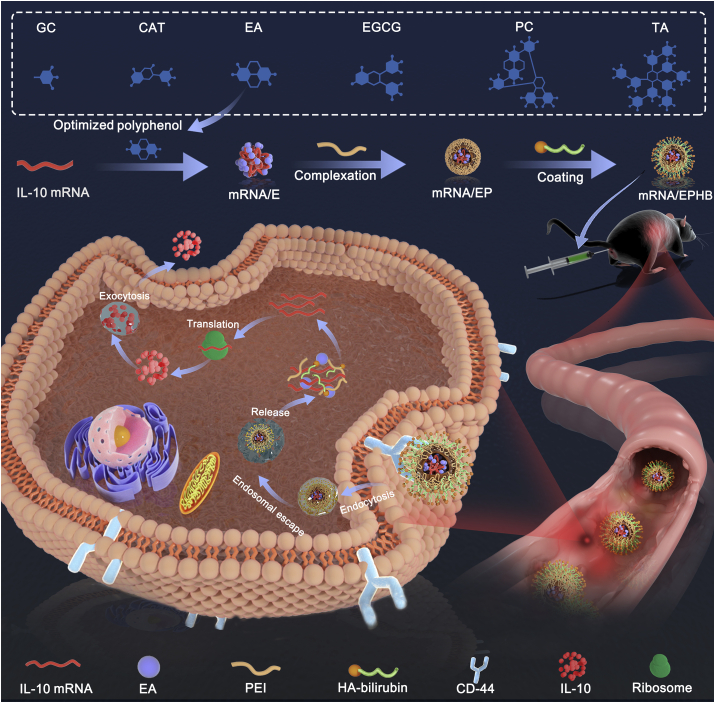
Abstract
With the development of synthesis technology, modified messenger RNA (mRNA) has emerged as a novel category of therapeutic agents for a broad of diseases. However, effective intracellular delivery of mRNA remains challenging, especially for its sensitivity to enzymatic degradation. Here, we propose a polyphenol-assisted handy delivery strategy for efficient in vivo delivery of IL-10 mRNA. IL-10 mRNA binds to polyphenol ellagic acid through supramolecular binding to yield a negatively charged core, followed by complexing with linear polyetherimide and coating with bilirubin-modified hyaluronic acid to obtain a layer-by-layer nanostructure. The nanostructure specifically up-regulated the level of IL-10, effectively inhibited the expression of inflammatory factors, promoted mucosal repair, protected colonic epithelial cells against apoptosis, and exerted potent therapeutic efficacy in dextran sulfate sodium salt-induced acute and chronic murine models of colitis. The designed delivery system without systemic toxicity has the potential to facilitate the development of a promising platform for mRNA delivery in ulcerative colitis treatment.
KEY WORDS: Polyphenol, mRNA, IL-10, Colitis, Inflammatory bowel disease, Drug delivery, mRNA delivery, Supramolecular binding
Graphical abstract
The polyphenol-assisted IL-10 mRNA delivery system with self-protection and active targeting mechanisms. Through rectal administration, this delivery system achieved conspicuous therapeutic effect on dextran sulfate sodium salt-induced ulcerative colitis model.

1. Introduction
Inflammatory bowel disease (IBD) is a chronic pathological disorder that causes a long-term inflammatory response of the gastrointestinal tract, characterized by the complex gene dysregulation and immune response1. Crohn's disease (CD) and ulcerative colitis (UC) are the principle types of IBD, which affects at least 3.6 million patients worldwide2, 3, 4. At present, glucocorticoids, immunosuppressant, and 5-aminosalicylic acid (5-ASA) are the first-line intervention agents of IBD. However, nontargeted traditional drug intervention commonly has multiple side effects because of their nonspecific systemic circulation characteristics5, 6, 7. Interleukin 10 (IL-10) is a central anti-inflammatory cytokine involved in maintaining intestinal immunity. Despite the potential, clinical trials using recombinant IL-10 failed to demonstrate a prominent therapeutic effect due to the short half-life. Recently, extensive studies have demonstrated the pivotally facilitative role of mRNA delivery for IBD treatment8, 9, 10. Therefore, through artificial delivery of IL-10 messenger ribonucleic acid (mRNA) to the inflamed colon, the level of IL-10 can be effectively improved to relieve colitis, which is an effective strategy for IBD treatment by regulating IL-10 level11.
However, the effective translation of IL-10 mRNA into bioactive proteins in cells is hindered due to inherent shortcomings of mRNA, such as the easy enzymatic degradation, poor efficiency of batch transcription due to the large molecules in vitro, and immunogenicity in vivo12, 13, 14, 15. The recently developed structural modification of mRNA allows the preparation of mRNA with unique biological activity, which has higher expression efficiency and lower immunogenicity in host cells. At present, the adjustment strategies mainly include the incorporation of m7G(5′)ppp(5′)G (mCAP), m7G(5′)ppp(5′) (2′OMeA)pG (EZ Cap™), and anti-reverse cap analog (ARCA) structures into transcriptional mRNA fragments to improve translation efficiency, adding modified nucleotides and poly(A) tails to reduce host cell immune response and enhance stability16, 17, 18, 19. Nevertheless, to realize the strategy of intervening IBD through manipulating the expression of IL-10 mRNA, the modification of mRNA structure alone is insufficient. The effective protection and specifically targeted delivery in vivo are the core challenges surrounding the utility of IL-10 mRNA as a gene therapy strategy.
To achieve targeted delivery of mRNA, bioactive materials with mature theoretical supports and technical practices, including cationic lipid, cationic polymers, and proteins, have been employed20, 21, 22, 23. However, these common delivery strategies have two major defects. First, although a ribonuclease (RNase)-free environment is mostly ensured in the preparation process, the preparation and delivery of mRNA delivery system is a complex process, which holds the possible risk of mRNA degradation due to unprotected exposure in vivo and in vitro. Traditional mRNA delivery systems cannot provide an active protective mechanism for mRNA, resulting in a substantial discount of the loaded mRNA in delivery systems. Second, for delivery systems constructed of single carrier material, including cationic lipid or cationic polymer, the main targeting mechanism is the passive targeting effect of nanopreparations10,11. However, in the process of mRNA delivery, due to the instability of the single chain macromolecular structure, an active targeting strategy can more effectively achieve accurate delivery.
Polyphenols are excellent antioxidants with dense ortho and meta-phenolic hydroxyls, which can realize complexation with nucleic acids by intermolecular forces24,25. Recently, studies have demonstrated that polyphenols mixed with nucleic acids achieved supramolecular binding and effectively reduced the enzymatic hydrolysis risk of nucleic acids10,26. Therefore, to achieve effective protection of IL-10 mRNA, six frequently used polyphenols were screened. After combining with optimized polyphenol ellagic acid (EA), the negatively charged nanocore (mRNA/E) was prepared, and the enzymatic degradation of IL-10 mRNA by RNase was reduced. Next, mRNA/E was complexed with linear polyetherimide (PEI) to form positively charged mRNA/EP. Subsequently, to realize the active and accurate targeted delivery of IL-10 mRNA, we introduced HA to coat the outside of the mRNA/EP. Although HA is a biocompatibility and biodegradability polysaccharide which can effectively target cluster of differentiation 44 (CD44)27,28, it is easy to be inactivated by hyaluronidase-mediated degradation and harsh oxidative conditions in the inflamed colon, which limits the combination of HA and CD4429,30. Therefore, we grafted bilirubin (BR), characterized by good antioxidant activity, to the molecular structure of HA (HA-BR) to protect HA from intestinal degradation30,31. By coating HA-BR on the outermost layer of mRNA/EP, we realized the construction of a novel IL-10 mRNA delivery system (mRNA/EPHB) with self-protection and active targeting mechanisms. Through rectal administration, this drug delivery system achieved conspicuous therapeutic effect on the dextran sulfate sodium salt (DSS)-induced acute and chronic UC models (Scheme 1). The research and development of mRNA/EPHB have the potential to provide more innovative and constructive strategies for the efficient delivery of mRNA agents for the treatment of IBD.
Scheme 1.
The polyphenol-assisted IL-10 mRNA delivery system with self-protection and active targeting mechanisms. Through rectal administration, this delivery system achieved conspicuous therapeutic effect on dextran sulfate sodium salt-induced ulcerative colitis model.
2. Materials and methods
2.1. Materials
Polyadenylated and capped modified IL-10 mRNA and Cy5-labeled modified IL-10 mRNA (mRNA-Cy5) were synthesized by APExBIO Technology LLC (Shanghai, China). Tannic acid (TA), punicalagin (PC), epigallocatechin gallate (EGCG), EA, catechin (CAT), and gallic acid (GC) were purchased from Aladdin Biochemical Technology Co., Ltd. (Shanghai, China). Linear polyetherimide (PEI), N-hydroxysuccinimide (NHS), dimethyl sulfoxide (DMSO), ethylenediamine, and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) were obtained from Sigma‒Aldrich. BR, ethidium bromide (EB), and diethyl pyrocarbonate (DEPC) were purchased from Macklin (Shanghai, China). Hyaluronic acid (HA) was obtained from Lifecore™ biomedica Biomedical, LLC (Boston, USA). Enzyme linked immunosorbent assay (ELISA) kits for mice IL-10, IL-1β, TNF-α and IL-6 were manufactured by BioLegend, Inc. (San Diego, USA). All other reagents were analytical grade without further purification.
