Abstract
Platelets buoy up cancer metastasis via arresting cancer cells, enhancing their adhesion, and facilitating their extravasation through the vasculature. When deprived of intracellular and granular contents, platelet decoys could prevent metastatic tumor formation. Inspired by these, we developed nanoplatesomes by fusing platelet membranes with lipid membranes (P-Lipo) to restrain metastatic tumor formation more efficiently. It was shown nanoplateletsomes bound with circulating tumor cells (CTC) efficiently, interfered with CTC arrest by vessel endothelial cells, CTC extravasation through endothelial layers, and epithelial-mesenchymal transition of tumor cells as nanodecoys. More importantly, in the mouse breast tumor metastasis model, nanoplateletsomes could decrease CTC survival in the blood and counteract metastatic tumor growth efficiently by inhibiting the inflammation and suppressing CTC escape. Therefore, nanoplatelesomes might usher in a new avenue to suppress lung metastasis.
KEY WORDS: Nanoplateletsome, Breast cancer, Tumor metastasis, Circulating tumor cells, Nanodecoy, Immunosuppressive micrometastatic environment, Lung pre-metastasis
Graphical abstract
Nanoplateletsomes were inaugurated to restrain metastatic tumor formation efficiently through decoy and active targeting, serving as a platform to orchestrate the inhibition of circulating tumor cell survival and metastasis.
1. Introduction
Though surgical excision and adjuvant therapy can cure well-confined primary tumors, metastatic disease is almost remediless owing to its systemic nature and the resistance of disseminated tumor cells to existing therapeutic agents, accounting for over 90% of cancer-associated mortalities1. The progression of much fatal metastasis is spawned by the vascular dissemination of tumor cells, which are also termed as circulating tumor cells (CTCs) and act as harbingers of this process2,3. CTCs that have been shed into the vasculature may contribute to the aggressive metastasis and decreased time of relapse, generating micro-metastasis with markedly different biology from that of the primary tumor. However, compared with the primary tumor, the pathophysiological characteristics of metastatic tumors are less informative and the corresponding treatment strategies are less available due to the heterogeneity within the CTCs. In this context, there is an urgent demand to propose specific preclinical or clinical strategies targeting the early treatment of metastasis in real time4.
To date, CTCs have been explored as predictive biomarkers for early prognosis, recurrence risk assessment, and therapeutic monitoring of metastasis5, 6, 7. Although several immunocytological technologies, molecular technologies, along with functional assays, have been proposed for CTC detection to indicate the effectiveness of cancer treatment8,9, limited studies based on CTC elimination or inactivation in vivo at the pre-metastasis stage were reported, which is an overarching challenge to be resolved. Hence, it is of great significance to eliminate CTCs or blunt their metastatic ability, yielding a lower possibility of tumor recurrence and metastasis10,11.
Platelets play a profound role in the whole tumor metastasis cascade, including the detachment and dissemination of tumor cells into the blood, the facilitation of the epithelial-to-mesenchymal transition, the generation of the CTC cluster, along with the subsequent metastatic niche12, 13, 14. Equipped with a plethora of adhesion receptors, when cancer cells detach and invade into the bloodstream as CTCs, platelets can be activated and attach to cancer cells to generate a protective cloak that helps cancer cells to escape immune surveillance and high shear stress from the bloodstream15,16, seed in distant organs and provides aid to metastatic lesion growth. Platelet membranes express many components that regulate cancer cell adhesion, immune evasion and metastasis, such as P-selectin, PECAM-1, GPIIb/IIIa, GPIb/IX, TLR4, etc17,18. Therefore, drugs or nanomedicines interrupting platelet-CTC interactions may weaken the metastatic ability of CTCs and prevent the formation of tumor metastasis11,19.
Recently, cell membrane-bioinspired nanotherapeutics based on cell‒cell interaction have been extensively developed and harnessed for various biomedical applications20, 21, 22, 23, 24, 25, 26, such as red blood cell-derived nanoparticles for immune evasion and antigen delivery27 or detoxication28, leukocyte-derived nanovesicles for anti-inflammatory29 or tumor-targeted therapy30, and platelet membrane-coated nanoparticles for targeted antitumor drug delivery31, 32, 33. Cell membrane-bioinspired nanotherapeutics essentially mimic the properties of the source cells from which their membrane originates, conferring a wide range of functions, such as long circulation and disease-targeting. To this end, these biomimetic nanotherapeutics have opened up new avenues to overcome the limitations of current nanotherapeutics, enhance their targeting capability without inducing immune responses. Inspired by the intrinsic properties of platelets and their interactions with CTCs, platelet membrane-based biomimetic nanotechnology has been explored for CTC detection34,35 and tumor metastasis treatment36, 37, 38. For instance, platelet membrane-cloaked microchips34 and platelet-leukocyte hybrid membrane-coated magnetic cores39 have been employed for CTC separation and detection. In general, two biomimetic strategies have been explored to restrain the metastatic formation or treat tumor metastasis. One strategy is eliminating CTCs by using platelet membrane-coated nanovehicles or microparticles loaded with drugs such as TRAIL36,37, doxorubicin (Dox)22. These biomimetic drug-loaded nanovehicles could actively target CTCs, kill them and inhibit metastatic tumor formation. The other strategy is platelet decoy strategy by disrupting the interactions between CTCs and platelets. Recently, Ingber's group19 has employed platelet decoys by denuding the platelet innards to interfere with platelet-mediated CTC aggregation and inhibit metastatic tumor growth after coinjection with tumor cells and platelets. Nevertheless, the anti-metastasis efficacy is moderate and a large amount of platelet decoys have to be coinjected with platelets to retard metastatic formation. In addition, the preparation of platelet decoys without activating platelets is challenging. Moreover, the stripped-down platelets are plagued by the limited half-time, thus decreasing their usefulness in the patients. Therefore, biomimetic decoys with high anti-metastasis efficacy, long circulation, fewer platelet resources, and easy production process are urgently needed to blunt CTCs for metastasis treatment.
In this work, we leveraged this platelet-CTC interaction to constitute nanoplateletsomes to actively target CTCs and treat metastasis as nano-platelet decoys. The nanoplateletsomes were developed by fusing natural platelet membranes with synthetic lipid membranes (P-Lipo) through extrusion (Scheme 1) and taking advantage of both biomaterials. We hypothesized that P-Lipo could provide functional cell surface receptors, which retained their binding capabilities with CTCs or pre-metastasis but were deprived of their intrinsic functional abilities, such as aggregation and activation, to facilitate CTC colonization and extravasation. Featuring the large surface-volume ratio and large quantity, nanoplateletsomes would greatly encase CTCs while not offering a shield against the immune cells. The subversion of the host immune responses was prevented, leading to a remarkable inhibition effect toward the generation of premetastatic tumor cascade. The feasibility of our bioinspired nano-design to achieve enhanced CTC binding and micrometastasis targeting, and restrain CTCs to metastasize in distant tissues as nano-platelet decoys was validated. The anti-metastasis efficacy and underlying mechanism of nanoplatesomes were investigated in a preclinical metastasis mouse model.
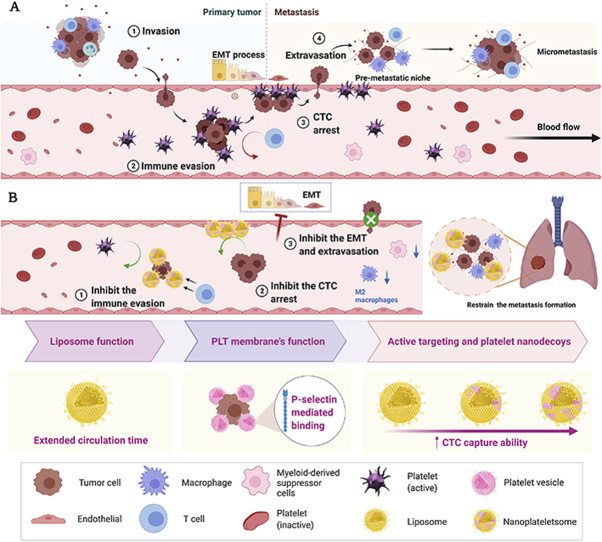
Scheme 1.
Schematic illustration of the role of platelet in the whole tumor metastasis cascade. (A) Platelets-promoted tumor metastasis cascade. Platelets can be activated and attach to cancer cells to generate a protective cloak that helps cancer cells to escape immune surveillance and high shear stress from the bloodstream. Additionally, activated platelets could help circulating tumor cells (CTC) arrest by vessel endothelial cells, and CTC extravasation through blood vessels, and provide aid to metastatic lesion growth. (B) Nanoplateletsomes (P-Lipo) inhibit metastasis formation through active targeting and act as a decoy to inhibit the function of platelets. The designed P-Lipo could bind with CTCs, interfere with CTC arrest by vessel endothelial cells, CTC extravasation through endothelial layers, and epithelial-mesenchymal transition of tumor cells. More importantly, P-Lipo could decrease CTC survival in the blood and counteract metastatic tumor growth efficiently by inhibiting the inflammation and suppressing CTC escape.
2. Materials and methods
2.1. Materials
1,2-Distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (DSPE-PEG2000) and egg yolk lecithin (EPC, PL-100M) were purchased from AVT Pharmaceutical Tech Co., Ltd. (China). Prostaglandin E1 (PGE1) was purchased from MedChemExpress (USA). Protease inhibitor mini-tablets, Dulbecco's modified Eagle medium (DMEM), Roswell Park Memorial Institute (RPMI) 1640 medium, and penicillin–streptomycin (10,000 U/mL) were purchased from Thermo Fisher (USA). CD41/CD61 antibody (362802) was from BioLegend (USA). GPVI antibody (STJ192229) was from StJohnsLabs (UK). CD29 antibody (UUY0319031), CD49b antibody (CBDM011909A), and CD42b antibody (CANBO41909A) were from BD Biosciences (USA). CD62P antibody (AB6631) and CD63 antibody (ab134045) were from Abcam (UK). Fluorescence dyes 3,3′-dioctadecy-loxacarbocyanine perchlorate salt (DiO), 1,1′-dioctadecyl-3,3,3′,3′-tetra-methylindodicarbocyanine, 4-chlorobenzenesulfonate salt (DiD), 4ʹ,6-diamidino-2-phenylindole (DAPI) were purchased from Meilunbio (China), and CFSE was from eBioscience (USA). Coumarin 6 and Rhodamine B were purchased from Sigma–Aldrich (USA). Rhodamine-conjugated dextran was purchased from Shanxi Xinyan Bomei Biotechnology Co., Ltd. (China). Omni-Easy™ Instant BCA protein assay kit was from EpiZyme (USA). EDTA, cholesterol, chloroform, and ethanol were purchased from Sinopharm (China).
