Abstract
Introduction
Common variants in the UMOD gene are considered an evolutionary adaptation against urinary tract infections (UTIs) and have been implicated in kidney stone formation, chronic kidney disease (CKD), and hypertension. However, differences in UMOD variant-phenotype associations across population groups are unclear.
Methods
We tested associations between UMOD/PDILT variants and up to 1528 clinical diagnosis codes mapped to phenotype groups in the Million Veteran Program (MVP), using published phenome-wide association study (PheWAS) methodology. Associations were tested using logistic regression adjusted for age, sex, and 10 principal components of ancestry. Bonferroni correction for multiple comparisons was applied.
Results
Among 648,593 veterans, mean (SD) age was 62 (14) years; 9% were female, 19% Black, and 8% Hispanic. In White patients, the rs4293393 UMOD risk variant associated with increased uromodulin was associated with increased odds of CKD (odds ratio [OR]: 1.22, 95% CI: 1.20–1.24, P = 5.90 × 10−111), end-stage kidney disease (OR: 1.17, 95% CI: 1.11–1.24, P = 2.40 × 10−09), and hypertension (OR: 1.03, 95% CI: 1.05–1.05, P = 2.11 × 10−06) and significantly lower odds of UTIs (OR: 0.94, 95% CI: 0.92–0.96, P = 1.21 × 10−10) and kidney calculus (OR: 0.85, 95% CI: 0.83–0.86, P = 4.27 × 10−69). Similar findings were observed across UMOD/PDILT variants. The rs77924615 PDILT variant had stronger associations with acute cystitis in White female (OR: 0.73, 95% CI: 0.59–0.91, P = 4.98 × 10−03) versus male (OR: 0.99, 95% CI: 0.89–1.11, P = 8.80 × 10−01) (P interaction = 0.01) patients. In Black patients, the rs77924615 PDILT variant was significantly associated with pyelonephritis (OR: 0.65, 95% CI: 0.54–0.79, P = 1.05 × 10−05), whereas associations with UMOD promoter variants were attenuated.
Conclusion
Robust associations were observed between UMOD/PDILT variants linked with increased uromodulin expression and lower odds of UTIs and calculus and increased odds of CKD and hypertension. However, these associations varied significantly across ancestry groups and sex.
Keywords: genetic renal disease, hypertension, kidney stones, multiethnic, urinary tract infection, uromodulin
Uromodulin (Umod) is an 85-kilodalton glycoprotein that is exclusively synthesized in the kidney by epithelial cells of the thick ascending limb of the loop of Henle and the distal convoluted tubule.1, 2, 3 It is the most abundant protein in healthy human urine.4,5 Umod is encoded by the UMOD gene on chromosome 16p12.3.6, 7, 8
Prior studies have suggested several physiological roles including regulation of ion (sodium ion and potassium ion) transport by interactions with the NKCC2 cotransporter and the ROMK channel9,10; defense against UTIs by binding uropathogenic Escherichia coli11, 12, 13; preventing formation of calculi by impairing aggregation of calcium oxalate crystals14; and playing a role in innate immunity by binding immunoglobulins and cytokines or activating monocytes and dendritic cells by Toll-like receptor 4.15,16
Recent genetic studies revealing common UMOD variants as important contributors to the architecture of renal traits (across multiple populations)17, 18, 19, 20, 21 and blood pressure,22 have reignited interest in Umod physiology. Because UMOD risk variants directly increase Umod expression,23,24 assessing their effect on clinically relevant phenotypes is paramount. More importantly, there may be relevant differences in the association between UMOD risk variants and clinical phenotypes across different ancestry groups given documented heterogeneity in the genetic basis of complex traits across population groups.25 However, this has been underexplored. We performed a PheWAS of common UMOD variants in non-Hispanic White, Black, Asian, and Hispanic patients in the MVP to investigate the pleiotropic effects of increased Umod expression on clinically relevant phenotypes and to evaluate differences in UMOD-phenome associations between population groups.
Methods
The data that support the findings of this study are available from the corresponding author on reasonable request.
Study Design
This study was designed as a PheWAS, which is a bioinformatics approach that enables the scanning of a broad range of clinical phenotypes (the clinical phenome) available in the electronic health record (EHR) of patients for associations with a genetic variant so as to evaluate the pleiotropic effects in several clinical phenotypes that share genetic architecture.26, 27, 28, 29
Study Population for Primary Analysis
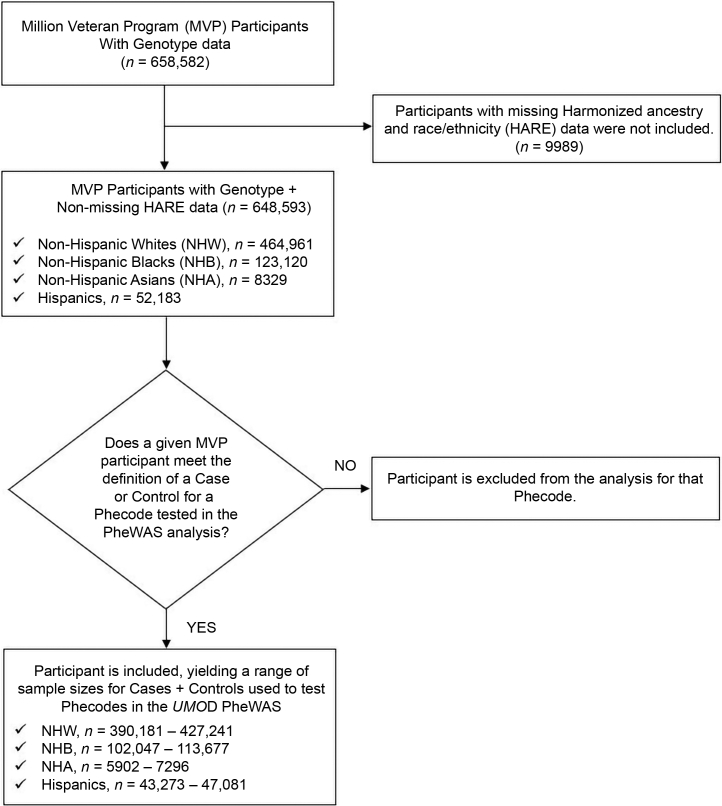
The main study population for this study was participants enrolled in the MVP, which is a large multiethnic longitudinal cohort and mega biobank designed to investigate the genetic underpinnings of common medical conditions among US veterans.30 The MVP data combine EHR data with genomic data to facilitate bioinformatic investigations. Full details of the MVP design and methods have been published elsewhere.30 Briefly, participants were recruited from 63 Veterans Affairs clinics beginning in 2011. At enrolment, participants provided blood samples for genotyping and biomarker studies and completed baseline questionnaires for demographic and lifestyle data. Participants also agreed for their medical records to be accessed. The study was approved by the Veterans Affairs Central Institutional Review Board, and patients signed informed consent. For the current study, 648,593 MVP participants (Table 1 and Figure 1) who had available genotyping and phenotypic data at the time of this study were included in this PheWAS.
Table 1.
Demographic characteristics and clinical phenotypes captured by phecodes in Million Veteran Program participants
| Characteristics | Non-Hispanic Whites na = 464,961 | Non-Hispanic Blacks na = 123,120 | Hispanics na = 52,183 |
|---|---|---|---|
| Demographics | |||
| Age, mean (SD) | 64 (14) | 58 (12) | 55 (16) |
| Female, n (%) | 34,438 (7.4) | 16,993 (13.8) | 5142 (9.9) |
| Years of follow-up in the EHR, median (25th, 75th) | 13.2 (8.0, 17.7) | 15.0 (9.3, 18.9) | 12.3 (6.5, 17.1) |
| General comorbidities | % | Cases | Allb | % | Cases | Allb | % | Cases | Allb |
|---|---|---|---|---|---|---|---|---|---|
| Hypertension | 74.7 | 303,313 | 406,067 | 80.1 | 87,895 | 109,752 | 62.8 | 28,089 | 44,710 |
| Type 2 diabetes | 35.1 | 144,376 | 411,196 | 43.5 | 47,407 | 109,088 | 39.2 | 17,827 | 45,496 |
| Gout | 9.3 | 38,796 | 417,943 | 11.9 | 13,193 | 111,141 | 6.0 | 2789 | 46,404 |
| Proteinuria | 2.2 | 9030 | 420,081 | 3.9 | 4357 | 110,957 | 3.4 | 1544 | 46,086 |
| Kidney disease | |||||||||
| Hypertensive heart and/or renal disease | 13.3 | 53,251 | 399,633 | 18.6 | 19,685 | 106,142 | 10.3 | 4625 | 44,866 |
| Hypertensive CKD | 7.4 | 30,565 | 411,933 | 11.6 | 12,633 | 109,157 | 6.2 | 2842 | 45,956 |
| Type 2 diabetes with renal manifestations | 5.2 | 21,706 | 417,549 | 7.6 | 8393 | 110,764 | 6.1 | 2817 | 45,597 |
| CKD | 13.8 | 56,757 | 411,325 | 17.8 | 19,540 | 109,540 | 10.4 | 4795 | 45,904 |
| CKD, stage I or II | 2.0 | 8236 | 419,746 | 4.1 | 4583 | 110,618 | 1.8 | 829 | 46,453 |
| CKD, stage III | 8.9 | 37,099 | 416,227 | 10.7 | 11,929 | 111,092 | 6.7 | 3083 | 46,295 |
| CKD, stage IV | 2.1 | 8918 | 425,070 | 3.5 | 3953 | 113,406 | 2.4 | 1133 | 47,003 |
| End-stage kidney disease | 1.2 | 5334 | 426,352 | 3.4 | 3873 | 113,748 | 2.1 | 1011 | 47,056 |
| Complications of CKD | |||||||||
| Anemia in CKD | 1.7 | 7330 | 423,505 | 3.9 | 4347 | 112,692 | 2.3 | 1056 | 46,817 |
| Anemia of chronic disease | 3.3 | 13,675 | 416,793 | 6.0 | 6641 | 110,416 | 3.4 | 1593 | 46,254 |
| Other anemias | 19.6 | 76,540 | 390,181 | 28.9 | 29,742 | 102,918 | 16.4 | 7150 | 43,696 |
| Hyperpotassemia | 3.8 | 15,510 | 411,143 | 4.1 | 4517 | 110,276 | 3.4 | 1567 | 45,792 |
| Secondary hyperparathyroidism (renal) | 1.1 | 4716 | 426,008 | 2.5 | 2860 | 113,514 | 1.5 | 686 | 47,045 |
| UTI complications | |||||||||
| UTI | 10.7 | 42,133 | 394,057 | 13.6 | 13,886 | 102,047 | 10.4 | 4488 | 43,273 |
| Pyelonephritis | 0.7 | 2816 | 424,496 | 0.7 | 797 | 113,362 | 0.8 | 364 | 46,780 |
| E. coli infection | 0.5 | 2217 | 422,520 | 0.6 | 711 | 112,696 | 0.5 | 231 | 46,584 |
| Urolithiasis-related phenotypes | |||||||||
| Hematuria | 9.0 | 36,418 | 404,436 | 10.3 | 11,005 | 106,978 | 8.4 | 3746 | 44,558 |
| Gross hematuria | 3.1 | 12,904 | 418,551 | 3.5 | 3922 | 111,742 | 2.5 | 1175 | 46,339 |
| Urinary calculus | 8.8 | 36,238 | 412,276 | 4.8 | 5323 | 111,169 | 7.2 | 3289 | 45,729 |
| Calculus of kidney | 7.3 | 30,249 | 413,504 | 4.0 | 4435 | 111,594 | 5.9 | 2713 | 45,798 |
| Calculus of ureter | 2.1 | 8817 | 422,749 | 1.1 | 1201 | 113,737 | 1.7 | 775 | 46,716 |
| Calculus of lower urinary tract | 0.6 | 2784 | 425,792 | 0.4 | 394 | 114,151 | 0.4 | 209 | 47,081 |
| Hydronephrosis | 1.5 | 6559 | 422,000 | 1.2 | 1354 | 113,350 | 1.1 | 530 | 46,632 |
| Stricture/obstruction of ureter | 0.6 | 2478 | 427,241 | 0.5 | 534 | 113,677 | 0.5 | 223 | 46,860 |
CKD, chronic kidney disease; E. coli, Escherichia coli; EHR, electronic health record; PheWAS, phenome-wide association study; UTI, urinary tract infection.
