Abstract
NOD-like receptor pyrin domain-containing 3 (NLRP3) has been considered to play a crucial role in triggering the host's immune and inflammatory responses. Genetic variants are critical determinants of interindividual variances in inflammatory responses and clinical outcomes. The role of NLRP3 gene variations in bipolar I (BPI) disorder, which is known to include genetic factors in its aetiology, has not been previously reported, at least to the best of our knowledge. The present study aimed to determine the role and frequency ofta exon 2 and exon 3 variants of NLRP3 in BPI disorder and to evaluate the association between different phenotypic traits. A case-control study with 123 patients and 107 healthy controls was conducted to investigate the association of variants identified in the exon 2 and exon 3 regions of NLRP3, with the risk of BPI. Regions of interest were sequenced using a PCR-based Sanger sequencing method. Three BPI-related variants were identified. The genotype Q705K CA was detected more frequently in BPI patients, as compared to the control group [P<0.001; odds ratio (OR), 0.202; 95% confidence interval (CI), 0.080-0.508]. In addition, two novel splice-site variants (c.393G>A and c.278_2A>G) that, to the best of our knowledge, have not been previously reported in any database, were detected only in the BPI patient group [P<0.001; OR, 0.846; 95% CI, 0.784-0.912; P<0.001; OR, 0.886; 95% CI, 0.832-0.944, respectively]. There was no significant association between the Q795K variant and phenotypic traits (P>0.05). However, there was a significant association between those carrying the heterozygous c.393G>A variant and a positive family history (P=0.043). It was also observed that those with the heterozygous c.278-2A>G variant presented with a significantly early-onset (P=0.003). On the whole, the data of the present study suggested that NLRP3 plays a crucial role in the pathogenesis of BPI and may be a potential risk factor. However, further functional studies and repeated studies in other populations are required to properly comprehend the roles of the NLRP3 variants in the risk of developing BPI.
Keywords: bipolar disorder, NOD-like receptor pyrin domain–containing 3, splice-site variant, polymorphism, neuroinflammation
Introduction
Bipolar disorder (BD) is a common chronic and cyclical psychiatric disorder characterized by unusual mood swings between mania/hypomania and depression (1). Although the etiology of BD is not precisely known, a distinct genetic component is considered to participate in its pathogenesis (2). Due to the complex and multifactorial nature of the disease, a predisposition to BD is believed to arise from the interaction of multiple low-impact genes, and in combination with environmental factors, may lead to a bipolar phenotype (3). Classic genetic epidemiological studies of family, twin, and adopted children have yielded substantial evidence that genes might cause a predisposition to bipolar disorder (4). In twin studies, it has been reported that the heritability of BD varies between 37 and 69%, and the risk of developing the disorder in first-degree biological relatives of patients with BD is seven-fold higher, as compared to the general population (5). However, a monozygotic concordance of <100% suggests that genes alone do fully explain this phenomenon (4).
Physical and mental traumas, mitochondrial dysfunction, neuroinflammation, altered-neurogenesis and apoptosis, oxidation, endoplasmic reticulum stress, and epigenetic changes, including histone alterations and methylation are the biological factors considered operative in BD (1,6). In a previously published study, it has been suggested that the NOD-like receptor protein 3 (NLRP3) inflammasome may mediate the association between mitochondrial dysfunction and inflammatory system activation (7). In addition, it has been postulated that the nucleotide-binding oligomerization domain-like receptor (NLR) family consists of intracellular pattern recognition receptors that play a critical role in triggering the host's immune and inflammatory responses (8). A number of NLRs (>20) have been identified in humans, with NLRP3 being the most familiar. The NLRP3 gene is expressed primarily in peripheral blood leukocytes (9).
The NLRP3 gene has a size of 32.9 kb, is located on chromosome 1q44, and contains nine exon regions (10). NLRP3 gene variants have been reported to increase the activation levels of inflammasomes and IL-1β and IL-18 by causing changes in their functions (10,11). To date, 60 single-nucleotide polymorphisms (SNPs) have been identified in the NLRP3 gene (11). These genetic variants of the NLRP3 gene appear to be a major predictor of the amplitude of autoinflammatory response (12). The most prevalent of these variants are rs35829419, rs10754558, rs4612666, rs4925648 and rs10925019. It has been stated in the literature that NLRP3 variants are associated with several inflammatory diseases, including rheumatoid arthritis, psoriatic arthritis, type 1 diabetes, multiple sclerosis, preeclampsia and mood disorders (13–18). However, in a meta-analysis study, certain variants were shown to exert a protective effect (11). Thus, the association between the NLRP3 gene variants and autoinflammatory diseases remains largely unknown, due to the use of a relatively small sample size, insufficient statistical power, and/or heterogeneity of diseases (11). To the best of our knowledge, no genetic study has been conducted to date to establish the role of the NLRP3 gene in the development of BD. It was hypothesized that the NLRP3 gene may lead to a susceptibility to bipolar I (BPI) disorder. Accordingly, the present study aimed to determine the frequency of genetic variants in exon 2 and exon 3 of the NLRP3 gene in BPI patients, to reveal their role in its pathogenesis, and to determine the association between the clinical findings.
