Abstract
A germination-specific amidase of bacilli is a major spore-lytic enzyme that is synthesized with a putative signal sequence and hydrolyses spore cortex in situ. The sleB gene encoding this amidase in Bacillus subtilis and Bacillus cereus was expressed in the forespore compartment of sporulating cells under the control of ςG, as shown by Northern blot and primer extension analyses. The forespore-specific expression of B. subtilis sleB was further indicated by the forespore-specific accumulation of a SleB-green fluorescent protein fusion protein from which a putative secretion signal of SleB was deleted. Immunoelectron microscopy with anti-SleB antiserum and a colloidal gold-immunoglobulin G complex showed that the enzymes from both Bacillus species are located just inside the spore coat layer in the dormant spore, and in the dormant spore, the amidases appear exist in a mature form lacking a signal sequence. These results indicate that SleB is translocated across the forespore’s inner membrane by a secretion signal peptide and is deposited in cortex layer synthesized between the forespore inner and outer membranes. The peripheral location of the spore-lytic enzymes in the dormant spore suggests that spore germination is initiated at the exterior of the cortex.
The cortex, a thick layer of peptidoglycan specific to the bacterial spores produced by the genera Bacillus and Clostridium, is responsible for maintenance of the highly dehydrated state of the core, contributing to the extreme dormancy and heat resistance of spores (5, 16). Bacterial spore germination, which is a series of interrelated degradation events triggered by specific germinants, leads to the irreversible loss of spore dormancy and the rehydration of the core. Once triggered, this process proceeds in the absence of germinant and germinant-stimulated metabolism (6, 16). This indicates that germination is a process controlled by the sequential activation of a set of preexisting germination-related enzymes but not by protein synthesis. However, little is known about the expression and localization of the germination-related enzymes and the mechanism and construction of the germination apparatus.
One of key enzymes involved in spore germination is a cortex-lytic enzyme. A germination-specific N-acetylmuramyl-l-alanine amidase (an amidase) has been identified in spores of Bacillus megaterium KM (4, 5), Bacillus cereus IFO13597 (11, 18), and Bacillus subtilis 168 AJ12866 (17) and is thought to be a major cortex-lytic enzyme. The genes for B. subtilis and B. cereus amidases (sleBs) were cloned, and the deduced amino acid sequences of SleBs indicated that the enzymes are synthesized in a form with a possible secretion signal at the N terminus (17, 18). In B. subtilis, it was also demonstrated that the amidase responds to germination triggered by l-alanine (17), the most universal germinant for spores of different species (16). In this article, we show that the B. subtilis and B. cereus amidases are synthesized in the forespore compartment of sporangia under the control of ςG, a sporulation-specific sigma factor, and that these amidases are located inside of the spore coat layer in a mature form. These results lead to a hypothesis that spore germination is initiated in the outer region of the cortex, which is not in accord with proposed models suggesting that the initial events of spore germination occur in the inner membrane and/or spore core (8, 24).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli XL1-Blue (Stratagene, La Jolla, Calif.) was used as the host for plasmid construction, and E. coli BL21(DE3) (Novagen, Madison, Wis.) was used for the expression of recombinant proteins. The strains of B. subtilis used in this study are listed in Table 1. B. subtilis was transformed as previously described (1). B. subtilis and E. coli were grown in LB medium (5 g of yeast extract, 10 g of polypeptone, and 10 g of NaCl per liter [pH 7.2]) at 37°C. For sporulation of B. subtilis and B. cereus, Schaeffer medium (21) was used. If necessary, ampicillin and tetracycline were added to final concentrations of 50 and 20 μg/ml, respectively.
TABLE 1.
B. subtilis used in this study
| Strain | Description or genotype | Reference or source |
|---|---|---|
| AJ12866 | Wild-type strain 168 | 17 |
| 1S38 | trpC2 spoIII94 | BGSCa |
| 1S60 | leuA8 tol-1 spoII G41 | BGSC |
| OD8603 | trpC2 pheA1 OR3Δ | 12 |
| SL1 | sleB::Cmr | 17 |
BGSC, Bacillus Genetic Stock Center, Ohio State University.
DNA manipulation.
Plasmids pBluescript SKII(−), pET22(+), pEGFP-C1, and pHY300PLK were purchased from Stratagene, Novagen, Clontech (Palo Alto, Calif.), and Takara Shuzo (Kyoto, Japan), respectively. Plasmid DNA was extracted from E. coli by the standard alkaline lysis procedure (19). Restriction enzymes, T4 DNA ligase, and T4 polynucleotide kinase were used as recommended by the manufacturers. The DNA restriction fragments were purified from agarose gels by using the Prep A Gene DNA Purification Matrix kit (Bio-Rad, Hercules, Calif.). Nucleotide sequences were determined using the dideoxy-chain termination method (20) with double-stranded DNA as the template and BigDye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems, Foster City, Calif.).