Represents the group-specific sample size with genotype and phenotype data.
Represents the total number of cases + controls for each phenotype after the PheWAS algorithm but excluded patients not meeting the phecode-specific case or control definition.
Figure 1.
Flowchart for MVP Participants included in the UMOD PheWAS. HARE, harmonized ancestry and race/ethnicity; MVP, Million Veteran Program; PheWAS, phenome-wide association study.
MVP Genotyping
Genotyping was performed using a customized Affymetrix Axiom Biobank Array using DNA extracted from whole blood31 with content added to provide coverage of African and Hispanic haplotypes. Details of the MVP genotyping methods have been described elsewhere.30,32,33 Briefly, standard QC and genotype calling algorithms were applied to the data using the Affymetrix Power Tools Suite. Duplicate samples and samples with an excess of missing genotype calls or a discordance of genetically inferred sex versus self-report were excluded. Related individuals as measured by the KING software34 were also excluded. Before imputation, poorly called variants or single nucleotide polymorphisms (SNPs) deviating from their expected allele frequency based on the 1000 genomes reference panel (1000G)35 were excluded.33 After prephasing using EAGLE version 2,36 genotypes from the 1000G37 reference panel were imputed into MVP participants by Minimac3 software.38 Principal component analysis was performed using the FlashPCA,39 to generate the top 30 principal components explaining the greatest variability in ancestry. After the imputation, variant-level quality control was performed using the EasyQC R package,40 and the following exclusion benchmarks were applied41: ancestry-specific Hardy–Weinberg equilibrium42 P < 1 × 10−20, posterior call probability <0.9, imputation quality (r2) or INFO score <0.3, minor allele frequencies (MAFs) <0.03%, call rate <97.5% for common variants (MAF >1%), and call rate <99% for rare variants (MAF <1%). Variants were also excluded if they deviated by >10% from their expected allele frequency based on the 1000 genomes reference data.43
Race/Ethnicity/Ancestry in MVP
Racial/ethnic groups in the MVP were assigned using a harmonized ancestry and race/ethnicity variable based on an algorithm that integrates genetically inferred ancestry based on the top 30 principal components with self-identified race/ethnicity. Details have been described elsewhere.25 On the basis of the harmonized ancestry and race/ethnicity variable, MVP participants with genotype data are assigned to 1 of the following 4 nonoverlapping groups: non-Hispanic White, non-Hispanic Black, non-Hispanic Asian, and Hispanic patients.
UMOD/PDILT Genetic Variants
The common UMOD variants referenced in this study are synonymous SNPs identified in genome-wide association studies (GWASs) of renal traits, blood pressure, and urinary uromodulin that in individuals of European ancestry are all within the same linkage disequilibrium block spanning the UMOD promoter.44 The ancestral alleles of these UMOD SNPs have allele frequencies of approximately 80% in the 1000G among persons of European ancestry (CEU). These variants include rs4293393 (A/G), rs12917707 (G/T), rs12922822 (C/T), rs13333226 (A/G), and rs13329952 (T/C).44 The squared correlation (r2) between these variants ranges from 0.91 to 1.0 for the 1000G CEU population (Supplementary Figure S1).
The linkage disequilibrium patterns of UMOD variants in 1000G African Americans (Supplementary Figure S2), 1000G Mexican ancestry (Supplementary Figure S3), and 1000G Han Chinese individuals (Supplementary Figure S4) are considerably different to that in 1000G CEU, but the frequencies of the major alleles are all >70%. We also investigated the effects of PDILT (upstream of the UMOD locus) variants, including rs12446492 (T/A), rs77924615 (G/A), and rs11864909 (C/T), that have been reported in GWAS of renal traits.
Definition of Outcomes for PheWAS Analysis in the MVP
In the MVP, the outcomes for the PheWAS analysis were clinical phenotypes (phecodes) derived from International Classification of Disease, ninth revision (ICD-9) and International Classification of Disease, tenth revision (ICD-10) diagnosis codes.45 ICD-9–based phenotypes were defined by mapping ICD-9 codes occurring in the EHR of patients to PheWAS codes, as previously described by Denny et al.26,46 Meanwhile, ICD-10 codes were first mapped to ICD-9 codes using a crosswalk and then subsequently mapped to the phecodes. Additional information on the mapping between phecodes and both ICD-9 and ICD-10 codes (including exclusion phecodes for each phecode) are publicly available at: https://phewascatalog.org/phecodes and https://phewascatalog.org/phecodes_icd10.
As described in previous studies, for each phenotype, a patient was defined as being a case if they had ≥2 phecodes on 2 different dates.46 Meanwhile, controls for the phenotype in question were individuals who had no phecodes for that phenotype and did not have phecodes for related phenotypes for that disease grouping (exclusion phecodes).26,46 Furthermore, individuals with 1 phecode for a given phenotype were not considered a case or a control and were thus excluded from the analysis for that specific phenotype (Figure 1). A list of all phecodes tested in the MVP is included in Supplementary Table S1.
Statistical Methods for PheWAS Analysis in the MVP
Clinical phenotypes with fewer than 200 cases were excluded a priori from the analysis given that ORs for rare events are unreliable.47 After these exclusions, the number of phenotypes available for PheWAS analyses in non-Hispanic White, White female, White male, Black, Asian, and Hispanic patients was 1528, 853, 1439, 1237, 256, and 930, respectively.
In each population, the primary analyses were conducted using the rs4293393 UMOD and rs77924615 PDILT variants as predictor variables. For each variant, a batch of logistic regression models (1 per phenotype totaling 256 to 1528 depending on the number of phecodes meeting the a priori criteria for a given population) was fit to investigate the association with clinical phenotypes. All models were adjusted for age, sex (except for sex-stratified analysis), and 10 principal components.
ORs (95% CI) for significant SNP-phenotype associations were reported. Bonferroni correction (0.05/number of phenotypes tested) was used to correct for multiple testing. Hence, statistical significance was defined as a P value < 0.05/1528 (or 3.27 × 10−05) for non-Hispanic White patients. Similarly, the thresholds for statistical significance in non-Hispanic Black, Hispanic, Asian, White female, and White male patients were 4.04 × 10−05, 5.38 × 10−05, 1.95 × 10−04, 5.86 × 10−05, and 3.47 × 10−05, respectively. In White female patients, we also reported ORs for phenotypes at a nominal significance level of 5.0 × 10−03 considering the smaller sample. We investigated variant-and-sex and variant-and-ancestry interactions by leveraging the difference in the stratum-specific log ORs and their variances to perform 2-sample Z-tests of interaction assuming asymptotic normality of the test statistics for large samples, as described elsewhere.48,49 P interaction < 0.05 was considered significant.
In sensitivity analyses, the PheWAS analyses were repeated using the other UMOD/PDILT variants (in high linkage disequilibrium with SNPs used in the primary analyses) to assess the consistency of the findings. In MVP, all UMOD/PDILT variants used in the PheWAS were either directly genotyped or imputed with imputation quality (Supplementary Table S2). All analyses were performed using the R PheWAS package.
Replication Cohorts
Replication of the MVP PheWAS results was conducted in the Vanderbilt University Medical Center biorepository (BioVU) and the UK Biobank.
The BioVU resource is a DNA biobank at Vanderbilt University Medical Center linked to the synthetic derivative, a deidentified mirror image of their EHR containing inpatient and outpatient data with a goal to investigate links between genetics and health outcomes.50 The BioVU replication was performed using individual-level data for 77,550 individuals who were genotyped on the Infinium Multiethnic Genotyping Array (MEGAchip). Details have been published elsewhere.50 Excluded DNA samples were those with per-individual call rate < 95%, wrongly assigned sex, cryptic relatedness closer than a third-degree relative, or unexpected duplication. Whole genome imputation was performed using the Michigan Imputation Server with the Haplotype Reference Consortium version r1.1 as reference. The list of phecodes tested in BioVU is listed in Supplementary Table S1.
The UK Biobank is a prospective cohort study of the effects of genetic, lifestyle, and environmental factors on disease outcomes. The study recruited >500,000 volunteers from the general population of the United Kingdom aged 40 to 69 years from 2006 to 2010.51 Details have been published elsewhere.51 Briefly, the phenotypes available in the UK Biobank are derived from diverse sources, including ICD-10 codes. Although Supplementary Table S1 lists the phecodes tested in the UK Biobank, there were 178 phenotypes tested in MVP that were not tested in the UK Biobank (Supplementary Table S3).
Replication Analyses in BioVU and UK Biobank
Clinical phenotypes that had significant associations with the candidate UMOD/PDILT variants in MVP were further evaluated in the BioVU repository. In BioVU, the UMOD/PDILT variants used in the PheWAS were also directly genotyped or imputed with high imputation quality (Supplementary Table S4). PheWAS regression analysis was performed adjusting for age, sex, 10 principal components, and length of EHR. For the replication studies, a less conservative nominal significance level was chosen, P < 0.05. All analyses for the BioVU data were performed on Vanderbilt’s Computing cluster.