2.2. Cells and animals
The 4T1 murine breast cancer cell line was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Human umbilical vein endothelial cell (HUVEC), mouse 4T1 breast tumor cells, and 4T1 cells expressing green fluorescent protein (4T1-GFP) or luciferase (4T1-Luc) were bought from American Type Culture Collection (ATCC, USA). 4T1 and HUVEC cells were cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS), streptomycin (100 U/mL), and penicillin (100 U/mL) at 37 °C in 95% air/5% CO2. 4T1-Luc and 4T1-GFP cell lines were cultured in Roswell Park Memorial Institute RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), HEPES (25 mmol/L), streptomycin (100 U/mL), penicillin (100 U/mL) at 37 °C in 95% air/5% CO2.
Female BALB/c mice aged 6 weeks were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. and housed under specific-pathogen free conditions. All animal protocols were approved by the Animal Ethics Committee of Fudan University (Shanghai, China).
2.3. P-Lipo preparation
The platelet membrane was derived by a repeated freeze-thawing method as previously described40,41. Briefly, the whole blood collected from healthy BALB/c mice was centrifuged at 100 × g for 20 min (Tx-400, Thermo, USA) to separate platelet-rich plasma (PRP) from red blood cells and white blood cells. Afterward, PBS buffer containing 1 mmol/L EDTA and 2 mmol/L PGE1 was added to the PRP to prevent platelet activation during the extraction process. Platelets were obtained by centrifuging 1 mL of PRP at 800 × g for 20 min (Thermo) and resuspended in 1 mL of PBS containing 1 mmol/L of EDTA and mixed with protease inhibitor mini-tablets. Platelet membranes were derived by a repeated freeze-thaw process three times, collected by centrifugation at 10,000 × g for 10 min (Thermo), were suspended in 1 mL of water (1 U) for spare.
P-Lipo were prepared by the thin-film hydration method as described previously with minor modifications42, 43, 44. Briefly, 4 mg of EPC, 0.4 mg of DSPE-PEG2000, and 0.8 mg of cholesterol were dissolved in 5 mL of chloroform/ethanol mixture (1:1, v/v). The organic solvent was removed through a rotary evaporator at 45 °C for 60 min to form a dry, thin film. The film was hydrated with 0.4 U (PL-Lipo) or 1.6 U (PH-Lipo) of platelet membranes dispersed in 2 mL of PBS buffer. The suspensions were frozen and thawed for ten cycles. Afterward, the suspension was successively extruded through 800, 400, and 200 nm polycarbonate filters (Nuclepore™, Whatman, US) repeatedly by using a Liposome Extruder (HandExtruder™, Genizer, USA). Thus PL-Lipo (0.1 U of platelet membranes per mg of lipid membranes) and PH-Lipo (0.4 U of platelet membranes per mg of lipid membranes) were obtained. Liposomes were prepared as a control with the same method except that no platelet membranes were input.
2.4. Characterization of P-Lipo
Dynamic light scattering (DLS) was used to determine the particle size and zeta potential of P-Lipo using a Zetasizer Nano ZS (Malvern, UK). The morphology of P-Lipo was visualized using a transmission electron microscope (TEM, Tecna G2S-Twin, FEI, USA) after staining with 2% uranyl acetate and cryogenic transmission electron microscopy (cyro-EM, 200 KV, FEI, USA) as previously described45,46.
2.5. Membrane fusion assay
To test the fusion of platelet membranes and artificial lipid membranes, a Förster resonance energy transfer (FRET) pair lipophilic dyes Coumarin 6 (C6, Ex/Em 460/510 nm) and Rhodamine B (RhB, Ex/Em 565/590 nm) were employed. Fluorescence C6-labeled lipid membranes were prepared according to previously reported47. Briefly, EPC, DSPE-PEG, and cholesterol in chloroform/ethanol were spiked with 5.2 μg of C6 before evaporation to form a lipid film. Platelet membranes were labeled with RhB by incubation with 2 mg/mL of RhB for 3 min, washed twice to remove free dyes, and then resuspended in PBS buffer. C6-stained lipid membranes (2 mg) were added to 0, 0.25, 0.5, 0.75, 1 and 1.25 U of RhB-labeled platelet membranes, respectively, followed by hydration, freeze-thaw process, and extruding through 800, 400, and 200 nm polycarbonate filters to facilitate the membrane fusion. The fluorescence emission spectrum of each sample was recorded from 450 to 700 nm at an excitation wavelength of 430 nm on a Cary Eclipse fluorescence spectrophotometer (Agilent, USA). FRET efficiency, defined as the proportion of the donor C6 that has transferred their excess energy to the acceptor RhB, was calculated using the following equation as previously described as Eq. (1),48:
| (1) |
where FDA and FD were the fluorescence intensities of the donor C6 in the presence and absence of the acceptor RhB, respectively.
To visualize the membrane fusion more vividly, the colocalization of platelet membranes and lipid membranes was investigated at the single nanoparticle level and cellular levels, respectively. Two lipophilic dyes, DiO (Ex/Em 488/501 nm) and DiD (Ex/Em 643/665 nm) were employed to label platelet membranes and lipid membranes, respectively. Platelet membranes were labeled with 5 μmol/L of DiO according to the manufacturer's protocol. The DiD-labeled lipid membranes were prepared as described above. The double-stained P-Lipo was prepared and visualized under a super-resolution microscope system (N-SIM, Nikon, Japan) with a 100 × oil objective. To obtain stable images, the samples were immobilized in 2% agarose to decrease their mobility. The mixture of DiD-labeled lipid membranes and DiO-labeled platelet membranes was treated as a control.
To further ascertain the colocalization of platelet membranes and lipid membranes in the nanoplateletsome, the double-stained P-Lipo were co-cultured with 4T1 cells for 1 h in an ice bath, washed with PBS, stained with DAPI, and then imaged under the confocal laser scanning microscope with a 63 × oil objective (CLSM, Zeiss LSM 710, Germany). The fluorescence colocalization was analyzed by Image J software.
2.6. Analysis of membrane proteins in P-Lipo
The protein content in P-Lipo was measured by BCA protein assay kit. The protein loading efficiency in P-Lipo after fusion was calculated as follows as Eq. (2):
| (2) |
Membrane proteins in P-Lipo were analyzed via sodium dodecyl sulfate-polyacryla mide gel electrophoresis (SDS-PAGE)28. In brief, samples with the same protein content were heated at 90 °C for 5min, and separated on a 10% SDS-polyacrylamide gel at 120 V for 1h, followed by Coomassie blue staining and imaging. Typical membrane proteins on P-Lipo were validated by Western blotting. Briefly, samples were separated by SDS-PAGE as described above, transferred to nitrocellulose membranes (PALL, USA), probed with primary antibodies for CD41/61, GPVI, CD42b, CD29, CD49b, CD62P, CD63, incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (R&D systems, USA), and detected using Immobilon™ Western Chemiluminescent HRP Substrate (Millipore, USA).
Proteomic analysis of P-Lipo was performed by label-free quantification proteomics technology to further illustrate the protein composition as previously described28,46 (refer to methods in the Supporting Information). To determine the orientation of membrane proteins on P-Lipo, P-Lipo was incubated with anti-CD61-gold conjugates (bs-0342R-Gold, Bioss, 1:500) for 1 h at 37 °C using CD61 as a typical platelet membrane protein, passed through a Sepharose CL-4B column to remove free anti-CD61-gold conjugates, and then visualized under TEM (Tecna G2S-Twin, FEI, USA) after staining with 2% uranyl. Fluorescent CD42b antibody was also explored to probe the membrane protein orientation on P-Lipo. Briefly, 0.5 mg/mL of P-Lipo suspended in PBS containing 1% BSA was incubated with anti-CD42b-APC (551061, BD, 1:5000) for 30 min at room temperature, washed through dialysis (MWCO 3500 kDa) to remove free anti-CD42b-APC, and analyzed with flow cytometry (FCM, CytoFlex S, Beckman Coulter, USA). Lipo without staining with anti-CD42b-APC was used as a control.
2.7. In vitro CTC capture with P-Lipo
In vitro CTC capture with P-Lipo was investigated as previously described39. Approximately 1 × 106 4T1-GFP cells were resuspended in PBS or in the whole blood diluent to obtain an artificial CTC-containing blood sample. 100 μL of DiD-labeled P-Lipo or Lipo was added to this sample and incubated for 2 h at 4 °C under 100 rpm. The cells were washed three times with PBS and resuspended in PBS. Then, the CTC capture was observed by CLSM (LSM710, Carl Zeiss, Germany) or analyzed on a flow cytometer (CytoFlex S, Beckman Coulter). 4T1-GFP cells without treatment were treated as a control.
To investigate the binding mechanism of P-Lipo with CTCs, PH-Lipo was pre-incubated with P-selectin inhibitor heparin (20 U/mL49) and fucoidan (50 μg/mL50), respectively. Afterward, pre-treated DiD-labeled PH-Lipo were incubated with 4T1-GFP cells in the presence of these inhibitors for 2 h. After washing the cells with PBS twice, the fluorescence intensity of DiD in the cells was measured via FCM (CytoFlex S, Beckman Coulter).