Northern hybridization.
B. cereus and B. subtilis cells sporulating in Schaeffer medium (50 mg [packed weight]) were collected by centrifugation (5,000 × g for 5 min at 4°C), frozen, and ground with pestle in mortar under liquid nitrogen. Nucleic acids were extracted with phenol-chloroform and chloroform-isoamyl alcohol and resuspended with 10 mM Tris-Cl (pH 8.0) containing 1 mM EDTA. RNA was precipitated at 4°C, with 8 M LiCl added to a final concentration of 2 M. The RNA pellet (15 μg) was separated in 1% agarose-formamide gels, transferred to Hybond nylon membranes, and hybridized at 65°C with a 32P-labeled DNA probe of the sleB gene. The sleB probe for B. cereus corresponded to nucleotides (nt) 623 to 1158 of D63645 (18) and was synthesized as a PCR product; the probe for B. subtilis was prepared as a HincII-SmaI fragment from plasmid pBS45H carrying D79978 (17).
Primer extension.
A 15-μg amount of RNA was annealed for 5 min at 65°C to 0.2 pmol of 32P-5′-labeled oligonucleotide (bcts 1 [5′-TAAAGACAGTCCTATGAACGCA-3′; nt 532 to 553 of D63645] for B. cereus and bsts1 [5′-GAAAAAAGGATGAGACATGCCATAATCG-3′; nt 626 to 646 of D79978] for B. subtilis) in 20 μl of 1× reverse transcriptase buffer (Amersham, Buckinghamshire, England) containing 0.5 mM deoxynucleoside triphosphates and extended for 1 h at 42°C with avian myeloblastosis virus transcriptase (Amersham). The cDNA products were loaded on a 6% polyacrylamide–6 M urea gel, together with sequencing reactions performed with the same primers and plasmid pBS45H or pBC15E carrying D63645 as the template, and bands were detected by autoradiography.
Construction of sleB-gfp fusion.
The gfp gene (encoding green fluorescent protein [GFP]) was obtained by PCR amplification from pEGFP-C1, using as primers oligonucleotides that created a SalI site at the 5′ end and an EcoRI site at the 3′ end. The PCR fragment was digested with SalI and EcoRI and then ligated to pHY300 PLK that had been digested with SalI and EcoRI, yielding pHYG1.
B. subtilis sleB was obtained by PCR amplification from pBS45H as a DNA segment encoding Met-109 to Glu-305 of SleB, using as primers oligonucleotides that created an NdeI site at 5′ end and a SalI site at 3′ end. The ends of the PCR fragment were rendered flush with T4 DNA polymerase, and the fragment was ligated to pBluescript SKII(−) that had been cut with SmaI. A plasmid in which the 5′ end of sleB is on the BamHI side of pBluescript SKII(−) was designated pBSL1. The 5′-upstream region of B. subtilis sleB was obtained as a DNA segment from positions −232 to +37 relative to the sleB transcription start site by PCR amplification from pBS45H, using as primers oligonucleotides that created NdeI sites at both ends. The fragment was digested with NdeI and then ligated to pBSL1 that had been cut with NdeI. A plasmid in which the Shine-Dalgarno sequence for sleB was adjacent to the codon for Met-109 of B. subtilis SleB, pBdSL3, was digested with XbaI and SalI, giving a fragment containing the sleB promoter, Shine-Dalgarno sequence, and partial sleB. This fragment was ligated to pHYG1 that had been digested with XbaI and SalI, yielding pHYdSLG, which contained the sleB-gfp in-frame fusion lacking the first 108 codons for SleB.
Preparation of antisera against B. subtilis and B. cereus SleBs.
To prepare the antibodies against B. subtilis SleB, parts of B. subtilis sleB encoding the proposed mature enzyme (from Phe-30 to Glu-305) and of E. coli pelB encoding a signal peptide were fused in the expression plasmid pET22(+). The recombinant protein was expressed in E. coli BL21(DE3) as described previously (18). Expression of two major proteins with molecular weights of approximately 33,000 and 31,000 (see Fig. 1, lane 2) were induced in an insoluble form with 2 mM isopropyl-β-d-thiogalactopyranoside, and N-terminal sequence analysis confirmed that these proteins were mature SleB with or without a PelB signal peptide, respectively. Recombinant B. subtilis SleB without a PelB signal peptide was separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and eluted from gels, and antibody was raised against the recombinant protein in mice as described previously (15). Antibody against B. cereus SleB was raised similarly against the purified SleB from spore germination exudate (11).
