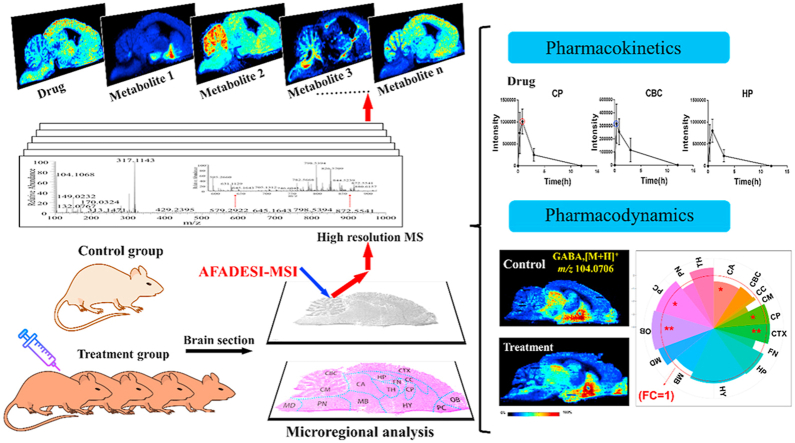
Abstract
The brain is the most advanced organ with various complex structural and functional microregions. It is often challenging to understand what and where the molecular events would occur for a given drug treatment in the brain. Herein, a temporo-spatial pharmacometabolomics method was proposed based on ambient mass spectrometry imaging and was applied to evaluate the microregional effect of olanzapine (OLZ) on brain tissue and demonstrate its effectiveness in characterizing the microregional pharmacokinetics and pharmacodynamics of OLZ for improved understanding of the molecular mechanism of drugs acting on the microregions of the brain. It accurately and simultaneously illustrated the levels dynamics and microregional distribution of various substances, including exogenous drugs and its metabolites, as well as endogenous functional metabolites from complicated brain tissue. The targeted imaging analysis of the prototype drug and its metabolites presented the absorption, distribution, metabolism, and excretion characteristics of the drug itself. Moreover, the endogenous functional metabolites were identified along with the associated therapeutic and adverse effects of the drug, which can reflect the pharmacodynamics effect on the microregional brain. Therefore, this method is significant in elucidating and understanding the molecular mechanism of central nervous system drugs at the temporo and spatial metabolic level of system biology.
KEY WORDS: Pharmacometabolomics, Pharmacokinetics, Pharmacodynamics, Mass spectrometry imaging, Antipsychotic drug
Graphical abstract
An effective temporo-spatial pharmacometabolomics method based on mass spectrometry imaging has been proposed for pharmacokinetic and pharmacodynamic characterization of brain microregions to elucidate the molecular mechanism of central nervous system (CNS) drugs.
1. Introduction
The central nervous system (CNS) has an intricate and fragile structure with high interconnectivity and interaction among numerous microregions of the brain. The brain has the most complicated and finest structure and function, which regulates all aspects of behavior, language, thinking, memory, movement, and emotions1. Based on the neural structure, chemical characteristics, and connectivity, it can be subdivided into numerous microregions, such as the cerebral cortex, midbrain, pons, medulla oblongata, hippocampus, and hypothalamus2. The functional differences in metabolic enzymes, receptors, ligands, transporters, and blood flow in the brain microregions3 cause differences in spatial distribution and efficacy of drugs. Additionally, the brain contains a variety of endogenous functional metabolites that are unevenly distributed in the different microregions4. Therefore, it is very challenging to develop CNS drugs and elucidate what and where the molecular events would occur for a given drug treatment, as most CNS drugs exert effects only after entering the brain. For drug development, it is crucial to understand the pharmacokinetics (absorption, distribution, metabolism, and excretion) and pharmacodynamics of the drug5. Developing novel technologies may provide new opportunities and promote the research of innovative drugs6,7.
In the recent years, liquid chromatography mass spectrometry (LC‒MS) based pharmacometabolomics has been proven to be a powerful tool in identifying an organism's metabolic response following drug administration through an investigation of the overall changes of metabolites in the body. LC‒MS can provide information on drug metabolism, drug action-associated alteration in the endogenous metabolic pathway, and can correlate drug pharmacokinetics, pharmacodynamics, and in vivo effects to elucidate the molecular mechanisms of drug effects and toxicity8, 9, 10. LC‒MS-based pharmacometabolomics analyzes body fluids, such as blood and urine, and reflects the average metabolic level of various tissues and organs in the body. Tissues and organs are the focal site of most diseases as well as drug action, with the brain being the target site for CNS diseases. Thus, information about the in situ distribution of drugs in the brain is significant to evaluate drug efficacy, toxicology, and pharmacokinetics11.
At present, the common techniques used to study the brain include several types of functional brain imaging, such as functional magnetic resonance imaging (fMRI)12 and positron emission tomography (PET)13,14, and structural brain imaging such as magnetic resonance imaging (MRI)15, computerized tomography (CT)16, single photon emission computed tomography (SPECT)17, and other CT technologies. Although structural and functional imaging technology plays an increasingly important role in disease research and CNS drug development, these technologies only provide images of the structure of brain tissue without analysis at the molecular level18, with low spatial resolution and with limited types of substances that can be monitored19. On the other hand, intracerebral drug analysis often uses tissue homogenates or microdialysis sampling based high-performance liquid chromatography mass spectrometry (HPLC‒MS) technology. The pre-processing of tissue sample is invasive and can damage the brain tissue microregions. The obtained results thus reflect only the average metabolic level of the sampled microregion20,21 with no information on the spatial distribution of the molecule throughout the brain.
Mass spectrometry imaging (MSI) has received great attention and has been rapidly developed in recent years. Compared with other imaging technologies (histochemical labeling, immunofluorescence, MRI, PET, whole body autoradiography, etc.), it is a molecular imaging technology and does not require complicated pre-processing and specific chemical labeling22, 23, 24, 25. MSI has become a powerful tool with high-throughput, high-sensitivity, and high-resolution that detects the qualitative and spatial distribution information of known or unknown molecules including peptides, proteins, endogenous metabolites, or exogenous drugs26, 27, 28. MSI has been applied to reveal asymmetrical spatial distribution of lipid metabolites from bisphenol S induced nephrotoxicity29, aristolochic acid-induced nephrotoxicity, tissue-specific metabolic reprogramming in diabetic nephropathy30,31, to determine glucose metabolism in different regions of the brain32, and to reveal the biologically relevant correlation between cholesterol and other metabolites in the brain33. In particular, we developed air flow-assisted desorption electrospray ionization (AFADESI)-MSI technology with high sensitivity, wide metabolites coverage, which can map thousands of structure-specific and functional metabolites in an untargeted analysis34. This technology is applied in many areas, such as in situ biomarker discovery for cancer diagnosis35, tumor metabolism research36, and elucidating mechanism of drug action37. A series of MSI techniques such as matrix-assisted laser desorption ionization (MALDI) MSI38,39, liquid extraction surface analysis tandem mass spectrometry (LESA‒MS/MS)40, and desorption electrospray ionization (DESI) MS41 have been reported to quantify OLZ from bio-tissue. There is still an urgent need for an imaging application method that can simultaneously characterize the pharmacokinetics and pharmacodynamics of brain microregions to facilitate CNS drug development.
