Abstract
Atherosclerosis (AS) remains the leading cause of mortality throughout the world, and vascular endothelial cell dysfunction is one of the key events leading to this pathology. In recent years, there has been an increased interest in the role of circulating RNAs in various diseases; these noncoding RNAs can regulate gene products by acting as microRNA (miR) sponges. Furthermore, it has been shown that foam cells exhibit high expression levels of hsa_circ_0005699 (circ_0005699); however, to the best of our knowledge, no previous study has investigated the role of circ_0005699 in the regulation of vascular endothelial function. The present study employed human umbilical vein endothelial cells (HUVECs), which have been widely used to study vascular endothelial cell function. In addition, apolipoprotein E (ApoE)-deficient mice were used, which have been shown to rapidly develop AS and are widely used as a model of this disease. Cellular and biochemical techniques were performed, including gene transfection and short hairpin RNA-mediated gene silencing for cell transfection, luciferase reporter gene assay to confirm predicted genes, Cell Counting Kit-8 assay and flow cytometry to assess cell viability and apoptosis, and reverse transcription-quantitative PCR and western blotting for detection of mRNA and protein expression. In the present study, the expression levels of circ_0005699 were increased by oxidized low-density lipoprotein in a time- and dose-dependent manner in HUVECs; this was also associated with increased apoptosis of these cells. In addition, the expression levels of circ_0005699 were elevated, along with increased levels of inflammatory cytokines, in ApoE-deficient mice. An RNA pull-down assay indicated that circ_0005699 can bind miR-450b-5p to decrease its expression, whereas silencing of circ_0005699 resulted in increased expression of miR-450b-5p. In addition, the online bioinformatics tool starBase predicted NFKB1 as a target gene of miR-450b-5p, which was further confirmed by the luciferase reporter gene assay. Notably, knockdown of circ_0005699 resulted in the increased survival of HUVECs, which was associated with decreased protein expression levels of NFKB1 and inflammatory cytokines. By contrast, the effects of circ-0005699 silencing on survival were reversed by miR-450b-5p inhibition or NFKB1 overexpression. In conclusion, knockdown of circ_0005699 may ameliorate endothelial cell injury through regulation of the miR-450b-5P/NFKB1 signaling axis.
Keywords: circ_0005699, microRNA-450b-5p, NFKB1, human umbilical vein endothelial cell, apolipoprotein E
Introduction
Atherosclerosis (AS) is one of the most common vascular lesions and is also considered the main pathological basis of cardiovascular disease (1). Several cell types and molecules are involved in the pathogenesis of AS, mainly including endothelial cells, smooth muscle cells and inflammatory cells (2). Furthermore, it has been reported that the pathophysiology of AS originates from endothelial cell injury; however, the specific pathogenesis has not yet been clarified (3).
As a class of endogenous non-coding RNA, circulating RNAs (circRNAs) do not have a 5′ cap and a 3′ poly tail, and exist in their characteristic covalent closed-loop form (4). Previous studies have indicated that circRNAs are widely present in vivo, are stably expressed in plasma, serum and tissues, and are involved in regulating gene expression in eukaryotes (5–7). Notably, circRNAs can adsorb microRNAs (miRs/miRNAs) by acting as molecular sponges, thus removing the negative regulatory effect of miRNAs on their downstream target genes, so as to achieve their own biological function (8). In addition, circRNAs have been shown to be involved in the development and progression of various cardiovascular diseases (9). For example, circ_0010283 has been reported to be highly expressed in oxidized low-density lipoprotein (ox-LDL)-induced vascular smooth muscle cells, and to promote cell proliferation and migration by regulating the miR-370-3P/HMGB1 signaling axis (10). Another study indicated that circ_0124644 was highly expressed in ox-LDL-induced human umbilical vein cells, and PAAP-A expression was upregulated via sponge adsorption of miR-149-5p, thus further worsening endothelial cell injury (11).
High expression levels of circ_0005699 in ox-LDL-stimulated foam cells have previously been reported (12). Formation of macrophage foam cells is known to be a major hallmark of the initiation of AS, and uncontrolled uptake of ox-LDL has been confirmed as one of the contributing factors leading to foam cell formation (13); however, the role of circ_0005699 in cardiovascular disease is still unclear. Therefore, the present study mainly investigated the role of circ_0005699 in regulating endothelial cell function and further explored its possible molecular mechanism. The online bioinformatics tool starBase was used to predict the miRNA target of circ_0005699 and the target gene of the identified miRNA, and the findings were confirmed by molecular and cellular biological assays. Furthermore, ApoE-deficient mice, which exhibit high ox-LDL levels, were used for in vivo studies. ox-LDL was also applied to human umbilical vein endothelial cells (HUVECs) in vitro to investigate its effects on the regulation of cell survival and circ_0005699 expression.
Materials and methods
Cell culture and treatment
The immortalized HUVEC line was obtained from American Type Culture Collection (cat. no. CRL-1730). HUVECs were cultured as previously described (14). The cells were cultured in a nutrient solution containing F-12 K basic medium, 10% fetal bovine serum and 1% penicillin-streptomycin (all from Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. Ox-LDL can act on endothelial cells and is the main risk factor of AS (15). Therefore, the present study treated HUVECs with different concentrations (0, 25, 50 and 100 µg/ml) of ox-LDL (Union-Biol) for different durations (0, 12, 24 and 48 h) at 37°C to establish the AS cell model. For experiments, the cells used were between passages 3 and 6.
Cell transfection
pLKO.1-puro was used as the backbone for the shRNA constructs. circ_0005699 short hairpin RNA (sh-circ_0005699#1/sh-circ_0005699#2), negative control shRNA (sh-NC), miR-450b-5p mimics and negative control (mimics-NC), miR-450b-5p inhibitor and negative control (inhibitor-NC) and pcDNA3.1-NFKB1 plasmid were constructed by Shanghai GenePharma Co., Ltd. Empty pcDNA3.1 was used as a control for pcDNA3-1-NFKB1. Cell (4×105) transfection was performed using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 48 h and cells were collected 72 h after transfection for subsequent experimentation. The concentration of shRNAs, mimics and inhibitors were 50 nM/1×105 cells; the concentration of overexpression plasmid was 0.8 µg/1×105 cells. The sequences were as follows: miR-450b-5p mimics, 5′-UUUUGCAAUAUGUUCCUGAAUA-5′; miR-NC, 5′-TTCTCCGAACGTGTCACGT-3′; miR-450b-5p inhibitor, 5′-UAUUCAGGAACAUAUUGCAAAA-3′; inhibitor NC: 5′-UUCUCCGAACGUGUCACGUTT-3′; sh-NC, 5′-GGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTTG-3′; sh-circ_0005699#1, 5′-CACCGCCACAAGGTCTATGGATTTCCGAAGAAATCCATAGACCTTGTGGC-3′; and sh-circ_0005699#2, 5′-CACCGCTTGGAAAGTCATCACTAAGCGAACTTAGTGATGACTTTCCAAGC-3′.