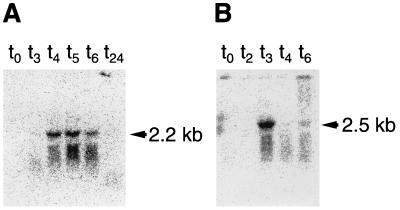
FIG. 1.
Northern blot analysis of sleB mRNAs from B. subtilis (A) and B. cereus (B) during sporulation. Total RNAs isolated from the cells at times (hours) indicated after the onset of sporulation (t0) at 37°C were separated in 1% agarose-formamide gels and transferred to nylon membranes. The filters were hybridized with 32P-labeled sleB probes for each species, as described in Materials and Methods. Each lane contains 15 μg of RNA.
Preparation of SDS extracts and germination exudate from B. subtilis spores.
Dormant spores of B. subtilis (0.1 g [packed weight]) were disrupted at 4°C with a bead beater in a 5-ml centrifuge tube containing 2 ml of 0.25 M potassium phosphate (pH 7.0) and 2 g of glass beads (diameter, 0.1 mm). After removal of glass beads with a no. 2 glass filter, cell debris and supernatant were separated by centrifugation (5,000 × g for 5 min at 4°C), and the debris was extracted with a 400 μl of 1% SDS at 95°C for 30 min. The supernatant fluid was also made 1% in SDS and heated at 95°C for 5 min. Aliquots of the SDS-treated supernatant and debris were subjected to SDS-polyacrylamide gel electrophoresis followed by immunoblot analysis (see below).
To prepare germination exudate, packed spores (0.1 g) were germinated at 30°C for 30 min in 10 volumes of germination buffer (10 mM l-alanine, 0.2 M KCl, 20 mM Tris-HCl [pH 7.0]) (17). After centrifugation (8,000 × g, 10 min, 4°C), the supernatant fluid was subjected to SDS-polyacrylamide gel electrophoresis followed by immunoblot analysis.
GFP visualization procedures.
An Olympus BX60 microscope was used with a PM-30 exposure control unit and a UplanApo universal objective (magnification, ×100; numerical aperture, 0.50 to 1.35). For visualization of GFP, a dichroic mirror cube unit with a narrow-band-pass (470- to 490-nm) excitation filter and a narrow-band-pass (515- to 550-nm) barrier filter (U-MNIBA; Olympus) for fluorescein isothiocyanate visualization was used. At various times of sporulation, cells in the same field were photographed by both fluorescence and differential interference contrast microscopy, using Fuji Fujichrome PROVIA (ASA 1600) film. Photo images were digitized with a Nikon LS-1000 film scanner, and image overlays and micrograph figures were prepared with Adobe Photoshop software.
Immunoelectron microscopy.
Thin sections of B. cereus and B. subtilis dormant spores immunolabeled with mouse anti-SleB antiserum and colloidal gold (10-nm particle diameter)-conjugated goat anti-mouse immunoglobulin G (IgG) (Zymed, San Francisco, Calif.) were prepared as described previously (13). The sections were observed with a JEM-1200EX electron microscope operating at 80 kV.
Analytical methods.
SDS-polyacrylamide gel electrophoresis was done on 12% (wt/vol) slab gels, using a Laemmli buffer system (10) at a constant current of 20 mA. Immunoblot analysis was performed as described previously (14). N-terminal amino acid sequence analysis was done on a protein sequencer (model 477A/120A; PE Applied Biosystems). Autoradiography was performed with a Fujix Bioimage analyzer BAS 2000II system.
RESULTS
Time of expression of sleB during sporulation.
Northern blot analysis using total RNA isolated before and at different times after the onset of sporulation (t0) indicated that B. subtilis sleB mRNA appeared as a 2.2-kb band at t4 (4 h after sporulation), t5, and t6 (Fig. 1A). B. cereus sleB mRNA was detected as a 2.5-kb band at t3 (Fig. 1B). RNAs isolated from exponentially growing cells and dormant spores (t24) gave no signal (data not shown), suggesting that transcription of sleB is sporulation specific.