For White British participants in the UK Biobank, data for clinical phenotypes associated with a given genetic variant including P values and number of samples per phenotype are available online at pheweb.org/UKB-SAIGE/. PheWAS analyses were conducted using SAIGE (a generalized mixed model), adjusting for genetic relatedness, sex, birth year, and the first 4 principal components.
Results
The mean (SD) age of the 648,593 MVP participants included in this UMOD PheWAS was 62 (14) years; 9% were female, 19% Black, 8% Hispanic, and 1.3% Asian. Table 1 illustrates the frequency of clinical phenotypes captured using phecodes. Hypertension and type 2 diabetes were common with case frequencies ranging from 62.8 to 74.7 and 35.1 to 43.5, respectively, across population groups. Supplementary Table S5 illustrates the characteristics of non-Hispanic White (n = 63,029) and non-Hispanic Black patients (n = 14,521) who were included in the BioVU replication. The proportion of female patients was 56% and 62%, respectively, in both groups, and the mean (SD) age was 57 (22) and 47 (21) years. Supplementary Table S6 illustrates the population characteristics of the UK Biobank participants. The mean age was 56 (8) years, 54.4% were female, 27.1% had hypertension, and >5.3% had self-reported diabetes.
In MVP, among non-Hispanic White patients, the MAFs of the rs4293393 and rs77924615 variants were 18% and 20%, respectively (Supplementary Table S7). The corresponding MAFs for these variants in non-Hispanic Black patients were 21% and 6%. In Hispanic and non-Hispanic Asian patients, the MAFs for these variants were 21% and 20% and 10% and 20%, respectively.
PheWAS of UMOD and PDILT Variants in Non-Hispanic Whites—Discovery and Replication
In non-Hispanic White patients, the rs4293393 UMOD promoter variant was significantly associated with 29 clinical phenotypes at the 3.27 × 10−05 threshold (Table 2).
Table 2.
Significant associations of the rs4293393 (A/G) UMOD variant with clinical phenotypes among non-Hispanic White patients in the Million Veteran Program and Replication results from BioVU (Vanderbilt’s Biobank) and UK Biobank
| Clinical phenotypes | Million Veteran Program |
Replication cohorts |
|||||
|---|---|---|---|---|---|---|---|
| PheWAS code description | Cases | Controls | OR (95% CI) per copy of the A allele | P value | BioVU | UK Biobank | |
| Increased risk | |||||||
| CKD | CKD | 56,757 | 354,568 | 1.22 (1.20–1.24) | 5.90 × 10−111 | Yesa | Yes |
| CKD, stages I–II | 8236 | 411,510 | 1.11 (1.07–1.19) | 3.29 × 10−05 | Nob | No | |
| CKD, stage III | 37,099 | 379,128 | 1.24 (1.21–1.26) | 2.79 × 10−86 | Yes | No | |
| CKD, stage IV | 8918 | 416,152 | 1.23 (1.19–1.29) | 1.33 × 10−23 | Yes | No | |
| Diabetic kidney disease | Type 2 diabetes with renal manifestations | 21,706 | 395,843 | 1.11 (1.08–1.13) | 2.76 × 10−14 | No | No |
| Proteinuria | Proteinuria | 9030 | 411,051 | 1.10 (1.05–1.14) | 4.31 × 10−06 | Yes | No |
| End-stage kidney disease and related disorders | End-stage renal disease | 5334 | 421,018 | 1.17 (1.11–1.24) | 2.40 × 10−09 | No | Yes |
| Renal failure, NOS | 5950 | 415,270 | 1.16 (1.10–1.22) | 4.20 × 10−09 | No | Yes | |
| Secondary hyperparathyroidism of renal origin | 4716 | 421,292 | 1.21 (1.14–1.28) | 1.69 × 10−11 | No | No | |
| Anemia of CKD | 7330 | 416,175 | 1.20 (1.14–1.25) | 7.31 × 10−15 | No | Yes | |
| Disorders resulting from impaired renal function | 6104 | 418,597 | 1.17 (1.12–1.23) | 1.79 × 10−10 | No | No | |
| Acute kidney injury | Acute renal failure | 37,825 | 367,058 | 1.06 (1.04–1.09) | 5.50 × 10−10 | No | Yes |
| Electrolyte imbalance | Electrolyte imbalance | 51,136 | 338, 927 | 1.04 (1.02–1.05) | 2.33 × 10−05 | No | No |
| Hyperkalemia | Hyperpotassemia | 15,510 | 395,633 | 1.07 (1.04–1.10) | 2.62 × 10−05 | No | Yes |
| Hypertension and complications | Hypertension | 303,313 | 102,754 | 1.03 (1.02–1.05) | 2.11 × 10−06 | No | Yes |
| Essential hypertension | 301,525 | 104,191 | 1.03 (1.02–1.05) | 3.08 × 10−06 | No | Yes | |
| Hypertensive CKD | 30,565 | 381,368 | 1.20 (1.17–1.22) | 1.89 × 10−53 | No | Yes | |
| Hypertensive heart or renal disease | 53,251 | 346,382 | 1.10 (1.09–1.12) | 5.11 × 10−29 | No | Yes | |
| Gout | Gout | 38,796 | 379,147 | 1.04 (1.02–1.06) | 1.94 × 10−05 | Yes | No |
| Anemia | Anemia of chronic disease | 13,675 | 403,118 | 1.13 (1.09–1.16) | 9.05 × 10−13 | No | No |
| Other anemias | 76,540 | 313,641 | 1.03 (1.02–1.05) | 2.00 × 10−05 | No | Yes | |
| Decreased risk | |||||||
| UTIs | UTI | 42,133 | 351,924 | 0.94 (0.92–0.96) | 1.21 × 10−10 | Yes | No |
| Urinary calculi | Urinary calculus | 36,238 | 376,038 | 0.85 (0.83–0.86) | 1.45 × 10−64 | Yes | Yes |
| Calculus of the kidney | 30,249 | 383,255 | 0.83 (0.81–0.85) | 4.27 × 10−69 | Yes | Yes | |
| Calculus of the ureter | 8817 | 413,932 | 0.81 (0.78–0.84) | 1.77 × 10−29 | Yes | Yes | |
| Calculus of the lower urinary tract | 2784 | 423,008 | 0.81 (0.76–0.87) | 4.93 × 10−10 | No | Yes | |
| Hydronephrosis | 6559 | 415,441 | 0.88 (0.84–0.92) | 9.20 × 10−09 | No | Yes | |
| Hematuria | Hematuria | 36,418 | 368,018 | 0.95 (0.93–0.97) | 4.40 × 10−07 | No | Yes |
| Gross hematuria | 12,904 | 405, 647 | 0.94 (0.91–0.97) | 3.26 × 10−05 | No | No | |
BioVU, biorepository; CKD, chronic kidney disease; NOS, not otherwise specified; OR, odds ratio; PheWAS, phenome-wide association study; UTI, urinary tract infection.
All associations listed were significant at the Bonferroni-corrected significance level of 3.27 × 10−05.
Yes means that the association was replicated in BioVU or the UK Biobank.
No means that the association was not replicated in BioVU or the UK Biobank.
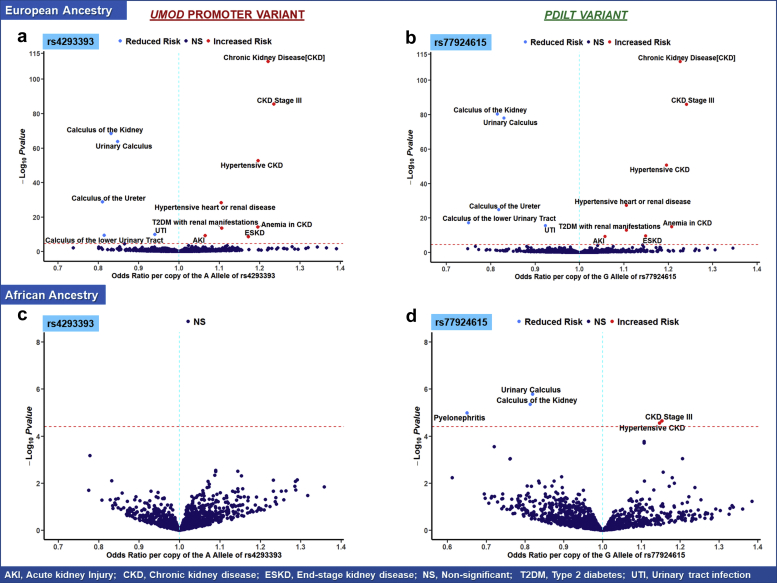
Each copy of the A allele of rs4293393 (that correlates with increased Umod expression) was associated with a significant 22% higher odds (OR: 1.22, 95% CI: 1.20–1.24) of CKD, 11% higher odds (OR: 1.11; 95% CI: 1.08–1.13) of diabetic kidney disease, 17% higher odds (OR: 1.17, 95% CI: 1.17–1.24) of end-stage kidney disease, and 6% higher odds (OR: 1.06, 95% CI: 1.04–1.09) of acute kidney injury (AKI). The A allele of rs4293393 was also associated with increased odds of proteinuria (OR: 1.10, 95% CI: 1.05–1.14), anemia of CKD (OR: 1.20, 95% CI: 1.14–1.25), secondary hyperparathyroidism of renal origin (OR: 1.21, 95% CI: 1.14–1.21), hypertension (OR: 1.03, 95% CI: 1.02–1.05), hypertensive CKD (OR: 1.20, 95% CI: 1.17–1.22), hypertensive heart or renal disease (OR: 1.10, 95% CI: 1.09–1.12), hyperkalemia (OR: 1.07, 95% CI: 1.04–1.10), and gout (OR: 1.04, 95% CI: 1.02–1.06). Conversely, the A allele of rs4293393 was significantly associated with lower odds of UTI (OR: 0.94, 95% CI: 0.92–0.96), urinary calculus (OR: 0.85, 95% CI: 0.83–0.86), calculus of the kidney (OR: 0.83, 95% CI: 0.81–0.85), calculus of the ureter (OR: 0.81, 95% CI: 0.78–0.84), and hematuria (OR: 0.95, 95% CI: 0.93–0.97). These findings were consistent with those observed for the other primary analysis using the rs77924615 PDILT variant (Figure 2a and b) and further confirmed in supplementary analyses using other UMOD promoter and PDILT variants (Supplementary Tables S8–S13 and Supplementary Table S5).
Figure 2.
