Abstract
Drought is one of the most critical abiotic stresses, which significantly impair rapeseed (Brassica napus L.) productivity. Several factors can regulate the stress response, including changes in gene expression in biological pathways, extensive protein interaction networks, and post-translational regulatory factors like microRNAs. External factors can also affect the intensity of the stress response. Therefore, this study investigated protein–protein interactions of some essential genes involved in abscisic acid (ABA) production, antioxidant system, and Krebs cycle. The expression of phyton synthase (PSY), 9-cis-epoxycarotenoid dioxygenase (NCED3), aldehyde oxidase (AAO3), thioredoxin reductase (NTRC), and glutathione reductase (GR) genes in two rapeseed genotypes, i.e., Hyola308 (drought-sensitive) and SLM046 (drought-tolerant) were evaluated using qRT-PCR technique under 72 h of drought stress and methanol foliar application. In the SLM046 (tolerant) genotype, the expression levels of PYS, NCED, AAO3, and GR genes were increased after 8 h of foliar application. The expression level of the NTR gene was increased 8 and 24 h after stress and methanol treatment. In the Hyola308 genotype, PYS, AAO3, NTR, and GR genes' expression level was increased 8 h after methanol foliar application, and the NCED gene was increased 24 h after stress with methanol treatment. In general, methanol foliar application increased the expression levels of several genes. Particularly, the gene expression was considerably higher in the SLM046 genotype than in Hyola308. Bioinformatics prediction of microRNAs targeting PSY, NCED, GR, NTRC, and AAO3 genes was performed, and 38, 38, 13, 11, and 11 microRNAs were predicted for these genes, respectively. The study of effective microRNAs showed that sometimes more than one type of microRNA could affect the desired gene, and in some cases, a conserved family of microRNAs caused the main effect on gene expression. Overall, our results lay the foundation for functional characterization of these genes or gene-miRNA modules in regulating drought stress tolerance in rapeseed.
Keywords: Carotenoids, Gene expression, microRNA, Protein interaction, STRING
Introduction
Rapeseed (Brassica napus. L) is one of the most valuable oilseeds crops belonging to Brassicaceae family. Studies have shown that water scarcity in the various stages of growth affects the quantity and the quality of oil production (Raza et al. 2020). Many genes are induced under drought stress in plants, and different organs are affected. Drought stress in plants leads to inhibition of photosynthesis and respiration in organs such as chloroplasts and mitochondria, accumulation of reactive oxygen species (ROS), and reprogramming of gene expression (Liu et al. 2021; Meng et al. 2020; Raza et al. 2020; Rezaie et al. 2020). Productions from these genes appear to be involved in stress tolerance, regulation of gene expression, and signal response to stress response (Ito et al. 2006).
Under dehydration conditions, abscisic acid (ABA) plays an essential role in expressing stress-responsive genes and stomatal closure. Plants use the carotenoid pathway as an indirect pathway for ABA synthesis. ABA biosynthesis begins with zeaxanthin, and zeaxanthin epoxidase (ZEP) converts zeaxanthin into violaxanthin (Ali et al. 2020). The phyton synthase gene (PSY3) also performs a specialized role in controlling ABA biosynthesis in response to abiotic stress (Li et al. 2007). The enzyme 9-cis-epoxycarotenoid dioxygenase (NCED) is critical in ABA biosynthesis. In Arabidopsis, the enzyme aldehyde oxidase (AAO3) accelerates the finishing stage of ABA biosynthesis (Seo et al. 2004). AAO3 was found to perform an essential role in ABA biosynthesis (Mittler et al. 2004). Thioredoxin (TRX) plays a key role in plant tolerance to oxidative stress (Dos Santos and Rey 2006). TRX uses Ferredoxin-dependent heterodimeric thioredoxin reductase (FTR) or NADPH-dependent thyrotoxin reductases (NTR) to reduce itself (Reichheld et al. 2005). NTRs are vital regulatory enzymes that modulate the redox state of the Trx system in plants. NTRs directly reduce ROS and lead to stress tolerance in plants. Arabidopsis has three NTR isoforms, NTRA and NTRB, found in the cytoplasm and mitochondria, and NTRC in chloroplasts. The important role of NTRC has been proven in plant growth and development and response to environmental conditions (Danaeipour and Haddad 2020; Pérez-Ruiz et al. 2006). The essential role of NTRC in response to abiotic stresses was also reported (Da et al. 2017). The study on Arabidopsis showed that the higher expression of the NTR gene increased tolerance to drought and photo-oxidative stresses. Increased NTR expression helped maintain the homeostasis of ROS under stress conditions (Kim et al. 2017).
Antioxidant defense mechanisms have been developed in plants to reduce the damaging effects of ROS on plant cells (Kalisz et al. 2016; Rezaie et al. 2020). Vital members of this defense system include superoxide dismutase (SOD), ascorbate peroxidase (APX), peroxidase (POD), glutathione reductase (GR), catalase (CAT), etc. The study on cassava showed that the antioxidant response under drought stress might be associated with increased accumulation of ascorbate (AsA) and glutathione (GSH) and higher activities of SOD and CAT. However, in some of its genotypes, it mainly depended on total phenolic accumulation (TP) and increased GR activity (Chen et al. 2020). SOD catalyzes O2 and H2O2 from superoxide (O2−) and consequently reduces its adverse effects. CAT is an antioxidant enzyme found in all aerobic organisms that catalyze H2O2 to water and oxygen in cells (Raza et al. 2021). Another enzyme, GR, plays a significant role in adapting to plants' oxidative stress (Salin 1991). This enzyme is responsible for converting oxidized glutathione to reduced glutathione and maintaining a high ratio of GSH to GSSG (Noctor and Foyer 1998). The alternative oxidase pathway is critical to reducing ROS production in chloroplast and mitochondrial organs (Mittler 2002). This process is carried out by the alternative oxidase enzyme (AOX). The lack of the AOX1 gene in the plant reduces the ability to grow or tolerate stresses (Mohsenzadeh Golafazani et al. 2017). Alternative oxidase also plays a role in reducing cell damage under various environmental stresses. Therefore, many recent studies have focused on its role in response to salinity and drought stress (Li et al. 2013). One of the pathways affected by drought stress is the Krebs cycle (TCA), located in the mitochondrial matrix (Ramezanzadeh Bishegahi et al. 2021). TCA cycle enzymes are susceptible to oxidative stress (Fernie et al. 2004). Recent study showed that drought stress could affect the expression of pyruvate dehydrogenase 1 (PDH1), isocitrate dehydrogenase 1 (IDH1), and fumarase 1 (FUM1) genes involved in the Krebs cycle (Ramezanzadeh Bishegahi et al. 2021).
Foliar applications of aqueous methanol have been reported to increase yield, accelerate maturity, and reduce drought stress and irrigation requirements in crops grown in dry environments (Nonomura and Benson 1992; Ramezanzadeh Bishegahi et al. 2021). The increased growth and yield were attributed to the action of methanol as a C nutrient and as an inhibitor of photorespiration (Nonomura and Benson 1992). In another study, various crops such as tomato, bean, sugar beet, and oil seed rape treated with 20–30% methanol solutions showed an increase in yield (Zbieć et al. 2003). Methanol treatment on Arabidopsis and tobacco plants increased their fresh and dry weight (Ramírez et al. 2006). Plants can easily absorb methanol sprayed on leaves and utilize it as a carbon source (Hemming et al. 1995; Khalilzadeh et al. 2020). Rapid oxidation to CO2 combined with methanol foliar application delays leaf aging by acting on stimulants of ethylene production in the plant. Also, it increases leaf surface durability and increases the active photosynthetic period (Zbieć et al. 2003). Recent study on drought stress on two rapeseed genotypes, Hyola308 and SLM046, revealed that methanol treatment significantly affected the expression profiles of IDH1, PDH, FUM, TRX, and AOX genes and was able to modulate the effect of drought stress (Ramezanzadeh Bishegahi et al. 2021). Microarray-based gene expression study of 484 transcripts in leaves of Arabidopsis thaliana harvested at 1, 24, and 72 h after methanol treatment revealed that only two transcripts were significantly down-regulated, and the remainder were up-regulated. These included cytochrome P450’s, glucosyl transferases, and the ABC transporter family members. Also, the analysis revealed an increase in the number of ABA- and auxin-related transcripts, aldehyde oxidase, and glutathione S-transferase. Therefore, methanol treatment led to the expression of hundreds of genes, and multiple detoxifications and signaling pathways were activated (Downie et al. 2004).
One of the most useful, standard, and widespread types of biomolecular networks is the protein–protein network. This network contains all the genes encoding proteins in a particular genome and highlights their functional relationship (Szklarczyk et al. 2021). Since proteins act primarily as interacting networks, protein–protein interaction networks can be considered as a valuable indicator for understanding cellular processes (Taghvaei et al. 2019). Examining these relationships and pathways of protein interactions provides researchers with very beneficial information. MicroRNAs are post-transcriptional gene regulatory elements influencing the expression of proteins. If these 21 to 22 nucleotide sequences bind to the gene sequence, they can regulate the activity of that gene. In other words, if the binding to the target region is complete, it leads to cleavage. On the other hand, if the binding is incomplete, it prevents the translation of the target gene (Bartel 2004).
Bioinformatics study of the genes network and microRNAs affecting the expression of specific genes cause removing extra data. Therefore, the desired data can be obtained more quickly and reduce the cost and time required in experimental tests. Accurately identifying the genes associated with drought stress provides the basis for improving drought tolerance in rapeseed. Expression data of mRNA transcripts under drought stress and non-stress conditions in sensitive and tolerant genotypes using qRT-PCR technique; data analysis of protein–protein interaction networks and microRNAs can provide a proper basis for improving drought stress tolerance in rapeseed as well as other similar plants. Therefore, in this study, both tolerant and sensitive rapeseed genotypes were studied to investigate their response to drought stress and treatment with methanol.