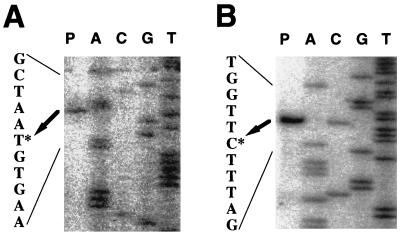
The promoter sequences involved in sleB expression were identified by primer extension analysis using total RNAs isolated at t4 from B. subtilis and t3 from B. cereus. The unique transcriptional start sites were located 34 bases upstream of the ATG start codon of B. subtilis sleB and 32 bases upstream of that of B. cereus sleB (Fig. 2). The transcriptional start site and the size of the B. subtilis sleB transcript indicated that B. subtilis sleB (918 bp) and the following gene ypeB (1,353 bp) are polycistronically transcribed. Similarly, it appears that in B. cereus sleB is the first gene in a two-gene operon with orf2, which is 15 bp away from sleB. B. subtilis ypeB and B. cereus orf2 encode putative homologous proteins (17, 18).
FIG. 2.
Mapping of the 5′ end of the sleB mRNA from B. subtilis (A) and B. cereus (B) by primer extension. Fifteen micrograms of total RNA isolated from cells at t4 for B. subtilis (A) or t3 for B. cereus (B) was annealed to the oligonucleotide of the sleB gene of the relevant origin and extended with avian myeloblastosis virus reverse transcriptase as described in Materials and Methods (lane P). Lanes T, C, G, and A contain a dideoxy sequencing ladder obtained with the same primer and plasmid pBS45H (A) or pBC15E (B) as described in Materials and Methods. The potential start point is marked by an asterisk.
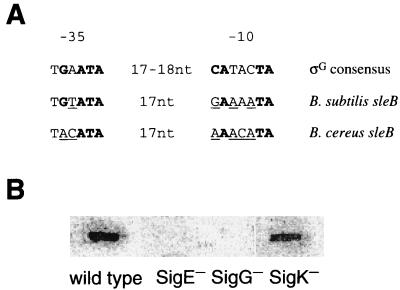
Comparison of sequence upstream of the sleB transcription start points with the consensus −10 and −35 sequences for ςG-dependent genes (Fig. 3A) shows that sleB exhibits four of (B. subtilis) or three (B. cereus) of six matches in the −35 region, and four (B. subtilis) or three (B. cereus) of seven matches in the −10 region. In addition, at all positions where both sleB sequences differ from the ςG consensus sequence, the residues found in sleBs are present in at least one other ςG-dependent promoter (Fig. 3A, underlined residues). The ςG dependency of sleB genes was confirmed by the slot blot analysis of RNAs obtained from various ς-deficient B. subtilis strains at t4. As shown in Fig. 3B, there was no detectable sleB transcript in t4 RNAs from both ςE and ςG null mutants, while a ςK null mutation had no effect on sleB transcription. These results suggest that sleB and ypeB (orf2 in B. cereus) are polycistronically transcribed by E-ςG.
FIG. 3.
Comparison of the ςG consensus promoter sequence with the sleB promoter sequences of B. subtilis and B. cereus and slot blot analysis of sleB mRNAs from various ς factor-deficient strains of B. subtilis. (A) The consensus ςG promoter sequence in the −10 and −35 regions is from Corfe et al. (3). Boldface bases are conserved in >80% of all B. subtilis promoters transcribed by E-ςG. Underlined bases in the sleB promoters that differ from the consensus sequence are found at this position in at least one other B. subtilis ςG-dependent promoter. (B) Five micrograms of total RNA from B. subtilis 168 AJ12866 (wild type), 1S60 (spoIIG SigE−), 1S38 (spoIIIC SigK−), or OD8603 (OR3Δ SigG−) at t4 was applied per lane and hybridized with the same 32P-labeled sleB probe as used for Fig. 1A. Use of an increased amount (up to 50 μg per lane) of RNA or RNA from t5 or t6 gave the same results (data not shown).
Forespore-specific expression of sleB.
