Abstract
Mycobacterium tuberculosis (MTB) utilizes multiple mechanisms to obtain antibiotic resistance during the treatment of infections. In addition, the biofilms, secreted by MTB, can further protect the latter from the contact with drug molecules and immune cells. These self-defending mechanisms lay a formidable challenge to develop effective therapeutic agents against chronic and recurring antibiotic-tolerant MTB infections. Although several inexpensive and effective drugs (isoniazid, rifampicin, pyrazinamide and ethambutol) have been discovered for the treatment regimen, MTB continues to cause considerable morbidity and mortality worldwide. Antibiotic resistance and tolerance remain major global issues, and innovative therapeutic strategies are urgently needed to address the challenges associated with pathogenic bacteria. Gratifyingly, the cell wall synthesis of tubercle bacilli requires the participation of many enzymes which exclusively exist in prokaryotic organisms. These enzymes, absent in human hepatocytes, are recognized as promising targets to develop anti-tuberculosis drug. In this paper, we discussed the critical roles of potential drug targets in regulating cell wall synthesis of MTB. And also, we systematically reviewed the advanced development of novel bioactive compounds or drug leads for inhibition of cell wall synthesis, including their discovery, chemical modification, in vitro and in vivo evaluation.
Key words: Mycobacterium tuberculosis, Small molecule inhibitor, Cell wall synthesis, Antibiotic resistance
Graphical abstract
The cell wall biosynthesis of Mycobacterium tuberculosis (MTB) requires the participation of many enzymes, which are becoming potential targets to develop anti-tuberculosis drugs with little toxicity.

1. Introduction
Tuberculosis (TB) is a chronic but fatal infectious disease and is one of the top 10 causes of death worldwide. Based on the latest “Global Tuberculosis Report 2020” from World Health Organization (WHO), 10 million people fell ill with TB and 1.2 million died in 2019. In particular, TB is a leading cause of death in HIV-positive patients. Multiple MTB complex members can cause TB, including MTB itself, Mycobacterium pinnipedii, Mycobacterium microti, Mycobacterium canettii, Mycobacterium bovis, Mycobacterium africanum and Mycobacterium caprae1. They may invade many organs in humans, but most often affect the lungs, which is called phthisical disease. TB can spread from person to person through the air and the main source of infection is patients with active pulmonary TB, but many patients acquire MTB as asymptomatic latent TB infection2. Although there is an effective and cheap drug regimen that is four times as effective as it was 40 years ago, the continuous increase in TB worldwide, most of whom are at risk of secondary injury3.
TB occurs in every country of the world and affects all age groups. According to the 2020 “Global Tuberculosis Report”, among the 1.2 million people worldwide who died of TB, 20,800 were HIV-positive. In addition, the best estimate from WHO is that 10.0 million (range, 8.9–11.1 million) people developed TB disease in 2019, including 5.6 million men, 3.2 million women and 1.2 million children. The most affected countries in Asia include India with 26% of cases, China with 8.4%, Indonesia with 8.5% and the Philippines with 6%. Multidrug-resistant (MDR)-TB began to significantly increase in the 1990s. About 620,000 people had MDR-TB, and the development of bacterial resistance needs more attention. It is the emergence of first-line drug resistance to TB that has caused panic in the healthcare field, and researchers are making more effort to develop innovative lead molecules that are effective against drug-resistant mycobacteria.
There are several challenges for the current treatment, including drug resistance and toxicity. Once these occur, the treatment needs to be interrupted and changed as soon as possible. Another challenge is pharmacokinetic drug‒drug interactions, in particular with antiretroviral therapy in patients with TB and HIV co-infection. One of the necessary conditions for achieving nonrecurrent therapy is that patients remain on treatment for a longer period of time. The current regimen has various drugs and fixed-dose combinations, which are divided into the following types: (1) two drugs (isoniazid and rifampicin); (2) three drugs (rifampicin, isoniazid and pyrazinamide); and (3) four drugs (ethambutol, pyrazinamide, rifampicin and isoniazid)4. Because most of the world's TB burden is caused by drug-resistance strains of MTB, use of the above three types of drug combinations are intended to reduce the emergence of drug resistance. Although researchers have invested a lot of time and effort in developing sensitive drugs for TB treatment, the existing standard of care is still a challenge and a hindrance to global TB treatment5.
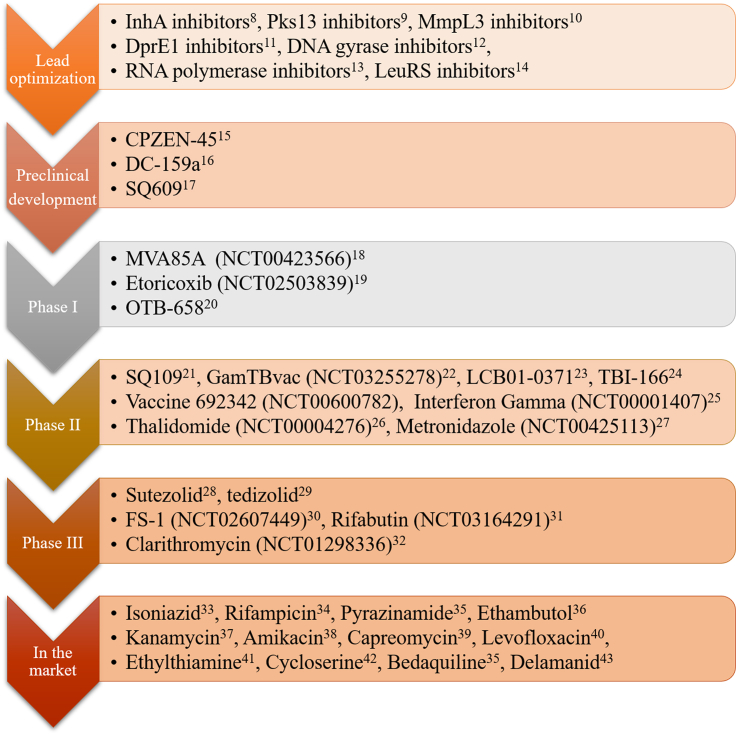
When experts encounter MDR-TB, the available drugs are limited, including only combination of clofazimine, amoxicillin and clavulanic acid, thioacetazone and linezolid. During the last 50 years, only three drugs were discovered and approved for treatment of MDR-TB. Bedaquiline was approved by the US Food and Drug Administration (FDA) in 2012, delamanid was accredited by the European Medicines Agency in 2014 and pretomanid was developed by the Global Alliance in 20196,7. Many new drugs are in the stage of clinical trials, lead compounds with many targets are in a lead-optimization state, such as InhA inhibitors8, PKS13 inhibitors9, MmpL3 inhibitors10, DprE1 inhibitors11, DNA gyrase inhibitors12, RNA polymerase inhibitors13, and LeuRS inhibitors14. Three compounds are in preclinical development, such as CPZEN-4515, DC-159a16, and SQ60917. Three drugs in phase I clinical trials include MVA85A18, etoricoxib19, and OTB-65820. Eight drugs in phase II clinical trials include SQ10921, GamTBvac22, LCB01-037123, TBI-16624, vaccine 692342, Interferon Gamma25, thalidomide26, and metronidazole27. Drugs in phase III clinical trials include sutezolid28, tedizolid29, FS-130, rifabutin31, and clarithromycin32. And 12 drugs have been approved in the market including isoniazid33, rifampicin34, pyrazinamide35, ethambutol36, kanamycin37, amikacin38, capreomycin39, levofloxacin40, ethylthiamine41, cycloserine42, bedaquiline35, and delamanid43 (Fig. 1).
Figure 1.
Current global pipeline of new anti-TB drugs. Representative agents at different research stages are listed for the treatment of TB.
2. M. tuberculosis cell wall: Drug targets and therapeutic agents
More effective TB drugs with novel scaffolds are being developed by scientists around the world. In drug discovery, the first step is to identify the drug targets. Advances in molecular biology technology, genetic tools and availability of the whole genome sequence of MTB in the post-genome era have created a new way to find chemotherapeutic biological targets6. Around 20 targets now have been identified for the new drug discovery process map in MTB (Fig. 2)6. Isocitrate lyase (Lcl) is essential role in the fatty acid metabolism, virulence, and growth regulation in active and dormant phases44, cytochrome bc1 complex (QcrB) regulates mycobacterial energy metabolism45, DNA gyrase regulates bacterial replication and transcription46, DosR (DevR) acts as dormancy regulator and is necessary for physiological maintenance47. Ribosome is involved in protein synthesis48, while proteasome is important for preventing reactive nitrogen forms and regulating proteins49. Then, InhA, FadD32, MmpL3, Ddl, and PKS13 are involved in mycolic acid synthesis and several other enzymes responsible for bacterial cell wall biosynthesis could be drug targets50, 51, 52. Because most of the previous studies focused on known targets, these efforts have not accelerated the pace of discovery of novel drugs. Therefore, finding novel targets and developing its inhibitors have become a major challenge in the treatment of TB.
Figure 2.
Diagrammatic representation of MTB targets within the cell structure.
The cell wall is the interface between the internal and external environments, and it performs many important basic processes, including structural definition, protection, and transmission53. Unlike animal cells, many enzymes responsible for bacterial cell wall biosynthesis are becoming attractive targets for development of novel drugs with unique mechanisms. Among the known drugs, ethambutol and isoniazid interfere with cell wall biosynthesis through interacting with enzymes on the cell wall. Therefore, we review the potential targets with critical roles in regulating four pathways in cell wall synthesis: mycolic acid biosynthesis pathway, peptidoglycan biosynthetic pathway, arabinogalactan (AG) biosynthesis pathway and other drug targets. We also systematically summarize the advanced development of novel drugs for inhibition of cell wall synthesis, including their discovery, chemical modification, and in vitro and in vivo evaluation.
2.1. Mycolic acid biosynthesis pathway
MTB consists of a variety of lipid components that both survive and cause disease, and has a special back-wall complex. It is worth mentioning that mycoplasma acid is a characteristic fatty acid that constitutes the integrity of the branched membrane, namely a large 3-hydroxyl, 2-alkyl long-chain fatty acid that can affect cell wall permeability and overall integrity of the cell. It is also a type of trehalose dimycolate, which is the main component of the outer membrane.
Mycolic acid is a long-chain (C70–90) α-alkyl-β-hydroxy fatty acid and the outermost component of the cell wall of MTB. Mycolic acid can be free or esterified to the underlying AG of the AG‒PG complex and responsible for maintaining the fluidity and permeability of the mycobacterial envelope. Mycolic acid is composed of highly saturated fatty acids that can be classified into three subsets: α-mycolic acids containing cyclopropane rings, which are only seen in the cis-configuration, while methoxy-and keto-mycolic acids contain cyclopropane rings in cis or trans configuration. The biosynthesis of mycolic acid consists of five distinct stages and requires synergistic action of more than 20 enzymes, which are essential for MTB reproduction and survival54. The enzymes involved in biosynthesis of mycolic acid may be ideal targets for development of new drugs.
