Abstract
The Czc system of Alcaligenes eutrophus mediates resistance to cobalt, zinc, and cadmium through ion efflux catalyzed by the CzcCB2A cation-proton antiporter. DNA sequencing of the region upstream of the czcNICBADRS determinant located on megaplasmid pMOL30 revealed the 5′ end of czcN and a gene for a MgtC-like protein which is transcribed in the orientation opposite that of czc. Additional open reading frames upstream of czc had no homologs in the current databases. Using oligonucleotide-probed Northern blotting experiments, a 500-nucleotide czcN message and a 400-nucleotide czcI message were found, and the presence of 6,200-nucleotide czcCBA message (D. Van der Lelie et al., Mol. Microbiol. 23:493–503, 1997) was confirmed. Induction of czcN, czcI, czcCBA, and czcDRS followed a similar pattern: transcription was induced best by 300 μM zinc, less by 300 μM cobalt, and only slightly by 300 μM cadmium. Reverse transcription-PCR gave evidence for additional continuous transcription from czcN to czcC and from czcD to czcS, but not between czcA and czcD nor between czcS and a 131-amino-acid open reading frame following czcS. The CzcR putative response regulator was purified and shown to bind in the 5′ region of czcN. A reporter strain carrying a czcNIC-lacZ-czcBADRS determinant on plasmid pMOL30 was constructed, as were ΔczcR and ΔczcS mutants of this strain and of AE128(pMOL30) wild type. Experiments on (i) growth of these strains in liquid culture containing 5 mM Zn2+, (ii) induction of the β-galactosidase in the reporter strains by zinc, cobalt, and cadmium, and (iii) cDNA analysis of czcCBA mRNA synthesis under inducing and noninducing conditions showed that the CzcRS two-component regulatory system is involved in Czc regulation.
Alcaligenes eutrophus CH34 contains at least seven determinants encoding resistance to toxic heavy metals, located either on the bacterial chromosome or on one of the two indigenous plasmids, pMOL28 (180 kb [34]) and pMOL30 (238 kb [4, 10]). The czc determinant of plasmid pMOL30 mediates inducible resistance to Co2+, Zn2+, and Cd2+ in A. eutrophus (6, 17, 20). The products of the genes czcA, czcB, and czcC form a membrane-bound protein complex catalyzing an energy-dependent efflux of these three metal cations (22, 23). The mechanism of action of the CzcCB2A complex (26) is that of a proton-cation antiporter (18). The CzcCB2A complex is composed of the proton-cation antiporter CzcA located in the cytoplasmic membrane, the putative membrane fusion protein CzcB in the periplasm, and CzcC, which is probably attached to the outer membrane (18, 26). CzcC contains a leader sequence, and its preprotein is processed during transport across the cytoplasmic membrane (26).
Possible Czc regulatory genes are arranged in regions upstream and downstream of the structural genes czcCBA (Fig. 1). The downstream regulatory region contains the genes czcD, czcR, and czcS, with CzcS (histidine kinase) and CzcR (response regulator) forming a two-component regulatory system (21, 35). CzcD is a membrane-bound protein required to sense metals by an unknown mechanism (19). The czcS gene is followed by an open reading frame (ORF), ORF131, encoding a possible product of 131 amino acids (aa) (35). The upstream regulatory region (19) contains the genes czcN and czcI (3) and another ORF, ORF69a, with a possible product of 69 aa. The predicted products of both ORFs have no known function and no homologs in current databases; however, the upstream regulatory region is essential for czc expression (19).
FIG. 1.
The czc determinant and its transcripts. A physical map of the czcNICBADRS determinant, mgtC, and ORF131 is shown; differences in shading indicate the different genes. The czc genes and the two ORFs (shown above the line) are transcribed from left to right; mgtC (below the line) is transcribed from right to left. A scaled-up view of the czcN upstream region is shown at the upper left. K, E, and X indicate sites for the restriction endonucleases KpnI, EcoRI, and XhoI, respectively. An arrow in front of czcN points to the binding site of the CzcR response regulator; the three lines under czcN indicate positions of the DNA fragments a, b, and c used in gel retardation experiments with CzcR. The figure is drawn to the scale given in kilobase pairs; the left scale represents the 2.879-kb czcN upstream region (gpAJ001159), and the right scale represents the previously published 11.3-kb czc sequence (gbX98451 [35]). The open circles above the physical map are potential rho-independent terminators (stem-loops followed by U stretches) with free energies of −60 kJ/mol (czcIt), −97 kJ/mol (czcAt), and −66 kJ/mol (czcSt), respectively. Closed circles below the line are the 5′ ends of the czcI-, czcC-, and czcD-specific mRNAs (primer extension results). The bar labeled “T7 fragment” shows the size, position, and orientation compared to the T7 promoter (black arrowhead) of the czc region used for T7 expression. The four black horizontal arrows pointing from left to right mark positions of the four czc-specific mRNAs (oligonucleotide-probed Northern blots); the arrow for the czcDRS-specific message is dashed because the size of the transcript could not be measured. The lines with two arrowheads mark positions of RT-PCR-generated fragments; two dashed lines with crosses are in regions where no RT-PCR fragment could be obtained. The dumbbell below czcA shows the position of the 529-bp fragment resulting from the competitive (Comp.) RT-PCR experiments. Finally, the dashed arrows at the bottom indicate all hypothetical transcripts of czc which yield the mRNAs observed in the Northern and RT-PCR experiments: czcNICBA (from czcNp to czcAt), czcICBA (from czcIp to czcAt), czcCBA (from czcCp to czcAt), czcDRS (from czcDp to czcSt), czcNI (from czcNp to czcIt), and czcI (from czcIp to czcIt), with the four assumed promoters czcNp, czcIp, czcCp, and czcDp.
It has not been shown if the CzcR-CzcS two-component system is indeed required for czc regulation (35) or, due to its homology to copper regulators (21), possibly for regulation of an associated copper resistance. Transcription of the upstream regulatory region of czc has been shown (35). Since a double-stranded DNA fragment was used as a probe for the Northern-type RNA-DNA hybridization experiments in this study (35), it was unclear if an ORF previously designated czcR (referred to as old czcR in this report) and divergent to czcCBA (19) or czcI on the same strand as czcCBA (3) was transcribed. Moreover, the old czcR contains sequence errors which interrupt the reading frame (changes in X67305 as published in reference 19: insertion of an additional C upstream of T-495, CA-600 to AC-600, and deletion of G-778; corrected sequence gbX98451 published in reference 35). Therefore, transcription of the czc upstream regulatory region was investigated again, using oligonucleotides as strand-specific probes. Moreover, the published czcN gene sequence was not complete, and it was not clear if czcN is really part of czc. In this report, the transcriptional organization of czc and the function of the CzcRS two-component system are clarified.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Tris-buffered mineral salts medium (10) containing 2 g of sodium gluconate/liter was used to cultivate A. eutrophus AE128(pMOL30) and its plasmid-free derivative AE104 (10). Analytical grade salts of CdCl2 · H2O, ZnCl2, CoCl2 · 6H2O, and CuSO4 · 5H2O were used to prepare 1 M stock solutions, which were sterilized by filtration. Lead acetate was used similarly in a 0.1 M stock solution. Solid Tris-buffered medium contained 20 g of agar/liter. β-Galactosidase activity was determined in permeabilized cells as published previously (19), with 1 U defined as the activity forming 1 nmol of o-nitrophenol per min at 30°C.
Genetic techniques.
Standard molecular genetic techniques were used (17, 28). For conjugal gene transfer, overnight cultures of donor strain Escherichia coli S17/1 (30) and of the A. eutrophus recipient strains grown at 30°C in complex medium were mixed (1:1) and plated onto nutrient broth agar. After overnight growth, the bacteria were suspended in saline (9 g of NaCl/liter), diluted, and plated onto selective media as previously described (17). PCGENE was used as standard computer program for the analysis of DNA sequences obtained with an A.L.F. sequencer (Pharmacia, Uppsala, Sweden). Total RNA of A. eutrophus was isolated as published elsewhere (24, 35). The amount and quality of total RNA were determined spectrophotometrically at 260 and 280 nm.