Volcano plot illustrating key clinical phenotypes significantly associated with UMOD promoter (rs4293393) and PDILT variants (rs77924615) in (a, b) non-Hispanic White and (c, d) non-Hispanic Black patients in the Million Veteran Program. In each plot, the red line indicates the significance threshold for Bonferroni correction as a reference. Phenotypes in the right upper quadrant have increased odds per copy of the allele associated with increased Umod expression, whereas phenotypes in the left quadrant have decreased odds. In Black patients, no significant variant-phenotype associations were observed for the rs4293393 UMOD promoter variant. Five significant variant-phenotype associations were observed for the PDILT variant rs4293393. In White patients, a wide range of significant variant-phenotype associations were observed corroborating the pleiotropic effects of Umod on human physiology. In addition, there were no noticeable differences in the patterns of variant-phenotype associations between UMOD promoter and PDILT variants. AKI, acute kidney injury; CKD, chronic kidney disease; ESKD, end-stage kidney disease; NS, nonsignificant; T2DM, type 2 diabetes mellitus; UTI, urinary tract infection.
Overall, the findings for most of these clinical phenotypes were replicated in the UK Biobank and/or BioVU at α = 0.05 (Table 2 and Supplementary Tables S14 and S15). For example, in BioVU, the OR (95% CI) per copy of the A allele of rs4293393 for calculus of the kidney, UTI, and CKD stage 3 was 0.85 (95% CI: 0.79–0.92, P = 4.02 × 10−05), 0.94 (95% CI: 0.90–0.98, P = 1.00 × 10−02), and 1.13 (95% CI: 1.05–1.22, P = 1.5 × 10−03), respectively.
PheWAS of UMOD/PDILT Variants in Non-Hispanic Black Patients
In Black patients, there were significant differences in the patterns of association between clinical phenotypes and both UMOD promoter variants (rs4293393 and rs12917707) and the PDILT variant (77924615; Figure 2c and 2d). The rs77924615 PDILT variant was significantly associated with phenotypes in 4 domains of interest (CKD, hypertension, calculus, and UTI). Each copy of the G allele of the rs77924615 variant was associated with increased odds of CKD stage III (OR: 1.15, 95% CI: 1.08–1.23, P = 2.27 × 10−05) and hypertensive CKD (OR: 1.15, 95% CI: 1.08–1.22, P = 2.78 × 10−06) and lower odds of urinary calculus (OR: 0.82, 95% CI: 0.76–0.89, P = 1.68 × 10−06), calculus of the kidney (OR: 0.81, 95% CI: 0.74–0.89, P = 4.52 × 10−06), and pyelonephritis (OR: 0.65, 95% CI: 0.54–0.79, P = 1.05 × 10−05) (Figure 2). However, no phenome-wide significant associations were observed between the rs4293392 UMOD promoter variant and any of the 4 domains of clinical phenotypes observed in non-Hispanic White patients, despite the number of cases for these phenotypes varying between 5323 and 87,895 (Table 3). The P values for differential effects between non-Hispanic White and Black patients for the association of the rs4293393 UMOD promoter variant and the 4 domains of clinical phenotypes observed in non-Hispanic Whites were 1.47 × 10−25 for CKD, 1.59 × 10−02 for hypertension, 2.15 × 10−05 for UTI, and 2.35 × 10−05 for urinary calculus (Supplementary Table S16). For the rs12917707 UMOD promoter variant, the G allele was significantly associated with 18% lower odds (OR: 0.82, 95% CI: 0.76–0.89, P = 6.0 × 10−06) of urinary calculus (Supplementary Table S5).
Table 3.
Effect estimates in non-Hispanic Blacks for clinical phenotypes that were significantly associated with the rs4293393 (A/G) UMOD variant in non-Hispanic White patients in the Million Veteran Program
| Clinical phenotypes | Million Veteran Program |
||||
|---|---|---|---|---|---|
| PheWAS code description | Cases | Controls | OR (95% CI) per copy of the A allele | P value | |
| CKD | CKD | 19,540 | 90,000 | 1.02 (0.99–1.05) | 1.14 × 10−01 |
| CKD, stages I–II | 4583 | 106,035 | 1.03 (0.98–1.09) | 2.61 × 10−01 | |
| CKD, stage III | 11,929 | 99,163 | 1.03 (0.99–1.07) | 9.57 × 10−02 | |
| CKD, stage IV | 3953 | 109,453 | 1.06 (1.00–1.12) | 4.58 × 10−02 | |
| Diabetic kidney disease | Type 2 diabetes with renal manifestations | 8393 | 102,371 | 1.01 (0.97–1.05) | 7.76 × 10−01 |
| Proteinuria | Proteinuria | 4357 | 106,600 | 1.02 (0.96–1.07) | 5.38 × 10−01 |
| End-stage kidney disease and related disorders | End-stage kidney disease | 3873 | 109,875 | 1.04 (0.98–1.10) | 1.91 × 10−01 |
| Renal failure, NOS | 3205 | 108,717 | 1.02 (0.96–1.08) | 5.82 × 10−01 | |
| Secondary hyperparathyroidism of renal origin | 2860 | 110,654 | 1.03 (0.96–1.10) | 3.92 × 10−01 | |
| Anemia of CKD | 4347 | 108,345 | 1.04 (0.98–1.04) | 2.00 × 10−01 | |
| Disorders resulting from impaired renal function | 3478 | 109,570 | 1.02 (0.97–1.09) | 3.57 × 10−01 | |
| Acute kidney injury | Acute renal failure | 13,954 | 92,382 | 1.01 (0.98–1.05) | 3.90 × 10−01 |
| Electrolyte imbalance | Electrolyte imbalance | 17,004 | 86,178 | 1.01 (0.98–1.04) | 4.66 × 10−01 |
| Hyperkalemia | Hyperpotassemia | 4517 | 105,759 | 1.05 (1.00–1.11) | 4.94 × 10−02 |
| Hypertension and complications | Hypertension | 87,895 | 21,857 | 0.99 (0.96–1.02) | 6.96 × 10−01 |
| Essential hypertension | 87,503 | 22,156 | 0.99 (0.97–1.02) | 7.26 × 10−01 | |
| Hypertensive CKD | 12,633 | 96,524 | 1.03 (1.00–1.06) | 7.60 × 10−02 | |
| Hypertensive heart or renal disease | 19,685 | 86,457 | 1.03 (1.00–1.06) | 3.68 × 10−02 | |
| Gout | Gout | 13,193 | 97,948 | 1.02 (0.98–1.05) | 3.62 × 10−01 |
| Anemia | Anemia of chronic disease | 6641 | 103,775 | 1.03 (0.98–1.05) | 3.62 × 10−01 |
| Other anemias | 29,742 | 73,176 | 1.01 (0.98–1.03) | 5.57 × 10−01 | |
| UTIs | UTI | 13,886 | 88,161 | 1.02 (0.98–1.05) | 2.45 × 10−01 |
| Urinary calculi | Urinary calculus | 5323 | 105,846 | 0.95 (0.90–0.99) | 2.49 × 10−02 |
| Calculus of the kidney | 4435 | 107,159 | 0.95 (0.91–1.01) | 7.83 × 10−01 | |
| Calculus of the ureter | 1201 | 112,536 | 0.91 (0.83–1.01) | 6.87 × 10−01 | |
| Calculus of the lower urinary tract | 394 | 113,757 | 0.91 (0.77–1.07) | 2.59 × 10−01 | |
| Hydronephrosis | 1354 | 111,996 | 0.94 (0.86–1.03) | 2.18 × 10−01 | |
| Hematuria | Hematuria | 11,005 | 95,973 | 1.02 (0.98–1.05) | 3.52 × 10−01 |
| Gross hematuria | 3922 | 107,820 | 1.04 (0.98–1.10) | 2.08 × 10−01 | |
CKD, chronic kidney disease; NOS, not otherwise specified; OR, odds ratio; PheWAS, phenome-wide association study.
PheWAS of UMOD and PDILT Variants in Hispanic and Non-Hispanic Asian Patients
In Hispanic patients, both UMOD promoter and PDILT variants had phenome-wide significant associations with renal phenotypes that were observed among non-Hispanic White patients. For example, each copy of the A allele of the rs4293393 UMOD promoter variant was associated with increased odds of CKD (OR: 1.15, 95% CI: 1.09–1.22, P = 7.10 × 10−07) and hypertensive CKD (OR: 1.16, 95% CI: 1.08–1.24, P = 4.27 × 10−05) and lower odds of calculus of the kidney (OR: 0.84; 95% CI: 0.79–0.90, P = 1.55 × 10−07; Supplementary Figure S6). These effect estimates were similar for the rs12917707 UMOD promoter variant and the rs77924615 PDILT variant (Supplementary Table S5). However, no significant association was observed with UTI despite a considerable number of observed cases, n = 4428 (Supplementary Tables S17 and S18). In non-Hispanic Asian patients, no phenome-wide significant phenotypes were observed for the rs4293393 UMOD promoter variant. However, for the rs77924615 PDILT variant, 1 phenotype namely CKD stage III (OR: 1.90, 95% CI: 1.34–2.04, P = 3.25 × 10−5) reached phenome-wide significance.
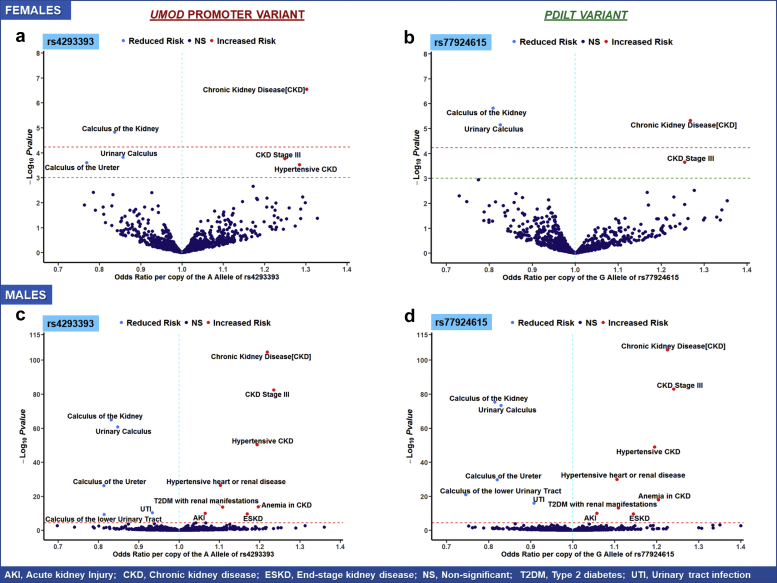
Sex Differences in the PheWAS of UMOD and PDILT Variants in Non-Hispanic White Patients
In non-Hispanic White male patients, the findings for the UMOD promoter and PDILT variants were consistent with the patterns observed in the total pool of non-Hispanic White patients with similar effect sizes and P values (Figure 3c and d). In White female patients, there were 3 clinical phenotypes reaching phenome-wide significance for the rs77924615 PDILT variant and 2 for the UMOD promoter variants rs12917707 and rs4293393 (Figure 3a and b). For example, each copy of the A allele of the rs4293393 variant was associated with 27% increased odds of CKD (OR: 1.27, 95% CI: 1.14–1.40) and 19% lower odds (OR: 0.81, 95% CI: 0.74–0.88) of calculus of the kidney (Table 4). Six other phenotypes reached nominal significance (α = 5 × 10−3), specifically hypertensive CKD, hypertensive heart or renal disease, CKD stage 3, elevated white blood cell count, urinary calculus, and calculus of the ureter. Overall, the difference in effect sizes for these phenotypes was minimal between the sexes. However, for acute cystitis, the effect was considerably stronger among White female patients with an OR (95% CI) per copy of the G allele of the rs77924615 PDILT variant of 0.73 (95% CI: 0.59–0.91, P = 4.98 × 10−03) compared with 0.99 (95% CI: 0.89–1.11, P = 8.80 × 10−01) among White male patients (P interaction = 0.01) (Supplementary Tables S16 and S19). The G allele of the rs77924615 variant was associated with increased odds of heart failure with preserved ejection fraction (OR: 1.28, 95% CI: 1.09–1.50, P = 3.01 × 10−03) among White female patients. Among White male patients, the effect estimate was 1.00 (95% CI: 0.97–1.03, P = 0.89).
