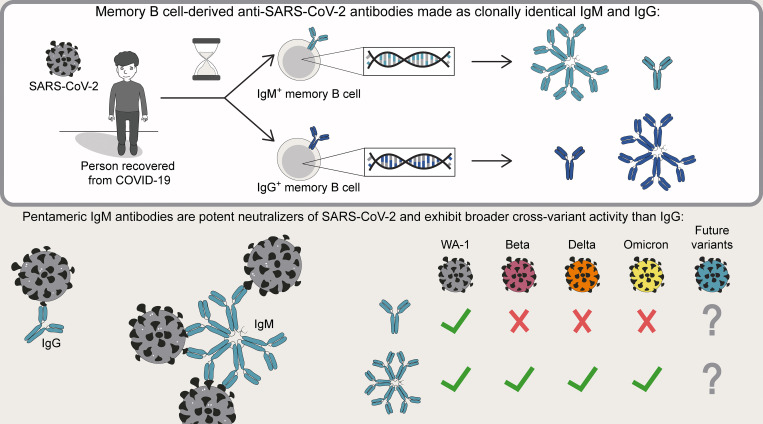
Using monoclonal antibodies built from SARS-CoV-2–specific memory B cells, Hale et al. demonstrate that IgM antibodies outperform IgG in neutralization assays against mutated variants. The authors suggest that IgM antibodies may be useful therapeutically, and that IgM memory B cells may be underappreciated protectors against rapidly evolving pathogens.
Abstract
Humoral immunity to SARS-CoV-2 can be supplemented with polyclonal sera from convalescent donors or an engineered monoclonal antibody (mAb) product. While pentameric IgM antibodies are responsible for much of convalescent sera’s neutralizing capacity, all available mAbs are based on the monomeric IgG antibody subtype. We now show that IgM mAbs derived from immune memory B cell receptors are potent neutralizers of SARS-CoV-2. IgM mAbs outperformed clonally identical IgG antibodies across a range of affinities and SARS-CoV-2 receptor-binding domain epitopes. Strikingly, efficacy against SARS-CoV-2 viral variants was retained for IgM but not for clonally identical IgG. To investigate the biological role for IgM memory in SARS-CoV-2, we also generated IgM mAbs from antigen-experienced IgM+ memory B cells in convalescent donors, identifying a potent neutralizing antibody. Our results highlight the therapeutic potential of IgM mAbs and inform our understanding of the role for IgM memory against a rapidly mutating pathogen.
Graphical Abstract
Introduction
Antibody products such as convalescent plasma or engineered mAbs provide passive immunity to SARS-CoV-2 and can protect vulnerable individuals from severe COVID-19 illness or death. IgM antibodies play a major and outsized role in SARS-CoV-2–neutralizing capacity based on studies of pooled convalescent plasma, despite comprising only ∼5% of the total antibody pool (Gasser et al., 2021; Kober et al., 2022). IgM is a natural pentamer with 10 epitope binding sites and in vivo effector functions well suited to neutralizing an invading virus, including activating complement and triggering activation of immune cells (Matsumoto, 2022; Zhang et al., 2022a). However, anti–SARS-CoV-2 mAbs in clinical use predominantly employ the monomeric, bivalent IgG structure most extensively developed for treatment of cancer and autoimmunity (Kreuzberger et al., 2021).
Emerging variants of SARS-CoV-2 have incorporated new mutations within the immunodominant receptor-binding domain (RBD) of the spike protein, leading to escape from neutralizing mAbs (Greaney et al., 2022; Harvey et al., 2021; Starr et al., 2021). While the identification of novel cross-variant neutralizing antibodies is one path forward, an alternative strategy would be to modify existing mAbs to be more tolerant of mutations. We previously found that expression as naturally multimerized IgM or engineered hexameric IgG enhanced the protective function of two malaria-specific mAbs via the cumulative binding strength of multiple interactions with an antigen-coated surface due to avidity (Thouvenel et al., 2021). Multimerization might therefore expand the functional range of an anti–SARS-CoV-2 mAb by compensating for reductions in affinity to a mutated RBD via enhanced avidity. Supporting this concept, artificial multimers can neutralize human immunodeficiency virus escape mutants, provided that the antigen-binding domains are arranged such that cross-linking could occur across viral spike proteins (Galimidi et al., 2015). Notably, SARS-CoV-2 spike proteins are distributed with a density such that inter- and intravirion crosslinking could theoretically be performed by IgM (Czajkowsky and Shao, 2009; Tai et al., 2021; Zhang et al., 2020).
The role of IgM and IgM-expressing cells in the human immune system must be better understood to maximize the therapeutic potential of IgM antibodies. IgM antibodies and IgM+ B cells are most appreciated in the early plasmablast response, providing a low-affinity humoral stopgap until higher-affinity antibodies of other isotypes are generated. This later response includes class-switched antibody-secreting plasmablasts, memory B cells (MBCs), and plasma cells that emerge from germinal centers. Reactivation of antigen-specific MBCs results in antibody production that can rapidly control pathogen propagation (Victora and Nussenzweig, 2012). A predominant pool of SARS-CoV-2–specific IgG+ MBCs and significantly smaller population of IgM+ MBCs are formed following infection or vaccination (Kim et al., 2022; Lederer et al., 2022; Zhang et al., 2022b). Despite intensive study of IgG+ MBCs, relatively little is known about the role for IgM+ MBCs in protection. Indirect evidence suggests that RBD-specific IgM+ MBCs may be important contributors to protective immunity, as well as a potentially underappreciated source of B cell receptor (BCR) sequences with therapeutic utility as neutralizing mAbs (Bullen et al., 2021; Callegari et al., 2022; Lenti et al., 2020; Newell et al., 2021; Piepenbrink et al., 2021; Purtha et al., 2011). However, descriptions of the SARS-CoV-2 IgM+ MBC repertoire are limited by the small number detectable in the blood at late time points and the technical challenge of producing high-quality pentameric IgM to study the BCR in its native isotype (Dan et al., 2021; Keyt et al., 2020; Wang et al., 2021).
Our group has established robust protocols for BCR sequencing of rare, antigen-specific MBCs and production of purified IgM mAbs (Krishnamurty et al., 2016; Rodda et al., 2021; Thouvenel et al., 2021). We used these methodologies to compare the relative functional activity of an array of SARS-CoV-2–specific sequences expressed as multimeric IgM vs. monomeric IgG that targeted a broad range of RBD epitopes. We then assessed the panel of clonally identical IgM vs. IgG mAbs for cross-variant neutralization. We also sought to determine whether IgM+ MBCs present in convalescent individuals encode antibodies that bind and neutralize SARS-CoV-2. Our combined findings suggest that IgM antibodies may play an important and underappreciated role in protection against SARS-CoV-2 variants when the protective capacity of serum IgG and IgG+ MBCs alone may prove inadequate.
Results and discussion
Pentameric IgM increases the potency of IgG+ MBC-derived neutralizing antibodies
For initial investigation into the effect of multimerization on SARS-CoV-2–specific antibodies, we employed eight neutralizing mAbs derived from RBD-specific IgG+ MBCs isolated from convalescent individuals (Rodda et al., 2021). Building on methods for studying the repertoire of antigen-specific MBCs, BCRs from individual RBD-specific B cells were sequenced, and the specificity-determining heavy and light chain variable regions were cloned into expression plasmids as γ1 (for the heavy chain) or κ/λ (light chain) constructs (Thouvenel et al., 2021), and IgG mAbs derived from each BCR were purified from cotransfected cells. Each heavy-chain variable sequence was also cloned into a plasmid upstream of the µ (IgM) constant region to prepare for multimerization studies.