2.2. Screening of polyphenols
mRNA solutions (1 mg/mL, 20 μL) were mixed and stirred with polyphenol solutions (TA, PC, EGCG, EA, CAT, and GC) at a weight ratio of 4:1 for 30 min to obtain mRNA/polyphenol complexes10. The Tyndall effect of mRNA/polyphenol complexes was observed under light beam irradiation, and the transmittance of the complex solution was determined at 450 nm by an ultraviolet‒visible (UV‒Vis) spectrophotometer (HACH, USA). The size and zeta potential of the mRNA/polyphenol complexes were measured by a dynamic light scattering (DLS) instrument, which was equipped with a Nanosizer ZS90 (Malvern, UK), at 25 °C32. All mRNA/polyphenols were separately mixed with RNase for 5 min. Then, 20 μL of the mRNA/polyphenol complex solutions treated with RNase were sampled to determine the degradation degree of mRNA by nucleic acid gel electrophoresis. Furthermore, the morphology of mRNA/E was scanned by transmission electron microscopy (TEM, JEM 1200X, JEOL, Japan).
2.3. Ethidium bromide competitive binding assay
The intermolecular force of the complexation of mRNA with EA was characterized by the EB competitive assay. Briefly, 1 mL EB solution (5 μmol/L) was mixed with 10 μL mRNA solution (1 mg/mL) to form the mRNA/EB. Next, 2.5 μg EA was added to the above complex solution10. The solution was further maintained at room temperature (RT) for 15 min before measurement by a fluorescence spectrometer (Thermo Fisher Scientific, USA) (excitation at 470 nm, emission at 525–725 nm).
2.4. Synthesis and characterization of HA-BR
AE-BR was synthesized as described in the reference30. In brief, 0.06 g of NHS and 0.44 g bilirubin were added to 8 mL of DMSO containing 0.225 μL of trimethylamine. Subsequently, 0.065 g of EDC was added to the mixture. After stirring for 15 min, 0.3 mL ethylenediamine was added to the mixture, and the reaction was conducted under nitrogen gas with stirring for 5 h at RT. Subsequently, the reaction product solution was mixed with 50 mL chloroform and then washed with 45 mL of 0.1 mol/L HCl, 0.1 mol/L NaHCO3, and then water, and the process was repeated. After evaporating the chloroform, the reaction mixture was mixed with 45 mL methanol and then centrifuged at 3000×g for 15 min. AE-BR was obtained by evaporating the supernatant.
The HA was desalted to obtain the product HA-BR. Briefly, the acidic form of HA was prepared by dialyzing HA sodium salt solution against 0.01 mol/L HCl overnight, followed by lyophilization. Then, 40 mg of the acidic form of HA and 1 mg of NHS were added to 5 mL of DMSO. Next, 8 mg EDC was added to the mixture and stirred for 10 min at RT. Subsequently, 4 mg of AE-BR was added to the mixture, and the reaction was conducted overnight under nitrogen gas at RT. Next, the reaction mixture was slowly dropped into 30 mL of 0.01 mol/L NaOH, and then, the dialysis process was performed against 0.01 mol/L NaOH for 5 h, followed by dialysis against a 1:1 ratio of water/acetonitrile solution for 12 h and then dialysis against distilled water for 12 h. The resulting solution was lyophilized to obtain HA-BR. Next, 1H NMR spectra were obtained via a Bruker Ultra Shield 600 PLUS NMR spectrometer (Bruker, Germany), and the UV‒Vis spectra were obtained (HACH, USA).
2.5. Preparation and physicochemical characterization of mRNA/EPHB
Briefly, 20 μL of 1 mg/mL mRNA solution was mixed with 200 μL of 25 μg/mL EA solution, and the mixture was stirred for 30 min at RT to obtain the mRNA/E solution. Subsequently, the mRNA/E solution was slowly dropped into 25 μL of 1.04 mg/mL PEI solution, and the mixture was stirred for another 30 min at RT to prepare the mRNA/EP solution. Next, the mRNA/EP solution was slowly added to 50 μL of 4 mg/mL HA-BR solution, and the mixture was stirred for 30 min to obtain the mRNA/EPHB solution. Tools soaked with 1‰ DEPC aqueous solution were used throughout the whole process, and water was RNase-free33. The size distribution, zeta potential, and polydispersity index (PDI) of mRNA/EP and mRNA/EPHB were determined by DLS. The particle morphologies of mRNA/EP and mRNA/EPHB were scanned using TEM. The N, O, and P element distributions of the formed mRNA/EPHB consisting of PEI, EA, and mRNA were analyzed by energy-dispersive X-ray spectroscopy (EDX) element mapping10.
2.6. In vitro stability of mRNA/EPHB
20 μL of 1 mg/mL mRNA solution was diluted to 295 μL with DEPC water to obtain the naked mRNA solution. The mRNA/E, mRNA/EP, and mRNA EPHB solutions were prepared as outlined above and diluted to the same volume as the naked mRNA solution. The mRNA/EPH solution was prepared by altering the HA-BR solution to 50 μL of 4 mg/mL HA. Then, the naked mRNA, mRNA/E, mRNA/EP, mRNA/EPH, and mRNA/EPHB solutions were separately mixed with RNase and hyaluronidase for 10 min (RNase 0.5 ng/mL, hyaluronidase 2.0 U/mL). Then, solutions were sampled to determine the degradation degree of mRNA by nucleic acid gel electrophoresis10. Additionally, storage stability experiments were performed on mRNA/EPHB over a storage period of 15 days. The size distribution and zeta-potential of mRNA/EPHB were observed and recorded on Days 0, 1, 3, 5, 7, 9, 11, 13, and 15.
2.7. CD44 receptor-mediated cellular uptake experiment in vitro
The cellular uptake characteristic of the mRNA delivery system was investigated by confocal laser scanning microscopy (CLSM, LEICA TCS SP8 SM, Germany). Briefly, mRNA-Cy5 was encapsulated in mRNA-Cy5/EPHB according to the method of mRNA/EPHB preparation. The Raw264.7 and NCM460 cells were cultured in 15-mm cell culture dishes (NEST Biotechnology, Wuxi, China) at 1 × 106 cells/well with fresh cell culture medium mixed with lipopolysaccharide (LPS, 1 μg/mL). The next day, the cells were treated with mRNA-Cy5 or mRNA-Cy5/EPHB (mRNA-Cy5: 5 μg/mL) for 6 h. To further investigate the active targeting mechanism of the delivery system, in the HA-pretreated group, both cell lines were incubated with 1 mg/mL of HA for 4 h before treatment with mRNA-Cy5/EPHB. Untreated Raw264.7 and NCM460 cells were used as the control groups. Subsequently, the cells were stained with Hoechst 33342 (Thermofisher), and then washed with PBS. After that, the cells were fixed paraformaldehyde solution. All samples were immediately visualized using a CLSM imaging system.
Quantitative analysis of cellular uptake was performed using flow cytometry (BD LSRFortessa, USA). Briefly, Raw264.7 cells and NCM460 cells were evenly seeded into 12-well plates at 2 × 106 cells/well and cultured. After 12 h, cells were treated with mRNA-Cy5 or mRNA-Cy5/EPHB (mRNA-Cy5: 10 μg/mL) and pretreated with HA, as described above. Then, cells were washed by PBS, blown repeatedly with a pipette to prepare cell suspension, and tested by flow cytometry.
2.8. In vitro expression of IL-10
NCM460 and Raw264.7 cells were seeded in 24-well plates at 5 × 105 cells/well, separately. The next day, cells were cultured with fresh culture medium mixed with lipopolysaccharide (LPS, 1 μg/mL). Then, the cells were treated with EA, EPHB, and mRNA/EPHB (taking EA content as the standard), separately. Cells without any treatment were used as negative controls. After 24 h of treatment, the cell supernatant of NCM460 and Raw264.7 cells in each well was collected. The concentrations of IL-10 were determined by commercial ELISA kits (Biolegend, USA), separately.
2.9. In vivo adhesion experiment
An experimental UC model induced by DSS was established in C57BL/6 mice to evaluate the in vivo adhesion ability of mRNA/EPHB. Briefly, DiR (50 μg/mL) was loaded in the delivery system to obtain DiR@mRNA/EPHB. Fluorescence images were scanned by an in vivo imaging system (IVIS, Lumina XR III, USA). Briefly, male mice were randomly divided into two groups (water + DiR@mRNA/EPHB and DSS + DiR@mRNA/EPHB group) with three mice per group. The water + DiR@mRNA/EPHB group was treated with normal distilled water as a control group, and the DSS + DiR@mRNA/EPHB group was treated with DSS solution (3%, w/v) for 6 days to establish the UC model. Next, DiR@mRNA/EPHB was administered rectally to the mice in the water + DiR@mRNA/EPHB and DSS + DiR@mRNA/EPHB groups. The mice were scanned at three consecutive time points after administration (6, 12, and 24 h). After 24 h, mice were sacrificed immediately, and the colon was completely separated by dissection.