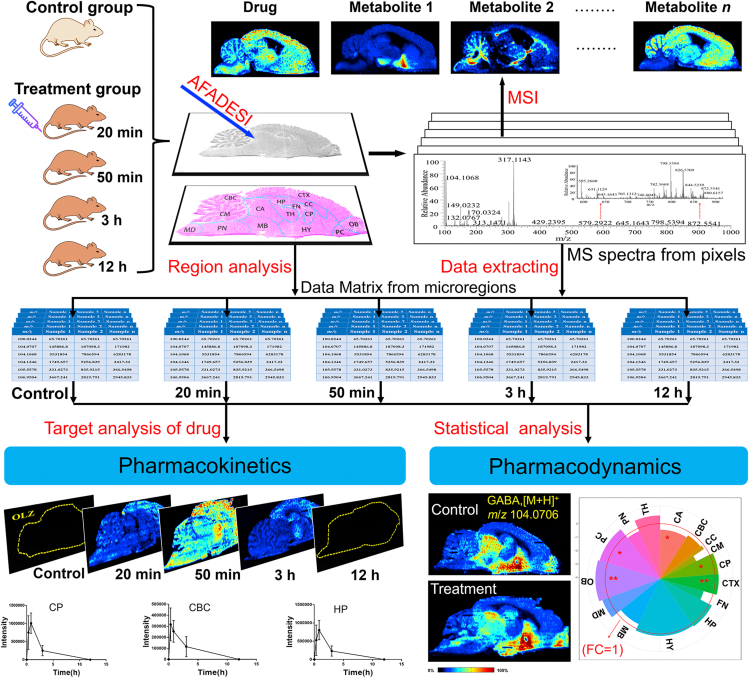
Herein, we developed a temporo-spatial pharmacometabolomics method, based on the air flow-assisted desorption electrospray ionization mass spectrometry imaging (AFADESI-MSI) system coupled with a high-resolution mass spectrometer, which can provide high-coverage molecular profile and microregional distribution information. The research strategy is shown in Scheme 1. This method was validated with a case study on the temporo-spatial changing of the levels of olanzapine (OLZ), a CNS drug used in clinical practice, and the endogenous metabolites in the rat brain over the period of administration and the analysis of pharmacokinetics and pharmacodynamics of the drug in the brain microregions. Overall, this method was successful at demonstrating the temporo-spatial characteristics of OLZ and its action-related metabolites and provided new insights into the molecular mechanism of action of CNS drugs.
Scheme 1.
Strategy and workflow of the proposed temporo-spatial pharmacometabolomics method to characterize the microregional pharmacokinetics and pharmacodynamics of CNS drugs in the brain.
2. Materials and methods
2.1. Chemicals and reagents
HPLC-grade acetonitrile was purchased from Merck (Darmstadt, Germany). Purified water was obtained from Wahaha (Hangzhou, China). Formic acid was provided by Millipore (Merck, Darmstadt, Germany), and olanzapine (OLZ) bulk drug powder (Supporting Information Fig. S1) was purchased from Shanghai Yuan Ye Biotechnology Co., Ltd., China.
2.2. Animal experiments
The animal experiments were conducted with the approval of the Animal Ethical Committee at the Institute of Materia Medical, Chinese Academy of Medical Science, and Peking Union Medical College (Beijing, China). Male Sprague–Dawley rats (weighing 180–200 g) were purchased from Vital River Laboratory Animal Technology Company (Beijing, China). The animals were maintained for a week with unlimited standard food and water at a constant room temperature of 22 ± 2 °C and 45%–55% humidity under a 12 h light/dark cycle. The rats were randomly divided into two groups: treatment group and control group. The treatment group was further divided into 4 groups of 3 animals each, which were orally administered OLZ solutions (50 mg/mL) prepared by mixing OLZ powder with saline. Rats were euthanized with high concentrations of ether at 20 min, 50 min, 3 h, and 12 h after administration (n = 3 at each time point). The control group (n = 3) were given an equal volume of saline by the gavage route. The whole brain of each animal was collected and stored at −80 °C until sectioning. The general brain perfusion with saline for blood‒brain barrier (BBB) permeable drugs was not performed to keep the original metabolic status for metabolic analysis and to avoid the loss of unstable endogenous metabolites, such as neurotransmitters (NTs).
2.3. Preparation of brain tissue sections
The brain tissue sections were sliced to consecutive sagittal slices of 15 μm using a Leica CM1860 Cryostat Microtome (Leica Microsystem Ltd., Germany) at −20 °C, and the tissue sections were thaw-mounted on positively charged desorption plates (Thermo Scientific, CA, USA) and stored in closed slice boxes at −80 °C until further analysis. The frozen slices were dried in a vacuum desiccator at −20 °C for 1 h and then kept at room temperature for another 1 h before AFADESI-MSI analysis. Meanwhile, the serial brain tissue sections were fixed with 4% paraformaldehyde and subjected to hematoxylin‒eosin (H&E) staining.
2.4. AFADESI-MSI analysis
The MSI experiments were performed using an AFADESI-MSI platform, which consisted of a home-built ambient AFADESI ion source and a Q Orbitrap Mass Analyzer (Q Exactive, Thermo Fisher Scientific, CA, USA)42,43. The key parameters included spray voltage, ion transfer tube voltage, solvent flow rate, and transporting gas flow rate. The SC100 Series Stepper Motor (Beijing Optical Century Instrument Co., Beijing, China) was used to control the movement of the brain tissue sections. The parameters were set in the self-designed control platform program. The experiments were achieved by continuously scanning the brain tissue surface in the x direction at a constant velocity of 160 μm/s, with a vertical step separation of 200 μm between adjacent lines in the y direction. A mixed solution of acetonitrile:water (8:2, v/v) was used as the optimized spray solvent with a flow rate of 5 μL/min. The MSI analysis was performed in positive and negative ion modes on a Q-Exactive mass spectrometer. A detailed parameter setting is shown in Supporting Information Table S1.
2.5. Data processing and statistical analysis
The raw data from the AFADESI-MS analysis was converted to cdf format files using Xcalibur 2.3 (Thermo Scientific, USA) and then imported into the custom-developed graphical software MassImager (Version 1.0, Chemmind Technologies Co., Ltd., Beijing, China) for ion image reconstruction44. After background subtraction, each section was normalized to the sample total ion chromatograms (TIC) value using MassImager, and the ion images were finally presented using the TIC normalized intensity threshold. MS profiles from the microregions of brain were precisely extracted from the MSI data by matching ion images with high-spatial resolution H&E staining images of the adjacent brain section, and each brain microregion profile was delineated and calculated 3 times from the subregions to reduce error. The average ion intensity of the microregion was calculated to generate separated two-dimensional data matrixes (m/z, ion intensity) in the.txt format. The mass tolerance for peak pick and background subtraction was set to 0.005 Da, and the proportional coefficient k was set to 1. Thereafter, the.txt file was imported into Markerview 1.2.1 (AB SCIEX, USA) for peak alignment and isotope ion deletion. Multivariate statistical analysis was carried out using SIMCA-P 14.0 (Umetrics AB, Umeå, Sweden), which included principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA). The metabolic profiles of the control and treatment groups were compared by performing supervised multivariate OPLS-DA to achieve the maximal separation. The variable importance in projection (VIP) value was calculated for each ion to indicate their contribution to the OPLS-DA model. Analysis of variance (ANOVA) was used to compare the differences between the control and treatment groups. P < 0.05 indicates a significant difference relative to the control group. Therefore, variables, which satisfied the following conditions, were considered to be potential metabolites: variables with a VIP value ≥ 1.0 and P < 0.05 in independent t-test. Finally, the isotope and adduct ions were deleted according to the corresponding extracted ion chromatograms (XICs). The area under the curve (AUC) was calculated using GraphPad Prism 6.0. software.
2.6. Analyte identification
The adducted ions of potential biomarkers were compared with the public databases including the Human Metabolome Database (www.hmdb.ca), Metlin (https://metlin.scripps.edu), MassBank (http://www.massbank.jp/), and LIPID MAPS (www.lipidmaps.org), using exact molecular weights with a mass error of less than 5 ppm and isotope patterns, combined with the isotopic abundance of high-resolution mass spectra that provide the elemental composition and likelihood list of the endogenous metabolites45. Then, the high-resolution MS/MS spectra were used for further identification. The high-resolution tandem MS experiments were performed on an orbitrap mass spectrometer (Q Exactive, Thermo Scientific, Bremen, Germany) and conducted directly from brain sections to obtain structural information based on the interpretation of the metabolites’ fragmentation patterns and database searches. For in situ AFADESI-MS/MS analysis, the resolution of the MS/MS acquisition was set to 70,000 with an AGC value of 5e6 and a maximum injection time of 200 ms. The ions of interest were considered as targets, and the NCE values were set to 25%, 35%, and 45% in the targeted MS2 scan mode.