For expression of the region upstream of czcCBA under control of the phage T7 promoter (Fig. 1), a 4-kb SacI (position 2259 in gbAJ001159)-PstI fragment containing czcNICB′ was cloned from pECD162 (19) into plasmid pT7-5 (32a). Expression in E. coli was done as published previously (16) and visualized by autoradiography.
Strain constructions.
In plasmid pMOL30-6 of A. eutrophus DN172, the ATG start codon of the ORF designated old czcR (19) was mutated to a TAG stop codon by PCR using an overlap extension method (11). The 1.6-kb fragment from positions 1 to 1595 (gbX98451 [Fig. 1, right scale]) was mutated. The 1.2-kb 5′ part of the DNA fragment was amplified from plasmid pECD277, which was constructed by cloning the 1.8-kb KpnI/PstI fragment of plasmid pECD177 (19) into pBluescript SK+ (Pharmacia). A primer corresponding to the DNA sequence of the KpnI site at the 5′ end of gbX98451 (Fig. 1, position 0) was used, and the primer additionally contained an XbaI site at the end to facilitate cloning (primer 641; 5′-AAA TCT AGA GGT ACC TGG GTG-3′ [XbaI site underlined]). The second primer for the amplification of the 1.2-kb DNA fragment corresponded to the DNA sequence of the mutated region (positions 1098 to 1100 of gbX98451 [Fig. 1, right scale]; primer 642; 5′-CTC GGA CGA TAG TTG AGT AT-3′ [mutated region underlined]). The 0.4-kb 3′ end of the 1.6-kb DNA fragment was similarly amplified with a primer corresponding to the mutated region (primer 643; 5′-ATA CTC AAC TAT CGT CCG AG-3′ [mutated region underlined]) and a primer corresponding to the region around bp 1595 (primer 640; AAA TCT AGA CCG CGG CAT TGA-3′ [XbaI site underlined]). The 1.2- and 0.4-kb PCR fragments were used as templates for another PCR with primers 640 and 641; the resulting mutated 1.6-kb PCR fragment was purified, digested with XbaI, cloned into pUC19 (37) to yield plasmid pECD425, and sequenced to verify the correct mutation.
The 1.6-kb XbaI fragment was cloned into the kanamycin-resistant suicide vector pLO2 (7), leading to plasmid pECD426. This plasmid was conjugated into A. eutrophus AE128(pMOL30), and kanamycin-resistant derivatives were selected. Since pLO2 derivatives do not replicate in A. eutrophus, kanamycin-resistant strains should contain a cointegrate between pMOL30 and pECD426 which was formed by homologous recombination of the 1.6-kb XbaI fragment with its counterpart on megaplasmid pMOL30. One kanamycin-resistant strain was selected and cultivated several times subsequently in liquid Tris-gluconate medium containing 1 mM Cu2+ and 0.1% (wt/vol) yeast extract to select for the presence of plasmid pMOL30 (copper resistance is not linked to czc in A. eutrophus). Sucrose-resistant and kanamycin-sensitive derivatives were isolated. These strains should have lost the pLO2 derivative (with a sacB gene mediating sucrose sensitivity) by a second recombination event; they should either be wild-type revertants or carry the desired mutation. Using primers 640 and 641, we amplified the 1.6-kb DNA fragment from those strains, and the resulting PCR fragments were used for a second PCR with primers 640 and 643, using an annealing temperature of 58°C. Due to the different annealing temperatures of primer 643 to wild-type and mutant DNA fragments, only mutant fragments yielded a 0.4-kb DNA fragment as a product, which was cloned into pUC19 (37) and sequenced for verification. Thus, strain DN172(pMOL30-6) was identified.
To generate A. eutrophus DN174(pMOL30-8) with a lacZ insertion between czcC and czcB, a 2.9-kb fragment containing czcC and the 5′ end of czcB was amplified from plasmid pECD110 (22), using primers with BamHI sites at their 5′ ends (primers 165 [5′-AAA AGG ATC CTT CTC CTG GTC ACA T-3′] and 166 [5′-AAA AGG ATC CTT CAA ACA TTG CTG T-3′]; BamHI sites underlined). The PCR fragment was cloned as a BamHI fragment in pUC9 (37), leading to plasmid pECD429. Using a Quick Change kit (Stratagene, Heidelberg, Germany), we inserted an XbaI site between czcC and czcB. A promoterless lacZ gene (25), amplified from transposon Tn5-B20 (31), was inserted into the XbaI site, and the resulting czcC-lacZ-czcB region was cloned into pLO2 (7), leading to plasmid pECD432. Plasmid pECD432 was conjugated into A. eutrophus DN172(pMOL30-6), and kanamycin-resistant derivatives forming blue colonies on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were selected. Again, after repeated cultivation in liquid Tris-gluconate medium containing copper and yeast extract, sucrose-resistant and kanamycin-sensitive colonies were isolated. The mutated region from those strains still able to form blue colonies on X-Gal was amplified again, using primers 165 and 166, and the PCR fragments were digested with several restriction endonucleases to verify the correct pattern. To construct strain DN175(pMOL30-9), a parallel approach was used to clone plasmid pECD432 into A. eutrophus AE128(pMOL30) wild type.
Construction of knockout mutants in czc genes.
To prevent polar effects mediated by deletions of single czc genes, the gene of interest was exchanged for a small ORF encoding a polypeptide of 20 aa. The first 9 aa coded by the small ORF were identical with the 9 amino-terminal aa of the product of the gene to be mutated, and the last 9 aa were identical with the last, carboxy-terminal amino acids of that gene. Positions 10 and 11 were E and L, coded by the hexanucleotide recognition sequence CAATTG of the restriction endonuclease MunI. Thus, the 500 bp upstream of the gene to be deleted were amplified by PCR, and this fragment ended with the 27-bp coding sequence for the first 9 aa of the gene, followed by a MunI hexanucleotide. Second, the 500 bp downstream of the gene were amplified by PCR, and this fragment started with the MunI recognition sequence and the last 27 bp of the gene. Both fragments were fused by MunI restriction and ligation, cloned, verified by DNA sequencing, and finally cloned into pLO2 (7). The resulting plasmid was used for mutation in A. eutrophus as described above. The mutant genotype was verified by PCR and DNA sequencing.
Oligonucleotide-probed Northern DNA-RNA hybridization.
Northern (RNA) blot analysis was performed by fractionation of RNA samples on a 1.5% agarose-formaldehyde gel, followed by transfer to a positively charged QIABRANE nylon filter (Qiagen, Hilden, Germany), using a pressure blot (Posi Blot; Stratagene). For quantitative analysis, the same amount of total RNA (40 μg) was loaded into each well of the same gel, and this was verified by ethidium bromide staining after electrophoresis. After prehybridization for 3 h at 42°C in hybridization buffer (5), the filters were hybridized for at least 14 h at 42°C in the same buffer. The filters were probed with oligonucleotides which were strand specific for czcN (5′-GAA TTC CGG GAA GGC GGA A-3′), czcI (5′-AGA TGG CAT ACC CCG CAG TC-3′), czcCBA (5′-ATG CTA CCG CCA GCC CGA-3′), or czcDRS (5′-GTC GTT GAG TGA CCT GCG CCC A-3′). The oligonucleotides were labeled with digoxigenin (DIG) by using terminal transferase and a Boehringer (Mannheim, Germany) DIG-oligonucleotide 3′-end labeling kit. After several washing steps, the filters were developed with a Boehringer DIG luminescence detection kit.
RT.
