Abstract
In recent years, studies have demonstrated that vascular endothelial growth factor B (VEGFB) can affect the metabolism of fatty acids and glucose, and it is expected to become a target for the diagnosis and treatment of metabolic diseases such as obesity and diabetes. At present, the specific mechanism that VEGFB regulates lipid and glucose metabolism balance is not completely understood. The present study used systemic VEGFB gene-knockout mice to investigate the effects of downregulation of the VEGFB gene on lipid metabolism and insulin secretion, and to explore the mechanism of the VEGFB pathway involved in the regulation of glucose and lipid metabolism. The morphological changes in the liver and pancreas of mice after VEGFB gene deletion were observed under a light microscope and a scanning electron microscope, and the effects of VEGFB gene deletion on lipid metabolism and blood glucose balance were detected by a serological technique. The detection indexes included total cholesterol (TC), triglyceride (TG), low-density lipoprotein cholesterol (LDL-C) and high-density lipoprotein cholesterol. Simultaneously, fasting blood glucose, glycosylated hemoglobin A1c (HbA1c), fasting insulin and glucagon were measured. Insulin sensitivity was assessed by using the insulin tolerance tests and glucose tolerance tests, and function of β-cell islets was evaluated by using the insulin resistance index (HOMA-IR) and pancreatic β-cell secretion index (HOMA-β). Τhe protein expression changes of vascular endothelial growth factor receptor 1 (VEGFR1) and vascular endothelial growth factor receptor 2 (VEGFR2) in mouse islets were detected by western blotting and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) after the VEGFB gene was knocked down to analyze the mechanism of VEGFB that may be involved in glucose and lipid metabolism. It was observed that after VEGFB was knocked down, mouse hepatocytes exhibited steatosis and increased secretory vesicles in islet cells. The lipid metabolism indexes such as TG, TC and LDL increased significantly; however, the levels of FBS, postprandial blood glucose and HbA1c decreased, whereas the glucose tolerance increased. Serum insulin secretion increased and HOMA-IR decreased since VEGFB was knocked down. Western blotting and RT-qPCR results revealed that the expression levels of VEGFR1 and neuropilin-1 decreased after the VEGFB gene was knocked down, while the expression levels of VEGFA and VEGFR2 increased. The absence of VEGFB may be involved in the regulation of glucose and lipid metabolism in mice by activating the VEGFA/VEGFR2 signaling pathway. VEGFB is expected to become a new target for the treatment of metabolic diseases such as obesity and diabetes. At present, the mechanism of VEGFB involved in regulating lipid metabolism and glucose metabolism is not completely clear. It was identified that downregulating VEGFB improved lipid metabolism and insulin resistance. The role of VEGFB/VEGFR1 pathway and other family members in regulating glucose and lipid metabolism was detected, which provided a theoretical and experimental basis for VEGFB to affect the regulation of glucose and lipid metabolism balance.
Keywords: vascular endothelial growth factor B, glucose and lipid metabolism, blood sugar balance, lipid metabolism, insulin resistance, vascular endothelial growth factor receptor 1
Introduction
An imbalance in metabolic homeostasis is the primary cause of various chronic diseases. The number of patients experiencing various chronic diseases caused by metabolic disorders has increased significantly in various countries worldwide. According to the prediction of the World Health Organization, obesity, hyperlipidaemia, fatty liver disease and other diseases dominated by lipid metabolism disorders have affected more than one third of the global population (1,2). The number of patients with diabetes and glucose metabolism disorders will reach 642 million by 2040, and more than 90% of these patients will have type 2 diabetes mellitus (T2DM) (3). Lipid metabolism disorder is a common metabolic disorder leading to targeted organ damage. For example, lipid metabolism disorder is one of the risk factors for cardiovascular disease, which can cause atherosclerosis, unstable plaque rupture and accelerated thrombosis (4). Disorder of lipid metabolism can also cause renal injury. High levels of triglycerides (TG) and low-density lipoprotein cholesterol (LDL-C) are deposited in the arterial wall and glomerular basement membrane, which may cause renal endothelial cell injury. In addition, chylomicrons and other residual lipoproteins in plasma can affect the development of diabetic nephropathy by damaging the vascular endothelial barrier and activating the platelets. Abnormal lipid deposition in the liver can cause hepatocyte steatosis and non-alcoholic steatohepatitis, accompanied by hepatocyte injury, inflammation, angiogenesis and varying degrees of fibrosis. Simultaneously, it can affect the insulin signal pathway and cause insulin resistance in the liver (5,6). Lipid and glucose metabolisms interact and regulate each other. High levels of TG can promote the disorder of insulin signal transduction, resulting in a decrease in glucose utilization. Furthermore, persistent hyperglycemia in patients with T2DM can promote fatty acid (FA) synthesis and TG accumulation, resulting in abnormal lipid metabolism and deposition. Long-term hyperglycemia will not only lead to diabetes but also increase the risk of metabolic diseases such as cardiovascular and cerebrovascular and non-alcoholic fatty liver diseases (7,8). The research on the influence of the regulation mechanism of blood glucose homeostasis and lipid metabolism on the development of chronic diseases is one of the hot issues concerned for the scholars.
VEGFB (vascular endothelial growth factor B) is a glycoprotein that induces a series of reactions via its receptors VEGFR1 and neuropilin-1 (NRP1) through the paracrine pathway (9). VEGFR1 is highly expressed in mitochondrial-rich tissues such as the heart, liver, muscle and brown adipose tissue. Previously, it was suggested that VEGFB is a survival molecule involved in the regulation of FA metabolism and glucose homeostasis (10,11). In 2010, Hagberg et al (12) first reported in Nature that VEGFB can regulate FA absorption in endothelial cells, transfer excess FAs to tissues with high energy metabolism and improve lipid transportation in endothelial cells. Targeting VEGFB may be a novel approach to preventing pathological lipid deposition (12). In 2012, Hagberg et al (10) reported that reducing VEGFB signal improves insulin sensitivity and glucose tolerance in T2DM animal models. In 2014, Wagenmakers et al (13) suggested that VEGFB controls the expression of FA transporters in capillary endothelial cells, which may prevent the accumulation of lipotoxic FAs. In 2017, Falkevall et al (14) revealed that VEGFB gene deletion can not only prevent pathological lipid deposition but also improve glucose homeostasis and insulin sensitivity by targeting the lipid transport properties of endothelial cells. In 2020, Moessinger et al (15) established a VEGFB gene knockout mouse model and observed that the VEGFB signal pathway can reduce endothelial cell glucose transport and cardiac glucose utilization. In the same year, Jensen et al (16) observed that islet β-cell specific VEGFB deficiency increased insulin secretion by upregulating the insulin gene. Therefore, numerous scholars have proposed that VEGFB may be a target for the treatment of metabolic diseases such as obesity and diabetes.
As one of the members of the VEGFs family, VEGFB mainly plays a biological role after binding to the membrane receptor VEGFR1. VEGFR1 consists of seven extracellular immunoglobulin homologous regions, one transmembrane structural region and one intracellular tyrosine kinase region (17). VEGFR1 is primarily expressed in endothelial cells, osteoblasts, monocytes/giant cells, phage cells, blastoderm trophoblast cells, renal interstitial cells and certain haematopoietic stem cells (18). In 2016, Robciuc et al (11) reported in Cell that in VEGFB transduced adipose tissue and VEGFB transgenic mouse models, fat vasodilation induced by VEGFB/VEGFR1 can counteract obesity and related metabolic complications (11). Shen et al (19) observed that VEGFB can reduce myocardial lipid accumulation and hypertrophy through VEGFR1 and its related downstream signaling pathways. In 2021, Hu et al (20) also proved that VEGFB/VEGFR1 could reduce lipid accumulation by activating the CaMKK/AMPK/ACC/CPT1 pathway.
At present, studies have reported that VEGFB can participate in the regulation of lipid metabolism. However, there are limited reports on whether it can cause changes in glucose metabolism while regulating lipid metabolism. Whether VEGFB can participate in the regulation of glucose and lipid metabolism via the VEGFB/VEGFR1 pathway remains unclear. In the present study, a VEGFB gene knockout mouse was established to study the effects of VEGFB gene deletion on glucose and lipid metabolism. In addition, the role of VEGFB in the signal pathway that regulates glucose and lipid metabolism was analyzed, and its findings provided evidence for the pathogenesis of metabolic diseases.
