Abstract
Multiple myeloma (MM) is still an incurable hematologic malignancy, which is eagerly to the discovery of novel therapeutic targets and methods. N-acetyltransferase 10 (NAT10) is the first reported regulator of mRNA acetylation that is activated in many cancers. However, the function of NAT10 in MM remains unclear. We found significant upregulation of NAT10 in MM patients compared to normal plasma cells, which was also highly correlated with MM poor outcome. Further enforced NAT10 expression promoted MM growth in vitro and in vivo, while knockdown of NAT10 reversed those effects. The correlation analysis of acetylated RNA immunoprecipitation sequencing (acRIP-seq) and ribosome profiling sequencing (Ribo-seq) combined with RIP-PCR tests identified centrosomal protein 170 (CEP170) as an important downstream target of NAT10. Interfering CEP170 expression in NAT10-OE cells attenuated the acceleration of cellular growth caused by elevated NAT10. Moreover, CEP170 overexpression promoted cellular proliferation and chromosomal instability (CIN) in MM. Intriguingly, remodelin, a selective NAT10 inhibitor, suppressed MM cellular growth, induced cellular apoptosis in vitro and prolonged the survival of 5TMM3VT mice in vivo. Collectively, our data indicate that NAT10 acetylates CEP170 mRNA to enhance CEP170 translation efficiency, which suggests that NAT10 may serve as a promising therapeutic target in MM.
KEY WORDS: Multiple myeloma, Target, NAT10, Acetylation, CEP170, Chromosomal instability, Translation, Remodelin
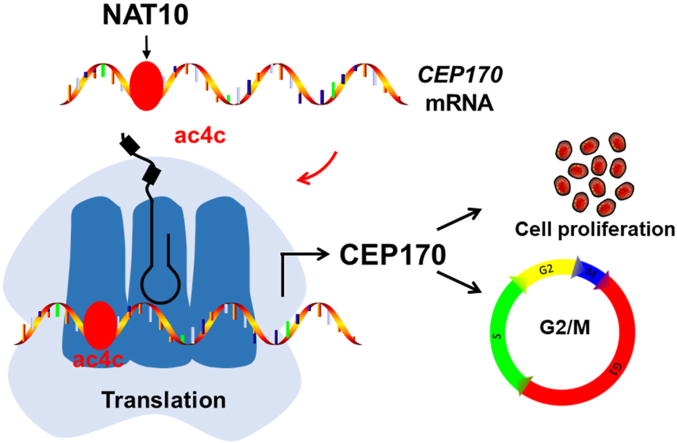
Graphical abstract
NAT10 regulates cell proliferation and cell cycle distribution via acetylating CEP170 mRNA to enhance translation efficiency in multiple myeloma.
1. Introduction
Multiple myeloma (MM) is still an incurable hematological malignancy, especially the high-risk subgroup patients are characterized by high-proliferation feature and they have no significant improvement of outcome even treated with the advanced novel drugs including proteasome inhibitors, immune modulators et al1,2. Furthermore, the prognostic stratification and therapeutic evaluation systems for MM also lack specific molecular indicators. Thus, seeking for novel diagnostic marker and potential therapeutic target for MM, especially for high-risk MM is of extreme urgency. Increasing studies demonstrate the importance of epigenetic regulation on MM malignancy, such as DNA methylation, histone acetylation and Epi-microRNAs3. Here we focus on N4-acetylcytidine (ac4C) mRNA modification, which is originally presented in tRNA and rRNA and recently investigated in multiple types of cancer mRNA, not yet in MM4, 5, 6.
N-Acetyltransferase 10 (NAT10) is the first identified acetyltransferase on ac4C mRNA modification containing 1025 amino acids with a molecular weight of 116 kD. There are three conserved domains in NAT10 protein, including the N-terminal acetylase domain (GNAT family), an ATP/GTP binding motif and an ATPase domain7. The acetylation site of NAT10 is lysine residue at 426 (K426), and acetylation of K426 is necessary for NAT10 activating rRNA transcription8. NAT10 catalyzes mRNA acetylation within coding sequences (CDS) to improve mRNA stability and increase translation efficiency6. It has been reported that NAT10 is involved in multiple cancers, such as epithelial ovarian cancer9, hepatocellular carcinoma10, 11, 12, 13, 14, breast cancer15, colorectal cancer16, 17, 18, acute myeloid leukemia19 et al. However, a large fraction of these studies describes other functions of NAT10 out of non-mRNA acetyltransferase. For instance, NAT10 acetylates p53 at K120 and stabilizes p53 at protein level by counteracting MDM2 action leading to inhibition of cellular proliferation in human colorectal carcinomas16. NAT10 and SIRT1 mediate the acetylation of CCDC84 protein at lysine 31 in Hela cells, and the acetylated CCDC84 promotes HsSAS-6 binding to APC/CCdh1 to regulate centrosome duplication20. The function of NAT10 in MM and the specific association between mRNA ac4C modification and MM pathogenesis are still poorly elaborated.
In this study, we first demonstrated the contributing role of NAT10 to MM growth in clinical cohorts and primary samples combined with laboratory works in vitro and in vivo. Furthermore, we discovered NAT10 catalyzing ac4C modification to enhance translation efficiency in MM cells and identified CEP170 as the major and functional downstream target of NAT10. In addition, inhibition of NAT10 by remodelin suppressed MM growth in vitro and in vivo. Taken together, we disclose that NAT10 is a potentially promising diagnostic marker and therapeutic target for MM.
2. Materials and methods
2.1. Gene expression profiling
The Gene expression profiling (GEP) cohorts were collected from GEO database as previous described21,22. Total therapy 2 (TT2, GSE2658), TT3 (GSE2658), and the assessment of proteasome inhibition for extending remission (APEX, GSE9782) were employed. The Dutch-Belgian Cooperative Trial Group for Hematology Oncology Group-65 (HOVON65) trials was collected from GSE19784.
2.2. Antibodies and reagents
The antibodies were used as following, NAT10 (13365-1-AP, Proteintech; DF12427, Affinity), CDK4 (11026-1-AP, Proteintech), CDK6 (14052-1-AP, Proteintech), CEP170 (18899-1-AP, Proteintech), PARP (9542 S, Cell Signaling Technology), cleaved caspase-3 (9661 S, Cell Signaling Technology), ac4C (ab252215, Abcam), rabbit IgG (7074, Cell Signaling Technology), β-actin (60008-1-Ig, Proteintech), Ki67 (AF0198, Affinity), α-tubulin (ab7291, Abcam), BrdU mAb (MI-11-3, MBL), secondary antibodies included goat anti-rabbit IgG(H+L) HRP (FMS-Rb01, Fcmacs) or mouse (S0002, Affinity). Remodelin was purchased from CSNpharm. Puromycin was from AEPxBIO (B7587), and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, M8180) was purchased from Solarbio. Actinomycin D was purchased from MCE (MedChemExpress, HY-17559). 5′-Bromo-uridine (BrU) was purchased from Sigma–Aldrich (850187). Lipofectamine Transfection Reagent (40802ES02), TRIeasy Total RNA Extraction Reagent (19201ES60), complementary DNA synthesis superMix (11123ES10), and SYBR Green Master Mix (11198ES03) were purchased from Yeasen.
2.3. Cell lines and culture
Human MM cell lines, CAG, KMS28PE, OPM2 and 293 T cells, were donated by Prof. Siegfried Janz (Division of Hematology and Oncology, Medical College of Wisconsin, Milwaukee, WI, USA) and 5TMM3VT murine myeloma cell was provided by Prof. Wen Zhou (Central South University, Changsha, China). The cells were cultured in RPMI-1640 (01-100-1ACS, Biological Industries, Israel). 293 T cells were cultured in Dulbecco's modified Eagle's medium (DMEM, 01-052-1ACS, Biological Industries, Israel). All media were supplemented with 10% fetal bovine serum (FBS, 04-001-1ACS, Biological Industries, Israel), 100 U/mL penicillin and 100 μg/mL streptomycin (03-031-1B, Biological Industries, Israel). All cells were cultured in humidified 5% CO2 incubators at 37 °C.