Materials and methods
Plant materials and treatment conditions
Drought-sensitive (Hayola308) and drought-tolerant (SLM046) genotypes’ seeds were obtained from the Seed and Plant Improvement Institute (SPII), Iran (Mirzai et al. 2013). The seeds were placed 4 days in a petri dish using water and filter paper for germination in the biotechnology laboratory of Guilan University, Iran. Then at least three seedlings were planted in the soil in each 1 kg plastic pot (90 pots in total). The pots were placed in a growth chamber under a 16-h light (25 °C)/8-h dark (22 °C) photoperiod (photo intensity 6000 Lx) and were watered daily until sampling. Statistically, it was performed as a completely randomized factorial design in three replications with three factors, including methanol, genotype, and time. When the tolerant and sensitive plants reached the 3 to 4-leaf stage, they were divided into three groups, including: Control plants (CI) that were regularly irrigated, drought-stressed plants (DR) that were exposed to drought stress for 72 h (30% of field capacity) and methanol-sprayed (DM) plants under 72-h drought stress. The single-step spraying of methanol solution 20% v/v (Methanol, 99.9%, Thermo Scientific™) was performed after 72-h drought stress on the DM plants with a hand-held sprayer at a rate of 20 ml per pot, with the nozzle approximately 20 cm above the leaf surface (Armand et al. 2016; Ramírez et al. 2006). The CI, DR, and DM plants’ leaves were harvested at 0 (72 h after water cut-off), 8, 24 h, and immediately frozen in liquid nitrogen, and stored in a freezer at −80° C for further analysis, RNA extraction and cDNA synthesize.
RNA extraction and cDNA synthesize
Total RNA was extracted using a total RNA isolation kit (DENAzist, IRAN) according to the manufacturer’s instructions. The extracted RNA’s quantity and quality were examined by Nanodrop® One (Thermo Fisher Scientific®) and 1% agarose gel electrophoresis. RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA) was used to synthesize cDNA following the manufacturer’s instructions. cDNA was stored at – 80 C freezer.
Analysis of protein–protein interactions
The genes involved in the production of ABA, antioxidant system, and Krebs cycle, were selected to investigate their relationship and analyze their protein–protein interactions. Information and genetic identity of selected genes were taken using the UniProt database (Table 1). Afterward, it was utilized in the STRING database to identify protein–protein interactions (PPIs). This database contains information from various sources, including experimental data, computational forecasting methods, and it is updated continuously. In this database, each interaction is given a score from 0 to 1, and the minimum required interaction score was set to medium (0.4) (Szklarczyk et al. 2019; Taghvaei et al. 2022).
Table 1.
Selected important genes involved in the production of ABA, antioxidant system, Krebs cycle using Arabidopsis thaliana as a reference plant
| Gene names | Identifier | UniProt | |
|---|---|---|---|
| Glutathione reductase | AT3G54660 | GR | https://www.uniprot.org/uniprot/P42770 |
| NADPH-dependent thioredoxin reductase 3 | AT2G41680 | NTRC | https://www.uniprot.org/uniprot/O22229 |
| Aldehyde oxidase 3 | AT2G27150 | AAO3 | https://www.uniprot.org/uniprot/Q7G9P4 |
| 9-cis-epoxycarotenoid dioxygenase | AT3G14440 | NCED3 | https://www.uniprot.org/uniprot/Q9LRR7 |
| Phytoene synthase | AT5G17230 | PSY | https://www.uniprot.org/uniprot/P37271 |
| Isocitrate dehydrogenase | AT4G35260 | IDH1 | https://www.uniprot.org/uniprot/Q8LFC0 |
| Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex | AT3G52200 | LTA3 | https://www.uniprot.org/uniprot/Q0WQF7 |
| Fumarate hydratase | AT2G47510 | FUM1 | https://www.uniprot.org/uniprot/P93033 |
| Thioredoxin | AT5G42980 | TRX3 | https://www.uniprot.org/uniprot/Q42403 |
| Alternative oxidase | AT3G22370 | AOX1A | https://www.uniprot.org/uniprot/Q39219 |
| Zeaxanthin epoxidase, chloroplastic | AT5G67030 | ZEP | https://www.uniprot.org/uniprot/Q9FGC7 |
| Catalase-2 | AT4G35090 | CAT | https://www.uniprot.org/uniprot/P25819 |
| Superoxide dismutase [Cu–Zn] 1 | AT1G08830 | CSD1 | https://www.uniprot.org/uniprot/P24704 |
| Fumarate hydratase 2 | AT5G50950 | FUM2 | https://www.uniprot.org/uniprot/Q9FI53 |
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit 2, mitochondrial | AT2G18450 | SDH1-2 | https://www.uniprot.org/uniprot/Q9ZPX5 |
| Malate dehydrogenase 1, mitochondrial | AT1G53240 | mMDH1 | https://www.uniprot.org/uniprot/Q9ZP06 |
| NADPH-dependent thioredoxin reductase 2 | AT2G17420 | NTRA | https://www.uniprot.org/uniprot/Q39242 |
| NADPH-dependent thioredoxin reductase 1 | AT4G35460 | NTRB | https://www.uniprot.org/uniprot/Q39243 |
| External alternative NAD(P)H-ubiquinone oxidoreductase B2, mitochondrial | AT4G05020 | NDB2 | https://www.uniprot.org/uniprot/Q94BV7 |
| External alternative NAD(P)H-ubiquinone oxidoreductase B4, mitochondrial | AT2G20800 | NDB4 | https://www.uniprot.org/uniprot/Q9SKT7 |
| Internal alternative NAD(P)H-ubiquinone oxidoreductase A2, mitochondrial | AT2G29990 | NDA2 | https://www.uniprot.org/uniprot/O80874 |
Design of specific primers
After examining the proteins' interaction and their functional relationships with each other, the candidate genes such as PSY, NCED3, GR, NTRC, and AAO3 genes were selected to evaluate the expression using qRT-PCR. The actin gene was used as a reference gene to normalize the data (Wang et al. 2011). Sequences of the desired genes PSY, NCED3, GR, NTRC, and AAO3 genes were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/). The primers were designed using Tcoffee (http://tcoffee.crg.cat/apps/tcoffee/do:mcoffee) and primer3 (http://primer3.ut.ee) (Table 2). Finally, the primers were checked for specificity in NCBI Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast).
Table 2.
Specifications of primers used in real-time PCR reaction
| Gene | Primer sequence | Melt ( °C) | PCR product | NCBI accession number |
|---|---|---|---|---|
|
Actin-F Actin-R |
5-TCCCGAGTATTGTTGGTCGT-3´ 5-TCCATGTCATCCCAGTTGCT-3´ |
54 | 157 | AF111812 |
|
AAO3-F AAO3-R |
5-GAAGCAACCCACACGAGTTT-3´ 5-AACCTCAGCCTCAAGACTCC-3´ |
54 | 175 | XM_013862421.2 |
|
PSY-F PSY-R |
5-TCGCTGATACCGTTGCTAGA-3´ 5-TGTCGTTGCTTTGGACTTGG-3´ |
54 | 197 | XM_013827623.2 |
|
NCED-F NCED-R |
5-AAGCAGTCCTCAACCTCTCC-3´ 5-TCTGAACACTAGGATCGGCG-3´ |
54 | 178 | XM_013788802 |
|
GR -F GR -R |
5´-GAGCAGATGTCTTTAAGAGGC-3´ 5´-CTTTGTATTAGGCTTGCGAC-3´ |
53.3 | 159 | XM_013855797.1 |
|
NTRC -F NTRC-R |
5-GCTGTAACTGCTGCTGGATC-3´ 5-ACTGTCCCCGATGCTTTGTA-3´ |
53.8 | 178 | XM_013892363.1 |
Investigation of gene expression by real-time PCR
Expression of selected genes was performed by qRT-PCR in cultivars SLM046 (tolerant) and Hyola308 (sensitive). The reactions were carried out in a LightCycler 96 real-time PCR machine (Roche, Basel, Switzerland) with the following amplification conditions: activation at 50 °C for 2 min; 95 °C for 2 min; followed by 40 cycles at 95 °C for 15 s and 58 °C for the 20 s; and 72 °C for 15 s and final holding at 4 °C. All reactions were performed in three biological replicates, each with three technical replicates. The Livak equation was used to compare the relative expression of the desired genes in real-time PCR (Livak and Schmittgen 2001). Graphs were constructed using Excel 2016. The standard error (SE) was used to compare the columns, and the relative expression rate was considered double (Mohsenzadeh Golfazani et al. 2019).
Identification of microRNAs targeting the studied genes
All microRNA sequences belong to the Brassicaceae family plants, including Arabidopsis thaliana, Arabidopsis lyrata, Brassica oleracea, Brassica rapa, Brassica napus, and Camelina sativa were obtained from the miRbase database (https://www.mirbase.org) (Release 22.1, October 2018). To identify microRNAs targeting PSY, NCED, GR, NTRC, and AAO3 genes, MicroRNA prediction was performed with psRNATarget database (expected value parameter 5) using CDS ( https://www.zhaolab.org/psRNATarget/) (Dai et al. 2018). Cytoscape software version 3.8 was additionally used to map the relationship between the identified microRNAs and the studied genes (Shannon et al. 2003).
Results and discussion
Phenotypic changes of rapeseed after drought stress and methanol foliar spray
The Hyola308 genotype showed signs of gradual wilting after drought stress compared to the control. In contrast, the SLM046 genotype wilting symptoms were seen in the early hours under drought stress, and then in the following hours of 72-h drought stress, the wilting symptoms gradually disappeared, the wilting symptoms gradually disappeared. Examination of both genotypes after methanol foliar application under drought stress showed that the plants’ freshness recovered again. It seems that methanol treatment increased plant tolerance to drought stress (Fig. 1A, B).
Fig. 1.

The treated plants were separated into two groups: drought-stressed plants (DR) that were exposed to drought stress for 72 h (30% of field capacity) and methanol-sprayed (DM) plants which sprayed with methanol solution 20% v/v after 72-h drought stress. A Hyola308 (drought-sensitive) genotype: DM and DR plants. B SLM046 (drought-tolerant) genotype: DM and DR plants
Protein–protein interaction network
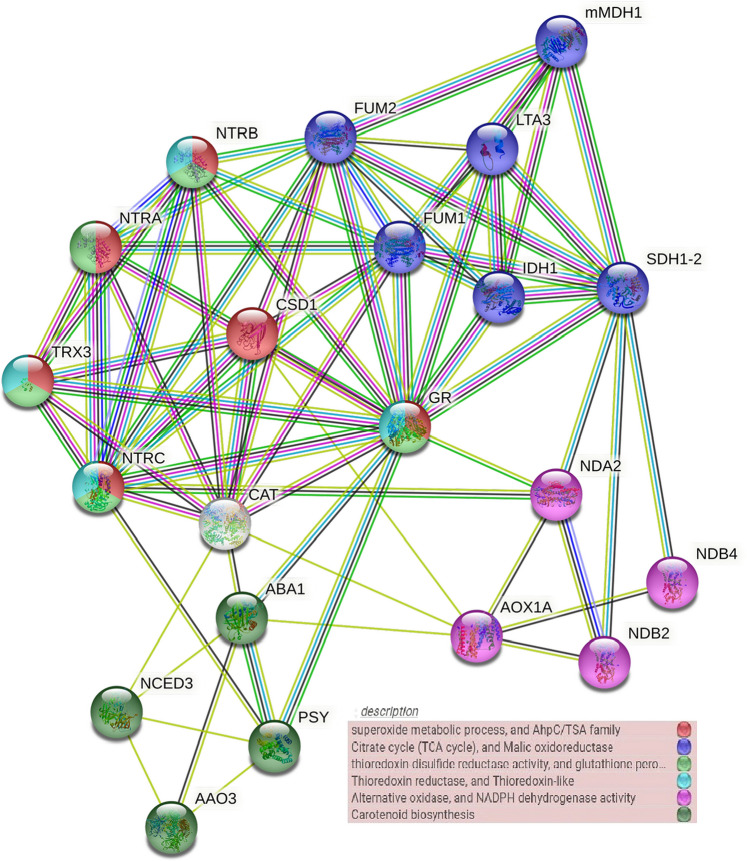
In this study, the STRING platform was utilized to investigate protein interactions in which nodes represent proteins and lines represent known or predicted and direct or indirect interactions. The relationship between the nodes was based precisely on the present information extracted from relevant databases and articles. Accordingly, the results of the interactions between the genes (Table 1) indicated 21 nodes and 72 edges (Fig. 2), and all of them were located in the network.