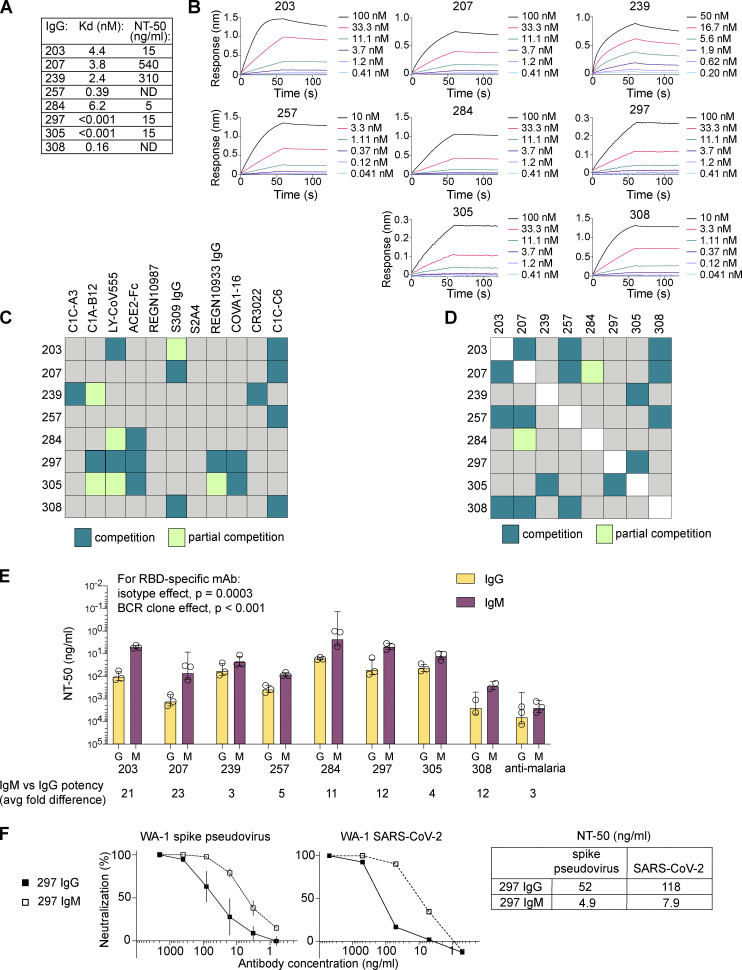
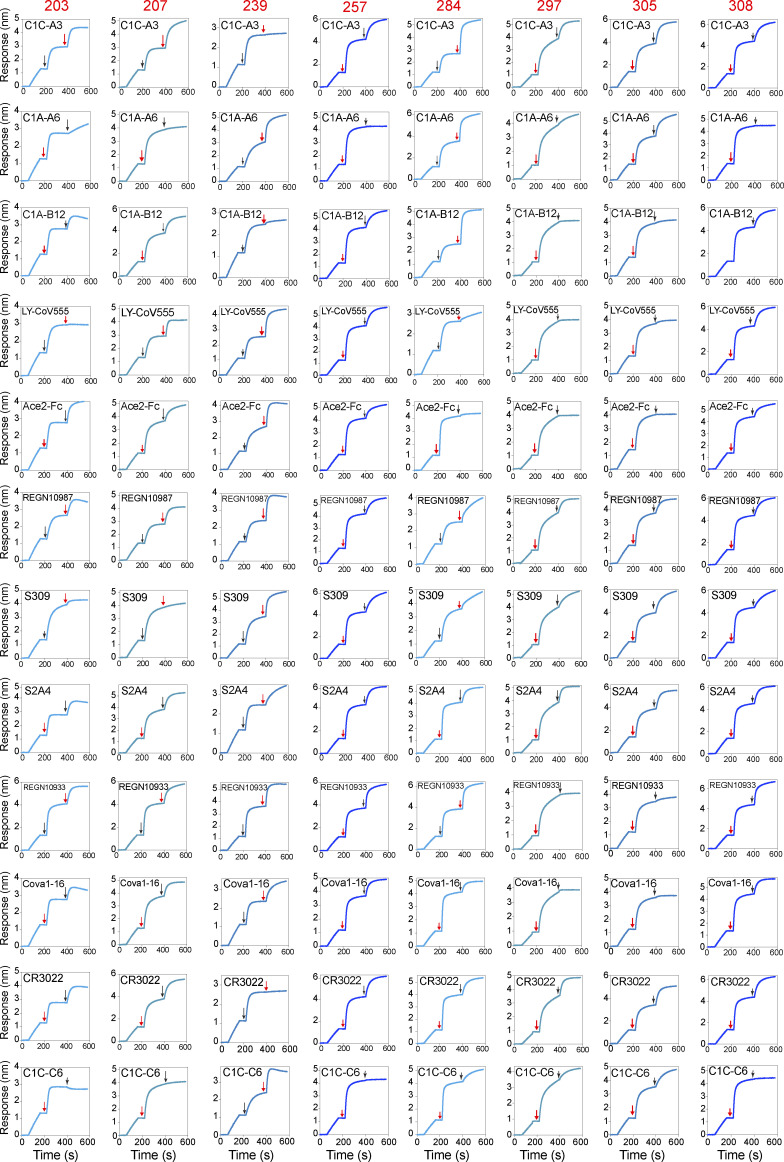
To build our panel of candidates for multimerization, we chose six IgG mAbs that had been previously tested in SARS-CoV-2 plaque neutralization tests (Rodda et al., 2021), with a range of half-maximal neutralization (NT50) titers (5–540 ng/ml; Fig. 1 A). We selected two additional IgG mAbs (257 and 308) that had failed to block RBD from binding ACE2 in a plate-bound assay, with the goal of also testing the impact of multimerization across a range of epitopes, including antibodies targeting regions outside of the ACE2:RBD interface (Rodda et al., 2021). Each mAb was affixed to a biosensor to assess RBD binding and dissociation by biolayer interferometry (BLI) to assess affinity. Affinities for RBD ranged from sub-picomolar (mAbs 297 and 305) to 6.2 nM (284; Fig. 1, A and B). To confirm that these mAbs represented a range of RBD target sites, we measured competition for RBD binding by BLI using a panel of Fabs and IgG mAbs with known epitopes (Figs. S1 and S2 A, summarized in Fig. 1, C and D). From the combined results, we predicted that three of eight mAbs (284, 297, and 305) bound at the ACE2 interface with likely class 1 or 2 RBD-specific binding moieties, using a classification scheme described by Barnes et al. (2020). mAb 203 did not appear to bind at the ACE2 interface, but competed strongly with LY-CoV555, a class 2 antibody, and partially with S309, the class 3 antibody that is the basis for sotrovimab, suggesting binding near but not directly blocking the ACE2 binding site (Pinto et al., 2020). mAb 207 was predicted to be a class 3–like antibody. mAb 239 competed with the tight-binding but nonneutralizing class 4 CR3022 antibody (Yuan et al., 2020), and with C1C-A3, a recently described class 4 antibody that binds outside of the ACE2 footprint at the conserved RBD core (Nabel et al., 2022). Contributing to evidence of binding outside the ACE2 footprint, 257 and 308 competed only with C1C-C6, a class 4–like neutralizing antibody (based on competition with CR3022) that also does not directly block ACE2:RBD interactions in protein-based assays (Nabel et al., 2022). Together, the predicted target sites within RBD, affinities, and neutralization potencies demonstrate that this panel’s MBC-derived mAbs encompass a diversity of immunologically relevant neutralizing epitopes.
Figure 1.
Diverse IgG+ MBC-derived RBD-specific antibodies gain potency when expressed as IgM. (A) Panel of eight IgG+ MBC-derived IgG mAbs indicating affinity for RBD and NT50 of WA-1 SARS-CoV-2 in PRNT (Rodda et al., 2021). Samples were analyzed in duplicate in at least two separate experiments. (B) Binding kinetics for individual mAbs as determined by BLI using sensor-bound IgG mAbs and serial dilution of soluble RBD protein. Each mAb was tested with six dilutions of RBD. (C and D) Summary of epitope-mapping experiments showing relative competition for RBD between the indicated MBC-derived IgG and well-characterized Fabs (C) or each other (D); individual data are provided in Figs. S1 A and S2 A. (E) Comparison of neutralizing potency of clonally identical IgG and IgM mAbs against pseudovirus. Bars indicate mean and SD for three independent experiments; individual symbols indicate the average of internal duplicates for each experiment. Average fold difference in NT50 potency for IgG vs. IgM for each BCR clone is shown below the graph. (F) Representative results for 297 IgM vs. IgG in neutralization assays using the pseudovirus (left panel; average and SD are shown for three independent experiments, each performed in duplicate) or a WA-1 SARS-CoV-2 PRNT (middle panel; representative of three independent experiments) and summary of NT50 data (right table).
Figure S1.
BLI competition assays. Competition with well-characterized mAbs for the panel of eight IgG+ MBC-derived IgG mAbs. The timing of the exposure to each antibody is indicated with a black arrow for the well-characterized mAb and a red arrow for the MBC-derived mAb.
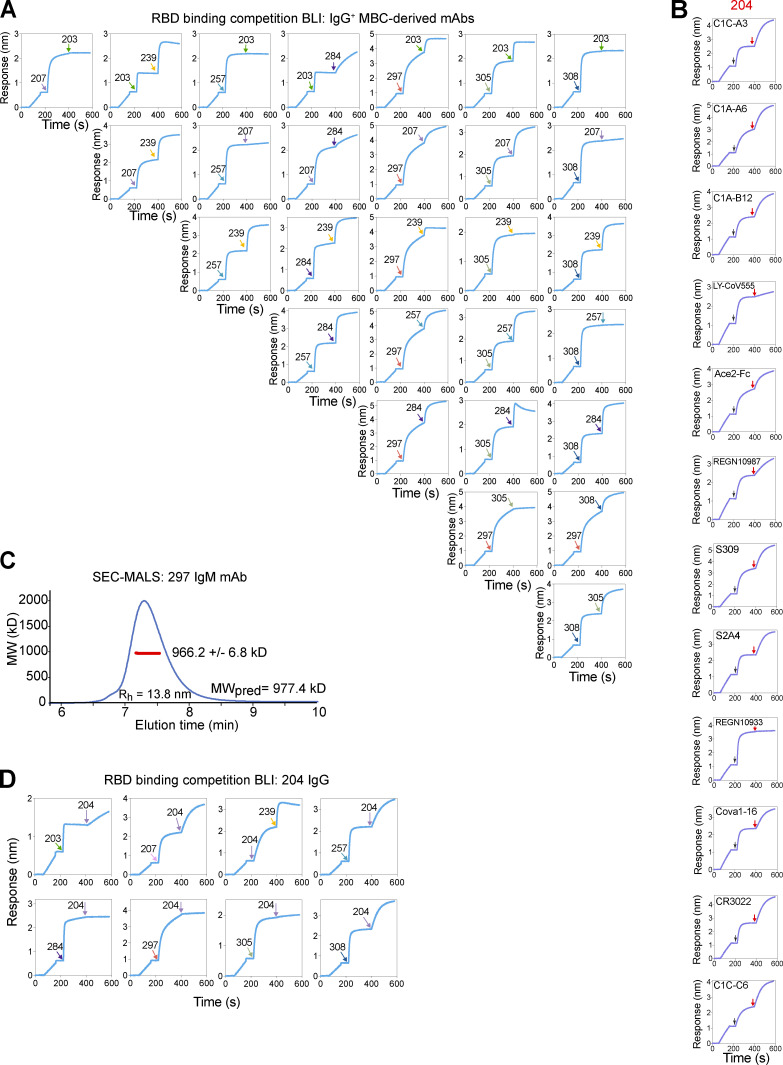
Figure S2.
BLI competition assays and SEC-MALS demonstrating molecular weight of IgM pentamer. (A) Intrapanel competition BLI plots displaying sensor-detected RBD binding after addition of the indicated IgG+ MBC-derived IgG mAb. (B) Competition with well-characterized mAbs and the IgM+ MBC-derived 204 IgG mAb for RBD by BLI. (C) SEC-MALS traces for the purified 297 IgM mAb. The light scattering trace is in blue, and the molecular weight (MW) calculated from the combination of scattering, refractive index, and UV absorbance is shown in red across the peak elution window. The average calculated MW is shown in the inset, along with the predicted MW (MWpred). The error is based on the SD from the 56 scans along the peak elution profile; however, the expected uncertainty can be as large as 10% due to assumptions about molar extinction coefficients and glycan occupancy. The hydrodynamic radius (Rh) was estimated from dynamic light scattering, which was also measured online and indicated under the peak. (D) Competition of mAb 204 against the panel of eight IgG+ MBC-derived mAbs.
