j.bbr.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 68.Smith KE, Pollak SD. Early life stress and development: potential mechanisms for adverse outcomes. J Neurodev Disord. 2020;12:1–15. doi: 10.1186/s11689-020-09337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Averill LA, Abdallah CG, Fenton LR, Fasula MK, Jiang L, Rothman DL, et al. Early life stress and glutamate neurotransmission in major depressive disorder. Eur Neuropsychopharmacol. 2020;35:71–80. doi: 10.1016/j.euroneuro.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Çalışkan G, Müller A, Albrecht A. Long-term impact of early-life stress on hippocampal plasticity: spotlight on astrocytes. Int J Mol Sci. 2020;21:1–19. doi: 10.3390/ijms21144999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gee DG, Casey BJ. The impact of developmental timing for stress and recovery. Neurobiol Stress. 2015;1:184–94. doi: 10.1016/j.ynstr.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orthner DK, Jones-Sanpei H, Williamson S. The resilience and strengths of low-income families. Fam Relat. 2004;53:159–67. doi: 10.1111/j.0022-2445.2004.00006.x. [DOI] [Google Scholar]
- 73.Levine S. Psychological and social aspects of resilience: a synthesis of risks and resources. Dialogues Clin Neurosci. 2003;5:273. doi: 10.31887/DCNS.2003.5.3/slevine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alyamani RAS, Murgatroyd C. Epigenetic programming by early-life stress. In: Progress in Molecular Biology and Translational Science. (Elsevier B.V., 2018) 133-50. [DOI] [PubMed]
- 75.McGowan PO, Szyf M. The epigenetics of social adversity in early life: Implications for mental health outcomes. Neurobiol Dis. 2010;39:66–72. doi: 10.1016/j.nbd.2009.12.026. [DOI] [PubMed] [Google Scholar]
- 76.Matthews SG, McGowan PO. Developmental programming of the HPA axis and related behaviours: epigenetic mechanisms. J Endocrinol. 2019;242:T69–T79. doi: 10.1530/JOE-19-0057. [DOI] [PubMed] [Google Scholar]
- 77.Lewis CR, Olive MF. Early-life stress interactions with the epigenome: Potential mechanisms driving vulnerability toward psychiatric illness. Behavioural Pharmacol. 2014;25:341–51. doi: 10.1097/FBP.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonté B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–8. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oakley RH, Cidlowski JA. Cellular processing of the glucocorticoid receptor gene and protein: new mechanisms for generating tissue-specific actions of glucocorticoids. J Biol Chem. 2011;286:3177–84. doi: 10.1074/jbc.R110.179325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carrillo-De Sauvage MÁ, Maatouk L, Arnoux I, Pasco M, Sanz Diez A, Delahaye M, et al. Potent and multiple regulatory actions of microglial glucocorticoid receptors during CNS inflammation. Cell Death Differ. 2013;20:1546–57. doi: 10.1038/cdd.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, Wang H, Zhang C, Liu B, Peng Z, Li Y, et al. Mifepristone attenuates depression-like changes induced by chronic central administration of interleukin-1β in rats. Behavioural Brain Res. 2018;347:436–45. doi: 10.1016/j.bbr.2018.03.033. [DOI] [PubMed] [Google Scholar]
- 82.Groeneweg F, Karst H, de Kloet E, Joëls M. Rapid non-genomic effects of corticosteroids and their role in the central stress response. J Endocrinol. 2011;209:153–67. doi: 10.1530/JOE-10-0472. [DOI] [PubMed] [Google Scholar]
- 83.Lappalainen T, Greally JM. Associating cellular epigenetic models with human phenotypes. Nat Rev Genet. 2017;18:441–51. doi: 10.1038/nrg.2017.32. [DOI] [PubMed] [Google Scholar]
- 84.Teissier A, Le Magueresse C, Olusakin J, Andrade da Costa BLS, De Stasi AM, Bacci A, et al. Early-life stress impairs postnatal oligodendrogenesis and adult emotional behaviour through activity-dependent mechanisms. Mol Psychiatry. 2020;25:1159–74. doi: 10.1038/s41380-019-0493-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Babenko O, Kovalchuk I, Metz GAS. Stress-induced perinatal and transgenerational epigenetic programming of brain development and mental health. Neurosci Biobehav Rev. 2015;48:70–91. doi: 10.1016/j.neubiorev.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 86.Bale TL. Epigenetic and transgenerational reprogramming of brain development. Nat Rev Neurosci. 