Fig. 2.
Protein–protein interaction network (PPIs) using the STRING database. Colored nodes represent proteins, and each color represents a cluster. Colored lines represent their interactions with each other based on the references in the database
Based on STRING clustering (Fig. 2 and Table 3), the studied genes were classified into clusters including, superoxide metabolic process (CL: 10,321), Citrate cycle (TCA cycle) (CL: 9184), thioredoxin-disulfide reductase activity (CL: 10,322), thioredoxin reductase (CL: 10,344), alternative oxidase (CL: 38,330), carotenoid biosynthesis (CL: 14,843), respectively. These results indicated that there were direct and indirect associations and interactions between Krebs cycle enzymes (FUM, SDHI, and IDH1), effective enzymes in oxidation–reduction reactions and scavenging ROS (CAT, CSD, NTR, and GR), alternative oxidase pathway (AOX1A), and ABA production processes (NCED, ABA1, AAO3, and PSY3) (Fig. 2 and Table 3).
Table 3.
Local network cluster based on STRING database
| #term ID | Term description | Observed gene count | Background gene count | Strength | False discovery rate |
|---|---|---|---|---|---|
| CL:10,321 | Superoxide metabolic process, and AhpC/TSA family | 6 | 31 | 2.4 | 1.13E-11 |
| CL:9184 | Citrate cycle (TCA cycle) and malic oxidoreductase | 6 | 85 | 1.96 | 7.17E-10 |
| CL:10,322 | Thioredoxin-disulfide reductase activity and glutathione peroxidase activity | 5 | 20 | 2.51 | 1.49E-10 |
| CL:10,344 | Thioredoxin reductase and Thioredoxin-like | 4 | 6 | 2.94 | 7.17E-10 |
| CL:38,330 | Alternative oxidase and NADPH dehydrogenase activity | 4 | 11 | 2.68 | 2.07E-09 |
| CL:14,843 | Carotenoid biosynthesis | 4 | 28 | 2.27 | 3.04E-08 |
| CL:38,332 | Alternative oxidase and extrinsic component of mitochondrial inner membrane | 3 | 5 | 2.89 | 6.86E-08 |
| CL:9306 | Malate metabolic process | 3 | 11 | 2.55 | 3.98E-07 |
| CL:9186 | Citrate cycle (TCA cycle) | 3 | 57 | 1.84 | 2.57E-05 |
| CL:14,881 | Abscisic acid biosynthetic process | 2 | 5 | 2.72 | 2.43E-05 |
| CL:14,848 | Carotenoid biosynthesis | 2 | 13 | 2.3 | 0.0001 |
| CL:9188 | Citrate cycle (TCA cycle) | 2 | 34 | 1.89 | 0.00058 |
ABA is involved in various abiotic stresses like drought, cold, salinity, and growth and development processes such as seed development, dormancy, germination, and stomatal movement (Li et al. 2021). Therefore, observing the interaction between effective enzymes involved in ABA biosynthesis, ROS scavenging and oxidative stress-reducing was expected. According to Fig. 2, a direct relationship was seen between ABA1 (ZEP), NCED3, PSY3, GR, CAT, and NTRC enzymes. Previous research reported that under drought stress conditions, the expression of Arabidopsis NCED gene (AtNCED3) was induced and controlled internal ABA levels (Bartel 2004; Iuchi et al. 2001). PSY3 was also found to induce ABA production under drought and salinity stress in Zea mays (Li et al. 2007). GR is an NADPH-dependent antioxidant enzyme and effectively promotes GSH reduction and tolerance to oxidative stress (Gill et al. 2013). According to Fig. 2, we observed direct interactions between GR and NTR enzymes and Krebs cycle enzymes (LTA3, FUM, IDH1, and SDH1) and enzymes affecting ABA production (PSY, and ABA1). Overall, based on strong interactions and genes expression, our results provide strong evidence for future characterization of the genes.
Expression changes of the studied genes
The biosynthesis of carotenoid and ABA hormone
In plants, carotenoids serve as vital antioxidant pigments in stress tolerance. These pigments control ROS and free radicals and maintain redox balance. Plants, under stress, manage the accumulation and degradation of carotenoids by regulating genes involved in carotenoid metabolism. Accumulation and degradation of carotenoids affect a complex process of gene regulation (Wang et al. 2021). Based on STRING clustering, PSY, NCED, and AAO3 genes were classified as carotenoid biosynthesis (CL: 14,843) (Table 3). The results of the gene network study (Fig. 2) revealed a direct association between the genes involved in ABA synthesis.
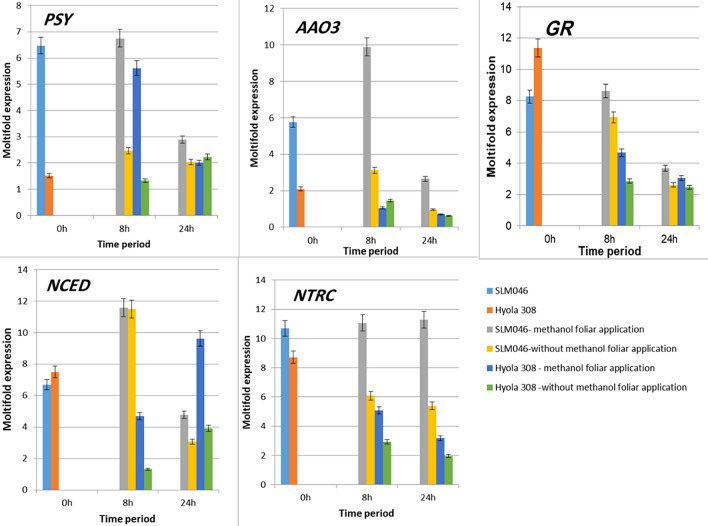
At the beginning of sampling (72 h without irrigation), an increase in the expression of PSY gene was observed in the drought-tolerant genotype (SLM046) (Fig. 3). Gene expression of DM samples showed that the highest gene expression was seen at 8 h after stress (6.7 times), and then it decreased at 24 h after stress. Nevertheless, PSY gene expression in DR samples had a decreasing trend at 8 and 24 h after stress. However, both cases of DM samples (SLM046 and Hyola308) showed higher gene expression than DR samples. In the SLM046 genotype, PSY gene expression in DM samples was about 6.7 times at 8 h after drought stress and almost three times at 24 h after stress. PSY gene expression in the drought-sensitive genotypes (Hyola308) increased slightly in the early hours of sampling compared to the tolerant genotype. In DM samples 8 h after stress, the highest gene expression (5.5 times) was observed and expression level was decreased at 24 h. PSY showed the lowest gene expression level (equivalent to 0 h) in DR samples at 8 h after drought stress and gradually increased over 24 h. Therefore, it seems that in sensitive plants, methanol foliar application has accelerated the response to drought stress. The PSY highest expression level was observed at 8 h in both DM genotypes under drought stress. PSY is the first reactive enzyme that limits the rate of carotenoid production, and the plant indirectly produces the ABA through this pathway (Li et al. 2020). This hormone plays a vital role as a regulator of development and in responding to drought stress (Ali et al. 2020). In both genotypes, PSY gene expression increased, leading to the production of the ABA. Also, PSY gene expression was increased by methanol application 8 h after stress. Methanol as an anti-stress agent (Hemming et al. 1995) increased the expression level of this gene, but it also decreased the expression level of PSY after 24 h. A recent study reported that salt stress and ABA could induce DcPSY2 expression by binding the AREB transcription factor (possibly DcAREB3) to ABRE cis-element in the promoter. Therefore, the expression of DcPSY2 in carrots increases carotenoid production, thereby increasing ABA levels and protecting the plant from abiotic stress (Simpson et al. 2018).
Fig. 3.
Expression pattern of PSY, NCED, GR, NTRC, AAO3 genes using real-time PCR under drought stress and methanol foliar application in seedling SLM046 and Hyola308
The NCED gene is a critical enzyme in ABA biosynthesis. NCED converts Neoxanthin into Xanthoxin, followed by the production of the phytohormone ABA (Wang et al. 2021). NCED gene expression increased in the tolerant genotype at 72 h without irrigation (Fig. 3). In both DM and DR samples in the drought-tolerant genotypes (SLM046), the NCED gene had the highest expression level at 8 h (11.5 and 11.4 times, respectively), and after 24 h, showed a decreasing trend. The expression pattern did not change in the DM samples, but the expression level in 24 h increased more than the DR samples. However, this expression pattern was similar to the PSY gene expression pattern in the tolerant plant. The tolerant plant reacts more rapidly with higher expression rates, and methanol spraying increases overall expression level. At 72 h without irrigation, the drought-sensitive genotypes (Hyola308) had higher expression of the NCED gene than DR samples (7.4 times). However, a similar expression pattern was observed in both DM and DR samples. In this way, initially, the expression decreased in the 8th hour and then increased in the 24th hour. However, methanol foliar application in DM samples increased the expression level to a large extent (5 times at 8 h and 9 times at 24 h). In both genotypes, methanol increased the expression of this gene as an anti-stress factor. Increased expression of this gene would be directly related to increasing ABA as a stress-responsive plant hormone. The production of ABA would control drought stress and produce responsive genes and closed pores in response to stress. The effect of methanol was various on gene expression in two genotypes at different hours. The drought-sensitive genotype had the highest expression level 24 h after methanol foliar application, while the drought-tolerant genotype had the highest expression level 8 h after methanol treatment. These results also suggest NCED is involved in drought tolerance. Differences in the expression of this gene under drought stress conditions may be related to differences in cultivars, degree of drought, and other factors. In Arabidopsis, drought stress induced the expression of the NCED gene (AtNCED3), and this gene adjusted the level of endogenous ABA (Iuchi et al. 2001). On the other hand, overexpression of AtNCED3 in transgenic plants increased the level of endogenous ABA. The results showed AtNCED3 expression played a significant role in ABA biosynthesis in drought stress in Arabidopsis. In another study, the expression of DcNCED1 and DcNCED2 was significantly increased in three carrot cultivars under drought stress (Wang et al. 2021).