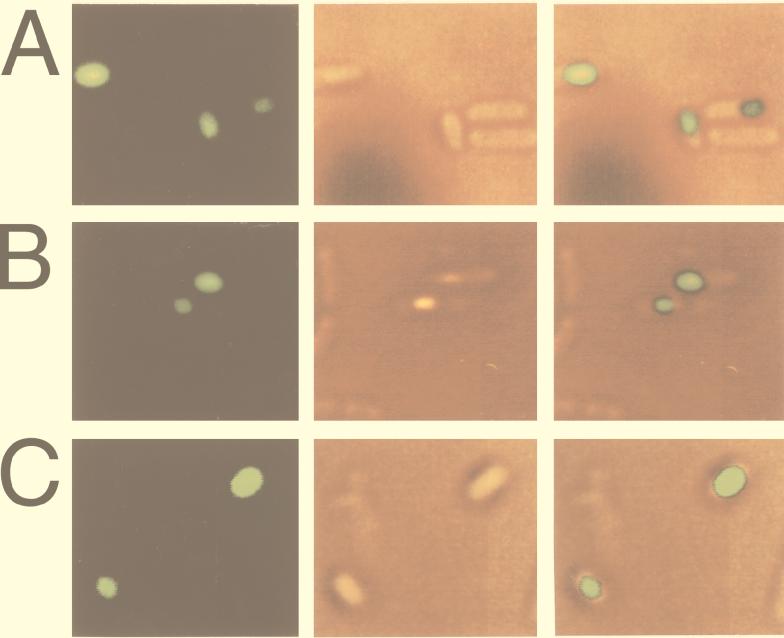
The ςG dependence of sleB genes of B. subtilis and B. cereus suggests that these germination-specific amidases are synthesized in the forespore compartment of sporulating cells. This was further examined by visualization of the site of expression of B. subtilis sleB by use of a SleB-GFP fusion. A 863-bp fragment of DNA containing the promoter and 197 codons of B. subtilis sleB was cloned into plasmid pHYG1 to generate an in-frame fusion to gfp (see Materials and Methods). Codons encoding the likely signal peptide of SleB were deleted in order to observe the accumulation of the fusion protein at its site of expression. The resultant plasmid, pHYdSLG, was transformed into B. subtilis, with selection for tetracycline resistance. When the transformant, designated strain SG109, was sporulated in Schaeffer medium containing tetracycline (20 μg/ml) at 37°C, no fluorescence was observed in sporangia, possibly because of impaired folding of the GFP moiety at high temperature. However, when this strain was sporulated at 30°C, there was significant fluorescence. As shown in Fig. 4, when cells were viewed by fluorescence microscopy at intervals after the onset of sporulation, small, roughly spherical areas of fluorescence, situated close to the cell pole, appeared at t15, which was just before phase-bright forespores became visible (Fig. 4A). Phase-contrast microscopic observation suggested that sporulation stage of t15 cells under this condition corresponded to that of B. subtilis t5 cells and B. cereus t3 cells which were sporulated at 37°C without antibiotic and used for Northern blot analysis (Fig. 1). About 8% of sporangia examined fluoresced at the point. The population of fluorescent cells increased as sporulation continued, and at t18 about 80% of forespores fluoresced (Fig. 4B). Fluorescence could be observed in free spores as well at t24 (Fig. 4C), and it persisted for over 48 h. Among hundreds of sporangia examined, there were a few (<3%) cells in which the entire sporangium exhibited fluorescence as early as t10, but fluorescence of this kind gradually disappeared with development of cells. In ςG-deficient cells carrying the sleB-gfp fusion, fluorescence was never observed throughout sporulation (data not shown).
FIG. 4.
Fluorescence of sporangia bearing a sleB-gfp fusion. Sporulation was induced by the Shaeffer medium nutrient exhaustion method at 30°C, and sporangia were photographed as described in Materials and Methods. Fluorescence photographs (left panels) of sporangia of strain SG109 bearing sleB-gfp were taken at approximately t15 (A), t18 (B), and t24 (C). Differential interference contrast micrographs of the same fields (middle panels) and those overlaid with corresponding fluorescence photographs (right panels) are also shown.
Subcellular location of SleB in dormant spores.
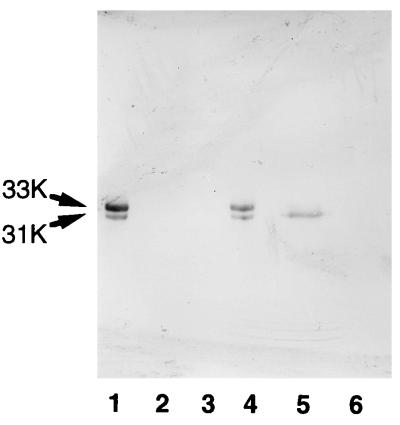
The germination-specific amidase of B. cereus spores is released into the germination exudate during germination (11). This enzyme was present in a mature form in dormant spores, as shown by the release of active enzyme from dormant spores disrupted in 0.25 M potassium phosphate (pH 7.0) at 25°C for 30 min (18). On the other hand, neither amidase activity nor a protein cross-reactive with anti-B. subtilis SleB antiserum was detected in the germination exudate of B. subtilis spores and the extract with 0.25 M potassium phosphate (pH 7.0) from disrupted dormant spores of B. subtilis (Fig. 5, lanes 2 and 3). However, the antiserum cross-reacted with a component of 31-kDa mass in disrupted spores extracted with 1% SDS at 90°C for 30 min (Fig. 5, lane 5). The size of this 31-kDa band coincided with that of the recombinant SleB without a signal peptide (Fig. 5, lanes 1 and 4), and this 31-kDa band was not detected in the extract made from disrupted dormant spores of the sleB-deficient strain B. subtilis SL1 (Fig. 5, lane 6). These data suggest that the germination-specific amidase of B. subtilis spores exists as a mature form in dormant spores like its counterpart of B. cereus, but that the B. subtilis protein interacts strongly but noncovalently with spore components. B. subtilis SleB could not be detected with anti-B. cereus SleB antiserum and vice versa.