2015;16:332–44. doi: 10.1038/nrn3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Franklin TB, Russig H, Weiss IC, Grff J, Linder N, Michalon A, et al. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry. 2010;68:408–15. doi: 10.1016/j.biopsych.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 88.Kundakovic M, Champagne FA. Early-life experience, epigenetics, and the developing brain. Neuropsychopharmacol. 2014;40:141–53. doi: 10.1038/npp.2014.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McKenzie AT, Wang M, Hauberg ME, Fullard JF, Kozlenkov A, Keenan A, et al. Brain cell type specific gene expression and co-expression network architectures. Sci Rep. 2018;8:1–19. doi: 10.1038/s41598-018-27293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cuevas‐Diaz Duran R, Wei H, Wu JQ. Single‐cell RNA‐sequencing of the brain. Clin Transl Med. 2017;6:20. doi: 10.1186/s40169-017-0150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen R, Wu X, Jiang L, Zhang Y. Single-cell RNA-Seq reveals hypothalamic cell diversity. Cell Rep. 2017;18:3227–41. doi: 10.1016/j.celrep.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol. 2018;36:70–80. doi: 10.1038/nbt.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, et al. Single-cell genomics identifies cell type–specific molecular changes in autism. Science. 2019;364:685–9. doi: 10.1126/science.aav8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Azevedo FAC, Carvalho LRB, Grinberg LT, Farfel JM, Ferretti REL, Leite REP, et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J Comp Neurol. 2009;513:532–41. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- 95.von Bartheld CS, Bahney J, Herculano-Houzel S. The search for true numbers of neurons and glial cells in the human brain: a review of 150 years of cell counting. J Comp Neurol. 2016;524:3865–95. doi: 10.1002/cne.24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Herculano-Houzel S, Mota B, Lent R. Cellular scaling rules for rodent brains. Proc Natl Acad Sci USA. 2006;103:12138–43. doi: 10.1073/pnas.0604911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jiang S, Postovit L, Cattaneo A, Binder EB, Aitchison KJ. Epigenetic modifications in stress response genes associated with childhood trauma. Front Psychiatry. 2019;10. 10.3389/fpsyt.2019.00808. [DOI] [PMC free article] [PubMed]
- 98.Sale A, Berardi N, Maffei L. Environment and brain plasticity: towards an endogenous pharmacotherapy. Physiol Rev. 2014;94:189–234. doi: 10.1152/physrev.00036.2012. [DOI] [PubMed] [Google Scholar]
- 99.Gartstein MA, Skinner MK. Prenatal influences on temperament development: the role of environmental epigenetics. Dev Psychopathol. 2018;30:1269–303. doi: 10.1017/S0954579417001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Blaze J, Roth TL. Caregiver maltreatment causes altered neuronal DNA methylation in female rodents. Dev Psychopathol. 2017;29:477–89. doi: 10.1017/S0954579417000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blaze J, Scheuing L, Roth TL. Differential methylation of genes in the medial prefrontal cortex of developing and adult rats following exposure to maltreatment or nurturing care during infancy. Developmental Neurosci. 2013;35:306–16. doi: 10.1159/000350716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boku S, Toda H, Nakagawa S, Kato A, Inoue T, Koyama T, et al. Neonatal maternal separation alters the capacity of adult neural precursor cells to differentiate into neurons via methylation of retinoic acid receptor gene promoter. Biol Psychiatry. 2015;77:335–44. doi: 10.1016/j.biopsych.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Monteleone MC, Adrover E, Pallarés ME, Antonelli MC, Frasch AC, Brocco MA. Prenatal stress changes the glycoprotein gpm6a gene expression and induces epigenetic changes in rat offspring brain. Epigenetics. 2014;9:152–60. doi: 10.4161/epi.25925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. Cell-type-based analysis of MicroRNA profiles in the mouse brain. Neuron. 2012;73:35–48. doi: 10.1016/j.neuron.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pomper N, Liu Y, Hoye ML, Dougherty JD, Miller TM. CNS microRNA profiles: a database for cell type enriched microRNA expression across the mouse central nervous system. Sci Rep. 2020;10:1–8. doi: 10.1038/s41598-020-61307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lu XC, Zheng JY, Tang LJ, Huang BS, Li K, Tao Y, et al. MiR-133b promotes neurite outgrowth by targeting RhoA expression. Cell Physiol Biochem. 2015;35:246–58. doi: 10.1159/000369692. [DOI] [PubMed] [Google Scholar]