Patients and methods
Patients
The present study was initiated following the approval of Süleyman Demirel University Faculty of Medicine Clinical Research Ethics Committee (date, 02.11.2020; reference no. 345). All procedures performed involving human participants complied with the ethical standards of the institutional and/or national research committee and the 1964 Helsinki declaration and any subsequent amendments or comparable ethical standards. All participants signed a written informed consent. The patient group consisted of 123 (62 females and 61 males) patients with bipolar I disorder and 107 (55 females and 52 males) healthy controls (matched by age and sex) who were recruited between January and July, 2021 from general psychiatric clinics of Süleyman Demirel University Hospital. All participants were ≥18 years of age. The phenotyping of tge patients was based on clinical interviews, medical records and a family history method according to DSM-5 diagnostic criteria using The Structured Clinical Interview for DSM-5 (SCID-5-CV). The clinical evaluation of the patients was performed by an experienced psychiatrist. The sociodemographic and clinical characteristics of the patients were recorded using the Mood Disorders Patient Registration Form (SKIP-TURK) (19). In the BPI patient group, those with any concomitant chronic/malignant disease were excluded. The control group consisted of healthy volunteers, blood donors, hospital staff, medical students with no family history of mood disorders and good social functions were selected. All patients with BPI were in the euthymic period. The Young mania rating scale (YMRS), Hamilton depression rating scale (HAM-D), functioning assessment short test (FAST) and global assessment of functioning (GAF) were administered to the participants by the clinician. Those with a HAM-D score <7 and a YMRS score <5 were defined as being in remission.
Evaluation instruments
SCID-5-CV, a semi-structured interview guide, was used for DSM-5 diagnoses (20). In previous studies, Turkish validity and reliability study of this scale have been conducted (21–25). YMRS, developed by Young et al (22), was used to evaluate mania symptoms. The HAM-D, developed by Hamilton was used to evaluate depressive symptoms (24). The GAF scale and FAST were used to measure global function (26). The Turkish validity and reliability of the assessment tools used were studied (27).
DNA extraction
A volume of 5 ml peripheral blood samples were obtained from patients and controls in EDTA-containing tubes. Genomic DNA was isolated from collected peripheral blood samples of the subjects using a DNA Isolation kit (cat. no. 11796828001; Roche Applied Science) following the manufacturer's instructions. After extraction, all DNA samples were stored at 20°C. The DNA concentration in dissolved solution was evaluated through optical density (OD) measurement at 260 nm and its 260/280 ratio using a NanoDrop (Thermo Fisher Scientific, Inc.).
Sanger sequencing and data analysis
Genetic analyses for NLRP3 gene (GenBank Accession no. NM_001243133.2) exons 2 and 3 were performed using Sanger-based DNA sequencing. Sequencing was performed from polymerase chain reaction (PCR)-amplified DNA using conventional protocols. PCR was performed using the following primers: for exon 2 forward, 5′-GTCTCCTCTCTCATGCCAAATA-3′ and reverse, 5′-CAAGAGCCACACAAACATGAA-3′; for exon 3 forward, 5′-CGTGACAGTCCTTCTGGAAA-3′ and reverse, 5′-CCCATTTATCCACCTACCATACA-3′. Various web tools were used for primer synthesis [NCBI (USA gov.), Ensembl database (EMBL-EBI), Primer3web (Whitehead Institute for Biomedical Research) (https://www.ncbi.nlm.nih.gov/, https://www.ensembl.org/index.html, https://primer3.ut.ee/]. In detail, the following protocol was applied: 25 µl Master mix, containing 2.5 µl of 10X Buffer, 0.5 µl PCR grade nucleotide mix (10 mM), 0.5 µl forward primer (10 µM), 0.5 µl reverse primer (10 µM), 3 µl DNA sample, 0.25 µl FastStart Taq DNA polymerase and 0.5 µl DMSO [FastStart High Fidelity PCR System, dNTPack (Roche Diagnostic)]. The amplification conditions were as follows: (94°C, 10 min) 1 cycle, (94°C, 2 min, 55°C 30 sec, 72°C 1 min) 35 cycles, (72°C 7 min) 1 cycle, and 4°C permanent storage (SimpliAmp Thermal Cycler; Applied Biosystems; Thermo Fisher Scientific, Inc.). At the end of the PCR analysis, agarose gel electrophoresis was performed to observe the band formation in the PCR samples. Subsequently, the gel was visualized with a UV Transilluminator (ECX-F20; Vilber Lourmat). The PCR product was then purified: 2 ml exosap were added to 5 ml of the PCR product and incubated at 37°C for 30 min and at 85°C for 15 min. The amplification products were sequenced in both directions using BigDye Terminator v3.1 (Thermo Fisher Scientific, Inc.) and specific primers for each region, in the ABI3500 Genetic Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Sequences were analyzed with the SeqScape Software v3.0 (Thermo Fisher Scientific, Inc.) using the GRCh37/hg19 sequence as reference. Each SNP was assessed for deviation from Hardy-Weinberg equilibrium (HWE) using a HWE calculator web tool (https://wpcalc.com/en/equilibrium-hardy-weinberg/).
Statistical analysis
Categorical variables were analyzed using the Chi-squared test or Fisher's exact test, where appropriate. Normality was tested using the Kolmogorov-Smirnov and Shapiro-Wilk W test. An independent samples (unpaired) t-test was used for age distribution evaluation. To evaluate the relative risk conferred by a particular allele or genotype, the odds ratio (OR) and 95% confidence interval (95% CI) were calculated. Deviation from HWE was analyzed using the Chi-squared test. Statistical analysis was performed using SPSS 18.0 software (SPSS, Inc.). Power analysis was performed by the GPower 3.1 software (https://g-power.apponic.com/). It was determined that it would be appropriate to analyze a total of 123 patients and 107 healthy individual controls, with a medium effect size of 0.30, type 1 error rate of 0.05, and power of the test as 87%. P<0.05 was considered to indicate a statistically significant difference.