2.8. In vivo CTC capture with P-Lipo
The CTC capture ability of P-Lipo in the blood vessels was monitored by in vivo FCM (IVFC, custom-built by Med-X Research Institute and School of Biomedical Engineering, Shanghai Jiao Tong University)51, 52, 53, 54. In brief, 106 CFSE-labeled 4T1 cells were injected intravenously (i.v.) via the tail vein to female BALB/c mice to mimic the CTC mouse model. Ten min later, the mice received an intravenous injection of DiD-labeled P-Lipo or Lipo (0.2 mg) and were anesthetized with 1% amobarbital by intraperitoneal injection. Afterward, the mouse was fixed on a glass plate, and the ear was glued to the glass panel with a maximum plane by glycerin. Major veins and arteries of the mouse ear were visualized under a CCD (charge-coupled device) camera, and an artery of 50–70 μm in diameter was selected for data acquisition. During acquisition, the fluorescence of CFSE-labeled 4T1 (510 ± 10 nm) and DiD-labeled P-Lipo (670 ± 20 nm) was excited by two lasers, 488 nm diode-pumped solid-state laser and 633 nm HeNe laser, respectively. When CFSE-labeled 4T1 or DiD-labeled Lipo or P-Lipo was passing through the slit of excitation light, the emitted fluorescence was collected by a photomultiplier tube (PMTs) and presented in the form of a peak. The data was analyzed using DT Measure Foundry. Each mouse was recorded for 10 min.
To further illustrate that P-Lipo could capture CTCs in the blood flow in vivo, real-time imaging of CTC capture was acquired with a spinning disk confocal microscope (SDC, SpinSR Olympus), which can provide fast, dual-color imaging to tract CTCs in the vessel at a high flow rate. In brief, 106 CFSE-labeled 4T1 cells were i.v. injected via the tail vein to female BALB/c mice. After 10 min, the mice received an intravenous injection of DiD-labeled P-Lipo (0.2 mg) and were anesthetized with 1% amobarbital by intraperitoneal injection. The mesenteric blood vessel was selected to track CTC capture and avoid the disturbing of the hair autofluorescence on the body surface. Two lasers (495 and 578 nm) were harnessed for fluorescence excitation, and two photomultiplier tube (PMT) detectors were used for the detection of CTC (519 nm) and P-Lipo (603 nm) fluorescence, respectively. The scanning speed was 4000 rpm with a 20 × objective lens. To avoid excessive burning of blood vessels by the laser, each mouse was recorded for 10 min.
2.9. In vitro CTC arrest inhibition by P-Lipo
To investigate the inhibition effect of P-Lipo on the CTC arrest by endothelial cells, Focht Chamber System 2 (FCS2®, Bioptechs Inc, USA) was built for flow cell imaging on inverted microscopes. To mimic the flowing CTC arrest in distant capillary beds, 50 × 106 HUVECs were seeded in the lower coverslip and were exposed to the fluid flow containing 10 ng/mL of TNF-α for 12 h. 4T1-GFP cells were collected using trypsin digestion and resuspended in HBSS. Afterward, 4T1-GFP (1 × 104/mL) and P-Lipo (or Lipo, 20 μg/mL) were filled into the syringes and connected to the FCS2 chamber via a three-way valve. A constant flow rate was generated by a syringe pump. Once the flow was generated, a high-speed video of moving 4T1-GFP cells was recorded using a trinocular inverted fluorescence phase-contrast microscope (Olympusix71, Japan). After 10 min perfusion, the picture of 4T1-GFP latched onto HUVECs was also gathered and the number of 4T1-GFP was analyzed using ImageJ.
2.10. In vitro CTC extravasation inhibition by P-Lipo
CTC extravasation across the vascular endothelial cells in vitro was evaluated by a transwell culture system55,56. Briefly, 30 μL of ice-cold matrigel (Corning) was added to a 24-well transwell insert and solidified at 37 °C for 30 min to form a thin gel layer. 5 × 104 HUVECs were added into the pre-coated insert, cultured overnight to form an endothelial monolayer, and pretreated with 10 ng/mL of TNF-α for 12 h. Afterward, 104 4T1-LUC cells and P-Lipo (or Lipo) (20 μg/mL) suspended in 200 μL of serum-free medium were added in the upper chamber. 600 μL of RPMI-160 medium containing 20% calf bovine serum was added carefully in the lower chamber. After 12 h coincubation, upper chambers were washed with PBS and scraped twice to remove the remaining cells. The transwell insert was immersed in d-luciferin (150 μg/mL), and bioluminescence imaging of the transwells was recorded using an In Vivo Imaging System (PerkinElmer, USA). Then the insert was immersed in 70% ethanol for 10 min to fix the migrating cells through the insert. After drying, the cells were stained with 0.1% crystal violet for 30 min followed by washing with PBS to remove excess dye. Images of cells were taken by an Olympusix71 inverted microscope (Japan). The cells in the donor wells were fixed and stained with crystal violet, as described above. Afterward, the crystal violet in the donor well was eluted using 33% acetic acid and quantified by measuring the OD570 nm with a microplate reader (Biorad, USA).
2.11. Wound healing inhibition by P-Lipo
To investigate the inhibition effect of P-Lipo on platelet-promoted wound healing of tumor cells, 4T1 cells were seeded into 6-well plates at a density of 106 cells per well and cultured at 37 °C in 5% CO2 overnight. Confluent cells were scratched and incubated with PBS, Lipo, PL-Lipo, and PH-Lipo (20 μg/mL) in the presence or absence of platelets (5 × 106/mL) for 12 h at 37 °C. Then the cells were imaged at different time points using an Olympus Ixpolor Standard inverted microscope (Japan) and percentage of the scratch area repopulated was calculated using ImageJ software.
To investigate the effect of P-Lipo on platelet-promoted epithelial-mesenchymal transition (EMT) in tumor cells, 4T1 cells were not scratched but treated with PBS, Lipo, PL-Lipo, and PH-Lipo in the presence or absence of platelets as described above. The morphology of 4T1 cells was observed under an Olympus Ixpolor Standard inverted microscope (Japan). Afterward, the cells were collected and lysed in a lysis buffer for 30 min on ice. The protein concentration in these samples was measured by a BCA kit. 20 μg of protein samples were separated by SDS-PAGE, transferred to PVDF membranes (Merck) as described above. The membranes were then blocked with protein-free rapid blocking buffer (1×) for 10 min and probed with primary antibodies specific for E-cadherin (GB11868, Servicebio, 1:500), N-cadherin (GB111273, Servicebio, 1:500), Vimentin (GB111192, Servicebio, 1:500), and β-actin (GB12001, Servicebio), followed by staining with horseradish peroxidase (HRP)-conjugated secondary antibodies (GB23303, Servicebio, 1:5000). The signals were visualized using ECL Plus reagents (EpiZyme) under the FluorChem™ M imaging system (ProteinSimple, USA) and quantified using ImageJ software.
2.12. In vivo imaging of P-Lipo in lung pre-metastasis
In vivo imaging of P-Lipo (or Lipo) in pre-metastasis niches was investigated as previously reported57,58. Each female BALB/c mouse (18–20 g) was injected with 106 4T1-LUC cells (200 μL) via the tail vein. Fifteen minutes later, mice were intravenously injected with DiD-labeled P-Lipo (or Lipo) at the lipid dose of 10 mg/kg. The ventral hair was removed using hair removal cream. Two hours later, each mouse received an intraperitoneal injection of 100 μL of d-luciferin at the dose of 150 mg/kg. In vivo bioluminescence imaging (BLI) and fluorescence imaging (FLI) were acquired using the IVIS Spectrum (PerkinElmer, USA) 10 min post d-luciferin injection. Afterward, the mice were sacrificed and major organs including the heart, the liver, the spleen, the lung, the kidney, and the brain of mice were harvested. BLI and FLI were performed using the IVIS Spectrum as described above. All quantitative measurements of BLI and FLI signals were obtained using the Living Image v. 4.3 software (PerkinElmer, USA).
2.13. Ex vivo imaging of P-Lipo in lung pre-metastasis
The ex vivo imaging of lung metastasis was created as previously reported59. Briefly, 106 4T1-GFP cells (200 μL) were injected into each BALB/c mouse (18–20 g) via the tail vein. Fifteen minutes later, the mice were intravenously injected with DiD-labeled P-Lipo (or Lipo) at the lipid dose of 10 mg/kg (equal to 10 μg/kg of DiD). To visualize lung capillaries, 100 μL of 70 kDa rhodamine-conjugated dextran (4 mg/mL) was injected via the tail vein. Afterward, the mice were injected with a lethal dose of 1% pentobarbital sodium (i.p.) and mounted on anatomical plates. Surgical scissors were utilized to cut the skin and expose the tracheal. 500 μL of 37 °C 2% low-melting-temperature agarose was injected into the lungs via the trachea until the lungs were inflated. After agarose cooling, the inflated lungs were separated from the mouse and immersed in warm RPMI-1640 to wash off excessive blood. Later on, inflated lungs were immersed into DAPI solution (2 μg/mL in PBS) for 15 min, washed with PBS three times at room temperature. The lobes were separated with scissors and placed into the 24-well imaging plate with the flat surface down to maximize the imaging surface. 100 μL of 37 °C RPMI-1640 was added on top of the lobes, and 15-mm circular microscope cover slides were placed on top of the lobes to prevent it from floating. Imaging was acquired with a spinning-disk confocal laser scanning microscopy (LSCM, A1R HD25, Nikon, Japan).