For expression as IgM mAbs, in addition to the light-chain and µ heavy-chain plasmids, we included a plasmid encoding the human joining (J) chain that enables self-assembly into pentameric IgM, the predominant form in humans (Hughey et al., 1998; Matsumoto, 2022; Thouvenel et al., 2021). We also confirmed appropriate size, hydrodynamic radius, and purity of pentameric IgM using size exclusion chromatography with multiple angle light-scattering (SEC-MALS; Fig. S2 C).
To compare the properties of IgM vs. IgG mAbs with identical specificities, we first measured neutralization potencies against a luciferase-encoding SARS-CoV-2 spike–pseudotyped lentiviral vector (pseudovirus; Crawford et al., 2020). Pseudovirus was preincubated with IgM or IgG mAbs and applied to ACE2-expressing 293T cells. Strikingly, multimerization as IgM lowered the concentration required to achieve NT50 (50% reduction in infection relative to untreated controls) for all eight IgG+ MBC-derived mAbs with impacts ranging from 3- to 23-fold (Fig. 1 E). There is a large size difference between monomeric IgG (∼150 kD) and pentameric IgM (∼970 kD; Keyt et al., 2020). Thus, considered on a molar basis, neutralizing activity increased ∼20–150-fold.
Relative effects for IgG/IgM pairs were not attributable solely to their RBD epitope. In particular, if, as has been speculated, steric hindrance of ACE2 binding by the bulky IgM molecule were the primary mediator of increased neutralization (Ku et al., 2021), then mAbs that competed with the ACE-2-Fc chimera might have been predicted to display the greatest enhancement with multimerization. However, while mAbs 297 and 305 each competed with ACE2-Fc and exhibited similar sub-picomolar affinity to RBD and 15-ng/ml potency as IgG, multimerization as IgM effected 12- vs. 4-fold increases in activity, respectively. One explanation for variance in multimerization impact is differing ability to cross-link spike proteins/virions as assembled pentamers, a geometric or paratope property that would increase potency in neutralization assays but not be detected via techniques used to quantify binding. The limited impact of multimerization on clone 239, which binds at the RBD core, may be due to relative steric hindrance for IgM in accessing the base of RBD, reducing the potential for cross-linking (Samsudin et al., 2020). While our studies are limited by reliance on competition assays rather than direct structural studies to define the RBD epitope, our combined findings suggest that the impact of multimerization is influenced by factors other than proximity to the ACE2 footprint.
In our study of MBC-derived mAbs targeting malaria merozoite surface proteins, enhanced avidity by multimerization as pentameric IgM or engineered hexameric IgG drove increased activity in parasite-blocking assays (Thouvenel et al., 2021). Anti–SARS-CoV-2 mAbs with lower affinities for RBD might therefore be predicted to benefit most from multimerization. Indeed, two of the lowest-affinity clones, 203 and 207, showed the greatest benefit of multimerization (21–23-fold).
Although neutralization assays that use a spike pseudotyped lentivirus have been validated for IgG mAbs, differences between pseudovirus and true SARS-CoV-2 might impact the assessment of IgM/multimer activity. We therefore assessed one mAb pair using a live viral assay. The results for clone 297 IgG and IgM mAbs in a plaque assay that employed a SARS-CoV-2 isolate were nearly identical to the data generated using the pseudovirus neutralization assay (Fig. 1 F). In summary, multimerization as IgM increased potency across an unexpectedly broad range of target epitopes and affinities, including for mAbs that exhibited sub-picomolar affinity and potent neutralizing activity as IgG monomers.
SARS-CoV-2–specific IgM+ B cells encode BCRs that primarily bind RBD when expressed as IgM
Having shown that the activities of IgG+ MBC-derived neutralizing antibodies are enhanced by expression as IgM, we next sought to investigate the characteristics of RBD-specific IgM+ MBCs present in the same donors (Rodda et al., 2021). We therefore determined BCR sequences from antigen-experienced RBD-specific IgM+ B cells collected but not previously evaluated in our prior study (Table S1). BCRs from IgM-expressing cells with a classic MBC surface phenotype (CD21+CD27+) exhibited evidence of somatic mutation. The extent of mutation was less than observed in malaria-specific IgM+ MBCs (Krishnamurty et al., 2016), likely reflecting the different kinetics of the immune responses to these pathogens, the time between exposure and sample collection, and/or the likelihood of multiple exposures in a malaria-endemic region (Muller et al., 2021). Of note, near-germline sequences can encode potent SARS-CoV-2–neutralizing antibodies (Brouwer et al., 2020; Bullen et al., 2021; Kreer et al., 2020). Consistent with data that the fraction of RBD-specific MBCs that are IgM+ declines in the months following a first infection in unvaccinated individuals, most IgM+ MBC BCR sequences were isolated at early time points after infection (Dan et al., 2021; Rodda et al., 2021; Sakharkar et al., 2021). While our data are limited by the duration of the study, other work suggests that individuals infected 12 mo before analysis can retain long-lived IgM+ MBCs (Ruggiero et al., 2022; Viant et al., 2021).
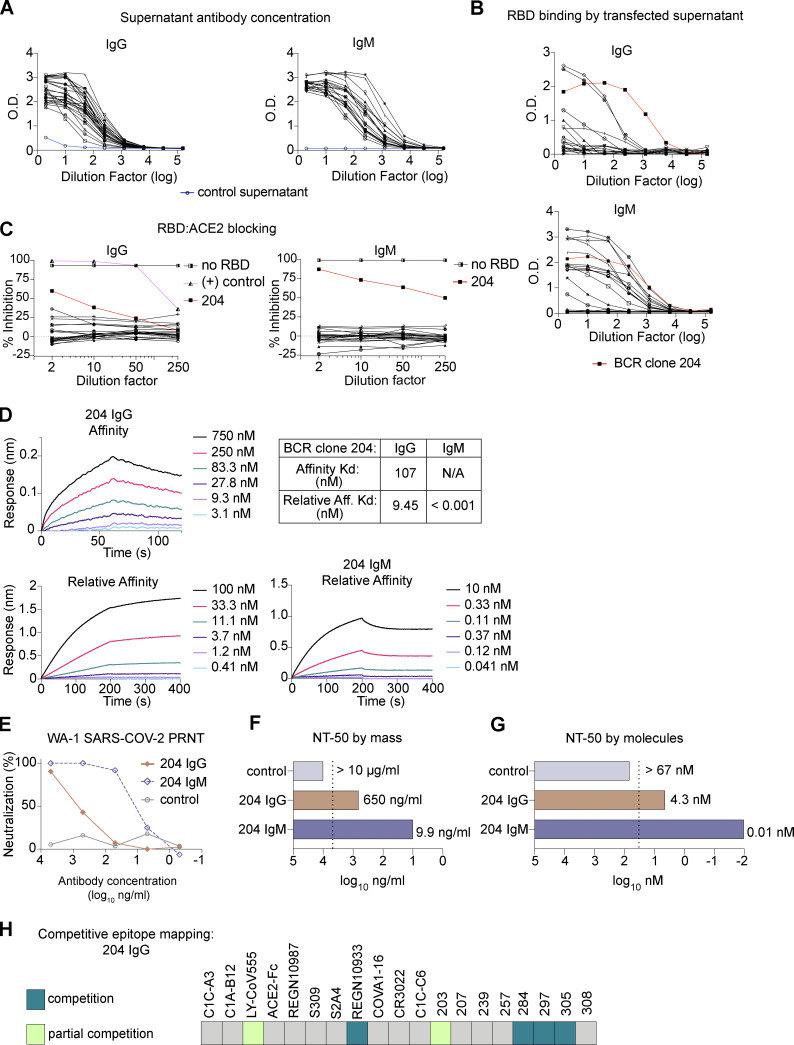
To understand the potential contribution of IgM memory to the antibody pool, we built a library of plasmids from the antigen specificity-determining V(D)J sequences of each BCR. Heavy chains were cloned into alternative plasmids to allow us to directly compare the binding and functional properties when expressed as the native IgM isotype vs. as IgG. As an initial screen, we tested supernatants from cells cotransfected with a plasmid encoding the BCR-derived light chain and a plasmid encoding its paired heavy-chain variable region upstream of the γ1 constant region (to make IgG) or the µ constant region (IgM). In this manner, 26 IgM-encoded BCRs were expressed as IgG and IgM antibodies. Despite achieving similar concentrations of IgG or IgM as assessed using anti-isotype ELISA, RBD binding was detected primarily for supernatants containing IgM (Fig. 2, A and B). Clones that bound RBD as IgM, but not IgG, likely represent low-affinity receptors that benefit from enhanced avidity provided by IgM’s multimeric structure. Our results demonstrate the value of studying IgM BCRs as IgM antibodies and highlight the ability of tetramer-based enrichment to identify even very-low-affinity antigen-specific MBCs (Taylor et al., 2012). Notably, supernatant from cells producing an IgM antibody derived from clone 204 prevented RBD from binding to human ACE2, a surrogate for neutralization (Fig. 2 C; Tan et al., 2020).
Figure 2.
IgM MBCs encode RBD-specific antibodies, including a neutralizing mAb with improved activity when expressed as IgM vs. IgG. (A and B) Supernatants from cells transfected with plasmids encoding IgM+ MBC-derived mAbs as IgG vs. IgM screened by ELISA for anti-IgG (left) or -IgM (right), with untransfected supernatant as a negative control (blue; A); or binding to SARS-CoV-2 spike RBD protein (B). (C) Supernatants’ ability to block RBD from binding to plate-bound human ACE2. BCR clone 204 is highlighted (in red) in B and C. IgG+ MBC-derived antibody, 297 IgG, is included as a positive control (in pink) in C. Data are representative of two independent experiments performed in triplicate. (D) BLI data showing binding and dissociation kinetics of BCR clone 204 as purified IgG vs. IgM. (E) SARS-CoV-2 PRNT on Vero cells at the indicated concentrations of 204 IgG vs. IgM or an anti-malaria mAb (negative control; Thouvenel et al., 2021). (F and G) NT50 titers are shown based on mass (F) or molar (G) concentrations. For conditions that did not approach an NT50, an arbitrary NT50 was assigned of 2× the maximum concentration tested (dashed line). The assay was performed in duplicate and repeated at least twice. (H) Epitope mapping of mAb 204 IgG by BLI against well-characterized Fabs/mAbs and the other mAbs in our panel; individual data are provided in Fig. S2, B and D.