AAO3 gene expression increased in the tolerant genotype (SLM046) in the early hours of sampling (Fig. 3). However, AAO3 gene expression decreased in DR samples at the 8th and 24th hours of drought stress. On the other hand, in DM samples, the highest expression was observed in the 8th hour (9.8 times), and then a decreasing trend was observed in the 24th hour. However, DM samples had higher expression levels than DR samples in the 8th and 24th hours. It seems that methanol changed the expression pattern in DM samples compared to DR samples. Also, the expression pattern of the AAO3 gene observed in the tolerant plants in DM samples was similar to PSY and NCED genes. A study on peanuts showed that the expression of AhZEP, AhNCED1, AhAAO2, and AhABA3 genes were significantly increased in response to drought stress (Long et al. 2019).
AAO3 gene expression was highest (2 times) in drought-sensitive genotype (Hyola308) in the first hours of sampling. In DR drought stress samples, gene expression decreased gradually at 8 and 24 h after drought stress. However, the expression level of the AAO3 gene in DM samples did not change at 8 and 24 h. Therefore, it seems that methanol foliar application had little effect on gene expression in DM samples of sensitive plants and even reduced the expression to some extent in the 8th hour. In Arabidopsis, the enzyme aldehyde oxidase (AAO3) is an enzyme that accelerates the finishing stage of ABA biosynthesis. According to the results, AAO3 performs a significant role in ABA biosynthesis in leaves (Seo et al. 2004).
The results showed that the expression of the AAO3 gene increased with methanol application. As a result, it will cause an increasing effect on ABA followed by controlling and coping with drought stress. However, this gene had the highest expression in the drought-tolerant genotype after 8 h in drought stress with methanol treatment. As mentioned, methanol changed the expression pattern in DM samples compared to DR samples, so it seems that methanol spraying was effective in stress-tolerant plants. In contrast, methanol did not significantly affect sensitive genotypes and even reduced gene expression. So the effectiveness of methanol was different in both sensitive and drought-tolerant genotypes. In Arabidopsis, AtAAO3 is one of four abscisic aldehyde oxidases (AtAAO1 to 4) that actively use abscisic aldehyde as a substrate. It is most likely the sole AAO involved in ABA biosynthesis (Long et al. 2019).
Drought is an essential factor in the biosynthesis and metabolism of carotenoids. Drought stress was reported affecting carotenoid content in many crops, including strawberries, corn, soybeans, etc. (Shafiq et al. 2015). The results showed that drought stress in the tolerant plant increased genes’ expression, affecting the biosynthesis of carotenoids and ABA. In DR samples, the expression of PSY, NCED, AAO3 genes increased at the beginning of stress and decreased after 24 h. However, the sensitive plant’s expression increase was less than in the tolerant plant. It seems that the application of methanol on the aerial parts of the plant was effective in reducing the influence of drought stress and water requirement. Therefore, spraying methanol on the leaves led to easier absorption by the plant and reduced stress. Methanol foliar application in DM samples in the tolerant plants significantly increased the expression level of PSY, NCED, and AAO3 genes. However, DM samples in the sensitive plants displayed an increasing effect of methanol only on PSY and NCED genes and had little impact on AAO3 gene expression.
Expression changes in genes involved in the antioxidant defense system
The GR gene expression was elevated in the tolerant genotype (SLM046) at the initial hours of sampling (Fig. 3). However, gene expression decreased in DR drought stress samples at the 8th and 24th hours of drought stress. Also, it was less than the GR expression level at the beginning of stress. Methanol spraying appears to have altered the expression pattern in DM samples compared with DR samples. Since, in DM samples, the highest expression rate was observed in the 8th hour (8.6 times), which was the highest expression rate between both DM and DR groups. However, a decreasing trend was observed again in the 24th hour. Nevertheless, at both 8th and 24th h, DM samples had higher expression levels than DR samples (8.6 times at 8 h and 3.6 times at 24 h).
The GR gene expression in the drought-sensitive genotype (Hyola308) had the highest expression (11.3 times) in the early hours of sampling, which was even higher than the expression in the tolerant plant. In DR drought stress samples, gene expression decreased significantly at 8 and 24 h after drought stress. It seems that the sensitive plant could not maintain the expression of the GR effective gene in regulating oxidative stress. However, GR gene expression decreased to a lesser extent in DM samples at the 8th and 24th hours. Therefore, it seems that methanol treatment had some compensatory effect on DM samples of sensitive plants.
GR is an NADP(H)-dependent antioxidant enzyme and effectively promotes GSH reduction and oxidative stress tolerance (Gill et al. 2013). The glutathione–ascorbate cycle is one of the ROS control cycles in most cell organs such as mitochondria, chloroplasts, and cytoplasm. GR is responsible for converting oxidized glutathione to reduced glutathione and maintaining a high ratio of GSH to GSSG (Noctor and Foyer 1998). In a study on Nicotiana tabacum, it was found that tolerance to photo-oxidative stress in plants is associated with the high activity of GR chloroplasts, so increasing the expression of the GR gene could prevent oxidative stress (Aono et al. 1993). Increased glutathione reductase gene expression was reported by drought stress in sensitive bean cultivars. They also stated the expression level of this gene remained constant in drought-resistant cultivars (Torres-Franklin et al. 2008). Other studies have reported increased glutathione reductase activity during drought stress compared to control plants (Chen et al. 2020; Ratnayaka et al. 2003; Yousuf et al. 2012).
Methanol application increased the expression of the GR gene in both tolerant and susceptible genotypes 8 h after stress, but this amount was higher in the tolerant plant. Increased expression of this gene has been reported in drought stress compared to controls (Ratnayaka et al. 2003; Yousuf et al. 2012). In the current study (72 h after final irrigation), GR gene expression was increased in both genotypes. The activity of the glutathione-ascorbate cycle increased by exposing the plant to drought stress and increasing ROS production. The amount of GR expression gradually decreases over time due to the activation of other antioxidant mechanisms. Unlike the susceptible plant, decreasing expression levels occurred to a lesser extent due to the tolerant plant’s more robust stress tolerance mechanism. It also seems that methanol spraying was able to increase the intensity of the impact of this defensive mechanism in both genotypes. However, this increasing impact was higher in the tolerant plant.
At the beginning of sampling, NTRC gene expression increased in the tolerant genotypes (SLM046) (Fig. 3). The expression level of the NTRC gene decreased gradually after 8 h and reached its lowest level in DR samples in the 24th hour (5.3 times). Therefore, we observed an upward and then a downward pattern in DR samples. In contrast, methanol spray significantly increased the expression level of this gene at the 8th and 24th hour in DM samples compared to the same time in DR samples (11 times at 8 h and 11.2 times at 24 h).
NTRC gene expression in drought-sensitive genotypes (Hyola308) increased in the early hours of sampling but was less than the amount expressed in the tolerant plant. Similar to the tolerant plants, the expression level of the NTRC gene in DR samples of sensitive plants decreased at 8 h after stress and reached its lowest level after 24 h (1.9 times). Therefore, the expression pattern was first upward and then downward, similar to drought-tolerant plants. However, unlike the DM samples of the tolerant plant, methanol treatment did not alter the expression pattern. It merely increased the expression level at the 8th and 24th hours compared to the same time in the sensitive plant.
NTRC is located inside the chloroplast and is a significant player in the chloroplast redox detoxification system. Plants without NTRC showed susceptibility to bacterial pathogens, leading to a high accumulation of ROS (Mata-Pérez and Spoel 2019). As noticed, drought stress increased GR gene expression, and this factor could increase glutathione–ascorbate cycle activity. On the other hand, the activity of this cycle is dependent on NADPH, and one of its sources is NTRC. Therefore, the expression of the NTRC gene increases with increased antioxidant activity. As the activity of this system decreased, the expression level of the NTRC gene also gradually reduced. Prolonged stress causes ROS to accumulate in the plant and induces oxidative damage at the cellular level. Therefore, increasing antioxidant inhibitors is essential to lessen ROS accumulation. Plants cope with stress by increasing gene expression like NTRC (Cha et al. 2014). Based on the results (Fig. 2), there is a direct relationship between NTRs and TRX. TRX plays an essential role in the tolerance of plants to oxidative stress. TRX uses FTR or NTR to reduce itself (Reichheld et al. 2005). Drought stress increased the expression of the NTRC gene in both sensitive and tolerant genotypes; because of the more robust defense system of the tolerant plant against drought stress, the expression level of the NTRC gene was higher in the tolerant genotype. Moreover, methanol application effectively increased the expression of the NTRC gene. In both genotypes, increased expression was observed in 8 and 24 h after stress.
According to the results of the gene network, GR and NTRC genes were classified as superoxide metabolic processes (CL: 10,321) (Table 3). A direct interaction was observed between GR and NTR enzymes. It revealed the involvement of both of these enzymes in the antioxidant system. For example, ntra and ntrb mutants were able to survive but were highly sensitive to GSH reduction (Marty et al. 2009). The expression of GR and NTRC genes effective in regulating ROS activity increased during drought stress. A complex antioxidant defense system was activated in both sensitive and tolerant genotypes by exposing the plant to drought stress and increasing ROS production. Hence, the expression of GR and NTRC genes increased as part of this defense system. A downward trend in the expression level of GR and NTRC genes was observed at the 8th and 24th hours, caused by the activation of other elements of the antioxidant defense system. A severe effect of stress was observed because of the greater reduction in gene expression level in the sensitive plant and possibly its weaker antioxidant defense system. On the other hand, in both genotypes, methanol foliar application increased the expression level of both genes. The difference was that less expression was seen in sensitive genotypes at the 8th and 24th hours.
In comparison, an increasing trend was observed in the drought-tolerant genotype in the expression level of GR and NTRC genes. The treatment likely caused an enhancing effect on other genes involved in the antioxidant system, especially in the tolerant plant. This issue can be investigated in future research. Overall, drought stress increased ROS levels, followed by activation of the antioxidant system. Methanol was able to increase gene expression significantly. As a result, it could enhance anti-stress properties and support the plant cope with drought stress.