2.4. The patient sample collection and immunohistochemistry analysis (IHC)
The posterior superior iliac spine of MM patients was chosen as the biopsy point, followed by disinfecting the skin, spreading a drape and anesthetizing with 2% lidocaine. Next, a bone marrow biopsy needle was used to pierce vertically and inserted the needle clockwise for 1–2 mm. Then the needle tube clockwise was withdrawn, the bone marrow tissue was obtained and placed in the fixative followed by the process of pathology filming. IHC was performed as described previously23. Immunostaining was performed on paraffin tissue sections. The main process was as follows. Slides were incubated with anti-NAT10 or anti-Ki67 overnight at 4 °C. Afterwards, the secondary antibody was applied and kept for 45 min at 37 °C, followed by dropping SABC at 37 °C for 30 min, 3,3′-diaminobenzidine (DAB) coloring, and finally counterstaining with hematoxylin. The study protocol was approved by the Human Research Ethics Committees of the Affiliated Hospital of Nanjing University of Chinese Medicine (Ethics number: KY2018005). All patients provided written informed consent for their bone marrow tissue samples to be used for research.
2.5. Plasmids and transfection
The plasmids containing the Homo NAT10 cDNA were purchased from TranSheepBio. The NAT10 coding sequence (NM_024662.3) was cloned into the lentiviral vector, pTSB carrying Flag tag. Lentiviruses containing the cDNA were obtained by co-transfection of the pTSB-NAT10 with packaging vectors (PSPAX.2 and PMD2.G) into 293 T cells in accordance with the protocol of Lipofectamine Transfection Reagent. The virus supernatant was harvested after 48 h, concentrated, and stored at −80 °C. MM cells were transfected with lentivirus containing NAT10 cDNA to yield NAT10-OE MM cells. Transfected cells were selected with puromycin treatment. Transduction efficiency was determined by Western blotting.
2.6. Transient transfection
Small interfering RNA (siRNA) was transfected into MM cells by using electrotransmitter (BTX). The specific process was as follows. BTXpress Cytoporation Media T4 (47-0003, BTX) was used to resuspend cells in 1 × 106/mL, added siRNA to a final concentration of 100 nmol/L, and transferred to electric shock cup after mixed and transfected finally. Electric transfer parameters were determined as following: square wave, voltage 960 V, duration 0.1 ms, number of pulses 2, pulse interval 1.0 s and electrode gap 4 mm siRNAs were synthesized by Shanghai Genepharma. Sequences of siRNA were as following:
Negative control: sense 5′-UUCUCCGAACGUGUCACGUTT-3′, anti-sense 5′-ACGUGACACGUUCGGAGAATT-3′;
NAT10: sense 5′-GCAUGGACCUCUCUGAAUATT-3′, anti-sense 5′-UAUUCAGAGAGGUCCAUGCTT-3′;
CEP170: sense 5′-GCAUGAGAAGUUUACCAUUTT-3′, anti-sense 5′-AAUGGUAAACUUCUCAUGCTT-3′.
The sequence of siNAT10 was designed for the coding sequences (CDS) targeted exon in site of 3049 base and the sequence of siCEP170 was target exon in site of 616 base.
2.7. Western blotting
Western blotting assay was performed as described previously23. Anti-β-actin was used as a loading control for total protein.
2.8. Cell proliferation and viability assay
Cell viability was detected as described previously23 by using MTT assay according to the manufacturer's protocol (Solarbio).
2.9. MM xenografts
CAG wild type (WT) cells and CAG NAT10-OE cells (2 × 106) were injected subcutaneously into the left and right flank abdominal, respectively, of 6–8-weeks old NOD/SCID mice. In all cases, tumor diameter was measured with calipers. Once the tumor diameter reached 15 mm, mice were sacrificed, and tumor tissues were collected, weighted, and photographed. All animal work was performed in accordance with government-published recommendations for the Care and Use of laboratory animals and guidelines of Institutional Ethics Review Boards of Nanjing University of Chinese Medicine (Ethics Registration No. 201905A003).
2.10. 5TMM3VT myeloma mouse model
Approximately 1 × 106 5TMM3VT cells were injected through the orbital vein into C57BL/KaLwRij mice from Harlan Laboratories (n = 8 per group). The mice were randomly divided into two groups (Control and Remodelin). On Day 3 after cell injection, intraperitoneal injection of remodelin (5 mg/kg) was performed twice weekly in remodelin group.
2.11. Flow cytometry analysis of cell cycle and apoptosis
Flow cytometry analysis of cell cycle and apoptosis was performed as described previously23. Flow cytometry equipped with Guava easyCyte System (Merck Millipore, Darmstadt, Germany) was applied to detect cell cycle and apoptosis.
2.12. ac4C detection by dot blot
Dot blot was performed using anti-ac4C antibody as described previously24. At first, 5 μg of RNA with denaturation solution (deionized formamide:37% formaldehyde:MOPS buffer = 66:21:13, v/v/v) was denatured in 65 °C for 5 min, followed by immediately placed on ice for 1 min and loaded onto Hybond-N+ membranes. The membranes were crosslinked twice with 150 mJ/cm2 in the UV254 nm Stratalinker 2400 (Stratagene). Afterwards, the membranes were stained with 0.02% methylene blue solution for 10 min, rinsed the background with DEPC water and scaned as an internal reference.
2.13. NAT10 immunoprecipitation
CAG NAT10-OE cells were utilized for NAT10 immunoprecipitation analysis. The procedure was performed as previously described6. Per each IP, 2.5 μg of anti-NAT10 antibody or 2.5 μg of rabbit IgG control were used.
2.14. Real-time PCR
Total RNA was extracted by using TRIeasy. Complementary DNA was synthesized by using reverse transcription kit according to instruction. Real-time quantitative PCR was performed with SYBR Green master Mix. Primer sequences were listed at Supporting Information Table S1.
2.15. Acetylated RNA immunoprecipitation and sequencing (acRIP-seq)
CAG WT and CAG NAT10-OE cells were used for acRIP-seq analysis. The procedure of acRIP-seq was performed as previously described6. All data analysis and processing were performed by Guangzhou Epibiotek Co., Ltd. (Guangzhou, China).
2.16. Ribosome profiling and sequencing (Ribo-seq)
CAG WT and CAG NAT10-OE cells were used for Ribo-seq analysis. The procedure of Ribo-seq was performed as previously described6. All data analysis and processing were applied by Guangzhou Epibiotek Co., Ltd.
2.17. RNA decay assay
MM cells were treated with mRNA transcription inhibitor actinomycin D (5 μg/mL) (HY-17559, MCE) for 0, 1, 2, and 3 h. Then, the total mRNA was isolated and used for qRT-PCR to quantify the relative abundance of CEP170 mRNA (relative to 0 h), and 18 S rRNA was used as internal control.
2.18. 5′-Bromo-uridine (BrU) immunoprecipitation chase-deep RT-qPCR (BRIC RT-qPCR)
BRIC RT-qPCR was performed as described previously6,25. Briefly, cells were incubated at 37 °C in the presence of 150 μmol/L 5′-bromo-uridine (BrU; 850187, Sigma–Aldrich) for 24 h in a humidified incubator with 5% CO2. The cells were collected at indicated time points after replacing BrU containing medium with BrU-free medium. Total RNA was isolated by using TReasy. BrU-labeled total RNA (12 μg) was denatured by heating at 80 °C for 1 min and then added to the anti-BrdU mAb conjugated beads containing 2 μg of anti-BrdU mAb (MI-11-3, MBL). The mixture was incubated at 4 °C overnight with rotation. After immunoprecipitation, elution of RNA was carried out by adding 300 μL of TReasy directly to the mixture. BrU-labeled RNA was extracted by the TReasy method and then used for RT-qPCR.
2.19. Immunofluorescent staining and confocal microscopy
The cells were fixed with 4% paraformaldehyde, permeabilized with PBS containing 0.1% Triton X-100, and blocked with 4% BSA. After overnight incubation with primary antibodies at 4 °C, the slides were incubated with corresponding secondary antibodies. The images were captured by using a confocal microscope (TCS SP8, Leica, Germany).
2.20. Giemsa staining
Giemsa staining was conducted by using Wright–Giemsa stain (Wuhan Servicebio technology Co., Ltd., Wuhan, China) according to the manufacturer's instructions. Briefly, the slides were stained for 5 min with solution A and next 5 min with solution B. Then the slides were washed with phosphate buffer (pH 6.8) and air dried.