The early steps in the biosynthesis of mycolic acid are extension of acetyl-CoA to produce malonyl-ACP and carbon chain elongation mediated by the type I fatty acid synthetase (FAS-I) system. Extension of acetyl-CoA: the acetyl moiety of acetyl-CoA is transformed by acetyl-CoA carboxylase (AccA3/AccD6) to produce malonyl-CoA, and then malonyl CoA/ACP transacylase (FabD) catalyzes the malonyl moiety of malonyl-CoA transfer to holo-ACP, resulting in malonyl-ACP. The FAS-I system is a single multienzyme complex that is responsible for utilizes malonyl-ACP and acetyl-CoA synthesis the Acyl-CoA54. The first step in the FAS-I system is transfer of acetyl-CoA and malonyl-CoA to the FAS-I complex to form acetyl-S-Enz and malonyl-S-Enz intermediates. Then, a condensation reaction catalyzed by the enzyme complex utilizing both types of the above intermediate yields a β-ketoacyl-C4-S-Enz intermediate. Subsequently, the intermediate is successively subjected to β-ketoacyl reduction, dehydration and enoyl reduction, resulting in butyryl-S-Enz, and a cyclical reaction is continued until the acyl chain reaches C16–18. This acyl-S-Enz is transformed into acyl-CoA and subsequently to generate the α-alkyl moiety (C24) or enter the type II fatty acid synthetase (FAS-II) system, resulting in production of the mero-chain54.
The FAS-II is an enzyme complex that consists of at least four enzymes: β-ketoacyl-ACP reductase (MabA), β-hydroxyacyl-ACP dehydrase (HadAB/BC), 2-trans-enoyl-ACP reductase (InhA), and β-ketoacyl-ACP synthases (KasA/B)54. β-Ketoacyl-ACP synthase-III (FabH) catalyzes condensation of malonyl-ACP and palmitoyl-CoA to form a β-ketoacyl-Acp intermediate. Subsequently, MabA utilizes NADPH as a reductant to reduce β-ketoacyl-Acp, resulting in β-hydroxyacyl-ACP, which is dehydrated by HadAB/BC to yield trans-2-enoyl-Acp. InhA utilizes NADPH to reduce trans-2-enoyl-Acp to form acyl-Acp. Compared with the above palmitoyl-CoA, Acyl-Acp is elongated by two carbons. The newly formed Acyl-Acp is catalyzed by KasA/B to form β-ketoacyl-ACP and continues involvement in the FAS-II circulating system for elongation of its carbon chain, or continues with further modification depending on whether the acyl chain reaches C42–62.
After the acyl chain reaches C42–62, the carbon chain is further modified to form three types of meroacid: α-meroacid, methoxy-meroacid and keto-meroacid, which undergo further processing to form three types of mycolic acid. In the primary modification stages, MmaA2 is responsible for introduction of the distal cyclopropane ring, cis-cyclopropane ring and proximal cis-cyclopropane ring to α-meroacid, methoxy-meroacid and keto-meroacid, respectively. MmaA1 and MmaA4 methylate the keto-meroacids. MmaA3 is responsible for introduction of the O-methyl group to the methoxy-meroacids. In addition, PcaA/UmaA2 are responsible for introduction of the proximal cyclopropane ring to α-meroacid, and CmaA2 is responsible for introduction of the cis- and trans-cyclopropane rings to methoxy-meroacid and keto-meroacid, respectively. These modifications of the meromycolate chain can adjust the fluidity and permeability of the mycobacterial envelope and improve its ability to resist the host immune system during infection.
Progression of Claisen-type condensation is catalyzed by AccD4/5 and an AccA3-AccD4-FadD32-PKS13 enzyme complex. Acyl-CoA, a product of the FAS-I system, is catalyzed by acyl-CoA carboxylase (AccA3/AccD4) to produce carboxyacyl-CoA. Fatty acyl-AMP ligase (FadD32) catalyzes the meromycolic acid chain (C42–62) to generate meromycolyl-AMP. Polyketide synthase 13 (PKS13) catalyzes the Claisen type condensation reaction of carboxyacyl-CoA and meromycolyl-CoA to yield an α-alkyl-β-keto-mycolic acid intermediate, which is then converted into a mature mycolic acid through a reduction reaction catalyzed by mycolyl reductase.
Numerous key enzymes, such as those involved in fatty acid synthesis, fatty-acid-modifying enzymes, fatty-acid-activating, condensing enzymes, as well as transporter and transferase are potential drug targets in the mycolic acid biosynthesis pathways (Fig. 3). In the background of MDR-TB, extremely drug resistant (XDR)-TB and fully drug resistant TB, mycolic acid biosynthesis pathways provide a valuable source for the development of potential targets for new anti-TB drugs.
Figure 3.
Mycolic acid biosynthesis pathway.
2.1.1. FadD32 inhibitors
One of the most important enzymes is FadD32, plays a significant role in the biosynthesis of mycolic acid. There is a long meromycolate chain (C55‒C63) and a shorter α-alkyl chain (C22‒C24), which consists of branched-chain fatty acids, that are the main components of the specific mycobacterial cell wall. FadD32 produces β-keto-alkyl mycolic acid, which is required to activate meromycolic acid, to facilitate condensation with shorter-chain fatty acids. FadD32 is an ideal target for the development of novel antimicrobial agents against MTB because there are two enzyme functions in FadD32: the ACP domain in the middle of the transfer activity polyketide synthase 13 (Pks13) is formed in the FadD32 fat acyl-amp ligase (FAAL) activity of the first adenosine meromycolic acid to produce fatty acid synthase (FAS) second biological synthesis pathway and its fatty acylate ACP synthase (FAAS) activity results.
The 4,6-diaryl-5,7-dimethyl has shown that FadD32 is a target for antibiotic development because it effectively blocked bacterial replication in vitro and in animal models of TB. It acts in the same way as isoniazid, an established antibiotic, and Stanley et al.55 reported in 2013 that a family of 4,6-diaryl-5,7-dimethyl killed MTB by inhibiting FadD32. In their report, compound 1 was the most powerful series of compounds showing antibacterial activity against MTB (H37Rv) with an MIC of 0.24 μmol/L (Supporting Information Fig. S1). Not long after this, Kawate et al.56 delved into the structure–activity relationships (SARs) of coumarin derivatives and found that the two methyl and aryl groups are essential for the activity. In their effort, 43 derivatives have been synthesized and the most powerful compound 2 exhibits strong inhibition ability for H37Rv with an IC90 of 0.5 μmol/L (Fig. S1). Unfortunately, the chemical instability of coumarin derivatives has greatly limited the development of this series of FadD32 inhibitors. To overcome this fatal defect, Fang et al.57 investigated other heterocyclic replacement of the coumarin ring, such as benzofuran, quinoline and 2-quinolone derivatives. Among these heterocyclic, only quinoline derivatives, such compound 3 (MIC = 0.8 μmol/L), have exhibits similar activity with compounds 1 and 2. Except the anti-H37Rv activity, compound 3 had minimal cytotoxicity (IC50 = 60 μmol/L HepG2) and high cross-species microsomal stability (19.4% R at 60 min in hLM and 19.1% R at 60 min in mLM) (Fig. S1). For structural optimization of lead compound, the case of coumarin scaffold development to quinoline scaffold has high reference significance.
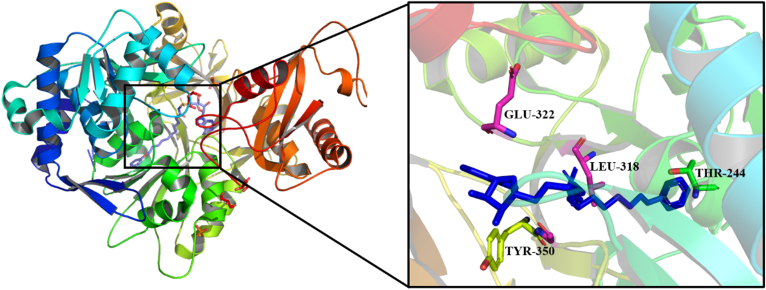
In 2016, Kuhn et al.58 found some of the best ligands of FadD32, such as 5′-o-[N-(11-phenoxyundecanoyl)-sulfamoyl]-adenosine (compound 4), which has an IC50 of 6.8 μmol/L, and compound 4 is safe (Fig. S1). A crystallization experiment was conducted to understand the mechanism by which complex inhibits this protein, in which compound 4 was co-crystallized with FadD32 protein (Fig. 4). There were 18 hydrophobic interactions between the compound 4 and the protein. The key residues include Thr244, Tyr350, Leu318 and Glu322. There are five H-bonds between compound 4 and the protein FadD32, which form the strong interactions.
Figure 4.
Co-crystal structure (PDB ID 5HM3) of compound 4 bound to FadD32 of MTB (compound 4 showed as blue color).
2.1.2. InhA inhibitors
InhA (enoyl-ACP reductase) is a key enzyme (FAS II) that catalyzes NADH-dependent reduction of 2-trans-enoyl-ACP (acyl-carrier protein), giving up NAD+ and reducing enoyl thioester-ACP substrate, and promoting synthesis of mycophenolic acid, and the synthesis of type II fatty acids (FAS II). In addition, InhA is an effective target of the most developed anti-TB drug59. Many compounds (compound A) tested had potent submicromolar IC50 values against purified MTB InhA (Supporting Information Table S1). For a comprehensive review of InhA inhibitors, please see the recent review from the Tonge Laboratory60.
Compounds 5 and 6 were reported by Sriram et al.61 in 2020. The synthesized compounds carry all structural moieties required to interact with InhA enzyme; formation of hydrogen bonds to NAD and Tyr and two hydrophobic rings. Compounds 5 and 6 had IC50 values of 17.7 ± 1.3 and 15.6 ± 1.6 μmol/L and MICs of 2.42 and 4.26 μmol/L, respectively (Supporting Information Fig. S2). It can be concluded that InhA is an important target for these compounds that exhibit antimycobacterial activities.
In 2020, Sarıpınar et al.62 synthesized new thiadiazolylhidrazones and increased the lipophilicity of these compounds by introducing two (substituted) phenyl rings to obtain better antimycobacterial activity. Compound 7 had an MIC of 2.52 μmol/L, and 75% inhibition of InhA at 50 μmol/L (Fig. S2). Introduction of a halogen to the N-phenyl ring led to an increase in the mentioned activity. However, the most active compound had a methyl group at the same locus, suggesting that the interaction with the binding site of the enzyme played a more important role than the electronic features of the substituents.