Cells of A. eutrophus AE128(pMOL30) were cultivated in Tris-gluconate medium in the presence of 300 μM Zn2+ to induce czc. Total RNA was isolated from those cells and digested with 1 U of RNase-free DNase I (Boehringer) per μg of RNA for 2 h at 37°C. Then 20 pmol of the 3′ antisense primer was mixed with 5 μg (reverse transcription [RT]-PCR) or 10 μg (primer extension) of total RNA in 12 μl of water, heated for 10 min at 70°C, and quickly cooled on ice. Buffer (4 μl; 250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2), 2 μl of 100 mM dithiothreitol, and 1 μl containing 10 mM each of the four deoxynucleostide triphosphates were added. After incubation at 50°C for 2 min, 200 U of Superscript II reverse transcriptase (Gibco BRL, Eggenstein, Germany) was added, and incubation was continued for 60 min at 50°C. The reaction was stopped by heating at 95°C for 5 min.
Primer extension experiments.
Primer extension analysis was performed by a modification of a standard protocol (28) using fluorescein-labeled oligonucleotides and an automated A.L.F. DNA sequencer (Pharmacia) as described previously (36). The fluorescein-labeled 3′ antisense primers (for czcN, 5′-CCC CCA ACA TCC CCA GCG TCA-3′ and 5′-TTA CGA CAA CCC GCC AAA CGC-3′; for ORF69a, 5′-CGC CTG CTC GTT TTG GGG TGC-3′; for czcI, 5′-GTG CCC AAG GTG CCA AGT G-3′; for czcC, 5′-ATT GCT TCC TGC CGC CA-3′; for czcD, 5′-CAC TTC GGC AAT CAG GA-3′; for czcR, 5′-GGT AAT CGT CTG CCC CCA-3′; and for czcS, 5′-CGA GCG AGT AGT AAG CA-3′) were complementary to the corresponding gene regions. After phenol-chloroform extraction, the cDNA was precipitated with ethanol, vacuum dried, and suspended in 4 μl of H2O and 4 μl of A.L.F. stop solution (Pharmacia). Following heat denaturation, the sample was loaded on a 7% polyacrylamide sequencing gel. In parallel, a sequencing reaction was performed with the same fluorescein-labeled primer and a DNA fragment containing the gene region of interest. The transcription start site was determined by comparison of the retention time of the primer extension reaction with that of the sequencing reaction. Primer extension experiments with total RNA from plasmid-free AE104 cells and a control reaction carried out without reverse transcriptase were used as negative controls.
RT-PCR experiments.
RT was carried out with the 3′ antisense primers 5′-AGA TGG CAT ACC CCG CAG TC-3′ for czcI, 5′-AAA TCT AGA CCG CGG CAT TGA-3′ for czcC, 5′-CAC TTC GGC AAT CAG GA-3′ for czcD, 5′-GGT AAT CGT CTG CCC CCA-3′ for czcR, 5′-TGC CCG TGC GAA ACA AAA G-3′ for czcS, and 5′-GCT TCG CCA TCG CCT GC-3′ for ORF131. PCR amplification was performed with 0.1 volume of the RT reaction mixture in a mixture containing 50 mM Tris-HCl (pH 9.0), 20 mM ammonium sulfate, 20 pmol of 5′ sense primer (for czcN, 5′-AAA TCT AGA GGT ACC TGG GTG-3′; for ORF69a, 5′-GTG GTC GTG GAG TCG TTT GAT-3′; for czcI, 5′-GAC TGC GGG GTA TGC CAT CT-3′; for czcA, 5′-TAT CGA CTT GCT CAC CGC A-3′; czcD, 5′-CAG GAG GAA ACT AAG GTG CG-3′; for czcR, 5′-AGG TCT GGG GCG TCA-3′; and for czcS, 5′-AGC GTC GTA ATC AGG GTA3′), 0.1 mM (each) deoxynucleoside triphosphate, 1.5 mM MgCl2, and 1 U of Tfl polymerase (Biozym; Hessisch Oldendorf) in a final volume of 100 μl. The amplification profile used consisted of 35 cycles of denaturation at 95°C for 30 s, annealing at 50 to 70°C for 1 min, and extension at 72°C for 90 s, followed by a final extension at 72°C for 10 min. The amplification was performed with a mineral oil overlay in a Trio-Thermoblock (Biometra, Göttingen, Germany).
Negative controls were RT-PCR of total RNA from plasmid-free metal-sensitive AE104 cells, experiments with a 3′ antisense primer at the position of the 5′ sense and a 5′ sense primer at the position of the 3′ antisense primer, DNA templates, no templates, PCR without previous RT reactions, and reactions with total RNA treated with DNase-free RNase. Additionally, Southern-type DNA-DNA hybridization experiments were performed with various primers as probes to determine the specificity of the RT-PCR products.
Purification of CzcR.
CzcR was heterologously expressed in E. coli as a MalE fusion. The czcR gene was amplified (primers 5′-CCC GAA TTC GTG CGG GTA CTT GT-3′ and 5′-CCC GGA TCC TTA TCG AAG TCC CG-3′ [EcoRI and BamHI site, respectively, underlined]) as a PCR fragment from plasmid pECD277 (19) and cloned as an EcoRI/BamHI fragment into plasmid pMAL-c2 (New England Biolabs, Schwalbach, Germany), leading to plasmid pECD529. A two-stage purification procedure was used to isolate CzcR as instructed by the manufacturer (New England Biolabs) with a Bio-Rad (Hempel Hempstead, United Kingdom) HRLC MA7Q anion-exchange column as the second stage. Fractions containing the CzcR protein, as monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, were pooled and concentrated with Vivaspin concentrator columns (Vivascience; Binbrook, Lincoln, United Kingdom). Aliquots of purified CzcR were stored in Tris-HCl buffer (20 mM, pH 8.0) containing 50% (vol/vol) glycerol at −20°C. The approximate protein concentration was determined by Bradford (1) assay, using bovine serum albumin as a standard.
Gel retardation assays.
Gel retardation assays were performed by the method of Mills et al. (12), with modifications. Purified restriction fragments or PCR products were labeled with 32P by a random-priming procedure. Approximately 100 ng of labeled DNA and 200 to 500 ng of purified CzcR protein were mixed and incubated for 40 min at 28°C in 30 μl of binding buffer (100 mM KCl, 20 mM Tris-HCl [pH 8.0], 3 mM MgCl2, 1 mM dithiothreitol, 100 μM EDTA, 100 μg of bovine serum albumin per ml) to form the DNA-protein complex. The reaction was analyzed on a 6% polyacrylamide gel at 4°C and 35 mA for 2 to 3 h.
Competitive RT-PCR (14).
DNase-treated total RNA was isolated from cells of A. eutrophus AE128(pMOL30), DN178(pMOL30-10 ΔczcR), and DN179(pMOL30-11 ΔczcS), either without or with induction for 10 min with 300 μM Zn2+. A sample (2 μg) of this RNA was reverse transcribed by using 200 U of Superscript II reverse transcriptase (Gibco BRL) and 50 pmol of random primer in a total volume of 20 μl. To determine the amount of czcCBA mRNA-specific cDNA for each strain and induction regimen, 10-fold dilutions of an internal DNA standard (10−7 to 102 pg) were added to 1.5 μl of the cDNA, and the mixture was amplified with 100 μM (each) deoxynucleoside triphosphate, 10 pmol of primers (3′ antisense primer B [ATGCCACCGATTACCACCGTTGCGA, positions 7144 to 7120] [gbX98451; Fig. 1, right scale] in czcA [positions 4092 to 7283]); 5′ sense primer A [ATTGGTTCATTCGTGCCCG, positions 6615 to 6633 in czcA; Fig. 1]) and 1 U of Taq polymerase (Qiagen) in a total volume of 50 μl, using the following PCR program: 1 cycle of 2.5 min at 94°C, 1 min at 60°C, and 1 min at 72°C and 28 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C, with final extension for 5 min at 72°C. A 10-μl aliquot of each PCR product was analyzed on a 1.5% agarose gel stained with ethidium bromide. The relative amounts of czcCBA cDNA (529-bp) and internal DNA standard (237-bp) products were quantified after densitometric analysis using ScanPack 2.0 software (Biometra). For each lane, which represents the cDNA with one dilution of the internal DNA standard, the resulting band intensity of the czcCBA cDNA was divided by 529 bp and the intensity of the internal standard was divided by 237 bp, and the ratio of the corrected intensities was calculated. For all 10 lanes, this ratio was plotted (as a log-log plot) against the amount of internal standard used in the competition experiment. A linear regression was calculated for these points, and the intercept on the x axis was defined the point at which the amount of the internal standard was the same as that of the czcCBA-specific cDNA in the RT probe. This value was used to calculate the czcCBA-specific cDNA per μg of total RNA used. Controls were (i) a complete assay without template and RT reaction, (ii) a complete assay with RNA template but without RT, and (iii) a complete assay with total RNA isolated from the plasmid-free A. eutrophus AE104.