3. Results and discussion
3.1. Mapping the drug and endogenous metabolites in the rat brain
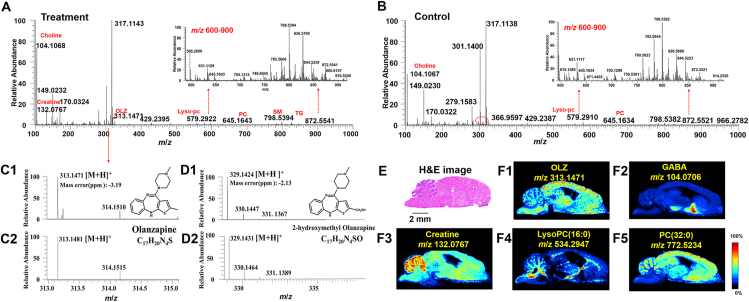
AFADESI-MSI is a powerful imaging tool used to map the exogenous compounds and endogenous metabolites from the brain tissue section. The representative mass spectra acquired using AFADESI-MSI from treatment and control groups brain tissue sections in positive ion mode is shown in Fig. 1A, B and that in negative ion mode is shown in Fig. S1A and B. The average mass spectrum showed that the endogenous metabolite ions detected in different brain microregions are diverse and the intensities differed greatly. In the low mass range of 100–500 Da, amino acids, nucleosides, nucleotides, small molecular organic acids, and some background ions could be detected; in the high mass range of 500–1000 Da, some lipid components, including sphingomyelin (SM), phosphatidylethanolamine (PE), phosphatidylcholine (PC), and some lysophosphatidylcholine (LysoPC) were detected. Components such as γ-aminobutyric acid (GABA, m/z 104. 0706), creatine (m/z 132.0765), carnitine (m/z 162.1120), acetylcarnitine (m/z 204.1231), and phosphatidylcholine (m/z 786.5258) were detected in the positive ion mode, while glutamic acid (Glu, m/z 146.0461), fatty acid (18:1) (m/z 281.2486), fatty acid (20:4) (m/z 303.2330), ascorbic acid (m/z 175.0276), and phosphatidylinositol (PI, m/z 885.5502) were detected in the negative ion mode. Ion images of these metabolites could be observed visually with their uneven microregional distribution information and the prominent structural profile of brain tissue (Fig. 1F1–F5, Fig. S1D1–S1D5) was characterized. The prototype drug OLZ and its metabolite 2-hydroxymethyl OLZ were detected in the positive ion mode, which was confirmed by their accurate mass assignment and identical isotopic peak shape. The results are shown in Fig. 1C1 and D1, and the theoretical matching isotopic peak is shown in Fig. 1C2 and D2. These results indicated that this non-targeted MSI method can simultaneously map the exogenous drugs and endogenous metabolites in a single experiment and can obtain their spatial distribution characteristics and microregional abundance variation.
Figure 1.
Imaging results for exogenous drug and endogenous metabolites obtained using AFADESI-MSI from brain tissue section. The representative mass spectra acquired using AFADESI-MSI from treatment (A) and control groups (B) brain tissue section in positive ion mode. C1 and D1 are experimental mass spectra of olanzapine (OLZ) and its metabolite, 2-hydroxymethyl OLZ, respectively. C2 and D2 are the calculated isotopic peaks of OLZ and 2-hydroxymethyl OLZ, respectively. (E) represents the hematoxylin-eosin (H&E) staining of sections of the brain. Scale bar = 2 mm. (F1)–(F5) show ion images of OLZ, γ-aminobutyric acid (GABA), Creatine, LysoPC (16:0), PC (32:0), respectively.
3.2. Temporo-spatial changes in OLZ and its metabolite in the brain
OLZ is a drug used to treat schizophrenia, with the brain being its main target organ. To explore the distribution of drugs in various functional microregions of the brain over the time of administration, the rat brain tissues of the treatment and control rats were collected at 20 min, 50 min, 3 h, and 12 h after drug administration for MSI analysis. As shown in Fig. 2A, 15 microregions of interests were focused on the caudate putamen (CP), cerebral cortex (CTX), hippocampus (HP), hypothalamus (HY), thalamus (TH), cerebellar cortex (CBC), cerebellar medulla (CM), medulla (MD), pons (PN), cerebral aqueduct (CA), middle brain (MB), fornix (FN), piriform cortex (PC), olfactory bulb (OB), and corpus callosum (CC). The microstructure of the rat brain samples is shown in Supporting Information Fig. S2.
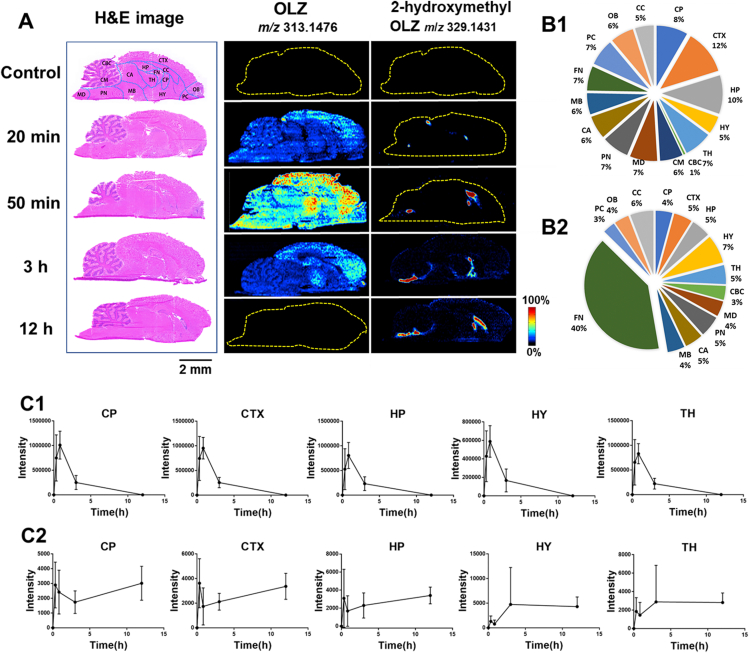
Figure 2.
Temporo-spatial alteration of OLZ and 2-hydroxymethyl OLZ in brain microregions. (A) MS images of OLZ and 2-hydroxymethyl OLZ in brain tissue section acquired using AFADESI-MSI. (B1, B2) The percentage of AUC in OLZ and 2-hydroxymethyl OLZ in different brain microregions. (C1, C2) The relative intensity changes of OLZ and 2-hydroxymethyl OLZ in the different brain microregions with the time. Data are presented as means ± standard deviation (SD), n = 3. CP: caudate putamen, CTX: cerebral cortex, HP: hippocampus, HY: hypothalamus, TH: thalamus, CBC: cerebellar cortex, CM: cerebellar medulla, MD: medulla, PN: pons, CA: cerebral aqueduct, MB: middle brain, FN: fornix, PC: piriform cortex, OB: olfactory bulb, and CC: corpus callosum.
From the MS images of the brain sections (Fig. 2A), the spatial distributions of the prototype drug OLZ and its metabolite 2-hydroxymethyl OLZ were detected. These results showed that OLZ can readily penetrate the BBB, largely dispersed in the ventricles and brain parenchymal tissue but not uniformly distributed in all microregions of the brain, to act on the CNS. It was found that OLZ is distributed mainly in the CTX 20 min after administration. After 50 min, the levels of OLZ increased significantly. With the progression of time, the drug signal in the brain quickly dropped below the imaging detection limit. Simultaneously, it was found that 2-hydroxymethyl OLZ, mainly distributed in the FN, had a different distribution pattern in the various microregions than that of OLZ. There was a significant increase in abundance of 2-hydroxymethyl OLZ 20 min after administration, and the metabolite was still present in high concentrations after 12 h of administration. Furthermore, the percentage of AUC of OLZ (Fig. 2B1) was similar in different microregions of the brain, except in CBC that was smaller, which indicates that the degree of drug absorption is similar in brain microregions, and less in the CBC. The percentage of AUC of 2-hydroxymethyl OLZ (Fig. 2B2) in FN was the largest, indicating that the exposure was the largest in the FN microregion. The metabolic exposure was similar in the other microregions. Additionally, the changes in relative intensity of OLZ in the different brain microregions with time (Fig. 2C1 and Supporting Information Fig. S3A) showed that in the CBC, CM, MD, PN, CA, MB, PC, OB, and CC, the highest level of OLZ was reached at 20 min after administration and then gradually decreased, while in the CP, CTX, HP, HY, TH, and FN, OLZ reached the highest level at 50 min after administration. Fig. 2C2 and Fig. S3B show that the level of 2-hydroxymethyl OLZ increased with time lapsed after administration in 12 of the microregions of the brain and increased dramatically in the PN.