Compounds with the methyl thiazole scaffold have been reported as direct InhA inhibitors, and the most active of these compounds is 863. Compound 8 had an IC50 of 0.003 μmol/L against InhA and an MIC of 0.19 μmol/L against MTB (Fig. S2). It has been shown that compound 8 is selective for bacteria. Mammalian cytotoxicity of 78 μmol/L also indicated that compound 8 was inactive in eukaryotes when targeting the A549 cell line. The crystal structure of InhA in complex with compound 8 is shown in Supporting Information Fig. S3. When intermolecular interaction of compound 8 was observed with the Y158, M103, and M98 residues, activity of InhA was also inhibited.
2.1.3. PKS13 inhibitors
Polyketide synthase (PKS), is one of the important enzymes that has not been used as a drug target for any microbial pathogen. There are about 24 PKS coding genes in the MTB H37Rv genome. A number of mycobacterial PKSs and complex lipid structures in MTB have been linked to genetic and biochemical studies of synthetic pathways. The special components of MTB, which has a lipid-rich and complex cell wall, are lipid metabolites from PKS.
In a study, some thiophenes killed MTB by targeting the N-terminal N-ACP domain of PKS13. The compounds function by obstructing the interaction between N-ACP and the FadD32 protein that transmits the meromycolyl chain. These results confirmed that PKS13 could be used as a drug target for MTB and highlight its potential to develop novel anti-TB drugs that interfere with the key pathway of MTB acid synthesis.
Some experts believe that PKS13 inhibitors are also useful compounds. Compounds 9 and 10, as examples of the Alland group, have been used to demonstrate that PKS13 inhibitors of thiophene have bactericidal effects in cells, and that they acquire bactericidal activity when used with isoniazid. Whole-genome sequencing of thiophene-resistant mutants has shown that the functional domain of PKS13 complex N-ACP was inhibited by compounds 9 and 10 with MICs of 1.0 and 0.5 μmol/L, respectively (Supporting Information Fig. S4)64. According to Thanna et al.65, one of the most active compounds, compound 11, in the 42 2-aminothiophenes Sucheck group library has an MIC ranging from 0.2 to 0.4 μmol/L, and MTB is resistant to isoniazid and rifampin (Fig. S4). However, this series of PKS13 inhibitors contain a high lipophilicity pentafluoroaryl group and a thiophene group, which may encounter difficult to enter the clinical stage due to the fact that most anti-TB drugs used clinically are hydrophilic.
Compounds 12 (TAM1) and 13 (TAM16), with IC50 of 0.29 and 0.19 μmol/L, and MIC of 2.3 and 0.09 μmol/L, respectively, have been identified (Fig. S4)66. The crystal structure of PKS13 in complex with compound 12 (TAM1) is shown in Supporting Information Fig. S5. The catalytic triad was identified as Ser1533, Ala1561, and His1699, and the oxygen anion holes were formed by amide N-atoms of Leu1534 and Ala1477. The benzofuran core of compound 12 wedges between Phe1670 and Asn1640. Compound 12 has strong in vitro bactericidal activity, is active against drug resistance and clinical isolates of MTB, and is a benzofuran inhibitor of PKS13. Compound 13 (TAM16) was effective against all MDR and XDR MTB clinical isolates with an MIC range of 0.05–0.42 μmol/L. In multiple mouse models of TB infection, compound 13 (TAM16), either alone or in combination with rifampicin, was as effective as isoniazid, the previous first-line anti-TB drug. Compound 13 (TAM16) could be developed as a new drug against acute TB. The toxicity of compound 13 (TAM16) is 100 times lower than that of isoniazid, with a good pharmacological and safety profile. Compound 13 shows low protein binding in mouse and human plasma and showing very low clearance in MLM and high grade (artificial liver microsomes, clint <0.5 mL/min/g liver).
Yu et al.67 discovered novel tetracyclic analogs that were synthesized and evaluated for their anti-TB activity against MTB H37Rv in 2020. Compound 14 demonstrated potent activity against MTB H37Rv with an MIC between 0.0313 and 0.0625 μg/mL (Fig. S4), and inhibited the PKS13-TE activity with an IC50 of 0.58 μmol/L, with high selectivity in Vero cells (64‒128 fold). Compound 14 also showed excellent selectivity against actinobacteria; therefore, it is unlikely that nonpathogenic bacteria will develop drug resistance. However, whether it is benzofuran or coumarin scaffold, all compounds among these series have a basic lipophilic amine, which might contribute to the off-target of hERG signal and induce cardiotoxicity. Therefore, there is still a long way to develop PKS13 inhibitors for the treatment of MTB.
2.1.4. MmpL3 inhibitors
There are 13 genes encoding RND protein MmpL (MTB membrane protein) in the MTB genome, and MmpL mediates translocation or extrusion of various physiological compounds. There are 12 membrane proteins in the MTB genome sequence, and the membrane proteins are thought to play a role in lipid transport. However, only one of these genes, MmpL, is important for survival, and insertion and inactivation of the other 11 genes have also indicated this phenomenon. Twelve transmembrane structures constitute MmpL3 and it contains 994 amino acids. MmpL3 is a membrane transporter that is responsible for the transport of branched fatty acids to the external side. In in vitro and in vivo infection models, inhibition of MmpL3 weakens the cell wall of mycobacteria and ultimately leads to cell death10. According to Li et al.68, the analog of the new TB drug compound 15 (SQ109) works by inhibiting MmpL and cell wall biosynthesis. In their opinion, compound 15 (SQ109) has a strong inhibitory effect on MTB with an MIC of 0.1–0.2 μg/mL (Supporting Information Fig. S6). N-Geranyl ethanolamines, compounds 16 and 17, are more effective (MIC = 0.02–0.05 μg/mL), and compound 16 is 4‒5 times more active than SQ109 (Fig. S6). Among these ethylenediamine derivatives, the center nitrogen is essential for activity, replacing the nitrogen with oxygen will result decreased activity. In addition, Lun et al.69 has reported some indoleamides with high activity against MDR and XDR MTB H37Rv strain.
The MICs for compounds 18, 19 and 20 are 0.125–0.25, 0.0156–0.0313, and 0.0039 μg/mL, respectively (Fig. S6). They assessed the MIC value of each of indoleamide compounds against IAR2 (indoleamide-resistant, compound 2) mutants, and it was found that the activity was much higher than the parental H37Rv strain. The MICs of compounds 19 and 20 were increased 32–64 times, which indicated that the target of indoleamide compounds was MmpL3. The SAR study of these series of indoleamide derivatives showed that the cycloalkane unit has extremely contribution to activity and replaced by phenyl ring or nitrogen-containing heterocyclic ring will significantly reduce the anti-TB activity. In addition, the size of the cycloalkane unit also plays a crucial role in the activity, small ring leads to the complete disappearance of activity while bulkier substituents yield potent activity.
Other researchers have also reported new anti-TB drugs that target MmpL3 protein. Grzegorzewicz et al.70 have reported that compound 21, 1-(2-adamantyl)-3-(2,3,4-trifluorophenyl) urea, is an effective bactericidal anti-TB agent with an MIC of 0.1 μg/mL (Fig. S6). It retains high activity against strains resistant to front-line anti-TB agents and lacks MIC potency against other bacteria, suggesting a novel and specific mechanism of action. The actual target of compound 21 was determined by genetic analysis of spontaneous drug-resistant mutants, and a single nucleotide polymorphism (G758A) was found in the gene encoding MmpL3.
In 2021, Tanya et al.71 found that compound 22 had potent activity against MTB H37Rv (MIC = 0.66 μmol/L) and low toxicity (Fig. S6). Compound 22 showed a 10-fold decrease in activity against the MmpL3F255L strain of MTB, indicating that it retained an MmpL3-related mechanism of action. An impact on this pathway was also indicated using a hypomorphic strain (P606-5C-mmpL3) that underexpressed MmpL3 when grown in the absence of anhydrotetracycline.
2.2. Peptidoglycan biosynthetic pathway
Peptidoglycan (PG), a complex macromolecule, is a component of bacteria cell walls and maintains cell wall rigidity and osmotic stability. PG of MTB consists of two glycan backbones that are crosslinked to each other by short tripeptide or tetrapeptide side chains to form a mesh-like structure. The components of the glycan backbone are conserved in all bacteria and formed by N-acetylglucosamine (GlcNAc) as well as N-acetylmuramic acid (MurNAc) crosslinked by α,β (1 → 4) linkages. The biosynthesis of PG requires many unique enzymes that are absent in human metabolic pathways. So, those enzymes have potential for new drug development.
The primary step in the biosynthesis of PG is catalysis of GlcN-1-P acetylation and uridylation by bifunctional enzyme GlmU (Rv1018c) to yield UDP-GlcNAc. UDP-GlcNAc is catalyzed by a sequential series of Mur ligases (MurA-F) as well as NamH, resulting in the Park's nucleotide intermediate (UDP-MurNAc/Glyc-l-Ala-d-isoGlu-m-DAP-d-Ala-d-Ala). MurA catalyzes enoyl pyruvate addition of UDP-GlcNAc to yield UDP-enoylpyruvyl-GlcNAc, and MurB utilizes NADPH to reduce the enoyl pyruvate group of UDP-enoylpyruvyl-GlcNAc to a lactoyl ether moiety and generate UDP-MurNAc. Subsequently, NamH (Rv3808) catalyzes UDP-MurNAc hydroxylation, resulting in the UDP-MurNGlyc intermediate, which is a unique structure in MTB compared with other bacteria. However, this enzyme may not be a suitable target for drug discovery because NamH is not essential in MTB. Lack of NamH results in hyper susceptibility of MTB to β-lactam antibiotics; therefore, inhibitors of NamH can be combined with β-lactam antibiotics for treatment of TB. Then, four ATP-dependent Mur ligases are responsible for addition of four types of amino acid residues to UDP-MurNAc/Glyc. The first step is addition of l-alanine to the MurNAc/Glyc residues, which is catalyzed by MurC (Rv2151c). Subsequently, MurD (Rv2155c) catalyzes d-isoglutamate binding of l-alanine residues; MurE (Rv2158c) catalyzes m-DAP attachment of d-isoglutamate residues; and MurF (Rv2157c) is responsible for addition of d-alanyl-d-alanine to the m-DAP residues, resulting in Park's nucleotide. MurX/MraY (Rv2156c) is a phospho-MurNAc-pentapeptide translocase that catalyzes decaprenyl phosphate transfer to Park's nucleotide at the expense of cleavage between UDP and MurNAc/Glyc glycosidic bond, to produce Lipid I. MurG (Rv2153), a glycosyltransferase, catalyzes transfers of GlcNAc from UDP-GlcNAc to Lipid I to form Lipid II. The formation of Lipid II marks the end of PG biosynthesis within the cytoplasm. Lipid II crosses the plasma membrane, which requires Lipid II flippase, and was recently recognized as MurJ or FtsW. Lipid II monomers are the final monomeric units involved in the polymerization of PG. Here, we describe the biosynthetic progress of PG and discuss some enzymes in the PG biosynthetic pathway, which have attracted the most attention for anti-TB drug discovery.