Figure 3.
Volcano plot illustrating key clinical phenotypes significantly associated with UMOD promoter (rs4293393) and PDILT variants (rs77924615) in non-Hispanic White female (a, b) and male (c, d) patients in the Million Veteran Program. In each plot, the red line indicates the significance threshold for Bonferroni correction as a reference whereas the green line indicates nominal significance at α = 5 × 10−3. Phenotypes in the right upper quadrant have increased odds per copy of the allele associated with increased Umod expression, whereas phenotypes in the left quadrant have decreased odds. All clinical phenotypes typically associated with Umod expression had significant associations with UMOD promoter and PDILT variants in White male patients. In White female patients, significant variant-phenotype associations were observed for CKD, urinary calculus, and calculus of the kidney. Nominally significant associations were observed with acute cystitis and heart failure with preserved EF. AKI, acute kidney injury; CKD, chronic kidney disease; EF, ejection fraction; ESKD, end-stage kidney disease; NS, nonsignificant; T2DM, type 2 diabetes mellitus; UTI, urinary tract infection.
Table 4.
Association of the rs4293393 (A/G) UMOD variant with key clinical phenotypes across sex groups in non-Hispanic White patients in the MVP
| Females | |||||
|---|---|---|---|---|---|
| Clinical phenotypes | PheWAS code description | Cases | Controls | OR (95% CI) per copy of the A allele | P value |
| CKD | CKD | 1685 | 28,924 | 1.30 (1.18–1.44) | 2.87 × 10−07,a |
| CKD, stage III | 1154 | 29,725 | 1.25 (1.10–1.45) | 2.39 × 10−04 | |
| CKD, stage IV | 263 | 31,007 | 1.62 (1.24–2.11) | 4.00 × 10−04 | |
| Hypertensive complications | Hypertensive CKD | 774 | 30,108 | 1.28 (1.11–1.49) | 7.50 × 10−04 |
| Hypertensive heart or renal disease | 1567 | 28,773 | 1.17 (1.06–1.30) | 2.20 × 10−03 | |
| UTIs | UTI | 7840 | 18,560 | 0.98 (0.98–1.05) | 4.03 × 10−01 |
| Cystitis | 977 | 28,707 | 0.87 (0.78–0.98) | 2.01 × 10−02 | |
| Acute cystitis | 245 | 30,151 | 0.84 (0.68–1.05) | 1.31 × 10−01 | |
| Pyelonephritis | 584 | 30,061 | 0.92 (0.79–1.07) | 2.62 × 10−01 | |
| Urinary calculi | Urinary calculus | 1990 | 28,328 | 0.86 (0.79–0.93) | 1.73 × 10−04 |
| Calculus of the kidney | 1710 | 28,751 | 0.84 (0.77–0.91) | 4.98 × 10−05,a | |
| Calculus of the ureter | 518 | 30,514 | 0.77 (0.66–0.89) | 5.37 × 10−04 | |
| Males | |||||
|---|---|---|---|---|---|
| Clinical phenotypes | PheWAS code description | Cases | Controls | OR (95% CI) per copy of the A allele | P value |
| CKD | CKD | 55,072 | 325,644 | 1.22 (1.20–1.24) | 2.09 × 10−105,a |
| CKD, stage III | 35,945 | 349,403 | 1.23 (1.21–1.26) | 2.82 × 10−83,a | |
| CKD, stage IV | 8655 | 385,145 | 1.23 (1.18–1.28) | 1.54 × 10−21,a | |
| Hypertension complications | Hypertensive chronic kidney disease | 29,791 | 351,260 | 1.19 (1.17–1.22) | 4.31 × 10−51,a |
| Hypertensive heart or renal disease | 51,684 | 317,609 | 1.10 (1.08–1.12) | 3.68 × 10−27,a | |
| UTIs | UTI | 34,293 | 333,364 | 0.93 (0.92–0.95) | 4.35 × 10−11,a |
| Cystitis | 3638 | 385,756 | 0.97 (0.92–1.03) | 3.44 × 10−01 | |
| Acute cystitis | 1096 | 391,828 | 0.92 (0.83–1.02) | 1.26 × 10−01 | |
| Pyelonephritis | 2232 | 391,619 | 0.91 (0.84–0.98) | 9.18 × 10−03 | |
| Urinary calculi | Urinary calculus | 34,248 | 347,710 | 0.85 (0.83–0.86) | 1.45 × 10−61,a |
| Calculus of the kidney | 28,539 | 354,504 | 0.83 (0.81–0.85) | 4.20 × 10−65,a | |
| Calculus of the ureter | 8299 | 383,418 | 0.81 (0.78–0.84) | 5.22 × 10−27,a | |
CKD, chronic kidney disease; MVP, Million Veteran Program; OR, odds ratio; PheWAS, phenome-wide association study; UTI, urinary tract infection.
Represents phenotypes reaching PheWAS significance among non-Hispanic White female (5.86 × 10−05) and male patients (3.47 × 10−05) in the MVP.
Discussion
In this PheWAS of common UMOD/PDILT gene variants using data from large multiethnic biobanks, we report significant associations of UMOD risk variants with increased risk of CKD and hypertension and lower risk of UTIs and kidney stones. Importantly, we observed differential patterns in the pleiotropic effects of UMOD risk variants on clinical phenotypes related to Umod physiology across ancestry groups, which have not been previously reported.
In non-Hispanic White patients, there were consistent SNP-phenotype associations across UMOD promoter and PDILT variants. UMOD promoter and PDILT risk variants (alleles associated with increased Umod expression) were significantly associated with increased risk of kidney disease phenotypes (CKD, diabetic kidney disease, and end-stage kidney disease), hypertension, and complications of hypertension (hypertensive heart or renal disease) while being protective for UTIs and urolithiasis. Conversely, in Black patients, clear differences in SNP-phenotype associations were observed between variants in the UMOD promoter and PDILT locus with variants in the latter having significant effects with the aforementioned Umod-related phenotypes. In particular, the rs77924615 PDILT variant had strong protective effects for pyelonephritis. The variant-phenotype associations in Hispanic patients were mostly consistent with those observed in non-Hispanic White patients but the associations with UTIs were null (ORs close to 1) raising potential questions regarding the causal variants in Hispanic patients and the genetic architecture of UTIs in this population. The findings in non-Hispanic Asian patients revealed a particularly strong association of the rs77924615 variant with CKD stage 3 (90% increased odds per copy of the G allele that increases Umod expression) despite a null effect for the rs4293393 UMOD promoter variant. The fact that these patterns are in clear contrast with those observed in non-Hispanic White patients suggests the needed for additional investigation into differential patterns in Asian versus European ancestry populations using larger samples in future studies. Sex differences in the ORs for the aforementioned phenotypes were minimal except for stronger effects of the rs7724615 PDILT variant on acute cystitis and heart failure with preserved ejection fraction in White female versus male patients which needs to be investigated further in future work.
Previous GWAS have established the association between common UMOD variants and CKD in cohorts from multiple populations, including European,17,18 Icelandic,52 and multiethnic.21,53 Case-control data have also revealed significant associations with hypertension and incident cardiovascular disease independently of eGFR.22 Our PheWAS findings extend these prior data by revealing significant associations with both early and advanced stages of CKD and both renal and cardiovascular complications of hypertension in clinical settings, for UMOD promoter risk variants among non-Hispanic White patients. Our findings corroborate those previously reported by Shang et al.54 who found significant associations for the rs28544423 UMOD variant (in complete linkage disequilibrium with rs4293393) with CKD stage 3 in 78,638 European ancestry individuals in the eMERGE network. However, in our multiethnic PheWAS, these associations with CKD and hypertension and hypertensive complications were not observed for UMOD promoter risk variants in non-Hispanic Black patients. Notably, in a previous GWAS of kidney traits conducted exclusively in African Americans in the CARE consortium, the rs4293393 UMOD promoter variant did not have a significant association with CKD and urinary albumin-to-creatinine ratio and only reached nominal significance for eGFR.55 These findings contrast with those from large multiethnic studies,21,53 whose results are likely driven by the larger European cohorts in the meta-analysis. Our multiethnic PheWAS suggests differences in the effect of UMOD promoter risk variants on advanced CKD and hypertensive renal and/or heart disease between patients of European and African ancestry, and this observed heterogeneity in allelic effects for UMOD promoter risk variants is novel and may portend potential clinical relevance. Prior data from Trudu et al.24 had established Umod as a potential therapeutic target for blood pressure lowering and nephroprotection. Trudu et al.24 reported that increased Umod expression in transgenic mice led to salt-sensitive hypertension (owing to Umod-mediated hyperactivation of NKCC2 in the thick ascending limb) and the development of age-dependent lesions in the kidney that were similar to those observed in older patients that are homozygous for high-risk alleles of UMOD promoter variants, such as the carriers of the AA genotype of the rs4293393 variant reported in the current study. Trudu et al.24 also reported that pharmacologic inhibition of the NKCC2 cotransporter using furosemide was more effective on blood pressure lowering in patients with hypertension who are homozygous for UMOD promoter risk variants than in other patients with hypertension revealing a link between genetic susceptibility to hypertension and CKD to Umod expression levels and Umod-mediated reabsorption of sodium chloride in the thick ascending limb. Devuyst et al.44 thus highlighted a potential personalized medicine recommendation for furosemide which may be more efficacious in patients with hypertension who are homozygous for UMOD promoter risk variants. Our findings further extend this potential precision medicine recommendation by highlighting that the possible benefit of SNP-dependent furosemide effects may also be ancestry dependent. Although patients with hypertension of European ancestry who are homozygotes for UMOD promoter variants may have more potent furosemide effects, this may not be the case for patients of African ancestry. However, the observed effect of the rs77924615 PDILT variant on hypertensive CKD in patients of African ancestry does raise the question whether this variant maybe a better potential candidate—compared with the UMOD promoter variants—to explore for precision medicine approaches in this patient population.