For construction of an internal standard specific for czcCBA cDNA, the 529-bp part of czcA was amplified from czcCBA cDNA with primers A and B. This fragment was purified and used again as the template in a PCR experiment with a loop-out primer (ATTGGTTCATTCGTGCCCGGGCGGTGCTCAATGGTCTG) and primer B. The loop-out primer was identical in the first 19 nucleotides (nt) (underlined) with primer A, the remaining 19 nt (bold) were the base pairs in position 6926 to 6944 (gbX98451 [Fig. 1]) of czcA, located between the positions of primers A and B. Thus, we amplified a 237-bp PCR product which could be clearly differentiated from the 529-bp czcCBA cDNA product, but with the same ends as the 529-bp fragment. The 237-bp PCR product was isolated, cleaned with QIAquick, quantified (Ultrospec II; Pharmacia), diluted and used for competitive RT-PCR.
RESULTS
The czcI and czcN genes are induced by zinc.
Figure 2 shows the 400-nt transcript of the czcI gene (Fig. 2A), which is different in size (Fig. 2B) from the 500-nt transcript of the czcN gene (Fig. 3C). Both messages were induced best by 300 μM Zn(II) (lanes 2 and 10) and less well by 300 μM Co(II) (lanes 3 and 11). Smears in the zinc- and cobalt-induced lanes may indicate the presence of larger transcripts (lanes 2, 3, 6, 10, and 11). In the presence of 300 μM Cd(II) (lanes 1 and 9), the levels of both mRNAs were close to those in uninduced cells. No czcI or czcN message was detected in cells of the plasmid-free control strain AE104 (Fig. 2 lanes 5 and 13). In contrast, no transcript of the proposed old czcR (19) was found under any induction conditions (data not shown).
FIG. 2.
Oligonucleotide-probed Northern blot analysis of transcription of the czc determinant. Total RNA was separated by electrophoresis in 1.5% agarose, transferred to a nylon filter, and hybridized with a czcI-specific probe (panel A, all lanes; panel B, lane 6 [for simplicity, lane B6]) or a czcN-specific probe (lane B8; panel C, all lanes). Total RNA was isolated from the czc-free, metal-sensitive strain A. eutrophus AE104 (lanes A5 and C13) and the czc-containing strain AE128 which was cultivated without toxic concentrations of heavy-metal cations (lanes A4 and C12), or induced with metal cations for 10 min: 300 μM Cd2+ (lanes A1 and C9), 300 μM Zn2+ (lanes A2, B6, B8, and C10), or 300 μM Co2+ (lane A3 and C11). Lane B7 is empty. In panel B, the RNA for future hybridization with the czcI- or czcN-specific probe was run on the same gel for direct size comparison. The sizes of RNA molecular size markers are marked with closed arrows; the transcripts are marked with dashed arrows. Blebs and compressions at 1,541 and 2,904 nt are due to the 16S and 23S rRNAs; positions of the rRNAs are marked by large arrowheads. The original photograph was scanned with Ofoto 2.0 (Light Source Computer Images, Inc.) and processed with Adobe Photoshop 3.0 (Adobe Systems, Inc.).
FIG. 3.
Synthesis of [35S]methionine-labeled polypeptides determined by the czcNICB′ region. The upstream region of czc containing czcNICB′ (grey bar labeled “T7 fragment” in Fig. 1) was cloned into plasmid pT7-5 (32a) and expressed in E. coli K38(pGP1-2) (33). (A) Control without the region; (B) czcNICB′ region. Size markers are given in kilodaltons on the left; the resulting polypeptides are indicated by arrows on the right. The original photograph was scanned with Ofoto 2.0 (Light Source Computer Images, Inc.) and processed with Adobe Photoshop 3.0 (Adobe Systems, Inc.).
To confirm that the ORF designated old czcR was not functional in czc expression, the proposed ATG start codon of the gene (positions 1098 to 1100 in EMBL/GeneBank accession no. X67305) on plasmid pMOL30 was changed into a TAG stop codon (pMOL30-6). This mutation in the old czcR led to a H94L mutation in the czcI gene encoded on the other DNA strand. There was no difference in growth on solid or in liquid media in the presence or absence of Zn(II), Co(II), or Cd(II) between AE128(pMOL30) and DN172(pMOL30-6) (data not shown). Finally, a promoterless lacZ gene was inserted between czcC and czcB on plasmid pMOL30-6, yielding plasmid pMOL30-8, and on the wild-type plasmid pMOL30, yielding plasmid pMOL30-9. Both strains, DN174(pMOL30-8) and DN175(pMOL30-9), were resistant to 2.5 mM Zn(II) (data not shown), and induction of β-galactosidase activity by 100 μM Zn(II) was identical in both strains (data not shown). Thus, the old czcR (19) does not exist. Instead, the czcI and czcN genes on the other strand were shown to be transcribed, and their transcription was inducible by the czc substrates. Moreover, a H94L mutation in czcI did not influence induction of czc.
Using different oligonucleotides, we also investigated transcription of the czcCBA and czcDRS regions. Transcription of both regions followed the same pattern as induction of czcN or czcI, with zinc and cobalt as good inducers and cadmium as a poor inducer (data not shown). The existence of the 6,200-nt czcCBA mRNA (35) was confirmed with an oligonucleotide as the probe, but the message(s) of the czcDRS region gave no discrete signal in the Northern experiments (data not shown).
czcN and its upstream region.
Sequencing of czcN, now demonstrated to be part of the czc system, was completed. The region upstream of the previous 5′ limit of czc (gbX98451) was cloned from plasmid pECD162 (19), and a 2,788-bp sequence of each DNA strand was determined (GenBank accession no. AJ001159 [Fig. 1, left scale]). The KpnI site at the beginning of gbX98451 starts at position 2789 in gbAJ001159 (Fig. 1, position 0).
Two potential translational start sites of czcN are at A2471TG (with a single G as the ribosome binding site [Fig. 1, left scale]) or at A2642TG (GA as the ribosome binding site). The corresponding CzcN protein would have size of 274 aa (30.1 kDa) or 217 aa (23.7 kDa), respectively. If the first start site is used, CzcN has additional 60 aa at its amino terminus compared to NccN from the nickel-cobalt-cadmium resistance of Alcaligenes xylosoxidans (29); if the second site is used, the additional N-terminal region is only 3 aa. The CzcN and NccN proteins are 67% identical in an overlap of 212 aa.
On the opposite DNA strand, a 125-aa ORF starts at an ATG (position 2311 [Fig. 1, left scale]) and ends at a TGA stop codon (position 1934 [Fig. 1, left scale]). The predicted protein is 42% identical to SrpB from Synechococcus sp. strain PCC 7952 (15) and between 37 and 42% identical to other MgtC-like proteins. Thus, the 125-aa ORF is probably an MgtC-like protein (32), which interacts in an unknown manner with inducible P-type ATPases responsible for magnesium uptake. It was designated mgtC.