Animal handling
Six male C57BL/6 wild-type (WT) mice and six ApoE-knockout mice with a C57BL/6 background were obtained from Beijing HFK Bioscience Co., Ltd. Mice were maintained under the following standard laboratory conditions: Temperature, 22°C; humidity, 55%; 12-h light/dark cycle; and ad libitum access to food and water. Handling of mice was performed according to the Institutional Animal Care of The Affiliated Ganzhou Hospital of Nanchang University (Gangzhou, China) (16). After 12 weeks, the mice were sacrificed under excessive anesthesia using sodium pentobarbital (120 mg/kg; intraperitoneal injection). The research team and veterinary staff monitored the mice twice daily. Health was monitored by measuring their weight (twice weekly), assessing food and water intake, and general assessment of animal activity, panting and fur condition. Before the end of the experiment, the mice were euthanized once they had fasted for 12 h. The mice were euthanized by sodium pentobarbital. The drug was injected intraperitoneally at a dose of 120 mg/kg, and death was confirmed by the lack of a heartbeat. Serum samples were obtained via cardiac puncture and centrifugation of blood samples for 10 min at 1,000 × g at 4°C, and were stored at −80°C until further use. All of the procedures were reviewed and approved by the Ethical Committee for Animal Experimentation, The Affiliated Ganzhou Hospital of Nanchang University (Ganzhou, China; approval no. KY-E-2019-11-19).
ELISA
ELISA kits for the determination of serum and cell concentrations of TNF-α (cat. no. ab208348), IL-6 (cat. no. ab222503) and IL-1β (cat. no. ab197742) were purchased from Abcam. Prior to ELISA, cells were treated with 100 µg/ml ox-LDL for 48 h. The experiments were conducted according to the manufacturer's protocols.
Annexin V/PI assay
The Annexin V/PI assay was used to determine the apoptosis of HUVECs according to a previously described protocol (17). After treatment (100 µg/ml ox-LDL for 48 h), cells were harvested and washed three times with PBS. Subsequently, cells (1×104) were incubated in the dark at room temperature with 5 µl Annexin V-FITC and PI (cat. no. 331200; Thermo Fisher Scientific, Inc.) for 15 min. Cells undergoing apoptosis (early + late) were detected by flow cytometry (Accuri C6; BD Biosciences) and cell apoptosis was determined using FlowJo 7.6.1 software (FlowJo LLC).
Cell counting kit-8 (CCK-8) assay
The CCK-8 assay was used to assess the proliferation of cells. Briefly, HUVECs were seeded in 96-well plates at a density of 1×103 cells/well, and 10 µl CCK-8 solution (Dojindo Laboratories, Inc.) was added to each well after 0, 24, 48 and 72 h. Subsequently, the cells were cultured at 37°C for a further 120 min, followed by the measurement of optical density at 450 nm using a plate reader (Tecan Group, Ltd.).
Western blot analysis
Briefly, cells were lysed using RIPA buffer (Wuhan Sanying Biotechnology) containing protease inhibitors, and total protein was extracted. The BCA method was used to determine protein concentration in the samples. Subsequently, SDS-PAGE was used for separation of proteins (40 µg) on 10% gels, which were transferred onto polyvinylidene difluoride membranes (MilliporeSigma). Membranes were then blocked with 4% BSA (Cell Signaling Technology, Inc.) at 4°C for 2 h and incubated with the following primary antibodies (Abcam): Anti-B-cell lymphoma-2 (Bcl-2; cat. no. ab196495; 1:1,000), anti-Bax (cat. no. ab32503; 1:1,000), anti-NFKB1 (cat. no. ab32360; 1:1,000) and anti-GAPDH (cat. no. ab8245; 1:3,000; Abcam) at 4°C overnight; anti-GAPDH was used as the internal reference. The primary antibodies were then removed and the membranes were further incubated with HRP-conjugated secondary antibodies (anti-mouse IgG(H+L); cat. no. ab205719; anti-rabbit IgG (H+L); cat. no. ab205718; 1:5,000; Abcam) for 1 h at room temperature. Protein bands were visualized using an ECL assay kit (Bio-Rad Laboratories, Inc.).
Bioinformatics analysis
The online bioinformatics tool starBase database (http://starbase.sysu.edu.cn/), which predicts the target miRNA of the circRNA (18) was used in the present study. This software was also used to predict the miRNA and target gene interaction.
Dual-luciferase reporter gene assay
WT and mutant (Mut) sequences of circ_0005699 and NFKB1 were inserted into the BamHI and SalI sites downstream of the luciferase gene in the pGL3-control vector (Promega Corporation); the constructs were named circ_0005699 WT/circ_0005699 Mut and NFKB1 WT/NFKB1 Mut, respectively. The final concentration of 20 nM for each plasmid was subsequently co-transfected into HUVECs (lx106) along with miR-NC and miR-450b-3p mimics using Lipofectamine 3000 for 48 h at 37°C according to the manufacturer's protocol. Relative firefly luciferase activity was normalized against Renilla luciferase activity. The Dual-Glo Luciferase Reporter Assay System (Promega Corporation) was used for measurement of luciferase activity according to the manufacturer's protocol.