Materials and methods
Animals
All animal experiments were approved by the ethics committee of Binzhou Medical University (Yantai, China) and strictly abided by the animal ethics code. All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee of the Medical Ethics Committee of Binzhou Medical University (IACUC protocol no. 2021-300). C57BL/6N male mice were selected as experimental animals. The VEGFB gene knockout mouse model was constructed by Saiye (Guangzhou) Biological Technology Co., Ltd. using Crispr/cas9 system. VEGFB gene ID is 22340, locates on chromosome 19, and NM_ 011697.3 transcript was selected as reference transcript. This transcript has 7 exons, ATG is on exon 1 and TAG is on exon 6. In the present experiment, 90% of the protein coding region of exon 2~6 of VEGFB gene was knocked out in a large fragment manner, and the knockout fragment was ~2,000 bp. VEGFB gene retains the first 30 amino acids of exon 1 (Fig. 1A). The level of VEGFB knockdown in fertilized eggs was verified by PCR amplification and sequencing. Two pairs of primers were used for PCR cycle and the primer 1 sequence was: forward, 5′-TCTCAAGGTTGGCGGAAGTGG-3′ and reverse,: 5′-CAAACTCACCATGTCACCAAGGAG-3′. Primer 2 sequence was: forward, 5′-TCTCAAGGTTGGCGGAAGTGG-3′ and reverse, 5′-TTGGGATCACGCAAGATAAGGG-3′. The gRNA vector and cas9 mRNA were co-injected into in vitro fertilized eggs and transplanted into the fallopian tubes of C57BL/6N strain surrogate mice. Positive VEGFB heterozygous mice (VEGFB+/−) mice of generation 0 (F0 mice) were obtained after inoculation. After sexual maturation, F0 mice were bred and propagated with C57BL/6N mice, and F1 hybrid knockdown mice were identified. The F1 generation of heterozygous knockdown mice were selfed to obtain homozygous knockdown mice (VEGFB−/−), heterozygous knockdown mice and wild mice (VEGFB+/+). Healthy male mice, aged 4 weeks and weighing 18–22 g were randomly selected for the experiment. In total 9 mice were selected for each of the VEGFB+/+ and VEGFB−/− groups. All animals were kept in the SPF animal room of the Medical Research Center of Binzhou Medical College. Conditions were maintained at 20°C and a relative humidity of 50±20%, with food and water obtained freely and a 12-h light/dark cycle. During animal experiment, 6 mice of each group were selected for anesthesia with 3% isoflurane, and then the mice were sacrificed by cervical dislocation after blood collection from the eyeball. The liver and pancreas tissue were received in 4°C condition for molecular biology experiments. Meanwhile, 3 mice of each group were sacrificed by cervical dislocation to obtain liver and pancreas for morphological experiments.
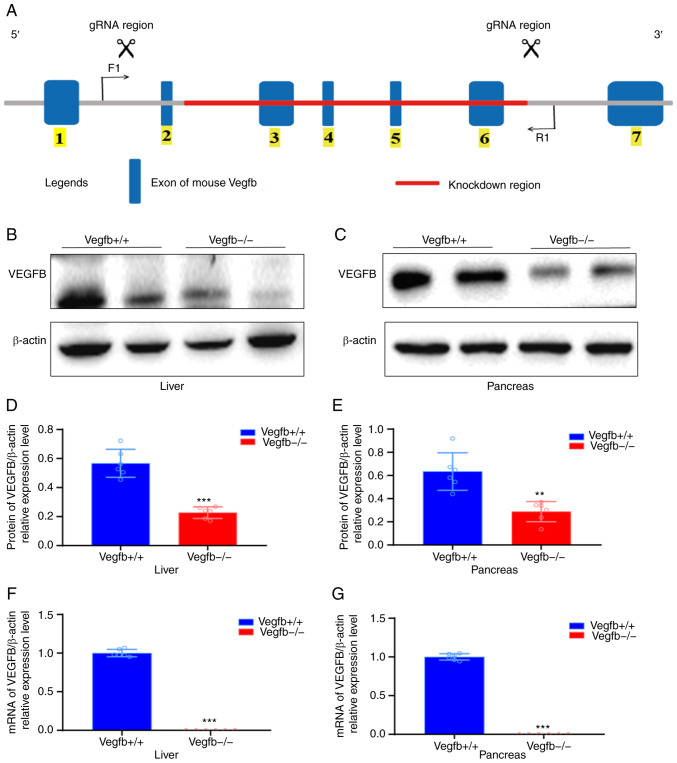
Figure 1.
Systemic VEGFB-knockdown mouse model construction and genetic identification. (A) VEGFB gene structure, gRNA target site and deletion region. Boxes and lines respectively represent exons and introns. F1 and R1 arrows indicate the position of primers for PCR genotyping. 1–7 shows the VEGFB gene sequence, where the red line represents the VEGFB knockout gene sequence. (B-E) Western blot analysis of VEGFB protein in liver tissues and pancreas. (F and G) mRNA analysis in the liver tissues and pancreas. **P<0.01 vs. VEGFB+/+ group and ***P<0.001 vs. VEGFB+/+ group. VEGFB, vascular endothelial growth factor B.
Isolation of mouse islets
Mice in VEGFB+/+ and VEGFB−/− groups were received after fasting for 12 h, with 6 mice in each group. After anesthesia, they were disinfected with alcohol, the abdominal cavity was opened, then the pancreas was extracted and placed in the culture dish. The adipose tissue was picked out in Hank's buffer, and then 0.5 mg/ml collagenase P was poured into the pancreas. After the pancreas was completely expanded, it was digested in a 37°C water bath for 10 min. The digested pancreas was shaken quickly until it reached the shape of sediment; then it was placed into the petri dish and precooled (4°C) Hank's buffer was added to stop digestion. Under the ×100 visual field of a stereomicroscope, isolated round and smooth islet cell masses were selected.
Islet identification
The same amount of dithizone (DTZ) solution was added to 50 µl islet cell suspension, and staining was performed at room temperature for 10 min. The islet cell mass was identified under a ×200 visual field of a fluorescence microscope.
Western blot analysis
Mouse tissue lysates were analyzed by western blotting to detect changes in related protein expression. The liver tissue and islets from six mice in each group were selected for organ extraction. Protease inhibitor (1% PMSF; cat. no. PO100) and tissue/cell high-efficiency lysate (RIPA; cat. no. R0010; both from Beijing Solarbio Science & Technology Co., Ltd.) were added into tissue and then tissue was lysed on ice in 4°C for 30 min using ultrasonication (Ultrasonic Cell Crusher XC-CD). The concentration of the extracted proteins was measured using a BCA kit. The supernatant was collected after centrifugation at 15,300 × g and 4°C for 20 min. Sample loading buffer (D1020-5; Beijing Solarbio Science & Technology Co., Ltd.) was added, boiled in a pan (Tu-100C) at 95°C for 10 min, and 20 µg sample protein was dissolved in gel after cooling and then transfered to a polymer PVDF membrane. VEGFB (22 kDa) and VEGFA (27 kDa) are small molecular weight proteins and the concentration of gel was 12%. VEGFR1 (180 kDa), VEGFR2 (151 kda) and NRP1 (103 kDa) are high molecular weight proteins and the gel concentration was 10%. The membrane was blocked with 5% skimmed milk at room temperature for 1 h, and then incubated overnight with primary antibody (Table I) at 4°C. The next day, incubation was performed with anti-rabbit IgG HRP-conjugated antibody (cat. no. 111-035-003) and anti-mouse IgG HRP-conjugated antibody (cat. no. 115-035-003) (1:5,000; Jackson ImmunoResearch Laboratories, Inc.) at room temperature for 2 h. Protein band Biosharp ECL prime Western blot Reagent (Biosharp Life Sciences; cat. no. BL520b) was used and detected in the enhanced chemiluminescence system (Tanon-5200; Tanon Science & Technology). ImageJ software v1.8.0 (National Institutes of Health) was used to analyze the gray value of protein.
Table I.
Antibody information.
| Antibody name | Dilution | Species/source | Company | Cat. no. |
|---|---|---|---|---|
| VEGFB | 1:1,000 | Rabbit | Affinity Biosciences | AF5250 |
| VEGFA | 1:1,000 | Rabbit | Affinity Biosciences | DF7470 |
| VEGFR1 | 1:1,000 | Rabbit | Affinity Biosciences | AF6204 |
| VEGFR2 | 1:1,000 | Rabbit | Affinity Biosciences | AF6281 |
| NRP1 | 1:1,000 | Rabbit | Affinity Biosciences | DF7877 |
| β-actin | 1:1,000 | Mouse | Affinity Biosciences | AF7018 |
Reverse transcription-quantitative (RT-q) PCR
A total of six mice in each group of VEGFB+/+ group and VEGFB−/− group were selected to extract liver and islet tissues; total RNA was extracted with TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RT was performed according to the manufacturer's instructions using an RNA Easy Isolation Reagent (Vazyme Biotech Co., Ltd.). qPCR was performed according to the manufacturer's instructions using TB Green Premix Ex Taq II (Takara Bio USA, Inc.) fluorescence quantitative kit. PCR amplification was performed on PCR machine QuantStudio 3 (Thermo Fisher Scientific, Inc.). qPCR was performed using the following thermocycling conditions: Initial denaturation at 95°C for 30 sec; then 40 cycles were performed at 95°C for 5 sec and 60°C for 34 sec; finally, the dissolution process was carried out at 95°C, 60°C and 95°C for 15 sec, 1 min and 15 sec, respectively. The 2−ΔΔCq method was used to quantify the level of mRNA expression using β-actin as an internal reference gene (21). Primer sequences are provided in Table II.
Table II.
Primer sequences for quantitative PCR.
| Primer name | Primer sequence (5′-3′) |
|---|---|
| VEGFB | F: AGCCACCAGAAGAAAGTGGT |
| R: GCTGGGCACTAGTTGTTTGA | |
| VEGFA | F: GAGGCTGCTGTAACGATGAA |
| R: TATGTGCTGGCTTTGGTGAT | |
| VEGFR1 | F: TTGGTGGTGGCTGACTCTCA |
| R: TCTCCTTCGGCTGGCATCTT | |
| VEGFR2 | F: TGATTTCACCTGGCACTCTCC |
| R: CCTTGGTCACTCTTGGTCACA | |
| NRP1 | F: CAGGGTTTTCCATCCGCTATG |
| R: ACTCCAGTAGGTGCTGTATAGTT | |
| β-actin | F: CATCCGTAAAGACCTCTATGCCAAC |
| R: ATGGAGCCACCGATCCACA |
VEGFB, vascular endothelial growth factor B; NRP1, neuropilin-1; VEGFR, vascular endothelial growth factor receptor.