FIG. 5.
Immunological detection of SleB-related protein in dormant spores of B. subtilis. Spores were disrupted and extracted as described in Materials and Methods. The germination exudate and the proteins released or extracted from disrupted spores were subjected to SDS-polyacrylamide gel electrophoresis followed by immunoblot analysis. For comparison, aliquots of the lysate of E. coli cells producing recombinant B. subtilis SleB, of which a proposed mature region had been fused with signal peptide of E. coli PelB protein, was also electrophoresed. Approximately the same amount (∼100 μg) of protein except for E. coli lysate (∼10 μg for lane 1 and ∼5 μg for lane 4) was loaded on the gel. Lanes: 1, recombinant SleB expressed in E. coli; 2, germination exudate from B. subtilis 168 spores; 3, proteins released from disrupted spores of B. subtilis 168; 4, recombinant SleB; 5, extract made from disrupted spores of B. subtilis 168; 6, extract made from disrupted spores of B. subtilis SL1. Arrows labeled 31K and 33K indicate migration positions of recombinant SleB with or without E. coli signal peptide.
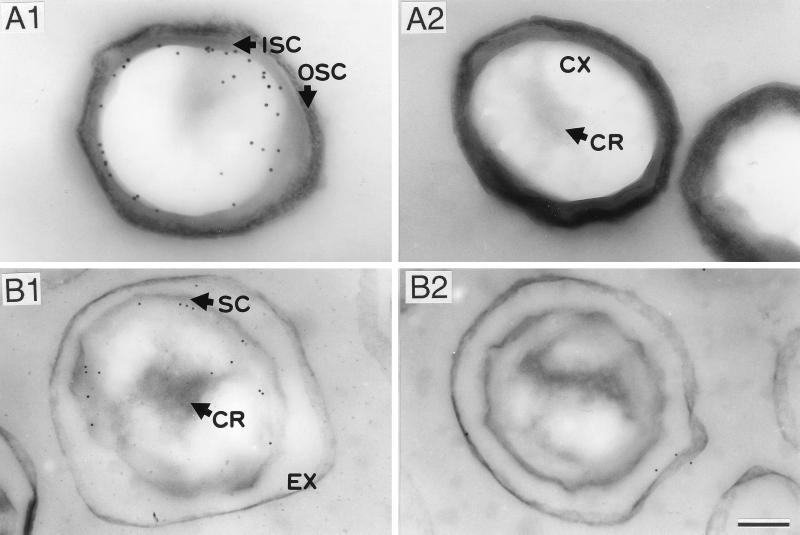
The localization of the amidases of B. subtilis and B. cereus in dormant spores was further examined by immunoelectron microscopy with anti-SleB antiserum and a colloidal gold-IgG complex. Electron microscopic observations of the immunolabeled sections indicated that the colloidal gold particles were located just inside the spore coat layer in both species (Fig. 6). These results suggest that the amidases interact with the outer region of cortex or the outer spore membrane. Although the core regions of the dormant spores fixed with 4% paraformaldehyde–0.5% glutaraldehyde were only faintly stained with uranyl acetate, it has been reported that components in the core regions can be fixed with paraformaldehyde alone (9).
FIG. 6.
Immunoelectron microscopic localization of germination-specific amidases in dormant spores of B. subtilis wild-type strain (A1) and SleB-deficient mutant strain (A2) and B. cereus wild-type strain (B1 and B2). Thin sections of the spores, which were fixed with 4% paraformaldehyde–0.5% glutaraldehyde, were stained with anti-SleB antiserum and colloidal gold (10 nm)-IgG complex (A1, A2, and B1) or with preimmune serum and colloidal gold (10 nm)-IgG complex (B2). EX, exosporium; SC, spore coat; ISC, inner spore coat; OSC, outer spore coat; CX, cortex; CR, core. Bar = 200 nm.