- 107.Jovičić A, Roshan R, Moisoi N, Pradervand S, Moser R, Pillai B, et al. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. Ann Intern Med. 2013;158:5127–37. doi: 10.1523/JNEUROSCI.0600-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zucchi FCR, Yao Y, Ward ID, Ilnytskyy Y, Olson DM, Benzies K, et al. Maternal stress induces epigenetic signatures of psychiatric and neurological diseases in the offspring. PLoS ONE. 2013;8:56967. doi: 10.1371/journal.pone.0056967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xie L, Korkmaz KS, Braun K, Bock J. Early life stress-induced histone acetylations correlate with activation of the synaptic plasticity genes Arc and Egr1 in the mouse hippocampus. J Neurochemistry. 2013;125:457–64. doi: 10.1111/jnc.12210. [DOI] [PubMed] [Google Scholar]
- 110.Pastuzyn ED, Day CE, Kearns RB, Kyrke-Smith M, Taibi AV, McCormick J, et al. The neuronal gene Arc encodes a repurposed retrotransposon Gag protein that mediates intercellular RNA transfer. Cell. 2018;172:275–288.e18. doi: 10.1016/j.cell.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the leg Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–51. doi: 10.1016/S0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 112.Albert PR. Transcriptional regulation of the 5-HT1A receptor: implications for mental illness. Philos Trans R Soc B: Biol Sci. 2012;367:2402–15. doi: 10.1098/rstb.2011.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mattern F, Post A, Solger F, O’Leary A, Slattery DA, Reif A, et al. Prenatal and postnatal experiences associated with epigenetic changes in the adult mouse brain. Behavioural Brain Res. 2019;359:143–8. doi: 10.1016/j.bbr.2018.10.037. [DOI] [PubMed] [Google Scholar]
- 114.Kefas B, Comeau L, Floyd DH, Seleverstov O, Godlewski J, Schmittgen T, et al. The neuronal microRNA miR-326 acts in a feedback loop with Notch and has therapeutic potential against brain tumors. J Neurosci. 2009;29:15161–8. doi: 10.1523/JNEUROSCI.4966-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, et al. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004;101:360–5. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Allen L, Dwivedi Y. MicroRNA mediators of early life stress vulnerability to depression and suicidal behavior. Mol Psychiatry. 2020;25:308–20. doi: 10.1038/s41380-019-0597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang Y, Wang Y, Wang L, Bai M, Zhang X, Zhu X. Dopamine receptor D2 and associated microRNAs are involved in stress susceptibility and resistance to escitalopram treatment. Int J Neuropsychopharmacol. 2015;18:1–10. doi: 10.1093/ijnp/pyu077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou JN, Fang H. Transcriptional regulation of corticotropin-releasing hormone gene in stress response. IBRO Rep. 2018;5:137–46. doi: 10.1016/j.ibror.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen J, Evans AN, Liu Y, Honda M, Saavedra JM, Aguilera G. Maternal deprivation in rats is associated with corticotrophin-releasing hormone (CRH) promoter hypomethylation and enhances CRH transcriptional responses to stress in adulthood. J Neuroendocrinol. 2012;24:1055–64. doi: 10.1111/j.1365-2826.2012.02306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Refojo D, Schweizer M, Kuehne C, Ehrenberg S, Thoeringer C, Vogl AM, et al. Glutamatergic and dopaminergic neurons mediate anxiogenic and anxiolytic effects of CRHR1. Science. 2011;333:1903–7. doi: 10.1126/science.1202107. [DOI] [PubMed] [Google Scholar]
- 121.Kim YS, Joh TH. Microglia, major player in the brain inflammation: their roles in the pathogenesis of Parkinson’s disease. Exp Mol Med. 2006;38:333–47. doi: 10.1038/emm.2006.40. [DOI] [PubMed] [Google Scholar]
- 122.Kaur C, Rathnasamy G, Ling EA. Biology of microglia in the developing brain. J Neuropathol Exp Neurol. 2017;76:736–53. doi: 10.1093/jnen/nlx056. [DOI] [PubMed] [Google Scholar]
- 123.Jurga AM, Paleczna M, Kuter KZ. Overview of general and discriminating markers of differential microglia phenotypes. Front Cell Neurosci. 2020;14:198. doi: 10.3389/fncel.2020.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Martins-Ferreira R, Leal B, Costa PP, Ballestar E. Microglial innate memory and epigenetic reprogramming in neurological disorders. Prog Neurobiol. 2021;200:101971. doi: 10.1016/j.pneurobio.2020.101971. [DOI] [PubMed] [Google Scholar]