Body weight, food intake and blood glucose measurement
The gene identification of mice started at the 3rd week, and the weight and food intake of wild and homozygote group were measured every 2 weeks from the 4th week to draw the growth and food intake curve. The mice were euthanized until the 32nd week for tissue section. Experiments in vivo lasted 7 months. Meanwhile, the fasting blood sugar and postprandial blood sugar of mice were examined every 2 weeks to draw the blood sugar curve from the 4th week to the 32nd week. After fasting for 12 h, blood was drawn from the tail vein by using a Roche blood glucose-meter for fasting blood glucose (FBG) measurement. Then food was provided for 2 h and the Roche blood glucose-meter was used to draw blood for postprandial blood glucose (PBG) measurement and the curve graph of blood glucose was drawn.
Organ coefficient ratio
Mice (32 weeks old) were sacrificed by cervical dislocation. The liver and pancreas were immediately dissected, the fatty tissue around the organ was removed and the organ surface and residual blood in the cavity were treated. The net weight of the organ was measured with an electronic balance (BS210S) according to the following formula: organ coefficient=net organ weight (g)/body weight (g).
Hematoxylin-eosin (H&E) staining
The liver and pancreas of the mice were fixed with 4% paraformaldehyde at room temperature for 24–48 h, then dehydrated with gradient alcohol, transparency was achieved with xylene, tissues embedded in paraffin and sectioned at a thickness of 4 µm. The tissue sections were incubated at 60°C in an oven for 2 h for H&E staining at room temperature. The protocol was as follows: the tissue sections were dewaxed with xylene twice for 10 min each time. Then they were treated with 100, 100, 95, 85 and 75% alcohol for 5 min each time. After washing with distilled water, tissue sections were immersed in 0.4% hematoxylin, stained for 5 min and washed with running water for 1 min. Subsequently, the tissue sections were immersed in 1% hydrochloric acid ethanol differentiation solution for 10 sec and washed with water until they turned into blue. Then, the tissue sections were immersed in 0.1% eosin, stained for 1 min, and dehydrated with 75, 85, 95, 100 and 100% inverse concentration alcohol for 5 min each time. After xylene transparency-treatment twice, the sections were sealed with neutral gum and observed under the microscope.
Transmission electron microscopy
A total of 3 mice (aged 32 weeks) in each group were fasted for 12 h and sacrificed under anesthesia. The liver and pancreas of the mice were isolated and fixed with 2.5% glutaraldehyde solution at 4°C for 2 h. After rinsing with buffer solution 3 times at room temperature, the tissues were immediately fixed with 1% osmic acid at 4°C for 90 min, and rinsed with buffer solution 3 times again. Sequential gradient ethanol solution dehydration followed by acetone replacement treatment twice for 15 min each time. The samples were embedded in different proportions of acetone (V1/V2=1/2; V1/V2=2/1) and mixed and processed for 1 and 2 h, and then embedded with epoxy resin overnight at 70°C. The samples were sliced with a Reichert ultrathin microtome (70 nm). The sections were stained with lead citrate solution for 15 min at 25°C and then stained with uranyl acetate 50% ethanol saturated solution for 15 min at 25°C. Then observed and photographed through a transmission electron microscope (JEM-1400, Japan), and used Image-Pro Plus version 6.0 (media cybernetics, Inc.) to analyze.
ELISA
A total of 6 mice (aged 32 weeks) were selected from each of the VEGFB+/+ and VEGFB-/- groups. After 12 h of fasting, they were deeply anesthetized with 3% isoflurane. Eyeball blood was taken and then the mice were killed by cervical dislocation. Whole blood was taken in each group. The supernatant was collected using 3,800 × g centrifugation at 4°C for 20 min, and a microplate reader was used to measure the optical density value. A standard curve was established based on the measured values of the standard products, and the level of each group of samples was calculated from the standard curve obtained. Data results were statistically analyzed. The mouse ELISA kits used for the detection of TG (cat no: a110-1-1), total cholesterol (TC; cat. no.: a111-1-1), LDL (cat. no.: 113-1-1) and high-density lipoprotein (HDL; cat. no.: 112-1-1) were purchased from Nanjing Jiancheng Bioengineering Institute. The mouse ELISA kits used for the detection of glycosylated hemoglobin (HbA1c; cat. no.: ml063816), glucose (cat. no.: ml076701), insulin (cat. no.: ml001983) and glucagon (cat. no.: ml057708) were all purchased from Shanghai Enzyme-linked Biotechnology Co., Ltd. The ELISA tests were performed according to the manufacturers' protocol and the OD value was measured at 510 or 450 nm on the microplate reader (BioTek China).
Oral glucose tolerance test (OGTT) and intraperitoneal insulin tolerance test (IPITT)
In VEGFB+/+ group and VEGFB−/− group, 9 mice in each group were selected for OGTT and IPITT experiments. In the OGTT experiment, mice were fed with 40% glucose at the dose of 2 g/kg after fasting for 12 h, and then the venous blood glucose was measured with a blood glucose-meter at 0, 15, 30, 60, 90 and 120 min, and the OGTT curve was drawn. In the IPITT experiment, the mice fasted for 6 h, were injected with insulin intraperitoneally at the rate of 0.5 µg/kg, the venous blood glucose of the mice was measured at 0, 15, 30, 60, 90 and 120 min and the IPITT curve was drawn.
Insulin resistance index (HOMA-IR) and pancreatic β cell function index (HOMA-β) measurement
Calculating mouse insulin resistance index (HPMA-IR) and pancreatic β-cell function index (HOMA-β) were determined by FBG and fasting insulin (FINS). The formula is as follows: HOMA-IR=FBG × FINS)/22.5; HOMA-β=FINS × 20/(FBG-3.5).
Statistical analysis
All data were analyzed statistically with SPSS 22.0 statistical software (IBM Corp.). The results were expressed as the mean ± SD. One-way ANOVA followed by Dunnet's post hoc test was used, while comparisons between two groups were assessed using paired Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Systemic VEGFB-knockdown mouse model construction and genetic identification
In order to explore the regulatory effect of VEGFB on glucose and lipid metabolism and its mechanism, systemic VEGFB-knockdown mice were constructed through CRISPR/cas9-mediated genomic engineering. The VEGFB gene is located on mouse chromosome 19. A total of 7 exons were identified. The ATG start codon was in exon 1, the tag stop codon was in exon 6 and exons 2–6 were selected as targeted sites (Fig. 1A). To determine whether the VEGFB gene was knocked out, liver and pancreatic tissues with high VEGFB expression were selected for western blotting and gene transcription level analysis. Compared with VEGFB+/+ mice, the protein and mRNA levels of VEGFB in liver and pancreas tissues of VEGFB−/− mice were significantly downregulated (P<0.05; Fig. 1B-G).
The effect of VEGFB downregulation on the liver and pancreas
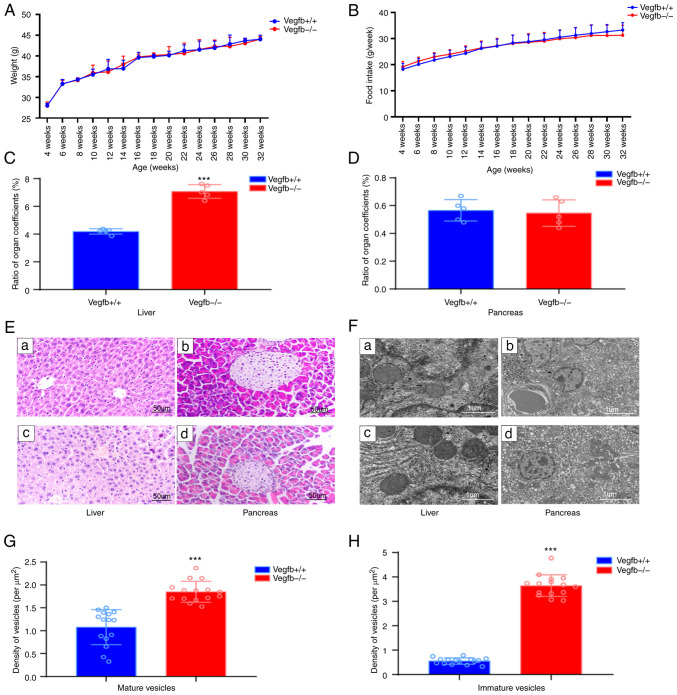
Mice in both groups were in favorable mental health, lively and active, with bright coat colors, normal diet and water intake and urine output and dry litter. There was no significant change in weight gain or average food intake in VEGFB−/− mice from week 4 to week 32 compared with VEGFB+/+ mice under the same feeding conditions (P>0.05; Fig. 2A and B). Compared with that of VEGFB+/+ mice, the liver-to-viscera ratio of VEGFB−/− mice increased (P<0.001), but the pancreas-to-viscera ratio was not significantly different (P>0.05; Fig. 2C and D). The results of H&E staining showed that the structure of liver lobules in VEGFB+/+ mice was normal and the liver cells were arranged neatly. The liver tissue of VEGFB−/− mice showed obvious steatosis, the volume of hepatocytes was increased, and circular lipid droplets of varying numbers and sizes were observed in the cytoplasm. The nuclei of liver cells were squeezed from the centre to the periphery (Fig. 2E). Compared with that of VEGFB+/+ mice, the pancreatic tissue of VEGFB−/− mice showed no obvious changes. Under the microscope, spherical islet structures scattered among spicules of different sizes and stains were observed (Fig. 2E). To further observe the effects of VEGFB gene deletion on mouse liver and pancreas, sections of 32-week-old VEGFB+/+ mice and VEGFB−/− mice were received to observe ultrastructural changes of liver and islet cells by transmission electron microscopy. The liver cells of VEGFB−/− mice had a large number of lipid droplets, up to 5-µm in diameter, irregular concave nuclei and abnormal inhomogeneous chromatin in the nuclei. The islet nuclei of VEGFB+/+ mice and VEGFB−/− mice were round, the nuclear membrane was clear and complete, the perinuclear space was normal and the organelles such as mitochondria, ribosomes and rough endoplasmic reticulum could be clearly observed in the cytoplasm (Fig. 2F). In addition, more endocrine granules were observed, most of which were secretory granules of pancreatic islet β-cells. Insulin-secreting vesicles were present in two different forms: immature vesicles with a uniform grey color within and mature vesicles with a dense black core (Fig. 2F). Compared with VEGFB+/+ mice, VEGFB−/− mice had significantly more insulin vesicles. The numbers of mature and immature vesicles were also significantly higher than those of VEGFB+/+ mice (P<0.05; Fig. 2G and H).