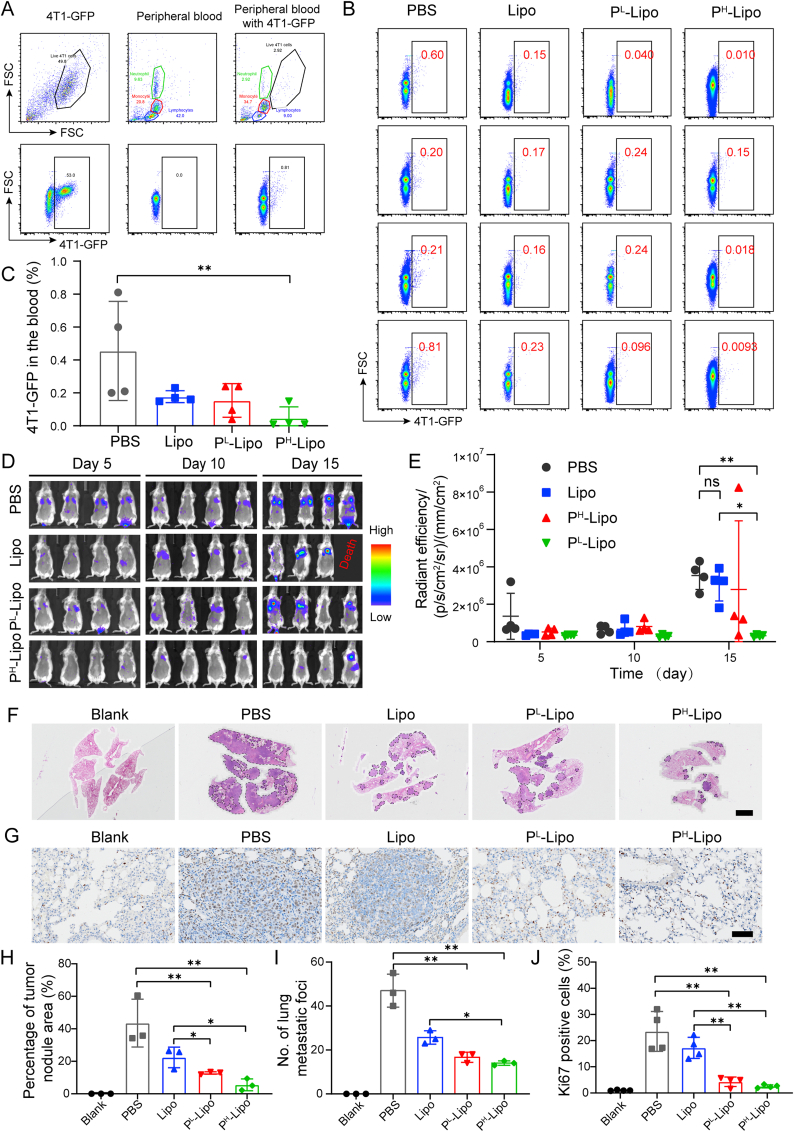
2.14. Anti-metastasis efficacy of P-Lipo
To test the efficacy of P-Lipo on the progression of metastasis, 104 4T1-LUC or 4T1-GFP cells suspended in 100 μL of PBS were injected into BALB/c female mice via the tail vein. Then 100 μL of P-Lipo or Lipo (0.2 mg) were intravenously injected via the tail vein at 15 min after 4T1-LUC or 4T1-GFP injection. For 4T1-GFP mouse models, blood was sampled from the mouse eyes on Day 3 after treatment. The blood was treated with red blood cell (RBC) lysis buffer for 5 min on ice to lyse RBCs, and subjected to FCM (CytoFlex S, Beckman Coulter) to monitor 4T1-GFP cells. For 4T1-Luc mouse models, in vivo bioluminescence imaging was conducted every 5 days for 15 days to monitor tumor metastasis growth with the IVIS Spectrum (PerkinElmer, USA). BLI was recorded at 10 min after intraperitoneal injection of d-luciferin (150 mg/kg) every time. Fifteen days after treatment, blood was sampled from mouse eyes and blood routine examination was performed by an auto hematology analyzer (BC-2800vet, Mindray, China). Afterward, the mice were sacrificed, and lungs were collected. Lungs were sliced, subjected to hematoxylin-eosin (H&E) and Ki67 staining according to the manufactures' protocols, and observed using an Olympus VS200 Research Slide Scanner (Japan). Besides, main organs, including the heart, the liver, the spleen, and the kidney, were collected for H&E staining according to the routine protocols.
2.15. Anti-tumor immune response of P-Lipo
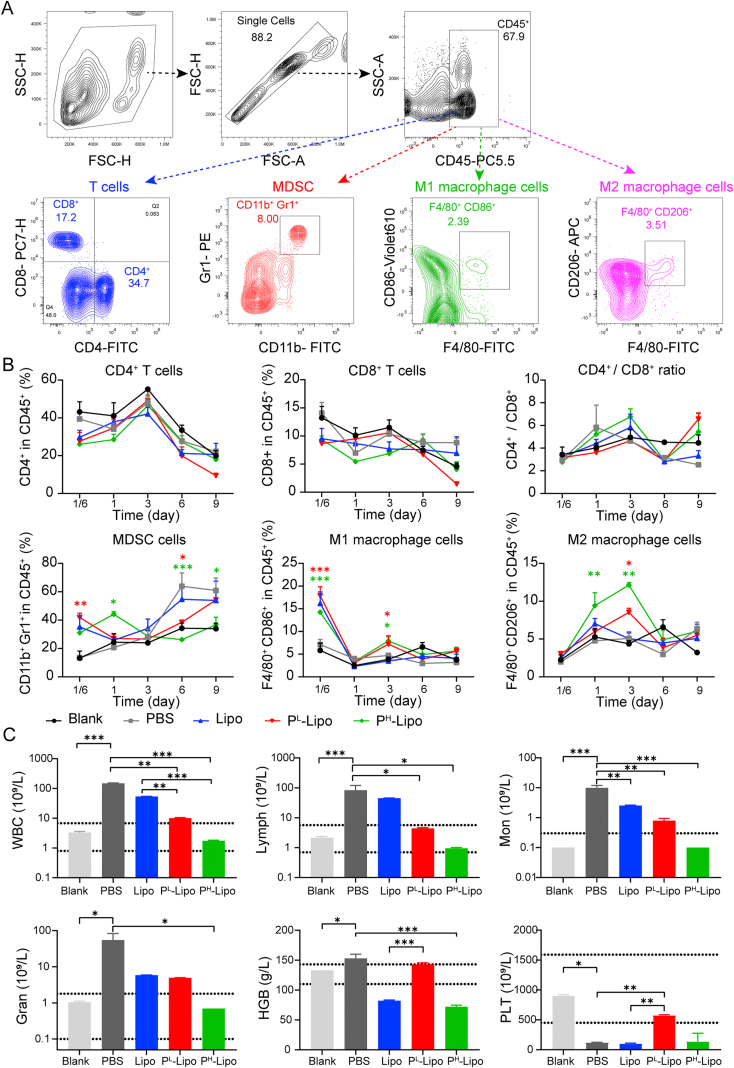
To test the anti-tumor immune response of P-Lipo treatment, 104 4T1-LUC cells were injected into BALB/c female mice and treated with P-Lipo as described above. Blood was sampled from the submaxillary veins at different time points after treatment, treated with RBC lysis buffer, blocked with 5% FBS followed by staining with fluorescent antibodies, and then subjected to a CytoFlex S flow cytometer (Beckman Coulter) for immune cell detection. For T cell detection, blood samples were stained with anti-CD45-PerCP-Cy5.5 (eBioscience, 45-0451-82, 1:500), anti-CD4-FITC (BD, 553650, 1:500), anti-CD8a-PE-Cy7 (eBioscience, 14-0081-82, 1:250) antibodies. For myeloid-derived suppressor cell (MDSC) detection, blood samples were stained with anti-CD45-PerCP-Cy5.5 (eBioscience, 45-0451-82, 1:500), anti-CD11b-FITC (eBioscience, 17-01120-81, 1:500) and anti- Gr1-PE (BD, 553128, 1:500) antibodies. For macrophage detection, blood samples were stained with anti-CD45-PerCP-Cy5.5 (eBioscience, 45-0451-82, 1:500), anti-F4-80-FITC (Biolegend, 123108, 1:500), anti-CD86-Super bright 600 (Invitrogen, 63-0862-80, 1:500), and anti-CD206-APC (Biolegend, 141708, 1:500) antibodies.
2.16. Statistical analysis
Data were presented as mean ± Standard deviation (SD). Unpaired Student's t-test was applied for statistical analyses between two groups, and one-way ANOVA with Tukey post-hoc analysis was used for multiple group comparison. Statistical differences were defined as ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. The abbreviation “ns” indicated no significant difference between two groups.
3. Results and discussion
3.1. Preparation and characterization of P-Lipo
P-Lipo was prepared by fusing platelet membranes with artificial lipid membranes, as illustrated in Fig. 1A. Platelets were extracted from the blood of BALB/c mice, which were pre-mixed with heparin to prevent clotting. The adding of prostacyclin E1 (PGE1) and protease inhibitor mini tablets could inhibit the activation of platelets and facilitate platelet resuspension during platelet isolation60,61. The optical microscopy images of the whole blood and extracted platelets were shown in Supporting Information Fig. S1. The extracted platelets had high purity and biconvex discoid structures, 2–3 μm in greatest diameter, and belonging to unactivated platelets. Platelet membranes were deviated by three freeze-thaw cycles and collected by high-speed centrifugation. Meanwhile, a thin artificial lipid film was formed in a round-bottom flask by the removal of organic solvent. The platelet membranes were then added into the flask to hydrate the lipid membranes and fused with the lipid membranes to generate homogenous P-Lipo after a freeze-thaw and extrusion process.
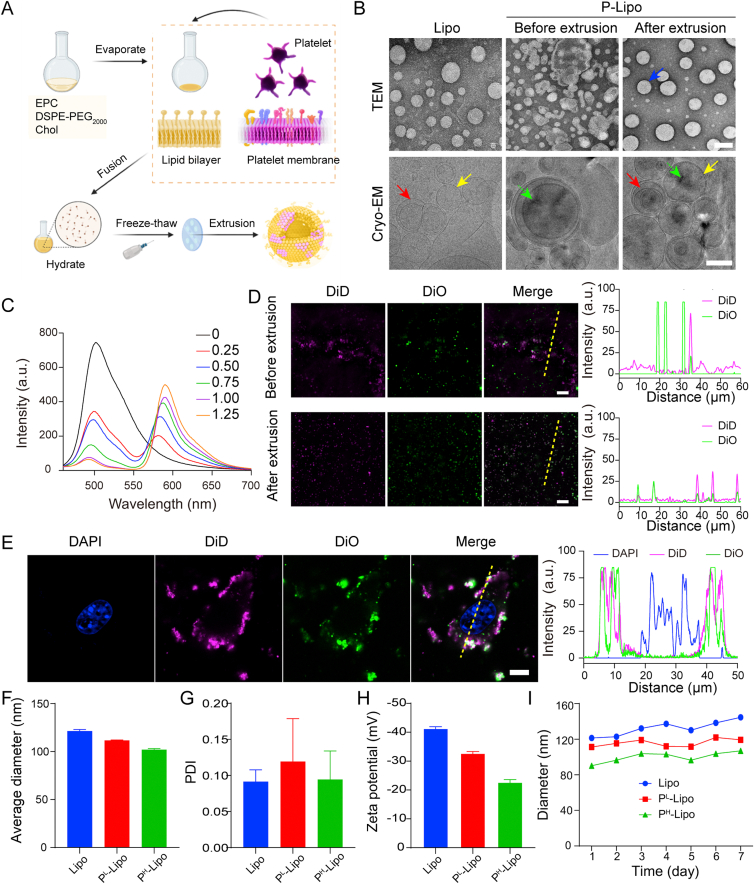
Figure 1.