These results indicated that the rate of drug absorption, distribution, and metabolism differ in various microregions of the brain, suggesting microregional effects on pharmacokinetics. It also demonstrated that the proposed temporo-spatial pharmacometabolomics method based on AFADESI-MSI has the ability to simultaneously illustrate changes in the levels of and spatial distribution of drugs and their metabolites in the complicated microregions of the brain.
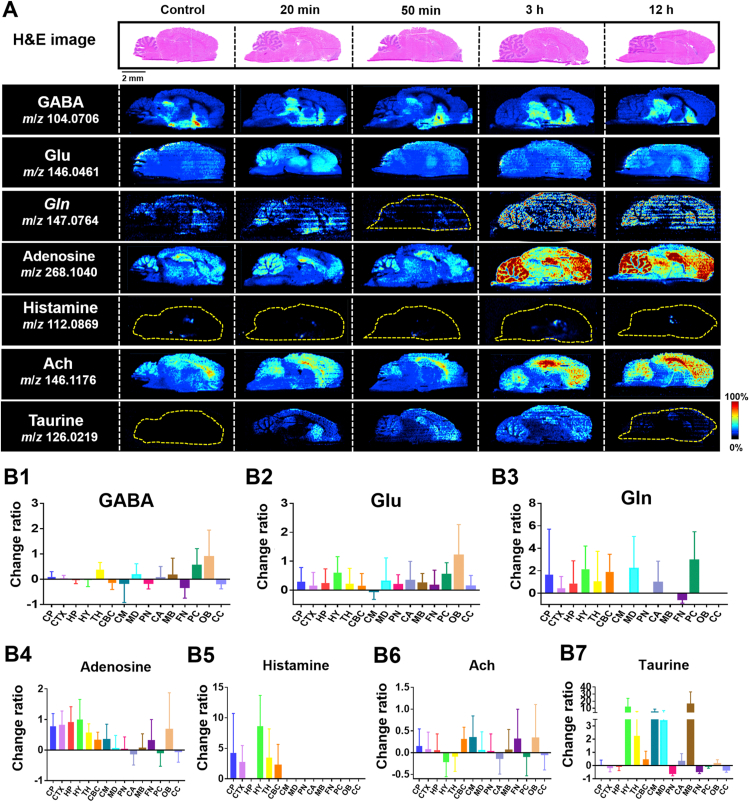
3.3. Microregional regulation of NTs by OLZ
OLZ is a first-line drug used in the clinical setting to treat schizophrenia. It acts mainly by regulating NTs. The known mechanism of action is by blocking the dopamine D2 receptor or the serotonin 2A receptor46,47. However, the microregional effect and the molecular mechanism of action of OLZ are still unclear. Therefore, we further analyzed the temporo-spatial changes of NTs that are closely related to the physiological activity of OLZ. As shown in Fig. 3A, a variety of NTs, such as GABA, Glu, glutamine (Gln), and adenosine, were detected. The AUC change ratio of NTs is shown in Fig. 3B1‒B7. Supporting Information Figs. S4‒S10 reflects the relative intensity changes of NTs in the different brain microregions with time. This mode of targeted analysis of functional metabolites from the non-targeted metabolic data has the advantage of not requiring predetermined parameters for the detection of metabolites.
Figure 3.
Drug effect on the distribution and AUC change ratio for NTs in brain. (A) represents the images of H&E-stained brain sections followed by the distribution of NTs in the brain acquired by AFADESI-MSI. (B1–B7) show the AUC change ratio of NTs. AUC change ratio = (AUC after drug‒AUC before drug)/AUC before drug. AUC change ratio >0 indicates an up-regulated status. AUC change ratio <0 indicates a down-regulated status. The larger absolute values of the AUC change ratio represent greater up-regulation or down-regulation with the drug action or effect.
GABA is a NT in the CNS that inhibits the nerve center, induces sleep, calmness, and prevents elevated body temperature by reducing neuronal activity to produce anti-anxiety effects48. The ion image showed that GABA (m/z 104.0706) was mainly distributed in the HY and was down-regulated after drug intervention. In the HY, the level of GABA decreased 20 min from drug administration and increased after 20 min up to 3 h. After 3 h from administration, the level of GABA gradually decreased (Fig. S4). The changed AUC ratio in the HY was −0.03-fold (Fig. 3B1). The minor variation in ratio indicates that GABA was lightly regulated after drug intervention in the HY. However, simultaneously, significant upregulation after drug intervention was observed in OB and PC, combined with the AUC changed ratio. Glu, a major neurotransmitter in the CNS, has an excitatory effect on nerve cells and can excite cells to death49. The temporal and spatial dynamic patterns of Glu and its metabolite, Gln, present a relatively consistent change trend in microregions after drug intervention (Figs. S5 and S6). Adenosine is widely distributed in the CNS and is considered to be an excitatory and inhibitory neurotransmitter in the brain50,51. It was unevenly distributed in various microregions, and the high levels of adenosine in the HP and HY increased significantly after 3 h from administration (Fig. S7), indicating that the up-regulation of adenosine would be more obvious when the drug accumulates. Histamine, as a central neurotransmitter, participates in the regulation of cognitive memory, sleep and wakefulness, diet, and other functions52,53. It was specifically distributed in the CP, CTX, HY, and TH and was up-regulated during drug action (Fig. S8). The effect of acetylcholine (Ach) on central neurons is mainly through the excitement and activation of brain nerve conduction function to enhance the brain's memory capacity54. Ach is highly distributed in the CTX and tends to increase with a prolonged administration (Fig. S9). Taurine promotes the growth and development of the nervous system and cell proliferation and differentiation55. It was observed that the levels of taurine increased after OLZ intervention (Fig. S10).
The results of the targeted imaging analysis of the above-mentioned NTs demonstrated that this method can detect the spatial distribution and variation in an abundance of prototype drugs, their metabolites, and endogenous metabolites related to the mechanism of action of CNS drugs. This is significant in elucidating the mechanism of action of CNS drugs and understanding schizophrenia and related diseases.
3.4. Microregional metabolic regulation with drug intervention by OLZ
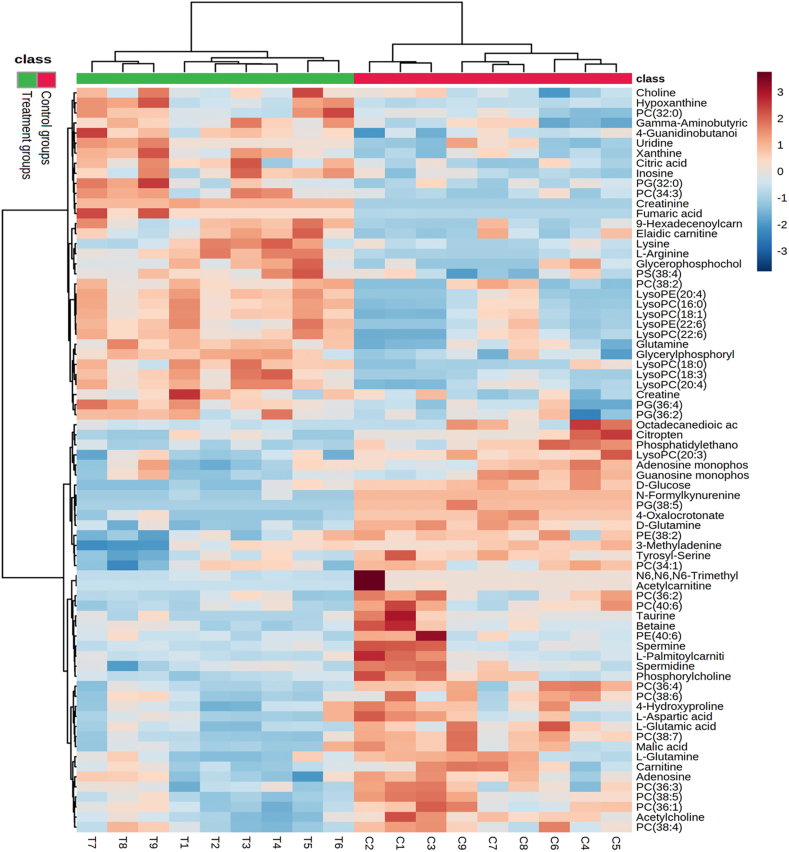
Endogenous metabolic changes in tissues and organs can reflect the effect of drug stimulation. To explore the microregional metabolic effects after drug intervention, the molecular profile of endogenous metabolites and the distribution information of their dynamic changes were investigated with pharmacometabolomic testing in various brain microregions. Three brain tissue samples from each group of treatment and control rats taken 50 min after OLZ and saline administration, respectively, were selected for microregional analysis. In order to verify the stability of the AFADESI-MSI analysis system, the blank area of the slice was selected as quality control, and data were analyzed by unsupervised PCA (Supporting Informaion Fig. S11A and S11B) and linear score graph (Fig. S11C‒S11F). The samples were in the 95% confidence interval, and the relative deviation was in the range of 2SD. Thereafter, OPLS-DA analysis was performed to observe the changes in the overall microregional metabolic profiles of brain metabolites in the control and treatment groups. The OPLS-DA results indicated that the control and treatment groups were clearly separated based on quantitative analysis in the brain microregions in positive and negative ion mode, respectively. The results are shown in Supporting Information Figs. S12 and S13. Discriminated metabolites were screened with variable importance for the projection (VIP) value ≥ 1, P < 0.05 in Student's t-tests, and further validated by the method described in section 2.6. A total of 90 differential endogenous metabolites were screened and identified to contribute greatly as drug action-associated effectors in the brain microregions (Supporting Information Table S2). Among them, 81 analytes were identified by MS2, while nine were identified by isotope pattern.