Figure 2.
Effect of VEGFB downregulation on liver and pancreas. (A) Body weight curve of mice (4–32 weeks). (B) Food intake curve of mice (4–32 weeks) (C and D) Organ coefficient ratio between liver and pancreas in mice. (E) Light microscopic structure of liver and pancreas; a and b: VEGFB+/+ group, c and d: VEGFB-/- group (scale bar, 50 µm; magnification, ×400). (F) Electron microscopic structure of liver and pancreas; a and b: VEGFB+/+ group, c and d: VEGFB-/- group (scale bar, 1 µm; images a and c, ×15,000 magnification; images b and d, ×8,000 magnification). (G and H) Density of mature and immature vesicles. ***P<0.001 vs. VEGFB+/+ group. VEGFB, vascular endothelial growth factor B.
The effect of VEGFB downregulation on serum glucose and lipid metabolism
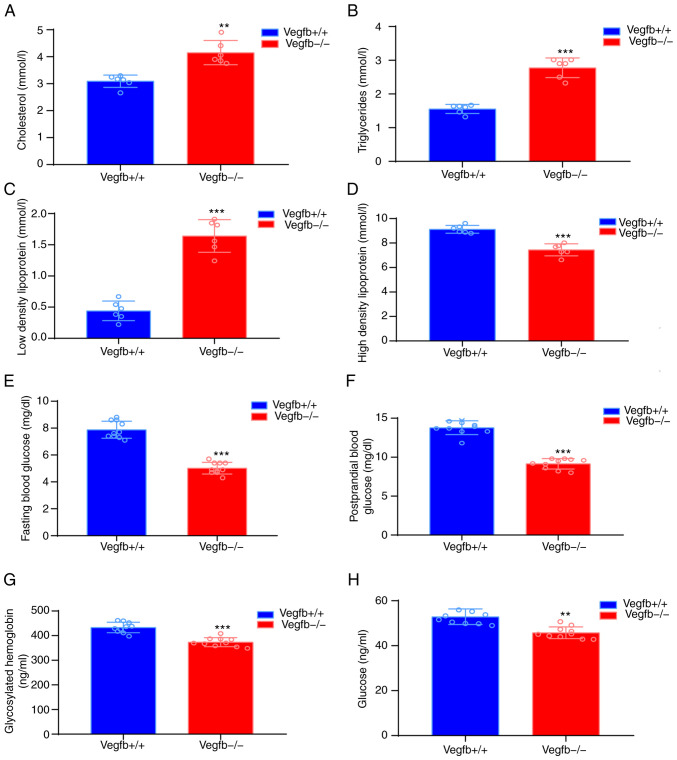
Serological analysis revealed that VEGFB−/− TC (Fig. 3A), TG (Fig. 3B) and LDL-C (Fig. 3C) levels were significantly increased, while HDL-C levels were significantly lower in VEGFB−/− mice (P<0.001; Fig. 3D). Compared with those of VEGFB+/+ mice, the FBG and PBF levels of VEGFB−/− mice were significantly lower (P<0.001; Fig. 3E and F, respectively). Serum ELISA results demonstrated that HbA1c and glucose metabolism in 32-week-old VEGFB−/− mice were significantly lower than those of VEGFB+/+ mice (P<0.01; Fig. 3G and H).
Figure 3.
Effect of VEGFB downregulation on serum glucose and lipid metabolism. (A) Total cholesterol. (B) Triglycerides. (C) Low density lipoprotein-cholesterol (D) High density lipoprotein-cholesterol. (E) Fasting blood glucose. (F) Postprandial blood glucose. (G) HbA1c. (H) Serum glucose level. **P<0.01 vs. VEGFB+/+ group and ***P<0.001 vs. VEGFB+/+ group. VEGFB, vascular endothelial growth factor B.
The effect of VEGFB downregulation on blood glucose balance and insulin resistance
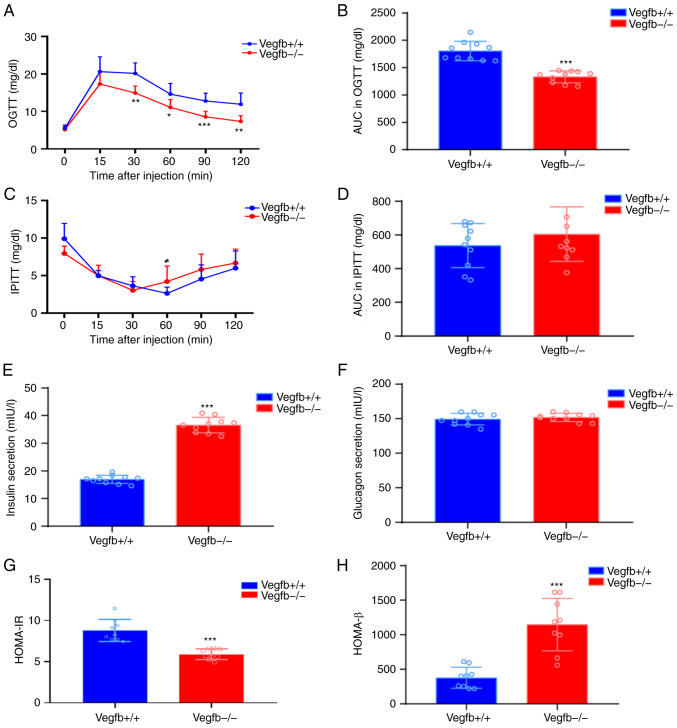
To determine whether loss of VEGFB affects glycemic homeostasis, OGTT and IPITT tests were performed on VEGFB+/+ and VEGFB−/− mice. The OGTT results revealed that compared with that of VEGFB+/+ mice, the blood glucose level of VEGFB−/− mice was significantly reduced at 30, 60, 90 and 120 min (P<0.05; Fig. 4A), and the area under the curve (AUC) was also significantly reduced (P<0.001; Fig. 4B). The IPITT results showed that after intraperitoneal injection of insulin, the blood glucose level of VEGFB−/− mice was significantly reduced at 0 min and 60 min (P<0.05; Fig. 4C). At the other times, compared with VEGFB+/+ mice, there was no significant change in blood glucose level (P>0.05; Fig. 4C), and there was no significant difference in AUC (P>0.05; Fig. 4D). The ELISA results demonstrated that compared with that in VEGFB+/+ mice, insulin secretion in VEGFB−/− mice increased (P<0.001; Fig. 4E), while VEGFB had no effect on the secretion of glucagon in mice (P>0.05; Fig. 4F). Compared with that of VEGFB+/+ mice, HOMA-IR of VEGFB−/− mice was significantly reduced (P<0.001; Fig. 4G), while HOMA-β of VEGFB−/− mice had significantly increased (P<0.001; Fig. 4H).
Figure 4.
Effect of VEGFB downregulation on blood glucose balance and insulin resistance. (A) OGTT. (B) Area under the curve in OGTT. (C) IPITT. (D) Area under the curve in IPTT. (E) Fasting insulin. (F) Serum glucagon. (G) HOMA-IR. (H) HOMA-β. *P<0.05 vs. VEGFB+/+ group, **P<0.01 vs. VEGFB+/+ group and ***P<0.001 vs. VEGFB+/+ group. HOMA-IR, insulin resistance index; HOMA-β, pancreatic β-cell secretion index. VEGFB, vascular endothelial growth factor B.