Characterization of P-Lipo. (A) Schematic showing the preparation of P-Lipo. (B) Transmission electron microscopy and cryo-electron microscopy analysis revealed a spherical homogeneous shape for bare liposomes and P-Lipo after extrusion (scale bar = 100 nm). (C) The fluorescence spectra of P-Lipo when different amounts of platelet membranes were input. C6-stained lipid membranes and RhB-labeled platelet membranes were fused to form P-Lipo. (D) Representative super-resolution fluorescence images showed the colocalization of DiD-labeled lipid membranes and DiO-labeled platelet membranes after fusion together. Scale bar = 20 μm. Fluorescence intensity traces (dotted yellow lines) from the left image were plotted at the right. Peak overlapping indicated the colocalization of artificial lipid membranes and platelet membranes. (E) Representative confocal laser scanning microscopy (CLSM) images of single 4T1 cell after incubation with P-Lipo (scale bar = 5 μm). Cell nuclei, artificial lipid membranes, and platelet membranes were labeled with DAPI (blue), DiO (pinkish-red), and DiO (green), respectively. Fluorescence intensity traces (dotted yellow line) from the left image were plotted at the right. Dynamic light scattering analysis displayed (F) the average diameter, (F) polydispersity index (PDI), and (H) zeta potential of Lipo, PL-Lipo, and PH-Lipo (n = 3). (I) Size stability of P-Lipo over 7 days in PBS at 4 °C (n = 3).
The morphology of P-Lipo under TEM and cryo-EM was shown in Fig. 1B. Compared with the mixture of platelet membranes and lipid membranes, P-Lipo showed a completely different morphology. Before the fusion process, the mixture had an irregular shape and was not of uniform size. After the fusion process, spherical vesicles with a size range of 90–120 nm were formed. Negatively stained P-Lipo appeared a dark halo (indicated with blue arrows) around it that was not present on the Lipo. This halo was similar to that of cell membrane-coated nanoparticles, and it was not present on the non-coated nanoparticles either62. Cryo-EM images reserve the most native state and avoid interference from staining artifacts associated with traditional TEM. From cryo-EM images, we could see spherical unilamellar (yellow arrow) and some multilamellar vesicles (red arrow) were present in P-Lipo after fusion. Compared with the Lipo, spherical P-Lipo were decorated with black dots (indicated with light green arrows) similar to those seen in the mixture, indicating that the membrane proteins were possibly embedded into the lipid bilayer of P-Lipo63. TEM and Cryo-EM results demonstrated that platelet membranes were composed of vesicles of different shapes and different sizes (Supporting Information Fig. S2). In the absence of extrinsic lipids, platelet membranes consisted of typical phospholipid bilayers and a particularly thick (15–20 nm) glycocaly64 were very difficult to be extruded through the polycarbonate membranes serially and most membrane proteins (>80%) were trapped in the membranes, especially after passing through 0.2 μm polycarbonate membranes. We could not obtain platelet membrane vesicles with similar size to P-Lipo. Thus, in subsequent experiments, platelet membrane vesicles were not included.
The fusion of platelet membranes and artificial lipid membranes was validated by FRET. Artificial lipid membranes and natural platelet membranes were labeled with FRET donor C6 and acceptor RhB probes, respectively. The donor C6 is excited at a maximal wavelength of 460 nm and emits fluorescence at 510 nm. The acceptor RhB is excited at 565 nm and emits at 590 nm. Energy transfer occurs due to the spectral overlap between the emission spectrum of the donor and the excitation spectrum of the acceptor and the spatial distance of the FRET pairs (RhB-C6) close enough once they are fused (Supporting Information Fig. S3A and S3B). FRET between donor and acceptor fluorophores decreases the donor fluorescence and increases the acceptor fluorescence. As shown in Fig. 1C, the fluorescence spectra of P-Lipo demonstrated an increase of the fluorescence signal at 590 nm (RhB) and a decrease at 495 nm (C6) when increasing the input of platelet membranes. The results suggested that the RhB-labeled platelet membranes were fused with C6-labeled lipid membranes successfully. To quantify the FRET effect, FRET efficiency was calculated. FRET efficiency in P-Lipo was increased with the input of platelet membranes (Fig. S3C and S3D). Approximately 90% of FRET efficiency was obtained when 1.25 U of platelet membranes was fused with lipid membranes. This observation confirmed the successful fusion of platelet membranes with lipid membranes.
To further confirm the membrane fusion, the colocalization of platelet membranes and lipid membranes in P-Lipo was observed by a super-resolution microscope which provided molecule-specific information at the single-nanoparticle level. As shown in Fig. 1D, DiO-labeled platelet membranes (green) and DiD-labeled lipid membranes (pinkish-red) largely stayed separately in the mixture before extrusion. After extrusion to form P-Lipo, almost all platelet membranes were overlayed with lipid membranes suggesting that platelet membranes were successfully fused into lipid membranes.
The fusion of platelet membranes and lipid membranes was also confirmed through the binding between P-Lipo and tumor cells (Fig. 1E). It was found that DiO-labeled platelet membranes were colocalized with DiD-labeled lipid membranes on the surface of 4T1 cells, indicating that P-Lipo was composed of both platelet membranes and lipid membranes. The apparent binding of P-Lipo to tumor cells also laid the foundation to explore the targeting of P-Lipo to CTCs.
The diameter size and zeta potential of P-Lipo were measured by DLS. The incorporation of platelet membranes into lipid membranes induced a slight reduction of the average diameter of the resulting P-Lipo, from 121.4 ± 1.4 nm (Lipo) to 111.7 ± 0.4 nm (PL-Lipo) and 102.1 ± 0.9 nm (PH-Lipo), respectively (Fig. 1F). The intensity-based particle size and the zeta potential of platelet membranes were 589 ± 35 nm and ‒27.1 ± 1.3 mV, respectively. The polydispersity index (PDI) of platelet membranes was 0.775 ± 0.040, indicating platelet membranes had a broad size distribution. This size change is different from cell membrane-coated PLGA nanoparticles, which always exhibit an increase in diameter after cell membrane coating on nanoparticles41,62,65, but is similar to cell membrane protein-incorporated lipid nanovesicles43,66. The possible cause of this result is that the artificial liposomes and platelet membranes are soft and deformable, and platelet membranes or cell membrane proteins could be easily incorporated into liposomes. On the contrary, PLGA nanoparticles have solid cores, and cell membranes could only be coated on them. The PDI values of both Lipo and P-Lipo were less than 0.2 (Fig. 1G), revealing a high size homogeneity for these liposomes. These results were consistent with TEM and cryo-EM analysis. Compared with Lipo (‒41.5 ± 0.4 mV), P-Lipo presented an increase in zeta potential, increasing to ‒32.8 ± 0.4 mV (PL-Lipo) and ‒22.9 ± 0.6 mV (PH-Lipo) (Fig. 1H). The increased zeta potential of P-Lipo was attributed to the shielding effect of the membrane proteins towards the negative charge of the phosphate groups43. When stored at 4 °C for up to 7 days, both P-Lipo and Lipo showed minor size change (Fig. 1I), indicating that P-Lipo had superb stability in PBS.
3.2. Analysis of membrane proteins in P-Lipo
To examine the platelet membrane content in P-Lipo, membrane protein content in P-Lipo was analyzed using a BCA assay. As shown in Supporting Information Fig. S4A, around 93% of platelet membranes were fused into PL-Lipo, while 63% of platelet membranes were found in PH-Lipo. Part of platelet membranes could be rejected by the polycarbonate filters during the extrusion process when the platelet membrane input increased. The retention of membrane proteins on P-Lipo was further investigated by SDS‒PAGE (Fig. 2A). Almost all of the protein bands of platelet membranes can be found in the P-Lipo group. No protein bands were found in the Lipo group. These results indicated most kinds of membrane proteins were retained in P-Lipo after platelet membranes were fused with lipid membranes.
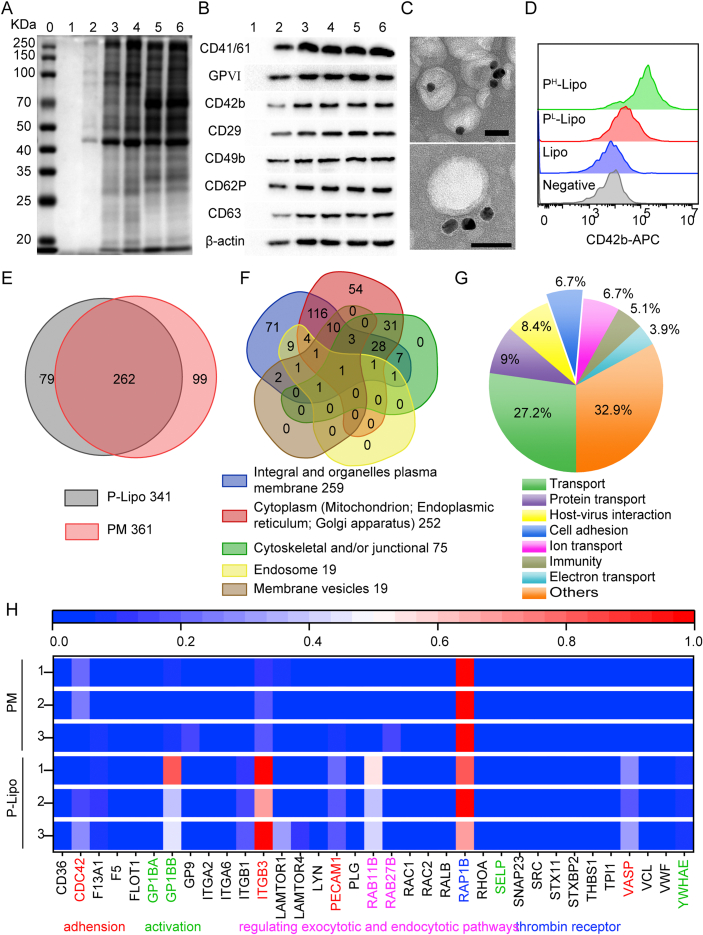
Figure 2.