The remaining DNA sequence upstream of czcN and mgtC contains only small ORFs encoding proteins up to 191 aa in size. None of these potential proteins has significant similarity to any other protein in the current databases. In the first 800 bp of the sequence, four stem-loop structures with free energies between −50 and −92 kJ/mol were found (data not shown). The 31-bp 1415-GCCTTAGCGTGCAATATTTTCCGTTTTCTGA-1445 are in 29 positions identical to inverted repeats of the mercury resistance transposon Tn21 and related to other mer transposon repeats. This may suggest that the czc determinant was formerly transposable, although it is not now flanked by inverted repeats. Thus, there is no evidence for additional czc-associated genes upstream of czcN with the exception of the mgtC-like gene (Fig. 1).
Heterologous expression of the czc upstream region.
No expression in E. coli of any protein was seen when the first 4.6 kb of the czc upstream region (positions 1 to 4600 in gbAJ001159 [Fig. 1, left scale]) were expressed in the direction of czc transcription under control of the phage T7 promoter (data not shown). Thus, the upstream region of czc containing czcNICB′ (grey bar labeled “T7 fragment” in Fig. 1) was cloned into plasmid pT7-5 (32a) and expressed in E. coli K38(pGP1-2) (33). As an internal control for successful expression, both bands of CzcC were visible, with sizes of 43.5 ± 1.3 kDa (sequence, 44.608 kDa) for the pre-CzcC and 39.2 ± 1.2 kDa (sequence, 42.224 kDa) for the mature CzcC (Fig. 3). A band corresponding to a size of 23.7 ± 0.7 kDa fits exactly the predicted size of CzcN if this protein starts at A2642TG. If CzcN starts earlier, it should have a size of 30.096 kDa; however, no band was observed in that region (Fig. 3). Another strong band corresponded to a polypeptide of about 9 kDa. This band was assigned to CzcI (predicted size, 13.257 kDa), which probably has a leader sequence and may be truncated to a mature protein of 10 kDa.
Start sites of the czc messages.
Using primer extension, we determined the start sites of all czc mRNAs present in zinc- induced AE128(pMOL30) cells (Fig. 4; Table 1). Primer extension experiments with RNA isolated from plasmid-free, metal-sensitive AE104 cells served as a negative control for the specificity of primer binding.
FIG. 4.
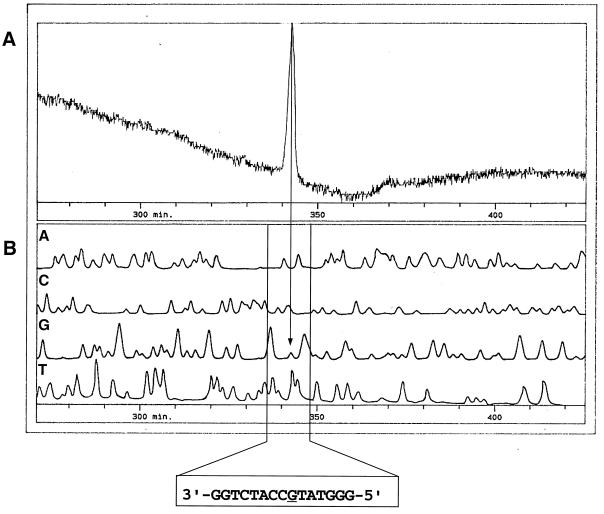
Determination of the transcription initiation site of czcC by primer extension analysis. A. eutrophus AE128(pMOL30) cells were induced for 10 min with 300 μM Zn2+, and total RNA was isolated. Primer extension analyses were performed on an automated fluorescence DNA sequencer using this total RNA and a fluorescently labeled oligonucleotide as the primer (A). Dideoxy sequencing was run as a size marker with the same primer (B); the raw data output is shown, with lines A, C, G, and T representing the successively detected dideoxynucleotide sequencing reaction products. The arrow marks the initiation site at a G. The DNA sequence shown is in the 3′-5′ orientation; the G initiation site is underlined.
TABLE 1.
5′ ends of the czc-specific mRNAs as determined by primer extension
Start sites of the mRNAs are underlined. Vertical lines join bases identical between the czcI/czcC and czcC/czcD mRNAs; asterisks mark bases identical between the czcI and czcD mRNAs; dashes indicate gaps introduced for better alignment.
One experiment shown in Fig. 4.
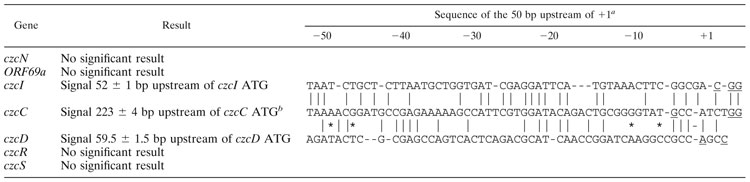
Even though a discrete 500-nt transcript of the czcN gene was visible in Northern experiments, the primer extension experiments gave no signals although several czcN-specific primers were used (data not shown). The 400-nt transcript of czcI starts at position 766 (gbX98451 [Fig. 1, right scale]) 52 ± 1 bp upstream of the czcI ATG (Table 1). Two additional signals in the primer extension experiments could not be reproduced or were also present in the negative control (data not shown). With the start site 52 bp upstream of czcI, the 400-nt transcript should end at a potential stem-loop structure downstream of czcI. The sequence of this region is 1155-CGTCTCGCTTGATCGGCGAGACGACGACTCTTTTTCT-1191 (gbX98451 [Fig. 1, right scale]). The inverted repeat is underlined; the boldfaced TGA is the potential stop codon of czcI. The calculated free energy of the stem-loop is −60 kJ/mol (25°C), and it is followed by a stretch of T’s. Thus, the position of the czcI transcript is clearly defined.
The 6,200-nt czcCBA transcript started at about position 1,019 ± 4 (gbX98451 [Fig. 1, right scale]), which is 223 ± 4 upstream of the czcC ATG and in the middle of czcI (Fig. 4). Thus, the 6,200-nt transcript fits between this start site and the potential terminator czcAt at position 7320 (gbX98451 [35]). The 50 bp upstream of the czcCBA start site were 52% identical with the 50 bp upstream of the czcI start site (Table 1).
The czcD transcript started at positions 7273 and 7276 (gbX98451 [Fig. 1, right scale]) 59 or 60 bp upstream of the czcD ATG and 20 bp upstream of the beginning of czcAt. The 50 bp upstream of this start site were about 26% identical to the respective czcI and czcC regions (Table 1). No start sites of the czcS and the czcR transcripts were found (Table 1).
Thus, defined 5′ ends of the discrete 400-nt czcI and the discrete 6,200-nt czcCBA mRNAs were found. The discrete 500-nt czcN mRNA had no defined 5′ end; however, a defined 5′ end was determined for the messages of the czcDRS region. Therefore, after specific zinc induction, there were up to four points of transcription initiation in the czc determinant (Fig. 1).
Binding sites of CzcR.
If these 5′ ends of the respective mRNAs are the result of zinc/cobalt-specific transcription initiation and CzcR is the activator of czc transcription, CzcR should bind to operators upstream of these four mRNA start sites. Thus, the czcR gene was fused to malE, overexpressed in E. coli, and purified to homogeneity.
To identify the binding site of CzcR to a possible operator upstream of czcN, the 734-bp EcoRI fragment harboring the 5′ end of czcN (Fig. 1, scaled-up view, gel retardation fragment c) was isolated. Since CzcR bound to this fragment (data not shown), we prepared a 501-bp (Fig. 1, gel retardation fragment b) and a 389-bp (Fig. 1, gel retardation fragment a) PCR fragment which contained a smaller part of the 734-bp EcoRI fragment; however, all three DNA fragments started at the EcoRI fragment upstream of czcN. CzcR bound to the two smaller fragments (Fig. 5) too, and a 100-fold (data not shown), 50-fold, or 10-fold excess (Fig. 5) of unlabeled, identical DNA competed for CzcR. Thus, binding was specific.