2.21. TUNEL assay
The tunnel assay was performed by using TUNEL Apoptosis Detection Kit (Alexa Fluor 640; 40308, Yeasen) according to the manufacturer's instructions. The images were captured by using a confocal microscope (TCS SP8, Leica, Germany).
2.22. Statistical analysis
All values were expressed as means ± standard deviation (SD). Two-tailed Student's t-test and one-way analysis of variance (ANOVA) (≥three groups) were used to determine significance between experimental groups. The Kaplan–Meier method was used to evaluate the correlation of NAT10 expression with myeloma patient survival. In all cases, significance was defined as P < 0.05.
3. Results
3.1. NAT10 expression is elevated in MM, which confers poor survival of MM patients
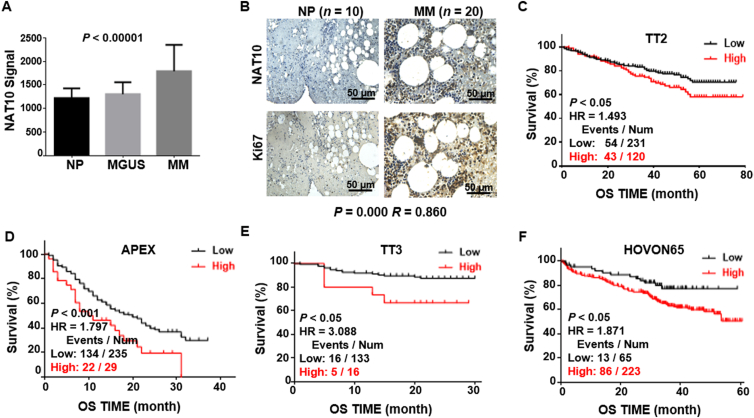
To examine the expression of NAT10 in MM, we interrogated the gene expression profiling (GEP) dataset of normal plasma (NP) cells, monoclonal gammopathy of undetermined significance (MGUS) and MM bone marrow plasma cells. The analysis showed that NAT10 expression was significantly higher in plasma cells from MM (n = 351) patients than MGUS (n = 44) and NP (n = 22) cells (GSE2658, Fig. 1A). We further detected NAT10 expression in MM patient bone marrow tissues (n = 20) compared with normal controls (n = 10) by IHC at protein level. Consistently, IHC staining intensity score presented elevated NAT10 in bone marrow from MM patients compared with normal controls (Supporting Information Fig. S1A). In addition, Ki67, a key marker of proliferation, was highly in accord with NAT10 expression (Fig. 1B and Fig. S1B). These results indicated that NAT10 expression was increased in MM cells and associated with abnormal proliferation of MM. In addition, we correlated NAT10 expression with patient outcome in four independent MM cohorts with over 1000 samples. It was worth noting that MM patients with increased NAT10 expression had poor survival in four cohorts, TT2 (Fig. 1C), APEX (Fig. 1D), TT3 (Fig. 1E) and HOVON65 (Fig. 1F). Taken together, NAT10 expression is elevated in MM patients and correlated with poor survival, indicating that NAT10 may be a biomarker and potential therapeutic target of MM.
Figure 1.
Elevated NAT10 expression is associated with poor outcomes in MM patients. (A) NAT10 mRNA levels were significantly increased in MM samples. The signal level of NAT10 (217884_at signaling) was shown on the y-axis (NP, n = 22; MGUS, n = 44; MM, n = 351). (B) IHC staining verified that NAT10 protein level was elevated in MM patient samples and NAT10 expression was positively correlated with Ki67 expression in MM samples (NP, n = 10; MM, n = 20). (C–F) High expression of NAT10 indicted poor overall survival (OS) in TT2 (C), APEX (D), TT3 (E), and HOVON65 (F) cohorts. All data are displayed as mean ± SD; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.2. NAT10 is a driver gene for MM malignancy in vitro and in vivo
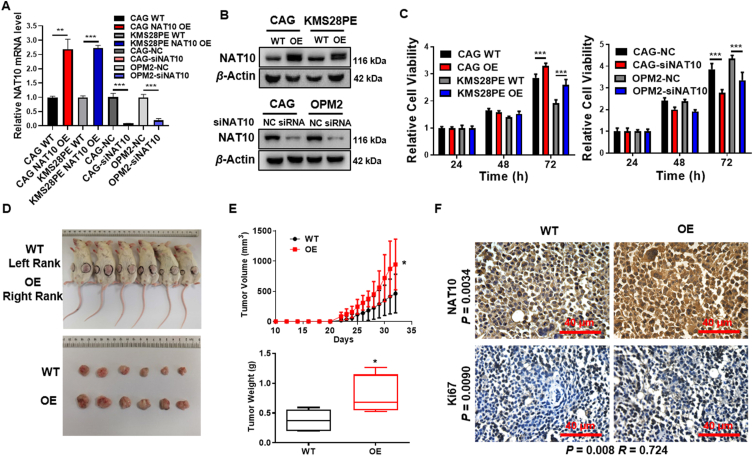
To verify NAT10 acting as a driver gene for MM malignancy, we overexpressed NAT10 in MM cells (CAG and KMS28PE) via lentivirus and knocked down NAT10 by siRNA, which were validated by RT-PCR (Fig. 2A) and Western blotting (Fig. 2B, Supporting Information Fig. S2A and S2B). MTT assay showed that cellular proliferation capacity was enhanced by elevated NAT10 relative to WT cells, and weakened by siNAT10 compared with negative control (NC, Fig. 2C), suggesting that NAT10 boosted MM cell proliferation in vitro. To further extend these observations in vivo, we injected 2 × 106 CAG WT cells (left flank) and CAG NAT10-OE cells (right flank) subcutaneously to NOD/SCID mice (n = 6). Thirty-two days later, NAT10-OE cells generated larger tumors visually compared with WT cells (Fig. 2D), while the mean volume and weight of NAT10-OE tumors were also significantly higher than that of WT tumors statistically (Fig. 2E). Western blotting and IHC assays confirmed that NAT10 was increased in NAT10-OE tumors compared with WT tumors (Supporting Information Fig. S3A). In addition, we examined the correlation of NAT10 and Ki67 in xenograft tumors. The results showed that the expressions of NAT10 and Ki67 in NAT10-OE tumor tissues were significantly higher than WT tumor tissues (Fig. S3B), and the expression of Ki67 was highly correlated with NAT10 expression (Fig. 2F and Fig. S3C). Collectively, we assume that NAT10 is a driver gene in MM and enforced expression of NAT10 promotes MM proliferation both in vitro and in vivo.
Figure 2.