Results
Description of the phenotype
The demographic and clinical characteristics of the 123 patients with BPI and the 107 healthy controls recruited for the sequence analysis are presented in Table I. The mean age was 41.20±11.86 years for the BPI group and 42.09±8.45 years for the healthy control group. The BPI prevalence was distributed equally between the two sexes. The BPI patient and control groups did not differ significantly as regards age and sex parameters (P>0.05). According to the DSM-5 criteria, age of onset was defined as the age at which probands first encountered symptoms, related to a manic or severe depressive episode. There was no authorized age of onset thresholds for the clinical and genetic study of the BPI. In the present study, occurrence at an age <22 years was accepted as early onset, based on the study of Hamshere et al (28). Patients with intermediate and late-onset occurrences were grouped as ‘late-onset’, generating a comparable subgroup sample size and accounting for genetic homogeneity (29). There was a significant difference between sexes at the age of onset (P=0.024). The age of onset in males [standard deviation (SD), 27.28±9.7] occurred considerably earlier than the age of onset in female (SD, 31.06±8.61). The mean values of the HAM-D, YMRS, GAF and FAST scales which were used for the classification between groups are presented in Table I.
Table I.
Characteristics of the controls and patients with bipolar disorder in the present study.
| Characteristic | BPI | Control | P-value |
|---|---|---|---|
| No. of subjects | 123 | 107 | - |
| Male/female (no. of subjects) | 61/62 | 52/55 | 0.896a |
| Age, years (mean ± SD) | 41.20±11.86 | 42.09±8.45 | 0.509b |
| Age at onset (mean ± SD) | - | ||
| Early-onset | 19.35±2.41 | ||
| Late-onset | 33.83±8.61 | ||
| Age at onset, n (%) | - | ||
| Early-onset | 37 (30.1) | ||
| Late-onset | 86 (69.9) | ||
| Age at onset (mean ± SD) | 0.024 | ||
| Male | 27.28±9.7 | ||
| Female | 31.06±8.61 | ||
| Illness duration, years | 11.98±9.2 | - | |
| Family history, n (%) | - | ||
| Positive | 58 (47.2) | ||
| Negative | 65 (52.8) | ||
| First illness period, n (%) | - | ||
| Euphoric mania | 89 (72.4) | ||
| Mixed episode | 12 (9.8) | ||
| Hypomania | 5 (4.1) | ||
| Depression | 17 (13.8) | ||
| Pharmacotherapy, n (%) | - | ||
| Mood stabilizers | |||
| Lithium | 58 (50.9) | ||
| Valproate | 54 (47.4) | ||
| Lamotrigine | 7 (6.1) | ||
| Carbamazepine | 2 (1.8) | ||
| Antipsychotic | 8 (6.5) | ||
| Combination | 77 (62.6) | ||
| No treatment | 3 (2.4) | ||
| HAM-D (mean ± SD) | 3.97±5.07 | 0.98±1.30 | <0.001 |
| YMRS (mean ± SD) | 1.89±5.37 | 0.09±0.29 | <0.001 |
| GAF (mean ± SD) | 80.69±11.29 | 91.81±4.73 | <0.001 |
| FAST (mean ± SD) | 6.87±7.03 | 0.92±1.07 | <0.001 |
Data are presented as the number (%) and mean ± SD. n, total number of individuals. P<0.05 was considered to indicate a statistically significant difference.
The Chi-squared test was used for statistical analysis.
An independent samples t-test was used for statistical analysis. BPI, bipolar I disorder; SD, standard deviation; HAM-D, Hamilton depression rating scale; YMRS, Young mania rating scale; GAF, global assessment of functioning; FAST, functioning assessment short test.
Genotype and allele distribution of variants
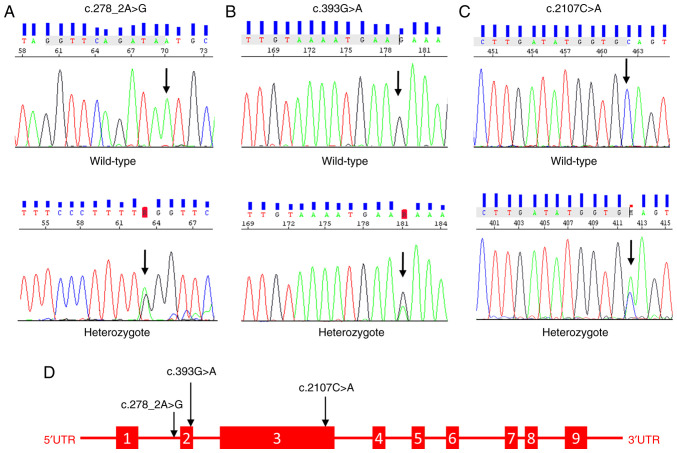
In both groups, Sanger-sequencing was used to genotype variations of the NLRP3 gene (NM_001243133.2) in exons 2 and 3. In addition to the missense-mutating variant rs35829419 (c.2107C>A, p.Q705K) identified after alignment with the reference genome (GRCh38.p13), two previously unreported in the database, novel variants (c.393G>A and c.278 2A>G) were discovered in the splice region, posing a risk for the development of BPI. These novel variants were absent in the control group. The schematic structure of the NLRP3 gene and the Sanger chromatogram of the detected variants are presented in Fig. 1. According to the statistical analysis, individuals carrying the CA genotype of rs35829419 (OR, 0.202; 95% CI, 0.080-0.508), the GA genotype of c.393G>A (OR, 0.846; 95% CI, 0.784-0.912) or the AG genotype of c.278_2A>G (OR, 0.886; 95% CI, 0.832-0.944) were associated with an increased risk of developing BPI (P<0.001). The genotype and allele frequencies and the estimated relative risks (OR) of the variants are presented in Table II. In addition to these variants, BPI patients had rs1178937944 (n=2), rs759389274 (n=2), rs763971331 (n=2), and rs770170892 (n=1) missense variants in exon 2.
Figure 1.