There were many types of differential metabolites including amino acids, fatty acids, glycerophospholipids, organic acids, polyamine, and acyl carnitines. First, hierarchical clustering analysis (HCA, Fig. 4) and correlation analysis (Supporting Information Fig. S14) were performed to visualize the change trend and association of different metabolites between the two groups using the MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/). The results indicated that the different metabolites have obvious clustering and grouping trends in the brain in the control and treatment groups.
Figure 4.
Hierarchical cluster analysis (HCA) of identified differential metabolites in control and treatment groups. The row on the right lists the metabolites, the column on the top indicates class (green: treatment groups and red: control groups), and the column on the bottom indicates sample ID (T: treatment groups sample and C: control groups sample). The metabolites were clustered, and shades of red and blue represent high expression levels and low expression levels, respectively.
Then, the discriminating metabolites were imported into MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/) and combined with Kyoto Encyclopedia of Genes and Genomes PATHWAY Database (https://www.genome.jp/kegg/pathway.html) to perform metabolic pathway matching analysis, facilitating the discovery of altered metabolic pathways56. This analysis suggested that multiple metabolic pathways were significantly dysregulated (Supporting Information Fig. S15 and Table S3). The impact-value threshold calculated from the pathway topology analysis was set to 0.10. Seven metabolic pathways significantly perturbed between treatment and control groups were identified, which included alanine, aspartate and glutamate metabolism, d-glutamine and d-glutamate metabolism, taurine and hypotaurine metabolism, starch and sucrose metabolism, glycerophospholipid metabolism, arginine and proline metabolism, arginine biosynthesis, purine metabolism, and citrate cycle (TCA cycle). To more accurately explore the changes in metabolic levels associated with the mechanism of action of OLZ, we focused on the abnormal metabolic pathways of alanine, aspartate, glutamate, and glycerophospholipid metabolism with parameters of large pathway impact (PI) and small Raw P.
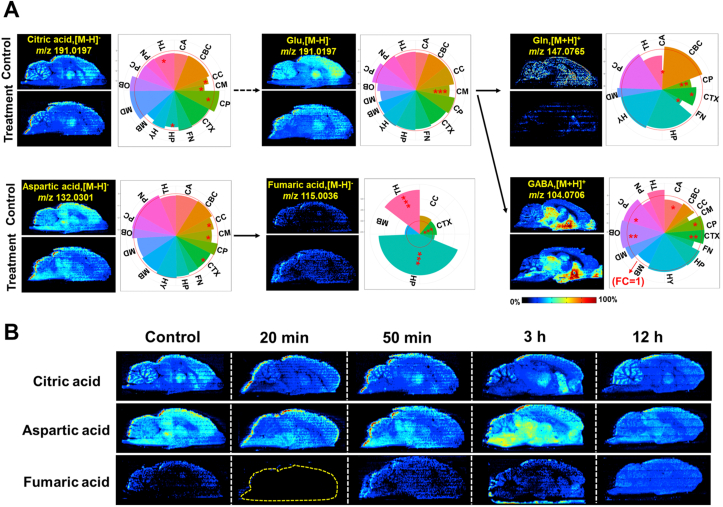
3.4.1. Disturbance in alanine, aspartate, and glutamate metabolism
Glu is synthesized from Gln by glutamine synthetase (GLS). Its clearance can prevent neuronal excitotoxicity caused by excessive activation of glutamate receptors57. Gln can generate Glu through GLS and enter the tricarboxylic acid (TCA) cycle to provide energy for cell life activities58. An abnormal Glu–Gln cycle plays an important role in the pathophysiological process of schizophrenia59. The temporo-spatial distributions of metabolites of the alanine, aspartate, and glutamate metabolism pathway are shown in Fig. 5. The AUC change ratio of metabolites is shown in Supporting Information Fig. S16. From the spatial distribution maps along with drug intervention, it was observed that the proportion of the key metabolites varied in different microregions of the brain and their fold change after drug intervention showed trends of up-regulation or down-regulation. Among them, citric acid was evenly distributed in most of the microregions of the brain; the level was significantly increased compared with those in control. It is well known that citric acid is an intermediate of the TCA cycle, which is the final metabolic pathway for the complete oxidation of sugar, fat, and protein in the body and is the main way for the body to obtain energy60,61. This result suggested that the level of citric acid was up-regulated after drug intervention, indicating that drug intervention accelerated the metabolism of TCA cycle and provided more energy to the body. Glu was also evenly distributed in various microregions, showing a down-regulation after drug intervention. Its metabolism produced Gln and GABA, which were up-regulated mainly in the HY and in multiple microregions of the brain, respectively.
Figure 5.
Temporo-spatial distribution of metabolites of the alanine, aspartate, and glutamate metabolism pathway. (A) Spatial distribution and changes of metabolites involved in the alanine, aspartate, and glutamate metabolism pathway in the treatment (50 min after dosing) and control groups. (B) MS images of metabolites of the alanine, aspartate, and glutamate metabolism pathways in brain tissue section acquired using AFADESI-MSI. Statistical analysis of the metabolome data extracted from control and treatment groups (50 min) in different brain microregions. (means ± SD), n = 3. ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001. The red circle in the pie chart represents the conversion of log2 (1) equal to 0 when the fold change of metabolite before and after administration is 1 (FC = 1). The sector outside the red circle represents the fold change greater than 1 (FC > 1) and indicates this metabolite with up-regulation after drug intervention in this microregion, while the sector inside the red circle represents the fold change less than 1 (FC < 1) and indicates this metabolite with down-regulation after drug intervention in this microregion. The distance of the radius of the sector from the red circle represents the degree of fold change. Outside the red circle, a larger radius of the sector represents more pronounced up-regulation, and within the red circle, a smaller radius of the sector represents more significant down-regulation. The size of the rounding angle represents the relative abundance in the microregion; the larger the rounding angle, the greater the relative abundance.
According to the above pathway analysis and the metabolic glutamate decarboxylase (GAD) enzyme reaction, it could be speculated that OLZ directly activated GAD to promote the synthesis of GABA62. GABA increases the activity of hexokinase in glycolysis, thereby accelerating the metabolism of glucose, increasing the supply of blood and oxygen to the brain, and promoting sleep63, 64, 65. The distribution of glucose in brain is presented in Supporting Information Fig. S17. The spatial distribution shows that glucose is distributed in all microregions of the brain but primarily in the OB and PC after administration. There was a significant increase in glucose levels 20 min after administration.