RNA pull-down assay and RNA immunoprecipitation (RIP)
The RNA pull-down assay was performed according to a previously published protocol (19,20). This assay was performed to confirm the interaction between circ_0005699 and miR-450b-3p. Biotin-labeled circ_0005699 probe and Bio-Oligo (Oligo probe) was purchased from Sangon Biotech Co., Ltd. HUVECs were trypsinized (Gibco; Thermo Fisher Scientific, Inc.) followed by cell lysis using RIP lysis buffer (Thermo Fisher Scientific, Inc.). The lysate alone served as the input control. The 0.7 ml lysate was incubated at 4°C overnight with 50 µl magnetic Dynabeads M-280 Streptavidin beads (Invitrogen; Thermo Fisher Scientific, Inc.). RNA complexes were subjected to centrifugation at 11,100 × g for 10 min at 37°C and then eluted by denaturation in 1X protein loading buffer for 10 min at 100°C. The enrichment of miR-450b-3p was detected using reverse transcription-quantitative PCR (RT-qPCR). Furthermore, the Magna RIP kit (cat. no. 17-704; MilliporeSigma) was used according to the manufacturer's guidelines and a previously described protocol (21). Briefly, cells were lysed using RIP lysis buffer (MedChemExpress,). Subsequently, 5 µg AGO2-specific antibody (cat. no. 2897; Cell Signaling Technology, Inc.) and a normal IgG antibody (cat. no. 58802; Cell Signaling Technology, Inc.) were conjugated to magnetic beads and mixed with the lysate (40 µg) for 4 h at 4°C. The magnetic beads were then harvested and incubated with 50 µl protein G at 37°C for 1 h. Finally, RNA was extracted from the beads and relative enrichment was analyzed using RT-qPCR.
RT-qPCR
RNAiso Plus reagent (Takara Bio, Inc.) was used to extract total RNA from HUVECs and 1 µg total RNA was reverse transcribed into cDNA using PrimeScript cDNA synthesis kit (Takara Bio, Inc.) according to manufacturer's protocol. The resulting cDNA was amplified using SYBR Premix Ex Taq™ II (Takara Bio, Inc.) in a real-time PCR detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). GAPDH was used as an internal reference for circ_0005699 and NFKB1, and U6 served as an endogenous control for miR-450b-5p. qPCR was performed as follows: Pre-denaturation at 95°C for 1 min; followed by 35 cycles of 95°C for 10 sec, 60°C for 20 sec and 72°C for 10 sec; and a final extension step at 72°C for 2 min. Gene expression was analyzed using the 2−ΔΔCq method (22). The primer sequences were as follows: circ_0005699 forward (F), 5′-TCCCCTTGTACGAAATCATTCCA-3′ reverse (R), 5′-ATTGAGACGTGTGAAGATGCCC-3′; miR-450b-5p F, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTATTCAGG-3′ R,5′-ACACTCCAGCTGGGTTTTGCAATATGTTCC-3′; NFKB1 F, 5′-GCAGCACTACTTCTTGACCACC-3′ R, 5′-TCTGCTCCTGAGCATTGACGTC-3′. GAPDH F, 5′-ACGGGAAGCTCACTGGCATGG-3′ R, 5′-GGTCCACCACCCTGTTGCTGTA-3′. U6 F, 5′-CTCGCTTCGGCAGCACA-3′ R, 5′-AACGCTTCACGAATTTGCGT-3′.
Statistical analysis
GraphPad Prism software version 6 (GraphPad Software, Inc.) was used to statistically analyze the results. The data are presented as the mean ± SD of at least three independent experiments. Unpaired Student's t-test was used to compare two groups, whereas the statistical analysis of more than two groups was performed using one-way ANOVA with Bonferroni's correction. P<0.05 was considered to indicate a statistically significant difference.
Results
circ_0005699 is upregulated in HUVECs induced by ox-LDL and in vivo
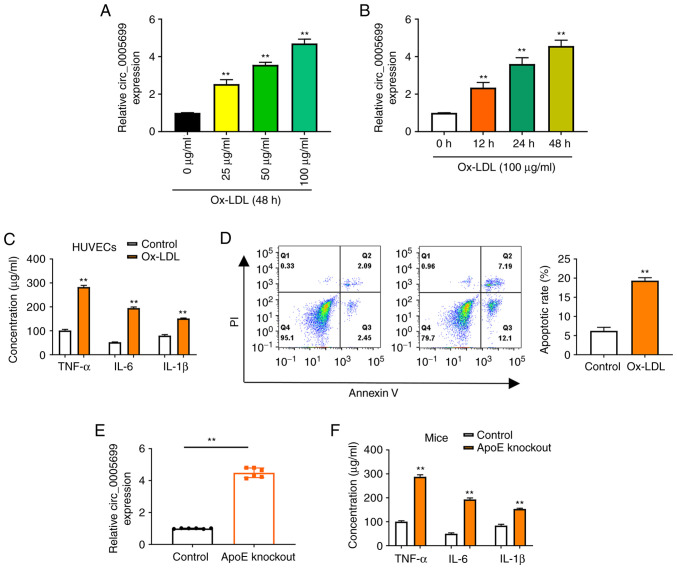
The present study observed that ox-LDL resulted in increased expression of circ_0005699 in HUVECs in a dose-dependent (Fig. 1A) and time-dependent manner (Fig. 1B). Notably, ox-LDL treatment was also associated with significantly increased concentrations of TNF-α, IL-6 and IL-1β in HUVECs compared with those in the control group (untreated cells) (Fig. 1C). Annexin V/PI double-staining and flow cytometry was used to detect the apoptotic rate of ox-LDL-treated HUVECs. It was observed that the apoptotic rate was significantly increased in the ox-LDL group compared with that in the control group (Fig. 1D). Increased ox-LDL has previously been reported in ApoE-knockout mice (23). In the present study, increased expression of circ_0005699 (Fig. 1E), and significantly increased serum concentrations of TNF-α, IL-6 and IL-1β were observed in ApoE-knockout mice (Fig. 1F).