NAT10 is a driver gene for MM malignancy in vitro and in vivo. (A, B) Confirmation of NAT10 expression in NAT10-OE and siNAT10 MM cells by RT-qPCR (n = 3) (A) and Western blotting (B). (C) MTT assay showed that increased NAT10 enhanced cellular proliferation and siNAT10 impeded cell growth in MM cells (n = 6). (D) Photographic images of harvested xenograft tumors (n = 6). (E) Mean tumor volume and weight from WT and NAT10-OE xenograft mice on Day 32. (F) IHC staining showed that NAT10 protein level was elevated in NAT10-OE xenograft tumor tissues and NAT10 expression was positively correlated with Ki67 expression in xenograft tumor tissues. All data are displayed as mean ± SD; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.3. NAT10 regulates cycle distribution and promotes cellular proliferation in MM cells
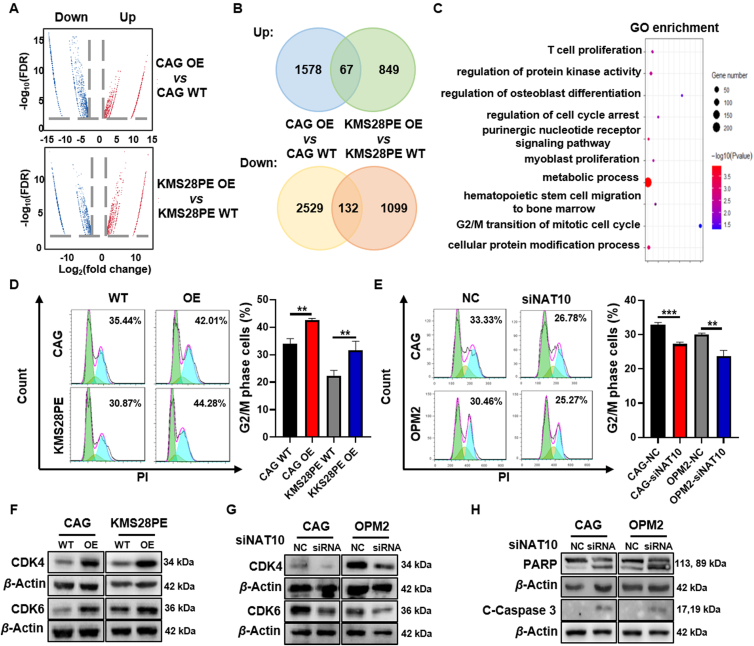
To identify the mechanism responsible for NAT10 promoting MM malignancy, we performed RNA-seq in CAG and KMS28PE cells. Volcano plots showed that there were 1578 upregulated and 2529 downregulated genes in CAG NAT10-OE cells compared with CAG WT cells, as well as 849 upregulated and 1099 downregulated genes in KMS28PE NAT10-OE cells compared with KMS28PE WT cells (Fig. 3A). The intersection of all up-regulated 67 genes or down-regulated 132 genes in the two pairs of cells was shown in the Venn diagrams (Fig. 3B). Further Gene ontology (GO) analysis presented that NAT10 was highly associated with cell proliferation and cell cycle signaling pathway (GSE155417, Fig. 3C), which was consistent with the data in Fig. 2. Next, we adopted flow cytometry to detect cell cycle distribution. An increase in G2/M phase fraction was observed with NAT10 overexpression (Fig. 3D) and correspondingly a decrease of cellular G2/M phase fraction was presented with siNAT10 (Fig. 3E) in MM cells. CDK4/6 are key initiators of the G1-to-S phase transition, while inhibition of CDK4/6 leads to G1 arrest of the cell cycle26,27. It was reported that inhibition of NAT10 induced cell cycle arrest in G1 phase and downregulated CDK4 expression28. Therefore, we tested whether NAT10 could affect the expression of CDK4 and CDK6 in MM cells. The results showed that the expressions of CDK4 and CDK6 were upregulated by increased NAT10 (Fig. 3F, Supporting Information Fig. S4A and S4B) and decreased by siNAT10 (Fig. 3G, Fig. S4C and S4D) at protein level. Additionally, siNAT10 upregulated the expression of cleaved-PARP and cleaved-caspase 3 (Fig. 3H, Fig. S4E and S4F). In summary, NAT10 regulates cell cycle distribution and promotes cellular proliferation in MM.
Figure 3.
NAT10 regulates cycle distribution and promotes cellular proliferation in MM cells. (A) Volcano plots showed genes with upregulated (red) and downregulated (blue) expression upon NAT10 overexpression. (B) Venn diagram of two pairs of MM cells with common upregulated or downregulated genes. (C) Pathway enrichment analysis of RNA-seq data revealed that NAT10 was correlated to cell proliferation and cell cycle. (D, E) Flow cytometry analysis displayed that NAT10 overexpression increased G2/M phase fraction (D) and siNAT10 decreased G2/M phase fraction (E) in MM cells (n = 3). (F, G) Western blotting assays indicated that NAT10 upregulated CDK4 and CDK6 expression (F) and siNAT10 decreased CDK4 and CDK6 expression (G) in MM cells. (H) Western blotting analysis confirmed that siNAT10 promoted apoptotic protein expression. All data are displayed as mean ± SD; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.4. NAT10 acetylates mRNA to modulate progression of MM cells
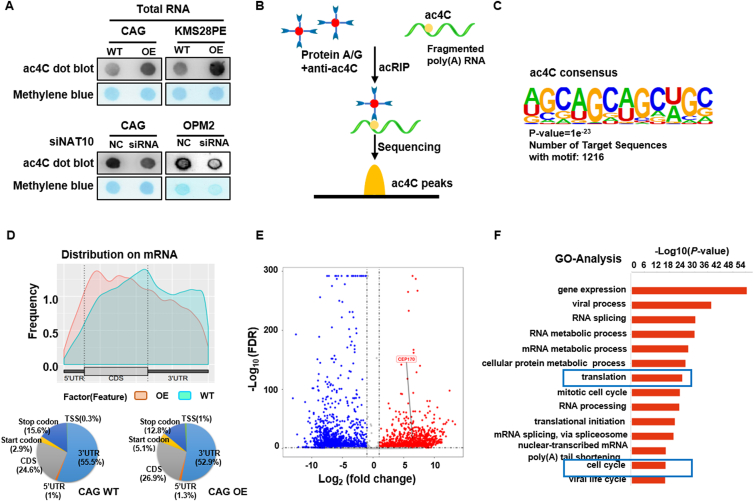
Since NAT10 is characterized as acetyltransferase, we continued to investigate whether NAT10 promoted MM cell proliferation depending on the acetyltransferase catalytic function. With the evidence showing that NAT10 acetylates mRNA6, we detected mRNA acetylation level in MM cells via dot blot assay. The results indicated that increased ac4C was in accordance with overexpressed NAT10, while silencing NAT10 expression resulted in decreased ac4C acetylation in MM cells (Fig. 4A). Therefore, it might be concluded that NAT10 enhanced mRNA acetylation in MM cells. As to determine the NAT10 targets of ac4C, we performed the acRIP-seq assay (Fig. 4B) in CAG WT and CAG NAT10-OE cells to assess the distribution and switch of ac4C in the transcriptome. Typical CXX motif of ac4C peaks is shown in Fig. 4C, indicating that the quality of acRIP-seq assay was guaranteed. Interestingly, ac4C peaks mainly appeared within CDS and 3′ untranslated region (3′UTR) in MM cells (Fig. 4D), suggesting that NAT10 might modulate the expression of mRNA directly. Volcano plot illustrated the genes of significant changes with upregulated (red) and downregulated (blue) mRNA acetylation under the condition of NAT10 overexpression (Fig. 4E). GO-analysis revealed that mRNA acetylation was associated with gene expression, translation, cell cycle and so on. (Fig. 4F). Here, we infer that NAT10 accelerates MM cells progression via catalyzing mRNA acetylation within CDS and promoting translation.
Figure 4.
NAT10 acetylates mRNA to regulate progression of MM cells. (A) Dot blot assay showed that increased NAT10 led to higher mRNA acetylation level and conversed result was observed in siNAT10 MM cells. (B) Schematic of acRIP-seq. (C) Typical CXX motif of ac4C peaks. (D) ac4C peaks mainly occurred within CDS and 3′UTR in MM cells. (E) Volcano plot described genes with upregulated (red) and downregulated (blue) mRNA acetylation levels upon NAT10 overexpression. (F) GO analysis of acRIP-seq. All data are displayed as mean ± SD; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.5. NAT10 acetylates CEP170 mRNA to enhance translation efficiency
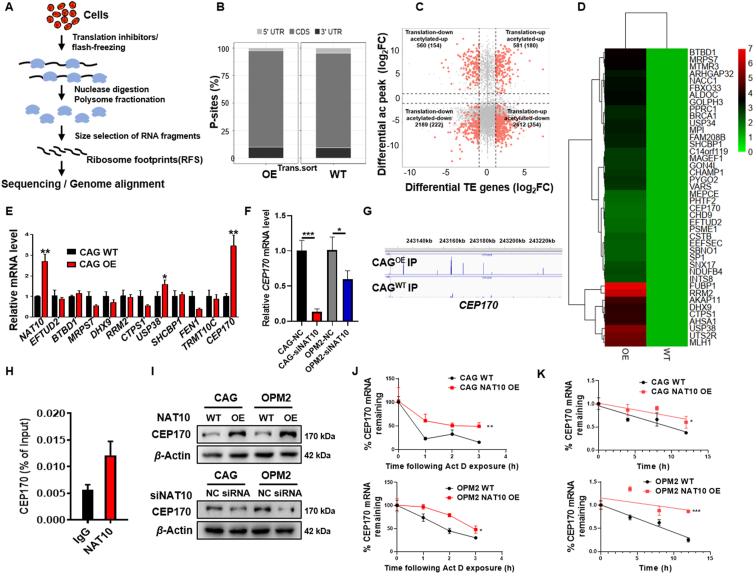
The decay of mRNA and translation are intricately associated with each other. For instance, the decrease in mRNA stability is manifested as the decrease in translation, while the decrease in translation weakens the stability of mRNA29. In this study, we focused on the signaling pathway of NAT10-regulated mRNA translation. To further explore the relationship between mRNA acetylation and translation, Ribo-seq (Fig. 5A), a translationomics research technology was utilized to detect the enhanced translational gene by NAT10-mediated mRNA acetylation in MM cells. According to the alignment position of ribosome footprints (RFs) on the genome, RFs were divided into four categories: CDS, 5′UTR, 3′UTR, Others. Generally, RFs were mostly distributed in the CDS area, but less in the UTR area (Fig. 5B). To screen the translation level of genes that undergo mRNA acetylation, we combined analyses of acRIP-seq and Ribo-seq, and the above four groups were divided based on mRNA acetylation level and translation level (GSE155917; Fig. 5C). Next, acRIP-seq and Ribo-seq analyses showed that 180 genes (45 genes in CDS region) were elevated at both mRNA acetylation and translation level in NAT10-OE cells compared with WT cells (Fig. 5D).