2.2.1. UDP-GlcNAc biosynthetic pathway
It has been predicted that the enzymes of UDP-GlcNAc biosynthesis are important for the growth of MTB in vitro. The UDP-GlcNAc biosynthetic pathway is located at the bifurcation point of two important biosynthetic pathways of PG and disaccharide ligand d-NGlcNAc-1-rhamnose, which is the key precursor molecule of the MTB cell wall. The significance of the UDP-GlcNAc pathway in bacteria is to make the enzymes of the pathway a substantial target for drug development. Heijenoort72 demonstrated the ability of bacteria, rather than human pathways, to identify drug targets that might reduce the toxicity of antimicrobial compounds by comparing human genomes with bacterial genomes. A new antibacterial drug identification method with a new mechanism is the enzyme of UDP-GlcNAc biosynthesis pathway.
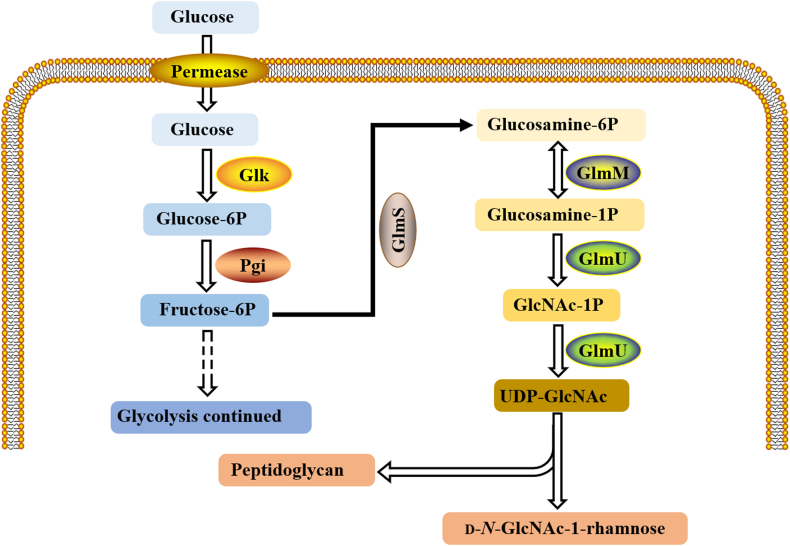
The biosynthesis of UDP-GlcNAc involves three enzymes in four steps (Fig. 5). In the first step, glutamine catalyzes the conversion of fructose-6-phosphate into glucosamine-6-phosphate (GlcN-6-P) by glucosamine-6-phosphate synthetase (GlmS). The GlmS on MTB is encoded by glms gene (rv3436c), which is homologous to E. coli glms. GlcN-6-P is then converted to glucosamine-1-phosphate (GlcN-1-P) by glucose phosphate mutase (GlmM). GlmM, similar to E. coli GlmM, is encoded by the GlmM gene (Rv3441c, recorded as mrsA on the TB server). In the process of converting GlcN-1-P into N-acetylglucosamine-1-phosphate, the last two sequential steps are acetyl and uridyl transfer reactions, and the final product is UDP-GlcNAc. These two steps are catalyzed by the single and double functional enzyme N-acetylglucosamine-1-phosphouridine transferase (GlmU) encoded by GlmU gene (Rv1018c), which has acetyltransferase and uridine transferase activities.
Figure 5.
UDP-GlcNAc biosynthetic pathway.
2.2.1.1. GlmU enzyme inhibitor
Some groups have confirmed that four homologs existed in the UDP-GlcNAc pathway in the MTB genome, and one of the potential and effective targets in TB drug development may be the GlmU enzyme. The inactivation of GlmU provides evidence for the necessity of the UDP-GlcNAc pathway and is an important gene. However, there is a lack of a bifunctional enzyme in eukaryotes, which corresponds to GlmU, acetyl transfer and uridyl transfer, which are sequentially related to GlmU and are accomplished by two different enzymes. In addition, the absence of the substrate required for GlmU acetyltransferase activity in humans means that a more important strategy for the development of new anti-TB drugs may be targeted at C-terminal enzyme activity of MTB GlmU.
There is a report that the chemical stents, as the specific inhibitors, play a significant role in the recognition of the GlmU acetyltransferase domain of MTB. A total of 1607 compounds were selected from among 20,000 in the ChemBridge Library, and assessed as having the potential to inhibit GlmU acetyltransferase activity in bioassays of MTB. The accumulation of glucosamine-1-P in the identified cells was tested experimentally and these cells were treated with inhibitors. The experimental results showed that the accumulation of glucosamine-1-P in the cell wall precursor was shown on compound 23, and this demonstrates its inhibitory effect on cell wall biosynthesis. The experiments suggest that GlmU may only be one of the possible targets of the compounds, the reason is that the correlation between in vitro antibodies TB data and in vitro enzymology data is weak, because of the proximity of the purified IC50 value of GlmU to the MIC value of its molecule (2 μg/mL) on compound 23 (Supporting Information Fig. S7).
The potential of MTB gene has not been fully exploited. Then, after its whole genome sequencing, genome-derived targeted therapy has attracted much attention. This method uses small molecules in the large chemical library to inhibit the growth of MTB in culture. This study suggests that action patterns and drug resistance are easy to identify, the reason is that the target, which based on chemical libraries, screening is superior to whole-cell screening. A total of 533 small molecules were found to be active in the acetyltransferase domain of MTB GlmU in PubChem Bioassay AID 1376. The IC50 values of 125 molecules ranged from 1 to 9999 μmol/L. It can be indicated that the activity under 30 μmol/L of the structure of the inhibitor is shown. In Chitra et al.73, the most efficient structures with multiple chemical structures with the best IC50 values were compounds 24, 25 and 26 (Fig. S7). They further cited the results of PubChem, and these compounds have undergone other tests besides GlmU activity assays. A variety of high throughput screening (HTS) assays, for example, histone lysine methyltransferase activity bioassays, T-RNA-2′ phosphotransferase (TPT1) assays, and RecQ-like DNA Helicase 1 assays have been reported.
2.2.1.2. GlmS enzyme inhibitor
The first decisive step in catalyzing hexose amine metabolism is glucosamine-6-P synthetase (GlmU), a dimer enzyme that belongs to the glutamine-dependent amide transferase family. l-Glutamine as a nitrogen source is used in this way, and this enzyme converts d-fructose-6-P into d-glusomine-6-P irreversibly. UDP-N-acetylglucosamine-6-P is the final product of this pathway and it also is a major component of bacterial and fungal cell walls. For this reason, various in-depth studies have been conducted in the past decade, and glucosamine-6-P synthase (GlmS) is also considered to be a pharmacological target. The latest X-ray structure of the bacterial GlmU has been reported by Floquet et al.74, and 50,000 possible compounds in the database are found around the active site. Researchers eventually tested 14 molecules as putative GlmS inhibitors, and three of them had significant inhibitory properties with a probability of 23%. In addition, compounds 27–29 inhibited GlmS significantly, with an IC50 of 70 μmol/L, and it was predicted that these three molecules would bind at the interface between these two GlmS monomers (Supporting Information Fig. S8).
2.2.2. Mur pathway
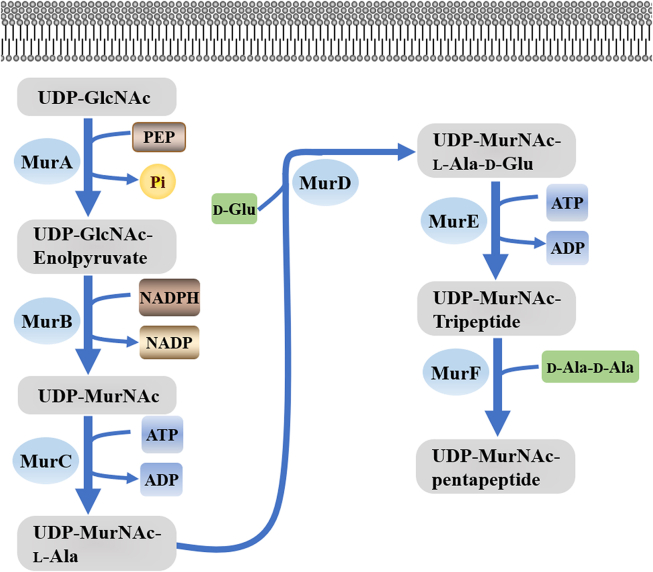
Mur enzymes have no counterparts in eukaryotes, and they are necessary to maintain cellular integrity and resistance to osmotic changes, and are highly conserved in bacteria. MurC and MurF (ATP-dependent ligases) are closely related to each other among Mur pathway enzymes, but MurA transferase and MurB reductase are obviously different from Mur ligase. MurA catalysis is the first step of the Mur pathway, in which polypyruvate of phosphoenolpyruvate (PEP) is partially transferred to UDP-N-acetylglucosamine (UDP-GlcNAc). Second, the alkyl pyruvate portion of MurB was reduced to the lactyl group by using NADPH as the cofactor, forming UDP-NAc-Mur cholic acid. Mur ligases (MurC-MurF) UDP-MurNAc pentapeptide was formed after the catalytic addition of l-alanine, d-glutamic acid, middle diamino-opiate and d-alanine-d-alanine successively into UDP-MurNAc, which is the following four steps (Fig. 6). These Mur enzymes have been found in several microorganisms, and knowledge of their structure, function, and regulation in MTB is largely incomplete.
Figure 6.
Mur pathway.
Tetrahydroisoquinoline drugs that inhibit the growth of MTB H37Rv have been experimentally studied and synthesized, and the whole cell phenotype is affected by such drugs, as well as the activity of ATP-dependent MurE ligase. Compound 30, which has an inhibitory effect on the growth of MTB H37Rv has low cytotoxicity, and its 50% growth inhibitory concentration is 28 μg/mL and its MIC is 30 μg/mL (Supporting Information Fig. S9). HPLC has been used to detect the compounds that inhibit MTB MurE synthetase75. The negative control, meanwhile, was isoniazid, a drug that has no inhibitory effect at any detectable concentration. The most effective inhibitor of MurE was compound 30, with IC50 less than 111 μmol/L.
Konduri et al.76 synthesized many new purine-linked piperazine derivatives. The synthesized compounds were screened for antibacterial activity, and it was found that six compounds had good antibacterial activity. They used computational molecular docking analysis to predict the correlation between these results and MurB. Among the six compounds, compound 31 (MIC = 5.08 μmol/L) had a strong hydrogen bond with the amino acid residues of MTB MurB (Fig. S9). The interaction between the binding energy and hydrogen bonds observed in the docking study is closely related to the anti-TB test.