Figure 1.
ox-LDL increases the expression of circ_0005699 in HUVECs. Expression levels of circ_0005699 were increased by ox-LDL in a (A) dose-dependent and (B) time-dependent manner. **P<0.01 vs. 0 µg/ml ox-LDL or 0 h ox-LDL. (C) ELISA indicated the effect of ox-LDL on the concentration of inflammatory factors in HUVECs. (D) Effect of ox-LDL on the apoptotic rate of HUVECs, as determined by Annexin V/PI double staining and flow cytometry. (E) circ_0005699 was highly expressed in ApoE-knockout mice. (F) Serum levels of circulating inflammatory cytokines in control and ApoE-knockout mice. **P<0.01 vs. control. ApoE, apolipoprotein E; HUVEC, human umbilical vein endothelial cell; ox-LDL, oxidized low-density lipoprotein.
circ_0005699 knockdown decreases ox-LDL-treated HUVEC apoptosis and inflammation
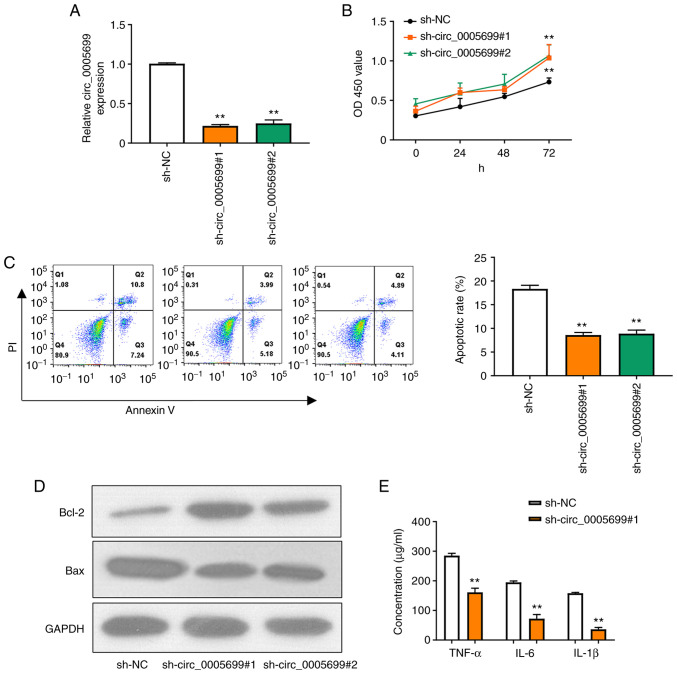
Two shRNAs were designed to target circ_0005699 and the knockdown efficiency was confirmed by RT-qPCR (Fig. 2A); as illustrated, the expression of circ_0005699 was significantly decreased following transfection with the two knockdown shRNAs and there was no significant difference between the two, thus one of them could be selected for subsequent experiments. Notably, circ_0005699 knockdown was associated with increased proliferation of ox-LDL-treated HUVECs (Fig. 2B). In addition, circ_0005699 knockdown significantly reduced the apoptotic rate in these cells, as indicated by Annexin V/PI double-staining and flow cytometry (Fig. 2C). The apoptosis-related proteins Bcl-2 and Bax were detected by western blotting. Compared with that in the control group, knockdown of circ_0005699 markedly increased the protein expression levels of Bcl-2, whereas Bax expression was decreased. Furthermore, knockdown of circ_0005699 significantly reduced the levels of inflammatory factors, including TNF-α, IL-6 and IL-1β in the supernatants of ox-LDL-treated HUVECs (Fig. 2E).
Figure 2.
Knockdown of circ_0005699 decreases the rate of HUVEC apoptosis and inflammation. (A) shRNAs were designed that targeted circ_0005699 and the knockdown efficiency was determined by reverse transcription-quantitative PCR. (B) Cell Counting Kit-8 assay was used to detect HUVEC proliferation. (C) Annexin V/PI double staining and flow cytometry were used to detect the apoptotic rate of HUVECs. (D) Western blot analysis of the expression levels of apoptosis-related proteins Bcl-2 and Bax. (E) Concentration of inflammation-related factors (TNF-α, IL-6 and IL-1β) in HUVECs, as determined by ELISA. **P<0.01 vs. sh-NC. Bcl-2, B-cell lymphoma 2; HUVEC, human umbilical vein endothelial cell; NC, negative control; sh/shRNA, short hairpin RNA.
circ_0005699 sponges miR-450b-5b
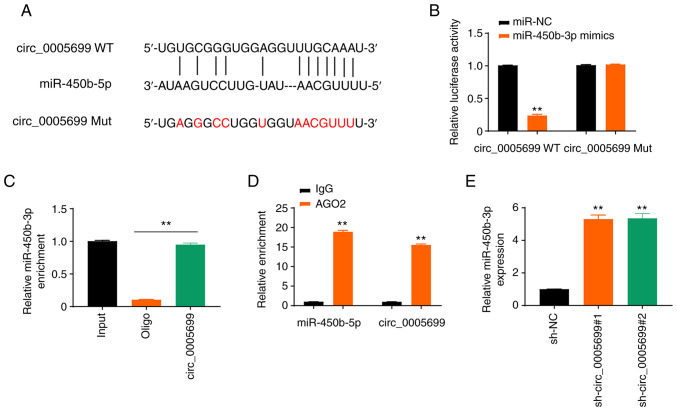
The starBase online database revealed that circ_0005699 possessed a binding site for miR-450b-5p (Fig. 3A). The expression levels of miR-450b-5p in ox-LDL-treated cells transfected with miR-450b-5p mimics were detected and it was revealed that the expression levels of miR-450b-5p were significantly increased (Fig. S1A). Notably, miR-450b-5p overexpression resulted in decreased luciferase activity in circ_0005699 WT ox-LDL-treated cells, but not in circ_0005699 Mut cells (Fig. 3B). In addition, the circ_0005699 probe pulled down more miR-450b-5p than the Oligo probe (Fig. 3C), which verified that circ_0005699 could bind to miR-450b-5p, whereas the beads coupled with AGO2 pulled down significantly greater circ_0005699 and miR-450b-5p than the IgG control (Fig. 3D). Furthermore, circ_0005699-knockdown via shRNA resulted in significantly increased expression of miR-450b-3p compared with that in the control group, as indicated by RT-qPCR analysis (Fig. 3E).
Figure 3.
circ_0005699 sponges miR-450b-5b. (A) circ_0005699 and miR-450b-5p binding sites were determined using the starBase online database. (B) Results of dual-luciferase reporter gene assay. **P<0.01 vs. miR-NC. (C) Results of RNA pull-down assay. (D) RNA immunoprecipitation assay using AGO2, with IgG used as a control. **P<0.01 vs. IgG. (E) Relative expression of miR-450b-5p after circ_0005699-knockdown. **P<0.01 vs. sh-NC. miR, microRNA; Mut, mutant; NC, negative control; sh, short hairpin RNA; WT, wild-type.