A mAb derived from an IgM+ MBC is a potent neutralizer of SARS-CoV-2
Next, we aimed to determine whether IgM BCR clone 204 encoded a neutralizing antibody. We therefore produced purified 204 IgG and IgM for use in advanced assays. To measure affinity, 204 IgG was immobilized onto sensor tips that were immersed into solutions with various concentrations of RBD. Next, we immobilized RBD on a sensor and allowed varying dilutions of IgG or IgM antibody to bind, such that a calculated dissociation constant (Kd) reflected relative affinity, or avidity. For 204 IgG, the Kd of the avidity assay was 9.45 nM, while the 204 IgM Kd could not be calculated (Fig. 2 D). Consistent with our observations of malaria-specific multimerized antibodies, IgM strongly resisted dissociation through increased numbers of binding events and/or the effective increase in local concentration of binding moieties.
To assess neutralizing activity, we performed plaque reduction neutralization tests (PRNTs) with SARS-CoV-2 and 204 IgM or IgG. 204 IgM, but not 204 IgG, exhibited potent neutralization (Fig. 2, E and F). In comparing the two isotypes, the NT50 was 66-fold lower for 204 IgM than the clonally identical IgG. Thus, ∼400-fold fewer molecules of IgM than IgG were required to achieve equivalent reduction in plaques vs. untreated controls (Fig. 2 G). Interestingly, while 204 IgG affinity and potency were lower than the IgG MBC-derived antibodies tested above, 204 IgM’s observed neutralizing potency was similar to the highly potent 297 IgM. To investigate whether a unique target site explained the robust effect of multimerization for clone 204, we again performed competitive BLI assays with well-characterized mAbs and the IgG+ MBC-derived mAbs (Fig. 2 H; and Fig. S2, B and D). Adding 204 IgG did not increase sensor-detected RBD binding after immersions into solution containing IgG mAbs 284, 297, or 305 or the REGN10933 IgG, and only moderately when 203 IgG or a LY-CoV555 Fab fragment was preloaded. Cumulatively, these results suggest that clone 204’s RBD target site is similar to others in our panel and challenge the hypothesis that a unique epitope is responsible for the larger effect size of multimerization. Instead, our results support a model wherein the avidity benefit of IgM enables antibodies of mediocre affinity to achieve equivalent binding strength to a high-affinity interaction with lower valency. For clones with high baseline affinity, additional avidity may contribute less to enhanced function than other attributes of IgM, such as the potential for crosslinking and greater steric hindrance for RBD:ACE2 interactions.
IgM mAbs maintain neutralizing activity against variants that escape clonally identical IgG
In settings of seasonal reexposure to a highly variable pathogen, a likely future scenario for SARS-CoV-2, an ideal therapeutic, like an ideal humoral memory response, would tolerate mutations in neutralizing target sites, without incurring significant risk for autoreactivity. We hypothesized that IgM antibodies could accomplish this goal through enhanced avidity.
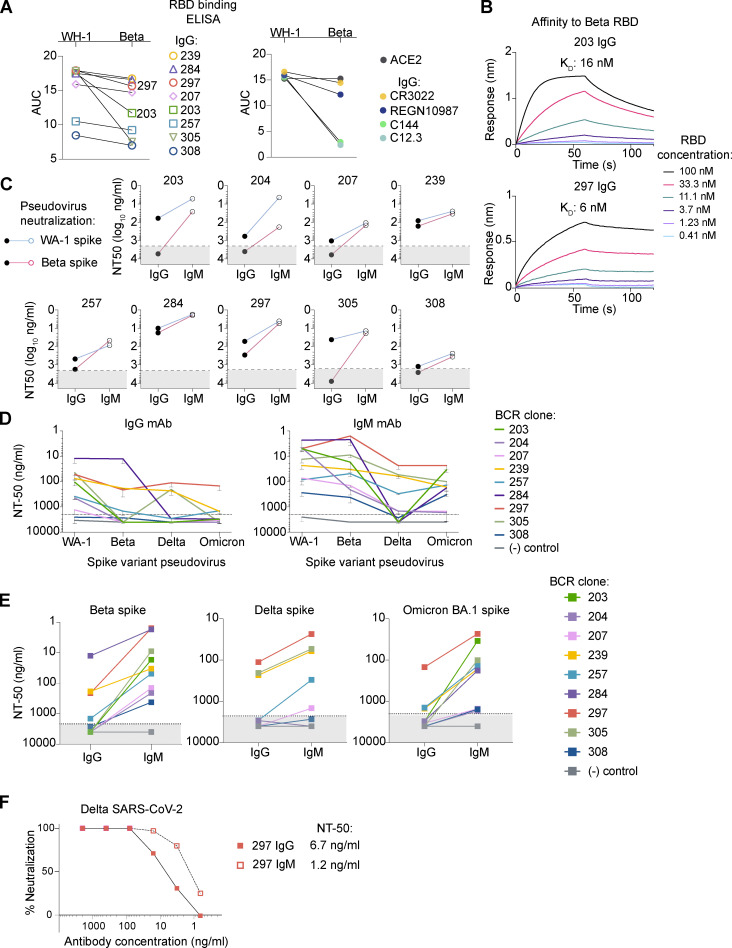
Based on observations that the Beta variant escapes neutralization by mAbs that are protective against earlier SARS-CoV-2 isolates, we compared binding to the Beta vs. Wuhan-Hu-1 RBD for each IgG mAb by ELISA (Sakharkar et al., 2021; Wang et al., 2021; Fig. 3 A). In parallel, we also tested well-characterized IgG mAbs from each epitope-defined class that are known to exhibit either limited (CR3022 and REGN10987) or substantial (C144 and C12.3) reductions in Beta vs. Wuhan-Hu-1 RBD binding (Barnes et al., 2020; Clark et al., 2021; Greaney et al., 2021; Starr et al., 2021). Together with an ACE2-Fc chimera, the four published antibodies exhibited the predicted RBD binding profile, validating the RBD proteins generated for this assay and contextualizing results for our in-house mAbs. The parental MBCs in our study were isolated in early 2020, when a Wuhan-Hu-1-like virus (WA-1 SARS-CoV-2) predominated (Muller et al., 2021). Several IgG mAbs exhibited reduced binding to the Beta variant RBD protein, most notably 203 and 305. Next, affinity to Beta RBD was quantified using BLI for 203 IgG, which exhibited significant loss of affinity in ELISA, and 297 IgG, for which binding appeared only slightly reduced (Fig. 3 B). The results were consistent with ELISAs: 16 nM to Beta vs. 4.4 nM to Wuhan-Hu-1 RBD for 203 IgG, and 6 nM to Beta vs. sub-picomolar to Wuhan-Hu-1 RBD for 297 IgG.
Figure 3.
IgM antibodies retain activity against viral spike variants that escape clonally identical IgG. (A) Binding to Wuhan-Hu-1 (WH-1) or Beta variant RBD proteins by purified IgG mAbs in ELISAs, quantified as the area under the curve (AUC) for a 10-dilution series. Results for the panel of eight IgG+ MBC-derived mAbs are shown (left). For clarity, simultaneously performed ELISAs testing well-characterized IgG mAbs, or an ACE2-Fc fusion protein (ACE2), are shown separately (right). Data are representative of two independent experiments performed in triplicate. (B) Affinity for Beta RBD of representative antibodies with significantly reduced binding, 203 (top), and moderate reduced binding, 297 (bottom). (C) Neutralization potencies for each mAb as IgG vs. IgM against WA-1 (blue) or Beta (pink) spike pseudovirus. Dashed line illustrates the maximum antibody concentration tested (2 μg/ml). For antibodies that did not approach an NT50, the NT50 is graphed arbitrarily in the shaded area as 4 μg/ml. Each IgG/IgM pair was tested in duplicate against both viruses in three independent experiments. (D) Varying neutralization potency against WA-1, Beta, Delta, and Omicron BA.1 spike pseudoviruses for IgG (left) vs. IgM (right) mAbs. Error bars illustrate mean ± SD for three or more experiments with internal duplicates. (E) Summary neutralization potencies of IgG vs. IgM mAbs for the indicated variants in experiments described in D. Control, malaria-specific IgG or IgM (MaliA01 mAbs; Thouvenel et al., 2021). (F) Neutralization in a Vero cell plaque reduction assay of Delta SARS-CoV-2 for 297 IgG vs. IgM. Representative plot of five independent experiments.
We next tested all nine IgG/IgM pairs for the ability to neutralize a Beta-spike pseudovirus. As expected, the poorest binders to Beta RBD failed to reduce pseudovirus infection as IgG mAbs, even at the highest concentration tested (2 μg/ml). In contrast, all IgM mAbs neutralized the WA-1 and Beta pseudoviruses with similar potencies (Fig. 3 C). These results expand on a study in which two IgM mAbs were tested for retained binding and neutralization activity against mutated RBD proteins and viruses (Ku et al., 2021). However, in that prior study, only one of the two IgM mAbs retained activity against the key mutations found in the Beta spike (NT50 >16 μg/ml). Our data contradict the authors’ conclusion that careful selection of targeted epitope is necessary to identify antibodies that overcome escape mutations when multimerized as IgM. All IgM mAbs in our panel overcame even dramatic losses in neutralization potency. Besides testing more IgM/IgG pairs, our study differs in that MBC-encoded BCRs were the source for antigen-binding domains, while the previous work used variable regions derived from a phage display library that were selected on the basis of binding to yeast-displayed protein. The ACE2-competing MBC-derived IgG mAbs are 10-fold more potent in neutralizing WA-1 SARS-CoV-2 than reported for the phage-derived IgG (Ku et al., 2021). These combined findings demonstrate that expression as IgM overcomes reduced affinity to the Beta variant for mAbs that target a range of RBD epitopes.
Next, we tested the previously unexplored question of whether IgM mAbs might also outperform clonally identical IgG against the Delta and Omicron BA.1 variants. In particular, the Omicron BA.1 variant is extensively mutated and partially or entirely escapes neutralization by all but one current Emergency Use Authorization mAb (Planas et al., 2021). Indeed, in neutralization assays using the Delta or Omicron BA.1 spike, eight of the nine IgG mAbs exhibited substantial reduction in activity (Fig. 3, D and E). In contrast, IgM mAbs retained activity at concentrations <2 μg/ml. Apparent exceptions were against the Delta strain, for 203, 284, and 308.