MicroRNAs targeting the studied genes
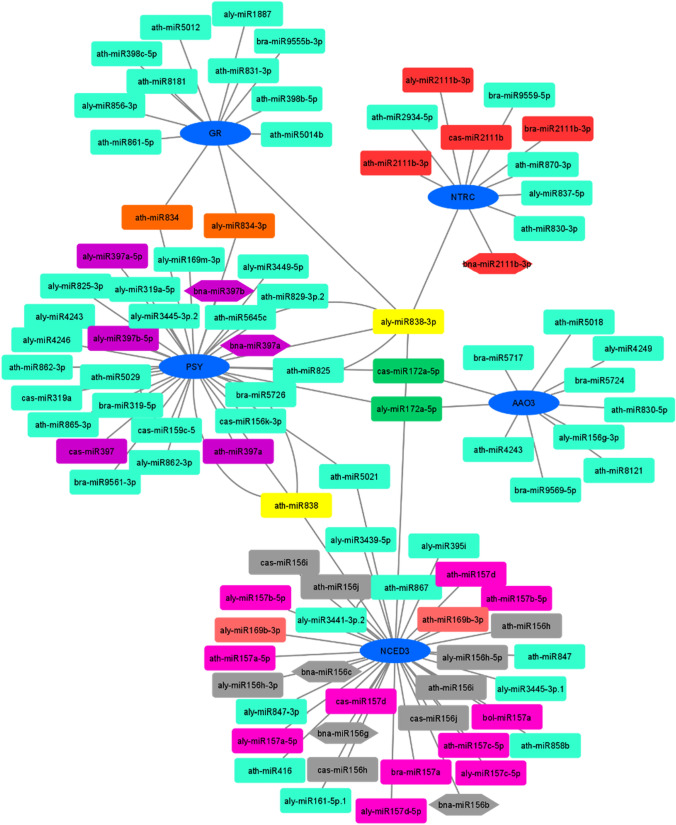
Examination of the Brassicaceae family microRNAs targeting PSY, NCED, GR, NTRC, and AAO3 was performed. The detailed information of the identified miRNAs is presented in Table 4. As a result, 38 microRNAs were identified for the PSY gene. Five microRNAs used the translation inhibition mechanism, and the rest used the cleavage system (Table 4). Several families of microRNAs were identified for the PSY gene, most of which belonged to the miR397 family with six numbers. Also, bna-miR397a and bna-miR397b were specifically identified in rapeseed (Fig. 4). A recent study indicated that bna-miR397a/b also targeted BnCSD10 in rapeseed and this genes’ expression level under different abiotic stress like drought stress was changed (Su et al. 2021). This microRNA is recognized as one of the factors in response to abiotic stresses (Huang et al. 2020). Recently, it has been observed that LAC2, a negative regulator of lignin deposition, was regulated by an increase in miR397b in Arabidopsis roots under water-deficient conditions (Khandal et al. 2020). Another study showed that miR397 expression increased in alfalfa (Medicago sativa) under drought stress, and by overexpression of alfalfa's miR397 in the tobacco plant, drought stress resistance occurred (Chen et al. 2020). According to the expectation index, the lowest value of the index belonged to miR159 (Table 4). The miR159 family is highly protected among monocotyledonous and dicotyledonous plants. However, the miR159’s relative abundance behaves differently in plants under drought stress because of the tissue and plant species (López-Galiano et al. 2019). For example, miR159 was reported to increase in Arabidopsis and maize under drought stress (Liu et al. 2008; Wei et al. 2009) but decreased in cotton and potato (Xie et al. 2015; Yang et al. 2014). While in barley and alfalfa, miR159 decreased in roots and increased in leaves in response to drought stress (Da et al. 2017; Hackenberg et al. 2015).
Table 4.
Identified microRNAs affecting PSY, NCED, GR, NTRC, AAO3 genes using psRNATarget software with the expected value 5
| miRNA_Acc | Target_Acc | Expectation | Target_start | Target_end | Inhibition | Target_Desc |
|---|---|---|---|---|---|---|
| aly-miR172a-5p | XM_013862421.2 | 3.5 | 1513 | 1533 | Cleavage | AAO3 |
| bra-miR5717 | XM_013862421.2 | 3.5 | 593 | 613 | Cleavage | AAO3 |
| cas-miR172a-5p | XM_013862421.2 | 3.5 | 1513 | 1533 | Cleavage | AAO3 |
| bra-miR9569-5p | XM_013862421.2 | 4 | 686 | 706 | Cleavage | AAO3 |
| ath-miR4243 | XM_013862421.2 | 4.5 | 1258 | 1278 | Cleavage | AAO3 |
| ath-miR8121 | XM_013862421.2 | 4.5 | 1729 | 1752 | Cleavage | AAO3 |
| ath-miR830-5p | XM_013862421.2 | 4.5 | 1419 | 1440 | Cleavage | AAO3 |
| bra-miR5724 | XM_013862421.2 | 4.5 | 854 | 874 | Cleavage | AAO3 |
| aly-miR156g-3p | XM_013862421.2 | 5 | 403 | 423 | Cleavage | AAO3 |
| aly-miR4249 | XM_013862421.2 | 5 | 1153 | 1173 | Cleavage | AAO3 |
| ath-miR5018 | XM_013862421.2 | 5 | 1389 | 1412 | Cleavage | AAO3 |
| ath-miR5014b | XM_022709985.1 | 3.5 | 1577 | 1597 | Cleavage | GR |
| aly-miR838-3p | XM_022709985.1 | 4 | 1858 | 1878 | Cleavage | GR |
| aly-miR834-3p | XM_022709985.1 | 4.5 | 284 | 304 | Cleavage | GR |
| aly-miR856-3p | XM_022709985.1 | 4.5 | 772 | 792 | Cleavage | GR |
| ath-miR5012 | XM_022709985.1 | 4.5 | 1629 | 1649 | Cleavage | GR |
| ath-miR834 | XM_022709985.1 | 4.5 | 284 | 304 | Cleavage | GR |
| aly-miR1887 | XM_022709985.1 | 5 | 1567 | 1590 | Translation | GR |
| ath-miR398b-5p | XM_022709985.1 | 5 | 1481 | 1501 | Translation | GR |
| ath-miR398c-5p | XM_022709985.1 | 5 | 1481 | 1501 | Translation | GR |
| ath-miR8181 | XM_022709985.1 | 5 | 249 | 268 | Cleavage | GR |
| ath-miR831-3p | XM_022709985.1 | 5 | 1461 | 1482 | Cleavage | GR |
| ath-miR861-5p | XM_022709985.1 | 5 | 960 | 980 | Cleavage | GR |
| bra-miR9555b-3p | XM_022709985.1 | 5 | 2025 | 2045 | Translation | GR |
| ath-miR2111b-3p | XM_013892363.2 | 4 | 483 | 503 | Cleavage | NTRC |
| aly-miR2111b-3p | XM_013892363.2 | 4.5 | 483 | 503 | Cleavage | NTRC |
| ath-miR830-3p | XM_013892363.2 | 4.5 | 910 | 930 | Cleavage | NTRC |
| bna-miR2111b-3p | XM_013892363.2 | 4.5 | 483 | 503 | Cleavage | NTRC |
| aly-miR837-5p | XM_013892363.2 | 5 | 1223 | 1243 | Cleavage | NTRC |
| aly-miR838-3p | XM_013892363.2 | 5 | 1546 | 1566 | Cleavage | NTRC |
| ath-miR2934-5p | XM_013892363.2 | 5 | 408 | 428 | Cleavage | NTRC |
| ath-miR870-3p | XM_013892363.2 | 5 | 1049 | 1069 | Cleavage | NTRC |
| bra-miR2111b-3p | XM_013892363.2 | 5 | 483 | 503 | Cleavage | NTRC |
| bra-miR9559-5p | XM_013892363.2 | 5 | 823 | 844 | Cleavage | NTRC |
| cas-miR2111b | XM_013892363.2 | 5 | 483 | 503 | Cleavage | NTRC |
| cas-miR159c-5 | XM_013827623.2 | 2.5 | 416 | 436 | Cleavage | PSY |
| ath-miR5029 | XM_013827623.2 | 3 | 345 | 365 | Cleavage | PSY |
| aly-miR838-3p | XM_013827623.2 | 3.5 | 1166 | 1186 | Cleavage | PSY |
| ath-miR5021 | XM_013827623.2 | 3.5 | 422 | 441 | Cleavage | PSY |
| aly-miR169m-3p | XM_013827623.2 | 4 | 1450 | 1468 | Cleavage | PSY |
| aly-miR834-3p | XM_013827623.2 | 4 | 1000 | 1020 | Cleavage | PSY |
| aly-miR838-3p | XM_013827623.2 | 4 | 33 | 53 | Cleavage | PSY |
| ath-miR834 | XM_013827623.2 | 4 | 1000 | 1020 | Cleavage | PSY |
| aly-miR319a-5p | XM_013827623.2 | 4.5 | 960 | 980 | Cleavage | PSY |
| aly-miR4246 | XM_013827623.2 | 4.5 | 949 | 969 | Cleavage | PSY |
| aly-miR838-3p | XM_013827623.2 | 4.5 | 198 | 218 | Cleavage | PSY |
| aly-miR862-3p | XM_013827623.2 | 4.5 | 240 | 260 | Translation | PSY |
| ath-miR825 | XM_013827623.2 | 4.5 | 154 | 174 | Cleavage | PSY |
| bra-miR319-5p | XM_013827623.2 | 4.5 | 960 | 980 | Cleavage | PSY |
| cas-miR159c-5 | XM_013827623.2 | 4.5 | 278 | 298 | Cleavage | PSY |
| cas-miR319a | XM_013827623.2 | 4.5 | 960 | 980 | Cleavage | PSY |
| aly-miR172a-5p | XM_013827623.2 | 5 | 584 | 603 | Cleavage | PSY |
| aly-miR3445-3p.2 | XM_013827623.2 | 5 | 351 | 371 | Cleavage | PSY |
| aly-miR3449-5p | XM_013827623.2 | 5 | 729 | 749 | Cleavage | PSY |
| aly-miR397a-5p | XM_013827623.2 | 5 | 880 | 900 | Cleavage | PSY |
| aly-miR397b-5p | XM_013827623.2 | 5 | 880 | 900 | Cleavage | PSY |
| aly-miR4243 | XM_013827623.2 | 5 | 98 | 118 | Cleavage | PSY |
| aly-miR825-3p | XM_013827623.2 | 5 | 154 | 174 | Cleavage | PSY |
| ath-miR397a | XM_013827623.2 | 5 | 880 | 900 | Cleavage | PSY |
| ath-miR5645c | XM_013827623.2 | 5 | 814 | 834 | Cleavage | PSY |
| ath-miR829-3p.2 | XM_013827623.2 | 5 | 623 | 643 | Cleavage | PSY |
| ath-miR838 | XM_013827623.2 | 5 | 1166 | 1186 | Translation | PSY |
| ath-miR838 | XM_013827623.2 | 5 | 39 | 58 | Translation | PSY |
| ath-miR838 | XM_013827623.2 | 5 | 198 | 217 | Translation | PSY |
| ath-miR862-3p | XM_013827623.2 | 5 | 240 | 260 | Translation | PSY |
| ath-miR865-3p | XM_013827623.2 | 5 | 958 | 978 | Cleavage | PSY |
| bna-miR397a | XM_013827623.2 | 5 | 879 | 900 | Cleavage | PSY |
| bna-miR397b | XM_013827623.2 | 5 | 879 | 900 | Cleavage | PSY |
| bra-miR5726 | XM_013827623.2 | 5 | 537 | 557 | Cleavage | PSY |
| bra-miR9561-3p | XM_013827623.2 | 5 | 338 | 360 | Cleavage | PSY |
| cas-miR156k-3p | XM_013827623.2 | 5 | 820 | 841 | Cleavage | PSY |