Characterization of platelet membrane proteins in P-Lipo. (A) Coomassie staining and (B) Western blot analysis of protein markers (0), Lipo (1), PL-Lipo (2), PH-Lipo (3), platelet membranes (4), platelet supernatant (5), and platelets (6) after SDS-PAGE separation. (C) Transmission electron microscopy images of P-Lipo probed with anti-CD61-gold, followed by negative staining with 2% uranyl acetate (scale bar = 50 nm). (D) Flow cytometry histograms of CD42b expression on the surface of P-Lipo. Unstained Lipo functioned as a negative control. (E) Venn diagram of identified proteins of P-Lipo and platelet membranes (PM) analyzed by liquid chromatography-tandem mass spectrometry. (F) Venn diagram of identified proteins extracted from P-Lipo by cellular component. (G) Functional characterization of the platelet membrane proteins identified in P-Lipo by biological process. (H) A heat map depicting the typical membrane protein levels (normalized to array reference) from P-Lipo and PM.
There are platelet surface cluster of differentiation (CD) antigens before activation, like CD41, CD42c, CD42b, and CD61. CD antigens such as CD31, CD62P (P-selectin), and CD63 appeared on the platelet surface during activation67. CD41/61, also named GPIIb-IIIa or αIIβ, could bind plasma proteins, such as fibrinogen, fibronectin, von Willebrand factor (vWF), and vitronectin, and plays a critical role in platelet aggregation and adhesion, promote tumor cell metastasis68. GPVI (GP6) and CD42 (GPIbα), are known as platelet membrane glycoproteins. GPVI participates in collagen-induced activation and aggregation. CD42b involves platelet adhesion and recovery from vascular injury, especially under high shear blood flow, and promotes tumor cell metastasis15. CD29 (ITGB1) and CD49b (ITGA2) are integrins, also known as platelet adhesion CD antigens, widely expressed on platelet surfaces. Western blot analysis confirmed the presence of the platelet-related surface proteins CD41/61, GPVI, CD42b, CD29, CD49b, CD62P, and CD63 on P-Lipo (Fig. 2B). The results indicate that platelet membranes were derived from partly activated platelets, and platelets were broken and partly activated during repeated freeze-thaw circles.
Immunolabelling with antibodies directed against the extracellular domain of membrane proteins was explored to confirm the orientation of platelet membrane proteins in P-Lipo. As shown in Fig. 2C, the dark solid gold nanoparticles (10 nm) were distributed randomly on the surface of round P-Lipo after it was labeled with a gold-labeled anti-CD61 antibody. When P-Lipo was incubated with anti-CD42b-APC, APC-labeled P-Lipo showed discernible populations compared with Lipo (Fig. S4B and S4C). It was found that CD42b was highly enriched in PL-Lipo and PH-Lipo (Fig. 2D). Utilizing CD61 and CD42b as typical platelet membrane proteins, both immune-TEM visualization and FCM analysis validated the right-side-out membrane orientation of membrane proteins on P-Lipo.
The protein composition of P-Lipo and platelet membranes (PM) was determined by label-free quantification proteomics technology, and 341 and 361 distinct proteins were identified from P-Lipo and PM, respectively. More than three-quarters of proteins (262) from P-Lipo overlapped with PM (Fig. 2E). According to the cellular component, proteins from P-Lipo could be classified into three types: membrane-associated (259), cytoplasm (Mitochondrion; Endoplasmic reticulum; Golgi apparatus) (252), cytoskeletal and/or junctional (75), endosome (19), and vesicular or secreted proteins (19) (Fig. 2F). From the perspective of the biological process, proteins from P-Lipo could be associated with transport (27.2%), protein transport (9%), host-virus interaction (8.4%), cell adhesion (6.7%), ion transport (6.7%), immunity (5.1%), electron transport (3.9%), and others (32.9%) (Fig. 2G). Further, 35 typical membrane proteins were shown in Fig. 2H according to their abundance. The results demonstrate that the protein abundance of P-Lipo was essentially in agreement with PM. The relative expression level of RAP1B, a key molecule involved in platelet activation/adhesiveness in the hemostatic process after injury69, was very high in P-lipo. ITGB3, also named Integrin β-3, mediating diverse processes including cellular adhesion, cancer metastases, and lymphocyte helper and killer70 was also highly expressed in P-lipo. Platelet glycoprotein Ib (GPIb, or CD42c) encoded by GP1BB, together with GP9 (CD42a) and GP1BA (CD42b), which are components of the GPIb-IX-V glycoprotein complex for VWF71, could be found in P-Lipo. Other cell adhesion proteins (ITGB3, SELP, PECAM1, and VASP), platelet activation markers (GP1BA, GPABB, SRC, and YWHAE), and thrombin receptor (PAP1B) could also be found in P-Lipo. These results indicated P-Lipo retained the same membrane protein composition and correct protein orientation as platelet membranes and might be endowed with biological functions similar to platelets.
3.3. Precise CTC capture by P-Lipo in vitro and in vivo
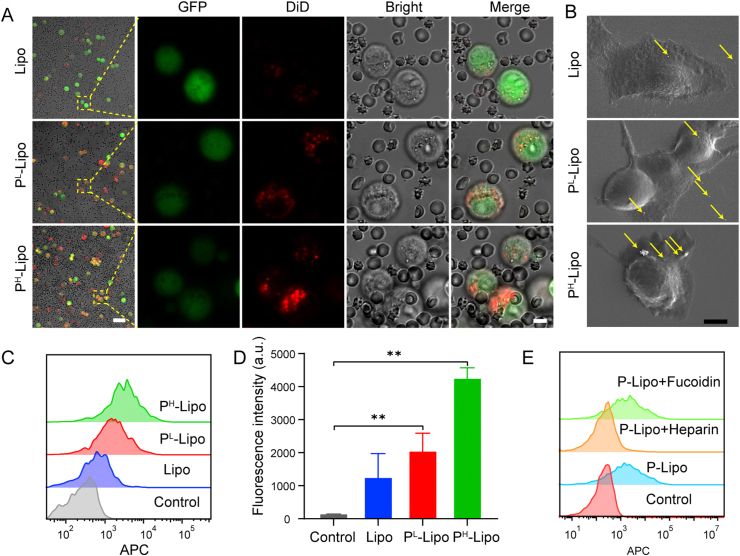
It is generally believed that platelets serve as physical shields from blood shear and natural killer (NK) cells attack, and enhance tumor metastasis by binding to CTCs. Therefore, we propose the hypothesis that P-Lipo may possess similar functionalities of CTC binding. CLSM and FCM were used to evaluate P-Lipo binding with cancer cells in vitro. As expected, significantly more P-Lipo bound with 4T1-GFP when compared with Lipo, whether in PBS (Supporting Information Fig. S5) or in the artificial CTCs-containing blood environment (Fig. 3A). In the artificial CTCs-containing blood, notable P-Lipo adhered to the surface of tumor cells, especially, and barely latched onto the surface of blood cells, including RBC, white blood cell (WBC), and platelets (Fig. 3A). Scanning electron microscopy (SEM) images revealed that numerous P-Lipo adhered onto the surface of 4T1 cells, but very few Lipo were observed on 4T1 cells (Fig. 3B). Further quantitative analysis by FCM showed that the mean fluorescence intensity of DiD-labeled PL-Lipo and PH-Lipo adhered on 4T1-GFP cells was 1.6- and 3.7-fold of that on Lipo (Fig. 3C and D). When DiD-labeled P-Lipo was incubated with 4T1-GFP cells in PBS, the fluorescence intensity of PL-Lipo and PH-Lipo bound on 4T1-GFP cells saw growth rates 2.9 and 3.8 times, respectively, compared with that of Lipo (Fig. S5). These results suggest that P-Lipo had a higher CTC capture ability than Lipo.
Figure 3.
In vitro CTC targeting of P-Lipo. (A) CLSM images of 4T1-GFP cells (green) captured by DiD-labeled P-Lipo (red) in the artificial CTCs-containing blood. (B) Scanning electron microscopy images of P-Lipo attached to the surface of 4T1 cell (scale bar = 5 μm). The yellow arrows indicated P-Lipo (or Lipo) adherent on the cell surface. (C) Flow cytometry histograms and (D) the corresponding fluorescence intensity of 4T1-GFP captured by DiD-labeled P-Lipo (or Lipo) from the artificial CTC-containing blood (n = 3). Untreated CTC-containing blood served as a control. ∗∗P < 0.01. (E) Flow cytometry histograms of 4T1-GFP in the artificial CTC-containing blood captured by DiD-labeled PH-Lipo, which were preincubated with heparin or fucoidin. Untreated CTC-containing blood served as a control.
To investigate which membrane proteins were involved in the adhesion between P-Lipo and CTCs72, heparin and fucoidan were applied to pretreat PH-Lipo before incubation with 4T1-GFP cells. Heparin is a known potent inhibitor of P-selectin and has been confirmed to efficiently inhibit metastasis in mouse models73,74. Fucoidan is known as a platelet agonist for the C-type lectin-like receptor 275. It was shown heparin attenuated the binding of PH-Lipo to 4T1-GFP cells while fucoidan had little impact on it (Fig. 3E), suggesting that P-Lipo binding with CTC was mainly mediated by P-selectin.