2.10. Therapeutic effects of mRNA/EPHB in DSS-induced acute UC mice
The drug intervention effect of mRNA/EPHB was evaluated using a DSS-induced acute UC model on C57BL/6 male mice. Briefly, adaptively reared mice were randomly divided into five groups (n = 6 per group), including control, model, 5-ASA, EPHB, and mRNA/EPHB groups. Then, the experimental mice in the model, 5-ASA, EPHB, and mRNA/EPHB groups were allowed to drink water containing DSS (3%, w/v) for 9 consecutive days to establish an acute UC model. The control group was given normal drinking water. The mice in the 5-ASA, EPHB, and mRNA/EPHB groups were rectally administered 5-ASA (100 mg/kg), EPHB (12.3 mg/kg), and mRNA/EPHB (13.3 mg/kg, totaling 1 mg/kg mRNA), respectively, on Days 2, 4, 6, and 8. The mice in the model group were rectally administered the equivalent amounts of normal saline. The body weight of the mice was recorded daily, and the disease activity index (DAI) was scored according to the summation of three indicators, including body weight loss, stool consistency, and fecal bleeding, and each indicator was scored from 0 to 4 according to the severity from low to high. On the last day of the experiment, mice were anesthetized, and blood was collected by eyeball extraction. The blood was centrifuged to acquire serum. The colon tissues, heart, liver, spleen, lungs, and kidneys were collected. Serum H2O2 and myeloperoxidase (MPO) levels were tested using commercial kits (Solarbio, Beijing). The expression level of cytokines in the colon tissues, including IL-10, IL-1β, TNF-α, and IL-6, was detected via corresponding ELISA kits (Biolegend, USA). The colon tissue was embedded in paraffin, cut into 5 μm sections, and stained with hematoxylin/eosin (H&E) and periodic acid Schiff (PAS), separately. H&E-stained sections of heart, liver, spleen, lungs, and kidneys were carry out to evaluate the toxicity of mRNA/EPHB.
Next, real-time quantitative polymerase chain reaction (qPCR), immunohistochemistry (IHC), and Western blot (WB) were employed to determine the expression level of IL-10 in the colon tissues of each group. For the qPCR experiments, RNA was extracted from cryopreserved colon tissues using the Animal Total RNA Isolation Kit (Foregene, Chengdu) according to the manufacturer's protocol. RNA was transcribed into cDNA by High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Next, qRT-PCR was performed using SYBR Green qPCR Kits (Yeasen, China). The relative mRNA expression was calculated using β-actin as the endogenous control. For the IHC experiment, tissue sections were incubated with rabbit anti-IL-10 antibody (primary antibody; Proteintech, Wuhan, China) and HRP-labeled anti-rabbit IgG (secondary antibody; Thermo Fisher Scientific) and stained with hematoxylin and DAB. The final stained sections were observed under a Nikon Eclipse Ts2R-FL microscope system (Nikon, Japan). For the WB assay, colonic tissues were homogenized using RIPA lysis buffer (Beyotime Biotechnology, Shanghai), with β-actin used as the control. Rabbit anti-IL-10 antibody (1:1000) and HRP-labeled anti-rabbit IgG were combined to display the signal to image IL-10.
To demonstrate the protective effect of mRNA/EPHB on intestinal barrier function, the upstream RNA levels of ZO-1 and occludin-1 were detected via qPCR. A corresponding immunofluorescence (IF) assay was also carried out. The specific operation flows of qPCR test of RNA levels of ZO-1 and occludin-1 were consistent with the detection of IL-10 mRNA. To acquire in vivo confocal microscopy images, colon tissue sections were stained with occludin-1 antibody conjugated with AlexaFluor-594 (Thermo Fisher Scientific) and anti ZO-1 antibody conjugated with AlexaFluor-488 (Thermo Fisher Scientific), and the nuclei were stained with DAPI. To investigate the regulatory effect of inflammatory drug intervention on intestinal cell apoptosis, the colon tissues were stained with BrdU monoclonal antibody (Thermo Fisher Scientific) and β-catenin antibody conjugated with Cy3 (Bioss, USA), and the nuclei were stained with DAPI. All the tissue sections were examined by IF microscopy (Nikon Eclipse C1, Nikon). The fluorescence intensity was determined using ImageJ software.
2.11. Therapeutic effects of mRNA/EPHB in mice with DSS-induced chronic UC
Briefly, C57BL/6 male mice were randomly divided into five groups same as in acute UC experiment. The control group received no treatment. To establish a chronic UC model, the other groups were allowed to freely drink DSS (1.5%, w/v) for 5 consecutive days and then water for a further 5 consecutive days; this cycle was repeated three times34. During each DSS drinking stage, the experimental mice in each group were rectally administered with different intervention substances, as in the acute UC model, on Days 1, 3, and 5. The body weight and DAI of the mice were recorded and assessed daily throughout the experiment. On the last day, the colon tissues, major visceral organs, and serum were collected. The serum H2O2 and MPO levels were tested using commercial kits (Solarbio, Beijing). The level of cytokines in the colon tissues, including IL-10, IL-1β, TNF-α, and IL-6, was detected via corresponding ELISA kits (Biolegend, USA). To determine the expression level of IL-10 in the colon tissues of each group, qPCR IHC, and WB were employed. The colon tissues were stained with H&E and picrosirius red (PSR) separately. Blood biochemistry analysis and H&E-stained sections of major organs were prepared to evaluate the toxicity of mRNA/EPHB. The RNA expression level of ZO-1 and occludin-1 was tested via qPCR, and the expression of ZO-1, occludin-1, β-catenin, and Apo-BrdU in the colon was detected by IF. After sealing, the images were observed and captured by a fluorescence microscope (Nikon Eclipse C1, Japan). The fluorescence intensity was determined using ImageJ software.
2.12. Ethics statement
All experiments involving animals were conducted according to the ethical policies and procedures approved by the Committee on Use and Care of Animals of University of Macau, Macau (Ethics No.: UM ARE-037-2018). C57BL/6 mice (male, 8 weeks) were obtained from Faculty of Healthy Science Animal Centre of University of Macau.
2.13. Statistical analysis
All experiment data are presented as the mean ± SD. Student's t-test or one-way analysis of variance (ANOVA) was used to analyze statistically significant differences. Statistical significance was indicated as ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3. Results and discussion
3.1. Screening polyphenols and the protective effect of EA on mRNA
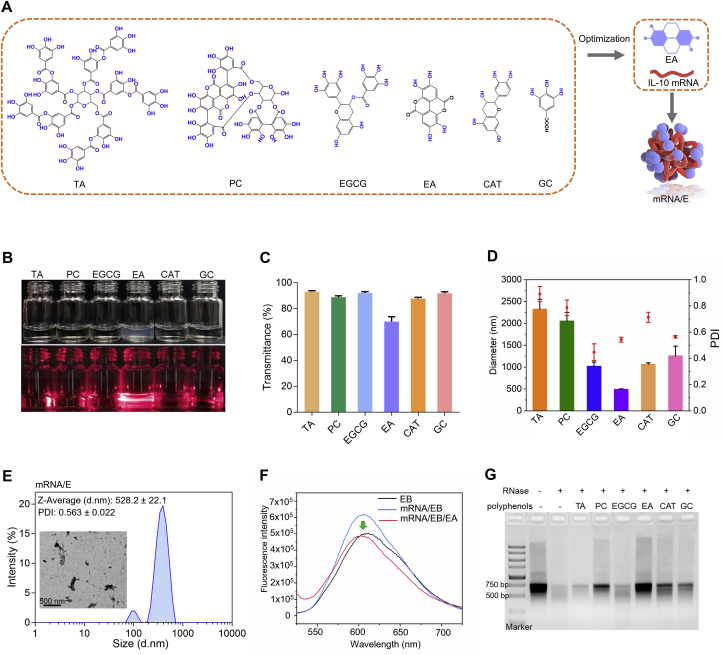
Although IL-10 mRNA has great potential in the clinical treatment of UC, the easy enzymatic hydrolysis of mRNA has been difficult to solve. Polyphenols possess dense ortho and meta-phenolic hydroxyl groups which can complex with nucleic acids through intermolecular forces24. Recently, studies have revealed that small interfering RNA (siRNA) mixed with EGCG effectively reduced enzymatic hydrolysis because the polyphenolic structure enabled strong affinity to siRNA via hydrogen bonds and hydrophobic interactions10,26. Inspired by this, we aimed to determine the polyphenol with the strongest protective effect on mRNA by screening common polyphenols including TA, PC, EGCG, EA, CAT, and GC (Fig. 1A). The polyphenols and mRNA were mixed to obtain self-assembled mRNA/polyphenol complexes. The Tyndall effect was especially obvious in the product comprising EA and mRNA (mRNA/E) (Fig. 1B). This observation was also corroborated by determining the transmittance at 450 nm, which was dominated by scattering light (Fig. 1C). In addition, compared to other mRNA/polyphenol complexes, the mRNA/E solution showed the smallest particle size (528.2 ± 22.1 nm) and a relatively lower PDI (0.563 ± 0.022) (Fig. 1D). Radiography research was performed via TEM, which demonstrated that EA and mRNA form complexes. Moreover, as revealed by DLS, the uniformity of the particle size distribution was low (Fig. 1E). The interaction of EA with mRNA was further confirmed by EB competitive binding. The results showed that EB intercalated into the grooves of mRNA, yielding a fluorescent complex. After adding EA into the complex solution, the fluorescence intensity was significantly decreased (Fig. 1F), suggesting competitive binding of mRNA with EA. Additionally, the results of RNase degradation assay showed that the mRNA/E complex efficiently prevented mRNA from enzymatic degradation (Fig. 1G), which might be attributed to the formation of mRNA/E nanoparticles to protect IL-10 mRNA from exposure to RNase. These results proved that polyphenol EA formed a complex with IL-10 mRNA and could effectively protect IL-10 mRNA from hydrolysis dominated by RNase.
Figure 1.