| cas-miR172a-5p | XM_013827623.2 | 5 | 584 | 603 | Cleavage | PSY |
| cas-miR397 | XM_013827623.2 | 5 | 880 | 900 | Cleavage | PSY |
| aly-miR838-3p | XM_013788802.2 | 3 | 2031 | 2051 | Cleavage | NCED3 |
| aly-miR157d-5p | XM_013788802.2 | 3.5 | 2107 | 2126 | Cleavage | NCED3 |
| ath-miR157d | XM_013788802.2 | 3.5 | 2107 | 2126 | Cleavage | NCED3 |
| cas-miR156i | XM_013788802.2 | 3.5 | 2104 | 2123 | Cleavage | NCED3 |
| aly-miR3445-3p.1 | XM_013788802.2 | 4 | 149 | 169 | Cleavage | NCED3 |
| ath-miR847 | XM_013788802.2 | 4 | 2032 | 2052 | Cleavage | NCED3 |
| cas-miR157d | XM_013788802.2 | 4 | 2104 | 2123 | Cleavage | NCED3 |
| aly-miR161-5p.1 | XM_013788802.2 | 4.5 | 946 | 966 | Cleavage | NCED3 |
| aly-miR169b-3p | XM_013788802.2 | 4.5 | 981 | 1002 | Cleavage | NCED3 |
| ath-miR169b-3p | XM_013788802.2 | 4.5 | 981 | 1002 | Cleavage | NCED3 |
| ath-miR5021 | XM_013788802.2 | 4.5 | 2114 | 2133 | Translation | NCED3 |
| ath-miR838 | XM_013788802.2 | 4.5 | 2031 | 2051 | Translation | NCED3 |
| ath-miR867 | XM_013788802.2 | 4.5 | 2311 | 2331 | Cleavage | NCED3 |
| aly-miR156h-3p | XM_013788802.2 | 5 | 2034 | 2054 | Translation | NCED3 |
| aly-miR156h-5p | XM_013788802.2 | 5 | 2107 | 2126 | Translation | NCED3 |
| aly-miR157a-5p | XM_013788802.2 | 5 | 2107 | 2127 | Cleavage | NCED3 |
| aly-miR157b-5p | XM_013788802.2 | 5 | 2107 | 2127 | Cleavage | NCED3 |
| aly-miR157c-5p | XM_013788802.2 | 5 | 2107 | 2127 | Cleavage | NCED3 |
| aly-miR3439-5p | XM_013788802.2 | 5 | 1139 | 1159 | Cleavage | NCED3 |
| aly-miR3441-3p.2 | XM_013788802.2 | 5 | 448 | 468 | Cleavage | NCED3 |
| aly-miR395i | XM_013788802.2 | 5 | 1153 | 1173 | Cleavage | NCED3 |
| aly-miR847-3p | XM_013788802.2 | 5 | 2032 | 2052 | Cleavage | NCED3 |
| ath-miR156h | XM_013788802.2 | 5 | 2107 | 2126 | Translation | NCED3 |
| ath-miR156i | XM_013788802.2 | 5 | 2107 | 2126 | Translation | NCED3 |
| ath-miR156j | XM_013788802.2 | 5 | 2107 | 2126 | Translation | NCED3 |
| ath-miR157a-5p | XM_013788802.2 | 5 | 2107 | 2127 | Cleavage | NCED3 |
| ath-miR157b-5p | XM_013788802.2 | 5 | 2107 | 2127 | Cleavage | NCED3 |
| ath-miR157c-5p | XM_013788802.2 | 5 | 2107 | 2127 | Cleavage | NCED3 |
| ath-miR416 | XM_013788802.2 | 5 | 650 | 670 | Cleavage | NCED3 |
| ath-miR858b | XM_013788802.2 | 5 | 1213 | 1233 | Cleavage | NCED3 |
| ath-miR867 | XM_013788802.2 | 5 | 1276 | 1296 | Cleavage | NCED3 |
| bna-miR156b | XM_013788802.2 | 5 | 2107 | 2127 | Cleavage | NCED3 |
| bna-miR156c | XM_013788802.2 | 5 | 2107 | 2127 | Cleavage | NCED3 |
| bna-miR156g | XM_013788802.2 | 5 | 2107 | 2127 | Cleavage | NCED3 |
| bol-miR157a | XM_013788802.2 | 5 | 2107 | 2127 | Cleavage | NCED3 |
| bra-miR157a | XM_013788802.2 | 5 | 2107 | 2127 | Cleavage | NCED3 |
| cas-miR156h | XM_013788802.2 | 5 | 2107 | 2126 | Translation | NCED3 |
| cas-miR156j | XM_013788802.2 | 5 | 2107 | 2126 | Translation | NCED3 |
Fig. 4.
Relationships between microRNAs affecting PSY, NCED3, GR, NTRC, AAO3 genes were mapped using Cytoscape software. The hexagon shapes represent rapeseed microRNAs, and the cyan color indicates individually located microRNAs, other groups with the same color represent the microRNA family
However, some microRNAs were mutual among various genes, including miR838 among PSY, NCED, GR, NTRC, and miR834 genes between PSY and GR genes, miR172 between PSY and AAO3 genes, and miR5021 between PSY and NCED genes (Fig. 4). 38 microRNAs were identified for the NCED gene, of which nine microRNAs used the translation inhibition mechanism, and the rest used the cleavage system (Table 4). Several microRNAs were identified for the NCED gene; most of them belonged to miR156, miR157 families, with 11 numbers. Also, bna-miR156b, bna-miR156c, bna-miR156g were specifically identified in rapeseed (Fig. 4). The miR156 family is highly conserved in plants and plays a significant role in responding to abiotic stresses (Feyissa et al. 2021). Studies on alfalfa (Medicago sativa) have shown that miR156 has a regulatory role in drought stress (Feyissa et al. 2019). The overexpression of miR156’s Oryza sativa affected the expression of genes associated with abiotic stress resistance and growth in alfalfa (Wang et al. 2021). The lowest expectations were for miR838 and miR157 (Table 4). It was observed that miR838 was effective in drought stress in wheat (Akdogan et al. 2016). The miR157, which belonged to the same miRNA family as that of miR156, showed the same expression trend.
Studies have shown that miR157 is a factor involved in response to drought stress and high temperatures (Chen et al. 2020; Li et al. 2013). 13 microRNAs were identified for the GR gene, of which four microRNAs used the translation inhibition mechanism, and the rest used the cleavage system. No microRNA was found in rapeseed for this gene. However, the lowest Expectations belonged to ath-miR5014b, aly-miR838-3p, and aly-miR834-3p, respectively (Table 4). Specifically, ath-miR5014b has been identified in Arabidopsis (Borges et al. 2011), and no evidence has been found for its involvement in stress regulation. aly-miR838-3p was in common with PSY and NTRC, and aly-miR834-3p was also in common with PSY. The miR838 and miR834 families are conserved in the Brassicaceae family, and their regulatory role in regulating abiotic stress has been identified (Çelik and Akdaş 2019; Srivastava et al. 2013). Examination of the Zanthoxylum bungeanum revealed that miR834 had a regulatory role in drought stress (Fei et al. 2020). miR838 was also influential in drought stress in Triticum aestivum (Akdogan et al. 2016).
Eleven microRNAs were identified for the NTRC gene, and all microRNAs used the cleavage mechanism (Table 4). Several families of microRNAs were identified for the NTRC gene, most of which belonged to the miR2111 family with five numbers. Bna-miR2111b-3p was also detected in rapeseed (Fig. 4). Studies have also indicated the role of this microRNA in drought stress (Ghorecha et al. 2017; Wang et al. 2011). MiR2111 has also been involved in phosphate homeostasis and plant survival (Li et al. 2013). Studies have shown that simulated drought stress increases miR2111 expression (Zheng et al. 2016). In soybean gma-miR2111b/c/e/f had increased in leaves, but its expression decreased in roots (Huang et al. 2020).
Eleven microRNAs were identified for the AAO3 gene, and all microRNAs used the cleavage mechanism (Table 4). Several families of microRNAs were identified for the AAO3 gene, most of which belonged to the miR172 family with two numbers (Fig. 4). The lowest expectation value belonged to aly-miR172a-5p, bra-miR5717 (Table 4). miR172 is involved in the development of anther in many species like Arabidopsis, cotton, and alfalfa miR172. It regulates flower organ recognition genes, such as the AP2 and AP2 genes. Its expression was significantly inhibited under high-temperature stress, while its target gene (TOE) expression was increased (Chen et al. 2020). A recent study demonstrated that bna-miR172a/b/c/d targeted BnCSD2 in rapeseed (Su et al. 2021). The role of this microRNA under drought stress has been clarified; mir172 expression was affected in Brassica juncea, Helianthus annuus, and Malus domestica (Ali et al. 2020; Bhardwaj et al. 2014; Liang et al. 2020).