Figure 5.
NAT10 acetylates CEP170 mRNA to enhance translation efficiency. (A) Schematic of Ribo-seq. (B) RFs were mostly distributed in the CDS area. (C) Quadrant diagram for combined analysis of acRIP-seq and Ribo-seq depicted the levels of mRNA acetylation and translation. (D) Hot map of genes with co-upregulated acetylation and translation efficiency in NAT10-OE cells. (E) qPCR tested the expression of the candidate genes in CAG WT and NAT10-OE cells (n = 3). (F) CEP170 mRNA level was downregulated in siNAT10 cells. (G) ac4C peaks of CEP170 in CAG WT and NAT10-OE IP samples (n = 3). (H) NAT10 immunoprecipitation followed by RT-qPCR disclosed that CEP170 interacted with NAT10 (n = 3). (I) CEP170 was positively correlated with NAT10 at protein level in MM cells. (J) RT-qPCR was performed to detect CEP170 mRNA stability in CAG and OPM2 cells with the addition of actinomycin D (5 μg/mL) (n = 3). (K) BRIC RT-qPCR was conducted to measure the half-life of CEP170 mRNA in CAG and OPM2 cells (n = 3). All data are displayed as mean ± SD; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
To further identify the downstream factors of NAT10, RT-PCR was performed to validate the upregulated candidate genes in both of the acRIP-seq and Ribo-seq analyses at the levels of acetylation and translation. CEP170 was increased evidently in NAT10-OE cells compared with WT cells (Fig. 5E) and decreased after NAT10 was interfered by siRNA in MM cells (Fig. 5F). In addition, ac4C peaks of CEP170 in group of NAT10-OE IP demonstrated significant increased enrichment compared with WT IP (Fig. 5G), indicating that NAT10 promoted CEP170 mRNA acetylation. Subsequently, RIP followed by RT-qPCR assay confirmed that NAT10 bound with CEP170 mRNA (Fig. 5H). Western blotting analysis showed that CEP170 expression was increased upon NAT10 overexpression and reduced by NAT10 siRNA at protein level (Fig. 5I, Supporting Information Fig. S5A and S5B). RNA decay assay presented a relatively higher stability of CEP170 transcripts in NAT10-OE cells compared with WT cells (Fig. 5J). BRIC RT-qPCR indicated that NAT10 significantly prolonged the half-life of CEP170 mRNA (Fig. 5K). Above data suggest that CEP170 may serve as an important target of NAT10, which upregulates CEP170 mRNA acetylation to stabilize CEP170 and enhances its translation efficiency.
3.6. NAT10 promotes chromosomal instability (CIN) to accelerate MM progression by interacting with CEP170
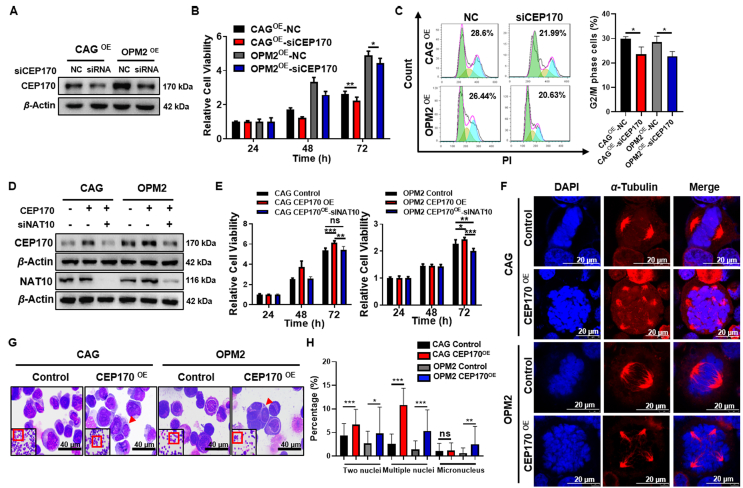
Even though we have identified that CEP170 is a downstream of NAT10, there is still no sufficient evidence showing that NAT10 influences MM cell proliferation via regulating CEP170. Therefore, we interfered CEP170 in NAT10-OE cells (Fig. 6A and Supporting Information Fig. S6) and overexpressed CEP170 in siNAT10 cells (Supporting Information Fig. S7A). MTT assays indicated that silencing CEP170 with siRNA significantly suppressed cellular proliferation in NAT10-OE cells (Fig. 6B) and CEP170 promoted cellular proliferation in siNAT10 cells (Fig. S7B). Consistently, inhibition of CEP170 expression decreased G2/M phase fraction in NAT10-OE cells (Fig. 6C). To further validate the function of CEP170 in MM cells, we overexpressed CEP170 in CAG and OPM2 cells and interfered NAT10 expression with siRNA in CEP170-OE cells, as indicated by Western blotting (Fig. 6D). MTT assay results showed that CEP170 indeed promoted MM cell proliferation and silencing NAT10 with siRNA inhibited this effect in CEP170-OE cells (Fig. 6E). CEP170 is a centrosome protein as a marker for mature centrioles that is responsible for centrosome microtubule anchoring30,31. Abnormal centrosome amplification leads to CIN32,33. Our previous work identified that CEP170 significantly increased chromosomal plate width and decreased mitotic bipolar spindle length, suggesting that CEP170 was associated with MM CIN34. Then, we conducted IF staining for α-tubulin and DAPI and found that elevated CEP170 compromised the integrity of the centrosome and bipolar shape of the spindle (Fig. 6F). Giemsa staining demonstrated that overexpression of CEP170 profoundly promoted an increase in the number of multiple nuclear cells as well as the separation error rate in MM cells (Fig. 6G and H). Here, we propose that NAT10 promotes CIN to accelerate MM progression by interacting with CEP170.
Figure 6.