Expression of VEGFR1 and NRP1 in the pancreas and liver after VEGFB downregulation
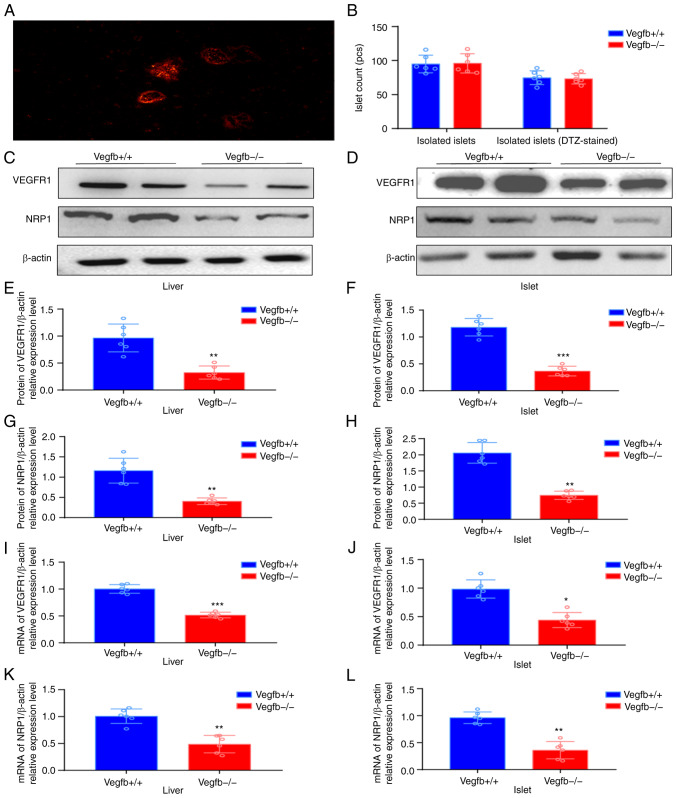
After the mouse islet tissue was stained with DTZ solution, the isolated and purified intact or incomplete islet cells were in the shape of red mass, and the peripheral pancreatic exocrine cells were not stained (Fig. 5A). After islet culture and isolation, each mouse could exchange 80–100 islet cell clusters (Fig. 5B). VEGFR1 and NRP1 protein expression was also downregulated in islets and liver tissues of VEGFB−/− mice (P<0.05; Fig. 5C-H). qPCR results revealed that the expression of VEGFR1 and NRP1 in VEGFB−/− mice was downregulated (P<0.05; Fig. 5I-L).
Figure 5.
Expression of VEGFR1 and NRP1 in the liver and islets after VEGFB downregulation. (A) Dithizone staining to identify islet cells (magnification, ×400). (B) Numbers of islets. (C-H) Western blot analysis of VEGFB and NRP1 protein in liver tissues and islets. (I-L) mRNA analysis of VEGFB and NRP1 in liver tissues and islets. *P<0.05 vs. VEGFB+/+ group, **P<0.01 vs. VEGFB+/+ group and ***P<0.001 vs. VEGFB+/+ group. VEGFB, vascular endothelial growth factor B; NRP1, neuropilin-1.
Expression of VEGFA and its receptor VEGFR2 in the pancreas and liver after VEGFB downregulation
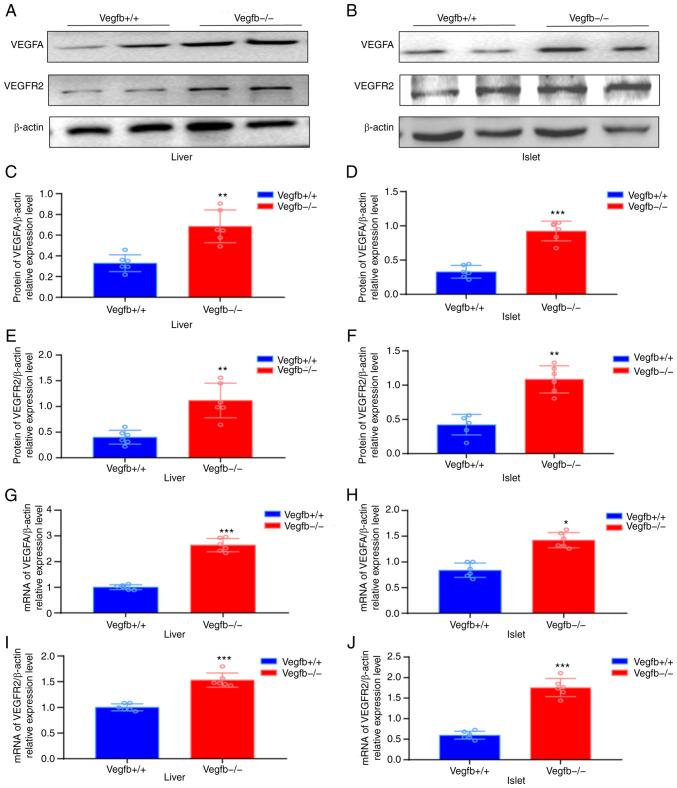
Western blot analysis showed that the expression levels of VEGFA and VEGFR2 in VEGFB−/− mice were higher than those in VEGFB+/+ mice (P<0.01; Fig. 6A-F). The qPCR results revealed that the expression levels of VEGFA and VEGFR2 in VEGFB−/− mice were higher than those in VEGFB+/+ mice (P<0.01; Fig. 6G-J).
Figure 6.
Expression of VEGFA and VEGFR2 in the liver and islets after VEGFB downregulation. (A-F) Western blot analysis of VEGFA and VEGFR2 protein in liver tissues and islets. (B) Numbers of islets. (G-J) mRNA analysis of VEGFA and VEGFR2 in liver tissues and islets. *P<0.05 vs. VEGFB+/+ group, **P<0.01 vs. VEGFB+/+ group and ***P<0.001 vs. VEGFB+/+ group.
Discussion
VEGFB is a glycoprotein that is mainly present in the heart, liver, skeletal muscle, brown fat and other tissues with high metabolic activity (22,23). VEGF family is composed of 7 members: VEGFA, VEGFB, VEGFC, VEGFD, VEGFE, VEGFF and PIGF (24,25). As a member of the VEGF family, VEGFB not only maintains the development of vascular and promotes neuroprotection and nutrition but also acts as a factor to maintain homeostasis. In recent years, the role of VEGFB in regulating lipid metabolism and blood glucose balance has attracted the attention of numerous scholars.
Hagberg et al (10,12) reported on Nature that VEGFB can regulate lipid uptake in endothelial cells and participate in lipid metabolism. And in the study of db/db mice, VEGFB gene knockdown prevented ectopic lipid deposition. Similar to the findings of Hagberg et al (12), Falkevall et al (14) also revealed that VEGFB gene deletion can prevent pathological lipid deposition. Mehlem et al (26) revealed that the deletion of the VEGFB gene reduced lipid metabolism and also prevented hyperglycaemia and hyperinsulinemia. In 2017, Wu et al (27) observed that high plasma VEGFB content causes abnormal glucose tolerance and elevates blood glucose, aggravating the condition of patients with T2DM. By contrast, Moessinger et al (15) reported that reducing the VEGFB signal pathway can increase cardiac glucose accumulation. Kivelä et al (28) revealed that in cardiomyocyte-specific VEGFB transgenic rats, enhancing the effect of VEGFB can alleviate glucose metabolism disorders. In 2021, Hu et al (20) injected VEGFB into high fat diet (HFD-induced obese mice for 10 weeks and observed that both the blood glucose level and the blood glucose AUC in the VEGFB treatment group were lower than that of the HFD group. Therefore, VEGFB can improve glucose tolerance and increase insulin sensitivity. An increasing number of scholars have discovered that the VEGFB signaling pathway is involved in the development of obesity and T2DM. It is predicted that VEGFB may become a key factor in regulating blood glucose regulation and lipid metabolism disorders. Altering the VEGFB signal pathway has certain therapeutic potential for obesity and T2DM (10,12).
Abnormal blood glucose and lipid regulation are closely related to T2DM, insulin resistance, non-alcoholic fatty liver disease, hypertension, diabetic nephropathy and other metabolic diseases. Abnormal deposition of lipids in non-fat tissues such as the skeletal muscle and the myocardium can interfere with the insulin signal pathway and indirectly affect key factors associated with tissue glucose uptake, leading to insulin resistance (29). Interfering with lipid and glucose metabolism in early diabetes and diagnosed patients with diabetes can help improve impaired insulin secretion in the early stage. Anti-aliphatic deposition and improving glucose intake may bring therapeutic benefits. However, drugs that improve the abnormal deposition of adipose and glucose metabolism in tissues are very rare at present. It has become one of the important impediments to the treatment of metabolic diseases such as obesity and diabetes. VEGFB has potential value as a target for treatment, and may benefit from diseases such as lipid metabolism disorders, diabetes, obesity and other diseases. Therefore, elucidating the mechanism by which VEGFB regulates blood lipid metabolism and blood glucose homeostasis has become the focus and hot-spot of medical research.
In the present study, a systemic VEGFB knockdown mouse model was constructed by CRISPR/cas9 genetic engineering to investigate the effect of VEGFB gene deletion on lipid metabolism and blood glucose homeostasis in mice and its mechanism. The nutritional research results on lipid metabolism in mice after VEGFB gene deletion are similar to those of Hagberg et al (10) and Robciuc et al (11). The research revealed that after VEGFB gene deletion, the levels of serum TG, TC and LDL in mice increased, suggesting that VEGFB gene deletion can lead to lipid accumulation in mice. Through the morphological detection of liver tissue, it was observed that following VEGFB gene deletion, mouse hepatocytes have obvious steatosis, suggesting that VEGFB gene deletion can accelerate the pathological progression of a mouse fatty liver. In the present study, the glucose metabolism indexes of mice with VEGFB gene deletion were determined. It was observed that the contents of blood glucose, serum glucose and HbA1c decreased after the VEGFB gene deletion. To further clarify the effect of VEGFB on blood glucose, an OGTT and IPITT were performed. It was observed that following the deletion of the VEGFB gene in mice, the glucose tolerance increased, and the insulin resistance index decreased significantly. This suggested that reducing the VEGFB signal can improve glucose metabolism and insulin resistance while aggravating the accumulation of lipids. By observing the expression changes of VEGFR1, VEGFA and VEGFR2 in the liver and islet tissues after the VEGFB gene deletion, it was concluded that the VEGFB gene deletion may participate in the regulation of glucose and lipid metabolism in mice by activating the VEGFA/VEGFR2 signal pathway.