- 125.Frank MG, Weber MD, Watkins LR, Maier SF. Stress-induced neuroinflammatory priming: a liability factor in the etiology of psychiatric disorders. Neurobiol Stress. 2016;4:62. doi: 10.1016/j.ynstr.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Harberts E, Datta D, Chen S, Wohler JE, Oh U, Jacobson S. Translocator protein 18 kDa (TSPO) expression in multiple sclerosis patients. J Neuroimmune Pharm. 2013;8:51. doi: 10.1007/s11481-012-9397-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu Y, Wang M, Marcora EM, Zhang B, Goate AM. Promoter DNA hypermethylation – implications for Alzheimer’s disease. Neurosci Lett. 2019;711:134403. doi: 10.1016/j.neulet.2019.134403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Galic MA, Riazi K, Pittman QJ. Cytokines and brain excitability. Front Neuroendocrinol. 2012;33:116–25. doi: 10.1016/j.yfrne.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vogel Ciernia A, Careaga M, LaSalle JM, Ashwood P. Microglia from offspring of dams with allergic asthma exhibit epigenomic alterations in genes dysregulated in autism. GLIA. 2018;66:505–21. doi: 10.1002/glia.23261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chastain LG, Franklin T, Gangisetty O, Cabrera MA, Mukherjee S, Shrivastava P, et al. Early life alcohol exposure primes hypothalamic microglia to later-life hypersensitivity to immune stress: possible epigenetic mechanism. Neuropsychopharmacology. 2019;44:1579–88. doi: 10.1038/s41386-019-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schwarz JM, Hutchinson MR, Bilbo SD. Early-life experience decreases drug-induced reinstatement of morphine CPP in adulthood via microglial-specific epigenetic programming of anti-inflammatory IL-10 expression. J Neurosci. 2011;31:17835–47. doi: 10.1523/JNEUROSCI.3297-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Carloni S, Favrais G, Saliba E, Albertini MC, Chalon S, Longini M, et al. Melatonin modulates neonatal brain inflammation through endoplasmic reticulum stress, autophagy, and miR-34a/silent information regulator 1 pathway. J Pineal Res. 2016;61:370–80. doi: 10.1111/jpi.12354. [DOI] [PubMed] [Google Scholar]
- 133.Pelvig DP, Pakkenberg H, Stark AK, Pakkenberg B. Neocortical glial cell numbers in human brains. Neurobiol Aging. 2008;29:1754–62. doi: 10.1016/j.neurobiolaging.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 134.Keller D, Erö C, Markram H. Cell densities in the mouse brain: a systematic review. Front Neuroanatomy. 2018;12. 10.3389/fnana.2018.00083. [DOI] [PMC free article] [PubMed]
- 135.Miller SJ. Astrocyte heterogeneity in the adult central nervous system. Front Cell Neurosci. 2018;12:15. doi: 10.3389/fncel.2018.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nagy C, Suderman M, Yang J, Szyf M, Mechawar N, Ernst C, et al. Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Mol Psychiatry. 2015;20:320–8. doi: 10.1038/mp.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brenner M, Kisseberth WC, Su Y, Besnard F, Messing A. GFAP promoter directs astrocyte-specific expression in transgenic mice. J Neurosci. 1994;14:1030–7. doi: 10.1523/JNEUROSCI.14-03-01030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ahmed OG, Shehata GA, Ali RM, Makboul R, Abd Allah ESH, Abd El-Rady NM. Folic acid ameliorates neonatal isolation-induced autistic like behaviors in rats: Epigenetic modifications of bdnf and gfap promotors. Appl Physiol Nutr Metab. 2021;46:964–75. doi: 10.1139/apnm-2020-0923. [DOI] [PubMed] [Google Scholar]
- 139.Lutz PE, Tanti A, Gasecka A, Barnett-Burns S, Kim JJ, Zhou Y, et al. Association of a history of child abuse with impaired myelination in the anterior cingulate cortex: Convergent epigenetic, transcriptional, and morphological evidence. Am J Psychiatry. 2017;174:1185–94. doi: 10.1176/appi.ajp.2017.16111286. [DOI] [PubMed] [Google Scholar]