FIG. 5.
Binding of CzcR to the DNA region upstream of czcN. Two PCR fragments (fr.) containing overlapping parts of the czcN upstream region were isolated, labeled with 32P, and used in gel retardation experiments with the CzcR protein. The left ends of both fragments, the 389-bp (lanes 1 to 5) and 501-bp (lanes 6 to 10) fragments were the EcoRI site between mgtC and czcN (Fig. 1). The DNA fragments were incubated without CzcR (negative control; lanes 1 and 6), with CzcR (lanes 2 and 7), with CzcR and acetyl phosphate (lanes 3 and 8), with CzcR and a 50-fold excess of unlabeled DNA fragment (lanes 4 and 9), and with CzcR and a 10-fold excess of unlabeled DNA fragment (lanes 5 and 10). The autoradiograms were scanned with Ofoto 2.0 (Light Source Computer Images, Inc.) and processed with Adobe Photoshop 3.0 (Adobe Systems, Inc.).
To identify CzcR operators upstream of czcI, czcC, or czcD, the 7,922-bp KpnI-BamHI fragment, which contains the region from the middle of czcN to the middle of czcD (Fig. 1), was isolated. The fragment was digested with AvaII, yielding 11 fragments. CzcR did not bind to the 984-bp AvaII fragment containing the upstream regions of czcI and czcC. Instead, there was some weak binding to one of the larger AvaII fragments, the 1,615- or 1,711-bp fragment containing the middle part of czcA or the region between czcC and czcB (data not shown).
To investigate CzcR binding upstream of czcR and czcS, the 2,148-bp BamHI-EcoRI fragment containing the region from the 3′ end of czcD to the middle of czcS (Fig. 1) was isolated and digested with BanII, yielding five fragments between 195 and 672 bp in size. CzcR bound to none of these fragments (data not shown).
Thus, the purified CzcR protein bound to a single site upstream of czcN, and this site is located on a 389-bp PCR fragment (Fig. 5).
Additional transcripts.
RT-PCR experiments were done to search for transcripts not visible in Northern-type experiments. RNA was isolated from zinc-induced AE128(pMOL30) cells, digested with DNase, and reverse transcribed by using a 3′ antisense primer. The resulting cDNA was amplified by using the same primer and a 5′ sense primer. To exclude artifacts, we performed a variety of control experiments, such as (i) use of RNA from the plasmid-free strain AE104, (ii) exchange of the sense and antisense PCR primers, (iii) no template, (iv) no RT reaction, and (v) addition of DNase-free RNase. Moreover, the sizes and restriction patterns of the resulting PCR products were verified.
With an RT reaction starting from a czcI-specific transcript, PCR products were obtained with 5′ sense primers in ORF69a and in czcN (Fig. 1 and 6). There was also a PCR product when a czcC-specific 3′ antisense primer and a czcI-specific 5′ sense primer were used. In contrast, there was never any product in case of a czcD-specific 3′ antisense primer and a czcA-specific 5′ sense primer. In the czcDRS region, RT-PCR indicated cotranscription of all three genes; however, no transcripts were found between czcS and ORF131, an ORF with a possible product of 131 amino acid residues located downstream of czc (Fig. 1) (35). Thus, there is evidence for additional transcripts which might span the complete region from upstream of czcN to czcAt (as a czcN-ORF69a-ICBA message), and for a tricistronic czcDRS mRNA (Fig. 1).
FIG. 6.
Additional transcripts of czc. DNA-free RNA was isolated from zinc-induced cells of A. eutrophus AE128(pMOL30). The RNA was reverse transcribed by using 3′ antisense (antis.) primers. The resulting cDNA was amplified with the same primer and a 5′ sense primer. The resulting PCR fragments were analyzed on an agarose gel. The genes or ORFs in which the primers are located are indicated above the gel image, and their positions are indicated in Fig. 1. The gel image was inverted with Adobe Photoshop 3.0 (Adobe Systems, Inc.); thus, the ethidium bromide-mediated fluorescence image is shown black on white. Lane 1, czcI-ORF69a; lane 2, czcI-czcN; lane 3, czcC-czcI; lane 4, czcD-czcA; lane 5, 100-bp ladder (sizes indicated on the left); lane 6, czcR-czcD; lane 7, czcS-czcR; lane 8, czcS-czcD; lane 9, ORF131-czcS. Locations of the antisense primers are given first for each pair.
Function of the CzcRS two-component regulatory system.
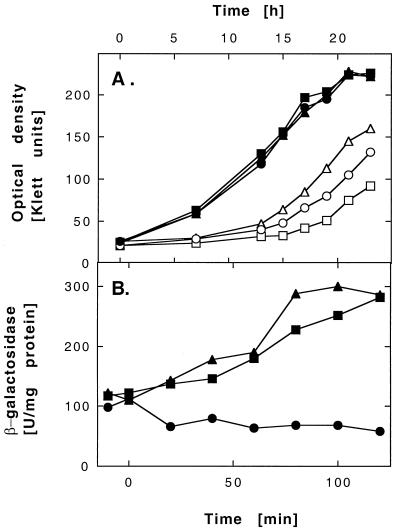
To elucidate the function of the CzcRS proteins in regulation of the Czc system, in-frame deletions of the czcR or czcS gene were constructed in plasmid pMOL30, leading to strains DN178(pMOL30-10 czcNICBAD ΔczcR czcS) and DN179(pMOL30-11 czcNICBADR ΔczcS). There was no significant difference in the MIC of zinc, cobalt, or cadmium between these two strains and AE128(pMOL30) wild type; however, growth of the deletion strains on solid media at metal ion concentrations close to the MIC was slower than growth of the wild type (data not shown). In liquid culture containing 5 mM Zn2+, all three strains grew identically when they had been induced for 24 h with 300 μM Zn2+; however, the uninduced ΔczcR mutant displayed an extended lag phase compared to the wild type, while the lag phase of the ΔczcS mutant was shorter (Fig. 7A).
FIG. 7.
Influence of CzcR and CzcS on growth and Czc expression in the presence of zinc. (A) Cells of strains AE128(pMOL30) (●, ○), DN178(pMOL30-10 ΔczcR) (■, □), and DN179(pMOL30-11 ΔczcS) (▴, ▵) were cultivated for 24 h in the presence of 300 μM Zn2+ (■, ●, ▴) or without inducer (□, ○, ▵) and diluted into fresh medium containing 5 mM Zn2+ up to an optical density of 10 Klett units. The cells were incubated with shaking at 30°C, and the increase in optical density was determined. (B) Cells of strains DN175(pMOL30-9 czcNIC lacZ czcBADRS) (●), DN180(pMOL30-12 czcNIC lacZ czcBAD ΔczcR czcS) (■), and DN181(pMOL30-13 czcNIC lacZ czcBADR ΔczcS) (▴) were cultivated for 48 h in Tris-buffered mineral salts medium containing 0.2% (wt/vol) sodium gluconate as the carbon source at 30°C with shaking. The cell suspension was diluted to an optical density of 35 Klett units, incubated with shaking until an optical density of 70 Klett units was reached, and induced with 100 μM Cd2+ at time zero. Incubation was continued with shaking at 30°C. The turbidity of the cells and the β-galactosidase activity of the reporter gene product over time were determined and used to calculate the specific activity in units per milligram (dry weight).