The observed deleterious effects of UMOD risk variants on diabetic kidney disease and proteinuria in patients of European ancestry may be an important finding that is relevant to ongoing research on therapies for diabetic kidney disease. In future work, it may be of interest to investigate whether the presence of UMOD risk variants modifies the beneficial effect of nephroprotective drugs (RAAS inhibitors,56,57 SGLT2 inhibitors,58, 59, 60, 61, 62 and mineralocorticoid antagonists63) in diabetic patients. In our eGFR GWAS, the observed UMOD effect on kidney function was greater in patients with diabetes versus patients without diabetes.18,64 The effect of extraluminal Umod may be attributed to the proximity of the thick ascending limb to the macula densa where Umod may interfere with the tubuloglomerular feedback increasing intraglomerular pressure.65
UMOD risk variants were also associated with increased AKI risk. The consistent findings across UMOD and PDILT variants and concurrent significant signals with accompanying signs of AKI such as hyperkalemia provide some corroboration to the observed UMOD-AKI association. However, this association may have been confounded by underlying CKD especially given that previous studies have suggested Umod as a potentially useful biomarker in models of AKI with higher levels being associated with better prognosis.66,67
The observed protective effects of the major alleles of the UMOD variants on nephrolithiasis and UTIs in this study among individuals of European ancestry corroborate previous data.11, 12, 13,68, 69, 70 Umod’s propensity to reduce kidney stone formation has been linked to its sialylated negatively charged glycans that inhibit aggregation of calcium oxalate and calcium phosphate crystals.70,71 Meanwhile, although Umod has been known to protect against UTIs by binding uropathogenic Escherichia coli,12,72 the precise mechanism was poorly understood until the landmark paper by Weiss et al.73 They revealed that the Umod filament consists of a zigzag-shaped backbone with laterally protruding arms. Specifically, Umod acts as a multivalent ligand for FimH on the bacterial pili, presenting several specific glycan epitopes which outcompete glycan receptors on the urothelium.11,73 Several Umod-uropathogen interactions occur between the multiple binding sites of the Umod filament and several pili on the bacterium causing bacterial aggregation and preventing attachment to the urothelium and clearance by micturition.73
In Black patients, no protective effects on urolithiasis were observed for the major allele of the rs4293393 UMOD promoter variant. These effects were observed for the rs12917707 and rs77924615 variants. The latter also had a strong protective effect on pyelonephritis. Similarly, in White female patients, stronger effects were observed for the rs77924615 PDILT variant on acute cystitis compared with UMOD promoter variants. In a transethnic eGFR GWAS in >1 million individuals, Wuttke et al.21 found that the rs77924615 PDILT variant had a strong causal regulatory role on the UMOD gene and Umod expression in the kidney by extension. This strong regulatory role of the rs77924615 variant on uromodulin production and urinary uromodulin levels may explain its greater effect on UTIs in female patients.
The strengths of our study include the large sample of patients with available genetic data, racial/ethnic diversity, and the presence of long follow-up time in the EHR, which increases the potential to capture clinical phenotypes using diagnostic codes. In addition, the Veterans Affairs is a closed health care system where patients have continuity of care which further minimizes ascertainment bias for patient outcomes. Furthermore, although the primary findings were obtained using MVP data, a replication phase was conducted in BioVU and the UK Biobank. Several limitations exist including the exclusive utilization of diagnostic codes to define outcomes. However, Cai et al.45 revealed PPVs of 80% to 100% for major cardiovascular phenotypes in their PheWAS of IL6R variants in the MVP. Requiring at least 2 codes to ascertain a case for each phenotype in our study reduces false positives. Furthermore, eliminating “uncertain” cases from the case-control pool makes the comparison groups more biologically disparate and likely increases the signal-to-noise ratio in this study. Moreover, assuming any potential outcome misclassification may likely be nondifferential across alleles of the UMOD variants; the ORs would be, at worst, biased toward the null. Dense phenotyping information on study outcomes (e.g., information on bacteriology for UTIs and antibiotic treatments) would have facilitated more in-depth analyses. However, as these extraneous factors are unlikely to have different distributions across alleles of the UMOD variants (which are antecedent by definition), they are unlikely to confound the ORs reported in this study. The absence of data on urinary uromodulin levels to supplement the reported genetic effects is another limitation. Although the PheWAS analyses in non-Hispanic White and Black patients were adequately powered for the main findings (Supplementary Table S20), our analysis in Hispanic patients (Supplementary Table S21) and non-Hispanic White female patients was limited by lower statistical power.
In summary, we found significant associations between UMOD/PDILT risk variants linked with increased Umod expression and increased risk of CKD and hypertension and lower odds of UTIs and nephrolithiasis. However, these associations varied significantly across ancestry groups and sex.
Implications
Using a PheWAS approach in multiethnic biobanks, we have highlighted significant heterogeneity in the pleiotropic effects of increased Umod expression on human disease between population groups—differences that may portend future relevance to personalized medicine for patients from diverse populations.
Disclosure
This work was supported by Veterans Affairs grant MVP Merit CX001897 (PI AMH) and is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration. The data analysis was supported using resources and facilities of the Veterans Affairs Informatics and Computing Infrastructure and the Veterans Affairs Genomic Information System for Integrative Science. EAA was supported by the American Heart Association Grant #20POST35210952/Akwo/2020. AG was funded by the Building Interdisciplinary Research Careers in Women’s Health program as a scholar (Grant number: 5K12HD043483-18; PI: KE Hartmann) and the National Institute of Diabetes and Digestive and Kidney Diseases (Grant number: 1K01DK120631-01A1; PI: AG) when this work was performed. The data used for the replication phase were obtained from Vanderbilt University Medical Center's resources, BioVU, and the synthetic derivative, which are supported by institutional funding, the 1S10RR025141 instrumentation award, and by the Vanderbilt National Center for Advancing Translational Science grant UL1TR000445 from National Center for Advancing Translational Sciences/National Institutes of Health. Existing genotypes in BioVU were funded by National Institutes of Health grants RC2GM092618 from National Institute of General Medical Sciences/OD and U01HG004603 from National Human Genome Research Institute/National Institute of General Medical Sciences. The authors acknowledge the expert technical support of the VANTAGE and VANGARD core facilities, supported in part by the Vanderbilt-Ingram Cancer Center (P30 CA068485) and Vanderbilt Vision Center (P30EY08126). The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. This publication does not represent the views of the Department of Veterans Affairs or the US Government.
Acknowledgments
This research is based on data from the Million Veteran Program, Veterans Health Administration's Office of Research and Development, Veterans Health Administration, and was supported by the CSR&D Merit award title Genetics of CKD and Hypertension—Risk Prediction and Drug Response in the MVP (#CX001897 to AMH). This publication does not represent the views of the Department of Veteran Affairs or the US Government. Acknowledgment of the Million Veteran Program leadership and staff contributions can be found in the supplementary material entitled “MVP core acknowledgements.”
Author Contributions
Study design: AMH and EAA. Data acquisition: AMH and CMS. Data analysis and interpretation: EAA, HC, GL, ZH, AG, QF, RT, AMH. Drafting of the manuscript: EAA. Critical revision of the manuscript for intellectual content: HC, CPC, AG, CR, TAI, CMS, EDS, QF, RT, AMH. All authors approve the final version of the manuscript.
Footnotes
Figure S1. Linkage disequilibrium patterns between UMOD promoter and PDILT variants among individuals of European ancestry (CEU) in the 1000 Genomes reference panel.
Figure S2. Linkage disequilibrium patterns between UMOD promoter and PDILT variants among African Americans (ASW) in the 1000 Genomes reference panel.
Figure S3. Linkage disequilibrium patterns between UMOD promoter and PDILT variants among individuals of Mexican ancestry (MXL) in the 1000 Genomes reference panel.
Figure S4. Linkage disequilibrium patterns between UMOD promoter and PDILT variants among individuals of Han Chinese ancestry (CHB) in the 1000 Genomes reference panel.
Figure S5. Volcano plot showing clinical phenotypes significantly associated with the rs4293393 UMOD promoter and rs77924614 PDILT variants in non-Hispanic Asian and Hispanic patients in the Million Veteran Program.
Figure S6. Volcano plot illustrating key clinical phenotypes significantly associated with the rs12917707 UMOD promoter in the 4 populations (non-Hispanic White, non-Hispanic Black, non-Hispanic Asian, and Hispanic patients) in the Million Veteran Program.
Table S1. List of Phecodes tested in MVP and BioVU participants.
Table S2. Imputation quality scores (R2) for UMOD/PDILT variants in MVP.
Table S3. List of Phecodes that were tested in MVP but not in the UK Biobank.
Table S4. Imputation quality scores (R2) for UMOD/PDILT variants in BioVU and UK Biobank participants.
Table S5. Characteristics of BioVU (Vanderbilt’s Biobank) participants that were included in the UMOD PheWAS.
Table S6. Baseline characteristics of UK Biobank participants (n = 502,650).
Table S7. Allele frequencies of UMOD and PDILT variants across population groups in the Million Veteran Program.
Table S8. Association of the rs12917707 (G/T) UMOD promoter variant with clinical phenotypes that were significant in the primary analysis using the rs4293393 (A/G) UMOD promoter variant in non-Hispanic White patients in the MVP.
Table S9. Association of the rs12922822 (C/T) UMOD promoter variant with clinical phenotypes that were significant in the primary analysis using the rs4293393 (A/G) UMOD promoter variant in non-Hispanic White patients in the MVP.
Table S10. Association of the rs13333226 (A/G) UMOD promoter variant with clinical phenotypes that were significant in the primary analysis using the rs4293393 (A/G) UMOD promoter variant in non-Hispanic White patients in the MVP.
Table S11. Association of the rs13329952 (T/C) UMOD promoter variant with clinical phenotypes that were significant in the primary analysis using the rs4293393 (A/G) UMOD promoter variant in non-Hispanic White patients in the MVP.
Table S12. Association of the rs12446492 (T/A) PDILT variant with clinical phenotypes that were significant in the primary analysis using the rs77924615 (G/A) PDILT variant in non-Hispanic White patients in the MVP.
Table S13. Association of the rs11864909 (C/T) PDILT variant with clinical phenotypes that were significant in the primary analysis using the rs77924615 (G/A) PDILT variant in non-Hispanic White patients in the MVP.