Schematic structure of the NLRP3 gene and Sanger chromatogram image of the detected variants. (A) Wild-type and heterozygous Sanger chromatogram image of novel splice-site c.278_2A>G variant, detected in the exon 2 of the NLRP3 gene. (B) Wild-type and heterozygous Sanger chromatogram image of novel splice-site c.393G>A variant, detected in the exon 2 of the NLRP3 gene. (C) Wild-type and heterozygous Sanger chromatogram image of the Q705K variant detected in the exon 3 of the NLRP3 gene. (D) Schematic representation of NLRP3 gene structure and identified variants in patients with BPI. NOD-like receptor pyrin domain-containing 3; BPI, bipolar I disorder.
Table II.
Allele and genotype frequencies of the NLRP3 variants.
| Variant | BPI (N=123) | Controls (N=107) | Pearson's χ2 test (df=1) | P-value | OR (95% CI) |
|---|---|---|---|---|---|
| rs35829419 | |||||
| Genotype | 13.371 | <0.001a | 0.202 (0.080-0.508) | ||
| CC | 95 (77.2%) | 101 (94.4%) | |||
| CA | 28 (22.8%) | 6 (5.6%) | |||
| AA | 0 | 0 | 4.324 (1.754-10.660) | ||
| Alelles | 11.746 | 0.001a | |||
| C | 218 (88.6%) | 202 (97.1%) | |||
| A | 28 (11.4%) | 6 (2.9%) | |||
| HWE P=0.154 | HWE P=0.765 | ||||
| c.393G>A | |||||
| Genotype | - | <0.001a | 0.846 (0.784-0.912) | ||
| GG | 104 (84.6%) | 107 | |||
| GA | 19 (15.4%) | 0 | |||
| AA | 0 | 0 | 1.091 (1.049-1.135) | ||
| Alelles | 18.718 | <0.001a | |||
| G | 208 (91.6%) | 214 | |||
| A | 19 (8.4%) | 0 | |||
| HWE P=0.353 | HWE - | ||||
| c.278-2A>G | |||||
| Genotype | - | <0.001a | 0.886 (0.832-0.944) | ||
| AA | 109 (88.6%) | 107 | |||
| AG | 14 (11.4%) | 0 | |||
| GG | 0 | 0 | 1.060 (1.028-1.093) | ||
| Alelles | 12.561 | <0.001a | |||
| A | 232 (94.3%) | 214 | |||
| G | 14 (5.7%) | 0 | |||
| HWE P=0.503 | HWE - |
(%), frequencies of alleles and genotypes; N, number of studies involved.
The Chi-squared test was used for statistical analysis. P<0.05 was considered to indicate a statistically significant difference. The P-value and OR of significant values are in presented in bold font. BPI, bipolar I disorder; OR, odds ratio; CI, confidence intervals; HWE, Hardy-Weinberg equilibrium.
HWE was tested in the population as it was a mandatory quality control step for population-based genetic association studies. The genotype distributions of the NLRP3 gene variants were observed to be in balance in both control and BPI populations. Therefore, it proved the absence of selection bias, population stratification, or genotyping errors in the present study (30). The frequencies and HWE values of the variations identified in the control group and patients with BPI disorder are listed in Table II.
Analysis of the genotype-phenotype association
The association between the genotypes and several phenotypic traits of the patients was studied. While the association between family history and c.393G>A variant was significant (P=0.043), no association was detected with other variants. Patients with c.278_2A>G variant AG genotype had significantly early onset of the disease (P=0.003). There was no significant association between the first disease period, lithium use, scales and variants (P>0.05). The association between phenotypic traits and variations is depicted in Table III.
Table III.
Association between some phenotypic traits and variants.
| Q705K | c.393G>A | c.278-2A>G | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Phenotype | CC | CA | P-value | GG | GA | P-value | AA | AG | P-value |
| Family history, n (%) | 0.930a | 0.043a | 0.426a | ||||||
| With | 45 (77.6) | 13 (22.4) | 45 (77.6) | 13 (22.4) | 50 (86.2) | 8 (13.8) | |||
| Without | 50 (23.1) | 15 (76.9) | 59 (90.8) | 6 (9.2) | 59 (90.8) | 6 (9.2) | |||
| Age of onset, n (%) | 0.460a | 0.351a | 0.003a | ||||||
| Early-onset | 27 (73) | 10 (27) | 33 (89.2) | 4 (10.8) | 28 (75.7) | 9 (24.3) | |||
| Late-onset | 68 (79.1) | 18 (20.9) | 71 (82.6) | 15 (17.4) | 81 (94.2) | 5 (5.8) | |||
| Lithium treatment, n (%) | 0.149a | 0.094a | 0.341a | ||||||
| With | 48 (82.8) | 10 (17.2) | 45 (77.6) | 13 (22.4) | 53 (91.4) | 5 (8.6) | |||
| Without | 40 (71.4) | 16 (28.6) | 50 (89.3) | 6 (10.7) | 48 (85.7) | 8 (14.3) | |||
| Scales (mean ± SD) | |||||||||
| HAM-D | 3.71±4.47 | 4.81±6.73 | 0.324b | 3.93±5.19 | 4.16±4.49 | 0.858b | 4.12±5.32 | 2.86±2.18 | 0.385b |
| YMRS | 2.11±5.99 | 1.15±2.14 | 0.416b | 2.06±5.82 | 1±1.2 | 0.432b | 1.98±5.69 | 1.21±1.48 | 0.618b |
| GAF | 81.31±11.3 | 78.63±11.09 | 0.281b | 80.66±11.19 | 80.84±12.09 | 0.950b | 80.52±11.47 | 81.93±10.14 | 0.664b |
| FAST | 6.63±7.18 | 7.67±6.59 | 0.505b | 7.13±7.52 | 5.53±3.36 | 0.364b | 7.01±7.37 | 5.86±3.71 | 0.567b |
Data are presented as the number (%) and mean ± SD. N, number of individuals.