2.3. Arabinogalactan (AG) biosynthesis pathway
AG is a heteropolysaccharide located between the MA and PG layers in the mycobacterial cell wall, and is a highly branched macromolecule composed mainly of arabinose (Ara) and galactose (Gal) sugar residues. Specifically, both Ara and Gal sugar residues are present in furanose (f) ring form77.
The process of AG biosynthesis can be divided into three components: linker unit biosynthesis, linear galactan biosynthesis and arabinan biosynthesis.
Linker unit biosynthesis is as follows: first, the GlcNAc transferase WecA catalyzes transfer of GlcNAc-1-P from UDP-GlcNAc to the lipid carrier and then the rhamnosyltransferase WbbL attaches the rhamnosyl (Rha) residue to the C50-P-P-GlcNAc, forming C50-P-P-GlcNAc-Rha, which is the linker unit between AG and PG. WecA serves as a promising drug target and has attracted much attention. Recently, Xu et al.78 have found that downregulation of WecA can enhance the sensitivity of MTB to first-line rifampin. Thus, with the development of inhibitors of WecA, a new combination treatment for TB may be established. Like MraY, WecA is a member of the polyprenyl-phosphate N-acetylhexosamine-1-phosphate transferase superfamily, which is essential for mycobacterial cell wall biosynthesis. In 2019, Yoshimasa et al.79 have found that caprazamycin (CPZ), an inhibitor of MarY, and its derivative CPZEN-45 have strong inhibitory activity against WecA. Subsequently, a new series of CPZ derivatives have been discovered through fluorescence-based assays for WecA activity80. Among these derivatives, UT-01320 has shown 100% inhibition at a concentration of 2 nmol/L (Supporting Information Fig. S10).
Among the enzymes involved in the biosynthesis of arabinan chains, DprE1 is the most popular target for research in the field of anti-TB drug development. From the discovery of the first DprE1 inhibitors in 2009 to the present time, more than 10 classes of different scaffolds have been found that can ultimately destroy MTB by inhibition of this enzyme. Chikhale et al.6 group published a review that has systematically summarized compounds involved in this field from 2009 to 2018, such as BTZ04381, PBTZ16982 and TCA183. Based on this work, our focus is to review new breakthrough of compounds in this field from 2018 to date.
Benzothiazinone (BTZ) is one of the first class of compounds that selectivity inhibits DprE1. Nitro-BTZ derivative BTZ043 and its analog 2-piperazinobenzothiazinone (PBTZ) PBTZ169 show a high potential for development of new anti-TB drugs6. Stimulated by this positive result, many BTZ derivatives have been synthesized their anti-TB activity has been determined either in vivo or in vitro in the past decade. Among these derivatives, the structural modifications are concentrated in the oxidation valence state of S atoms, linking units between the BTZ scaffold and amino groups and substituents, and transformation of the nitro group and modification of the scaffold84, 85, 86, 87. Based on the structure of the preclinical drug PBTZ169, Li et al.87, through a ring-opening strategy, designed and synthesized a series of benzamide derivatives. Among these derivatives, N-benzyl 3,5-dintrobenzamide analogs showed in vitro activity against H37Rv strain and two MDR strains with MICs of 0.0625 and 0.016–0.125 μg/mL, respectively (Supporting Information Fig. S11). Compared with positive compound PBTZ169, with similar activity, was found to have safer and better pharmacokinetic profiles.
Piton et al.88 have shown that logP and BTZ activity had a strong correlation and sulfonyl groups could increase the solubility and metabolic stability of many drugs. They also used the structure of PBTZ169 for structure-based rational design of a new series of sulfone–PBTZ derivatives. Most of these compounds exhibited high activity against MTB H37Rv strain with an MIC99 of 0.06–0.001 μg/mL, while that of PBTZ169 was 0.0001 μg/mL (Fig. S11). To determine whether addition of a sulfonyl group influenced the binding of compounds and protein pocket, they analyzed the crystal structure of complexes of sPBTZ169 and DprE1. However, compared with PBTZ169, the sulfonyl moiety of sPBTZ was not influenced by the binding mode. Although these derivatives exhibit decreased anti-TB activity compared with PBTZ169, they have better solubility profile and among these derivatives, 11626091 shows better stability in microsomal assays as originally hypothesized.
Regarding the structure of BTZ043, Li et al.89 used a scaffold-morphing strategy to design and synthesize three series of benzopyranone, benzoxazinone, and benzothiopyranone analogs. However, only the benzothiopyranone series exhibited good activity against MTB and had low cytotoxicity. Among these derivatives, compound 32 inhibited (MIC less than 0.016 μg/mL) drug-susceptible and MDR MTB in an in vitro anti-TB assay (Fig. S11). In an in vivo assay, compound 32 also demonstrated anti-TB activity in an acute mouse model. Since the BTZ scaffold was found in 2009, great efforts were devoted to discover the more druggable BTZ small compounds. Among these derivatives, nitro group was an essential group for activity and responsible for the formation of covalent bond to DprE1. The introduction of CF3 can significantly improve the anti-TB activity, while the alkyl chain was responsible for regulating the physical and chemical properties of compounds.
Besides the BTZ scaffold, Rogacki et al.90 have found a novel series of hydantoin-based scaffolds using a target-based, high-throughput screening strategy against DprE1. Based on compound 33 [pIC50(DprE1) = 7.0, MIC = 8.3 μmol/L], they synthesized >100 analogs to explore crude structure–activity relationships and development potential of this series of compounds. Combinations of these analogs had an inhibitory effect on DprE1. Modification of compound 33 at the A or B ring still resulted in significant inhibitory activity, with pIC50>6, while change in the length of the linker between the A and C rings, as well as carbonyl removal reduced the activity. In addition, replacement of the methyl side chain of the C ring by ethyl or another long aliphatic chain had no significant influence on the inhibitory activity. Among these analogs, some promising compounds are stable in both mouse and human microsomes, with intrinsic clearance values below 3 mL/(min·g) and 0.4 mL/(min·g), respectively. Although some compounds, including compound 33, exhibit measurable inhibitory activity for the hERG potassium channel, substitution of the A ring of compound 33 to a pyridine ring, forming compound 34, eliminated hERG inhibition, suggesting that suitable modification of this series results in no cardiotoxicity. To further understand DprE1 protein and this series, the authors tested the relationship between the concentration of compounds and their inhibitory effect vs. time. The linearity between concentration of product (catalyzed by DprE1) and time was not affected by compound concentration, suggesting that this series are reversible DprE1 inhibitors (Supporting Information Fig. S12).
In a further effort for optimization of hydantoins as selective DprE1 inhibitors, Rogacki et al.91 synthesized more than 80 analogs, with modification mainly around the A and B rings. The B ring was modified with a polar group, such as carboxylic acid, ester or a urea moiety. As expected, most of analogs showed significant in vitro enzymatic potency and whole-cell inhibitory activity with IC50 in the range 7–7.4 μmol/L and MIC from 0.6 to 0.9 μmol/L. In contrast, modification of the A ring showed no significant potency development. Compound 35 and similar compounds show excellent anti-TB activity with no cytotoxicity or cardiotoxicity (hERG), a reasonable physicochemical profile, and satisfactory metabolic stability. Nevertheless, this series still needs further preclinical development to explore their drug potential (Fig. S12).
PG and AG components are associated with l-rhamnose in MTB, and the essential genes in this bacterium are related to l-rhamnose biosynthesis. Glucose-1-phosphate and deoxidization thymidylic triphosphate (DTTP) are synthesized by DTDP-l-rhamnose, through the d-1-phosphate glucose-thymidine acyl transferase (RmlA), an enzyme encoded by gene RmlA-D, which was previously called rfbA-D62, dTDP-d-glucose-4,6-dehydratase (RmlB), DTDP-4-ketone-6-deoxidization glucose-3,5-epimerase (RmlC) and DTDP-6-deoxy-l-xylo-4-hexanone sugar reductase (RmlD). The synthesis of DTDP-d-glucose and pyrophosphate, using d-glucose-1-phosphate (G1P) in a reaction with DTTP, as catalyzed by RmlA, is independent of any other rhamnose-forming enzyme. DTDP-d-glucose is then oxidized by RmlB to form DTDP-6-deoxy-d-xylo-4-hexose. RmlC catalyzes DTDP-6-deoxy-l-xylo-4-hexose to DTDP-6-deoxy-d-xylo-4-hexose. It is worth mentioning that the RmlB (Rv3464) and RmlC (Rv3465) genes coexist in an operon. DTDP-rhamnose and NADP are produced by RmlD-catalyzed reactions of DTDP-6-deoxy-l-xylo-4-hexose with NADPH. d-n-Acetylglucosamine-1-phosphate is converted from DTDP-rhamnose residues by rhamnose transferase encoded by the wbbL gene, and then d-n-acetylglucosamine l-rhamnose disaccharide ligands are formed. However, the MTB Rmla-d gene was not located at a locus, but RmlD (Rv3266c) gene was present in the operon of wbbL (Rv3265c) and manB (Rv3264c)92 (Fig. 7). This could explain why RmlA, RmlB, RmlC, and RmlD are important targets for the development of new anti-TB drugs.
Figure 7.
DTDP-l-rhamnose pathway.
DTDP-d-glucose is obtained by condensation of G1P with DTTP catalyzed by RmlA (Supporting Information Fig. S13), which is homologous to other bacterial sugar nucleotide transferases such as G1P uridylyltransferase. The existence of a feedback mechanism, that is, inhibition of RmlA by DTDP-l-rhamnose as the final product of the pathway, occurs in competitive and noncompetitive ways, indicating that RmlA is the control point of the pathway. In addition, from the perspective of protein engineering, the application of RmlA in the glucuronization has attracted extensive attention.
A series of triazine-indole-benzimidazolone compounds have been reported by Sivendran et al.93, which inhibited the activity of an essential cell wall biosynthetic enzyme of MTB. The most preferred compound is 1-(3-(5-ethyl-5H[1,2,4]triazino[5,6-b]indol-3-ylthio)propyl)-1H-benzo[d]imidazole-2(3H)-one having an IC50 for inhibition of RmlC of 0.2 μmol/L (compound 36). This compound is nontoxic and the IC50 remains unchanged when the RmlD concentration is increased by a factor of 80-fold (Supporting Information Fig. S14). RmlC is inhibited by compound 36 and the IC50 changes as the RmlC concentration changes.
For DTDP-6-deoxy-l-xylo-4-hexose sugar reductase (RmlD), Wang et al.94 reported virtual screenings. Studies have shown that the RmlD inhibitor is compound 37 with an IC50 of 0.9 μmol/L, which binds to RmlD at the DTDP-l-rhamnose binding site in the presence of NADPH (Fig. S14). The formation of a hydrogen bond between the hydroxyl group and Asp105 and/or Thr104 is a structural feature of the compound. It was found that the enzymatic activity of RmlD72 was important, and Thr104 was part of the conserved catalytic triad95. RmlD is an enzyme that inhibits compound 37.