Louzier et al revealed that the functional status of mice lacking the VEGFB gene did not change significantly, and their lifespan was similar to that of normal mice (30,31). In 2014, Sun et al (32) reported that there was no significant difference between VEGFB−/− mice with increased body weight and increased food intake and control group mice. The weight of the constructed systemic VEGFB-knockdown mice gradually increased as the age of weeks. The deletion of the VEGFB gene did not affect the growth of mice, which was consistent with the findings of a previous study by Sun et al (32) in a VEGFB-deficient mouse model. In the present study, it was also found that although there was no significant change in body weight compared with VEGFB+/+ mice, the organ ratio of VEGFB−/− mice liver increased significantly, and there was steatosis in hepatocytes, suggesting that VEGFB gene deletion affected liver lipid accumulation. In 2021, Hu et al (20) proposed that VEGFB treatment can protect mice from liver lipid deposition induced by an HFD, which is consistent with our results. Furthermore, Shen et al revealed that VEGFB/IL22 fusion protein therapy can reverse hepatic lipid accumulation induced by diabetes mellitus (33).
In 2014, Sun et al (32) linked the VEGFB signal pathway with human lipid metabolism, glucose metabolism and islet resistance through genetic associations. Studies have shown that there was no change in peripheral VEGFB levels between healthy individuals and patients with diabetes, but there is a correlation between VEGFB levels in patients with diabetes and the levels of C-reactive protein, TC, TG and blood glucose levels (15,20,32). In 2015, Cheng et al (34) also observed an increase in serum VEGFB levels in insulin-resistant subjects in clinical trials, VEGFB was positively correlated with insulin resistance, and metformin treatment could reduce plasma VEGFB levels. These studies showed that there is a certain relationship between VEGFB levels and obesity and insulin resistance. In 2016, Mehlem et al (26) examined model mice and revealed that the TG diacylglycerol and PBG levels in the muscles of VEGFB−/− mice were lower than those in normal mice, and VEGFB−/− mice were not prone to hyperinsulinaemia. It has been suggested that VEGFB gene deletion can prevent hyperglycaemia and hyperinsulinaemia, reduce insulin resistance and improve dyslipidaemia (26). Robciuc et al (11) transfected AAV-B186 into obese mice and found that VEGFB not only significantly increased TG and cholesterol in mice but also improved the response to abdominal glucose and insulin tolerance tests and the metabolism of obese mice. The lipid metabolism indexes of VEGFB−/− mice were examined, and the results showed that after VEGFB gene deletion, the levels of TC, TG and LDL were increased, while HDL decreased. This finding revealed that VEGFB regulates lipid accumulation, which is consistent with the results of Hagberg et al (10), Robciuc et al (11) and other previous studies (12,19).
Disorders of lipid metabolism in the body and abnormal blood lipids can cause glucose metabolism disorders, which results in impaired glucose regulation, diabetes and other diseases. By detecting the glucose metabolism indexes of VEGFB−/− mice, it was observed that after 32 weeks of VEGFB knockdown, the FBG and PBG levels of mice decreased significantly, along with the contents of serum glucose and HbA1c when compared with that of VEGFB+/+ mice. According to the analysis of the research results, the deletion of the VEGFB gene can promote the glucose uptake of the body, which affects the levels of serum glucose, HbA1c, FBG and PBG. Wu et al (27) revealed that the level of human plasma VEGFB was positively correlated with FINS, HOMA-IR, blood glucose and HbA1c, indicating that VEGFB was closely related to insulin resistance and deterioration of glucose metabolism. In 2021, Hu et al (20) observed that the level of blood glucose in the VEGFB treatment group was lower than that in the HFD group and that the area under the curve of the blood glucose curve was also significantly lower, indicating that VEGFB could improve glucose tolerance and increase insulin sensitivity. According to the results of the present study, it was stated that glucose tolerance and level were improved after VEGFB deletion which was similar to the result by Hu et al. Unlike the aforementioned study, however, there was no significant effect on insulin sensitivity. This was controversial with the present findings.
Diabetes mellitus is the most common endocrine disorder characterized by chronic hyperglycaemia owing to relatively insufficient insulin secretion (35,36). Insulin or glucagon can impair blood glucose homeostasis and cause diabetes and other diseases. Currently, there have been no studies on the effect of VEGFB on the secretion of glucagon and its mechanism. By detecting the fasting serum insulin and glucagon levels of VEGFB−/− mice, it was observed that insulin secretion increased after VEGFB gene knockdown, while glucagon levels did not change significantly.
The detection of islet cell function is an important part of the pathophysiology of abnormal glucose metabolism, and it is important to clarify the β-cell functional status of abnormal glucose metabolism. The amount of insulin secretion is closely related to the number of vesicles in pancreatic islet cells (37). There are ~9,000-13,000 dense-core secretory vesicles in each rodent (mouse/rat) β-cell (38). In 2016, Pan et al (39) concluded that the pattern of insulin release corresponds to the number of cellular vesicles. Insulin-secreting vesicles can be divided into two types, mature and immature, according to their morphology. Mature vesicles are formed by insulin, zinc and calcium crystals and contain dense core particles. Immature vesicles are formed by the processing and packaging of proinsulin in the reverse Golgi apparatus. They are light grey and have low electron density (40). The maturation process of immature vesicles requires acidification by an ATP-dependent proton pump, and the coat protein must be removed for proinsulin to be converted into insulin and C peptide through proteolysis (41). It was observed that the islet vesicle density significantly increased in VEGFB−/− mice compared with VEGFB+/+ mice, and the number of mature and immature vesicles increased. The present study suggested that deletion of VEGFB can cause increased serum insulin levels by affecting the production of insulin secretory vesicles in islet cells.
Insulin resistance is the main link in the pathogenesis of T2DM. The effect of VEGFB gene deletion on insulin resistance index and islet cell function index was examined and it was found that VEGFB gene decreased insulin resistance index and cell function index (HOMA-) increased after VEGFB gene deletion. This finding suggested that the deletion of VEGFB gene can not only affect insulin secretion, but also improved insulin resistance. Robciuc et al (11) also found similar results to the present study in obese and insulin-resistant mice, suggesting that VEGFB can improve insulin secretion, insulin supply and signal transduction. These findings predicted the therapeutic potential of VEGFB in the treatment of disorders of glucose and lipid metabolism.
VEGFB plays a biological role primarily by binding to cell membrane receptors VEGFR1 and NRP1 (42). VEGFR1 was the first receptor in the VEGF family that was identified. In addition to binding to VEGFB, VEGFR1 has an increased affinity for VEGFA and placental growth factors. NRP1, as a coreceptor of VEGFR1, performs biological functions after binding to VEGFB (11,43,44). Falkevall et al (14) reported that VEGFB signal can enhance the uptake and exocytosis of FAs by endothelial cells through VEGFR1 and its coreceptor NRP1. In 2021, a study by Hu et al (20) also revealed that the effect of VEGFB on lipid metabolism depended on VEGFR1. In addition, it has been observed that VEGFB gene knockout and VEGFR1 deletion in the db/db mouse model can maintain metabolic homeostasis and restore lipid and glucose metabolism (45).
Anisimov et al (46) confirmed that VEGFB binding to VEGFR1 does not induce VEGFR1 downstream signal. By contrast, VEGFB is regarded to activate the VEGFA/VEGFR2 pathway by replacing VEGFR1 with VEGFR2, thereby promoting the normal angiogenesis pathway (47,48). In a study of the effect of VEGFB on insulin secretion, Robciuc et al (11) reported that VEGFB and VEGFA competitively bind VEGFR1. VEGFB and VEGFR1 binding promote the binding of VEGFA to VEGFR2, thereby increasing vascular remodeling and blood perfusion of adipose tissue and participating in metabolic regulation, insulin secretion and signal transduction. The expression of VEGFR1 and NRP1 in the liver and islet, respectively, was verified. The results revealed that the expression of VEGFR1 and NRP1 decreased significantly following VEGFB gene deletion, suggesting that VEGFB gene deletion reduced the expression of receptors VEGFR1 and NRP1 on the islet cell membrane. The expression of VEGFA and VEGFR2 in the liver and islet was evaluated and it was observed that their expression increased significantly following VEGFB deletion. VEGFA is a vascular permeability factor observed in tumor cells, which primarily plays its biological role by connecting with the extracellular receptor VEGFR2. The VEGFA/VEGFR2 signal pathway is involved in tumor angiogenesis, inflammation and oxidative stress; however, it also leads to endothelial cell dysfunction and lipid deposition (49,50). Ghorbanzadeh et al (51) reported that the VEGFA and VEGFR2 signals are closely related to glucose. In 2017, Jin et al (52) revealed that overexpression of VEGFA protected against obesity caused by high-fat foods and reduced insulin sensitivity.