The Chi-squared test was used for statistical analysis.
The Independent samples t-test was used for statistical analysis. P<0.05 was considered to indicate a statistically significant difference. SSD, standard deviation; HAM-D, Hamilton depression rating scale; YMRS, Young mania rating scale; GAF, global assessment of functioning; FAST, functioning assessment Short Test.
Discussion
The present study comparatively analyzed the effects of exon 2 and 3 NLRP3 variants on disease development, their frequency, and the association between various phenotypic traits in BPI patients and healthy individuals. To the best of our knowledge, the present study is the first to demonstrate an association between NLRP3 variants and BPI. The variant rs35829419 and two novel splice-site variants c.393G>A and c.278_2A>G identified in the NLRP3 gene demonstrated a significant association between BPI susceptibility and some phenotypic traits.
NLRP3 is the most intensively studied receptor in the NOD-like receptor family (31). Several studies have reported that the NOD-like receptor is a pattern recognition receptor that functions as a redox sensor in the inflammatory system. It has been reported that these recognition receptors can recognize distress signals, arising from physical and psychological stress (32,33). The variants in the NLRP3 gene can enhance the activation of the inflammasome, by causing changes in gene function. To date, ~60 SNPs have been identified in the NLRP3 gene (11). The gain-of-function variant rs35829419, which has been associated with a pro-inflammatory phenotype, has been demonstrated to promote the disease state by triggering NLRP3 inflammation activation (34). Therefore, it has been previously attempted to investigate the role of the rs35829419 variant in various inflammatory diseases. The association of the rs35829419 variant with several autoinflammatory disorders has been investigated, including celiac disease, Crohn's disease, psoriatic arthritis, myasthenia gravis, multiple sclerosis, type 1 diabetes, and systemic lupus erythematosus (15,16,35–38). In Parkinson's disease, in which inflammation is effective in pathogenesis, no significant association has been demonstrated between the frequency of the NLRP3 rs35829419 variant and the control group (39). In another study evaluating the effects of NLRP3 variants on the susceptibility and severity of rheumatoid arthritis (RA), no significant association was revealed between variants with joint damage and increased susceptibility (13). By contrast, in a study conducted to determine the association between inflammation-related genetic variants and susceptibility to psoriatic arthritis, a significant association was revealed between the destructive/deformative subset of psoriatic arthritis and the C allele of variant rs35829419 (14). In a previous study, a significant association was found between pediatric celiac disease and the NLRP3 variant rs358294199 in the Brazilian population (15). Similarly, allele and genotype frequencies of NLRP3 rs358294199 have been observed to be similar in case-control studies of patients with multiple sclerosis and ulcerative colitis in the Iranian population (16,40). As regards glucose homeostasis in patients with polycystic ovary syndrome, it has also been previously observed that the NLRP3 rs358294199 variant is significantly associated with the response to glycemic load (41). Moreover, it has been argued that the individuals with the A-risk allele of rs35829419 are more likely to develop the disease, whereas on the contrary, other researchers suggested a protective role; therefore, the clinical significance of this polymorphism has not been fully elucidated (11). The findings of the present study were consistent with other studies, suggesting that the rs35829419 variant may induce susceptibility to inflammatory diseases (14–16,40,41). In the present study, a substantial increase in BPI susceptibility was found in individuals carrying the A-risk allele of the rs35829419 variant, leading to the assumption that patients with BPI may have a higher inflammatory response than healthy individual controls. Therefore, it was suggested that the NLRP3 gene may play a critical role in the genetic etiology of BPI, and with the inflammation mediating this association.
The variants in the exon and splice regions of genes can alter the amino acid sequence, and thus affecting protein functions. Novel variants that cause amino acid changes in these regions are difficult to classify, since there are no entries in population databases, and segregation analyses cannot be performed. In the present study, two novel splice-site variants in the NLRP3 exon-2 region were identified. In total, 19 individuals (15.4%) in the single BPI patient group (MAF=0.084) presented with the novel c.393G>A variant, which resulted in synonymous alterations. There was a significant association with positive family history concerning the novel variant c.393G>A, as compared with phenotypic traits. There are only a few studies in the literature having addressed the family history of BPI patients. As previously reported, a family history of bipolar disorder could influence the course of the disorder and its prevalence rates (42). It has been revealed that patients with bipolar disorder who have a family history of depression have an early onset of the disease, experience more severe deterioration, experience multiple episodes during their lifetime, and are hospitalized more frequently (43). In their study involving data from patient families, Post et al (44) reported that the illness occurred early in adult patients with a family history of bipolar disorder and was associated with the occurrence of multiple psychiatric disorders in their children. In another study in patient family data, 22.5% of children with BD were observed to present with a higher incidence of depression, anxiety, conduct disorder, attention-deficit/hyperactivity disorder, and substance use disorders in comparison with their parents (45). In total, 14 individuals (11.4%) belonging to the BPI patient group were heterozygous for novel splice-site variant, c.278_2A>G. This variant was classified as ‘likely pathogenic’ in the VarSome and Franklin databases. The comparison of the novel c.278_2A>G variant with phenotypic features, revealed that there was an early onset of the disease in patients with the AG genotype. The early-onset BP has been generally associated with a worse prognosis than late-onset BP, including more psychotic features, substance addiction, comorbidity with panic and obsessive-compulsive disorders, a lower lithium response, and further suicide attempts (46–48). In a previous study, it has been suggested that early-onset BD has a high degree of homogeneity and a unique genetic etiology that has not yet been established in practice (49). In a study investigating the role of age at onset and family history in the clinic for BD, a significantly higher prevalence of family history and a higher number of attacks per year were observed in the early-onset group (50). Although a number of interesting preliminary studies have been conducted for the investigation of the basis of the different phenotypic characteristics of BD disease among individuals, the results obtained in genome-wide association studies (GWAS) with large samples need to be replicated to prove their actual validity. GWAS can identify genetic variations between early and late-onset BD, and the proportion of common SNPs that constitute each subtype can be estimated (49–51).