2.4. Other drug targets
Cell wall synthesis is inhibited by a cell wall inhibitor, compound 38 (teixobactin), which binds to highly conserved motifs of lipid II (a precursor of peptidoglycan) and lipid III (a precursor of cell wall phospho-teichoic acid). MTB mutants resistant to teixobactin have not been reported. However, a cell wall inhibitor that was developed from uncultured bacteria culled from in situ diffusion chambers has been reported Ling96. The development of antibiotic resistance is characteristic of compound 38. MTB H37Rv with an MIC of 0.125 μg/mL is inhibited by teixobactin. Teixobactin was not toxic to NIH/3T3 and HepG2 cells in mammals at 100 μg/mL, which was the highest dose tested. The compound does not bind to DNA, has no hemolytic activity, and the target of the compound is not a protein. Teixobactin inhibited the biosynthesis of PG in vitro, with lipid I, lipid II or undecaenyl pyrophosphate as a substrate in a dose-dependent manner. Teixobactin may bind conjugated lipid intermediates with PG and AG, and thus, it is active against MTB. The PD50 (a protective dose of 0.2 mg/kg for half the animals that survive) was determined in vivo to be equivalent to 2.75 mg/kg PD50 of vancomycin, a major antibiotic used to treat methicillin-resistant Staphylococcus aureus (Supporting Information Fig. S15)96.
MTB glutamine synthetase (GS) is a promising target for anti-TB drugs. Several types of GS inhibitors targeting ATP binding sites have been identified by Gising et al.97, and a class of 2-tert-butyl-4,5-diarylimidazoles has been identified, furthering the study of anti-TB drugs. The design, synthesis and X-ray crystallographic studies have demonstrated that GS inhibitors have sub-micromolar IC50 values and promising MIC values. The best GS inhibitor (compound 39) has an IC50 of 0.049 μmol/L and an MIC of 2 μg/mL (Supporting Information Fig. S16). The crystal structure of MTB GS is shown in Supporting Information Fig. S17, which is complexed with compound 39. Two hydrogen bonds are shown on the crystal structure; one is formed by the nitrogen of the pyridyl ring and Ser280, and the other by the amino group on the pyridyl ring and the Lys361 amino acid residue. In addition, it has been found that the nitrogen of imidazole interacts with one of the water molecules in the vicinity. The compound is stabilized by hydrophobic interactions with Tyr129, Phe232, and Trp282.
3. Conclusions and outlook
The discovery of high efficiency, low toxicity new anti-TB drugs is urgently needed to shorten the treatment time, especially after the increase in cases of MDR and XDR MTB infections. This paper reviewed the enzymes and inhibitors that are currently known; that is, those involved in cell wall biosynthesis of MTB.
Sequencing of the MTB genome92 has greatly assisted in the prevention and treatment of TB, and many potential therapeutic targets involving lipid biosynthesis and metabolism have been discovered. Many large chemical libraries contain phenotypic and targeted screening data, and there are many novel anti-TB agents to be identified98. Rapid whole-genome sequencing is included among modern molecular biology techniques, and may help with rapid and accurate identification of drugs. Therefore, in the past 10 years, techniques for the discovery of anti-TB drugs have greatly improved and have been applied successfully to the unique therapeutic targets of mycobacterial cell walls81.
Cell wall biosynthesis has been investigated by researchers as a target for novel antimicrobial agents and more-specific anti-TB drugs. New antibacterial drugs have been used in recent years with unsatisfactory results, so the methods have been questioned to some extent. In the case of anti-TB drugs, no specific enzyme has been targeted among the most promising candidates. However, the effectiveness of this method has been proven in treating acquired immunodeficiency virus (AIDS) proteases. Thus, only one or two successful treatments for a single bacterial enzyme is needed. We suggest that researchers may underestimate the value of screening bacteria for drug targets; therefore, now is not the right time to abandon this targeted approach. We hope that this review inspires researchers to think about which bacterial cell wall enzymes are the most effective drug targets, and to conduct studies to design more effective small molecule inhibitors.
Acknowledgments
This study was supported by National Natural Science Foundation of China (82073701, 31900687), Natural Science Foundation of Jiangsu Province (SBK2019040713, China) and the Project Program of State Key Laboratory of Natural Medicines, China Pharmaceutical University (SKLNMZZ202013). This study was also supported by Jiangsu Key Laboratory of Drug Design and Optimization, China Pharmaceutical University (No. 2020KFKT-5), and “Double First-Class” University Project (CPU2018GF04, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.04.014.
Author contributions
All the authors contributed to the writing and editing of this review article.
Conflicts of interest
The authors declare no competing financial interest.
Appendix A. Supporting information
The following is the Supporting data to this article:
References
- 1.Furin J., Cox H., Pai M. Tuberculosis. Lancet. 2019;393:1642–1656. doi: 10.1016/S0140-6736(19)30308-3. [DOI] [PubMed] [Google Scholar]
- 2.Fu H., Lewnard J.A., Frost I., Laxminarayan R., Arinaminpathy N. Modelling the global burden of drug-resistant tuberculosis avertable by a post-exposure vaccine. Nat Commun. 2021;12:424. doi: 10.1038/s41467-020-20731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harding E. WHO global progress report on tuberculosis elimination (vol 8, pg 19, 2020) Lancet Respir Med. 2020;8:19. doi: 10.1016/S2213-2600(19)30418-7. [DOI] [PubMed] [Google Scholar]
- 4.Lienhardt C., Cook S.V., Burgos M., Yorke-Edwards V., Rigouts L., Anyo G., et al. Efficacy and safety of a 4-drug fixed-dose combination regimen compared with separate drugs for treatment of pulmonary tuberculosis: the study c randomized controlled trial. JAMA. 2011;305:1415–1423. doi: 10.1001/jama.2011.436. [DOI] [PubMed] [Google Scholar]
- 5.Dooley K.E., Mitnick C.D., Ann De Groote M., Obuku E., Belitsky V., Hamilton C.D., et al. Old drugs, new purpose: retooling existing drugs for optimized treatment of resistant tuberculosis. Clin Infect Dis. 2012;55:572–581. doi: 10.1093/cid/cis487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chikhale R.V., Barmade M.A., Murumkar P.R., Yadav M.R. Overview of the development of DprE1 inhibitors for combating the menace of tuberculosis. J Med Chem. 2018;61:8563–8593. doi: 10.1021/acs.jmedchem.8b00281. [DOI] [PubMed] [Google Scholar]
- 7.Keam S.J. Pretomanid: first approval. Drugs. 2019;79:1797–1803. doi: 10.1007/s40265-019-01207-9. [DOI] [PubMed] [Google Scholar]
- 8.Prasad M.S., Bhole R.P., Khedekar P.B., Chikhale R.V. Mycobacterium enoyl acyl carrier protein reductase (InhA): a key target for antitubercular drug discovery. Bioorg Chem. 2021;115:105242. doi: 10.1016/j.bioorg.2021.105242. [DOI] [PubMed] [Google Scholar]
- 9.Lun S., Xiao S., Zhang W., Wang S., Gunosewoyo H., Yu L.F., et al. Therapeutic potential of coumestan PKS13 inhibitors for tuberculosis. Antimicrob Agents Chemother. 2021;65 doi: 10.1128/AAC.02190-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shao M., McNeil M., Cook G.M., Lu X. MmpL3 inhibitors as antituberculosis drugs. Eur J Med Chem. 2020;200:112390. doi: 10.1016/j.ejmech.2020.112390. [DOI] [PubMed] [Google Scholar]
- 11.Imran M., Alshrari A.S., Thabet H.K., Abida, Afroz Bakht M. Synthetic molecules as DprE1 inhibitors: a patent review. Expert Opin Ther Pat. 2021;31:759–772. doi: 10.1080/13543776.2021.1902990. [DOI] [PubMed] [Google Scholar]
- 12.Dighe S.N., Collet T.A. Recent advances in DNA gyrase-targeted antimicrobial agents. Eur J Med Chem. 2020;199:112326. doi: 10.1016/j.ejmech.2020.112326. [DOI] [PubMed] [Google Scholar]
- 13.Khan M.T., Kaushik A.C., Bhatti A.I., Zhang Y.J., Zhang S., Wei A.J., et al. Marine natural products and drug resistance in latent tuberculosis. Mar Drugs. 2019;17:549. doi: 10.3390/md17100549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouz G., Zitko J. Inhibitors of aminoacyl-tRNA synthetases as antimycobacterial compounds: an up-to-date review. Bioorg Chem. 2021;110:104806. doi: 10.1016/j.bioorg.2021.104806. [DOI] [PubMed] [Google Scholar]
- 15.Igarashi M., Nakagawa N., Doi N., Hattori S., Naganawa H., Hamada M. Caprazamycin B, a novel anti-tuberculosis antibiotic, from Streptomyces sp. J Antibiot (Tokyo) 2003;56:580–583. doi: 10.7164/antibiotics.56.580. [DOI] [PubMed] [Google Scholar]
- 16.Hoshino K., Inoue K., Murakami Y., Kurosaka Y., Namba K., Kashimoto Y., et al. In vitro and in vivo antibacterial activities of DC-159a, a new fluoroquinolone. Antimicrob Agents Chemother. 2008;52:65–76. doi: 10.1128/AAC.00853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogatcheva E., Hanrahan C., Nikonenko B., de los Santos G., Reddy V., Chen P., et al. Identification of SQ609 as a lead compound from a library of dipiperidines. Bioorg Med Chem Lett. 2011;21:5353–5357. doi: 10.1016/j.bmcl.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riste M., Marshall J.L., Satti I., Harris S.A., Wilkie M., Lopez Ramon R., et al. Phase I trial evaluating the safety and immunogenicity of candidate TB vaccine MVA85A, delivered by aerosol to healthy M.tb-infected adults. Vaccines (Basel) 2021;9:336. doi: 10.3390/vaccines9040396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.la Torre L.F., Franco-Gonzalez D.L., Brennan-Bourdon L.M., Molina-Frechero N., Alonso-Castro A.J., Isiordia-Espinoza M.A. Analgesic efficacy of etoricoxib following third molar surgery: a meta-analysis. Behav Neurol. 2021;2021:9536054. doi: 10.1155/2021/9536054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo S., Wang B., Fu L., Chen X., Zhang W., Huang H., et al. In vitro and in vivo activity of oxazolidinone candidate OTB-658 against Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2021;65 doi: 10.1128/AAC.00974-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B., Li J., Yang X., Wu L., Zhang J., Yang Y., et al. Crystal structures of membrane transporter MmpL3, an anti-TB drug target. Cell. 2019;176 doi: 10.1016/j.cell.2019.01.003. 636–648.e13. [DOI] [PubMed] [Google Scholar]