Notably, 297 IgG acted as a broad neutralizer across all tested variants, with NT50 values for the Delta and Omicron BA.1 pseudoviruses of 111 and 147 ng/ml, respectively. Multimerization as IgM further improved potency to 23 ng/ml against both variants. To confirm that increased tolerance for spike mutations by IgM and broad neutralization by clone 297 were not specific to the pseudovirus, we next performed assays with true Delta SARS-CoV-2. As predicted, 297 IgG retained neutralizing activity, and multimerization as IgM enhanced potency fivefold (Fig. 3 F).
IgM mAbs exhibit enhanced protection in human airway cultures
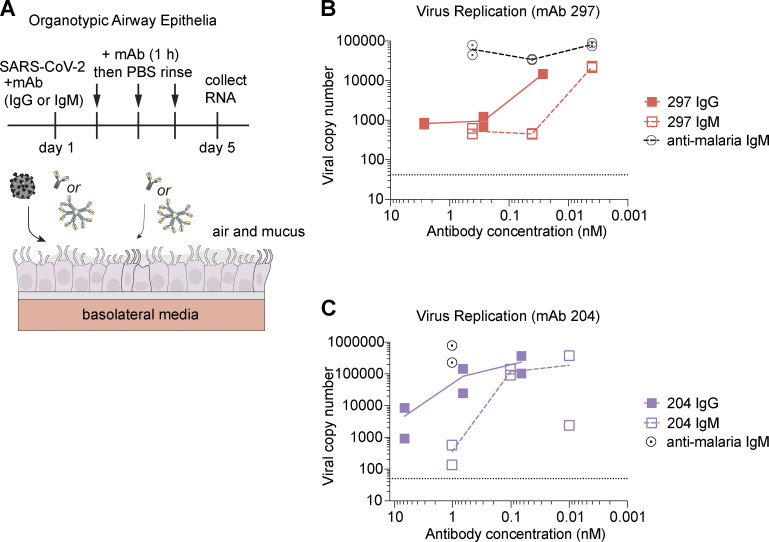
IgG antibodies do not easily penetrate the lung and respiratory epithelia, primary sites of SARS-CoV-2 infection. In contrast, pentameric IgM incorporates the J chain that allows transcytosis to mucosa (Brewer et al., 1994; Matsumoto, 2022). Evidence from subjects with immunoglobulin deficiencies suggests that IgM is important for long-term protection against respiratory pathogens (Micol et al., 2012). One potential challenge for an IgM therapeutic may be its reduced serum half-life relative to IgG (Keyt et al., 2020). However, a recent study showed that an IgM mAb delivered to the nasal passages protected mice subsequently challenged with SARS-CoV-2 (Ku et al., 2021). To test the effect of IgM mAbs in human respiratory epithelia, we used polarized organotypic airway epithelia cultures generated from primary human bronchial epithelial cells (Altman et al., 2018) and applied SARS-CoV-2 together with either IgG or IgM mAbs. For 1 h each subsequent day, the apical side of the culture was incubated with a small volume of mAb, and then rinsed (Fig. 4 A). Both the IgG and IgM antibodies reduced viral copy number in a dose-responsive manner (Fig. 4, B and C). Notably, a lower dose of IgM was required. As predicted by our experiments in cell lines, the enhanced performance of IgM was more striking for clone 204 than 297 due to the relatively lower potency of 204 IgG. These results demonstrate that IgM mAbs are protective in a model that recapitulates physiologic conditions of the human airway, including air exposure and mucus production.
Figure 4.
An anti-RBD IgM mAb protects against SARS-CoV-2 infection in differentiated human airway epithelia cultures. (A) Schematic of experimental design using airway epithelial cultures differentiated to an organotypic state at an ALI. (B and C) Quantification of viral RNA copy number in ALI cultures treated with the indicated antibody. Data represent independent experiments; cultures were generated from primary human bronchial epithelial cells derived from unique donors. Each condition was tested in independent duplicates. For B, duplicates were pooled as RNA before quantitative PCR.
Multimerization as the dimeric antibody isotype, IgA, has also been explored as a strategy to enhance SARS-CoV-2–neutralizing mAbs. Such reagents exhibit greater potency in vitro compared with clonally identical IgG, although not achieving that of IgM as demonstrated here or in other studies (Ku et al., 2021; Pisil et al., 2021; Wang et al., 2021). IgA is more efficiently transported to respiratory mucosa, while IgM is maintained in higher concentrations in the blood, where it controls hematogenous spread, in addition to accessing the mucosa (Keyt et al., 2020). Unlike IgA, IgM is a potent activator of complement, which both targets a pathogen directly and enhances the immune response. Delivery of IgM to respiratory mucosa might achieve both therapeutic effects (Zhang et al., 2022a). Interestingly, systemic delivery might offer enhanced stability of mAbs at the mucosal surface, as IgM acquires the protective secretory component during transcytosis (Michaud et al., 2020).
Artificial multimers also harness avidity to enhance the potency and/or breadth of SARS-CoV-2–binding domains. Among the most successful are trimeric nanobodies and a 24-valent structure of RBD-specific variable regions connected to an apoferritin scaffold (Hunt et al., 2022; Mast et al., 2021; Rujas et al., 2021; Xu et al., 2021). Interestingly, the hydrodynamic radius of the apoferritin construct is similar to IgM, potentially enabling similar cross-linking and steric hindrance (Fig. S3 in Rujas et al., 2021). However, despite potent in vitro properties and proof of principle in small mammal models, novel proteins face greater challenges in safety, pharmacokinetics, and antidrug immune responses. Moreover, their function is likely limited to direct neutralization of virus. In contrast, IgM antibodies can interface with the endogenous immune system (Keyt et al., 2020). Further, the use of variable domains derived from MBCs takes advantage of germinal center in vivo selection and refinement not feasible with phage display, including heavy and light chains that are selected in concert while, in parallel, screening against autoreactivity. Of note, a recent report described three spike-specific mAbs presumed to be derived from IgM+ MBCs (based on the dominant B cell subset present at the time of sample collection; Callegari et al., 2022). The two RBD-specific mAbs and one N-terminal domain–specific mAb exhibited increased neutralizing potency as IgM vs. IgG. However, expression of an IgG-derived, N-terminal domain–specific mAb as IgM did not statistically improve its activity. Our study expands upon these observations with characterization of >20 RBD-specific mAbs derived from IgM+ MBCs. Further, we provide a comprehensive assessment of the role of multimerization in enhancing neutralization and the impact of antibody isotype on resistance to viral escape.
The high per-molecule potency and enhanced durability of binding across mutant viral proteins demonstrated here for IgM mAbs may help to explain the remarkable evolutionary conservation of multimeric IgM in jawed vertebrates (Matz et al., 2021). Although our investigation of IgM+ MBCs encompassed a small number of cells, we identified a potent neutralizing antibody. These results contribute to growing evidence that IgM memory has an underappreciated role in protective immunity. IgM mAbs generated from diverse neutralizing antibodies are potent and broad neutralizers of SARS-CoV-2 and include candidate mAbs with likely therapeutic utility. Our results underscore the value of focused investigation into the role for IgM memory in immunity to SARS-CoV-2 and other evolving pathogens.
Materials and methods
MBC isolation
Peripheral blood samples from convalescent COVID-19 subjects were obtained with informed consent under the approval of the University of Washington Institutional Review Board (Gale Lab; 00009810) and isolated as previously described (Rodda et al., 2021). Briefly, peripheral blood mononuclear cells were thawed, washed, and stained with a decoy tetramer and then RBD tetramer; bound cells were magnetically enriched (Miltenyi Biotec) and stained with surface antibodies. Single tetramer-positive B cells were index-sorted using a FACS Aria II and collected in 96-well plates containing SMART-Seq v4 capture buffer (Takara Bio) for BCR sequencing.
BCR sequencing and cloning
Our methods for BCR sequencing of singly sorted B cells were described previously (Rodda et al., 2021). Briefly, after cDNA amplification using SMART-Seq v4 (Takara Bio) at half reaction volume, BCR sequences from each cell’s cDNA were amplified in a multiplex reaction using a universal 5′ primer for the template switch region combined with pooled 3′ primers for the µ, γ, α, κ, and λ constant regions. After gel electrophoresis to confirm amplification, amplicons were purified and sequenced by Sanger sequencing (GenBank IDs: ON886550–ON886835). Alignment of trimmed sequences was performed using IGMT/HighV-QUEST (Alamyar et al., 2012). Primer design and cloning into expression vectors followed the manufacturer’s protocol for in-fusion cloning (Takara Bio). Each light chain was cloned into vectors of its respective isotype, κ or λ. All heavy chains were cloned into IgG1 plasmids. Additionally, heavy chains from BCR clones 203, 207, 239, 257, 284, 297, 305, and 308 (Rodda et al., 2021) and any heavy-chain cDNA sequence using µ constant regions were also cloned into IgM plasmids. Cloned plasmids were sequenced and screened to ensure concordance with the parental cDNA.
Production of purified mAbs
Production of purified IgG and IgM mAbs was carried out by polyethylenimine transfection of heavy- and light-chain plasmids as previously described (Thouvenel et al., 2021), except that a plasmid encoding human J chain (GenBank ID: NM_144646.4) was included in IgM transfections at a µ:light:J chain plasmid ratio of 1:1:1, and supernatant was collected on days 3 and 6 after transfection. After 0.4-μm filtration, antibody purification was carried out following the manufacturer’s instructions using a HiTrap Protein G HP purification column for IgG antibodies (GE Healthcare), and a POROS CaptureSelect IgM Affinity Matrix Column (Thermo Fisher Scientific) for IgM. Antibodies were concentrated and buffer-exchanged to PBS at 1–3 mg/ml using Amicon Ultra Centrifugal filters (Millipore Sigma) and then filter-sterilized and stored at −80°C. Antibody concentration and purity were assessed by spectrophotometry and protein gel electrophoresis.