- 140.Fan HB, Chen LX, Qu XB, Ren CL, Wu XX, Dong FX, et al. Transplanted miR-219-overexpressing oligodendrocyte precursor cells promoted remyelination and improved functional recovery in a chronic demyelinated model. Sci Rep. 2017;7:1–18. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bruinsma IB, van Dijk M, Bridel C, van de Lisdonk T, Haverkort SQ, Runia TF, et al. Regulator of oligodendrocyte maturation, miR-219, a potential biomarker for MS. J Neuroinflammation. 2017;14:1–7. doi: 10.1186/s12974-017-1006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, et al. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther. 2019;4:1–39. doi: 10.1038/s41392-019-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kanherkar RR, Bhatia-Dey N, Makarev E, Csoka AB. Cellular reprogramming for understanding and treating human disease. Front Cell Developmental Biol. 2014;0:67. doi: 10.3389/fcell.2014.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu X, Wu H, Krzisch M, Wu X, Graef J, Muffat J, et al. Rescue of fragile X syndrome neurons by DNA methylation editing of the FMR1 gene. Cell. 2018;172:979–992.e6. doi: 10.1016/j.cell.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Millan M. An epigenetic framework for neurodevelopmental disorders: from pathogenesis to potential therapy. Neuropharmacology. 2013;68:2–82. doi: 10.1016/j.neuropharm.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 146.Schiele MA, Gottschalk MG, Domschke K. The applied implications of epigenetics in anxiety, affective and stress-related disorders - A review and synthesis on psychosocial stress, psychotherapy and prevention. Clin Psychol Rev. 2020;77:101830. doi: 10.1016/j.cpr.2020.101830. [DOI] [PubMed] [Google Scholar]
- 147.Moody L, Chen H, Pan Y-X. Early-life nutritional programming of cognition—the fundamental role of epigenetic mechanisms in mediating the relation between early-life environment and learning and memory. Process Adv Nutr Int Rev J. 2017;8:337–50. doi: 10.3945/an.116.014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Avila Cobos F, Alquicira-Hernandez J, Powell JE, Mestdagh P, De Preter K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat Commun. 2020;11:1–14. doi: 10.1038/s41467-020-19015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dong X, Chen K, Cuevas-Diaz Duran R, You Y, Sloan SA, Zhang Y, et al. Comprehensive identification of long non-coding RNAs in purified cell types from the brain reveals functional LncRNA in OPC fate determination. PLoS Genet. 2015;11:1005669. doi: 10.1371/journal.pgen.1005669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Arzua T, Jiang C, Yan Y, Bai X. The importance of non-coding RNAs in environmental stress-related developmental brain disorders: A systematic review of evidence associated with exposure to alcohol, anesthetic drugs, nicotine, and viral infections. Neurosci Biobehav Rev. 2021;128:633–47. doi: 10.1016/j.neubiorev.2021.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Clevers H. Cell Systems Voices What Is Your Conceptual Definition of “‘Cell Type’” in the Context of a Mature Organism? What Is an Adult Cell Type, Really? 2017 10.1016/j.cels.2017.03.006. [DOI] [PubMed]
- 152.Zeng H, Sanes JR. Neuronal cell-type classification: challenges, opportunities and the path forward. Nat Rev Neurosci. 2017;18:530–46. doi: 10.1038/nrn.2017.85. [DOI] [PubMed] [Google Scholar]
- 153.Mukamel EA, Ngai J. Perspectives on defining cell types in the brain. Curr Opin Neurobiol. 2019;56:61–68. doi: 10.1016/j.conb.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Assenov Y, Brocks D, Gerhäuser C. Intratumor heterogeneity in epigenetic patterns. Semin Cancer Biol. 2018;51:12–21. doi: 10.1016/j.semcancer.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 155.Hamilton PJ, Lim CJ, Nestler EJ, Heller EA. Neuroepigenetic editing. Methods Mol Biol. 2018;1767:113. doi: 10.1007/978-1-4939-7774-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yim YY, Teague CD, Nestler EJ. In vivo locus-specific editing of the neuroepigenome. Nat Rev Neurosci. 2020;21:471–84. doi: 10.1038/s41583-020-0334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu XS, Jaenisch R. Editing the epigenome to tackle brain disorders. Trends Neurosci. 2019;42:861–70. doi: 10.1016/j.tins.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 158.Hamilton P, Burek D, Lombroso S, Neve R, Robison A, Nestler E, et al. Cell-type-specific epigenetic editing at the Fosb gene controls susceptibility to social defeat stress. Neuropsychopharmacology. 2018;43:272–84. doi: 10.1038/npp.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther - Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]