Screening polyphenols and the protective effect of EA on mRNA. (A) Schematic illustration of polyphenol screening and mRNA/E complex preparation. (B) The Tyndall effect of different solutions obtained via the complexation of polyphenols and IL-10 mRNA. (C) The transmittance of mRNA/polyphenol solutions at 450 nm. (D) The size and PDI of mRNA/polyphenol solutions. (E) TEM image and size distribution of mRNA/E. (F) EB competitive binding experiment. (G) Nucleic acid degradation experiment of mRNA/polyphenol complexes. Values are represented as the means ± SD (n = 3).
3.2. Preparation and physicochemical characterization of mRNA/EPHB
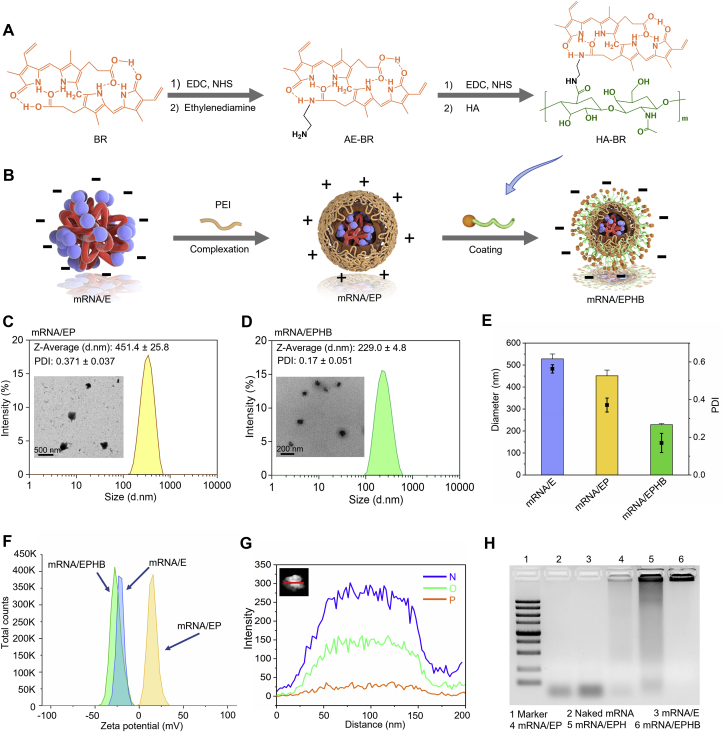
CD44 is highly expressed on inflammatory cells, including colon epithelial cells and colonic macrophages in IBD mice35. HA can selectively bind to CD44 and is commonly used in colon-targeted drug delivery36. Nevertheless, HA is unstable in a strong oxidative atmosphere and the complex intestinal enzyme environment of the inflamed colon29. Therefore, we grafted antioxidant BR to the molecular structure of HA through chemical synthesis technology to obtain HA-BR (Fig. 2A) and protected HA from intestinal degradation30. The chemically synthesized structure of HA-BR was confirmed by 1H NMR (Supporting Information Fig. S1A) and UV‒Vis spectrophotometry (Supporting Information Fig. S1B). Although BR was poorly soluble in water, HA-BR was readily dispersed in ultrapure water (Supporting Information Fig. S1C). The negatively charged mRNA/E complex was obtained by stirring the mixture of mRNA and EA for 30 min. Next, the positively charged PEI and negatively charged mRNA/E formed an mRNA/EP complex in aqueous solution by electrostatic interaction. Finally, negatively charged HA-BR macromolecules were coated on the outermost layer of the drug delivery system through electrostatic interaction (Fig. 2B). As revealed by TEM and DLS measurements, the hydrodynamic size of mRNA/EP was 451.4 ± 25.8 nm, and the PDI was 0.371 ± 0.037 (Fig. 2C); and the hydrodynamic size of mRNA/EPHB was 229.0 ± 4.8 nm, and the PDI was 0.17 ± 0.051 (Fig. 2D). Through horizontal comparison of the particle size and PDI of mRNA/E, mRNA/EP, and mRNA/EPHB, we found that layer-by-layer wrapping of materials with different charges led to an obvious decreasing trend in the particle size and PDI (Fig. 2E). This finding indicated that the whole particle structure realized an obvious compression process through electrostatic action among macromolecules, forming smaller and more uniform particles, which were more conducive to the efficient targeted delivery of nanoparticles. The potential measurement results revealed obvious conversions (from negative charge to positive charge, and then from positive charge to negative charge) in the potential of the three particles during the preparation process of mRNA/EPHB (Fig. 2F), confirming successful wrapping of polymer materials. Next, the structure of the formed mRNA/EPHB, consisting of mRNA, EA, PEI, and mRNA, was analyzed by EDX element mapping. The phosphorus (P, represents mRNA) was located in the interior of the mRNA/EPHB. Because HA-BR (containing large quantities of nitrogen from BR) was coated on the outermost layer of the nanoparticles, nitrogen (N) had the highest response strength (Fig. 2G), suggesting that mRNA/EPHB was a layer-by-layer core-shell-structured nanoparticle. Furthermore, the nucleic acid degradation experiment demonstrated that after wrapping the mRNA layer-by-layer with PEI and HA-BR, the mRNA/EPHB more effectively resisted the degradation by RNase compared to naked IL-10 mRNA and other IL-10 mRNA particles (Fig. 2H). Additionally, storage stability in vitro showed that the particle size and zeta potential of mRNA/EPHB did not change significantly during a 15-day storage period (Supporting Information Fig. S2A and S2B).
Figure 2.
Preparation and physicochemical characterization of mRNA/EPHB. (A) Flow chart of chemical synthesis of HA-BR. (B) Flow chart of preparation process of mRNA/EPHB. (C) TEM image and size distribution of mRNA/EP. (D) TEM image and size distribution of mRNA/EPHB. (E) Transverse comparison of size and PDI of mRNA/E, mRNA/EP, and mRNA/EPHB. (F) Zeta potentials of mRNA/E, mRNA/EP, and mRNA/EPHB. (G) High-angle annular dark-field TEM image and EDX element mapping of a single mRNA/EPHB consisting of IL-10 mRNA, EA, PEI, and HA-BR. Scale bar = 200 nm. (H) Stability of naked mRNA, semi-finished delivery systems, and mRNA/EPHB against the combination of RNase and hyaluronidase in vitro. Values are represented as the means ± SD (n = 3).
3.3. CD44-mediated effective cellular uptake in vitro
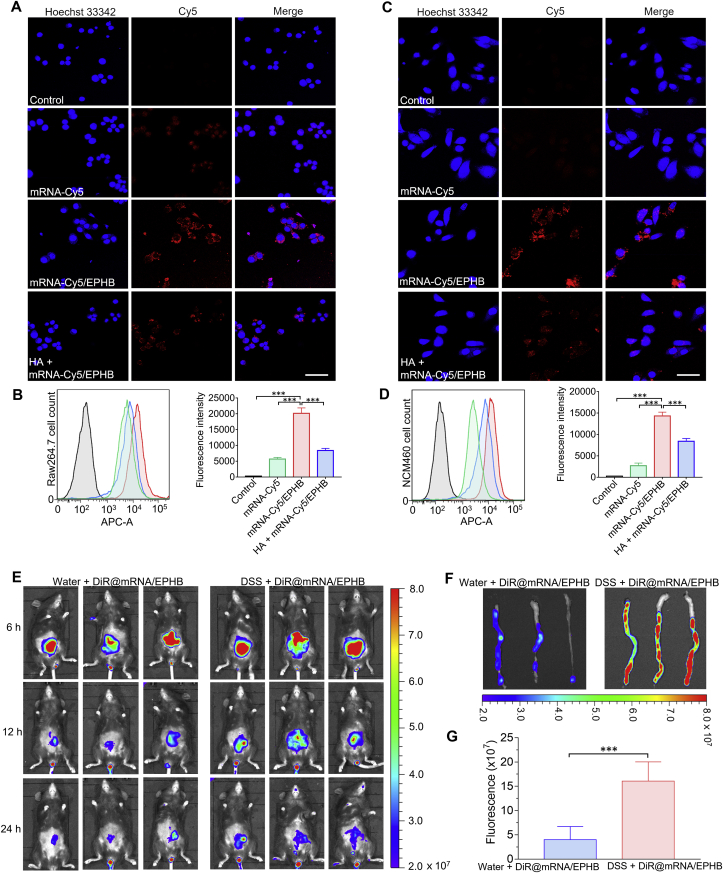
To investigate whether the better anti-inflammatory activity of mRNA/EPHB was related to the targeting of CD44 mediated by the outermost HA-BR, mRNA-Cy5 was encapsulated in the system to obtain mRNA-Cy5/EPHB. The in vitro uptake experiments were conducted on Raw264.7 and NCM460 cells. The fluorescence of Raw264.7 cells treated with free mRNA-Cy5 was weak and almost invisible, while cells showed obvious fluorescence following ingestion of mRNA-Cy5/EPHB 4 h after administration. However, pretreatment of 1 mg/mL HA obviously decreased the fluorescence intensity of mRNA-Cy5/EPHB in Raw264.7 cells (Fig. 3A). Correspondingly, quantitative analysis of mRNA-Cy5/EPHB in Raw264.7 cells by flow cytometry showed that the fluorescence intensity of cells treated with mRNA-Cy5/EPHB was significantly higher than that of free mRNA-Cy5 and HA pretreatment (Fig. 3B). Similar results were also confirmed on NCM460 cells (Fig. 3C and D). Furthermore, mRNA/EPHB showed significant lysosomal escape ability in Raw264.7 cells (Supporting Information Fig. S3). These results demonstrated that mRNA/EPHB was not only effectively ingested by intestinal epithelial cells and macrophages, also escaped from the lysosome to avoid the degradation of mRNA.