SEC-MALS
SEC-MALS was performed using an Agilent 1260 HPLC system coupled to a light scattering detector (miniDawn Treos; Wyatt Instruments) and a refractive index detector (TRex; Wyatt Instruments). 50 μl of each IgM construct (1 mg/ml) was injected and flowed over a Sepax SRTC SEC column (5 μm, 300 Å, 4.6 × 300 mm with a 4.6 × 50-mm matching guard column) at a flow rate of 0.35 ml/min in 150 mM sodium phosphate, pH 7.0, and 0.02% sodium azide. Chromatograms were aligned and integrated, and the MW was calculated using ASTRA (Wyatt Instruments). A refractive index of 0.181 was used for calculations reflecting a weighted combined contribution of protein 0.185 and glycan 0.146 (Arakawa and Wen, 2001). A glycan content of 8,635 daltons of N-linked glycans on each heavy chain and 2,155 daltons for the J-chain was based on the estimated average of the predominant glycoforms observed in previous site-specific glycan studies of human IgM (Moh et al., 2016).
ELISA for antibody expression and RBD binding
Cloned heavy- and light-chain plasmids derived from RBD-specific IgM+ B cells were initially screened in small-scale transfections. 293T cells (ATCC) were plated at 80% confluency and transfected with 0.5 μg each of paired heavy- and light-chain plasmids using polyethylenimine. 16 h later, medium was replaced with serum free-medium, and after 3–4 d, supernatants were harvested and cellular debris was removed by centrifugation. Antibody expression levels were determined using human IgG or IgM ELISA Antibody Pair Kit (Stemcell Technologies) according to the manufacturer’s instructions. RBD specificity was determined by ELISA as previously described (Rodda et al., 2021). Briefly, 96-well high-bind plates (Corning) were coated with 2 μg/ml SARS-CoV-2 RBD protein overnight at 4°C, washed with PBS and 0.05% Tween 20 (PBS-T), blocked with 3% milk in PBS-T, and incubated with serially diluted culture supernatants for 2 h at room temperature. Plates were washed, and bound antibodies were detected using anti-human IgG-HRP or anti-human IgM-HRP (Jackson ImmunoResearch) at a 1:3,000 dilution followed by 1× 3,3′,5,5″-tetramethylbenzidine (Invitrogen) and 1 M HCl. OD was measured on a spectrophotometer at 450 and 570 nm, and data were analyzed in Prism (v9.01; GraphPad).
ACE2:RBD blocking surrogate virus neutralization test
Antibodies’ ability to inhibit interaction with human ACE2 was assessed as previously described (Tan et al., 2020). 96-well plates (Corning) were coated with 5 μg/ml hACE2-Fc in 100 mM carbonate buffer overnight at 4°C. Plates were washed with PBS-T and blocked with 3% milk in PBS-T. mAbs were serially diluted and incubated with 18 ng of SARS-CoV2 RBD-HRP for 1 h at 37°C and added to the blocked plates for 1 h at room temperature. Binding was detected using 3,3′,5,5′-tetramethylbenzidine, reactions were quenched with 1 M HCl, and absorbance was measured at 450 and 570 nm. Percentage inhibition was calculated as (1 − Sample OD/Negative Control OD) × 100 and analyzed in Prism.
BLI
BLI assays for affinity and relative affinity/avidity were performed at ambient temperature with shaking set at 1,000 rpm using an Octet Red 96 System (Pall FortéBio/Sartorius). Individual IgG mAbs (for affinity assays) or purified RBD protein (for relative affinity/avidity assays) were diluted 10 µg/ml to in kinetics buffer (1× Hepes-EP+ [Pall FortéBio], 0.05% nonfat milk, and 0.02% sodium azide). Biosensors were hydrated in kinetics buffer for 10 min, and the diluted protein was immobilized onto Protein A biosensors (IgG mAbs in affinity assays) or anti-Penta-His biosensors (RBD in avidity assays) and then equilibrated in kinetics buffer for 60 s. For affinity assays, monomeric RBD was diluted to 5,000, 750, or 100 nM in kinetics buffer and serially diluted threefold for a final concentration of 20.6, 3.1, and 0.4 nM, respectively. For avidity assays, individual IgG or IgM mAbs were diluted to 100 or 10 nM in kinetics buffer and serially diluted threefold for a final concentration of 0.41 or 0.041 nM, respectively, in a black 96-well Greiner Bio-one microplate at 200 μl per well. To measure association, loaded biosensors were dipped into the diluted protein. Association and dissociation was performed for 60 s each in affinity assays. Association and dissociation times for avidity assays were extended to 200 s each. The data were baseline subtracted and plotted using Pall FortéBio/Sartorius analysis software (v12.0).
Protein production for BLI competition assays
For BLI competition assays, we produced SARS-CoV-2 spike protein RBD (GenBank ID: QHD43416.1, residues 319–541), as previously described (Nabel et al., 2022). Previously characterized antibodies were produced as either IgGs or Fabs for use in this assay. Antibodies REGN10933 and REGN10987 (Hansen et al., 2020), CR3022 (Yuan et al., 2020), C1A-B12, C1C-A3, C1C-C6, and C1A-A6 were prepared and purified as previously described (Clark et al., 2021). Antibodies S309, LY-CoV555, Cova1-16, and S2A4 were also generated as previously described (Nabel et al., 2022). An Fc-fusion protein comprising the human ACE2 ectodomain (GenBank ID: BAB40370.1, residues 18–740) was expressed and purified as previously described (Clark et al., 2021).
BLI competition assays
We performed competition experiments using an Octet RED 96e (Sartorius). First, we loaded SARS-CoV-2 spike protein RBD onto streptavidin sensors (FortéBio) at 1.5 μg/ml for 80 s in kinetics buffer (PBS containing 0.02% [vol/vol] Tween and 0.1% [vol/vol] BSA). For each pair of antibodies tested, we associated the first antibody (IgG, Fab, or ACE2-Fc fusion protein) at 250 nM for 180 s. We then associated the second analyte (IgG, Fab, or ACE2-Fc fusion protein) at 250 nM for 180 s. All antibodies were tested as Fabs unless only the IgG form was available. Antibodies tested as Fabs included 203, C1A-B12, C1C-A3, C1C-C6, Cova1-16, CR3022, LY-CoV555, REGN10987, and S2A4. Antibodies tested as IgGs included 204, 207, 239, 257, 284, 297, 305, 308, C1A-A6, S309, and REGN10933. We used FortéBio data analysis software and Prism to generate and analyze curves for competition assays. Antibodies were designated as competing if there was little to no change in the refractive index following association of the second protein.
Production of spike-variant pseudoviruses
To make Beta and Omicron BA.1 spike pseudotyping plasmids, sequence fragments encoding the variant spike protein were gene synthesized (Integrated DNA Technologies) and cloned into a digested plasmid (NR-53765; BEI Resources; National Institutes of Health) using NEBuilder HiFi DNA Assembly (NEB). To produce pseudotyped virus, 293T cells were transiently transfected with a vector plasmid encoding luciferase (NR-52516; BEI Resources), a pseudotyping plasmid encoding the Beta, Omicron BA.1, WA-1 (gift from David Veesler, University of Washington, Seattle, WA), or Delta (pLV-Spike-V8; InvivoGen) spike and a psPAX2 helper plasmid (Genscript). 48 h after transfection, culture supernatant was harvested and passed through a 0.22-μm filter. 100× concentrations of virus were achieved by overnight centrifugation at 8,500 g at 4°C and then resuspending the pellet in HBSS.
Pseudovirus neutralization assays
Pseudovirus neutralization assays were performed as previously described (Crawford et al., 2020; Rodda et al., 2022). Briefly, ACE-2–expressing 293T cells (BEI Resources; NR-52511) were seeded onto poly-L-lysine–coated 96-well plates and grown to 85–95% confluency. Pseudovirus was incubated with serially diluted IgG or IgM mAbs, or medium alone, for 1 h at 37°C and then gently applied to cells. 48 h after infection, cells were lysed following the manufacturer’s instructions using the Bright-Glo Luciferase Assay System reagent (E2610; Promega), and luminescence was measured in black-bottom plates using a Centro LB Microplate Luminometer (Berthold Technologies) with MikroWin 2000 software set to a 1-s exposure time. Percentage neutralization was calculated relative to the luminescence in control wells that had been transduced with virus coincubated with medium alone (internal for each plate, average of six wells), after subtracting background luminescence in virus-exposed 293T cells that lacked ACE2 expression (internal for each plate, average of six wells). NT50 was calculated by sigmoidal interpolation method in Prism. If a curve could not be fitted and 50% neutralization was not achieved at any dilution, where necessary for the purposes of data visualization, an arbitrary NT50 was assigned at 2× the highest dilution tested.
PRNT
PRNTs were performed as previously described (Erasmus et al., 2020; Rodda et al., 2021). Briefly, purified mAbs were diluted 1:10 followed by serial dilution. Diluted mAbs were mixed 1:1 with 600 PFU/ml SARS-CoV-2 WA-1 or Delta (BEI Resources; NR-52881 and NR-55612) virus in PBS + 0.3% cold water fish skin gelatin (Sigma-Aldrich) and incubated for 30 min at 37°C. Next, the mAb/virus mixture, or virus-only and mAb-only control solutions, was applied to duplicate wells of Vero cells in a 12-well plate and incubated for 1 h at 37°C, rocking every 15 min. Following incubation, plates were washed with PBS and overlaid with a 1:1 mixture of 2.4% Avicel RC-591 (FMC) and 2× MEM (Thermo Fisher Scientific) supplemented with 4% heat-inactivated FBS and penicillin/streptomycin (Thermo Fisher Scientific). The overlay was removed 2 d after infection, and plates were fixed by applying 10% formaldehyde (Sigma-Aldrich) in PBS and incubating for 30 min at room temperature and stained in a solution of 1% crystal violet (Sigma-Aldrich) in 20% ethanol (Thermo Fisher Scientific). Percentage neutralization was calculated as (1 − no. sample plaques/no. positive control plaques) × 100. Data was analyzed in Prism, and an NT50 for each condition was calculated by sigmoidal interpolation.