Table S14. Effect estimates in BioVU participants for the association of the rs4293393 (A/G) UMOD promoter variant with clinical phenotypes that were significant in MVP non-Hispanic White patients.
Table S15. Effect estimates in UK Biobank participants for the association of the rs4293393 (A/G) UMOD promoter variant with clinical phenotypes that were significant in MVP non-Hispanic White patients.
Table S16.P values for the test of differential effects of the rs4293393 (A/G) UMOD variant on key clinical phenotypes in non-Hispanic White versus Non-Hispanic Black patients in the Million Veteran Program.
Table S17. Association of the rs4293393 (A/G) UMOD variant with UTIs across population groups in the Million Veteran Program.
Table S18. Association of the rs77924615 (G/A) PDILT Variant with UTIs across population groups in the Million Veteran Program.
Table S19. Association of the rs77924615 (G/A) PDILT variant with key clinical phenotypes across sex groups in the Million Veteran Program.
Table S20. Power estimates for the association between the rs4293393 UMOD variant and key clinical phenotypes in non-Hispanic White and Black patients.
Table S21. Power estimates for the association between the UMOD/PDILT variants and key clinical phenotypes in Hispanic patients.
Supplementary VA Million Veteran Program: Core Acknowledgment for Publication.
STROBE Statement.
Supplementary Material
Figure S1. Linkage disequilibrium patterns between UMOD promoter and PDILT variants among individuals of European ancestry (CEU) in the 1000 Genomes reference panel.
Figure S2. Linkage disequilibrium patterns between UMOD promoter and PDILT variants among African Americans (ASW) in the 1000 Genomes reference panel.
Figure S3. Linkage disequilibrium patterns between UMOD promoter and PDILT variants among individuals of Mexican ancestry (MXL) in the 1000 Genomes reference panel.
Figure S4. Linkage disequilibrium patterns between UMOD promoter and PDILT variants among individuals of Han Chinese ancestry (CHB) in the 1000 Genomes reference panel.
Figure S5. Volcano plot illustrating clinical phenotypes significantly associated with the rs4293393 UMOD promoter and rs77924614 PDILT variants in non-Hispanic Asian and Hispanic patients in the Million Veteran Program.
Figure S6. Volcano plot illustrating key clinical phenotypes significantly associated with the rs12917707 UMOD promoter in the 4 populations (non-Hispanic White, non-Hispanic Black, non-Hispanic Asian, and Hispanic patients) in the Million Veteran Program.
Table S1. List of Phecodes tested in MVP and BioVU participants.
Table S2. Imputation quality scores (R2) for UMOD/PDILT variants in MVP.
Table S3. List of Phecodes that were tested in MVP but not in the UK Biobank.
Table S4. Imputation quality scores (R2) for UMOD/PDILT variants in BioVU and UK Biobank participants.
Table S5. Characteristics of BioVU (Vanderbilt’s Biobank) participants that were included in the UMOD PheWAS.
Table S6. Baseline characteristics of UK Biobank participants (n = 502,650).
Table S7. Allele frequencies of UMOD and PDILT variants across population groups in the Million Veteran Program.
Table S8. Association of the rs12917707 (G/T) UMOD promoter variant with clinical phenotypes that were significant in the primary analysis using the rs4293393 (A/G) UMOD promoter variant in non-Hispanic White patients in the MVP.
Table S9. Association of the rs12922822 (C/T) UMOD promoter variant with clinical phenotypes that were significant in the primary analysis using the rs4293393 (A/G) UMOD promoter variant in non-Hispanic White patients in the MVP.
Table S10. Association of the rs13333226 (A/G) UMOD promoter variant with clinical phenotypes that were significant in the primary analysis using the rs4293393 (A/G) UMOD promoter variant in non-Hispanic White patients in the MVP.
Table S11. Association of the rs13329952 (T/C) UMOD promoter variant with clinical phenotypes that were significant in the primary analysis using the rs4293393 (A/G) UMOD promoter variant in non-Hispanic White patients in the MVP.
Table S12. Association of the rs12446492 (T/A) PDILT variant with clinical phenotypes that were significant in the primary analysis using the rs77924615 (G/A) PDILT variant in non-Hispanic White patients in the MVP.
Table S13. Association of the rs11864909 (C/T) PDILT variant with clinical phenotypes that were significant in the primary analysis using the rs77924615 (G/A) PDILT variant in non-Hispanic White patients in the MVP.
Table S14. Effect estimates in BioVU participants for the association of the rs4293393 (A/G) UMOD promoter variant with clinical phenotypes that were significant in MVP non-Hispanic White patients.
Table S15. Effect estimates in UK Biobank participants for the association of the rs4293393 (A/G) UMOD promoter variant with clinical phenotypes that were significant in MVP non-Hispanic White patients.
Table S16.P values for the test of differential effects of the rs4293393 (A/G) UMOD variant on key clinical phenotypes in non-Hispanic White versus non-Hispanic Black patients in the Million Veteran Program.
Table S17. Association of the rs4293393 (A/G) UMOD variant with UTIs across population groups in the Million Veteran Program.
Table S18. Association of the rs77924615 (G/A) PDILT variant with UTIs across population groups in the Million Veteran Program.
Table S19. Association of the rs77924615 (G/A) PDILT variant with key clinical phenotypes across sex groups in the Million Veteran Program.
Table S20. Power estimates for the association between the rs4293393 UMOD variant and key clinical phenotypes in non-Hispanic White and Black patients.
Table S21. Power estimates for the association between the UMOD/PDILT variants and key clinical phenotypes in Hispanic patients.
References
- 1.Mount D.B. Thick ascending limb of the loop of Henle. Clin J Am Soc Nephrol. 2014;9:1974–1986. doi: 10.2215/CJN.04480413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rampoldi L., Scolari F., Amoroso A., et al. The rediscovery of uromodulin (Tamm-Horsfall protein): from tubulointerstitial nephropathy to chronic kidney disease. Kidney Int. 2011;80:338–347. doi: 10.1038/ki.2011.134. [DOI] [PubMed] [Google Scholar]
- 3.Tamm I., Horsfall F.L., Jr. Characterization and separation of an inhibitor of viral hemagglutination present in urine. Proc Soc Exp Biol Med. 1950;74:106–108. [PubMed] [Google Scholar]
- 4.Bachmann S., Dawnay A.B., Bouby N., Bankir L. Tamm-Horsfall protein excretion during chronic alterations in urinary concentration and protein intake in the rat. Ren Physiol Biochem. 1991;14:236–245. doi: 10.1159/000173411. [DOI] [PubMed] [Google Scholar]
- 5.Goodall A.A., Marshall R.D. Effects of freezing on the estimated amounts of Tamm-Horsfall glycoprotein in urine, as determined by radioimmunoassay. Biochem J. 1980;189:533–539. doi: 10.1042/bj1890533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rindler M.J., Naik S.S., Li N., et al. Uromodulin (Tamm-Horsfall glycoprotein/uromucoid) is a phosphatidylinositol-linked membrane protein. J Biol Chem. 1990;265:20784–20789. doi: 10.1016/S0021-9258(17)45284-7. [DOI] [PubMed] [Google Scholar]
- 7.Serafini-Cessi F., Malagolini N., Cavallone D. Tamm-Horsfall glycoprotein: biology and clinical relevance. Am J Kidney Dis. 2003;42:658–676. doi: 10.1016/s0272-6386(03)00829-1. [DOI] [PubMed] [Google Scholar]
- 8.Devuyst O., Dahan K., Pirson Y. Tamm-Horsfall protein or uromodulin: new ideas about an old molecule. Nephrol Dial Transplant. 2005;20:1290–1294. doi: 10.1093/ndt/gfh851. [DOI] [PubMed] [Google Scholar]
- 9.Mutig K., Kahl T., Saritas T., et al. Activation of the bumetanide-sensitive Na+,K+,2Cl− cotransporter (NKCC2) is facilitated by Tamm-Horsfall protein in a chloride-sensitive manner. J Biol Chem. 2011;286:30200–30210. doi: 10.1074/jbc.M111.222968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renigunta A., Renigunta V., Saritas T., et al. Tamm-Horsfall glycoprotein interacts with renal outer medullary potassium channel ROMK2 and regulates its function. J Biol Chem. 2011;286:2224–2235. doi: 10.1074/jbc.M110.149880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pak J., Pu Y., Zhang Z.T., et al. Tamm-Horsfall protein binds to type 1 fimbriated Escherichia coli and prevents E. coli from binding to uroplakin Ia and Ib receptors. J Biol Chem. 2001;276:9924–9930. doi: 10.1074/jbc.M008610200. [DOI] [PubMed] [Google Scholar]
- 12.Mo L., Zhu X.H., Huang H.Y., et al. Ablation of the Tamm-Horsfall protein gene increases susceptibility of mice to bladder colonization by type 1-fimbriated Escherichia coli. Am J Physiol Ren Physiol. 2004;286:F795–F802. doi: 10.1152/ajprenal.00357.2003. [DOI] [PubMed] [Google Scholar]
- 13.Ghirotto S., Tassi F., Barbujani G., et al. The uromodulin gene locus shows evidence of pathogen adaptation through human evolution. J Am Soc Nephrol. 2016;27:2983–2996. doi: 10.1681/ASN.2015070830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mo L., Huang H.Y., Zhu X.H., et al. Tamm-Horsfall protein is a critical renal defense factor protecting against calcium oxalate crystal formation. Kidney Int. 2004;66:1159–1166. doi: 10.1111/j.1523-1755.2004.00867.x. [DOI] [PubMed] [Google Scholar]
- 15.Saemann M.D., Weichhart T., Zeyda M., et al. Tamm-Horsfall glycoprotein links innate immune cell activation with adaptive immunity via a Toll-like receptor-4-dependent mechanism. J Clin Invest. 2005;115:468–475. doi: 10.1172/JCI22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Achkar T.M., Wu X.R., Rauchman M., et al. Tamm-Horsfall protein protects the kidney from ischemic injury by decreasing inflammation and altering TLR4 expression. Am J Physiol Ren Physiol. 2008;295:F534–F544. doi: 10.1152/ajprenal.00083.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Köttgen A., Glazer N.L., Dehghan A., et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet. 2009;41:712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pattaro C., Teumer A., Gorski M., et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun. 2016;7:10023. doi: 10.1038/ncomms10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devuyst O., Pattaro C. The UMOD locus: insights into the pathogenesis and prognosis of kidney disease. J Am Soc Nephrol. 2018;29:713–726. doi: 10.1681/ASN.2017070716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wuttke M., Kottgen A. Insights into kidney diseases from genome-wide association studies. Nat Rev Nephrol. 2016;12:549–562. doi: 10.1038/nrneph.2016.107. [DOI] [PubMed] [Google Scholar]