Conclusion
Morphological observation showed that rapeseed plants, both sensitive and tolerant genotypes, regained their freshness after methanol foliar application under irrigation cut-off stress. This study showed that the expression of PSY, NCED, GR, NTRC, AAO3 genes in rapeseed drought-tolerant genotype (SLM046) increased more than the sensitive genotype (Hyola308). Drought stress reduces plant growth by changing photosynthesis and water status. The cell also tries to prevent oxidative stress and ROS accumulation by implementing special measures (including altering gene expression). The protein–protein interaction results showed extensive correlations between enzymes in the Krebs cycle and the mechanisms involved in oxidation–reduction reactions effective in controlling ROS and the pathway of carotenoid synthesis and ABA synthesis. This study observed that the expression of GR and NTRC genes increased during drought stress. The plant tries to prevent the accumulation and damage of ROS and activates the antioxidant system. Also, the expression of PSY, NCED, AAO3 genes effective in the pathway of carotenoid synthesis and ABA synthesis during drought stress was increased. Therefore, this pathway was activated under drought stress, and the production of the ABA increased as one of the factors controlling stress. This study showed that after methanol treatment, the expression of PSY, NCED, GR, NTRC, AAO3 genes in rapeseed drought-tolerant genotype (SLM046) increased significantly compared to sensitive genotype (Hyola308). Apparently, methanol increased tolerance, especially in the SLM046 genotype. In the early hours of stress in the sensitive genotypes, the expression of these genes increased after methanol treatment compared to plants without foliar application; however, in the next hour, the amount decreased. Considering the effect of methanol on the expression of genes under drought stress, it seems that methanol causes a positive impact under drought stress. Carbon dioxide fixation occurred, and hydrogen peroxide production decreased by the application of methanol. The study of effective microRNAs showed that sometimes more than one type of microRNA could affect the desired gene, and in some cases, a conserved family of microRNAs caused the main effect on gene expression. Therefore, by examining and manipulating these main groups of microRNAs, the genes expression level can be affected. This issue can be further investigated in future experiments. Further research on rapeseed and the genes involved in stress resistance may reveal relevant molecular patterns for identifying drought resistance.
Author contributions
MMG, planned and designed the study, drafted the manuscript, and participated in all the experiments. SA, participated in all experiments. MMT, participated in bioinformatics study and data analysis. HSL. and AR, participated in study design and supervised the study. All authors participated in revising and finalizing the manuscript.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Mohammad Mohsenzadeh Golfazani, Email: mohsenzadeh.mohamad@guilan.ac.ir.
Mohammad Mahdi Taghvaei, Email: Mahdi.Taghvaei@gmail.com.
Habibollah Samizadeh Lahiji, Email: hsamizadeh@guilan.ac.ir.
Seddigheh Ashery, Email: ashery138@gmail.com.
Ali Raza, Email: alirazamughal143@gmail.com.
References
- Akdogan G, Tufekci ED, Uranbey S, Unver T. miRNA-based drought regulation in wheat. Funct Inte Genom. 2016;16:221–233. doi: 10.1007/s10142-015-0452-1. [DOI] [PubMed] [Google Scholar]
- Ali S, Hayat K, Iqbal A, Xie L. Implications of abscisic acid in the drought stress tolerance of plants. Agronomy. 2020;10:1323. doi: 10.3390/agronomy10091323. [DOI] [Google Scholar]
- Aono M, Kubo A, Saji H, Tanaka K, Kondo N. Enhanced tolerance to photooxidative stress of transgenic Nicotiana tabacum with high chloroplastic glutathione reductase activity. Plant Cell Physiol. 1993;34:129–135. doi: 10.1093/oxfordjournals.pcp.a078386. [DOI] [Google Scholar]
- Armand N, Amiri H, Ismaili A. The effect of methanol on photosynthetic parameters of bean (Phaseolus vulgaris L.) under water deficit. Photosynthetica. 2016;54:288–294. doi: 10.1007/s11099-015-0178-2. [DOI] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bhardwaj AR, Joshi G, Pandey R, Kukreja B, Goel S, Jagannath A, Kumar A, Katiyar-Agarwal S, Agarwal M. A genome-wide perspective of miRNAome in response to high temperature, salinity and drought stresses in Brassica juncea (Czern) L. PLoS ONE. 2014 doi: 10.1371/journal.pone.0092456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Pereira PA, Slotkin RK, Martienssen RA, Becker JD. MicroRNA activity in the Arabidopsis male germline. J Exp Bot. 2011;62:1611–1620. doi: 10.1093/jxb/erq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çelik Ö, Akdaş EY. Tissue-specific transcriptional regulation of seven heavy metal stress-responsive miRNAs and their putative targets in nickel indicator castor bean (R. communis L.) plants. Ecotoxicology Env Safety. 2019;170:682–690. doi: 10.1016/j.ecoenv.2018.12.006. [DOI] [PubMed] [Google Scholar]
- Cha J-Y, Kim JY, Jung IJ, Kim MR, Melencion A, Alam SS, Yun D-J, Lee SY, Kim MG, Kim W-Y. NADPH-dependent thioredoxin reductase A (NTRA) confers elevated tolerance to oxidative stress and drought. Plant Physiol Biochem. 2014;80:184–191. doi: 10.1016/j.plaphy.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Chen J, Pan A, He S, Su P, Yuan X, Zhu S, Liu Z. Different microRNA families involved in regulating high temperature stress response during cotton (Gossypium hirsutum L.) anther development. Int J Mol Sci. 2020 doi: 10.3390/ijms21041280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Q, Wang P, Wang M, Sun T, Jin H, Liu B, Wang J, Grimm B, Wang H-B. Thioredoxin and NADPH-dependent thioredoxin reductase C regulation of tetrapyrrole biosynthesis. Plant Physiol. 2017;175:652–666. doi: 10.1104/pp.16.01500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release) Nucleic Acids Res. 2018;46:W49–W54. doi: 10.1093/nar/gky316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaeipour Z, Haddad R. Influence of drought stress on photosynthetic characteristics and protective enzymes in plants. Iranian J Gene Plant. 2020;9:114–129. [Google Scholar]
- Dos Santos CV, Rey P. Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci. 2006;11:329–334. doi: 10.1016/j.tplants.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Downie A, Miyazaki S, Bohnert H, John P, Coleman J, Parry M, Haslam R. Expression profiling of the response of Arabidopsis thaliana to methanol stimulation. Phytochem. 2004;65:2305–2316. doi: 10.1016/j.phytochem.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Fei X, Li J, Kong L, Hu H, Tian J, Liu Y, Wei A, Biochemistry, miRNAs and their target genes regulate the antioxidant system of Zanthoxylum bungeanum under drought stress. Plant Physiol. 2020;150:196–203. doi: 10.1016/j.plaphy.2020.01.040. [DOI] [PubMed] [Google Scholar]
- Fernie AR, Carrari F, Sweetlove LJ. Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol. 2004;7:254–261. doi: 10.1016/j.pbi.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Feyissa BA, Arshad M, Gruber MY, Kohalmi SE, Hannoufa A. The interplay between miR156/SPL13 and DFR/WD40–1 regulate drought tolerance in alfalfa. BMC Plant Biol. 2019;19:1–19. doi: 10.1186/s12870-019-2059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyissa BA, Amyot L, Nasrollahi V, Papadopoulos Y, Kohalmi SE, Hannoufa A. Involvement of the miR156/SPL module in flooding response in Medicago sativa. Sci Rep. 2021;11:1–16. doi: 10.1038/s41598-021-82450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorecha V, Zheng Y, Liu L, Sunkar R, Krishnayya N. MicroRNA dynamics in a wild and cultivated species of Convolvulaceae exposed to drought stress. Physiol Mol Biol Plants. 2017;23:291. doi: 10.1007/s12298-017-0426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Anjum NA, Hasanuzzaman M, Gill R, Trivedi DK, Ahmad I, Pereira E, Tuteja N. Glutathione and glutathione reductase: a boon in disguise for plant abiotic stress defense operations. Plant Physiol Biochem. 2013;70:204–212. doi: 10.1016/j.plaphy.2013.05.032. [DOI] [PubMed] [Google Scholar]
- Hackenberg M, Gustafson P, Langridge P, Shi BJ. Differential expression of micro RNA s and other small RNA s in barley between water and drought conditions. Plant Biotechnol J. 2015;13:2–13. doi: 10.1111/pbi.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming DJ, Criddle RS, Hansen LD. Effects of methanol on plant respiration. J Plant Physiol. 1995;146:193–198. doi: 10.1016/S0176-1617(11)82040-7. [DOI] [Google Scholar]
- Huang S, Zhou J, Gao L, Tang Y. Plant miR397 and its functions. Funct Plant Biol. 2020;48:361–370. doi: 10.1071/fp20342. [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- Kalisz A, Pokluda R, Jezdinský A, Sękara A, Grabowska A, Gil J, Neugebauerová J. Chilling-induced changes in the antioxidant status of basil plants. Acta Physiol Plant. 2016;38:196. doi: 10.1007/s11738-016-2214-7. [DOI] [Google Scholar]
- Khalilzadeh R, Seid Sharifi R, Pirzad A. Mitigation of drought stress in pot marigold (Calendula officinalis) plant by foliar application of methanol. J Plant Physiol Breed. 2020;10:71–84. [Google Scholar]
- Khandal H, Singh AP, Chattopadhyay D. The MicroRNA397b-LACCASE2 module regulates root lignification under water and phosphate deficiency. Plant Physiol. 2020;182:1387–1403. doi: 10.1104/pp.19.00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MR, Khaleda L, Jung IJ, Kim JY, Lee SY, Cha J-Y, Kim W-Y. Overexpression of chloroplast-localized NADPH-dependent thioredoxin reductase C (NTRC) enhances tolerance to photo-oxidative and drought stresses in Arabidopsis thaliana. J Plant Bio. 2017;60:175–180. doi: 10.1007/s12374-016-0464-y. [DOI] [Google Scholar]
- Li F, Murillo C, Wurtzel ET. Maize Y9 encodes a product essential for 15-cis-zeta-carotene isomerization. Plant Physiol. 2007;144:1181–1189. doi: 10.1104/pp.107.098996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Liang D, Xu R, Li H, Zhang Y, Qin R, Li L, Wei P, Yang J. Overexpression of an alternative oxidase gene, OsAOX1a, improves cold tolerance in Oryza sativa L. Genet Mol. 2013;12:5424–5432. doi: 10.4238/2013.November.11.4. [DOI] [PubMed] [Google Scholar]