To examine the influence of CzcR and CzcS on czc expression, czcR and czcS were also deleted from the czc-lacZ plasmid pMOL30-9, leading to strains DN180(pMOL30-12 czcNIC lacZ czcBAD ΔczcR czcS) and DN181(pMOL30-13) (czcNIC lacZ czcBADR ΔczcS). There was no difference of β-galactosidase induction between those strains at high (millimolar) concentrations of zinc, cobalt, or cadmium; however, at 100 μM Cd2+, 300 μM Co2+, or 10 μM Zn2+, β-galactosidase in both mutant strains was induced, whereas the wild-type strain exhibited low-level expression of the reporter gene (shown for 100 μM Cd in Fig. 7B; results for cobalt and zinc not shown). Uninduced cells of wild-type and mutant strains remained at a level of about 60 U of β-galactosidase/mg of protein if zinc-containing trace element solution was omitted from the mineral salts medium used for the experiment (data not shown).
To evaluate the influence of CzcR and CzcS on the mRNA level, competitive RT-PCR experiments were performed with RNA isolated from cells of strains AE128(pMOL30) and its ΔczcR and ΔczcS mutant derivatives, uninduced or after induction with 300 μM Zn2+ for 10 min (Table 2). In uninduced cells without czcR, the czcCBA mRNA-dependent cDNA level was only half of that in wild-type cells, while in uninduced cells without czcS, the cDNA level was more than fourfold higher (Table 2). In cells induced with zinc, deletion of czcR or czcS yielded a higher amount of czcCBA mRNA which was 2-fold or 1.5-fold, respectively, higher than the control level.
TABLE 2.
Effects of czcRS mutations on the concentration of czcCBA mRNA
| Bacterial strain | Genotype | cDNA concn (ng/g of RNA; mean ± SD [n = 2])
|
Fold induction | |
|---|---|---|---|---|
| Uninduced | Induceda | |||
| AE128(pMOL30) | Wild type | 11 ± 5 | 804 ± 42 | 73.7 |
| DN178(pMOL30-10) | ΔczcR | 5 ± 0 | 1,420 ± 270 | 313 |
| DN179(pMOL30-11) | ΔczcS | 40 ± 3 | 1,150 ± 20 | 28.9 |
Cells were induced for 10 min with 300 μM Zn2+ before the RNA was isolated.
Thus, in uninduced ΔczcR cells, the czcCBA mRNA level was lower than in wild-type cells and ΔczcR cells needed a longer time to adjust when confronted with heavy metals. On the other hand, uninduced ΔczcS cells contained more czcCBA message and resumed growth faster when challenged. Third, the increase in mRNA after induction observed in both mutant types was higher than in the wild type.
DISCUSSION
Four major transcripts of the czcNICBADRS determinant, the czcN, czcI, czcCBA, and czcDRS mRNAs, were identified by Northern and/or RT-PCR experiments (35) (Fig. 1). The patterns of control of these four transcripts are very similar; all were optimally induced by zinc, less by cobalt, and only a little by cadmium. Since regulation of czcD expression judged from a czcD::lacZ fusion (35) and negative evidence from RT-PCR experiments excludes transcription from czcA into czcD, a huge czcNICBADRS mRNA, which is processed into the four observed mRNAs and which is transcribed from one promoter upstream of czcN, should not exist. Thus, there should be four promoters, czcNp, czcIp, czcCp, czcDp, serving as start sites for each of the four transcripts, and these promoters are apparently controlled in the same manner.
Two of the transcripts, the czcI and the czcCBA mRNAs, are stable enough to be seen in a Northern experiment. In contrast, transcription of czcDRS yields a smear, and continuous transcription of the respective region could be shown only by RT-PCR. The size of the czcN message, 500 nt, was not sufficient to cover the czcN gene, which has a size of at least 651 bp, and no 5′ end of the mRNA was found. Both results indicate a highly unstable mRNA of czcN, and thus the 500-nt transcript may be a metastable degradation product.
Computer analyses do not predict any potential terminator structure downstream of czcN (or of ORF69a following czcN), and RT-PCR experiments demonstrated continuous transcription from czcN into czcI. Thus, a czcNI dicistronic mRNA should exist. With DNA-probed Northern experiments, a 1,200-nt mRNA has been demonstrated (35), and in all Northern experiments done in the czcNI region (Fig. 2), smears above the discrete bands indicate the presence of larger transcripts.
The czcI mRNA with a size of 400 nt fits between the start site found with primer extension and the proposed terminator czcIt. However, RT-PCR indicates again that some transcription extends beyond czcIt into czcC, and smears in Northern experiments of the czcCBA region (35) could result from transcripts larger than the 6,200-nt czcCBA message.
Thus, the transcription initiation and termination events in the upstream regulatory region of czc seem to be quite complex: transcription starts from the czcNp, czcIp, and czcCp, there is probably termination at czcIt downstream of czcI and maybe some termination downstream of czcN. The resulting primary transcripts (the czcNICBA, czcNI, czcN, czcICBA, czcI, and czcCBA mRNAs) are then processed into the three observed mRNAs of 400, 500, and 6,200 nt.
All transcripts, however, are terminated at czcAt. The proposed promoter of the czcDRS region overlaps with the czcAt region, which may explain the regulation of czcD expression as examined with a czcD::lacZ fusion (35) and Northern experiments: expression of czcD::lacZ starts vigorously when zinc is present, however, under conditions of high expression of czcCBA, expression of czcD decreases (35). If the RNA polymerase pauses at czcAt after transcription of czcCBA, the czcDRS promoter may be blocked. Thus, the stronger the expression of czcCBA, the less available the czcD promoter may be for transcription initiation events. Expression of the czcDRS downstream regulatory region of the czc determinant could therefore be, in the long run, inversely controlled with respect to expression of the czcCBA structural gene region.
It is still unclear which proteins are actually responsible for control of the four proposed promoters czcNp, czcIp, czcCp, and czcDp. As shown by the knockout experiments, the two-component regulatory system CzcRS is not essential for Czc control. This was shown before (19), because deletion of all czc genes downstream of czcC did not abolish regulation of Czc. CzcRS, however, is involved in Czc regulation. In the absence of CzcS, CzcR should be present in an unphosphorylated state, and transcription of czcCBA, compared to the wild type, was fourfold higher in uninduced cells but only slightly higher in zinc-induced cells. In the absence of CzcR, czcCBA transcription decreases in uninduced cells but increases in induced cells. The CzcR response regulator, as purified, bound only upstream of czcN. Preliminary footprinting experiments (data not shown) indicated a protected region of 62 bp (positions 2483 to 2544 in gbAJ001159 [Fig. 1, left scale]) with some similarity to the cop box, which is required for regulation of copper resistance (27). Thus, CzcR probably acts only on czcNp; the CzcRS two-component regulatory system may therefore control the differential expression of czcN compared to the other czc genes.
There is little information available concerning CzcN. It is probably membrane bound, with four or five transmembrane α-helices; it is 67% identical to NccN, which is not essential for the function of the combined nickel-cobalt-cadmium resistance (mediated by the CzcCB2A-related NccCBA efflux system) from A. xylosoxidans (29); however, transposon insertions into nccN led to a threefold reduction in nickel resistance in this bacterium (29).
CzcN is not essential for a slow nonspecific Czc regulation, since a truncated czc determinant without the 5′ end of czcN shows some inducibility (19). If this residual upstream regulatory region is deleted, metal resistance is abolished but may be regained when a lac promoter is cloned upstream of czcCBA. That leaves the problem that of all potential Czc regulators identified so far: (i) CzcRS and CzcN are not essential for Czc regulation, (ii) CzcD is membrane bound, and (iii) CzcI is probably located in the periplasm. Thus, another, unknown regulator could exist.