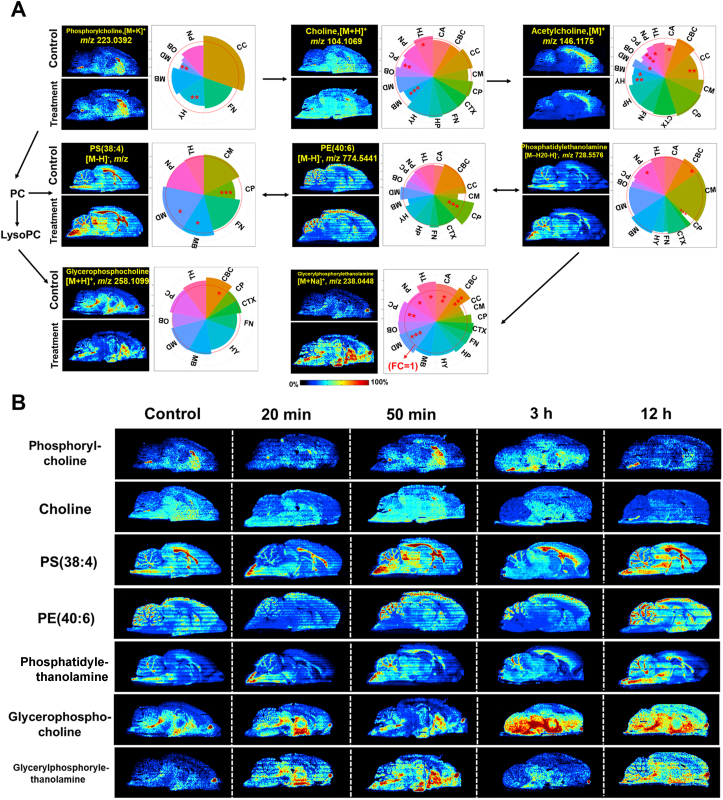
3.4.2. Disturbance of the glycerophospholipid metabolism pathway
Lipids have a variety of biological functions, including material transportation, energy metabolism, information recognition and transmission, cell development, differentiation, and apoptosis66. Glycerophospholipids help to control liver lipid metabolism, promote memory, enhance immunity, and delay aging67. Lipids may participate in the regulation of schizophrenia by regulating the integrity of the myelin sheath and neuronal function68. Lipid metabolism is associated with brain cognitive function69. Several studies have confirmed that independent phospholipase A2β (iPLA2β) is a novel ferroptosis regulator, and the associated mutations may be involved in the pathological process of Parkinson's disease, which is mainly related to the insufficient repair of oxidized phospholipids70,71. The temporo-spatial distribution of metabolites of the glycerophospholipid metabolism pathway is shown in Fig. 6. The AUC change ratio of metabolites is shown in Supporting Information Fig. S18. The highest levels of phosphorylcholine were seen in the CC and were significantly upregulated. PS (38:4) was significantly downregulated in the CP and upregulated in the CM, MB, and MD. Glycerophosphocholine showed an up-regulation in the CBC. Upregulated PE (40:6) was observed in the CP. Phosphatidylethanolamine was significantly downregulated in the CP, and the levels were the highest in the CM. Glycerylphosphorylethanolamine showed an upregulation in most microregions. The results of this study showed that after the administration of OLZ, most of the lipids showed an up-regulation in majority of the microregions. OLZ has metabolic side effects in clinical applications, such as weight gain, dyslipidemia, hypertriglyceridemia, and insulin resistance. These experimental results proved that the upregulation of lipid metabolism may contribute to these side effects during treatment with OLZ, and further confirmation is needed about the biological significance of more lipid substances.
Figure 6.
Temporo-spatial distribution of metabolites of the glycerophospholipid metabolism pathway. (A) Spatial distribution and changes of metabolites involved in the glycerophospholipid metabolism pathway in the treatment (50 min after dosing) and control groups. (B) MS images of metabolites of the glycerophospholipid metabolism pathway in brain tissue section acquired using AFADESI-MSI. Statistical analysis was performed using Student's t-test. ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001.
These results demonstrated that the temporo-spatial pharmacometabolomics method was effective in identifying the endogenous molecular effectors associated with drug action. The data revealed targeted and non-targeted metabolic alterations with variation in their abundance while also illustrating their spatial distribution information to show the precise location in the microregions targeted by the drug. This method is, therefore, significant in understanding the pharmacodynamics of CNS drugs.
4. Conclusions
We developed a temporo-spatial pharmacometabolomics method to characterize the pharmacokinetics and pharmacodynamics of CNS drugs in the brain using ambient MSI. Compared with LC‒MS based pharmacometabolomics, AFADESI-MSI based temporo-spatial pharmacometabolomics has the advantage of detecting the static level changes of endogenous and exogenous substances and providing dynamic time-dependency trends and spatial distribution information in the different microregions of the brain, accurately presenting in situ and microregional molecular events. Therefore, correlation of pharmacokinetics and pharmacodynamics with metabolic pathways at the level of systems biology is conducive for obtaining critical information for deeper understanding of the molecular mechanisms of drug action.
The temporo-spatial pharmacometabolomics method developed in this study was applied to evaluate the microregional effect of OLZ on brain tissue and demonstrate its effectiveness in characterizing the microregional pharmacokinetics and pharmacodynamics of CNS drugs crossing the blood–brain-barrier. This method clearly presented the pharmacokinetics of the prototype drug and its metabolite 2-hydroxymethyl OLZ in different microregions of the rat brain. Moreover, the microregional pharmacodynamics of OLZ was illustrated through mapping metabolic pathways. It was found that some NTs, including GABA, adenosine, and histamine, displayed multiple metabolic pathways related to drug intervention. Furthermore, the regulation of aspartate, glutamate, and the glycerophospholipid metabolism pathway could be associated with the therapeutic and adverse effects observed with the clinical usage of OLZ, which provides critical information for understanding the molecular mechanism of its action.
In summary, the temporo-spatial pharmacometabolomics method provided a comprehensive and effective tool for elucidating the in situ pharmacokinetics and pharmacodynamics of CNS drugs.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant Nos. 81773678 and 81974500) and the CAMS Innovation Fund for Medical Sciences (CIFMS, 2021-I2M-1-026, China).
Author contributions
Jiuming He conceived and designed the experiments and revised the manuscript. Dan Liu designed and performed experiments, analyzed and interpreted the data, and drafted the manuscript. Jianpeng Huang and Shanshan Gao helped with the animal experiments. Hongtao Jin helped with manuscript editing. All authors have read and approved the final manuscript before submission.
Conflicts of interest
The authors declare no conflict of interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.03.018.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Park H.J., Friston K. Structural and functional brain networks: from connections to cognition. Science. 2013;342:1238411. doi: 10.1126/science.1238411. [DOI] [PubMed] [Google Scholar]
- 2.Hassabis D., Maguire E.A. The construction system of the brain. Philos Trans R Soc Lond B Biol Sci. 2009;364:1263–1271. doi: 10.1098/rstb.2008.0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao R., Pollack G.M. Regional differences in capillary density, perfusion rate, and P-glycoprotein activity: a quantitative analysis of regional drug exposure in the brain. Biochem Pharmacol. 2009;78:1052–1059. doi: 10.1016/j.bcp.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Pang X., Gao S., Ga M., Zhang J., Luo Z., Chen Y., et al. Mapping metabolic networks in the brain by ambient mass spectrometry imaging and metabolomics. Anal Chem. 2021;93:6746–6754. doi: 10.1021/acs.analchem.1c00467. [DOI] [PubMed] [Google Scholar]
- 5.Shou W.Z. Current status and future directions of high-throughput ADME screening in drug discovery. J Pharm Anal. 2020;10:201–208. doi: 10.1016/j.jpha.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo H.H., Feng C.L., Zhang W.X., Luo Z.G., Zhang H.J., Zhang T.T., et al. Liver-target nanotechnology facilitates berberine to ameliorate cardio-metabolic diseases. Nat Commun. 2019;10:1981. doi: 10.1038/s41467-019-09852-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan N.U., Ni J., Ju X., Miao T., Chen H., Han L. Escape from abluminal LRP1-mediated clearance for boosted nanoparticle brain delivery and brain metastasis treatment. Acta Pharm Sin B. 2021;11:1341–1354. doi: 10.1016/j.apsb.2020.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayet-Robert M., Morvan D., Chollet P., Barthomeuf C. Pharmacometabolomics of docetaxel-treated human MCF7 breast cancer cells provides evidence of varying cellular responses at high and low doses. Breast Cancer Res Treat. 2010;120:613–626. doi: 10.1007/s10549-009-0430-1. [DOI] [PubMed] [Google Scholar]