- 21.Wuttke M., Li Y., Li M., et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet. 2019;51:957–972. doi: 10.1038/s41588-019-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padmanabhan S., Melander O., Johnson T., et al. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLOS Genet. 2010;6 doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olden M., Corre T., Hayward C., et al. Common variants in UMOD associate with urinary uromodulin levels: a meta-analysis. J Am Soc Nephrol. 2014;25:1869–1882. doi: 10.1681/ASN.2013070781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trudu M., Janas S., Lanzani C., et al. Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med. 2013;19:1655–1660. doi: 10.1038/nm.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang H., Hui Q., Lynch J., et al. Harmonizing genetic ancestry and self-identified race/ethnicity in genome-wide association studies. Am J Hum Genet. 2019;105:763–772. doi: 10.1016/j.ajhg.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denny J.C., Bastarache L., Ritchie M.D., et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–1110. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pendergrass S.A., Brown-Gentry K., Dudek S., et al. Phenome-wide association study (PheWAS) for detection of pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) network. PLOS Genet. 2013;9 doi: 10.1371/journal.pgen.1003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bush W.S., Oetjens M.T., Crawford D.C. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat Rev Genet. 2016;17:129–145. doi: 10.1038/nrg.2015.36. [DOI] [PubMed] [Google Scholar]
- 29.Rastegar-Mojarad M., Ye Z., Kolesar J.M., et al. Opportunities for drug repositioning from phenome-wide association studies. Nat Biotechnol. 2015;33:342–345. doi: 10.1038/nbt.3183. [DOI] [PubMed] [Google Scholar]
- 30.Gaziano J.M., Concato J., Brophy M., et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 31.Bick A.G., Akwo E., Robinson-Cohen C., et al. Association of APOL1 risk alleles with cardiovascular disease in Blacks in the Million Veteran Program. Circulation. 2019;140:1031–1040. doi: 10.1161/CIRCULATIONAHA.118.036589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giri A., Hellwege J.N., Keaton J.M., et al. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat Genet. 2019;51:51–62. doi: 10.1038/s41588-018-0303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunter-Zinck H, Shi Y, Li M, et al. Measuring genetic variation in the multi-ethnic Million Veteran Program (MVP). bioRxiv. Published online January 7, 2020. https://doi.org/10.1101/2020.01.06.896613
- 34.Manichaikul A., Mychaleckyj J.C., Rich S.S., et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auton A., Brooks L.D., Durbin R.M., et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loh P.R., Danecek P., Palamara P.F., et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Consortium G.P., Abecasis G.R., Altshuler D., et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das S., Forer L., Schonherr S., et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abraham G., Inouye M. Fast principal component analysis of large-scale genome-wide data. PLoS One. 2014;9:e93766. doi: 10.1371/journal.pone.0093766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winkler T.W., Day F.R., Croteau-Chonka D.C., et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9:1192–1212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klarin D., Damrauer S.M., Cho K., et al. Genetics of blood lipids among ∼300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet. 2018;50:1514–1523. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hyde C.L., Nagle M.W., Tian C., et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016;48:1031–1036. doi: 10.1038/ng.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.1000 Genomes Project Consortium. Auton A., Brooks L.D., et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Devuyst O., Olinger E., Rampoldi L. Uromodulin: from physiology to rare and complex kidney disorders. Nat Rev Nephrol. 2017;13:525–544. doi: 10.1038/nrneph.2017.101. [DOI] [PubMed] [Google Scholar]
- 45.Cai T., Zhang Y., Ho Y.-L., et al. Association of interleukin 6 receptor variant with cardiovascular disease effects of interleukin 6 receptor blocking therapy: a phenome-wide association study. JAMA Cardiol. 2018;3:849–857. doi: 10.1001/jamacardio.2018.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denny J.C., Ritchie M.D., Basford M.A., et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.King G., Zeng L. Logistic regression in rare events data. Pol Anal. 2001;9:137–163. doi: 10.1093/oxfordjournals.pan.a004868. [DOI] [Google Scholar]
- 48.Buckley J.P., Doherty B.T., Keil A.P., Engel S.M. Statistical approaches for estimating sex-specific effects in endocrine disruptors research. Environ Health Perspect. 2017;125 doi: 10.1289/EHP334. 067013-067013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paternoster R., Brame R., Mazerolle P., Piquero A. Using the correct statistical test for the equality of regression coefficients. Criminology (Beverly Hills) 1998;36:859–866. doi: 10.1111/j.1745-9125.1998.tb01268.x. [DOI] [Google Scholar]
- 50.Roden D.M., Pulley J.M., Basford M.A., et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sudlow C., Gallacher J., Allen N., et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLOS Med. 2015;12 doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sveinbjornsson G., Mikaelsdottir E., Palsson R., et al. Rare mutations associating with serum creatinine and chronic kidney disease. Hum Mol Genet. 2014;23:6935–6943. doi: 10.1093/hmg/ddu399. [DOI] [PubMed] [Google Scholar]
- 53.Mahajan A., Rodan A.R., Le T.H., et al. Trans-ethnic fine mapping highlights kidney-function genes linked to salt sensitivity. Am J Hum Genet. 2016;99:636–646. doi: 10.1016/j.ajhg.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shang N., Khan A., Polubriaginof F., et al. Medical records-based chronic kidney disease phenotype for clinical care and “big data” observational and genetic studies. NPJ Digit Med. 2021;4:70. doi: 10.1038/s41746-021-00428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu C.T., Garnaas M.K., Tin A., et al. Genetic association for renal traits among participants of African ancestry reveals new loci for renal function. PLOS Genet. 2011;7 doi: 10.1371/journal.pgen.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lewis E.J., Hunsicker L.G., Clarke W.R., et al. Renoprotective effect of the angiotensin-receptor antagonist irbesartan in patients with nephropathy due to type 2 diabetes. N Engl J Med. 2001;345:851–860. doi: 10.1056/NEJMoa011303. [DOI] [PubMed] [Google Scholar]
- 57.Brenner B.M., Cooper M.E., de Zeeuw D., et al. Effects of losartan on renal and cardiovascular outcomes in patients with type 2 diabetes and nephropathy. N Engl J Med. 2001;345:861–869. doi: 10.1056/NEJMoa011161. [DOI] [PubMed] [Google Scholar]
- 58.Perkovic V., Jardine M.J., Neal B., et al. Canagliflozin and renal outcomes in type 2 diabetes and nephropathy. N Engl J Med. 2019;380:2295–2306. doi: 10.1056/NEJMoa1811744. [DOI] [PubMed] [Google Scholar]
- 59.Heerspink H.J.L., Stefánsson B.V., Correa-Rotter R., et al. Dapagliflozin in patients with chronic kidney disease. N Engl J Med. 2020;383:1436–1446. doi: 10.1056/NEJMoa2024816. [DOI] [PubMed] [Google Scholar]
- 60.Wanner C., Inzucchi S.E., Lachin J.M., et al. Empagliflozin and progression of kidney disease in type 2 diabetes. N Engl J Med. 2016;375:323–334. doi: 10.1056/NEJMoa1515920. [DOI] [PubMed] [Google Scholar]
- 61.Neal B., Perkovic V., Mahaffey K.W., et al. Canagliflozin and cardiovascular and renal events in type 2 diabetes. N Engl J Med. 2017;377:644–657. doi: 10.1056/NEJMoa1611925. [DOI] [PubMed] [Google Scholar]
- 62.Perkovic V., de Zeeuw D., Mahaffey K.W., et al. Canagliflozin and renal outcomes in type 2 diabetes: results from the CANVAS Program randomised clinical trials. Lancet Diabetes Endocrinol. 2018;6:691–704. doi: 10.1016/S2213-8587(18)30141-4. [DOI] [PubMed] [Google Scholar]
- 63.Bakris G.L., Agarwal R., Anker S.D., et al. Effect of finerenone on chronic kidney disease outcomes in type 2 diabetes. N Engl J Med. 2020;383:2219–2229. doi: 10.1056/NEJMoa2025845. [DOI] [PubMed] [Google Scholar]
- 64.Hellwege J.N., Velez Edwards D.R., Giri A., et al. Mapping eGFR loci to the renal transcriptome and phenome in the VA Million Veteran Program. Nat Commun. 2019;10:3842. doi: 10.1038/s41467-019-11704-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Padmanabhan S., Graham L., Ferreri N.R., et al. Uromodulin, an emerging novel pathway for blood pressure regulation and hypertension. Hypertens (Dallas: Tex. 2014;64:918–923. doi: 10.1161/HYPERTENSIONAHA.114.03132. [DOI] [PubMed] [Google Scholar]
- 66.Yoshida T., Kurella M., Beato F., et al. Monitoring changes in gene expression in renal ischemia-reperfusion in the rat. Kidney Int. 2002;61:1646–1654. doi: 10.1046/j.1523-1755.2002.00341.x. [DOI] [PubMed] [Google Scholar]
- 67.El-Achkar T.M., McCracken R., Liu Y., et al. Tamm-Horsfall protein translocates to the basolateral domain of thick ascending limbs, interstitium, and circulation during recovery from acute kidney injury. Am J Physiol Ren Physiol. 2013;304:F1066–F1075. doi: 10.1152/ajprenal.00543.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parkkinen J., Virkola R., Korhonen T.K. Identification of factors in human urine that inhibit the binding of Escherichia coli adhesins. Infect Immun. 1988;56:2623–2630. doi: 10.1128/iai.56.10.2623-2630.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Conover M.S., Ruer S., Taganna J., et al. Inflammation-induced adhesin-receptor interaction provides a fitness advantage to uropathogenic E. coli during chronic infection. Cell Host Microbe. 2016;20:482–492. doi: 10.1016/j.chom.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y., Mo L., Goldfarb D.S., et al. Progressive renal papillary calcification and ureteral stone formation in mice deficient for Tamm-Horsfall protein. Am J Physiol Ren Physiol. 2010;299:F469–F478. doi: 10.1152/ajprenal.00243.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coe F.L., Evan A., Worcester E. Pathophysiology-based treatment of idiopathic calcium kidney stones. Clin J Am Soc Nephrol. 2011;6:2083–2092. doi: 10.2215/CJN.11321210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Serafini-Cessi F., Monti A., Cavallone D. N-Glycans carried by Tamm-Horsfall glycoprotein have a crucial role in the defense against urinary tract diseases. Glycoconj J. 2005;22:383–394. doi: 10.1007/s10719-005-2142-z. [DOI] [PubMed] [Google Scholar]
- 73.Weiss G.L., Stanisich J.J., Sauer M.M., et al. Architecture and function of human uromodulin filaments in urinary tract infections. Science. 2020;369:1005–1010. doi: 10.1126/science.aaz9866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.