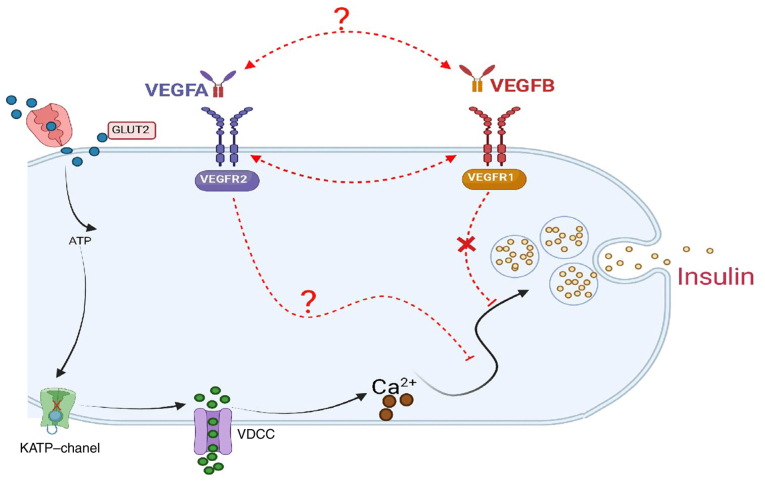
VEGFA and VEGFB participate in multiple signal pathways including cell proliferation and differentiation, apoptosis and metabolic balance in vivo. In 2011, Osawa et al (25) reported that VEGFB induced signal transduction of pericytes and other cells by binding to VEGFR1. Simultaneously, the binding of VEGFB to VEGFR1 increased the binding of VEGFA to VEGFR2 and promoted tumor growth (25). Jin et al (52) reported that VEGFA overexpression can resist obesity by increasing vascular density and browning of white adipose tissue (WAT). In addition, it was observed that VEGFA downregulation could upregulate the expression of VEGFB in WAT. It was concluded that VEGFB and VEGFA have a certain relationship in regulating adipose metabolism (52). Recent studies have reported that VEGFB played its metabolic role by indirectly activating the VEGFA/VEGFR2 pathway (11,28,47). As a result of the downregulation of VEGFB and VEGFR1 expression caused by the deletion of the VEGFB gene, it was concluded that VEGFB may regulate the blood glucose homeostasis and lipid metabolism in mice by activating the VEGFA/VEGFR2 signal pathway (Fig. 7), which provided a novel approach for VEGFB to participate in the pathogenesis of T2DM.
Figure 7.
Mechanism of VEGFB involved in insulin secretion of β-cells. VEGFB, vascular endothelial growth factor B.
The present study analyzed the effect of VEGFB gene knockdown on insulin secretion by observing the levels of glucose and lipid metabolism in systemic VEGFB knockdown mice, and revealed that VEGFB knockdown may participate in the regulation of glucose and lipid metabolism in mice by activating the VEGFA/VEGFR2 signal pathway. Since the mechanisms of glucose and lipid metabolism and insulin secretion are not consistent under physiological and pathological conditions, further exploration of the regulatory mechanism of VEGFB on blood glucose, lipid metabolism and insulin secretion is required via construction of animal models of obesity and diabetes.
In conclusion, the present study identified that the deletion of VEGFB gene could aggravate the lipid deposition in mice, affect glucose metabolism, increase insulin secretion and reduce blood glucose level. It was also found that the expression of VEGFR1 decreased after downregulation of VEGFB, which may affect the balance of glucose and lipid metabolism by activating VEGFA/VEGFR2 pathway. The present findings provided theoretical and experimental evidence for VEGFB in the diagnosis and treatment of obesity and diabetes.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the National Natural Science Foundation of China Youth Project (grant no. 31702024), the Shandong Province Higher Educational Science and Technology Plan Project (grant no. J17KA258) and the Shandong University Student Innovation Training Project (grant no. S202010440029).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YNL and WGJ designed the study, XL, RRL, YQL, HP and HNY performed the experiments, wrote and revised the manuscript. XL and YNL wrote the manuscript and analyzed the data. RRL searched the literature. All authors read and approved the final manuscript. YNL and XL confirm the authenticity of all the raw data. All authors agree to the publication of the manuscript.
Ethics approval and consent to participate
The present study was reviewed and approved by the Institutional Review Board of Binzhou Medical University (Yantai, China). All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee of the Medical Ethics Committee of Binzhou Medical University (IACUC protocol number: 2021-300).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: Principles of pathogenesis and therapy. Lancet. 2005;365:1333–1346. doi: 10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- 2.Trayhurn P. Hypoxia and adipocyte physiology: Implications for adipose tissue dysfunction in obesity. Annu Rev Nutr. 2014;34:207–236. doi: 10.1146/annurev-nutr-071812-161156. [DOI] [PubMed] [Google Scholar]
- 3.Lysaght J, van der Stok EP, Allott EH, Casey R, Donohoe CL, Howard JM, McGarrigle SA, Ravi N, Reynolds JV, Pidgeon GP. Pro-inflammatory and tumour proliferative properties of excess visceral adipose tissue. Cancer Lett. 2011;312:62–72. doi: 10.1016/j.canlet.2011.07.034. [DOI] [PubMed] [Google Scholar]
- 4.Bai T, Li M, Liu Y, Qiao Z, Wang Z. Inhibition of ferroptosis alleviates atherosclerosis through attenuating lipid peroxidation and endothelial dysfunction in mouse aortic endothelial cell. Free Radic Biol Med. 2020;160:92–102. doi: 10.1016/j.freeradbiomed.2020.07.026. [DOI] [PubMed] [Google Scholar]
- 5.Sinha RA, Bruinstroop E, Singh BK, Yen PM. Nonalcoholic fatty liver disease and hypercholesterolemia: Roles of thyroid hormones, metabolites, and agonists. Thyroid. 2019;29:1173–1191. doi: 10.1089/thy.2018.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santoleri D, Titchenell PM. Resolving the paradox of hepatic insulin resistance. Cell Mol Gastroenterol Hepatol. 2019;7:447–456. doi: 10.1016/j.jcmgh.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laakso M, Kuusisto J. Insulin resistance and hyperglycaemia in cardiovascular disease development. Nat Rev Endocrinol. 2014;10:293–302. doi: 10.1038/nrendo.2014.29. [DOI] [PubMed] [Google Scholar]
- 8.Khan RMM, Chua ZJY, Tan JC, Yang Y, Liao Z, Zhao Y. From pre-diabetes to diabetes: Diagnosis, treatments and translational research. Medicina (Kaunas) 2019;55:546. doi: 10.3390/medicina55090546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aase K, Lymboussaki A, Kaipainen A, Olofsson B, Alitalo K, Eriksson U. Localization of VEGF-B in the mouse embryo suggests a paracrine role of the growth factor in the developing vasculature. Dev Dyn. 1999;215:12–25. doi: 10.1002/(SICI)1097-0177(199905)215:1<12::AID-DVDY3>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 10.Hagberg CE, Mehlem A, Falkevall A, Muhl L, Fam BC, Ortsäter H, Scotney P, Nyqvist D, Samén E, Lu L, et al. Targeting VEGF-B as a novel treatment for insulin resistance and type 2 diabetes. Nature. 2012;490:426–430. doi: 10.1038/nature11464. [DOI] [PubMed] [Google Scholar]
- 11.Robciuc MR, Kivelä R, Williams IM, de Boer JF, van Dijk TH, Elamaa H, Tigistu-Sahle F, Molotkov D, Leppänen VM, Käkelä R, et al. VEGFB/VEGFR1-Induced expansion of adipose vasculature counteracts obesity and related metabolic complications. Cell Metab. 2016;23:712–724. doi: 10.1016/j.cmet.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagberg CE, Falkevall A, Wang X, Larsson E, Huusko J, Nilsson I, van Meeteren LA, Samen E, Lu L, Vanwildemeersch M, et al. Vascular endothelial growth factor B controls endothelial fatty acid uptake. Nature. 2010;464:917–921. doi: 10.1038/nature08945. [DOI] [PubMed] [Google Scholar]
- 13.Wagenmakers AJM, Strauss JA, Shepherd SO, Keske MA, Cocks M. Increased muscle blood supply and transendothelial nutrient and insulin transport induced by food intake and exercise: Effect of obesity and ageing. J Physiol. 2016;594:2207–2222. doi: 10.1113/jphysiol.2014.284513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falkevall A, Mehlem A, Palombo I, Heller Sahlgren B, Ebarasi L, He L, Ytterberg AJ, Olauson H, Axelsson J, Sundelin B, et al. Reducing VEGF-B signaling ameliorates renal lipotoxicity and protects against diabetic kidney disease. Cell Metab. 2017;25:713–726. doi: 10.1016/j.cmet.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Moessinger C, Nilsson I, Muhl L, Zeitelhofer M, Heller Sahlgren B, Skogsberg J, Eriksson U. VEGF-B signaling impairs endothelial glucose transcytosis by decreasing membrane cholesterol content. EMBO Rep. 2020;21:e49343. doi: 10.15252/embr.201949343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen N, Ning FC, Mi J, Lindström W, Balan M, Muhl L, Eriksson U, Nilsson I, Nyqvist D. VEGF-B ablation in pancreatic β-cells upregulates insulin expression without affecting glucose homeostasis or islet lipid uptake. Sci Rep. 2020;10:923. doi: 10.1038/s41598-020-57599-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibuya M. VEGF-VEGFR system as a target for suppressing inflammation and other diseases. Endocr Metab Immune Disord Drug Targets. 2015;15:135–144. doi: 10.2174/1871530315666150316121956. [DOI] [PubMed] [Google Scholar]
- 18.Zachary I, Gliki G. Signaling transduction mechanisms mediating biological actions of the vascular endothelial growth factor family. Cardiovasc Res. 2001;49:568–581. doi: 10.1016/S0008-6363(00)00268-6. [DOI] [PubMed] [Google Scholar]