- 9.Gao Y., Li W., Chen J., Wang X., Lv Y., Huang Y., et al. Pharmacometabolomic prediction of individual differences of gastrointestinal toxicity complicating myelosuppression in rats induced by irinotecan. Acta Pharm Sin B. 2019;9:157–166. doi: 10.1016/j.apsb.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D., Li D., Zhang Y., Chen J., Zhang Y., Liao C., et al. Functional metabolomics reveal the role of AHR/GPR35 mediated kynurenic acid gradient sensing in chemotherapy-induced intestinal damage. Acta Pharm Sin B. 2021;11:763–780. doi: 10.1016/j.apsb.2020.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willmann J.K., van Bruggen N., Dinkelborg L.M., Gambhir S.S. Molecular imaging in drug development. Nat Rev Drug Discov. 2008;7:591–607. doi: 10.1038/nrd2290. [DOI] [PubMed] [Google Scholar]
- 12.Logothetis N.K. What we can do and what we cannot do with fMRI. Nature. 2008;453:869–878. doi: 10.1038/nature06976. [DOI] [PubMed] [Google Scholar]
- 13.Lan Y., Bai P., Chen Z., Neelamegam R., Placzek M.S., Wang H., et al. Novel radioligands for imaging sigma-1 receptor in brain using positron emission tomography (PET) Acta Pharm Sin B. 2019;9:1204–1215. doi: 10.1016/j.apsb.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drake L.R., Brooks A.F., Stauff J., Sherman P.S., Arteaga J., Koeppe R.A., et al. Strategies for PET imaging of the receptor for advanced glycation endproducts (RAGE) J Pharm Anal. 2020;10:452–465. doi: 10.1016/j.jpha.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox M.D., Raichle M.E. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8:700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y., Zheng T., Fan L.I. Diagnosis of elderly brain atrophy patients with mental disorder according to their CT image. Chin J Geriatric Heart Brain and Vessel Diseases. 2017;19:1299–1301. [Google Scholar]
- 17.Kolla N.J., Houle S. Single-photon emission computed tomography and positron emission tomography studies of antisocial personality disorder and aggression: a targeted review. Curr Psychiatr Rep. 2019;21:24. doi: 10.1007/s11920-019-1011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brahimaj B.C., Kochanski R.B., Pearce J.J., Guryildirim M., Gerard C.S., Kocak M., et al. Structural and functional imaging in glioma management. Neurosurgery. 2021;88:211–221. doi: 10.1093/neuros/nyaa360. [DOI] [PubMed] [Google Scholar]
- 19.Ganesana M., Lee S.T., Wang Y., Venton B.J. Analytical techniques in neuroscience: recent advances in imaging, separation, and electrochemical methods. Anal Chem. 2017;89:314–341. doi: 10.1021/acs.analchem.6b04278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theodoridis G., Gika H.G., Wilson I.D. LC‒MS-based methodology for global metabolite profiling in metabonomics/metabolomics. Trac Trends Anal Chem. 2008;27:251–260. [Google Scholar]
- 21.Gika H.G., Theodoridis G.A., Plumb R.S., Wilson I.D. Current practice of liquid chromatography‒mass spectrometry in metabolomics and metabonomics. J Pharm Biomed Anal. 2014;87:12–25. doi: 10.1016/j.jpba.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 22.Castaing R., Slodzian G. Microanalyse par émission ionique secondaire. J Microsc. 1962:I960. [Google Scholar]
- 23.Castaing R., Slodzian G. Microanalysis using secondary ion emission. J Mass Spectrom. 2021;56 doi: 10.1002/jms.4800. [DOI] [PubMed] [Google Scholar]
- 24.Wiseman J.M., Ifa D.R., Venter A., Cooks R.G. Ambient molecular imaging by desorption electrospray ionization mass spectrometry. Nat Protoc. 2008;3:517–524. doi: 10.1038/nprot.2008.11. [DOI] [PubMed] [Google Scholar]
- 25.Takáts Z., Wiseman J.M., Gologan B., Cooks R.G. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 26.Swales J.G., Dexter A., Hamm G., Nilsson A., Strittmatter N., Michopoulos F., et al. Quantitation of endogenous metabolites in mouse tumors using mass-spectrometry imaging. Anal Chem. 2018;90:6051–6058. doi: 10.1021/acs.analchem.7b05239. [DOI] [PubMed] [Google Scholar]
- 27.Caprioli R.M., Farmer T.B., Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 28.Wang S.S., Wang Y.J., Zhang J., Sun T.Q., Guo Y.L. Derivatization strategy for simultaneous molecular imaging of phospholipids and low-abundance free fatty acids in thyroid cancer tissue sections. Anal Chem. 2019;91:4070–4076. doi: 10.1021/acs.analchem.8b05680. [DOI] [PubMed] [Google Scholar]
- 29.Zhao C., Xie P., Yong T., Wang H., Chung A.C.K., Cai Z. MALDI-MS imaging reveals asymmetric spatial distribution of lipid metabolites from bisphenol S-induced nephrotoxicity. Anal Chem. 2018;90:3196–3204. doi: 10.1021/acs.analchem.7b04540. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z., He B., Liu Y., Huo M., Fu W., Yang C., et al. In situ metabolomics in nephrotoxicity of aristolochic acids based on air flow-assisted desorption electrospray ionization mass spectrometry imaging. Acta Pharm Sin B. 2020;10:1083–1093. doi: 10.1016/j.apsb.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z., Fu W., Huo M., He B., Liu Y., Tian L., et al. Spatial-resolved metabolomics reveals tissue-specific metabolic reprogramming in diabetic nephropathy by using mass spectrometry imaging. Acta Pharm Sin B. 2021;11:3665–3677. doi: 10.1016/j.apsb.2021.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleinridders A., Ferris H.A., Reyzer M.L., Rath M., Soto M., Manier M.L., et al. Regional differences in brain glucose metabolism determined by imaging mass spectrometry. Mol Metabol. 2018;12:113–121. doi: 10.1016/j.molmet.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X., Hou Y., Hou Z., Xiong W., Huang G. Mass spectrometry imaging of brain cholesterol and metabolites with trifluoroacetic acid-enhanced desorption electrospray ionization. Anal Chem. 2019;91:2719–2726. doi: 10.1021/acs.analchem.8b04395. [DOI] [PubMed] [Google Scholar]
- 34.He J., Sun C., Li T., Luo Z., Huang L., Song X., et al. A sensitive and wide coverage ambient mass spectrometry imaging method for functional metabolites based molecular histology. Adv Sci. 2018;5:1800250. doi: 10.1002/advs.201800250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li T., He J., Mao X., Bi Y., Luo Z., Guo C., et al. In situ biomarker discovery and label-free molecular histopathological diagnosis of lung cancer by ambient mass spectrometry imaging. Sci Rep. 2015;5:14089. doi: 10.1038/srep14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun C., Li T., Song X., Huang L., Zang Q., Xu J., et al. Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc Natl Acad Sci U S A. 2019;116:52–57. doi: 10.1073/pnas.1808950116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He J., Luo Z., Huang L., He J., Chen Y., Rong X., et al. Ambient mass spectrometry imaging metabolomics method provides novel insights into the action mechanism of drug candidates. Anal Chem. 2015;87:5372–5379. doi: 10.1021/acs.analchem.5b00680. [DOI] [PubMed] [Google Scholar]
- 38.Hamm G., Bonnel D., Legouffe R., Pamelard F., Delbos J.M., Bouzom F., et al. Quantitative mass spectrometry imaging of propranolol and olanzapine using tissue extinction calculation as normalization factor. J Proteonomics. 2012;75:4952–4961. doi: 10.1016/j.jprot.2012.07.035. [DOI] [PubMed] [Google Scholar]
- 39.Taylor A.J., Dexter A., Bunch J. Exploring ion suppression in mass spectrometry imaging of a heterogeneous tissue. Anal Chem. 2018;90:5637–5645. doi: 10.1021/acs.analchem.7b05005. [DOI] [PubMed] [Google Scholar]
- 40.Swales J.G., Strittmatter N., Tucker J.W., Clench M.R., Webborn P.J.H., Goodwin R.J.A. Spatial quantitation of drugs in tissues using liquid extraction surface analysis mass spectrometry imaging. Sci Rep. 2016;6:37648. doi: 10.1038/srep37648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swales J.G., Tucker J.W., Strittmatter N., Nilsson A., Cobice D., Clench M.R., et al. Mass spectrometry imaging of cassette-dosed drugs for higher throughput pharmacokinetic and biodistribution analysis. Anal Chem. 2014;86:8473–8480. doi: 10.1021/ac502217r. [DOI] [PubMed] [Google Scholar]