Figure 3.
CD44-mediated effective cellular uptake in vitro and effective retention of mRNA/EPHB in the inflamed colon in vivo. (A) Fluorescence images of cellular uptake of free mRNA-Cy5, mRNA-Cy5/EPHB, and HA pretreatment administrated Raw264.7 cells. Scale bar = 50 μm. (B) Flow cytometric histogram profiles and histograms of the fluorescence intensities of free mRNA-Cy5, mRNA-Cy5/EPHB, and HA pretreatment treated Raw264.7 cells. (C) Fluorescence images of cellular uptake of free mRNA-Cy5, mRNA-Cy5/EPHB, and HA pretreatment administrated NCM460 cells. Scale bar = 50 μm. (D) Flow cytometric histogram profiles and histograms of the fluorescence intensities of free mRNA-Cy5, mRNA-Cy5/EPHB, and HA pretreatment treated NCM460 cells. (E) In vivo fluorescence images of UC mice and healthy mice rectally administrated with DiR@mRNA/EPHB. (F) Fluorescence images of isolated colons separated from UC mice and healthy mice after 24 h of administration. (G) Quantitative analysis of fluorescence intensity of the isolated colons. All the values are represented as the means ± SD (n = 3). Statistical significance was indicated as ∗∗∗P < 0.001.
3.4. Effective retention of mRNA/EPHB in the inflamed colon
Rectal administration can appreciably avoid the influence of the complex gastrointestinal environment on the drug delivery system, but therapeutic agents are easily excreted. Therefore, drugs should have definite retention characteristics in inflamed colon tissues to adapt to the requirements of rectal administration37. We chose to encapsulate DiR, with a maximum absorbance at 780 nm, in our drug delivery system (named DiR@mRNA/EPHB) to demonstrate the retention of mRNA/EPHB in vivo. The solution of DiR@mRNA/EPHB was administered rectally in both healthy mice (water + DiR@mRNA/EPHB) and UC mice (DSS + DiR@mRNA/EPHB). The average fluorescence intensity of DiR in the DSS + DiR@mRNA/EPHB group was higher than that in the water + DiR@mRNA/EPHB group at 6, 12, and 24 h after rectal administration (Fig. 3E). Moreover, mice from the two groups were sacrificed, and their colons were separately collected 24 h after rectal administration, and the fluorescence intensity of the colons was imaged (Fig. 3F). Quantitative analysis showed that the average fluorescence intensity of the inflamed colons was approximately 3-fold that of the healthy colons (Fig. 3G). It has been suggested that the positively charged proteins will accumulate on the ulcer surface during the formation of ulcer wounds in IBD, resulting in a positive charge on the entire ulcer surface38,39. HA is a biocompatible polysaccharide with a strong negative charge, which is appropriate for the HA-BR encapsulated drug delivery system. Thus, the designed mRNA delivery system displays excellent intestinal adhesion characteristics.
3.5. mRNA/EPHB promoted the expression of IL-10 in vitro
We conducted an intervention experiment of mRNA/EPHB on the expression of IL-10 in Raw264.7 and NCM460 cells. Briefly, the results of ELISA performed using the supernatant of Raw264.7 cells showed that after 24 h of treatment, mRNA/EPHB significantly improved the expression of IL-10 compared to the control, model, EA, and EPHB groups. The IL-10 expression level of the mRNA/EPHB group was approximately 8-fold higher than that of the control group (Supporting Information Fig. S4A). Similarly, the quantitative analysis of proteins in the supernatant of NCM460 cells showed that mRNA/EPHB administration obviously increased the concentration of IL-10 compared to the other groups and was approximately 4-fold higher than that of the control group (Fig. S4B). These results showed that IL-10 mRNA was effectively translated into IL-10 in macrophages and intestinal epithelial cells via the encapsulation of the novel drug delivery system. Besides, mRNA/EPHB showed low cytotoxicity in both RAW264.7 and NCM460 cells compare with mRNA/EP (Supporting Information Fig. S5).
3.6. Efficacy evaluation of the acute UC model
3.6.1. Therapeutic efficacy against acute UC
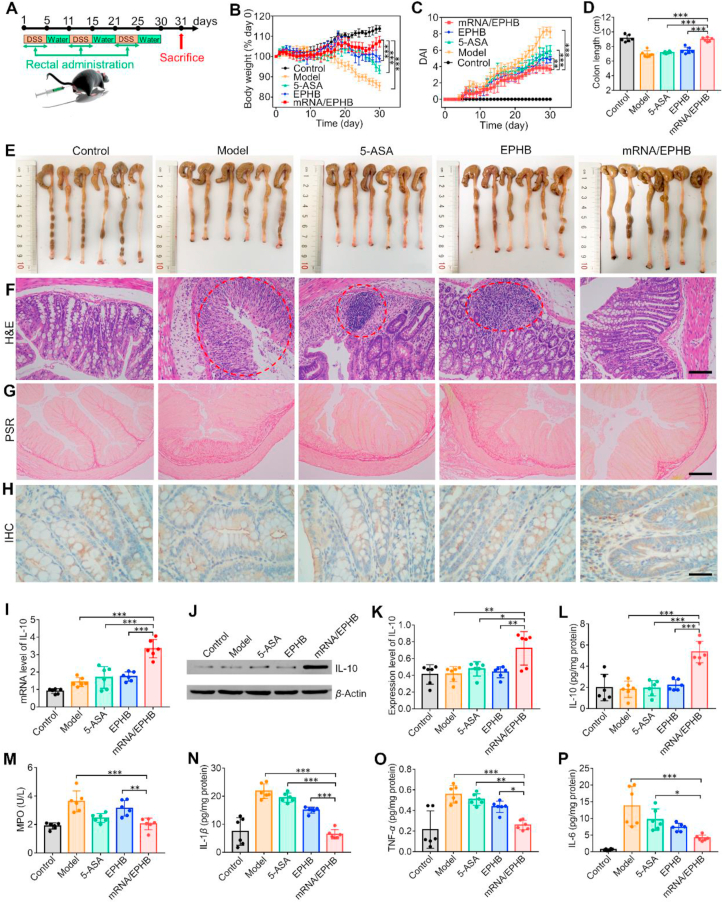
It is also necessary to conduct in vivo experiments to evaluate the therapeutic effect of mRNA/EPHB on DSS-induced UC model mice. We employed 5-ASA, one of the first-line therapeutic drugs for IBD, as the positive control agent. Briefly, all mice were randomly divided into five groups, including control, model, 5-ASA, EPHB, and mRNA/EPHB groups. Except for the control group, C57BL/6 mice in the model, 5-ASA, EPHB, and mRNA/EPHB groups were administered 3% DSS in drinking water for 9 days to establish the acute UC model, and various drug delivery formulations were correspondingly administered to experimental mice via rectal administration on Days 2, 4, 6, and 8 (Fig. 4A). Compared to the 5-ASA and EPHB treatments, the mRNA/EPHB treatment significantly protected the mice against DSS-induced body weight loss (Fig. 4B). The DAI score of the mRNA/EPHB group was significantly lower than that of the model, 5-ASA, and EPHB groups (Fig. 4C). Notably, on the final day of the experiment, only the mice treated with mRNA/EPHB presented no traces of hematochezia around the anus, which was comparable to that of the control group (Supporting Information Fig. S6A). Moreover, rectal administration of mRNA/EPHB significantly prevented the atrophy of colonic tissues caused by inflammation, and the colon length of the mRNA/EPHB group was significantly longer than that of the model, 5-ASA, and EPHB groups (Fig. 4D and E). H&E-stained sections showed that the overall structure of intestinal tissue of DSS-induced UC mice was abnormal. Local mucosal necrosis was obvious, and normal structure of intestinal tissue basically disappeared. Plenty of inflammatory cells existed in the whole layer of the tissue. The H&E-stained sections of the 5-ASA group were similar to those of the model group, which was characterized by full-thickness necrosis of local mucosa and disappearance of normal structure of intestinal tissue, indicating the unsatisfactory therapeutic activity of 5-ASA. Slight amelioration of the colon injury in EPHB treated mice were revealed, but inflammatory cell infiltration was still existed. After the treatment of mRNA/EPHB, cured colitis wounds was realized, characterized by a nearly normal histological microstructure. Typical areas of inflammatory infiltration were marked by red circles (Fig. 4F). Goblet cells are unicellular glands, the main function of which is to synthesize and secrete mucin and form a mucosal barrier to protect epithelial cells. The number of goblet cells is positively correlated with the health of the colon tissues. PAS staining revealed that the colon goblet cells were completely damaged in the UC model group. 5-ASA, EPHB, and mRNA/EPHB alleviated the apoptosis of goblet cells in the colon, and mRNA/EPHB showed the most significant protective effect on goblet cells. Goblet cells were marked by yellow arrows (Fig. 4G).
Figure 4.