Air–liquid interface (ALI) primary airway epithelial cultures
Airway epithelial cultures were differentiated to an organotypic state at an ALI as previously described (Altman et al., 2018). Briefly, bronchial epithelial cells were obtained under study #12490 approved by the Seattle Children’s Institutional Review Board and following the rules of the Declaration of Helsinki of 1975. Cells were differentiated for 21 d at an ALI on 12-well collagen-coated Corning plates with permeable Transwells in PneumaCult ALI medium (Stemcell). SARS-CoV-2 WA-01 was added to the apical surface of differentiated cultures at an MOI of 0.5 together with either IgG or IgM mAbs for 1 h, then removed. Every 24 h, mAb in 100 µl of PBS was again added to the apical surface of cultures for 1 h then removed. After 96 h of infection, RNA was extracted from cultures, and SARS-CoV-2 replication was assessed by measuring viral genome copy number by quantitative PCR, with duplicate assays of harvested RNA from each SARS-CoV-2–infected experimental condition completed (Genesig Coronavirus Strain 2019-nCoV Advanced PCR Kit, Primerdesign).
Online supplemental material
Figs. S1 and S2 A show the individual BLI curves for the epitope-mapping competition assays that are summarized in Fig. 1, C and D, respectively. Fig. S2, B–D, shows the individual BLI curves for epitope mapping of the IgM+ MBC-derived antibody, 204 IgG (summarized in Fig. 2 H), and the SEC-MALS trace with calculated molecular weight for 297 IgM. Table S1 contains the characteristics of the RBD-specific IgM+ MBCs from which BCRs were sequenced, cloned, and expressed as antibodies. The symbols in the leftmost column correspond with the functional data for antibody-containing supernatants shown in Fig. 2, A–C.
Supplementary Material
shows characteristics of singly sorted RBD-specific IgM+ B cells from which BCRs were cloned and tested.
Acknowledgments
We thank Jennifer Haddock for administrative support.
This work was supported by the following: Seattle Children’s Research Integration Hub Covid-19 Award (to D.J. Rawlings), Burroughs Wellcome Fund Pilot Grant (to J. Abraham and M. Pepper), the Bill & Melinda Gates Foundation grants OPP1156262 (to N.P. King) and OPP1126258 (M. Guttman), Burroughs Wellcome Fund Career Award for Medical Scientists (J. Abraham), and National Institutes of Health grants T32GM007266 (M. Hale), T32GM007753 (K.N. Smith), R01AI153191 (M. Guttman), R01AI163160 (J.S. Debley), and K24AI150991-01S1 (J.S. Debley). D.J. Rawlings holds the Tom Hansen Investigator for Pediatric Innovation Endowment at Seattle Children’s.
Author contributions: Conceptualization: M. Hale, J. Netland, Y. Chen, C.D. Thouvenel, M. Pepper, and D.J. Rawlings. Supervision/direction: N.P. King, M. Guttman, M. Gale Jr, J. Abraham, J.S. Debley, M. Pepper, and D.J. Rawlings. Funding acquisition: N.P. King, M. Guttman, J. Abraham, J.S. Debley, M. Pepper, and D.J. Rawlings. Investigation: M. Hale, J. Netland, Y. Chen, C.D. Thouvenel, K.N. Smith, L.M. Rich, E.R. Vanderwall, M.C. Miranda, J. Eggenberger, L. Hao, M.J. Watson, C.C. Mundorff, and L.B. Rodda. Data analysis: M. Hale, J. Netland, Y. Chen, C.D. Thouvenel, K.N. Smith, L.M. Rich, E.R. Vanderwall, M.C. Miranda, J. Eggenberger, L. Hao, M.J. Watson, C.C. Mundorff, L.B. Rodda, M. Guttman, J. Abraham, J.S. Debley, M. Pepper, and D.J. Rawlings. Writing—original draft: M. Hale. Writing—review and editing: M. Hale, J. Netland, K.N. Smith, M. Guttman, J. Abraham, J.S. Debley, M. Pepper, and D.J. Rawlings.
References
- Alamyar, E., Duroux P., Lefranc M.-P., and Giudicelli V.. 2012. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 882:569–604. 10.1007/978-1-61779-842-9_32 [DOI] [PubMed] [Google Scholar]
- Altman, M.C., Reeves S.R., Parker A.R., Whalen E., Misura K.M., Barrow K.A., James R.G., Hallstrand T.S., Ziegler S.F., and Debley J.S.. 2018. Interferon response to respiratory syncytial virus by bronchial epithelium from children with asthma is inversely correlated with pulmonary function. J. Allergy Clin. Immunol. 142:451–459. 10.1016/j.jaci.2017.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa, T., and Wen J.. 2001. Determination of carbohydrate contents from excess light scattering. Anal. Biochem. 299:158–161. 10.1006/abio.2001.5432 [DOI] [PubMed] [Google Scholar]
- Barnes, C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. 2020. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 588:682–687. 10.1038/s41586-020-2852-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer, J.W., Randall T.D., Parkhouse R.M., and Corley R.B.. 1994. Mechanism and subcellular localization of secretory IgM polymer assembly. J. Biol. Chem. 269:17338–17348. 10.1016/S0021-9258(17)32559-0 [DOI] [PubMed] [Google Scholar]
- Brouwer, P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. 2020. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 369:643–650. 10.1126/science.abc5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen, G., Galson J.D., Hall G., Villar P., Moreels L., Ledsgaard L., Mattiuzzo G., Bentley E.M., Masters E.W., Tang D., et al. 2021. Cross-reactive SARS-CoV-2 neutralizing antibodies from deep mining of early patient responses. Front. Immunol. 12:678570. 10.3389/fimmu.2021.678570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegari, I., Schneider M., Berloffa G., Mühlethaler T., Holdermann S., Galli E., Roloff T., Boss R., Infanti L., Khanna N., et al. 2022. Potent neutralization by monoclonal human IgM against SARS-CoV-2 is impaired by class switch. EMBO Rep. 23:e53956. 10.15252/embr.202153956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.A., Clark L.E., Pan J., Coscia A., McKay L.G.A., Shankar S., Johnson R.I., Brusic V., Choudhary M.C., Regan J., et al. 2021. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell. 184:2605–2617.e18. 10.1016/j.cell.2021.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. 2020. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 12:513. 10.3390/v12050513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowsky, D.M., and Shao Z.. 2009. The human IgM pentamer is a mushroom-shaped molecule with a flexural bias. Proc. Natl. Acad. Sci. USA. 106:14960–14965. 10.1073/pnas.0903805106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan, J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. 2021. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 371:eabf4063. 10.1126/science.abf4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erasmus, J.H., Khandhar A.P., O’Connor M.A., Walls A.C., Hemann E.A., Murapa P., Archer J., Leventhal S., Fuller J.T., Lewis T.B., et al. 2020. An Alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci. Transl. Med. 12:eabc9396. 10.1126/scitranslmed.abc9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimidi, R.P., Klein J.S., Politzer M.S., Bai S., Seaman M.S., Nussenzweig M.C., West A.P., and Bjorkman P.J.. 2015. Intra-spike crosslinking overcomes antibody evasion by HIV-1. Cell. 160:433–446. 10.1016/j.cell.2015.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser, R., Cloutier M., Prévost J., Fink C., Ducas É., Ding S., Dussault N., Landry P., Tremblay T., Laforce-Lavoie A., et al. 2021. Major role of IgM in the neutralizing activity of convalescent plasma against SARS-CoV-2. Cell Rep. 34:108790. 10.1016/j.celrep.2021.108790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney, A.J., Starr T.N., Barnes C.O., Weisblum Y., Schmidt F., Caskey M., Gaebler C., Cho A., Agudelo M., Finkin S., et al. 2021. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 12:4196. 10.1038/s41467-021-24435-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney, A.J., Starr T.N., Eguia R.T., Loes A.N., Khan K., Karim F., Cele S., Bowen J.E., Logue J.K., Corti D., et al. 2022. A SARS-CoV-2 variant elicits an antibody response with a shifted immunodominance hierarchy. PLoS Pathog. 18:e1010248. 10.1371/journal.ppat.1010248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. 2020. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 369:1010–1014. 10.1126/science.abd0827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., COVID-19 Genomics UK COG-UK Consortium, Peacock S.J., and Robertson D.L.. 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 19:409–424. 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughey, C.T., Brewer J.W., Colosia A.D., Rosse W.F., and Corley R.B.. 1998. Production of IgM hexamers by normal and autoimmune B cells: Implications for the physiologic role of hexameric IgM. J. Immunol. 161:4091–4097 [PubMed] [Google Scholar]
- Hunt, A.C., Case J.B., Park Y.-J., Cao L., Wu K., Walls A.C., Liu Z., Bowen J.E., Yeh H.-W., Saini S., et al. 2022. Multivalent designed proteins neutralize SARS-CoV-2 variants of concern and confer protection against infection in mice. Sci. Transl. Med. 14:eabn1252. 10.1126/scitranslmed.abn1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyt, B.A., Baliga R., Sinclair A.M., Carroll S.F., and Peterson M.S.. 2020. Structure, function, and therapeutic use of IgM antibodies. Antibodies. 9:53. 10.3390/antib9040053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, W., Zhou J.Q., Horvath S.C., Schmitz A.J., Sturtz A.J., Lei T., Liu Z., Kalaidina E., Thapa M., Alsoussi W.B., et al. 2022. Germinal centre-driven maturation of B cell response to mRNA vaccination. Nature. 604:141–145. 10.1038/s41586-022-04527-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kober, C., Manni S., Wolff S., Barnes T., Mukherjee S., Vogel T., Hoenig L., Vogel P., Hahn A., Gerlach M., et al. 2022. IgG3 and IgM identified as key to SARS-CoV-2 neutralization in convalescent plasma pools. PLoS One. 17:e0262162. 10.1371/journal.pone.0262162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreer, C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K., et al. 2020. Longitudinal isolation of potent near-Germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell. 182:843–854.e12. 10.1016/j.cell.2020.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzberger, N., Hirsch C., Chai K.L., Tomlinson E., Khosravi Z., Popp M., Neidhardt M., Piechotta V., Salomon S., Valk S.J., et al. 2021. SARS-CoV-2-neutralising monoclonal antibodies for treatment of COVID-19. Cochrane Database Syst. Rev. 9:CD013825. 