- 22.Tkachuk A.P., Bykonia E.N., Popova L.I., Kleymenov D.A., Semashko M.A., Chulanov V.P., et al. Safety and immunogenicity of the GamTBvac, the recombinant subunit tuberculosis vaccine candidate: a phase II, multi-center, double-blind, randomized, placebo-controlled study. Vaccines (Basel) 2020;8:652. doi: 10.3390/vaccines8040652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foti C., Piperno A., Scala A., Giuffre O. Oxazolidinone antibiotics: chemical, biological and analytical aspects. Molecules. 2021;26:4280. doi: 10.3390/molecules26144280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu H., Fu L., Wang B., Chen X., Zhao J., Huang H., et al. Activity of clofazimine and TBI-166 against Mycobacterium tuberculosis in different administration intervals in mouse tuberculosis models. Antimicrob Agents Chemother. 2021;65 doi: 10.1128/AAC.02164-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shih H.P., Ding J.Y., Yeh C.F., Chi C.Y., Ku C.L. Anti-interferon-gamma autoantibody-associated immunodeficiency. Curr Opin Immunol. 2021;72:206–214. doi: 10.1016/j.coi.2021.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Lu Y., Wei Z., Yang G., Lai Y., Liu R. Investigating the efficacy and safety of thalidomide for treating patients with β-thalassemia: a meta-analysis. Front Pharmacol. 2021;12:814302. doi: 10.3389/fphar.2021.814302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pramar Y.V., Mandal T.K., Bostanian L.A., Le G., Morris T.C., Graves R.A. Physicochemical and microbiological stability of compounded metronidazole suspensions in PCCA suspendIt. Int J Pharm Compd. 2021;25:169–175. [PMC free article] [PubMed] [Google Scholar]
- 28.Black T.A., Buchwald U.K. The pipeline of new molecules and regimens against drug-resistant tuberculosis. J Clin Tuberc Other Mycobact Dis. 2021;25:100285. doi: 10.1016/j.jctube.2021.100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bloem A., Bax H.I., Yusuf E., Verkaik N.J. New-generation antibiotics for treatment of gram-positive infections: a review with focus on endocarditis and osteomyelitis. J Clin Med. 2021;10:1743. doi: 10.3390/jcm10081743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sethiya J.P., Sowards M.A., Jackson M., North E.J. MmpL3 inhibition: a new approach to treat nontuberculous mycobacterial infections. Int J Mol Sci. 2020;21:6202. doi: 10.3390/ijms21176202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gisbert J.P. Rifabutin for the treatment of Helicobacter pylori infection: a review. Pathogens. 2020;10:15. doi: 10.3390/pathogens10010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morino Y., Sugimoto M., Nagata N., Niikiura R., Iwata E., Hamada M., et al. Influence of cytochrome P450 2C19 genotype on Helicobacter pylori proton pump inhibitor-amoxicillin-clarithromycin eradication therapy: a meta-analysis. Front Pharmacol. 2021;12:759249. doi: 10.3389/fphar.2021.759249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jhun B.W., Koh W.J. Treatment of isoniazid-resistant pulmonary tuberculosis. Tuberc Respir Dis. 2020;83:20–30. doi: 10.4046/trd.2019.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bahraminia F., Azimi T., Zangiabadian M., Nasiri M.J., Goudarzi M., Dadashi M., et al. Rifampicin-resistant tuberculosis in Iran: a systematic review and meta-analysis. Iran J Basic Med Sci. 2021;24:720–725. doi: 10.22038/ijbms.2021.47360.10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gils T., Lynen L., de Jong B.C., Van Deun A., Decroo T. Pretomanid for tuberculosis: a systematic review. Clin Microbiol Infect. 2022;28:31–42. doi: 10.1016/j.cmi.2021.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Mohammadi B., Ramazanzadeh R., Nouri B., Rouhi S. Frequency of codon 306 mutations in embB gene of Mycobacterium tuberculosis resistant to ethambutol: a systematic review and meta-analysis. Int J Prev Med. 2020;11:112. doi: 10.4103/ijpvm.IJPVM_114_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khademi F., Vaez H., Sahebkar A., Taheri R.A. Group a Streptococcus antibiotic resistance in iranian children: a meta-analysis. Oman Med J. 2021;36:e222. doi: 10.5001/omj.2020.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoy S.M. Amikacin liposome inhalation suspension in refractory Mycobacterium avium complex lung disease: a profile of its use. Clin Drug Invest. 2021;41:405–412. doi: 10.1007/s40261-021-01010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rybak L.P., Ramkumar V., Mukherjea D. Ototoxicity of non-aminoglycoside antibiotics. Front Neurol. 2021;12:652674. doi: 10.3389/fneur.2021.652674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rizzo S., Gambini G., De Vico U., Rizzo C., Kilian R. A one-week course of levofloxacin/dexamethasone eye drops: a review on a new approach in managing patients after cataract surgery. Ophthalmol Ther. 2022;11:101–111. doi: 10.1007/s40123-021-00435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu T., Makino K. Physiological action of ethylthiamine. J Vitaminol. 1961;7:202–208. doi: 10.5925/jnsv1954.7.202. [DOI] [PubMed] [Google Scholar]
- 42.Henter I.D., Park L.T., Zarate C.A., Jr. Novel glutamatergic modulators for the treatment of mood disorders: current status. CNS Drugs. 2021;35:527–543. doi: 10.1007/s40263-021-00816-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pecora F., Dal Canto G., Veronese P., Esposito S. Treatment of multidrug-resistant and extensively drug-resistant tuberculosis in children: the role of bedaquiline and delamanid. Microorganisms. 2021;9:1074. doi: 10.3390/microorganisms9051074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munoz-Elias E.J., McKinney J.D. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang J., Wang B., Wu T., Wan J., Tu Z., Njire M., et al. Design, synthesis, and biological evaluation of pyrazolo[1,5-a]pyridine-3-carboxamides as novel antitubercular agents. ACS Med Chem Lett. 2015;6:814–818. doi: 10.1021/acsmedchemlett.5b00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Disratthakit A., Prammananan T., Tribuddharat C., Thaipisuttikul I., Doi N., Leechawengwongs M., et al. Role of gyrB mutations in pre-extensively and extensively drug-resistant tuberculosis in Thai clinical isolates. Antimicrob Agents Chemother. 2016;60:5189–5197. doi: 10.1128/AAC.00539-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voskuil M.I., Schnappinger D., Visconti K.C., Harrell M.I., Dolganov G.M., Sherman D.R., et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe S., Matsumura K., Iwai H., Funatogawa K., Haishima Y., Fukui C., et al. A mutation in the 16S rRNA decoding region attenuates the virulence of Mycobacterium tuberculosis. Infect Immun. 2016;84:2264–2273. doi: 10.1128/IAI.00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gandotra S., Lebron M.B., Ehrt S. The Mycobacterium tuberculosis proteasome active site threonine is essential for persistence yet dispensable for replication and resistance to nitric oxide. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan S., Nagarajan S.N., Parikh A., Samantaray S., Singh A., Kumar D., et al. Phosphorylation of enoyl-acyl carrier protein reductase inhA impacts mycobacterial growth and survival. J Biol Chem. 2010;285:37860–37871. doi: 10.1074/jbc.M110.143131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tahlan K., Wilson R., Kastrinsky D.B., Arora K., Nair V., Fischer E., et al. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2012;56:1797–1809. doi: 10.1128/AAC.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sassetti C.M., Boyd D.H., Rubin E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 53.Rozwarski D.A., Grant G.A., Barton D.H., Jacobs W.R., Jr., Sacchettini J.C. Modification of the nadh of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science. 1998;279:98–102. doi: 10.1126/science.279.5347.98. [DOI] [PubMed] [Google Scholar]
- 54.Takayama K., Wang C., Besra G.S. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin Microbiol Rev. 2005;18:81–101. doi: 10.1128/CMR.18.1.81-101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stanley S.A., Kawate T., Iwase N., Shimizu M., Clatworthy A.E., Kazyanskaya E., et al. Diarylcoumarins inhibit mycolic acid biosynthesis and kill Mycobacterium tuberculosis by targeting fadd32. Proc Natl Acad Sci U S A. 2013;110:11565–11570. doi: 10.1073/pnas.1302114110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawate T., Iwase N., Shimizu M., Stanley S.A., Wellington S., Kazyanskaya E., et al. Synthesis and structure‒activity relationships of phenyl-substituted coumarins with anti-tubercular activity that target FadD32. Bioorg Med Chem Lett. 2013;23:6052–6059. doi: 10.1016/j.bmcl.2013.09.035. [DOI] [PubMed] [Google Scholar]
- 57.Fang C., Lee K.K., Nietupski R., Bates R.H., Fernandez-Menendez R., Lopez-Roman E.M., et al. Discovery of heterocyclic replacements for the coumarin core of anti-tubercular fadd32 inhibitors. Bioorg Med Chem Lett. 2018;28:3529–3533. doi: 10.1016/j.bmcl.2018.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuhn M.L., Alexander E., Minasov G., Page H.J., Warwrzak Z., Shuvalova L., et al. Structure of the essential mtb fadd32 enzyme: a promising drug target for treating tuberculosis. ACS Infect Dis. 2016;2:579–591. doi: 10.1021/acsinfecdis.6b00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flint L., Korkegian A., Parish T. InhA inhibitors have activity against non-replicating Mycobacterium tuberculosis. PLoS One. 2020;15 doi: 10.1371/journal.pone.0239354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pan P., Tonge P.J. Targeting InhA, the FasII enoyl-acp reductase: sar studies on novel inhibitor scaffolds. Curr Top Med Chem. 2012;12:672–693. doi: 10.2174/156802612799984535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dogan S.D., Gunduz M.G., Dogan H., Krishna V.S., Lherbet C., Sriram D. Design and synthesis of thiourea-based derivatives as Mycobacterium tuberculosis growth and enoyl acyl carrier protein reductase (InhA) inhibitors. Eur J Med Chem. 2020;199:112402. doi: 10.1016/j.ejmech.2020.112402. [DOI] [PubMed] [Google Scholar]