Efficacy evaluation of the acute UC model. (A) Schematic illustration of drug intervention plan for acute UC model. (B) The weight changes of mice in different groups during the 9-day treatment. (C) DAI changes in different groups of mice. (D) Average lengths of colons isolated from mice after 9-day drug intervention. (E) Photos reflecting the actual situation of colons isolated from mice after 9-day administration. (F) H&E-stained histological sections (scale bar = 50 μm), (G) PAS-stained histological sections (scale bar = 200 μm), and (H) IHC-stained histological sections (scale bar = 50 μm) of mice in different groups. (I) Histograms of IL-10 mRNA expression level in colon tissues. (J) WB analysis of IL-10 expression level and (K) histogram of IL-10 WB experiment of colon tissues. The levels of (L) IL-10, (M) MPO, (N) IL-1β, (O) TNF-α, and (P) IL-6 in colonic tissues isolated from healthy or diseased mice treated with different formulations. All the values are represented as the means ± SD (n = 6). Statistical significance was indicated as ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3.6.2. Promotion of the expression of IL-10 and inhibition of inflammation in acute UC
As a complex immune and inflammatory disease, the severity of UC is directly revealed by the expression level of inflammatory cytokines. Generally, high expression of IL-10 can significantly reduce the expression of proinflammatory factors and alleviate the deterioration of UC40. Radiography research on the IHC sections of the acute UC model suggested that the expression level of IL-10 in the mRNA/EPHB group was significantly elevated compared to that of the other groups (Fig. 4H). Moreover, the result of qPCR demonstrated that the mRNA level of IL-10 in the colon tissues of mRNA/EPHB treated mice was significantly higher than that of other groups (Fig. 4I). Besides, WB (Fig. 4J and K) and ELISA (Fig. 4L) showed that mRNA/EPHB significantly promoted the expression of IL-10 in the colon. Besides, mRNA/EPHB could also effectively reduce the levels of H2O2 (Supporting Information Fig. S6B) and MPO (Fig. 4M) in mouse serum. Additionally, the pathological overexpression of IL-1β, TNF-α, and IL-6 was significantly down-regulated in the mRNA/EPHB group (Fig. 4N–P).
3.6.3. Restoration and anti-apoptosis effects in the colonic epithelium in acute UC
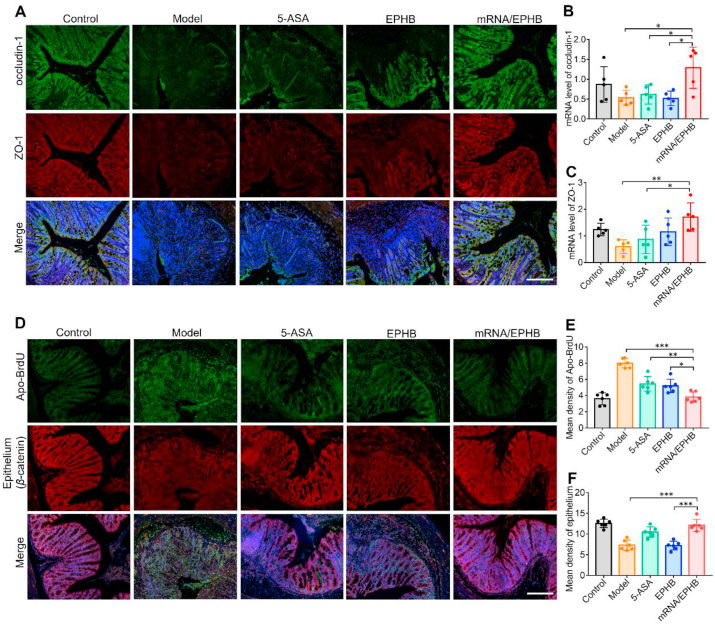
One of the main characteristics of IBD is the decrease of tight junction proteins, which results in an increase in intestinal permeability and further leads to the invasion of intestinal microbiota to the colon tissues, resulting in chronic inflammation and a complex immune response. ZO-1 and occludin-1 are the main types of intestinal epithelial tight junction proteins. Therefore, the expression levels of ZO-1 and occludin-1 were detected by IF and qPCR analysis. IF imaging analysis showed that mRNA/EPHB effectively maintained the tight distribution of occludin-1 and ZO-1 in the inflammatory colon as compared to the model, 5-ASA, and EPHB groups (Fig. 5A), which corresponded to the qPCR results (Fig. 5B and C). Besides, IF analysis showed that mRNA/EPHB treatment markedly maintained the normal level of β-catenin and reduced the level of apoptosis in the colon tissues, as shown by the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay (Fig. 5D). This finding was supported by the statistical results of the average fluorescence intensity of six randomly selected visual fields from the IF images of β-catenin and TUNEL assay (Fig. 5E and F). The above results proved that mRNA/EPHB played a positive role in treating acute UC by protecting the integrity of the epithelial cell barrier and inhibiting intestinal apoptosis.
Figure 5.
Restoration and anti-apoptosis effects in the colonic epithelium in acute UC. (A) IF of occludin-1 and ZO-1 in colon tissues. Scale bar = 200 μm. (B, C) The histogram of qPCR analysis of ZO-1 and occludin-1 mRNA expression levels. (D) Fluorescence photos of Apo-BrdU and β-catenin expression in colon tissues. Scale bar = 200 μm. (E, F) Quantitative analysis of fluorescence intensity of Apo-BrdU and β-catenin in colon tissues. All the values are represented as the means ± SD (n = 5 or 6). Statistical significance was indicated as ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3.7. Efficacy evaluation of the chronic UC model
3.7.1. Therapeutic efficacy against chronic UC
To establish a chronic UC model, the mice in the model, 5-ASA, EPHB, and mRNA/EPHB groups were allowed to freely drink DSS (1.5%, w/v) for 5 consecutive days, following which they were allowed to freely drink water for a further 5 consecutive days; this cycle was repeated three times (Fig. 6A). The mice in the experimental groups were administered different intervention agents rectally, as in the acute UC model, on Days 1, 3, and 5 during each DSS drinking period. Compared to the other groups, the mRNA/EPHB mice had significantly less body weight loss induced by DSS drinking (Fig. 6B). Moreover, the DAI score of the mRNA/EPHB treated mice was significantly lower than that of the mice in the model, 5-ASA, and EPHB groups (Fig. 6C). Following drug intervention, the spleen weight of mRNA/EPHB groups was the closest to that of control group (Supporting Information Fig. S7A and S7B). Furthermore, mRNA/EPHB prevented atrophy of colonic tissues caused by inflammation (Fig. 6D and E). The H&E-stained sections revealed that the overall structure of intestinal tissue of model group was abnormal. Intensive inflammatory cells existed in the whole layer of the tissue. The structure of intestinal tissue of 5-ASA group were similar to those of the model group, revealing the infinitesimal therapeutic effect of positive drug. Moreover, EPHB slightly attenuated colon destruction, and mRNA/EPHB cured the colitis, as shown by the presence of a nearly healthy histological microstructure. Typical areas of inflammatory infiltration were marked by red circles (Fig. 6F). The long-term inflammatory reaction in the colon will ultimately lead to colonic fibrosis which positively correlated with the severity of UC. The results of PSR staining showed that the fibrosis area of the colon was the largest in the model group, followed by the EPHB group and the 5-ASA group. The fibrosis of the colon tissues in mice treated with mRNA/EPHB was similar to that in healthy mice (Fig. 6G).
Figure 6.
Efficacy evaluation of the chronic UC model. (A) Schematic illustration of drug intervention plan for chronic UC model. (B) The weight changes of mice in different groups during the 30-day experiment. (C) DAI changes in different groups of mice. (D) Average lengths of colons isolated from mice after drug intervention. (E) Photos reflecting the actual situation of colons isolated from mice after 30 days of experiment. (F) H&E-stained histological sections (scale bar = 100 μm), (G) PSR-stained histological sections (scale bar = 200 μm), and (H) IHC-stained histological sections (scale bar = 50 μm) of mice in different groups. (I) Histograms of quantitative analysis of IL-10 mRNA expression level in colon tissues. (J) WB analysis of IL-10 expression level and (K) histogram of IL-10 WB experiment of colon tissues. The levels of (L) IL-10, (M) MPO, (N) IL-1β, (O) TNF-α, and (P) IL-6 in colonic tissues isolated from healthy or diseased mice treated with different formulations. All the values are represented as the means ± SD (n = 6). Statistical significance was indicated as ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3.7.2. Promotion of the expression of IL-10 and inhibition of inflammation in chronic UC
Image analysis of IHC revealed that the positive area of IL-10 in the sections of the mRNA/EPHB group was more remarkable compared to that of the other groups (Fig. 6H). The results of qPCR experiments showed that the mRNA expression level of IL-10 in the mRNA/EPHB group was significantly improved compared to that in the control, model, 5-ASA, and EPHB groups (Fig. 6I). WB (Fig. 6J and K) and ELISA (Fig. 6L) also demonstrated that mRNA/EPHB significantly promoted the expression of IL-10 in the colon. After the administration of mRNA/EPHB, the content of the oxidative stress-related molecular mediators H2O2 (Supporting Information Fig. S7C) and MPO (Fig. 6M) in serum was strikingly decreased, the expression levels of IL-1β, TNF-α, and IL-6 in the chronic inflammatory colon tissues were significantly inhibited (Fig. 6N‒P).