To the best of our knowledge, no genotype-phenotype correlation of NLRP3 gene variants has been described in BPI patients to date. The present study proved that NLRP3 gene variants, which may play an important role in modulating inflammatory responses, might genetically predispose to a chronic neuroinflammatory disease, including BPI. Furthermore, it was suggested that the rs35829419, c.393G>A, and c.278_2A>G NLRP3 variants may change the degree of systemic inflammation, and thus contribute to adverse inflammatory outcomes. However, there were some significant limitations to this study. For instance, there was a relatively small number of patients that comprised the study population. Another limitation was the inability to perform segregation analyses and functional studies, which were critical for demonstrating the effect of the identified variants on the disease.
In conclusion, the present study revealed that NLRP3 gene variants may be associated with an elevated risk of BPI. These findings need to be confirmed by family-based studies and replicated in different studies and large population samples. In addition, large-scale genetic studies, including GWAS, whole-exome sequencing, and whole-genome sequencing seem to be the most promising perspectives for future research.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the Scientific Research Projects Coordination Unit of Süleyman Demirel University with the project no. TSG-2020-8134.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
KHÖ and GÖÜ contributed to the conception and design of the study. KHÖ and GÖÜ performed experiments and samples collection. KHÖ performed data analysis and interpretation. KHÖ and GÖÜ drafted the manuscript. KHÖ and GÖÜ confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by Süleyman Demirel University Faculty of Medicine Clinical Research Ethics Committee (date, 02.11.2020, no. 345). All patients provided written informed consent prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Pereira AC, Oliveira J, Silva S, Madeira N, Pereira CM, Cruz MT. Inflammation in bipolar disorder (BD): Identification of new therapeutic targets. Pharmacol Res. 2021;163:105325. doi: 10.1016/j.phrs.2020.105325. [DOI] [PubMed] [Google Scholar]
- 2.Gubert C, Andrejew R, Moritz CE, Dietrich F, Vasconcelos-Moreno MP, Dos Santos BT, Fijtman A, Kauer-Sant'Anna M, Kapczinski F, da Silva Magalhães PV, Battastini AM. Bipolar disorder and 1513A>C P2RX7 polymorphism frequency. Neurosci Lett. 2019;16:143–147. doi: 10.1016/j.neulet.2018.11.055. [DOI] [PubMed] [Google Scholar]
- 3.Prata DP, Costa-Neves B, Cosme G, Vassos E. Unravelling the genetic basis of schizophrenia and bipolar disorder with GWAS: A systematic review. J Psychiatr Res. 2019;114:178–207. doi: 10.1016/j.jpsychires.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Gordovez FJ, McMahon FJ. The genetics of bipolar disorder. Mol Psychiatry. 2020;25:544–559. doi: 10.1038/s41380-019-0634-7. [DOI] [PubMed] [Google Scholar]
- 5.Gunderson JG, Zanarini MC, Choi-Kain LW, Mitchell KS, Jang KL, Hudson JI. Family study of borderline personality disorder and its sectors of psychopathology. Arch Gen Psychiatry. 2011;68:753–762. doi: 10.1001/archgenpsychiatry.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berk M, Post R, Ratheesh A, Gliddon E, Singh A, Vieta E, Carvalho AF, Ashton MM, Berk L, Cotton SM, et al. Staging in bipolar disorder: From theoretical framework to clinical utility. World Psychiatry. 2017;16:236–244. doi: 10.1002/wps.20441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sigitova E, Fišar Z, Hroudová J, Cikánková T, Raboch J. Biological hypotheses and biomarkers of bipolar disorder. Psychiatry Clin Neurosci. 2017;71:77–103. doi: 10.1111/pcn.12476. [DOI] [PubMed] [Google Scholar]
- 8.Mangan MSJ, Olhava EJ, Roush WR, Seidel HM, Glick GD, Latz E. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat Rev Drug Discov. 2018;17:688. doi: 10.1038/nrd.2018.97. [DOI] [PubMed] [Google Scholar]
- 9.Zhang AQ, Zeng L, Gu W, Zhang LY, Zhou J, Jiang DP, Du DY, Hu P, Yang C, Yan J, et al. Clinical relevance of single nucleotide polymorphisms within the entire NLRP3 gene in patients with major blunt trauma. Crit Care. 2011;15:R280. doi: 10.1186/cc10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villani AC, Lemire M, Fortin G, Louis E, Silverberg MS, Collette C, Baba N, Libioulle C, Belaiche J, Bitton A, et al. Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nat Genet. 2009;41:71–76. doi: 10.1038/ng.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Z, Wu S, Liang T. Association of NLRP3 rs35829419 and rs10754558 polymorphisms with risks of autoimmune diseases: A systematic review and meta-analysis. Front Genet. 2021;12:690860. doi: 10.3389/fgene.2021.690860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verma D, Lerm M, Julinder RB, Eriksson P, Söderkvist P, Särndahl E. Gene polymorphisms in the NALP3 inflammasome are associated with interleukin-1 production and severe inflammation: Relation to common inflammatory diseases? Arthritis Rheum. 2008;58:888–894. doi: 10.1002/art.23286. [DOI] [PubMed] [Google Scholar]