10.1002/14651858.CD013825.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurty, A.T., Thouvenel C.D., Portugal S., Keitany G.J., Kim K.S., Holder A., Crompton P.D., Rawlings D.J., and Pepper M.. 2016. Somatically hypermutated plasmodium-specific IgM+ memory B cells are rapid, plastic, early responders upon malaria rechallenge. Immunity. 45:402–414. 10.1016/j.immuni.2016.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, Z., Xie X., Hinton P.R., Liu X., Ye X., Muruato A.E., Ng D.C., Biswas S., Zou J., Liu Y., et al. 2021. Nasal delivery of an IgM offers broad protection from SARS-CoV-2 variants. Nature. 595:718–723. 10.1038/s41586-021-03673-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer, K., Bettini E., Parvathaneni K., Painter M.M., Agarwal D., Lundgreen K.A., Weirick M., Muralidharan K., Castaño D., Goel R.R., et al. 2022. Germinal center responses to SARS-CoV-2 mRNA vaccines in healthy and immunocompromised individuals. Cell. 185:1008–1024.e15. 10.1016/j.cell.2022.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenti, M.V., Aronico N., Pellegrino I., Boveri E., Giuffrida P., Borrelli de Andreis F., Morbini P., Vanelli L., Pasini A., Ubezio C., et al. 2020. Depletion of circulating IgM memory B cells predicts unfavourable outcome in COVID-19. Sci. Rep. 10:20836. 10.1038/s41598-020-77945-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast, F.D., Fridy P.C., Ketaren N.E., Wang J., Jacobs E.Y., Olivier J.P., Sanyal T., Molloy K.R., Schmidt F., Rutkowska M., et al. 2021. Highly synergistic combinations of nanobodies that target SARS-CoV-2 and are resistant to escape. Elife. 10:e73027. 10.7554/eLife.73027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, M.L. 2022. Molecular mechanisms of multimeric assembly of IgM and IgA. Annu. Rev. Immunol. 40:221–247. 10.1146/annurev-immunol-101320-123742 [DOI] [PubMed] [Google Scholar]
- Matz, H., Munir D., Logue J., and Dooley H.. 2021. The immunoglobulins of cartilaginous fishes. Dev. Comp. Immunol. 115:103873. 10.1016/j.dci.2020.103873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud, E., Mastrandrea C., Rochereau N., and Paul S.. 2020. Human secretory IgM: An elusive player in mucosal immunity. Trends Immunol. 41:141–156. 10.1016/j.it.2019.12.005 [DOI] [PubMed] [Google Scholar]
- Micol, R., Kayal S., Mahlaoui N., Beauté J., Brosselin P., Dudoit Y., Obenga G., Barlogis V., Aladjidi N., Kebaili K., et al. 2012. Protective effect of IgM against colonization of the respiratory tract by nontypeable Haemophilus influenzae in patients with hypogammaglobulinemia. J. Allergy Clin. Immunol. 129:770–777. 10.1016/j.jaci.2011.09.047 [DOI] [PubMed] [Google Scholar]
- Moh, E.S.X., Lin C.-H., Thaysen-Andersen M., and Packer N.H.. 2016. Site-specific N-Glycosylation of recombinant pentameric and hexameric human IgM. J. Am. Soc. Mass Spectrom. 27:1143–1155. 10.1007/s13361-016-1378-0 [DOI] [PubMed] [Google Scholar]
- Muller, N.F., Wagner C., Frazar C.D., Roychoudhury P., Lee J., Moncla L.H., Pelle B., Richardson M., Ryke E., Xie H., et al. 2021. Viral genomes reveal patterns of the SARS-CoV-2 outbreak in Washington State. Sci. Transl. Med. 13:eabf0202. 10.1126/scitranslmed.abf0202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel, K.G., Clark S.A., Shankar S., Pan J., Clark L.E., Yang P., Coscia A., McKay L.G.A., Varnum H.H., Brusic V., et al. 2022. Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain. Science. 375:eabl6251. 10.1126/science.abl6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell, K.L., Clemmer D.C., Cox J.B., Kayode Y.I., Zoccoli-Rodriguez V., Taylor H.E., Endy T.P., Wilmore J.R., and Winslow G.M.. 2021. Switched and unswitched memory B cells detected during SARS-CoV-2 convalescence correlate with limited symptom duration. PLoS One. 16:e0244855. 10.1371/journal.pone.0244855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenbrink, M.S., Park J.-G., Oladunni F.S., Deshpande A., Basu M., Sarkar S., Loos A., Woo J., Lovalenti P., Sloan D., et al. 2021. Therapeutic activity of an inhaled potent SARS-CoV-2 neutralizing human monoclonal antibody in hamsters. Cell Rep. Med. 2:100218. 10.1016/j.xcrm.2021.100218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. 2020. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 583:290–295. 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- Pisil, Y., Yazici Z., Shida H., and Miura T.. 2021. Is SARS-CoV-2 neutralized more effectively by IgM and IgA than IgG having the same Fab region? Pathogens. 10:751. 10.3390/pathogens10060751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas, D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. 2021. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 596:276–280. 10.1038/s41586-021-03777-9 [DOI] [PubMed] [Google Scholar]
- Purtha, W.E., Tedder T.F., Johnson S., Bhattacharya D., and Diamond M.S.. 2011. Memory B cells, but not long-lived plasma cells, possess antigen specificities for viral escape mutants. J. Exp. Med. 208:2599–2606. 10.1084/jem.20110740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda, L.B., Morawski P.A., Pruner K.B., Fahning M.L., Howard C.A., Franko N., Logue J., Eggenberger J., Stokes C., Golez I., et al. 2022. Imprinted SARS-CoV-2-specific memory lymphocytes define hybrid immunity. Cell. 185:1588–1601.e14. 10.1016/j.cell.2022.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda, L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. 2021. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 184:169–183.e17. 10.1016/j.cell.2020.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero, A., Piubelli C., Calciano L., Accordini S., Valenti M.T., Carbonare L.D., Siracusano G., Temperton N., Tiberti N., Longoni S.S., et al. 2022. SARS-CoV-2 vaccination elicits unconventional IgM specific responses in naïve and previously COVID-19-infected individuals. EBioMedicine. 77:103888. 10.1016/j.ebiom.2022.103888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujas, E., Kucharska I., Tan Y.Z., Benlekbir S., Cui H., Zhao T., Wasney G.A., Budylowski P., Guvenc F., Newton J.C., et al. 2021. Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers. Nat. Commun. 12:3661. 10.1038/s41467-021-23825-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar, M., Rappazzo C.G., Wieland-Alter W.F., Hsieh C.-L., Wrapp D., Esterman E.S., Kaku C.I., Wec A.Z., Geoghegan J.C., McLellan J.S., et al. 2021. Prolonged evolution of the human B cell response to SARS-CoV-2 infection. Sci. Immunol. 6:eabg6916. 10.1126/sciimmunol.abg6916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsudin, F., Yeo J.Y., Gan S.K., and Bond P.J.. 2020. Not all therapeutic antibody isotypes are equal: The case of IgM versus IgG in pertuzumab and trastuzumab. Chem. Sci. 11:2843–2854. 10.1039/C9SC04722K [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., and Bloom J.D.. 2021. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 371:850–854. 10.1126/science.abf9302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, L., Zhu G., Yang M., Cao L., Xing X., Yin G., Chan C., Qin C., Rao Z., Wang X., et al. 2021. Nanometer-resolution in situ structure of the SARS-CoV-2 postfusion spike protein. Proc. Natl. Acad. Sci. USA. 118:e2112703118. 10.1073/pnas.2112703118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, C.W., Chia W.N., Qin X., Liu P., Chen M.I.-C., Tiu C., Hu Z., Chen V.C.-W., Young B.E., Sia W.R., et al. 2020. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. 38:1073–1078. 10.1038/s41587-020-0631-z [DOI] [PubMed] [Google Scholar]
- Taylor, J.J., Pape K.A., and Jenkins M.K.. 2012. A germinal center–independent pathway generates unswitched memory B cells early in the primary response. J. Exp. Med. 209:597–606. 10.1084/jem.20111696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thouvenel, C.D., Fontana M.F., Netland J., Krishnamurty A.T., Takehara K.K., Chen Y., Singh S., Miura K., Keitany G.J., Lynch E.M., et al. 2021. Multimeric antibodies from antigen-specific human IgM+ memory B cells restrict Plasmodium parasites. J. Exp. Med. 218:e20200942. 10.1084/jem.20200942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viant, C., Wirthmiller T., ElTanbouly M.A., Chen S.T., Cipolla M., Ramos V., Oliveira T.Y., Stamatatos L., and Nussenzweig M.C.. 2021. Germinal center–dependent and –independent memory B cells produced throughout the immune response. J. Exp. Med. 218:e20202489. 10.1084/jem.20202489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora, G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Wang, Z., Lorenzi J.C.C., Muecksch F., Finkin S., Viant C., Gaebler C., Cipolla M., Hoffmann H.-H., Oliveira T.Y., Oren D.A., et al. 2021. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 13:eabf1555. 10.1126/scitranslmed.abf1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Xu K., Jung S., Conte A., Lieberman J., Muecksch F., Lorenzi J.C.C., Park S., Schmidt F., Wang Z., et al. 2021. Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants. Nature. 595:278–282. 10.1038/s41586-021-03676-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., and Wilson I.A.. 2020. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 368:630–633. 10.1126/science.abb7269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Zhang H., and Sun L.. 2022. a. Therapeutic antibodies for COVID-19: Is a new age of IgM, IgA and bispecific antibodies coming? MAbs. 14:2031483. 10.1080/19420862.2022.2031483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S., et al. 2020. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 11:6013. 10.1038/s41467-020-19808-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Mateus J., Coelho C.H., Dan J.M., Moderbacher C.R., Gálvez R.I., Cortes F.H., Grifoni A., Tarke A., Chang J., et al. 2022. b. Humoral and Cellular Immune Memory to Four COVID-19 Vaccines. Cell. 185:2434–2451.e17. 10.1016/j.cell.2022.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows characteristics of singly sorted RBD-specific IgM+ B cells from which BCRs were cloned and tested.