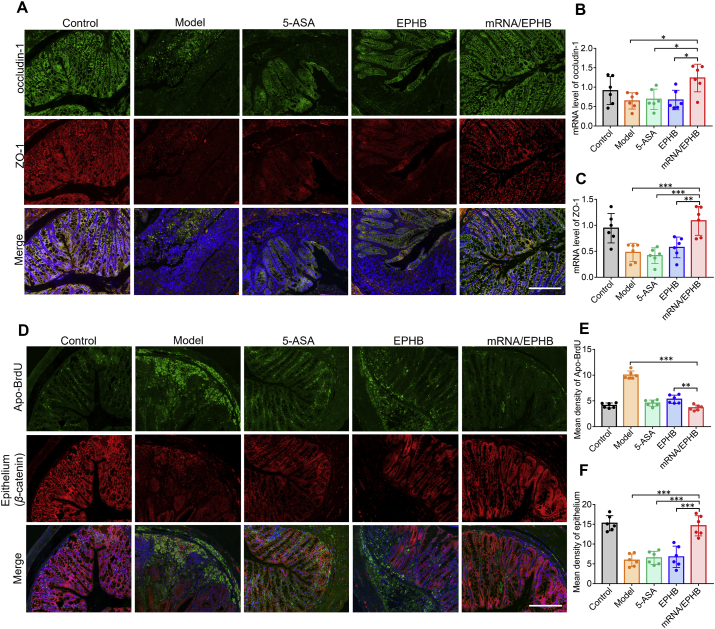
3.7.3. Restoration and anti-apoptosis effects in the colonic epithelium in acute UC
IF imaging analysis showed that mRNA/EPHB effectively increased the expression of occludin-1 and ZO-1 in the inflammatory colon compared to that in the model group, 5-ASA group, and EPHB group (Fig. 7A). Correspondingly, the qPCR analysis showed that mRNA/EPHB could significantly up-regulate the expression of occludin-1 and ZO-1 mRNA (Fig. 7B and C). In addition, IF analysis showed that mRNA/EPHB treatment maintained the normal level of intestinal epithelial cells and reduced the level of apoptosis in the colonic epithelium (Fig. 7D). This finding was supported by the statistical results of the average fluorescence intensity of six visual fields that were randomly selected from the IF images of β-catenin and Apo-BrdU (Fig. 7E and F). The above experimental results showed that mRNA/EPHB treatment protected the tight junction proteins in the model of chronic UC and significantly inhibited cell apoptosis in the colon.
Figure 7.
Restoration and anti-apoptosis effects in the colonic epithelium in chronic UC. (A) IF of occludin-1 and ZO-1 in colon tissues. Scale bar = 200 μm. (B, C) The histogram of qPCR analysis of ZO-1 and occludin-1 mRNA expression levels. (D) Fluorescence photos of Apo-BrdU and β-catenin expression in colon tissues. Scale bar = 200 μm. (E, F) Quantitative analysis of fluorescence intensity of Apo-BrdU and β-catenin in colon tissues. All the values are represented as the means ± SD (n = 6). Statistical significance was indicated as ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
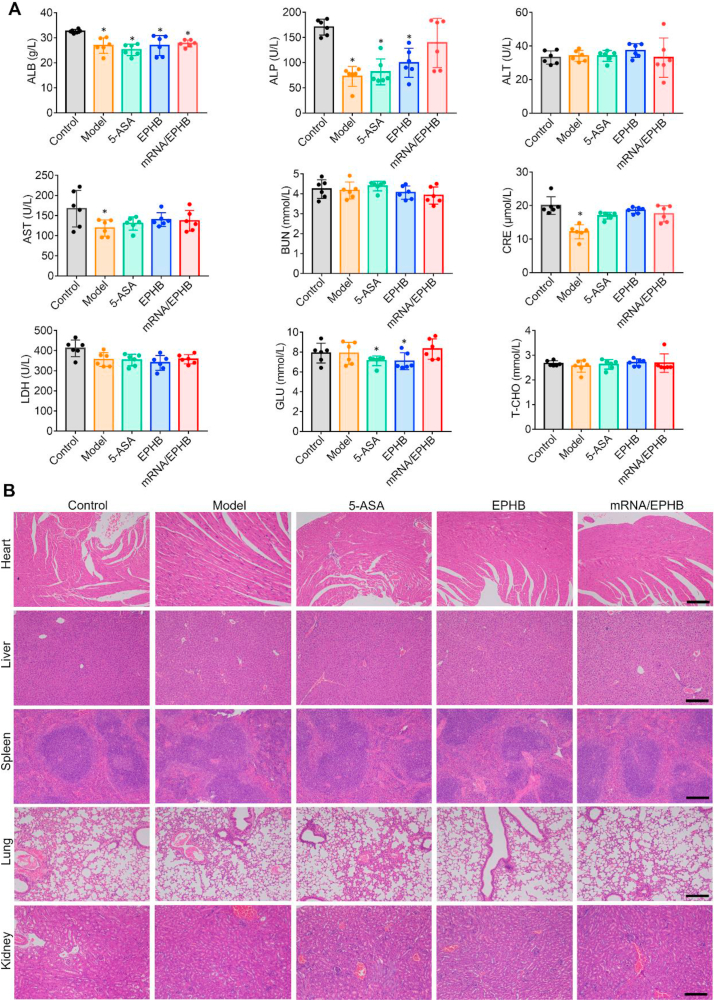
3.8. mRNA/EPHB exerted no significant toxicity
As mRNA/EPHB administered through the rectum are not affected by the first pass effect of the liver, the toxic risk of mRNA/EPHB is possible. Therefore, it is necessary to carry out the blood biochemistry analysis and organ pathological section. Fig. 8A indicates that no apparent enzyme activity differences of mRNA/EPHB treated chronic UC mice among all tested indicator enzyme (hepatotoxicity ALP, ALT, and AST; nephrotoxicity BUN, CRE; cardiotoxicity LDH; blood glucose GLU; blood fat T-CHO) compared with the control (normal mice). The sections of the main organs of acute (Supporting Information Fig. S8) and chronic UC model (Fig. 8B) mice were pathologically evaluated. After H&E staining, the histological results show no obvious abnormal lesions in the main internal organs of each group compared to the control group.
Figure 8.
Safety evaluation of mRNA/EPHB. (A) Blood biochemistry analysis of chronic UC model mice. (B) H&E staining of main internal organs (heart, liver, spleen, lungs, and kidneys) of chronic UC mice. Scale bar = 200 μm. All the values are represented as the means ± SD (n = 6). Statistical significance was indicated as ∗P < 0.05 vs. control group.
4. Conclusions
In summary, we prepared a layer-by-layer core–shell delivery system mRNA/EPHB using supramolecular binding. The stability of IL-10 mRNA was increased by adding natural polyphenol EA during the preparation process. Simultaneously, HA, which is easily degraded by the intestinal enzyme environment and superoxide, was grafted and modified by bilirubin. The obtained HA-BR not only had prominent biocompatibility but also had more reliable stability. HA-BR with good adhesion and stable negative charge endowed the system with colon retention characteristics. The drug delivery system can be effectively ingested by inflammatory cells in the colon and can smoothly enter the cells to release modified IL-10 mRNA by targeting CD44. Through the translation process in vivo, IL-10 mRNA successfully expressed the anti-inflammatory protein IL-10, which was further released from cells to play a broader anti-inflammatory role. Through the application of EA and HA-BR, the designed drug delivery system realized the active stability guarantee of mRNA and the active targeting assurance of the drug delivery system. Additionally, the drug delivery system prepared by the interaction between different molecules has a simple preparation process, high production efficiency, and great feasibility in clinical transformation, providing a new multifunctional design scheme for gene therapy of IBD.
Acknowledgments
This work was funded by the Science and Technology Development Fund, Macau SAR (00151/2020/A3 & SKL-QRCM(UM)-2020-2022, China), Major Basic and Applied Basic Research Projects of Guangdong Province of China (2019B030302005), Guangxi Science and Technology Research Project (GuiKeAA18242040, China), and the University of Macau (MYRG2020-00141-ICMS and EF013/ICMS-WYT/2019/GPU, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.03.025.
Contributor Information
Yitao Wang, Email: ytwang@um.edu.mo.
Shengpeng Wang, Email: swang@um.edu.mo.