The cell apoptosis of CTCs after incubation with P-Lipo in the whole blood diluent was performed by flow cytometry. As shown in Supporting Information Fig. S6, the percentages of the apoptotic cells, including early apoptosis, late apoptosis, and total apoptosis for the P-Lipo groups, were significantly higher than those for the Lipo group. In addition, the percentages of the apoptotic cells, including late apoptosis and total apoptosis for the PH-Lipo group were significantly higher than those for the PBS group. These results indicate that the PH-Lipo binding with CTCs enhanced their apoptosis, which might attribute to that PH-Lipo adherence onto the surface of 4T1 cells broken the protection from nature platelets and facilitated the attack of immune cells in the blood.
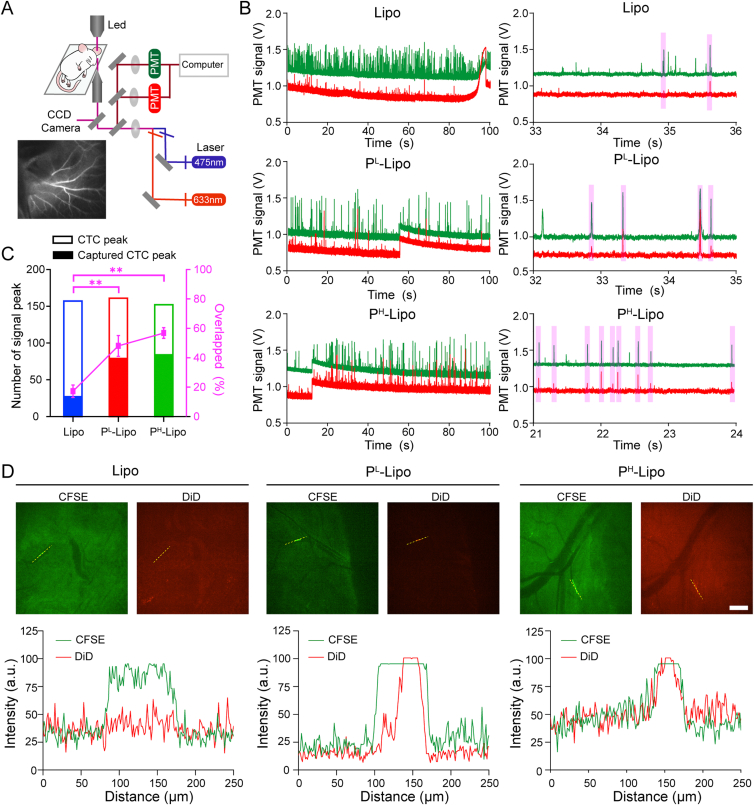
Inspired by encouraging CTC capture capability of P-Lipo in vitro, in vivo CTC capture ability of P-Lipo was future verified by IVFC51. Schematic of the custom-built IVFC system and the illustration of angiogram demonstrating ear vessels were shown in Fig. 4A. The advantage of IVFC was that cell populations could be measured continuously in real-time while the mouse was under anesthesia. When CFSE-labeled 4T1 cells or P-Lipo passed through the field-of-view, green and red signal amplitudes were recorded to identify CFSE-labeled CTCs and DiD-labeled P-Lipo (or Lipo), respectively. Two peaks overlapped represented the successful capture of CTC by P-Lipo (or Lipo) (purple rectangles in Fig. 4B). As shown in Fig. 4C, the bar chart revealed the number of CTC peak and captured CTC peak (CFSE and DiD double-positive peak). The percentage of double-positive peaks in CTC peaks increased from 17.2 ± 4.2% of Lipo to 48.0 ± 7% of PL-Lipo and 56.8 ± 3.6% of PH-Lipo, respectively. The result illustrated that the CTC capture ability of P-Lipo increased with the content of platelet membranes in P-Lipo.
Figure 4.
In vivo CTC targeting of P-Lipo. (A) Schematic illustration of the two-color, two-slit in vivo flow cytometer. (B) Visualization of digitized fluorescence signals of 4T1 cells (Green line) and DiD-labeled P-Lipo (or Lipo) (red line) by in vivo flow cytometry (IVFC). Each peak was created by the fluorescence burst resulting from tumor cells or P-Lipo traversing the excitation slit. Individual fluorescence peaks were shown in more detail on the right. (C) CTC capture efficiency in vivo (n = 3). The bar chart revealed the number of CTC peaks and captured CTC peaks, while the point plot depicted the percentage of dual-positive peaks in CTC peaks. ∗∗P < 0.01. (D) The images of CTC (green) captured by DiD-labeled P-Lipo or Lipo (Red) in the mesenteric vessels visualized by the spinning-disk confocal microscope (scale bar = 200 μm) and the corresponding fluorescence intensity profiles of P-Lipo or Lipo (red) and 4T1 cells (green) along the dotted yellow line crossing the representative cells.
To offer more intuitionistic information of CTC capture by P-Lipo in high-speed blood flow in vivo in real-time, SDC equipped with two cameras simultaneously to provide fast, two-color localization super-resolution imaging was applied to capture the dynamic processes76. To decrease the interference of spontaneous hair fluorescence, a peripheral mesentery vessel was chosen as the observation window. In the SDC image (Fig. 4D, and Supporting Movies 1‒3), individual CFSE-labeled CTC (green) could be visualized in the blood vessels once the CTC was captured by DiD-labeled P-Lipo (or Lipo), which appeared red. A good colocalization of the intensity profiles for DiD-labeled P-Lipo and CFSE-labeled 4T1 cells in the blood vessel further confirmed the capture of the circulating 4T1 cell by P-Lipo. The SDC imaging matched well with the results from the previous IVFC assay. In the P-Lipo group, most CTCs were stained red, while in the Lipo group, few red signals appeared simultaneously with green CTCs. These results indicated platelet membranes in P-Lipo strengthened the CTC capture ability of P-Lipo.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.apsb.2022.01.005
The following are the supplementary data related to this article:
3.4. In vivo imaging of P-Lipo in lung pre-metastasis
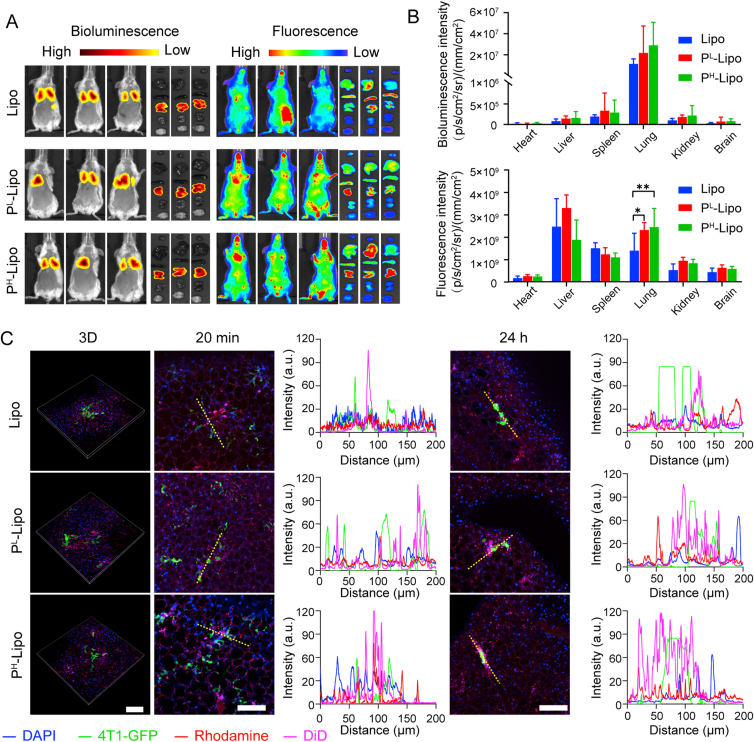
A murine model of lung pre-metastasis was adopted to track the distribution of cancer cells and P-Lipo. As shown in Fig. 5A and B, bioluminescence imaging revealed that 4T1-Luc mainly accumulated in the lung after intravenous injection. In vivo fluorescence imaging demonstrated that P-Lipo was mainly distributed in the liver, lung, and spleen at 2 h after injection. Compared with Lipo, significantly more P-Lipo was located in the lung where the pre-metastasis existed, suggesting platelet membranes assisted P-Lipo getting trapped in the pre-metastasis lung environment. The colocalization of P-Lipo with metastatic tumor cells was further visualized by ex vivo live imaging. 2% agarose was instilled into the lung to avoid lung collapse caused by the open-chest operation and preserved the maximum lung microenvironment ex vivo to achieve the SDC imaging of non-sectioned whole lung lobes. As shown in Fig. 5C and the Supporting Movies 4‒5, 4T1-GFP was arrested in the lung capillaries 20 min after injection, and much more P-Lipo surrounded 4T1-GFP, compared with Lipo (Supporting Information Fig. S7). More importantly, at 24 h after injection, although most 4T1-GFP were removed from the circulation, it could be found resident 4T1-GFP in the lung were still surrounded with P-Lipo while they were located separately and did not overlap with Lipo (Fig. 5C and Supporting Information Fig. S8). These results suggest P-Lipo could capture the arrested CTC in the pre-metastasis lung environment.
Figure 5.
In vivo targeting of P-Lipo in lung pre-metastasis. (A) Representative bioluminescence imaging (left) and fluorescence imaging (right) at 2 h after intravenous injection of DiD-labeled P-Lipo (or Lipo) in the pre-metastasis model. Ex vivo imaging of major organs (from top to down: heart, liver, spleen, lung, kidney, and brain) was to evaluate the distribution of P-Lipo (or Lipo) after injection. (B) Bioluminescence intensity and fluorescence intensity of major organs to reveal the distribution of P-Lipo (or Lipo) (n = 3). ∗P < 0.05, ∗∗P < 0.01. (C) Representative fluorescence images of 4T1-GFP cells colocalized with DiD-labeled P-Lipo (or Lipo) in the lung at 20 min and 24 h after intravenous injection and the corresponding fluorescence intensity profiles of yellow dotted lines shown on the right. Scale bar = 100 μm. Blue, nuclei; green, 4T1-GFP cells; red, vessels stained by rhodamine-conjugated dextran; rosy, DiD-labeled P-Lipo (or Lipo).