NFKB1 is the target of miR-450b-5p
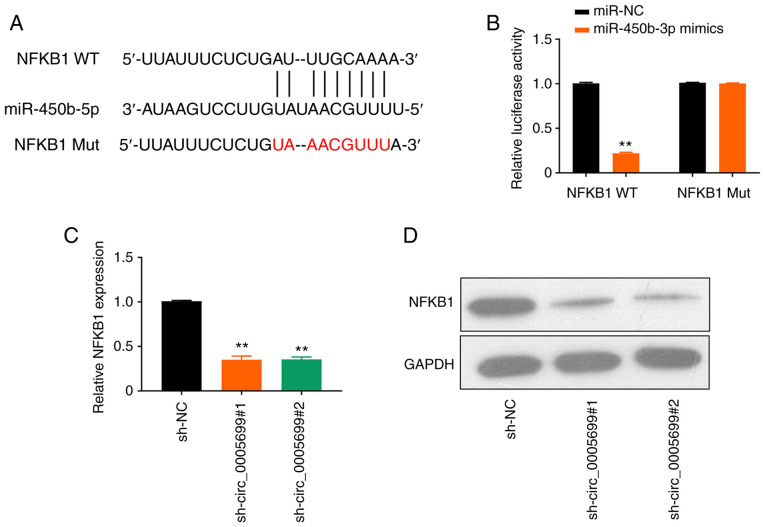
The online database starBase predicted that miR-450b-5p may target NFKB1 (Fig. 4A). It was observed that the overexpression of miR-450b-5p significantly decreased luciferase activity in NFKB1 WT ox-LDL-treated cells compared with the control; however, this effect was not observed following mutation in the binding site of NFKB1 (Fig. 4B). Furthermore, circ_0005699 silencing resulted in significantly decreased expression levels of NFKB1 in ox-LDL-treated cells compared with those in the control group, as determined using RT-qPCR (Fig. 4C) and western blot analysis (Fig. 4D).
Figure 4.
miR-450b-5p targets NFKB1. (A) StarBase online database analysis revealed that mir-450b-5p targets NFKB1. (B) Dual-luciferase reporter assay verified the targeting of miR-450b-5P to NFKB1. **P<0.01 vs. miR-NC. (C) mRNA and (D) protein expression levels of NFKB1 after circ_0005699 knockdown, as determined by reverse transcription-quantitative PCR and western blotting. **P<0.01 vs. sh-NC. miR, microRNA; Mut, mutant; NC, negative control; sh, short hairpin RNA; WT, wild-type.
circ_0005699 attenuates ox-LDL-treated HUVEC apoptosis and inflammation through the miR-450b-5p/NFKB1 axis
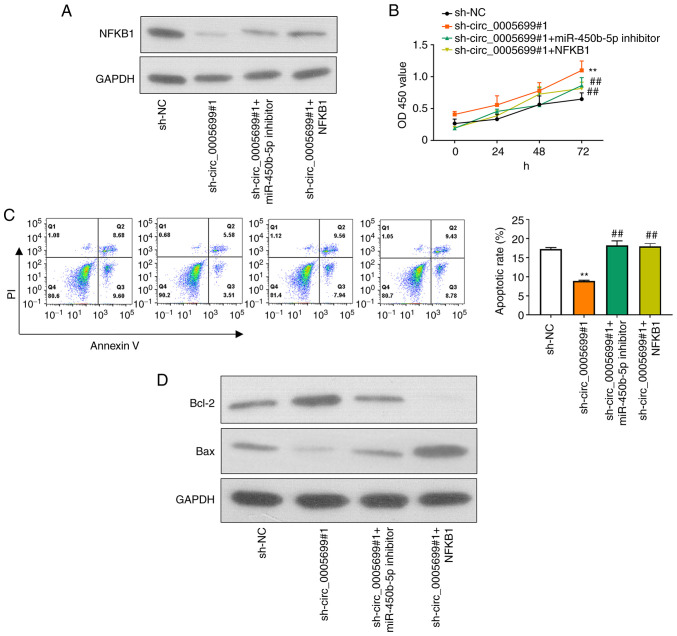
Western blotting and RT-qPCR were used to detect the protein and mRNA expression levels of NFKB1 in different groups of ox-LDL-treated HUVECs following different treatments. First, miR-450b-5p expression was markedly decreased in ox-LDL-treated HUVECs transfected with miR-450b-5p inhibitor (Fig. S1B). In addition, NFKB1 expression was significantly increased in ox-LDL-treated HUVECs transfected with NFKB1 plasmid (Fig. S1C). Compared with that in the control group, knockdown of circ_0005699 was associated with decreased NFKB1 protein expression (Fig. 5A). Notably, ox-LDL-treated HUVECs co-transfected with sh-circ_0005699#1 and miR-450b-5p inhibitor or NFKB1 overexpression plasmid exhibited partial restoration of NFKB1 expression compared with that in the circ_0005699 knockdown group (Fig. 5A). Furthermore, circ_0005699 knockdown was associated with increased cell proliferation (Fig. 5B) and significantly reduced apoptotic rate compared with sh-NC group (Fig. 5C); these effects were reversed when cells were co-transfected with sh-circ_0005699#1 and miR-450b-5p inhibitor or NFKB1 overexpression plasmid (Fig. 5C). In addition, the protein expression levels of Bcl-2 were markedly increased, whereas Bax protein expression was inhibited following knockdown of circ_0005699 (Fig. 5D). By contrast, when cells were co-transfected with sh-circ_0005699#1 and miR-450b-5p inhibitor or NFKB1 overexpression plasmid, Bcl-2 protein expression was reduced and Bax protein expression was partially increased compared with that in cells transfected with sh-circ_0005699#1 only (Fig. 5D).
Figure 5.
circ_0005699 attenuates HUVEC apoptosis and inflammation through the miR-450b-5p/NFKB1 axis. (A) Protein expression levels of NFKB1 in different groups of HUVECs, as detected by western blotting. (B) Cell Counting Kit-8 analysis of HUVEC proliferation following different treatments. (C) Apoptotic rate of HUVECs in different groups, as determined by Annexin V/PI double staining and flow cytometry. (D) Protein expression levels of apoptosis-related genes Bcl-2 and Bax. **P<0.01 vs. sh-NC, ##P<0.01 vs. sh-circ_0005699#1. Bcl-2, B-cell lymphoma 2; HUVEC, human umbilical vein endothelial cell; miR, microRNA; NC, negative control; sh, short hairpin RNA.