- 13.Kastbom A, Johansson M, Verma D, Söderkvist P, Rantapää-Dahlqvist S. CARD8 p.C10X polymorphism is associated with inflammatory activity in early rheumatoid arthritis. Ann Rheum Dis. 2010;69:723–726. doi: 10.1136/ard.2008.106989. [DOI] [PubMed] [Google Scholar]
- 14.Juneblad K, Kastbom A, Johansson L, Rantapää-Dahlqvist S, Söderkvist P, Alenius GM. Association between inflammasome-related polymorphisms and psoriatic arthritis. Scand J Rheumatol. 2021;50:206–212. doi: 10.1080/03009742.2020.1834611. [DOI] [PubMed] [Google Scholar]
- 15.Pontillo A, Brandao L, Guimaraes R, Segat L, Araujo J, Crovella S. Two SNPs in NLRP3 gene are involved in the predisposition to type-1 diabetes and celiac disease in a pediatric population from northeast Brazil. Autoimmunity. 2010;43:583–589. doi: 10.3109/08916930903540432. [DOI] [PubMed] [Google Scholar]
- 16.Imani D, Azimi A, Salehi Z, Rezaei N, Emamnejad R, Sadr M, Izad M. Association of nod-like receptor protein-3 single nucleotide gene polymorphisms and expression with the susceptibility to relapsing-remitting multiple sclerosis. Int J Immunogenet. 2018;45:329–336. doi: 10.1111/iji.12401. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Li S, Liu Z, Jiang S, Wang J, Guo M, Zhao X, Song W, Liu S. The NLRP3 rs10754558 polymorphism is a risk factor for preeclampsia in a Chinese Han population. J Matern Neonatal Med. 2019;32:1792–1799. doi: 10.1080/14767058.2017.1418313. [DOI] [PubMed] [Google Scholar]
- 18.Zhou XY, Fernando SM, Pan AY, Laposa R, Cullen KR, Klimes-Dougan B, Andreazza AC. Characterizing the NLRP3 inflammasome in mood disorders: Overview, technical development, and measures of peripheral activation in adolescent patients. Int J Mol Sci. 2021;22:12513. doi: 10.3390/ijms222212513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Özerdem A, Yazıcı O, Tunca Z, Tırpan K. Establishment of computerized registry program for bipolar illnes in turkey: SKIP-TURK. J Affect Disord. 2004;84:82–86. [Google Scholar]
- 20.First MB, Williams JB, Karg RS, Spitzer RL. User's guide for the SCID-5-CV Structured Clinical Interview for DSM-5® disorders: Clinical version: American Psychiatric Publishing, Inc. 2016 [Google Scholar]
- 21.Elbir M, Topbaş ÖA, Bayad S, Kocabaş T, Topak OZ, Çetin Ş, Özdel O, Ateşçi F, Aydemir Ö. Adaptation and reliability of the structured clinical interview for DSM-5-disorders-clinician version (SCID-5/CV) to the Turkish language. Turk Psikiyatri Derg. 2019;30:51–56. (In Turkish) [PubMed] [Google Scholar]
- 22.Young RC, Biggs JT, Ziegler VE, Meyer DA. A rating scale for mania: Reliability, validity and sensitivity. Br J Psychiatry. 1978;133:429–435. doi: 10.1192/bjp.133.5.429. [DOI] [PubMed] [Google Scholar]
- 23.Karadag F, Oral T, Yalcin FA, Erten E. Reliability and validity of Turkish translation of young mania rating scale. Turk Psikiyatri Derg. 2002;13:107–114. (In Turkish) [PubMed] [Google Scholar]
- 24.Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akdemir A, Türkçapar MH, Örsel SD, Demirergi N, Dag I, Özbay MH. Reliability and validity of the Turkish version of the hamilton depression rating scale. Compr Psychiatry. 2001;42:161–165. doi: 10.1053/comp.2001.19756. [DOI] [PubMed] [Google Scholar]
- 26.Rosa AR, Sánchez-Moreno J, Martínez-Aran A, Salamero M, Torrent C, Reinares M, Comes M, Colom F, Van Riel W, Ayuso-Mateos JL, et al. Validity and reliability of the functioning assessment short test (FAST) in bipolar disorder. Clin Pract Epidemiol Ment Health. 2007;3:5. doi: 10.1186/1745-0179-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aydemir O, Uykur B. Reliability and validity study of the Turkish version of functioning assessment short test in bipolar disorder. Turk Psikiyatri Derg. 2012;23:193–200. (In Turkish) [PubMed] [Google Scholar]
- 28.Hamshere ML, Gordon-Smith K, Forty L, Jones L, Caesar S, Fraser C, Hyde S, Tredget J, Kirov G, Jones I, et al. Age-at-onset in bipolar-I disorder: Mixture analysis of 1369 cases identifies three distinct clinical sub-groups. J Affect Disord. 2009;116:23–29. doi: 10.1016/j.jad.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 29.Grigoroiu-Serbanescu M, Martinez M, Nöthen MM, Grinberg M, Sima D, Propping P, Marinescu E, Hrestic M. Different familial transmission patterns in bipolar I disorder with onset before and after age 25. Am J Med Genet. 2001;105:765–773. doi: 10.1002/ajmg.10047. [DOI] [PubMed] [Google Scholar]
- 30.Namipashaki A, Razaghi-Moghadam Z, Ansari-Pour N. The essentiality of reporting hardy-weinberg equilibrium calculations in population-based genetic association studies. Cell J. 2015;17:187–192. doi: 10.22074/cellj.2016.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryant C, Fitzgerald KA. Molecular mechanisms involved in inflammasome activation. Trends Cell Biol. 2009;19:455–464. doi: 10.1016/j.tcb.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 33.Wei P, Yang F, Zheng Q, Tang W, Li J. The potential role of the NLRP3 inflammasome activation as a link between mitochondria ROS generation and neuroinflammation in postoperative cognitive dysfunction. Front Cell Neurosci. 2019;13:73. doi: 10.3389/fncel.2019.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Q, Fan HW, Zhang JZ, Wang YM, Xing HJ. NLRP3 rs35829419 polymorphism is associated with increased susceptibility to multiple diseases in humans. Genet Mol Res. 2015;14:13968–13980. doi: 10.4238/2015.October.29.17. [DOI] [PubMed] [Google Scholar]