Since the promoter regions of czc do not contain clear consensus sequences for typical ς70 promoters, an unknown sigma factor might be required for czc transcription. The cnr cobalt-nickel-resistance system (8), which is related to czc but located on plasmid pMOL28, is controlled by its own sigma factor, CnrH, of the extracellular function (9) sigma factor family. No such sigma factor has yet been identified for czc. Any of the genes whose functions are unknown (e.g., czcN, czcI, and even ORF69a) might encode products which, together with the CzcD sensor for extracellular cation, could be responsible for the control of a sigma factor by periplasmic signals, in a system similar to the Rse system (2) and many other extracellular function sigma factors (13).
ACKNOWLEDGMENTS
We thank Grit Schleuder for excellent technical assistance. C.G. thanks members of the N.L.B. laboratory, especially Ken Jakeman and Jon Wilson, for advice and help in protein purification.
This work was supported by grants from Forschungsmittel des Landes Sachsen-Anhalt and Fonds der Chemischen Industrie to D.H.N. and the Biotechnology and Biological Sciences Research Council (G07943) and Medical Research Council (G.9309093) to N.L.B.
REFERENCES
- 1.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.De Las Penas A, Connolly L, Gross C A. The sigmaE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol Microbiol. 1997;24:373–385. doi: 10.1046/j.1365-2958.1997.3611718.x. [DOI] [PubMed] [Google Scholar]
- 3.Diels L, Dong Q, van der Lelie D, Baeyens W, Mergeay M. The czc operon of Alcaligenes eutrophus CH34: from resistance mechanism to the removal of heavy metals. J Ind Microbiol. 1995;14:142–153. doi: 10.1007/BF01569896. [DOI] [PubMed] [Google Scholar]
- 4.Dressler C, Kües U, Nies D H, Friedrich B. Determinants encoding multiple metal resistance in newly isolated copper-resistant bacteria. Appl Environ Microbiol. 1991;57:3079–3085. doi: 10.1128/aem.57.11.3079-3085.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engler-Blum G, Meier M, Frank J, Müller G A. Reduction of background problems in nonradioactive Northern and Southern blot analysis enables higher sensitivity than 32P-based hybridizations. Anal Biochem. 1993;210:235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]
- 6.Kunito T, Kusano T, Oyaizu H, Senoo K, Kanazawa S, Matsumoto S. Cloning and sequence analysis of czc genes in Alcaligenes sp. strain CT14. Biosci Biotechnol Biochem. 1996;60:699–704. doi: 10.1271/bbb.60.699. [DOI] [PubMed] [Google Scholar]
- 7.Lenz O, Schwartz E, Dernedde J, Eitinger T, Friedrich B. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J Bacteriol. 1994;176:4385–4393. doi: 10.1128/jb.176.14.4385-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liesegang H, Lemke K, Siddiqui R A, Schlegel H-G. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J Bacteriol. 1993;175:767–778. doi: 10.1128/jb.175.3.767-778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lonetto M A, Brown K L, Rudd K E, Buttner M J. Analysis of the Streptomyces coelicolor sigF gene reveals the existence of a subfamily of eubacterial RNA polymerase ς factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mergeay M, Nies D, Schlegel H G, Gerits J, Charles P, van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985;162:328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michaelian I, Sergeant A. A general and fast method to generate multiple site directed mutations. Nucleic Acids Res. 1992;20:376. doi: 10.1093/nar/20.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mills S D, Lim C-K, Cooksey D A. Purification and characterization of CopR, a transcriptional activator protein that binds to a conserved domain (cop box) in copper-inducible promoters of Pseudomonas syringae. Mol Gen Genet. 1994;244:341–351. doi: 10.1007/BF00286685. [DOI] [PubMed] [Google Scholar]
- 13.Missiakas D, Raina S. The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 14.Navarrete Santos A, Kehlen A, Schütte W, Langner L, Riemann D. Regulation by transforming growth factor β1 of class II mRNA and protein expression in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Int Immunol. 1998;10:601–607. doi: 10.1093/intimm/10.5.601. [DOI] [PubMed] [Google Scholar]
- 15.Nicholson M L, Laudenbach D E. Genes encoded on a cyanobacterial plasmid are transcriptionally regulated by sulfur availability and CysR. J Bacteriol. 1995;177:2143–2150. doi: 10.1128/jb.177.8.2143-2150.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nies A, Nies D H, Silver S. Cloning and expression of plasmid genes encoding resistance to chromate and cobalt in Alcaligenes eutrophus. J Bacteriol. 1989;171:5065–5070. doi: 10.1128/jb.171.9.5065-5070.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nies D, Mergeay M, Friedrich B, Schlegel H G. Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J Bacteriol. 1987;169:4865–4868. doi: 10.1128/jb.169.10.4865-4868.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nies D H. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J Bacteriol. 1995;177:2707–2712. doi: 10.1128/jb.177.10.2707-2712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nies D H. CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc and cadmium (czc system) in Alcaligenes eutrophus. J Bacteriol. 1992;174:8102–8110. doi: 10.1128/jb.174.24.8102-8110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nies D H. Resistance to cadmium, cobalt, zinc and nickel in microbes. Plasmid. 1992;27:17–28. doi: 10.1016/0147-619x(92)90003-s. [DOI] [PubMed] [Google Scholar]
- 21.Nies D H, Brown N. Two-component systems in the regulation of heavy metal resistance. In: Silver S, Walden W, editors. Metal ions in gene regulation. London, England: Chapman & Hall; 1997. [Google Scholar]
- 22.Nies D H, Nies A, Chu L, Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci USA. 1989;86:7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nies D H, Silver S. Plasmid-determined inducible efflux is responsible for resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus. J Bacteriol. 1989;171:896–900. doi: 10.1128/jb.171.2.896-900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oelmüller U, Krüger N, Steinbüchel A, Friedrich C G. Isolation of prokaryotic RNA and detection of specific mRNA with biotinylated probes. J Microbiol Methods. 1990;11:73–84. [Google Scholar]
- 25.Peitzsch, N., and D. H. Nies. Unpublished data.
- 26.Rensing C, Pribyl T, Nies D H. New functions for the three subunits of the CzcCBA cation-proton antiporter. J Bacteriol. 1997;179:6871–6879. doi: 10.1128/jb.179.22.6871-6879.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rouch D A, Brown N L. Copper-inducible transcriptional regulation at two promoters in the Escherichia coli copper resistance determinant pco. Microbiology. 1997;143:1191–1202. doi: 10.1099/00221287-143-4-1191. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Schmidt T, Schlegel H G. Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans 31A. J Bacteriol. 1994;176:7045–7054. doi: 10.1128/jb.176.22.7045-7054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 31.Simon R, Quandt J, Klipp W. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in Gram-negative bacteria. Gene. 1989;80:161–169. doi: 10.1016/0378-1119(89)90262-x. [DOI] [PubMed] [Google Scholar]
- 32.Snavely M D, Miller C G, Maguire M E. The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J Biol Chem. 1991;266:815–823. [PubMed] [Google Scholar]
- 32a.Tabor, S. Personal communication.
- 33.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taghavi S, Mergeay M, Van der Lelie D. Genetic and physical map of the Alcaligenes eutrophus CH34 megaplasmid pMOL28 and it derivative pMOL50 obtained after temperature induced mutagenesis and mortality. Plasmid. 1997;37:22–34. doi: 10.1006/plas.1996.1274. [DOI] [PubMed] [Google Scholar]
- 35.Van der Lelie D, Schwuchow T, Schwidetzky U, Wuertz S, Baeyens W, Mergeay M, Nies D H. Two component regulatory system involved in transcriptional control of heavy metal homeostasis in Alcaligenes eutrophus. Mol Microbiol. 1997;23:493–503. doi: 10.1046/j.1365-2958.1997.d01-1866.x. [DOI] [PubMed] [Google Scholar]
- 36.Voss H, Wirkner U, Jakobi R, Hewitt N A, Schwager C, Zimmermann J, Ansorge W, Pyerin W. Structure of the gene encoding human casein kinase II subunit β. J Biol Chem. 1991;266:13706–13711. [PubMed] [Google Scholar]
- 37.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]