Discussion
Cardiovascular disease caused by ASAS remains the leading cause of mortality worldwide (24) and is considered a type of chronic inflammatory disease (25). It has previously been reported that ox-LDL may serve a critical role in the development of atherosclerotic plaque formation through various mechanisms. These mechanisms may include endothelial dysfunction (26–28), foam cell formation (29–31), smooth muscle cell migration and proliferation (32,33), and the induction of platelet adhesion and aggregation (34,35). Wang et al (12) previously indicated that ox-LDL-induced foam cells (an in vitro AS model) had increased expression of circ_0005699. To the best of our knowledge, there is no previous study available that investigates the role of circ_0005699 in the regulation of vascular endothelial cells. Therefore, HUVECs, endothelial cells isolated from the human umbilical cord vein, were used in the present study. These cells have been widely used as a model for the study of function and pathologies of endothelial cells, including AS and plaque formation (36–40). In addition, ApoE-deficient mice were used as an in vivo model of ASAS. These mice, first produced by Plump et al (41) exhibit atherosclerotic transformation even under a normal chow-diet (42).
The results of the present study indicated that ox-LDL resulted in increased expression of circ_0005699 in HUVECs in a time- and dose-dependent manner, which is in corroboration with a previous report (12). Notably, circ_0005699 silencing was associated with increased proliferation of HUVECs, along with a decreased rate of apoptosis in the present study. Furthermore, circ_0005699 was shown to sponge miR-450b-5b and NFKB1 was identified as a target gene of miR-450b-5p; therefore, circ_0005699 may attenuate HUVEC apoptosis and inflammation through the miR-450b-5p/NFKB1 axis.
The starBase database predicted that circ_0005699 could sponge miR-450b-5p. Furthermore, circ_0005699 silencing was associated with increased miR-450b-5p expression, thus indicating that miR-450b-5p was a target of circ_0005699. The role of miR-450b-5p in the regulation of various genes has previously been investigated, particularly in cancer. For example, miR-450b-5p downregulation has been reported to be associated with the progression of hepatocellular carcinoma (43), whereas miR-450b-5p expression was revealed to be upregulated in colorectal cancer cells (44). Furthermore, another study has indicated miR-450b-5p may be a potential biomarker of ischemic injury (45).
The starBase database further predicted that NFKB1 was a target gene of miR-450b-5p and this was confirmed by luciferase reporter gene assay. NFKB is a transcription factor that has been shown to regulate innate and adaptive immune responses, and can induce the transcription of various inflammatory mediators, including TNF-α, IL-1β, IL-6, IL-12p40 and cyclooxygenase-2 (46). Therefore, NFKB1 has been implicated in the pathogenesis of multiple inflammatory diseases, including AS (47–49). A recent report by Huang et al (50) revealed that miR-450b-5p inhibition could reduce ischemic injury in hepatic cells through an NFKB-dependent mechanism. Another report suggested that miR-148a-3p reduced NFKB signaling and thus proinflammatory gene expression in aortic valve cells (51), which further corroborates the present findings, which indicated that NFKB promoted the apoptosis of HUVECs and aggravated inflammation.
In conclusion, the findings of the present study indicated that circ_0005699 knockdown may have beneficial effects on the survival and function of endothelial cells through the regulation of miR-450b-5p/NFKB1.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
TC designed the project and collected data. BY and WC analyzed the data and drafted the manuscript. GZ performed almost all of the experiments. HX and YG performed some experiments, and LL was involved in data collection and analysis. TC and LL confirm the authenticity of all the raw data. All of the authors revised and corrected the manuscript, and read and approved the final manuscript.
Ethics approval and consent to participate
All of the procedures were reviewed and approved by the Ethical Committee for Animal Experimentation, The Affiliated Ganzhou Hospital of Nanchang University (Ganzhou, China; approval no. KY-E-2019-11-19).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Shah PK. Inflammation, infection and atherosclerosis. Trends Cardiovasc Med. 2019;29:468–472. doi: 10.1016/j.tcm.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Katakami N. Mechanism of development of atherosclerosis and cardiovascular disease in diabetes mellitus. J Atheroscler Thromb. 2018;25:27–39. doi: 10.5551/jat.RV17014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vidanapathirana AK, Psaltis PJ, Bursill CA, Abell AD, Nicholls SJ. Cardiovascular bioimaging of nitric oxide: Achievements, challenges, and the future. Med Res Rev. 2021;41:435–463. doi: 10.1002/med.21736. [DOI] [PubMed] [Google Scholar]
- 4.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 5.Holdt LM, Kohlmaier A, Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol Life Sci. 2018;75:1071–1098. doi: 10.1007/s00018-017-2688-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao L, Guo Y, Guo Y, Ji X, Fan D, Chen C, Yuan W, Sun Z, Ji Z. Effect and mechanism of circRNAs in tumor angiogenesis and clinical application. Int J Cancer. 2022;150:1223–1232. doi: 10.1002/ijc.33863. [DOI] [PubMed] [Google Scholar]