NAT10 promotes CIN to accelerate MM progression by interacting with CEP170. (A) Validation of CEP170 interference (siCEP170) relative to negative control (NC). (B) MTT assay showed that siCEP170 impeded cell growth in NAT10-OE cells (n = 6). (C) Flow cytometry analysis indicated siCEP170 decreased G2/M phase fraction in NAT10-OE cells (n = 3). (D) WB analysis of CEP170-OE and CEP170-OE treated with siNAT10 cells compared with vehicle-transfected control cells. (E) Elevated CEP170 facilitated cell proliferation and siNAT10 attenuated this effect in CAG and OPM2 cells (n = 6). (F) Increased the separation error rate in CEP170-OE cells compared with controls, as demonstrated by IF staining for α-tubulin and DAPI. (G, H) Giemsa staining revealed that elevated CEP170 increased numbers of multi-nuclear cells in CAG and OPM2 cells (n = 40). All data are displayed as mean ± SD; ns P > 0.05, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.7. Remodelin impedes MM cell growth in vitro and prolongs the survival of 5TMM3VT mice in vivo
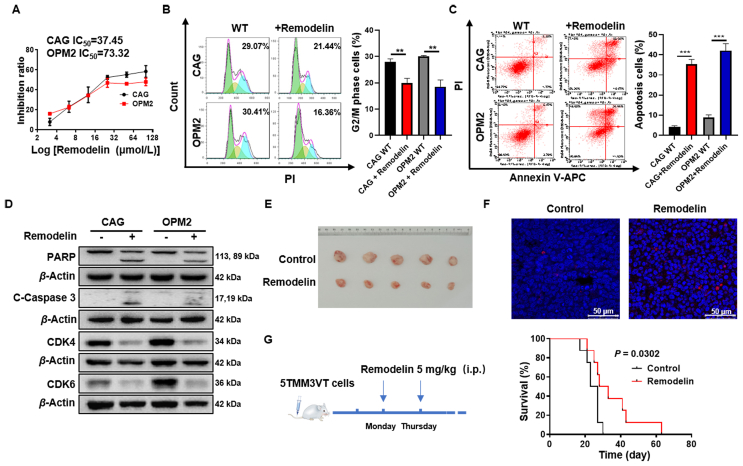
We further investigated if NAT10 could be a potent therapeutic target in MM by importing remodelin, a selective inhibitor of NAT10. Remodelin can correct nuclear architecture and attenuate senescence, which is lethal to ameliorat laminopathies35. We employed remodelin to evaluate the effect of NAT10 inhibition on MM cell growth, apoptosis and cell cycle. MTT assay showed that remodelin suppressed MM cell growth apparently compared with the untreated WT cells (Fig. 7A). Flow cytometry analysis indicated that remodelin strongly induced cell cycle arrest (Fig. 7B) and cell apoptosis (Fig. 7C). In addition, inhibition of NAT10 by remodelin increased the expression of cleaved-PARP and cleaved-caspase 3, and decreased the expression of CDK4 and CDK6 in MM cells (Fig. 7D, Supporting Information Fig. S8A–S8D). To further validate the effect of remodelin inhibiting NAT10 in vivo, we used the xenograft model of MM. It was showed that remodelin significantly inhibited the growth of xenograft tumors (Fig. 7E, Supporting Information Fig. S9A–S9C). Moreover, remodelin increased the expressions of cleaved-PARP and cleaved-caspase 3, and decreased the expressions of CDK4 and CDK6 in vivo (Fig. S9D). TUNEL assays also confirmed that remodelin induced cell apoptosis in xenograft tumors (Fig. 7F). In addition, 5TMM3VT mouse model presented that remodelin significantly extended the survival period of myeloma mice (Fig. 7G) relative to the untreated control mice in vivo. All the data above demonstrate that targeting NAT10 by remodelin can retard the development of MM in vitro and in vivo, indicating that NAT10 acts as a potential therapeutic target in MM.
Figure 7.
Remodelin impedes MM cell growth in vitro and prolongs the survival of 5TMM3VT mice in vivo. (A) Inhibition ratio of remodelin in MM cells (n = 4). (B) Remodelin induced cell cycle arrest (n = 3). (C) Flow cytometry analysis indicated that remodelin induced MM cell apoptosis (n = 3). (D) Western blotting analysis showed that remodelin promoted apoptotic protein expression and reduced CDK4 and CDK6 expression. (E) Photographic images of harvested xenograft tumors (n = 5). (F) Representative micrographs of TUNEL assay in tissues from xenograft mice treated with remodelin. (G) Remodelin extended the survival period of myeloma mice in vivo (n = 8). All data are displayed as mean ± SD; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
4. Discussion
Although the development of novel proteasome inhibitors and other treatment strategies have greatly improved the survival of MM patients, the acquired drug resistance and malignant proliferation eventually lead to relapse and poor outcome36. Our group is committed to focus on discovery and identification of novel MM therapeutic targets and developing corresponding target-therapies. In this study, we investigated a novel target NAT10 in MM, which could acetylate CEP170 mRNA to enhance the translation of CEP170 in regulating MM cell growth and survival.
NAT10 functions as an oncogene in human cancers and participates in cell proliferation and migration9,10,12, 13, 14. Originally, many studies of NAT10 focused on the protein-acetyltransferase activity. For instance, NAT10 acetylates MORC2 at K767 to regulate DNA damage-induced G2 checkpoint mediated by MORC2 in breast cancer37. NAT10 activates p53 via acetylating p53 at K120 and counteracting MDM2 action in colorectal cancer16. Recently, NAT10 is reported to acetylate mRNA6, while ac4C is the first acetylation event in mRNA, all of which are catalyzed by NAT10 or its homologs38. The ac4C is closely related to several human disease, as the ac4C content in the urine of patients is significantly higher than that of healthy people, including gestational diabetes39, interstitial cystitis40 and cancers41, 42, 43. Especially, ac4C is of great importance in diagnosis and treatment of cancers41, 42, 43. Analysis of ac4C function includes promoting protein translation, affecting RNA stability and alternative splicing, and regulating gene expression6,44,45. The ac4C peaks are enriched in the third codon encoding amino acid to improve the efficiency and accuracy of mRNA translation in human HeLa cells6,45. The modification of ac4C is involved in regulating gene expression via increasing stability and translation, possibly through its impact on mRNA decoding efficiency6. Additionally, NAT10 acetylates multiple cytidines on HIV-1 RNAs to ac4C and depletion of ac4C by drug treatment or mutagenesis decreases the stability of HIV-1 transcripts leading to the reduction of viral replication46. However, the role of ac4C modification and the function of NAT10 in either protein-acetyltransferase activity or acetylation of mRNA in MM warrant further research.
In present study, we first demonstrated that NAT10 expression was significantly correlated with poor survival of MM patients and increased NAT10 promoted MM growth in vitro and in vivo. Considering the crucial function of NAT10 in acetylating mRNA, we examined the mRNA-acetyltransferase activity of NAT10 in MM cells by using advanced high-throughput RNA sequencing techniques. The acRIP-seq was performed to delineate detailed mechanism on how NAT10 promoted MM cell proliferation. We found that ac4C regulated by NAT10 was associated with translation. To deeply explore the influence of ac4C on mRNA translation and the key downstream targets of NAT10, we combined the Ribo-seq data with acRIP-seq data to screen the potential targets with upregulated ac4C enrichment and translation efficiency. Finally, CEP170 was identified as the most significant downstream target of NAT10, and following studies confirmed that CEP170 expression was positively regulated by NAT10.
CEP170, localizing to centrosomes as well as spindle microtubules, is involved in microtubule organization and microtubule assembly47,48. CEP170 serves as a marker for centriole maturation during the cell cycle and contributes to distinguish distinct mechanisms causing centrosome amplification31, which is modulated by TBK1 to regulate microtubule dynamics and mitotic progression in cancer cells49. Centrosome abnormalities result in mitotic spindle defects to cause the formation of aneuploidy, which is the characteristic manifestation of CIN50,51. CIN is an important factor in tumor development52,53. As a centrosome protein, CEP170 abnormality contributes to CIN and then facilitates the occurrence and development of cancer34. Notably, NAT10 is associated with nuclear membrane and plays a vital role in mitosis54,55. Thus, we inferred that NAT10 might regulate CEP170 during cell mitosis to influence MM progression. Our data show that NAT10 directly bound with CEP170 mRNA and promoted CEP170 mRNA acetylation in MM cells. We also found that reducing CEP170 expression significantly attenuated the cellular growth acceleration and G2/M phase fraction caused by elevated NAT10. These results suggest that NAT10 acetylated CEP170 mRNA to enhance CEP170 translation efficiency and then promoted malignant progression of MM.
To validate that NAT10 might be targetable in MM, we used a selective inhibitor of NAT10 remodelin in this study. Remodelin has been reported to ameliorate Hutchinson–Gilford progeria syndrome (HGPS) cellular defects by inhibiting NAT1056 and restore nuclear shape of HGPS-derived patient cells via microtubule reorganization35. Remodelin inhibits cell proliferation and migration, and induces cell cycle arrest or apoptosis in various cancer cells12,13,15,57. In present study, we detected the effect of remodelin on MM in vitro and in vivo. Our data show that remodelin suppressed MM cell proliferation, induced cell cycle arrest and apoptosis in vitro and improved the survival of MM mice in vivo. Intriguingly, NAT10 is the only identified lysine acetyltransferase (KAT), and remodelin is the most potent and stable analog of KAT inhibitor 4-(4-chlorophenyl)-2-(2-cyclopentylidenehydrazinyl) thiazole to inhibit NAT10 KAT activity up to now35. Applying remodelin as the core for discovery or synthetization of more effective compounds to inhibit the activity of NAT10 is worthy of further exploration.