DISCUSSION
In this study, we demonstrated that the germination-specific amidases of B. subtilis and B. cereus which are crucial for spore cortex hydrolysis during l-alanine-induced germination are expressed in the forespore compartment of sporulating cells and localized on the outside of the cortex in the dormant spore. The B. subtilis amidase was also present in a form lacking the N-terminal 29 amino acid residues of SleB, in accordance with the observation that the B. cereus amidase exists as a mature enzyme without the N-terminal 32 residues (18), which have the characteristics of a cleavable signal sequence (22). This finding implies that SleB produced in the forespore compartment under control by ςG is transported across the inner forespore membrane with the aid of the secretion signal sequence. The SleB-GFP fusion protein appeared in sporangia before the refractivity of forespore is achieved, suggesting that SleB is synthesized prior to the deposition of cortex between spore membranes. However, the mechanism of the accumulation of SleB on the outside of the cortex layer during sporulation remains a topic for future study.
A number of genes are known to depend on ςG for their expression. From its known members including sleB, the ςG regulon appears to encode products that are synthesized within the forespore compartment during the later stages of sporulation, and whose function is to enhance spore survival and facilitate germination (7). Among them, gerA is the best-characterized cluster of germination genes, encoding a putative l-alanine receptor complex that senses l-alanine and transmits this information to the germination apparatus (16). Immunoelectron microscopic observation has demonstrated that GerA proteins are also localized just inside the spore coat layer in the dormant spore (23). Such a close location of GerA and SleB, both of which are involved in key events in germination process, is consistent with an effective transmission of initiation signal between germinant-sensor and cortex-degrading systems. This further suggests that spore germination is triggered at a rather peripheral site of dormant spore and that cortex hydrolysis during germination proceeds from the exterior to the core side of the cortex.
A remarkable difference in the amidases from B. subtilis and B. cereus is the tightness of the interaction of the enzyme with spore components. The homology of the mature enzymes between these two species is most notable in both the N-terminal (residues 33 to 99 of B. subtilis SleB and residues 30 to 97 of B. cereus SleB, in which 53 amino acid residues are identical) and C-terminal regions (residues 173 to 306 of B. subtilis SleB and residues 137 to 259 of B. cereus SleB, in which 95 residues are identical) (17, 18). However, the internal region linking these two regions differs in both length (74 amino acid residues in B. subtilis SleB versus 40 residues in B. cereus SleB) and polarity, and in this region, there are only eight residues which are identical between the two species. The internal region of the B. subtilis enzyme is notable in its high content of basic amino acids (eight Lys, three Arg, and one His). Possibly the excessive positive charge in this region of B. subtilis SleB cause a strong interaction between the enzyme and some spore component(s), such as the negatively charged spore peptidoglycan.
We have shown here that the amidases of B. subtilis and B. cereus exist as mature but inactive forms in the dormant spore. This finding suggests that regulation of the activity of these enzymes requires a mechanism different from the activation by proteolytic cleavage of an inactive proenzyme as observed in germination-specific amidases of Clostridium perfringens (14) and B. megaterium (6). As for B. subtilis and B. cereus amidases, a germination-specific muramidase of C. perfringens is present in a mature form in the dormant spore (2). However, there is a significant difference in substrate specificity between the amidases of bacilli and the C. perfringens muramidase. Germination-specific amidases have been indicated to cleave the cross bridge of in situ spore cortex, leading to the dissolution of the cortex structure (5, 6, 11, 15). On the other hand, the C. perfringens muramidase lyses only dissolved cortex (2), suggesting that the activity is tightly regulated by its requirement for disrupted cortex. It is apparent that this is not the case for the amidases of B. subtilis and B. cereus. Elucidation of the function of the products of B. subtilis ypeB and B. cereus orf2, which are polycistronically transcribed with sleB as possible germination-related proteins, may explain how the activity of the amidases are controlled.