- 7.Ye D, Gong M, Deng Y, Fang S, Cao Y, Xiang Y, Shen Z. Roles and clinical application of exosomal circRNAs in the diagnosis and treatment of malignant tumors. J Transl Med. 2022;20:161. doi: 10.1186/s12967-022-03367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Ryu J, Ahn Y, Kook H, Kim YK. The roles of non-coding RNAs in vascular calcification and opportunities as therapeutic targets. Pharmacol Ther. 2021;218:107675. doi: 10.1016/j.pharmthera.2020.107675. [DOI] [PubMed] [Google Scholar]
- 10.Ding P, Ding Y, Tian Y, Lei X. Circular RNA circ_0010283 regulates the viability and migration of oxidized low-density lipoprotein-induced vascular smooth muscle cells via an miR-370-3p/HMGB1 axis in atherosclerosis. Int J Mol Med. 2020;46:1399–1408. doi: 10.3892/ijmm.2020.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, Li Y, Liu Z, Ma X, Li M, Lu Q, Li Y, Lu Z, Niu L, Fan Z, Lei Z. Circular RNA circ_0124644 exacerbates the ox-LDL-induced endothelial injury in human vascular endothelial cells through regulating PAPP-A by acting as a sponge of miR-149-5p. Mol Cell Biochem. 2020;471:51–61. doi: 10.1007/s11010-020-03764-0. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Zheng Z, Feng X, Zang X, Ding W, Wu F, Zhao Q. circRNA/lncRNA-miRNA-mRNA network in oxidized, low-density, lipoprotein-induced foam cells. DNA Cell Biol. 2019;38:1499–1511. doi: 10.1089/dna.2019.4865. [DOI] [PubMed] [Google Scholar]
- 13.Yu XH, Fu YC, Zhang DW, Yin K, Tang CK. Foam cells in atherosclerosis. Clin Chim Acta. 2013;424:245–252. doi: 10.1016/j.cca.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Le Saux G, Plawinski L, Parrot C, Nlate S, Servant L, Teichmann M, Buffeteau T, Durrieu MC. Surface bound VEGF mimicking peptide maintains endothelial cell proliferation in the absence of soluble VEGF in vitro. J Biomed Mater Res A. 2016;104:1425–1436. doi: 10.1002/jbm.a.35677. [DOI] [PubMed] [Google Scholar]
- 15.Lu J, Mitra S, Wang X, Khaidakov M, Mehta JL. Oxidative stress and lectin-like ox-LDL-receptor LOX-1 in atherogenesis and tumorigenesis. Antioxid Redox Signal. 2011;15:2301–2333. doi: 10.1089/ars.2010.3792. [DOI] [PubMed] [Google Scholar]
- 16.Huang F, Chen W, Peng J, Li Y, Zhuang Y, Zhu Z, Shao C, Yang W, Yao H, Zhang S. LncRNA PVT1 triggers Cyto-protective autophagy and promotes pancreatic ductal adenocarcinoma development via the miR-20a-5p/ULK1 axis. Mol Cancer. 2018;17:98. doi: 10.1186/s12943-018-0845-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Zhu X, Li Z, Li T, Long F, Lv Y, Liu L, Liu X, Zhan Q. Osthole inhibits the PI3K/AKT signaling pathway via activation of PTEN and induces cell cycle arrest and apoptosis in esophageal squamous cell carcinoma. Biomed Pharmacother. 2018;102:502–509. doi: 10.1016/j.biopha.2018.03.106. [DOI] [PubMed] [Google Scholar]
- 18.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui J, Li W, Liu G, Chen X, Gao X, Lu H, Lin D. A novel circular RNA, hsa_circ_0043278, acts as a potential biomarker and promotes non-small cell lung cancer cell proliferation and migration by regulating miR-520f. Artif Cells Nanomed Biotechnol. 2019;47:810–821. doi: 10.1080/21691401.2019.1575847. [DOI] [PubMed] [Google Scholar]
- 20.Zhao W, Geng D, Li S, Chen Z, Sun M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018;7:842–855. doi: 10.1002/cam4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Peng L, Chen G, Zhu Z, Shen Z, Du C, Zang R, Su Y, Xie H, Li H, Xu X, et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung's disease. Oncotarget. 2017;8:808–818. doi: 10.18632/oncotarget.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 23.Kato R, Mori C, Kitazato K, Arata S, Obama T, Mori M, Takahashi K, Aiuchi T, Takano T, Itabe H. Transient increase in plasma oxidized LDL during the progression of atherosclerosis in apolipoprotein E knockout mice. Arterioscler Thromb Vasc Biol. 2009;29:33–39. doi: 10.1161/ATVBAHA.108.164723. [DOI] [PubMed] [Google Scholar]
- 24.Gisterå A, Hansson GK. The immunology of atherosclerosis. Nat Rev Nephrol. 2017;13:368–380. doi: 10.1038/nrneph.2017.51. [DOI] [PubMed] [Google Scholar]
- 25.Kattoor AJ, Pothineni NVK, Palagiri D, Mehta JL. Oxidative stress in atherosclerosis. Curr Atheroscler Rep. 2017;19:42. doi: 10.1007/s11883-017-0678-6. [DOI] [PubMed] [Google Scholar]
- 26.Frostegård J, Haegerstrand A, Gidlund M, Nilsson J. Biologically modified LDL increases the adhesive properties of endothelial cells. Atherosclerosis. 1991;90:119–126. doi: 10.1016/0021-9150(91)90106-D. [DOI] [PubMed] [Google Scholar]
- 27.Catapano AL, Maggi FM, Tragni E. Low density lipoprotein oxidation, antioxidants, and atherosclerosis. Curr Opin Cardiol. 2000;15:355–363. doi: 10.1097/00001573-200009000-00008. [DOI] [PubMed] [Google Scholar]
- 28.Forstermann U, Sessa WC. Nitric oxide synthases: Regulation and function. Eur Heart J. 2012;33:829–837. 837a–837d. doi: 10.1093/eurheartj/ehr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbieri SS, Cavalca V, Eligini S, Brambilla M, Caiani A, Tremoli E, Colli S. Apocynin prevents cyclooxygenase 2 expression in human monocytes through NADPH oxidase and glutathione redox-dependent mechanisms. Free Radic Biol Med. 2004;37:156–165. doi: 10.1016/j.freeradbiomed.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 30.Hansson GK, Robertson AK, Söderberg-Nauclér C. Inflammation and atherosclerosis. Annu Rev Pathol. 2006;1:297–329. doi: 10.1146/annurev.pathol.1.110304.100100. [DOI] [PubMed] [Google Scholar]