- 35.Pontillo A, Vendramin A, Catamo E, Fabris A, Crovella S. The missense variation Q705K in CIAS1/NALP3/NLRP3 gene and an NLRP1 haplotype are associated with celiac disease. Am J Gastroenterol. 2011;106:539–544. doi: 10.1038/ajg.2010.474. [DOI] [PubMed] [Google Scholar]
- 36.Roberts RL, Topless RKG, Phipps-Green AJ, Gearry RB, Barclay ML, Merriman TR. Evidence of interaction of CARD8 rs2043211 with NALP3 rs35829419 in Crohn's disease. Genes Immun. 2010;11:351–356. doi: 10.1038/gene.2010.11. [DOI] [PubMed] [Google Scholar]
- 37.Agah E, Nafissi S, Saleh F, Sarraf P, Tafakhori A, Mousavi SV, Saghazadeh A, Sadr M, Sinaei F, Mohebbi B, et al. Investigating the possible association between NLRP3 gene polymorphisms and myasthenia gravis. Muscle Nerve. 2021;63:730–736. doi: 10.1002/mus.27193. [DOI] [PubMed] [Google Scholar]
- 38.Pontillo A, Girardelli M, Kamada AJ, Pancotto JAT, Donadi EA, Crovella S, Sandrin-Garcia P. Polimorphisms in inflammasome genes are involved in the predisposition to systemic lupus erythematosus. Autoimmunity. 2012;45:271–278. doi: 10.3109/08916934.2011.637532. [DOI] [PubMed] [Google Scholar]
- 39.Redenšek S, Flisar D, Kojović M, Kramberger MG, Georgiev D, Pirtošek Z, Trošt M, Dolžan V. Genetic variability of inflammation and oxidative stress genes does not play a major role in the occurrence of adverse events of dopaminergic treatment in Parkinson's disease. J Neuroinflammation. 2019;16:50. doi: 10.1186/s12974-019-1439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanaei S, Sadr M, Rezaei A, Shahkarami S, Daryani NE, Bidoki AZ, Rezaei N. Association of NLRP3 single nucleotide polymorphisms with ulcerative colitis: A case-control study. Clin Res Hepatol Gastroenterol. 2018;42:269–275. doi: 10.1016/j.clinre.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 41.Herman R, Jensterle M, Janež A, Goričar K, Dolžan V. Genetic variability in antioxidative and inflammatory pathways modifies the risk for PCOS and influences metabolic profile of the syndrome. Metabolites. 2020;10:439. doi: 10.3390/metabo10110439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antypa N, Serretti A. Family history of a mood disorder indicates a more severe bipolar disorder. J Affect Disord. 2014;156:178–186. doi: 10.1016/j.jad.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 43.Zalar B, Blatnik A, Maver A, Klemenc-Ketiš Z, Peterlin B. Family history as an important factor for stratifying participants in genetic studies of major depression. Balkan J Med Genet. 2018;21:5–12. doi: 10.2478/bjmg-2018-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Post RM, Altshuler LL, Kupka R, McElroy SL, Frye MA, Rowe M, Grunze H, Suppes T, Keck PE, Jr, Leverich GS, Nolen WA. Multigenerational transmission of liability to psychiatric illness in offspring of parents with bipolar disorder. Bipolar Disord. 2018;21 doi: 10.1111/bdi.12668. doi: 10.1111/bdi.12668. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 45.Axelson D, Goldstein B, Goldstein T, Monk K, Yu H, Hickey MB, Sakolsky D, Diler R, Hafeman D, Merranko J, et al. Diagnostic precursors to bipolar disorder in offspring of parents with bipolar disorder: A longitudinal study. Am J Psychiatry. 2015;172:638–646. doi: 10.1176/appi.ajp.2014.14010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Studart-Bottó P, Bezerra-Filho S, Sarmento S, Miranda-Scippa Â. Social support in patients with bipolar disorder and differing ages at onset. Clin Psychol Psychother. 2022;29:351–359. doi: 10.1002/cpp.2617. [DOI] [PubMed] [Google Scholar]
- 47.Geoffroy PA, Etain B, Scott J, Henry C, Jamain S, Leboyer M, Bellivier F. Reconsideration of bipolar disorder as a developmental disorder: Importance of the time of onset. J Physiol. 2013;107:278–285. doi: 10.1016/j.jphysparis.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 48.Grigoroiu-Serbanescu M, Rietschel M, Hauser J, Czerski PM, Herms S, Sun X, Wickramaratne P, Elston RC. Commingling analysis of age-of-onset in bipolar I disorder and the morbid risk for major psychoses in first degree relatives of bipolar I probands. J Affect Disord. 2014;168:197–204. doi: 10.1016/j.jad.2014.06.054. [DOI] [PubMed] [Google Scholar]
- 49.Kennedy KP, Cullen KR, Deyoung CG, Klimes-Dougan B. The genetics of early-onset bipolar disorder: A systematic review. J Affect Disord. 2015;184:1–12. doi: 10.1016/j.jad.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soni A, Singh P, Kumar S, Shah R, Batra L, Verma M. Role of age at onset in the clinical presentation of bipolar disorder in Indian population. Ind Psychiatry J. 2021;30:41–46. doi: 10.4103/ipj.ipj_8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu LS, Huang MC, Fann CS, Lane HY, Kuo CJ, Chiu WC, Kwok PY, Cheng AT. Genome-wide association study of early-onset bipolar I disorder in the Han Taiwanese population. Transl Psychiatry. 2021;11:301. doi: 10.1038/s41398-021-01407-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.