- 19.Shen Z, Zhang Z, Wang X, Yang K. VEGFB-VEGFR1 ameliorates Ang II-induced cardiomyocyte hypertrophy through Ca2+-mediated PKG I pathway. J Cell Biochem. 2018;119:1511–1520. doi: 10.1002/jcb.26311. [DOI] [PubMed] [Google Scholar]
- 20.Hu L, Shan Z, Wang F, Gao X, Tong Y. Vascular endothelial growth factor B exerts lipid-lowering effect by activating AMPK via VEGFR1. Life Sci. 2021;276:119401. doi: 10.1016/j.lfs.2021.119401. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Claesson-Welsh L. VEGF receptor signal transduction-A brief update. Vascul Pharmacol. 2016;86:14–17. doi: 10.1016/j.vph.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 24.Bates DO. Vascular endothelial growth factors and vascular permeability. Cardiovasc Res. 2010;87:262–271. doi: 10.1093/cvr/cvq105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osawa T, Muramatsu M, Wang F, Tsuchida R, Kodama T, Minami T, Shibuya M. Increased expression of histone demethylase JHDM1D under nutrient starvation suppresses tumor growth via down-regulating angiogenesis. Proc Natl Acad Sci USA. 2011;108:20725–20729. doi: 10.1073/pnas.1108462109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehlem A, Palombo I, Wang X, Hagberg CE, Eriksson U, Falkevall A. PGC-1α coordinates mitochondrial respiratory capacity and muscular fatty acid uptake via regulation of VEGF-B. Diabetes. 2016;65:861–873. doi: 10.2337/db15-1231. [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Wei H, Qu H, Feng Z, Long J, Ge Q, Deng H. Plasma vascular endothelial growth factor B levels are increased in patients with newly diagnosed type 2 diabetes mellitus and associated with the first phase of glucose-stimulated insulin secretion function of β-cell. J Endocrinol Invest. 2017;40:1219–1226. doi: 10.1007/s40618-017-0677-z. [DOI] [PubMed] [Google Scholar]
- 28.Kivelä R, Bry M, Robciuc MR, Räsänen M, Taavitsainen M, Silvola JM, Saraste A, Hulmi JJ, Anisimov A, Mäyränpää MI, et al. VEGF-B-induced vascular growth leads to metabolic reprogramming and ischemia resistance in the heart. EMBO Mol Med. 2014;6:307–321. doi: 10.1002/emmm.201303147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samuel VT, Petersen KF, Shulman GI. Lipid-induced insulin resistance: Unravelling the mechanism. Lancet. 2010;375:2267–2277. doi: 10.1016/S0140-6736(10)60408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louzier V, Raffestin B, Leroux A, Branellec D, Caillaud JM, Levame M, Eddahibi S, Adnot S. Role of VEGF-B in the lung during development of chronic hypoxic pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol. 2003;284:L926–L937. doi: 10.1152/ajplung.00247.2002. [DOI] [PubMed] [Google Scholar]
- 31.Reichelt M, Shi S, Hayes M, Kay G, Batch J, Gole GA, Browning J. Vascular endothelial growth factor-B and retinal vascular development in the mouse. Clin Exp Ophthalmol. 2003;31:61–65. doi: 10.1046/j.1442-9071.2003.00602.x. [DOI] [PubMed] [Google Scholar]
- 32.Sun CY, Lee CC, Hsieh MF, Chen CH, Chou KM. Clinical association of circulating VEGF-B levels with hyperlipidemia and target organ damage in type 2 diabetic patients. J Biol Regul Homeost Agents. 2014;28:225–236. [PubMed] [Google Scholar]
- 33.Shen Y, Chen W, Han L, Bian Q, Fan J, Cao Z, Jin X, Ding T, Xian Z, Guo Z, et al. VEGF-B antibody and interleukin-22 fusion protein ameliorates diabetic nephropathy through inhibiting lipid accumulation and inflammatory responses. Acta Pharm Sin B. 2021;11:127–142. doi: 10.1016/j.apsb.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng F, Zhao L, Wu Y, Huang T, Yang G, Zhang Z, Wu Y, Jia F, Wu J, Chen C, Liu D. Serum vascular endothelial growth factor B is elevated in women with polycystic ovary syndrome and can be decreased with metformin treatment. Clin Endocrinol (Oxf) 2016;84:386–393. doi: 10.1111/cen.12950. [DOI] [PubMed] [Google Scholar]
- 35.Abdul Razak MK, Sultan AA. The importance of measurement of plasma fibrinogen level among patients with type-2 diabetes mellitus. Diabetes Metab Syndr. 2019;13:1151–1158. doi: 10.1016/j.dsx.2019.01.049. [DOI] [PubMed] [Google Scholar]
- 36.Lam DW, LeRoith D. The worldwide diabetes epidemic. Curr Opin Endocrinol Diabetes Obes. 2012;19:93–96. doi: 10.1097/MED.0b013e328350583a. [DOI] [PubMed] [Google Scholar]
- 37.Clayton EL, Evans GJ, Cousin MA. Bulk synaptic vesicle endocytosis is rapidly triggered during strong stimulation. J Neurosci. 2008;28:6627–6632. doi: 10.1523/JNEUROSCI.1445-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao A, Ohara-Imaizumi M, Brissova M, Benninger RK, Xu Y, Hao Y, Abramowitz J, Boulay G, Powers AC, Piston D, et al. Gαo represses insulin secretion by reducing vesicular docking in pancreatic beta-cells. Diabetes. 2010;59:2522–2529. doi: 10.2337/db09-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan JY, Yuan S, Yu T, Su CL, Liu XL, He J, Li H. Regulation of L-type Ca2+ channel activity and insulin secretion by huntingtin-associated protein 1. J Biol Chem. 2016;291:26352–26363. doi: 10.1074/jbc.M116.727990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wollam J, Mahata S, Riopel M, Hernandez-Carretero A, Biswas A, Bandyopadhyay GK, Chi NW, Eiden LE, Mahapatra NR, Corti A, et al. Chromogranin A regulates vesicle storage and mitochondrial dynamics to influence insulin secretion. Cell Tissue Res. 2017;368:487–501. doi: 10.1007/s00441-017-2580-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vakilian M, Tahamtani Y, Ghaedi K. A review on insulin trafficking and exocytosis. Gene. 2019;706:52–61. doi: 10.1016/j.gene.2019.04.063. [DOI] [PubMed] [Google Scholar]
- 42.Nash AD, Baca M, Wright C, Scotney PD. The biology of vascular endothelial growth factor-B (VEGF-B) Pulm Pharmacol Ther. 2006;19:61–69. doi: 10.1016/j.pupt.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 43.Gilbert M, Jung SR, Reed BJ, Sweet IR. Islet oxygen consumption and insulin secretion tightly coupled to calcium derived from L-type calcium channels but not from the endoplasmic reticulum. J Biol Chem. 2008;283:24334–24342. doi: 10.1074/jbc.M802097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gauthier BR, Wollheim CB. Synaptotagmins bind calcium to release insulin. Am J Physiol Endocrinol Metab. 2008;295:E1279–E1286. doi: 10.1152/ajpendo.90568.2008. [DOI] [PubMed] [Google Scholar]
- 45.Shang R, Lal N, Lee CS, Zhai Y, Puri K, Seira O, Boushel RC, Sultan I, Räsänen M, Alitalo K, et al. Cardiac-specific VEGFB overexpression reduces lipoprotein lipase activity and improves insulin action in rat heart. Am J Physiol Endocrinol Metab. 2021;321:E753–E765. doi: 10.1152/ajpendo.00219.2021. [DOI] [PubMed] [Google Scholar]
- 46.Anisimov A, Leppänen VM, Tvorogov D, Zarkada G, Jeltsch M, Holopainen T, Kaijalainen S, Alitalo K. The basis for the distinct biological activities of vascular endothelial growth factor receptor-1 ligands. Sci Signal. 2013;6:ra52. doi: 10.1126/scisignal.2003905. [DOI] [PubMed] [Google Scholar]
- 47.Bry M, Kivelä R, Leppänen VM, Alitalo K. Vascular endothelial growth factor-B in physiology and disease. Physiol Rev. 2014;94:779–794. doi: 10.1152/physrev.00028.2013. [DOI] [PubMed] [Google Scholar]
- 48.Rafii S, Carmeliet P. VEGF-B improves metabolic health through vascular pruning of fat. Cell Metab. 2016;23:571–573. doi: 10.1016/j.cmet.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 49.Feng S, Bowden N, Fragiadaki M, Souilhol C, Hsiao S, Mahmoud M, Allen S, Pirri D, Ayllon BT, Akhtar S, et al. Mechanical activation of hypoxia-inducible factor 1α drives endothelial dysfunction at atheroprone sites. Arterioscler Thromb Vasc Biol. 2017;37:2087–2101. doi: 10.1161/ATVBAHA.117.309249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu D, Lei L, Desir M, Huang Y, Cleman J, Jiang W, Fernandez-Hernando C, Di Lorenzo A, Sessa WC, Giordano FJ. Smooth muscle hypoxia-inducible factor 1α links intravascular pressure and atherosclerosis-brief report. Arterioscler Thromb Vasc Biol. 2016;36:442–455. doi: 10.1161/ATVBAHA.115.306861. [DOI] [PubMed] [Google Scholar]
- 51.Ghorbanzadeh V, Mohammadi M, Dariushnejad H, Chodari L, Mohaddes G. Effects of crocin and voluntary exercise, alone or combined, on heart VEGF-A and HOMA-IR of HFD/STZ induced type 2 diabetic rats. J Endocrinol Invest. 2016;39:1179–1186. doi: 10.1007/s40618-016-0456-2. [DOI] [PubMed] [Google Scholar]
- 52.Jin H, Li D, Wang X, Jia J, Chen Y, Yao Y, Zhao C, Lu X, Zhang S, Togo J, et al. VEGF and VEGFB play balancing roles in adipose differentiation, gene expression, and function. Endocrinology. 2018;159:2036–2049. doi: 10.1210/en.2017-03246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.