ACKNOWLEDGMENT
We thank Y. Kobayashi for kindly providing strain OD8603.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirement for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Miyata S, Makino S, Moriyama R. Molecular Characterization of a germination-specific muramidase from Clostridium perfringens S40 spores and nucleotide sequence of the corresponding gene. J Bacteriol. 1997;179:3181–3187. doi: 10.1128/jb.179.10.3181-3187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corfe B M, Moir A, Popham D, Setlow P. Analysis of the expression and regulation of the gerB spore germination operon of Bacillus subtilis 168. Microbiology (Reading) 1994;140:3079–3083. doi: 10.1099/13500872-140-11-3079. [DOI] [PubMed] [Google Scholar]
- 4.Foster S J, Johnstone K. Purification and properties of a germination-specific cortex-lytic enzyme from spores of Bacillus megaterium KM. Biochem J. 1987;242:573–579. doi: 10.1042/bj2420573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster S J, Johnstone K. Pulling the triggers: the mechanism of bacterial spore germination. Mol Microbiol. 1990;4:137–141. doi: 10.1111/j.1365-2958.1990.tb02023.x. [DOI] [PubMed] [Google Scholar]
- 6.Foster D J, Johnstone K. Germination-specific cortex-lytic enzyme is activated during triggering of Bacillus megaterium KM spore germination. Mol Microbiol. 1988;2:727–733. doi: 10.1111/j.1365-2958.1988.tb00083.x. [DOI] [PubMed] [Google Scholar]
- 7.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keynan A. Spore structure and its relation to resistance, dormancy and germination. In: Chambliss G, Vary J C, editors. Spores VII. Washington, D.C: American Society for Microbiology; 1978. pp. 43–53. [Google Scholar]
- 9.Kozuka S, Yasuda Y, Tochikubo K. Ultrastructural localization of dipicolinic acid in dormant spores of Bacillus subtilis by immunoelectron microscopy with colloidal gold particles. J Bacteriol. 1985;162:1250–1254. doi: 10.1128/jb.162.3.1250-1254.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1979;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Makino S, Ito N, Inoue T, Miyata S, Moriyama R. A spore-lytic enzyme released from Bacillus cereus spores during germination. Microbiology (Reading) 1994;140:1403–1410. doi: 10.1099/00221287-140-6-1403. [DOI] [PubMed] [Google Scholar]
- 12.Masuda E S, Anaguchi H, Yamada K, Kobayashi Y. Two developmental genes encoding factor homologs are arranged in tandem in Bacillus subtilis. Proc Natl Acad Sci USA. 1988;85:7637–7641. doi: 10.1073/pnas.85.20.7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyata S, Kozuka S, Yasuda Y, Chen Y, Moriyama R, Tochikubo K, Makino S. Localization of germination-specific spore-lytic enzymes in Clostridium perfringens S40 spores detected by immunoelectron microscopy. FEMS Microbiol Lett. 1997;152:243–247. doi: 10.1016/s0378-1097(97)00204-8. [DOI] [PubMed] [Google Scholar]
- 14.Miyata S, Moriyama R, Miyahara N, Makino S. A gene (sleC) encoding a spore cortex-lytic enzyme from Clostridium perfringens S40 spores; cloning, sequence analysis and molecular characterization. Microbiology (Reading) 1995;141:2643–2650. doi: 10.1099/13500872-141-10-2643. [DOI] [PubMed] [Google Scholar]
- 15.Miyata S, Moriyama R, Sugimoto K, Makino S. Purification and partial characterization of a spore cortex-lytic enzyme of Clostridium perfringens S40 spores. Biosci Biotechnol Biochem. 1995;59:514–515. doi: 10.1271/bbb.59.514. [DOI] [PubMed] [Google Scholar]
- 16.Moir A, Smith D A. The genetics of bacterial spore germination. Annu Rev Microbiol. 1990;44:531–553. doi: 10.1146/annurev.mi.44.100190.002531. [DOI] [PubMed] [Google Scholar]
- 17.Moriyama R, Hattori A, Miyata S, Kudoh S, Makino S. A gene (sleB) encoding a spore cortex-lytic enzyme from Bacillus subtilis and response of the enzyme to l-alanine-mediated germination. J Bacteriol. 1996;178:6059–6063. doi: 10.1128/jb.178.20.6059-6063.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moriyama R, Kudoh S, Miyata S, Nonobe S, Hattori A, Makino S. A germination-specific spore cortex-lytic enzyme from Bacillus cereus spores: cloning and sequencing of the gene and molecular characterization of the enzyme. J Bacteriol. 1996;178:5330–5332. doi: 10.1128/jb.178.17.5330-5332.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaeffer P, Millet J, Aubert J P. Catabolite repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simonen M, Palva I. Protein secretion in Bacillus species. Microbiol Rev. 1993;57:109–137. doi: 10.1128/mr.57.1.109-137.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasuda Y, Sakae Y, Tochikubo K. Immunological detection of the GerA spore germination proteins in the spore integuments of Bacillus subtilis using scanning electron microscopy. FEMS Microbiol Lett. 1996;139:235–238. doi: 10.1111/j.1574-6968.1996.tb08208.x. [DOI] [PubMed] [Google Scholar]
- 24.Vary J C. Glucose-initiated germination in Bacillus megaterium spores. In: Chambliss G, Vary J C, editors. Spores VII. Washington, D.C: American Society for Microbiology; 1978. pp. 104–108. [Google Scholar]