- Li X, Chen P, Xie Y, Yan Y, Wang L, Dang H, Zhang J, Xu L, Ma F, Guan Q. Apple SERRATE negatively mediates drought resistance by regulating MdMYB88 and MdMYB124 and microRNA biogenesis. Hortic Res. 2020;7:1–11. doi: 10.1038/s41438-020-0320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yang Y, Raza A, Yin S, Wang H, Zhang Y, Dong J, Wang G, Zhong C, Zhang H, et al. Heterologous expression of Arabidopsis thaliana rty gene in strawberry (Fragaria × ananassa Duch.) improves drought tolerance. BMC Plant Biol. 2021 doi: 10.1186/s12870-021-02839-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Wang W, Ma J, Wang J, Zhou F, Li W, Yu Y, Zhang L, Huang W, Huang X. Identification of differentially expressed microRNAs of sunflower seedlings under drought stress. Agronomy. 2020;112:2472–2484. doi: 10.1002/agj2.20254. [DOI] [Google Scholar]
- Liu H-H, Tian X, Li Y-J, Wu C-A, Zheng C-C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14:836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Liu H, Du H, Liu H, Kurtenbach R. Relationship between polyamines conjugated to mitochondrion membrane and mitochondrion conformation from developing wheat embryos under drought stress. J Biosci. 2021;46:1–11. doi: 10.1007/s12038-021-00155-5. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long H, Zheng Z, Zhang Y, Xing P, Wan X, Zheng Y, Li L. An abscisic acid (ABA) homeostasis regulated by its production, catabolism and transport in peanut leaves in response to drought stress. PLoS ONE. 2019;14:e0213963. doi: 10.1371/journal.pone.0213963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Galiano MJ, García-Robles I, González-Hernández AI, Camañes G, Vicedo B, Real MD, Rausell C. Expression of miR159 is altered in tomato plants undergoing drought stress. Plants. 2019;8:201. doi: 10.3390/plants8070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty L, Siala W, Schwarzländer M, Fricker MD, Wirtz M, Sweetlove LJ, Meyer Y, Meyer AJ, Reichheld J-P, Hell R. The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc Natl Acad Sci. 2009;106:9109–9114. doi: 10.1073/pnas.0900206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata-Pérez C, Spoel SH. Thioredoxin-mediated redox signalling in plant immunity. Plant Sci. 2019;279:27–33. doi: 10.1016/j.plantsci.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Meng C, Yang M, Wang Y, Chen C, Sui N, Meng Q, Zhuang K, Lv W. SlWHY2 interacts with SlRECA2 to maintain mitochondrial function under drought stress in tomato. Plant Sci. 2020;301:110674. doi: 10.1016/j.plantsci.2020.110674. [DOI] [PubMed] [Google Scholar]
- Mirzai M, Moeini A, Ghanati F. Effects of drought stress on the lipid peroxidation and antioxidant enzyme activities in two canola (Brassica napus L.) cultivars. J Agric. 2013;15:593–602. [Google Scholar]
- Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7:405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Mohsenzadeh Golafazani M, Samizadeh Lahiji H, Hassani Kumleh H. Patterns of mitochondrial gene expression in rapeseed leaves (Brassica napus L) at early growth stage in response to drought stress. Iranian J Plant Sci. 2017;48:67–77. [Google Scholar]
- Mohsenzadeh Golfazani M, Pasandideh Arjmand M, Hassani Kumleh H, Samizadeh Lahiji H, Vahedi R, Ramezanzadeh Bishegahi S. The effect of iron toxicity on some of morphological traits, relative gene expression of G6PDH and peroxidase enzyme activity in resistant and susceptible genotypes of Rice (Oryza sativa) Cereal Res. 2019;9:207–220. [Google Scholar]
- Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Nonomura AM, Benson AA. The path of carbon in photosynthesis: improved crop yields with methanol. Proc Natl Acad Sci U S A. 1992;89:9794–9798. doi: 10.1073/pnas.89.20.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Spínola MC, Kirchsteiger K, Moreno J, Sahrawy M, Cejudo FJ. Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell. 2006;18:2356–2368. doi: 10.1105/tpc.106.041541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezanzadeh Bishegahi S, Mohsenzadeh M, Samizadeh H. Effect of methanol foliar application on expression changes of some mitochondrial genes in canola under drought stress. Iranian J Plant Sci. 2021;52:113–128. [Google Scholar]
- Ramírez I, Dorta F, Espinoza V, Jiménez E, Mercado A, Peña-Cortés H. Effects of foliar and root applications of methanol on the growth of Arabidopsis, tobacco, and tomato plants. J Plant Growth Regul. 2006;25:30–44. doi: 10.1007/s00344-005-0027-9. [DOI] [Google Scholar]
- Ratnayaka HH, Molin WT, Sterling TM. Physiological and antioxidant responses of cotton and spurred anoda under interference and mild drought. J Exp Bot. 2003;54:2293–2305. doi: 10.1093/jxb/erg251. [DOI] [PubMed] [Google Scholar]
- Raza A, Charagh S, Razzaq A, Javed R, Khan RSA, Hasanuzzaman M. Brassicaceae plants response and tolerance to drought stress: physiological and molecular interventions. In: Hasanuzzaman M, editor. The plant family Brassicaceae: biology and physiological responses to environmental stresses. Springer Singapore: Singapore; 2020. pp. 229–261. [Google Scholar]
- Raza A, Su W, Gao A, Mehmood SS, Hussain MA, Nie W, Lv Y, Zou X, Zhang X. Catalase (CAT) gene family in rapeseed (Brassica napus L.): genome-wide analysis, identification, and expression pattern in response to multiple hormones and abiotic stress conditions. Int J Mol Sci. 2021 doi: 10.3390/ijms22084281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld J-P, Meyer E, Khafif M, Bonnard G, Meyer Y. AtNTRB is the major mitochondrial thioredoxin reductase in Arabidopsis thaliana. FEBS Lett. 2005;579:337–342. doi: 10.1016/j.febslet.2004.11.094. [DOI] [PubMed] [Google Scholar]
- Rezaie R, Mandoulakani BA, Fattahi M. Cold stress changes antioxidant defense system, phenylpropanoid contents and expression of genes involved in their biosynthesis in Ocimum basilicum L. Sci Rep. 2020;10:1–10. doi: 10.1038/s41598-020-62090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salin ML. Chloroplast and mitochondrial mechanisms for protection against oxygen toxicity. Free Radic Res Commun. 1991;13:851–858. doi: 10.3109/10715769109145867. [DOI] [PubMed] [Google Scholar]
- Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, Koshiba T. Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol. 2004;45:1694–1703. doi: 10.1093/pcp/pch198. [DOI] [PubMed] [Google Scholar]
- Shafiq S, Akram NA, Ashraf M. Does exogenously-applied trehalose alter oxidative defense system in the edible part of radish (Raphanus sativus L.) under water-deficit conditions? Sci Hortic. 2015;185:68–75. doi: 10.1016/j.scienta.2015.01.010. [DOI] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson K, Fuentes P, Quiroz-Iturra LF, Flores-Ortiz C, Contreras R, Handford M, Stange C. Unraveling the induction of phytoene synthase 2 expression by salt stress and abscisic acid in Daucus carota. J Exp Bot. 2018;69:4113–4126. doi: 10.1093/jxb/ery207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Srivastava AK, Suprasanna P, D’souza S. Identification and profiling of arsenic stress-induced microRNAs in Brassica juncea. J Exp Bot. 2013;64:303–315. doi: 10.1093/jxb/ers333. [DOI] [PubMed] [Google Scholar]
- Su W, Raza A, Gao A, Jia Z, Zhang Y, Hussain MA, Mehmood SS, Cheng Y, Lv Y, Zou X. Genome-wide analysis and expression profile of superoxide dismutase (SOD) gene family in rapeseed (Brassica napus L) under different hormones and abiotic stress conditions. Antioxidants (basel) 2021;10:1182. doi: 10.3390/antiox10081182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork, P.J.N.a.r. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghvaei MM, Samizadeh Lahiji H, Bakhtiarizadeh MR, Mohsenzadeh Golafazani M. Bioinformatics analysis of microRNAs related to cold Stress and their effects on proteins associated with fatty acids metabolism in rapeseed (Brassica napus L) J Crop Biotech. 2019;9:41. [Google Scholar]
- Taghvaei MM, Lahiji HS, Golfazani MM. Evaluation of expression changes, proteins interaction network, and microRNAs targeting catalase and superoxide dismutase genes under cold stress in rapeseed (Brassica napus L.) OCL. 2022 doi: 10.1051/ocl/2021051. [DOI] [Google Scholar]
- Torres-Franklin ML, Contour-Ansel D, Zuily-Fodil Y, Pham-Thi A-T. Molecular cloning of glutathione reductase cDNAs and analysis of GR gene expression in cowpea and common bean leaves during recovery from moderate drought stress. J Plant Physiol. 2008;165:514–521. doi: 10.1016/j.jplph.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Wang T, Chen L, Zhao M, Tian Q, Zhang WH. Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics. 2011;12:367. doi: 10.1186/1471-2164-12-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Liu Y, Teng F, Cen H, Yan J, Lin S, Li D, Zhang W. Heterogeneous expression of Osa-MIR156bc increases abiotic stress resistance and forage quality of alfalfa. Crop J. 2021;1:949–963. doi: 10.1016/j.cj.2020.11.009. [DOI] [Google Scholar]
- Wei L, Zhang D, Xiang F, Zhang Z. Differentially expressed miRNAs potentially involved in the regulation of defense mechanism to drought stress in maize seedlings. Int J Plant Sci. 2009;170:979–989. doi: 10.1086/605122. [DOI] [Google Scholar]
- Xie F, Wang Q, Sun R, Zhang B. Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J Exp Bot. 2015;66:789–804. doi: 10.1093/jxb/eru437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang N, Mi X, Wu L, Ma R, Zhu X, Yao L, Jin X, Si H, Wang D. Identification of miR159s and their target genes and expression analysis under drought stress in potato. Comput Biol Chem. 2014;53:204–213. doi: 10.1016/j.compbiolchem.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Yousuf, P.Y., Hakeem, K.U.R., Chandna, R., and Ahmad, P. (2012). Role of glutathione reductase in plant abiotic stress. In Abiotic Stress Responses in Plants (Springer), pp. 149–158
- Zbieć I, Karczmarczyk S, Podsiadło C. Response of some cultivated plants to methanol as compared to supplemental irrigation. Electron J Pol Agric Univ. 2003;6:1–7. [Google Scholar]
- Zheng Y, Hivrale V, Zhang X, Valliyodan B, Lelandais-Brière C, Farmer AD, May GD, Crespi M, Nguyen HT, Sunkar R. Small RNA profiles in soybean primary root tips under water deficit. BMC Syst Biol. 2016;10:1–10. doi: 10.1186/s12918-016-0374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]