- 31.Mietus-Snyder M, Friera A, Glass CK, Pitas RE. Regulation of scavenger receptor expression in smooth muscle cells by protein kinase C: A role for oxidative stress. Arterioscler Thromb Vasc Biol. 1997;17:969–978. doi: 10.1161/01.ATV.17.5.969. [DOI] [PubMed] [Google Scholar]
- 32.Mitra S, Goyal T, Mehta JL. Oxidized LDL, LOX-1 and atherosclerosis. Cardiovasc Drugs Ther. 2011;25:419–429. doi: 10.1007/s10557-011-6341-5. [DOI] [PubMed] [Google Scholar]
- 33.Shen CM, Mao SJ, Huang GS, Yang PC, Chu RM. Stimulation of smooth muscle cell proliferation by ox-LDL- and acetyl LDL-induced macrophage-derived foam cells. Life Sci. 2001;70:443–452. doi: 10.1016/S0024-3205(01)01428-X. [DOI] [PubMed] [Google Scholar]
- 34.Maiolino G, Rossitto G, Caielli P, Bisogni V, Rossi GP, Calò LA. The role of oxidized low-density lipoproteins in atherosclerosis: The myths and the facts. Mediators Inflamm. 2013;2013:714653. doi: 10.1155/2013/714653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Podrez EA, Byzova TV, Febbraio M, Salomon RG, Ma Y, Valiyaveettil M, Poliakov E, Sun M, Finton PJ, Curtis BR, et al. Platelet CD36 links hyperlipidemia, oxidant stress and a prothrombotic phenotype. Nat Med. 2007;13:1086–1095. doi: 10.1038/nm1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park HJ, Zhang Y, Georgescu SP, Johnson KL, Kong D, Galper JB. Human umbilical vein endothelial cells and human dermal microvascular endothelial cells offer new insights into the relationship between lipid metabolism and angiogenesis. Stem Cell Rev. 2006;2:93–102. doi: 10.1007/s12015-006-0015-x. [DOI] [PubMed] [Google Scholar]
- 37.Yamada T, Fan J, Shimokama T, Tokunaga O, Watanabe T. Induction of fatty streak-like lesions in vitro using a culture model system simulating arterial intima. Am J Pathol. 1992;141:1435–1444. [PMC free article] [PubMed] [Google Scholar]
- 38.Burns MP, DePaola N. Flow-conditioned HUVECs support clustered leukocyte adhesion by coexpressing ICAM-1 and E-selectin. Am J Physiol Heart Circ Physiol. 2005;288:H194–H204. doi: 10.1152/ajpheart.01078.2003. [DOI] [PubMed] [Google Scholar]
- 39.Kokura S, Wolf RE, Yoshikawa T, Granger DN, Aw TY. Molecular mechanisms of neutrophil-endothelial cell adhesion induced by redox imbalance. Circ Res. 1999;84:516–524. doi: 10.1161/01.RES.84.5.516. [DOI] [PubMed] [Google Scholar]
- 40.Zhang W, DeMattia JA, Song H, Couldwell WT. Communication between malignant glioma cells and vascular endothelial cells through gap junctions. J Neurosurg. 2003;98:846–853. doi: 10.3171/jns.2003.98.4.0846. [DOI] [PubMed] [Google Scholar]
- 41.Plump AS, Smith JD, Hayek T, Aalto-Setälä K, Walsh A, Verstuyft JG, Rubin EM, Breslow JL. Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell. 1992;71:343–353. doi: 10.1016/0092-8674(92)90362-G. [DOI] [PubMed] [Google Scholar]
- 42.Piedrahita JA, Zhang SH, Hagaman JR, Oliver PM, Maeda N. Generation of mice carrying a mutant apolipoprotein E gene inactivated by gene targeting in embryonic stem cells. Proc Natl Acad Sci USA. 1992;89:4471–4475. doi: 10.1073/pnas.89.10.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Shen S, Chen X, Ren Z, Li Z, Yu Z. miR-450b-5p loss mediated KIF26B activation promoted hepatocellular carcinoma progression by activating PI3K/AKT pathway. Cancer Cell Int. 2019;19:205. doi: 10.1186/s12935-019-0923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye YP, Wu P, Gu CC, Deng DL, Jiao HL, Li TT, Wang SY, Wang YX, Xiao ZY, Wei WT, et al. miR-450b-5p induced by oncogenic KRAS is required for colorectal cancer progression. Oncotarget. 2016;7:61312–61324. doi: 10.18632/oncotarget.11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo X, Wang W, Li D, Xu C, Liao B, Li F, Zhou X, Qin W, Liu J. Plasma exosomal miR-450b-5p as a possible biomarker and therapeutic target for transient ischaemic attacks in rats. J Mol Neurosci. 2019;69:516–526. doi: 10.1007/s12031-019-01341-9. [DOI] [PubMed] [Google Scholar]
- 46.Cartwright T, Perkins ND, L Wilson C. NFKB1: A suppressor of inflammation, ageing and cancer. FEBS J. 2016;283:1812–1822. doi: 10.1111/febs.13627. [DOI] [PubMed] [Google Scholar]
- 47.Fiordelisi A, Iaccarino G, Morisco C, Coscioni E, Sorriento D. NFkappaB is a key player in the crosstalk between inflammation and cardiovascular diseases. Int J Mol Sci. 2019;20:1599. doi: 10.3390/ijms20071599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hernández-Presa MA, Ortego M, Tuñón J, Martín-Ventura JL, Mas S, Blanco-Colio LM, Aparicio C, Ortega L, Gómez-Gerique J, Vivanco F, Egido J. Simvastatin reduces NF-kappaB activity in peripheral mononuclear and in plaque cells of rabbit atheroma more markedly than lipid lowering diet. Cardiovasc Res. 2003;57:168–177. doi: 10.1016/S0008-6363(02)00619-3. [DOI] [PubMed] [Google Scholar]
- 49.Kumar A, Takada Y, Boriek AM, Aggarwal BB. Nuclear factor-kappaB: Its role in health and disease. J Mol Med (Berl) 2004;82:434–448. doi: 10.1007/s00109-004-0555-y. [DOI] [PubMed] [Google Scholar]
- 50.Huang Z, Mou T, Luo Y, Pu X, Pu J, Wan L, Gong J, Yang H, Liu Y, Li Z, et al. Inhibition of miR-450b-5p ameliorates hepatic ischemia/reperfusion injury via targeting CRYAB. Cell Death Dis. 2020;11:455. doi: 10.1038/s41419-020-2648-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel V, Carrion K, Hollands A, Hinton A, Gallegos T, Dyo J, Sasik R, Leire E, Hardiman G, Mohamed SA, et al. The stretch responsive microRNA miR-148a-3p is a novel repressor of IKBKB, NF-κB signaling, and inflammatory gene expression in human aortic valve cells. FASEB J. 2015;29:1859–1868. doi: 10.1096/fj.14-257808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.