5. Conclusions
Our work demonstrates the carcinogenesis role of NAT10 in promoting MM malignancy and dug up a novel mechanism of NAT10 acetylating CEP170 mRNA to enhance CEP170 translation efficiency in MM. It may be concluded that NAT10 is a promising prognosis marker and potential therapeutic target for MM.
Acknowledgments
This work was supported by National Key R&D Program of China (2020YFA0509400) (to Ye Yang); National Natural Science Foundation of China 81970196 (to Chunyan Gu) and 82073885 (to Ye Yang); Natural Science Foundation of Jiangsu Province (China) BK20200097 (to Chunyan Gu); National Natural Science Foundation of China 82073888 (to Hongbo Wang); the Science and Technology Support Program for Youth Innovation in Universities of Shandong (China) (2019KJM009) (to Hongbo Wang); Bohai rim Advanced Research Institute for Drug Discovery (China) (LX211011) (to Hongbo Wang); Jiangsu Postgraduate Research and Practice Innovation Program (China) KYCX21_1769 (to Rongfang Wei).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.01.015.
Contributor Information
Chunyan Gu, Email: guchunyan@njucm.edu.cn.
Hongbo Wang, Email: hongbowangyt@gmail.com.
Ye Yang, Email: yangye876@sina.com.
Author contributions
Ye Yang, Chunyan Gu and Hongbo Wang designed and conceived the experiments. Rongfang Wei and Xing Cui developed methodology and conducted most of the experiments. Jie Min analyzed and processed the sequencing data. Zigen Lin and Yanyan Zhou developed all animal experiments. Mengjie Guo, Xiaojuan An and Hao Liu acquired the data. Rongfang Wei drafted the manuscript. Chunyan Gu and Ye Yang reviewed and edited the manuscript. Siegfried Janz and Hongbo Wang provided technical or material support.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Gu C., Lu T., Wang W., Shao M., Wei R., Guo M., et al. RFWD2 induces cellular proliferation and selective proteasome inhibitor resistance by mediating P27 ubiquitination in multiple myeloma. Leukemia. 2021;35:1803–1807. doi: 10.1038/s41375-020-01033-z. [DOI] [PubMed] [Google Scholar]
- 2.Gao M., Bai H., Jethava Y., Wu Y., Zhu Y., Yang Y., et al. Identification and characterization of tumor-initiating cells in multiple myeloma. J Natl Cancer Inst. 2020;112:507–515. doi: 10.1093/jnci/djz159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caprio C., Sacco A., Giustini V., Roccaro A.M. Epigenetic aberrations in multiple myeloma. Cancers (Basel) 2020;12:2996. doi: 10.3390/cancers12102996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sas-Chen A., Thomas J.M., Matzov D., Taoka M., Nance K.D., Nir R., et al. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature. 2020;583:638–643. doi: 10.1038/s41586-020-2418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van de Donk N., Pawlyn C., Yong K.L. Multiple myeloma. Lancet. 2021;397:410–427. doi: 10.1016/S0140-6736(21)00135-5. [DOI] [PubMed] [Google Scholar]
- 6.Arango D., Sturgill D., Alhusaini N., Dillman A.A., Sweet T.J., Hanson G., et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018;175:1872–1886. doi: 10.1016/j.cell.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lv J., Liu H., Wang Q., Tang Z., Hou L., Zhang B. Molecular cloning of a novel human gene encoding histone acetyltransferase-like protein involved in transcriptional activation of hTERT. Biochem Biophys Res Commun. 2003;311:506–513. doi: 10.1016/j.bbrc.2003.09.235. [DOI] [PubMed] [Google Scholar]
- 8.Cai S., Liu X., Zhang C., Xing B., Du X. Autoacetylation of NAT10 is critical for its function in rRNA transcription activation. Biochem Biophys Res Commun. 2017;483:624–629. doi: 10.1016/j.bbrc.2016.12.092. [DOI] [PubMed] [Google Scholar]
- 9.Tan T.Z., Miow Q.H., Huang R.Y., Wong M.K., Ye J., Lau J.A., et al. Functional genomics identifies five distinct molecular subtypes with clinical relevance and pathways for growth control in epithelial ovarian cancer. EMBO Mol Med. 2013;5:1051–1066. doi: 10.1002/emmm.201201823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan Y., Zheng J., Liu X., Lu M., Zhang C., Xing B., et al. Loss of nucleolar localization of NAT10 promotes cell migration and invasion in hepatocellular carcinoma. Biochem Biophys Res Commun. 2018;499:1032–1038. doi: 10.1016/j.bbrc.2018.04.047. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X., Liu J., Yan S., Huang K., Bai Y., Zheng S. High expression of N-acetyltransferase 10: a novel independent prognostic marker of worse outcome in patients with hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:14765–14771. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X., Chen J., Jiang S., He S., Bai Y., Zhu L., et al. N-Acetyltransferase 10 enhances doxorubicin resistance in human hepatocellular carcinoma cell lines by promoting the epithelial-to-mesenchymal transition. Oxid Med Cell Longev. 2019;2019:7561879. doi: 10.1155/2019/7561879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma R., Chen J., Jiang S., Lin S., Zhang X., Liang X. Up regulation of NAT10 promotes metastasis of hepatocellular carcinoma cells through epithelial-to-mesenchymal transition. Am J Transl Res. 2016;8:4215–4223. [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q., Liu X., Jin K., Lu M., Zhang C., Du X., et al. NAT10 is upregulated in hepatocellular carcinoma and enhances mutant p53 activity. BMC Cancer. 2017;17:605. doi: 10.1186/s12885-017-3570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J., Zhu H., Wu J., Chen W., Guan X. Inhibition of N-acetyltransferase 10 using remodelin attenuates doxorubicin resistance by reversing the epithelial–mesenchymal transition in breast cancer. Am J Transl Res. 2018;10:256–264. [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X., Tan Y., Zhang C., Zhang Y., Zhang L., Ren P., et al. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep. 2016;17:349–366. doi: 10.15252/embr.201540505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H., Hou W., Wang H.L., Liu H.J., Jia X.Y., Zheng X.Z., et al. GSK-3beta-regulated N-acetyltransferase 10 is involved in colorectal cancer invasion. Clin Cancer Res. 2014;20:4717–4729. doi: 10.1158/1078-0432.CCR-13-3477. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z., Liu X., Li Y., Ren P., Zhang C., Wang L., et al. miR-6716-5p promotes metastasis of colorectal cancer through downregulating NAT10 expression. Cancer Manag Res. 2019;11:5317–5332. doi: 10.2147/CMAR.S197733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang P., Hu R., Liu Z., Miao M., Jiang H., Li C. NAT10 upregulation indicates a poor prognosis in acute myeloid leukemia. Curr Probl Cancer. 2020;44:100491. doi: 10.1016/j.currproblcancer.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Wang T., Zou Y., Huang N., Teng J., Chen J. CCDC84 acetylation oscillation regulates centrosome duplication by modulating HsSAS-6 degradation. Cell Rep. 2019;29:2078–2091. doi: 10.1016/j.celrep.2019.10.028. [DOI] [PubMed] [Google Scholar]
- 21.Gu C., Yang Y., Sompallae R., Xu H., Tompkins V.S., Holman C., et al. FOXM1 is a therapeutic target for high-risk multiple myeloma. Leukemia. 2016;30:873–882. doi: 10.1038/leu.2015.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broyl A., Hose D., Lokhorst H., de Knegt Y., Peeters J., Jauch A., et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010;116:2543–2553. doi: 10.1182/blood-2009-12-261032. [DOI] [PubMed] [Google Scholar]