- 62.Dogan H., Dogan S.D., Gunduz M.G., Krishna V.S., Lherbet C., Sriram D., et al. Discovery of hydrazone containing thiadiazoles as Mycobacterium tuberculosis growth and enoyl acyl carrier protein reductase (InhA) inhibitors. Eur J Med Chem. 2020;188:112035. doi: 10.1016/j.ejmech.2020.112035. [DOI] [PubMed] [Google Scholar]
- 63.Shirude P.S., Madhavapeddi P., Naik M., Murugan K., Shinde V., Nandishaiah R., et al. Methyl-thiazoles: a novel mode of inhibition with the potential to develop novel inhibitors targeting InhA in Mycobacterium tuberculosis. J Med Chem. 2013;56:8533–8542. doi: 10.1021/jm4012033. [DOI] [PubMed] [Google Scholar]
- 64.Wilson R., Kumar P., Parashar V., Vilcheze C., Veyron-Churlet R., Freundlich J.S., et al. Antituberculosis thiophenes define a requirement for PKS13 in mycolic acid biosynthesis. Nat Chem Biol. 2013;9:499–506. doi: 10.1038/nchembio.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thanna S., Knudson S.E., Grzegorzewicz A., Kapil S., Goins C.M., Ronning D.R., et al. Synthesis and evaluation of new 2-aminothiophenes against Mycobacterium tuberculosis. Org Biomol Chem. 2016;14:6119–6133. doi: 10.1039/c6ob00821f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aggarwal A., Parai M.K., Shetty N., Wallis D., Woolhiser L., Hastings C., et al. Development of a novel lead that targets M. tuberculosis polyketide synthase 13. Cell. 2017;170 doi: 10.1016/j.cell.2017.06.025. 249–259.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang W., Liu L.L., Lun S., Wang S.S., Xiao S., Gunosewoyo H., et al. Design and synthesis of mycobacterial PKS13 inhibitors: conformationally rigid tetracyclic molecules. Eur J Med Chem. 2021;213:113202. doi: 10.1016/j.ejmech.2021.113202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li K., Schurig-Briccio L.A., Feng X., Upadhyay A., Pujari V., Lechartier B., et al. Multitarget drug discovery for tuberculosis and other infectious diseases. J Med Chem. 2014;57:3126–3139. doi: 10.1021/jm500131s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lun S., Guo H., Onajole O.K., Pieroni M., Gunosewoyo H., Chen G., et al. Indoleamides are active against drug-resistant Mycobacterium tuberculosis. Nat Commun. 2013;4:2907. doi: 10.1038/ncomms3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grzegorzewicz A.E., Pham H., Gundi V.A., Scherman M.S., North E.J., Hess T., et al. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat Chem Biol. 2012;8:334–341. doi: 10.1038/nchembio.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ray P.C., Huggett M., Turner P.A., Taylor M., Cleghorn L.A.T., Early J., et al. Spirocycle MmpL3 inhibitors with improved hERG and cytotoxicity profiles as inhibitors of Mycobacterium tuberculosis growth. ACS Omega. 2021;6:2284–2311. doi: 10.1021/acsomega.0c05589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Heijenoort J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat Prod Rep. 2001;18:503–519. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]
- 73.Rani C., Khan I.A. UDP-GlcNAc pathway: potential target for inhibitor discovery against M. tuberculosis. Eur J Pharmaceut Sci. 2016;83:62–70. doi: 10.1016/j.ejps.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 74.Floquet N., Richez C., Durand P., Maigret B., Badet B., Badet-Denisot M.A. Discovering new inhibitors of bacterial glucosamine-6P synthase (GlmS) by docking simulations. Bioorg Med Chem Lett. 2007;17:1966–1970. doi: 10.1016/j.bmcl.2007.01.052. [DOI] [PubMed] [Google Scholar]
- 75.Guzman J.D., Wube A., Evangelopoulos D., Gupta A., Hufner A., Basavannacharya C., et al. Interaction of N-methyl-2-alkenyl-4-quinolones with ATP-dependent MurE ligase of Mycobacterium tuberculosis: antibacterial activity, molecular docking and inhibition kinetics. J Antimicrob Chemother. 2011;66:1766–1772. doi: 10.1093/jac/dkr203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Konduri S., Prashanth J., Krishna V.S., Sriram D., Behera J.N., Siegel D., et al. Design and synthesis of purine connected piperazine derivatives as novel inhibitors of Mycobacterium tuberculosis. Bioorg Med Chem Lett. 2020;30:127512. doi: 10.1016/j.bmcl.2020.127512. [DOI] [PubMed] [Google Scholar]
- 77.McNeil M., Wallner S.J., Hunter S.W., Brennan P.J. Demonstration that the galactosyl and arabinosyl residues in the cell-wall arabinogalactan of Mycobacterium leprae and Mycobacterium tuberculosis are furanoid. Carbohydr Res. 1987;166:299–308. doi: 10.1016/0008-6215(87)80065-4. [DOI] [PubMed] [Google Scholar]
- 78.Xu L., Qian L., Kang J., Sha S., Xin Y., Lu S., et al. Down-regulation of N-acetylglucosamine-1-phosphate transferase (WecA) enhanced the sensitivity of Mycobacterium smegmatis against rifampin. J Appl Microbiol. 2016;121:966–972. doi: 10.1111/jam.13228. [DOI] [PubMed] [Google Scholar]
- 79.Ishizaki Y., Takahashi Y., Kimura T., Inoue M., Hayashi C., Igarashi M. Synthesis and biological activity of analogs of CPZEN-45, a novel antituberculosis drug. J Antibiot (Tokyo) 2019;72:970–980. doi: 10.1038/s41429-019-0225-5. [DOI] [PubMed] [Google Scholar]
- 80.Mitachi K., Siricilla S., Yang D., Kong Y., Skorupinska-Tudek K., Swiezewska E., et al. Fluorescence-based assay for polyprenyl phosphate-GlcNAc-1-phosphate transferase (WecA) and identification of novel antimycobacterial WecA inhibitors. Anal Biochem. 2016;512:78–90. doi: 10.1016/j.ab.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Makarov V., Manina G., Mikusova K., Mollmann U., Ryabova O., Saint-Joanis B., et al. Benzothiazinones kill Mycobacterium tuberculosis by blocking Arabinan synthesis. Science. 2009;324:801–804. doi: 10.1126/science.1171583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Makarov V., Lechartier B., Zhang M., Neres J., van der Sar A.M., Raadsen S.A., et al. Towards a new combination therapy for tuberculosis with next generation benzothiazinones. EMBO Mol Med. 2014;6:372–383. doi: 10.1002/emmm.201303575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang F., Sambandan D., Halder R., Wang J., Batt S.M., Weinrick B., et al. Identification of a small molecule with activity against drug-resistant and persistent tuberculosis. Proc Natl Acad Sci U S A. 2013;110:E2510–E2517. doi: 10.1073/pnas.1309171110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tiwari R., Miller P.A., Cho S., Franzblau S.G., Miller M.J. Syntheses and antituberculosis activity of 1,3-benzothiazinone sulfoxide and sulfone derived from BTZ043. ACS Med Chem Lett. 2015;6:128–133. doi: 10.1021/ml5003458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tiwari R., Mollmann U., Cho S., Franzblau S.G., Miller P.A., Miller M.J. Design and syntheses of anti-tuberculosis agents inspired by BTZ043 using a scaffold simplification strategy. ACS Med Chem Lett. 2014;5:587–591. doi: 10.1021/ml500039g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tiwari R., Miller P.A., Chiarelli L.R., Mori G., Sarkan M., Centarova I., et al. Design, syntheses, and anti-TB activity of 1,3-benzothiazinone azide and click chemistry products inspired by BTZ043. ACS Med Chem Lett. 2016;7:266–270. doi: 10.1021/acsmedchemlett.5b00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li L., Lv K., Yang Y., Sun J., Tao Z., Wang A., et al. Identification of N-benzyl 3,5-dinitrobenzamides derived from PBTZ169 as antitubercular agents. ACS Med Chem Lett. 2018;9:741–745. doi: 10.1021/acsmedchemlett.8b00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li P., Wang B., Zhang X., Batt S.M., Besra G.S., Zhang T., et al. Identification of novel benzothiopyranone compounds against Mycobacterium tuberculosis through scaffold morphing from benzothiazinones. Eur J Med Chem. 2018;160:157–170. doi: 10.1016/j.ejmech.2018.09.042. [DOI] [PubMed] [Google Scholar]
- 89.Piton J., Vocat A., Lupien A., Foo C.S., Riabova O., Makarov V., et al. Structure-based drug design and characterization of sulfonyl-piperazine benzothiazinone inhibitors of DprE1 from Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/AAC.00681-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rogacki M.K., Pitta E., Balabon O., Huss S., Lopez-Roman E.M., Argyrou A., et al. Identification and profiling of hydantoins-a novel class of potent antimycobacterial DprE1 inhibitors. J Med Chem. 2018;61:11221–11249. doi: 10.1021/acs.jmedchem.8b01356. [DOI] [PubMed] [Google Scholar]
- 91.Balabon O., Pitta E., Rogacki M.K., Meiler E., Casanueva R., Guijarro L., et al. Optimization of hydantoins as potent antimycobacterial decaprenylphosphoryl-beta-d-ribose oxidase (DprE1) inhibitors. J Med Chem. 2020;63:5367–5386. doi: 10.1021/acs.jmedchem.0c00107. [DOI] [PubMed] [Google Scholar]
- 92.Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 93.Sivendran S., Jones V., Sun D., Wang Y., Grzegorzewicz A.E., Scherman M.S., et al. Identification of triazinoindol-benzimidazolones as nanomolar inhibitors of the Mycobacterium tuberculosis enzyme TDP-6-deoxy-d-xylo-4-hexopyranosid-4-ulose 3,5-epimerase (RmlC) Bioorg Med Chem. 2010;18:896–908. doi: 10.1016/j.bmc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Y., Hess T.N., Jones V., Zhou J.Z., McNeil M.R., Andrew McCammon J. Novel inhibitors of Mycobacterium tuberculosis dTDP-6-deoxy-l-lyxo-4-hexulose reductase (RmlD) identified by virtual screening. Bioorg Med Chem Lett. 2011;21:7064–7067. doi: 10.1016/j.bmcl.2011.09.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blankenfeldt W., Kerr I.D., Giraud M.F., McMiken H.J., Leonard G., Whitfield C., et al. Variation on a theme of sdr. dTDP-6-deoxy-l-lyxo-4-hexulose reductase (RmlD) shows a new Mg2+-dependent dimerization mode. Structure. 2002;10:773–786. doi: 10.1016/s0969-2126(02)00770-0. [DOI] [PubMed] [Google Scholar]
- 96.Ling L.L., Schneider T., Peoples A.J., Spoering A.L., Engels I., Conlon B.P., et al. Erratum: a new antibiotic kills pathogens without detectable resistance. Nature. 2015;520:388. doi: 10.1038/nature14303. [DOI] [PubMed] [Google Scholar]
- 97.Gising J., Nilsson M.T., Odell L.R., Yahiaoui S., Lindh M., Iyer H., et al. Trisubstituted imidazoles as Mycobacterium tuberculosis glutamine synthetase inhibitors. J Med Chem. 2012;55:2894–2898. doi: 10.1021/jm201212h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ananthan S., Faaleolea E.R., Goldman R.C., Hobrath J.V., Kwong C.D., Laughon B.E., et al. High-throughput screening for inhibitors of Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2009;89:334–353. doi: 10.1016/j.tube.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.