- 42.He J., Tang F., Luo Z., Chen Y., Xu J., Zhang R., et al. Air flow assisted ionization for remote sampling of ambient mass spectrometry and its application. Rapid Commun Mass Sp. 2011;25:843–850. doi: 10.1002/rcm.4920. [DOI] [PubMed] [Google Scholar]
- 43.Lv Y., Li T., Guo C., Sun C., Tang F., Huang L., et al. A high-performance bio-tissue imaging method using air flow-assisted desorption electrospray ionization coupled with a high-resolution mass spectrometer. Chin Chem Lett. 2019;30:461–464. [Google Scholar]
- 44.He J., Huang L., Tian R., Li T., Sun C., Song X., et al. MassImager: a software for interactive and in-depth analysis of mass spectrometry imaging data. Anal Chim Acta. 2018;1015:50–57. doi: 10.1016/j.aca.2018.02.030. [DOI] [PubMed] [Google Scholar]
- 45.Palmer A., Phapale P., Chernyavsky I., Lavigne R., Fay D., Tarasov A., et al. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat Methods. 2017;14:57–60. doi: 10.1038/nmeth.4072. [DOI] [PubMed] [Google Scholar]
- 46.Lieberman J.A., Bymaster F.P., Meltzer H.Y., Deutch A.Y., Duncan G.E., Marx C.E., et al. Antipsychotic drugs: comparison in animal models of efficacy, neurotransmitter regulation, and neuroprotection. Pharmacol Rev. 2008;60:358–403. doi: 10.1124/pr.107.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amato D., Canneva F., Cumming P., Maschauer S., Groos D., Dahlmanns J.K., et al. A dopaminergic mechanism of antipsychotic drug efficacy, failure, and failure reversal: the role of the dopamine transporter. Mol Psychiatr. 2020;25:2101–2118. doi: 10.1038/s41380-018-0114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mombereau C., Kaupmann K., Froestl W., Sansig G., van der Putten H., Cryan J.F. Genetic and pharmacological evidence of a role for GABA(B) receptors in the modulation of anxiety- and antidepressant-like behavior. Neuropsychopharmacology. 2004;29:1050–1062. doi: 10.1038/sj.npp.1300413. [DOI] [PubMed] [Google Scholar]
- 49.Zhou Y., Danbolt N.C. Glutamate as a neurotransmitter in the healthy brain. J Neural Transm (Vienna) 2014;121:799–817. doi: 10.1007/s00702-014-1180-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romanowska M., Komoszyński M. Adenosine‒neurotransmitter and neuromodulator in the central nervous system. Postepy Biochem. 2002;48:230–238. [PubMed] [Google Scholar]
- 51.Fredholm B.B., Chen J.F., Cunha R.A., Svenningsson P., Vaugeois J.M. Adenosine and brain function. Int Rev Neurobiol. 2005;63:191–270. doi: 10.1016/S0074-7742(05)63007-3. [DOI] [PubMed] [Google Scholar]
- 52.Yu X., Franks N.P., Wisden W. Sleep and sedative states induced by targeting the histamine and noradrenergic systems. Front Neural Circ. 2018;12:4. doi: 10.3389/fncir.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parmentier R., Ohtsu H., Djebbara-Hannas Z., Valatx J.L., Watanabe T., Lin J.S. Anatomical, physiological, and pharmacological characteristics of histidine decarboxylase knock-out mice: evidence for the role of brain histamine in behavioral and sleep-wake control. J Neurosci. 2002;22:7695–7711. doi: 10.1523/JNEUROSCI.22-17-07695.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grossberg S. Acetylcholine neuromodulation in normal and abnormal learning and memory: vigilance control in waking, sleep, autism, amnesia and Alzheimer's disease. Front Neural Circ. 2017;11:82. doi: 10.3389/fncir.2017.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Q., Liu Y., Wang H., Ma L., Xia H., Niu J., et al. The preventive effects of taurine on neural tube defects through the Wnt/PCP-Jnk-dependent pathway. Amino Acids. 2017;49:1633–1640. doi: 10.1007/s00726-017-2462-x. [DOI] [PubMed] [Google Scholar]
- 56.Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hertz L., Zielke H.R. Astrocytic control of glutamatergic activity: astrocytes as stars of the show. Trends Neurosci. 2004;27:735–743. doi: 10.1016/j.tins.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 58.Bak L.K., Schousboe A., Waagepetersen H.S. The glutamate/GABA-glutamine cycle: aspects of transport, neurotransmitter homeostasis and ammonia transfer. J Neurochem. 2006;98:641–653. doi: 10.1111/j.1471-4159.2006.03913.x. [DOI] [PubMed] [Google Scholar]
- 59.Kumar J., Liddle E.B., Fernandes C.C., Palaniyappan L., Hall E.L., Robson S.E., et al. Glutathione and glutamate in schizophrenia: a 7T MRS study. Mol Psychiatr. 2020;25:873–882. doi: 10.1038/s41380-018-0104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernie A.R., Carrari F., Sweetlove L.J. Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol. 2004;7:254–261. doi: 10.1016/j.pbi.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 61.Akram M. Citric acid cycle and role of its intermediates in metabolism. Cell Biochem Biophys. 2014;68:475–478. doi: 10.1007/s12013-013-9750-1. [DOI] [PubMed] [Google Scholar]
- 62.Hashimoto K., Bruno D., Nierenberg J., Marmar C.R., Zetterberg H., Blennow K., et al. Abnormality in glutamine-glutamate cycle in the cerebrospinal fluid of cognitively intact elderly individuals with major depressive disorder: a 3-year follow-up study. Transl Psychiatry. 2016;6:e744. doi: 10.1038/tp.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zarrindast M., Rostami P., Sadeghi-Hariri M. GABA(A) but not GABA(B) receptor stimulation induces antianxiety profile in rats. Pharmacol Biochem Behav. 2001;69:9–15. doi: 10.1016/s0091-3057(01)00518-4. [DOI] [PubMed] [Google Scholar]
- 64.Michaeli S., Fait A., Lagor K., Nunes-Nesi A., Grillich N., Yellin A., et al. A mitochondrial GABA permease connects the GABA shunt and the TCA cycle, and is essential for normal carbon metabolism. Plant J. 2011;67:485–498. doi: 10.1111/j.1365-313X.2011.04612.x. [DOI] [PubMed] [Google Scholar]
- 65.Pastorino J.G., Hoek J.B. Hexokinase II: the integration of energy metabolism and control of apoptosis. Curr Med Chem. 2003;10:1535–1551. doi: 10.2174/0929867033457269. [DOI] [PubMed] [Google Scholar]
- 66.Fahy E., Subramaniam S., Brown H.A., Glass C.K., Merrill A.H., Jr., Murphy R.C., et al. A comprehensive classification system for lipids. J Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 67.Gibellini F., Smith T.K. The Kennedy pathway‒de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life. 2010;62:414–428. doi: 10.1002/iub.337. [DOI] [PubMed] [Google Scholar]
- 68.Gouvêa-Junqueira D., Falvella A.C.B., Antunes A.S.L.M., Seabra G., Brandão-Teles C., Martins-de-Souza D., et al. Novel treatment strategies targeting myelin and oligodendrocyte dysfunction in schizophrenia. Front Psychiatr. 2020;11:379. doi: 10.3389/fpsyt.2020.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bowers M., Liang T., Gonzalez-Bohorquez D., Zocher S., Jaeger B.N., Kovacs W.J., et al. FASN-dependent lipid metabolism links neurogenic stem/progenitor cell activity to learning and memory deficits. Cell Stem Cell. 2020;27:98–109. doi: 10.1016/j.stem.2020.04.002. [DOI] [PubMed] [Google Scholar]
- 70.Sun W.Y., Tyurin V.A., Mikulska-Ruminska K., Shrivastava I.H., Anthonymuthu T.S., Zhai Y.J., et al. Phospholipase iPLA2β averts ferroptosis by eliminating a redox lipid death signal. Nat Chem Biol. 2021;17:465–476. doi: 10.1038/s41589-020-00734-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luo X., Gong H.B., Gao H.Y., Wu Y.P., Sun W.Y., Li Z.Q., et al. Oxygenated phosphatidylethanolamine navigates phagocytosis of ferroptotic cells by interacting with TLR2. Cell Death Differ. 2021;28:1971–1989. doi: 10.1038/s41418-020-00719-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.