Author contributions
Zhejie Chen designed the research, carried out the experiments, performed data analysis, and wrote the manuscript. Wei Hao, Caifang Gao, and Yangyang Zhou participated part of the experiments. Shengpeng Wang and Yitao Wang designed and supervised the experiments and performed data analysis. Chen Zhang, Jinming Zhang, Ruibing Wang, and Shengpeng Wang revised the manuscript. All authors have read and approved the final manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bouma G., Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3:521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- 2.Peyrin-Biroulet L., Danese S., Argollo M., Pouillon L., Peppas S., Gonzalez-Lorenzo M., et al. Loss of response to vedolizumab and ability of dose intensification to restore response in patients with Crohn's disease or ulcerative colitis: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2019;17:838–846. doi: 10.1016/j.cgh.2018.06.026. [DOI] [PubMed] [Google Scholar]
- 3.Engel M.A., Neurath M.F. New pathophysiological insights and modern treatment of IBD. J Gastroenterol. 2010;45:571–583. doi: 10.1007/s00535-010-0219-3. [DOI] [PubMed] [Google Scholar]
- 4.Ng S.C., Tang W., Ching J.Y., Wong M., Chow C.M., Hui A.J., et al. Incidence and phenotype of inflammatory bowel disease based on results from the Asia-Pacific Crohn's and colitis epidemiology study. Gastroenterology. 2013;145:158–165. doi: 10.1053/j.gastro.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad A., Ansari M.M., Mishra R.K., Kumar A., Vyawahare A., Verma R.K., et al. Enteric-coated gelatin nanoparticles mediated oral delivery of 5-aminosalicylic acid alleviates severity of DSS-induced ulcerative colitis. Mater Sci Eng C. 2021;119:111582. doi: 10.1016/j.msec.2020.111582. [DOI] [PubMed] [Google Scholar]
- 6.De Iudicibus S., Franca R., Martelossi S., Ventura A., Decorti G. Molecular mechanism of glucocorticoid resistance in inflammatory bowel disease. World J Gastroenterol. 2011;17:1095–1108. doi: 10.3748/wjg.v17.i9.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu H., Luo R., Dong L., Pu X., Chen Q., Ye N., et al. pH/ROS dual-sensitive and chondroitin sulfate wrapped poly(β-amino ester)-SA-PAPE copolymer nanoparticles for macrophage-targeted oral therapy for ulcerative colitis. Nanomedicine. 2022;39:102461. doi: 10.1016/j.nano.2021.102461. [DOI] [PubMed] [Google Scholar]
- 8.Hajj K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat Rev Mater. 2017;2:17056. [Google Scholar]
- 9.Eygeris Y., Patel S., Jozic A., Sahay G. Deconvoluting lipid nanoparticle structure for messenger RNA delivery. Nano Lett. 2020;20:4543–4549. doi: 10.1021/acs.nanolett.0c01386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen W., Wang Q., Shen Y., Gao X., Li L., Yan Y., et al. Green tea catechin dramatically promotes RNAi mediated by low-molecular-weight polymers. ACS Cent Sci. 2018;4:1326–1333. doi: 10.1021/acscentsci.8b00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veiga N., Goldsmith M., Granot Y., Rosenblum D., Dammes N., Kedmi R., et al. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat Commun. 2018;9:4493. doi: 10.1038/s41467-018-06936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granot-Matok Y., Kon E., Dammes N., Mechtinger G., Peer D. Therapeutic mRNA delivery to leukocytes. J Control Release. 2019;305:165–175. doi: 10.1016/j.jconrel.2019.05.032. [DOI] [PubMed] [Google Scholar]
- 13.Ibba M.L., Ciccone G., Esposito C.L., Catuogno S., Giangrande P.H. Advances in mRNA non-viral delivery approaches. Adv Drug Deliv Rev. 2021;177:113930. doi: 10.1016/j.addr.2021.113930. [DOI] [PubMed] [Google Scholar]
- 14.Xiong Q., Lee G.Y., Ding J., Li W., Shi J. Biomedical applications of mRNA nanomedicine. Nano Res. 2018;11:5281–5309. doi: 10.1007/s12274-018-2146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weng Y., Li C., Yang T., Hu B., Zhang M., Guo S., et al. The challenge and prospect of mRNA therapeutics landscape. Biotechnol Adv. 2020;40:107534. doi: 10.1016/j.biotechadv.2020.107534. [DOI] [PubMed] [Google Scholar]
- 16.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granot Y., Peer D. Delivering the right message: challenges and opportunities in lipid nanoparticles-mediated modified mRNA therapeutics—an innate immune system standpoint. Semin Immunopathol. 2017;34:68–77. doi: 10.1016/j.smim.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Verbeke R., Lentacker I., Wayteck L., Breckpot K., Van Bockstal M., Descamps B., et al. Co-delivery of nucleoside-modified mRNA and TLR agonists for cancer immunotherapy: restoring the immunogenicity of immunosilent mRNA. J Control Release. 2017;266:287–300. doi: 10.1016/j.jconrel.2017.09.041. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto A., Kormann M., Rosenecker J., Rudolph C. Current prospects for mRNA gene delivery. Eur J Pharm Biopharm. 2009;71:484–489. doi: 10.1016/j.ejpb.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Islam M.A., Reesor E.K., Xu Y., Zope H.R., Zetter B.R., Shi J. Biomaterials for mRNA delivery. Biomater Sci. 2015;3:1519–1533. doi: 10.1039/c5bm00198f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Sun C., Wang C., Jankovic K.E., Dong Y. Lipids and lipid derivatives for RNA delivery. Chem Rev. 2021;121:12181–12277. doi: 10.1021/acs.chemrev.1c00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat Rev Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang T., Li C., Wang X., Zhao D., Zhang M., Cao H., et al. Efficient hepatic delivery and protein expression enabled by optimized mRNA and ionizable lipid nanoparticle. Bioact Mater. 2020;5:1053–1061. doi: 10.1016/j.bioactmat.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z., Farag M.A., Zhong Z., Zhang C., Yang Y., Wang S., et al. Multifaceted role of phyto-derived polyphenols in nanodrug delivery systems. Adv Drug Deliv Rev. 2021:113870. doi: 10.1016/j.addr.2021.113870. [DOI] [PubMed] [Google Scholar]
- 25.Zhou J., Lin Z., Ju Y., Rahim M.A., Richardson J.J., Caruso F. Polyphenol-mediated assembly for particle engineering. Acc Chem Res. 2020;53:1269–1278. doi: 10.1021/acs.accounts.0c00150. [DOI] [PubMed] [Google Scholar]
- 26.Shen W., Wang R., Fan Q., Li Y., Cheng Y. Natural polyphenol assisted delivery of single-strand oligonucleotides by cationic polymers. Gene Ther. 2020;27:383–391. doi: 10.1038/s41434-020-0151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., Gu H., Zhang H., Xian J., Li J., Fu C., et al. Oral core–shell nanoparticles embedded in hydrogel microspheres for the efficient site-specific delivery of magnolol and enhanced antiulcerative colitis therapy. ACS Appl Mater Interfaces. 2021;13:33948–33961. doi: 10.1021/acsami.1c09804. [DOI] [PubMed] [Google Scholar]
- 28.Wang S., Zhang J., Wang Y., Chen M. Hyaluronic acid-coated PEI-PLGA nanoparticles mediated co-delivery of doxorubicin and miR-542-3p for triple negative breast cancer therapy. Nanomedicine. 2016;12:411–420. doi: 10.1016/j.nano.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 29.Zheng L., Riehl T.E., Stenson W.F. Regulation of colonic epithelial repair in mice by toll-like receptors and hyaluronic acid. Gastroenterology. 2009;137:2041–2051. doi: 10.1053/j.gastro.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee Y., Sugihara K., Gillilland M.G., 3rd, Jon S., Kamada N., Moon J.J. Hyaluronic acid-bilirubin nanomedicine for targeted modulation of dysregulated intestinal barrier, microbiome and immune responses in colitis. Nat Mater. 2020;19:118–126. doi: 10.1038/s41563-019-0462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Z., Vong C.T., Gao C., Chen S., Wu X., Wang S., et al. Bilirubin nanomedicines for the treatment of reactive oxygen species (ROS)-mediated diseases. Mol Pharm. 2020;17:2260–2274. doi: 10.1021/acs.molpharmaceut.0c00337. [DOI] [PubMed] [Google Scholar]
- 32.Wang X., Yan J.J., Wang L., Pan D., Yang R., Xu Y., et al. Rational design of polyphenol-poloxamer nanovesicles for targeting inflammatory bowel disease therapy. Chem Mater. 2018;30:4073–4080. [Google Scholar]
- 33.Wang L.L., Feng C.L., Zheng W.S., Huang S., Zhang W.X., Wu H.N., et al. Tumor-selective lipopolyplex encapsulated small active RNA hampers colorectal cancer growth in vitro and in orthotopic murine. Biomaterials. 2017;141:13–28. doi: 10.1016/j.biomaterials.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 34.Li C., Zhao Y., Cheng J., Guo J., Zhang Q., Zhang X., et al. A proresolving peptide nanotherapy for site-specific treatment of inflammatory bowel disease by regulating proinflammatory microenvironment and gut microbiota. Adv Sci. 2019;6:1900610. doi: 10.1002/advs.201900610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puré E., Cuff C.A. A crucial role for CD44 in inflammation. Trends Mol Med. 2001;7:213–221. doi: 10.1016/s1471-4914(01)01963-3. [DOI] [PubMed] [Google Scholar]
- 36.Kwon M.Y., Wang C., Galarraga J.H., Puré E., Han L., Burdick J.A. Influence of hyaluronic acid modification on CD44 binding towards the design of hydrogel biomaterials. Biomaterials. 2019;222:119451. doi: 10.1016/j.biomaterials.2019.119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang S., Ermann J., Succi M.D., Zhou A., Hamilton M.J., Cao B., et al. An inflammation-targeting hydrogel for local drug delivery in inflammatory bowel disease. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aaa5657. 300ra128-300ra128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tirosh B., Khatib N., Barenholz Y., Nissan A., Rubinstein A. Transferrin as a luminal target for negatively charged liposomes in the inflamed colonic mucosa. Mol Pharm. 2009;6:1083–1091. doi: 10.1021/mp9000926. [DOI] [PubMed] [Google Scholar]
- 39.Courthion H., Mugnier T., Rousseaux C., Moller M., Gurny R., Gabriel D. Self-assembling polymeric nanocarriers to target inflammatory lesions in ulcerative colitis. J Control Release. 2018;275:32–39. doi: 10.1016/j.jconrel.2017.07.044. [DOI] [PubMed] [Google Scholar]
- 40.Gan J., Dou Y., Li Y., Wang Z., Wang L., Liu S., et al. Producing anti-inflammatory macrophages by nanoparticle-triggered clustering of mannose receptors. Biomaterials. 2018;178:95–108. doi: 10.1016/j.biomaterials.2018.06.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.