- 23.Li F., Liu Z., Sun H., Li C., Wang W., Ye L., et al. PCC0208017, a novel small-molecule inhibitor of MARK3/MARK4, suppresses glioma progression in vitro and in vivo. Acta Pharm Sin B. 2020;10:289–300. doi: 10.1016/j.apsb.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinclair W.R., Arango D., Shrimp J.H., Zengeya T.T., Thomas J.M., Montgomery D.C., et al. Profiling cytidine acetylation with specific affinity and reactivity. ACS Chem Biol. 2017;12:2922–2926. doi: 10.1021/acschembio.7b00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imamachi N., Tani H., Mizutani R., Imamura K., Irie T., Suzuki Y., et al. BRIC-seq: a genome-wide approach for determining RNA stability in mammalian cells. Methods. 2014;67:55–63. doi: 10.1016/j.ymeth.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 26.Li K., You J., Wu Q., Meng W., He Q., Yang B., et al. Cyclin-dependent kinases-based synthetic lethality: evidence, concept, and strategy. Acta Pharm Sin B. 2021;11:2738–2748. doi: 10.1016/j.apsb.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan K., Wang X., Dong H., Min W., Hao H., Yang P. Selective inhibition of CDK4/6: a safe and effective strategy for developing anticancer drugs. Acta Pharm Sin B. 2021;11:30–54. doi: 10.1016/j.apsb.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zi J., Han Q., Gu S., McGrath M., Kane S., Song C., et al. Targeting NAT10 induces apoptosis associated with enhancing endoplasmic reticulum stress in acute myeloid leukemia cells. Front Oncol. 2020;10:598107. doi: 10.3389/fonc.2020.598107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanson G., Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol. 2018;19:20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W., Yang S.L., Yang M., Herrlinger S., Shao Q., Collar J.L., et al. Modeling microcephaly with cerebral organoids reveals a WDR62–CEP170–KIF2A pathway promoting cilium disassembly in neural progenitors. Nat Commun. 2019;10:2612. doi: 10.1038/s41467-019-10497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guarguaglini G., Duncan P.I., Stierhof Y.D., Holmström T., Duensing S., Nigg E.A. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005;16:1095–1107. doi: 10.1091/mbc.E04-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Karim E.A., Hagos E.G., Ghaleb A.M., Yu B., Yang V.W. Krüppel-like factor 4 regulates genetic stability in mouse embryonic fibroblasts. Mol Cancer. 2013;12:89. doi: 10.1186/1476-4598-12-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Croessmann S., Wong H.Y., Zabransky D.J., Chu D., Mendonca J., Sharma A., et al. NDRG1 links p53 with proliferation-mediated centrosome homeostasis and genome stability. Proc Natl Acad Sci U S A. 2015;112:11583–11588. doi: 10.1073/pnas.1503683112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu C., Wang W., Tang X., Xu T., Zhang Y., Guo M., et al. CHEK1 and circCHEK1_246aa evoke chromosomal instability and induce bone lesion formation in multiple myeloma. Mol Cancer. 2021;20:84. doi: 10.1186/s12943-021-01380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larrieu D., Britton S., Demir M., Rodriguez R., Jackson S.P. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science. 2014;344:527–532. doi: 10.1126/science.1252651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda T., Tsubaki M., Kino T., Kawamura A., Isoyama S., Itoh T., et al. Mangiferin enhances the sensitivity of human multiple myeloma cells to anticancer drugs through suppression of the nuclear factor kappaB pathway. Int J Oncol. 2016;48:2704–2712. doi: 10.3892/ijo.2016.3470. [DOI] [PubMed] [Google Scholar]
- 37.Liu H.Y., Liu Y.Y., Yang F., Zhang L., Zhang F.L., Hu X., et al. Acetylation of MORC2 by NAT10 regulates cell-cycle checkpoint control and resistance to DNA-damaging chemotherapy and radiotherapy in breast cancer. Nucleic Acids Res. 2020;48:3638–3656. doi: 10.1093/nar/gkaa130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma S., Langhendries J.L., Watzinger P., Kötter P., Entian K.D., Lafontaine D.L. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015;43:2242–2258. doi: 10.1093/nar/gkv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law K.P., Han T.L., Mao X., Zhang H. Tryptophan and purine metabolites are consistently upregulated in the urinary metabolome of patients diagnosed with gestational diabetes mellitus throughout pregnancy: a longitudinal metabolomics study of Chinese pregnant women part 2. Clin Chim Acta. 2017;468:126–139. doi: 10.1016/j.cca.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 40.Parsons C.L., Shaw T., Berecz Z., Su Y., Zupkas P., Argade S. Role of urinary cations in the aetiology of bladder symptoms and interstitial cystitis. BJU Int. 2014;114:286–293. doi: 10.1111/bju.12603. [DOI] [PubMed] [Google Scholar]
- 41.Szymańska E., Markuszewski M.J., Markuszewski M., Kaliszan R. Altered levels of nucleoside metabolite profiles in urogenital tract cancer measured by capillary electrophoresis. J Pharm Biomed Anal. 2010;53:1305–1312. doi: 10.1016/j.jpba.2010.07.031. [DOI] [PubMed] [Google Scholar]
- 42.Zhang T., Wu X., Ke C., Yin M., Li Z., Fan L., et al. Identification of potential biomarkers for ovarian cancer by urinary metabolomic profiling. J Proteome Res. 2013;12:505–512. doi: 10.1021/pr3009572. [DOI] [PubMed] [Google Scholar]
- 43.Li H., Qin Q., Shi X., He J., Xu G. Modified metabolites mapping by liquid chromatography-high resolution mass spectrometry using full scan/all ion fragmentation/neutral loss acquisition. J Chromatogr A. 2019;1583:80–87. doi: 10.1016/j.chroma.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 44.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 45.Dominissini D., Rechavi G. N4-acetylation of cytidine in mRNA by NAT10 regulates stability and translation. Cell. 2018;175:1725–1727. doi: 10.1016/j.cell.2018.11.037. [DOI] [PubMed] [Google Scholar]
- 46.Tsai K., Jaguva Vasudevan A.A., Martinez Campos C., Emery A., Swanstrom R., Cullen B.R. Acetylation of cytidine residues boosts HIV-1 gene expression by increasing viral RNA stability. Cell Host Microbe. 2020;28:306–312. doi: 10.1016/j.chom.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Welburn J.P., Cheeseman I.M. The microtubule-binding protein Cep170 promotes the targeting of the kinesin-13 depolymerase Kif2b to the mitotic spindle. Mol Biol Cell. 2012;23:4786–4795. doi: 10.1091/mbc.E12-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazo G., Soplop N., Wang W.J., Uryu K., Tsou M.F. Spatial control of primary ciliogenesis by subdistal appendages alters sensation-associated properties of cilia. Dev Cell. 2016;39:424–437. doi: 10.1016/j.devcel.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pillai S., Nguyen J., Johnson J., Haura E., Coppola D., Chellappan S. Tank binding kinase 1 is a centrosome-associated kinase necessary for microtubule dynamics and mitosis. Nat Commun. 2015;6:10072. doi: 10.1038/ncomms10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lingle W.L., Barrett S.L., Negron V.C., D’Assoro A.B., Boeneman K., Liu W., et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A. 2002;99:1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iftekhar A., Berger H., Bouznad N., Heuberger J., Boccellato F., Dobrindt U., et al. Genomic aberrations after short-term exposure to colibactin-producing E. coli transform primary colon epithelial cells. Nat Commun. 2021;12:1003. doi: 10.1038/s41467-021-21162-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akimova E., Gassner F.J., Schubert M., Rebhandl S., Arzt C., Rauscher S., et al. SAMHD1 restrains aberrant nucleotide insertions at repair junctions generated by DNA end joining. Nucleic Acids Res. 2021;49:2598–2608. doi: 10.1093/nar/gkab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pihan G.A., Wallace J., Zhou Y., Doxsey S.J. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63:1398–1404. [PubMed] [Google Scholar]
- 54.Chi Y.H., Haller K., Peloponese J.M., Jr., Jeang K.T. Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in de-condensation of mitotic chromosomes. J Biol Chem. 2007;282:27447–27458. doi: 10.1074/jbc.M703098200. [DOI] [PubMed] [Google Scholar]
- 55.Gassmann R., Henzing A.J., Earnshaw W.C. Novel components of human mitotic chromosomes identified by proteomic analysis of the chromosome scaffold fraction. Chromosoma. 2005;113:385–397. doi: 10.1007/s00412-004-0326-0. [DOI] [PubMed] [Google Scholar]
- 56.Balmus G., Larrieu D., Barros A.C., Collins C., Abrudan M., Demir M., et al. Targeting of NAT10 enhances healthspan in a mouse model of human accelerated aging syndrome. Nat Commun. 2018;9:1700. doi: 10.1038/s41467-018-03770-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oh T.I., Lee Y.M., Lim B.O., Lim J.H. Inhibition of NAT10 suppresses melanogenesis and melanoma growth by attenuating microphthalmia-associated transcription factor (MITF) expression. Int J Mol Sci. 2017;18:1924. doi: 10.3390/ijms18091924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.