Supplementary video related to this article can be found at https://doi.org/10.1016/j.apsb.2022.01.005
The following are the supplementary data related to this article:
3.5. CTC arrest inhibition by P-Lipo in vitro
As described above, P-Lipo inherited the essential platelet adhesion proteins on their surfaces and retained some platelet functions, including CTC binding or capture. Then we evaluated the role of P-Lipo as platelet nanodecoys in CTC arrest and extravasation in the pre-metastasis environment. First, a co-culture cell model consisting of HUVECs and 4T1-GFP was built to examine the cell-binding ability of P-Lipo (Supporting Information Fig. S9). Compared with Lipo, P-Lipo showed higher cell-binding with both tumor cells and HUVECs, and PH-Lipo showed the most potent binding among these groups. However, in this co-culture cell model, there was no significant difference in P-Lipo binding between HUVEC and 4T1-GFP.
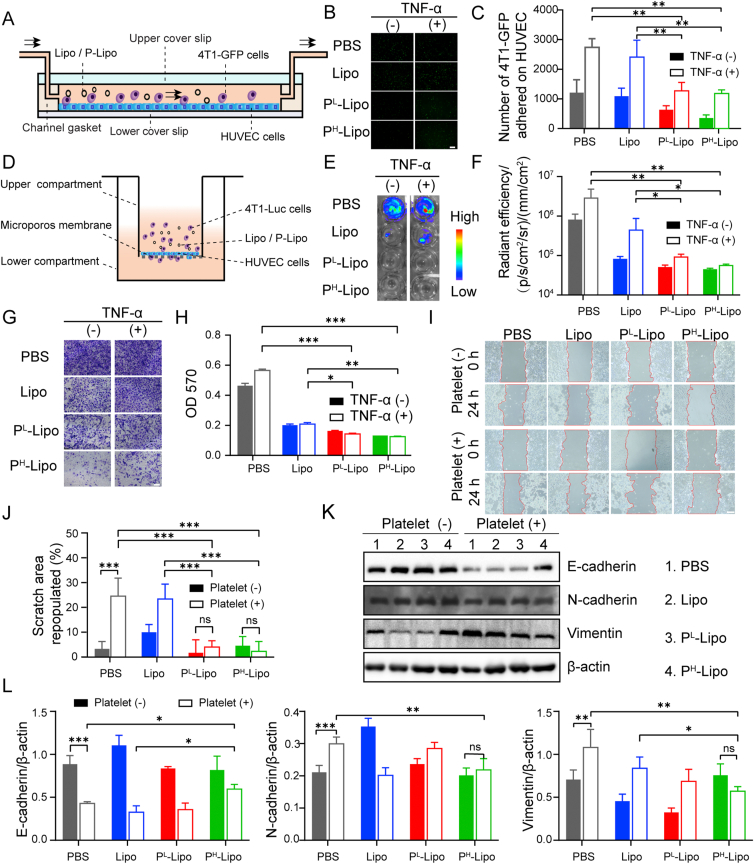
Platelets not only contribute to CTCs' survival by helping escape from immune surveillance but also enhance tumor-endothelium interactions77. To investigate whether P-Lipo impacts CTC arrest by vessel endothelial cells under flow conditions, flowing tumor cells adhered to HUVECs was recorded in the FCS2 chamber, whose structure was shown in Fig. 6A and Supporting Information Fig. S10. In the presence or absence of TNF-α, the amount of 4T1-GFP adhered to HUVECs increased over time, but the presence of P-Lipo made from 0.002 or 0.008 U/mL of platelets significantly decreased 4T1-GFP arrest on HUVECs (Fig. 6B and C). It was shown that platelet decoys made from approximately 0.039 U/mL of platelets could significantly decrease tumor cell arrest by HUVECs, considering there are 0.77 × 109 platelets per microliter of blood19. These results indicate P-Lipo inhibited CTC arrest on vessel endothelial cells with high efficiency.
Figure 6.
P-Lipo decreased tumor cell arrest, extravasation and epithelial-mesenchymal transition in vitro. (A) Schematic of CTC arrest on HUVEC investigated by the FCS2 chamber under the flow state. HUVEC cells were pre-seeded on the lower coverslip, and the mixture of tumor cells and P-Lipo (or Lipo) was perfused into the FCS2 chamber. (B) Representative fluorescence images of 4T1-GFP arrest by HUVEC, in the presence and absence of TNF-α (scale bar = 100 μm). (C) The number of 4T1-GFP arrested on HUVEC after 10 min of perfusion with the mixture of tumor cells and P-Lipo (or Lipo) in the presence and absence of TNF-α (n = 4). (D) Schematic of the transwell system to investigate CTC extravasation through the vascular endothelial layer. (E) Bioluminescence imaging and (F) the corresponding bioluminescence intensity of 4T1-luc cells migrating through the HUVEL monolayer (n = 3). (G) Crystal violet staining of 4T1-Luc was migrating through the HUVEC monolayer (scale bar = 100 μm). (H) Quantification of extravasated 4T1-Luc into the donor wells by measuring optical density (OD) at 570 nm (n = 3). (I) Representative wound field images of 4T1 cells at 0 h and 24 h after P-Lipo treatment in the presence of absence of platelets (scale bar = 200 μm). (J) The corresponding cell scratch area repopulated quantified by ImageJ (n = 3). (K) Representative Western blot images and (L) the corresponding quantification of E-cadherin, N-cadherin, and vimentin expression in 4T1 cells with different treatments (n = 3). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
3.6. CTC extravasation inhibition by P-Lipo in vitro
CTC extravasation inhibition by P-Lipo was studied in vitro using a transwell system under physiological and inflammatory conditions as previously reported56 (Fig. 6D). 4T1-Luc and P-Lipo were incubated with the HUVEC monolayer in the presence and absence of TNF-α. Bioluminescence imaging (Fig. 6E and F) and crystal violet staining (Fig. 6G) demonstrated that P-Lipo could significantly depress 4T1-Luc migration through the endothelial monolayer in both normal and inflammatory conditions, compared with PBS or Lipo. Importantly, it was also demonstrated that P-Lipo made from 0.002 or 0.008 U/mL of platelets could significantly inhibit 4T1-Luc extravasation through the endothelial monolayer into the donor wells in both normal and inflammatory conditions compared with PBS or Lipo (Fig. 6H). It was shown platelet decoys made from approximately 0.039 U/mL of platelets could significantly suppress tumor cell extravasation19. These results suggest P-Lipo could inhibit CTC to extravasate from the endothelial monolayer with high efficiency. A possible explanation is that P-Lipo could bind with CTC and vessel endothelial cells like platelets but interfere with CTC-platelet-endothelial cell interaction, disrupting CTC arrest on endothelial cells and CTC extravasation through vessels.
One of the precise mechanisms of the correlation between platelet-tumor cell interaction and metastasis is that platelets alter EMT in tumor cells and promote tumor invasion and metastasis78,79. To further validate whether P-Lipo could inhibit the platelet-induced EMT process, firstly, a wound healing test was carried out to evaluate the effect of P-Lipo on tumor cell migration in the presence of platelets. As shown in Fig. 6I and J, platelets significantly increased the scratch closure rate of 4T1 cells. However, compared with PBS and Lipo treatment, P-Lipo treatment significantly decreased the wound healing rate enhanced by platelets. Moreover, for both PL-Lipo and PH-Lipo groups, there were no significant differences in the percentage of scratch area repopulated between in the presence and absence of platelets. These outcomes indicated P-Lipo could inhibit platelet-enhanced wound healing of tumor cells. It was found that the co-incubation with platelets induced morphological alterations of tumor cells from a pebble-like shape to a spindle shape, which is a typical characteristic of EMT progress. However, P-Lipo treatment noticeably reduced the morphological changes elicited by platelets, indicating that the platelet-induced EMT could be suppressed by P-Lipo treatment (Supporting Information Fig. S11). Next, we further elucidated the tumor cells' EMT process by analyzing the expression of EMT markers (E-cadherin, N-cadherin, and vimentin) by Western blotting. As shown in Fig. 6K‒L, platelets significantly downregulated E-cadherin levels but significantly upregulated N-cadherin and vimentin levels in tumor cells, whereas PH-Lipo treatment could restore the protein levels of these markers in the presence of platelets. These results imply that platelet-induced EMT in 4T1 cells could be significantly weakened by P-Lipo.
3.7. P-Lipo decreased disseminated 4T1 invasion and metastasis in vivo
Before the anti-metastasis study, the pharmacokinetics of DiD-labeled P-Lipo containing different amounts of platelet membranes was investigated. As shown in Supporting Information Fig. S12, PL-Lipo, and PH-Lipo demonstrate similar long circulation profiles to Lipo. There were no significant differences in the pharmacokinetic parameters (e.g., MRT, AUC, t1/2 and Cl) between PL-Lipo and Lipo. However, the AUC and t1/2 for PH-Lipo were significantly lower than those for Lipo (Supporting Information Table S1), indicating a high platelet membrane content in P-Lipo could accelerate the blood clearance of P-Lipo to some extent. The long retention of P-Lipo in the blood could provide it more chance to catch CTCs or access pre-metastasis.
P-Lipo could not only capture both flowing CTCs and arrested CTCs, but also decrease CTC arrest and extravasation from the HUVEC monolayer. To test the ability of P-Lipo to reduce the formation of metastasis in vivo, 4T1-GFP cells and P-Lipo were injected into BALB/c mice successively. After 3 days, the percentage of 4T1-GFP in the blood after lysing the erythrocytes was monitored by FCM (Fig. 7A and B). It was found that PH-Lipo treatment significantly decreased the 4T1-GFP level in the peripheral blood compared with PBS treatment. Compared with the PBS group, 61%, and 66% and 89% reduction of disseminating 4T1-GFP in the blood was observed in the Lipo, PL-Lipo, and PH-Lipo treatment groups, respectively (Fig. 7C), indicating P-Lipo decreased CTC survival in the blood. These results might attribute to that PH-Lipo disrupted the CTC-platelet interaction and facilitated the attack of immune cells in the blood.
Figure 7.
Anti-tumor metastasis efficacy of P-Lipo. (A) Representative flow cytometry scatter plot of 4T1-GFP in the peripheral blood cells from heath mouse. (B) Flow cytometry scatter plot of peripheral blood cells from the pre-metastasis mouse models on Day 3 after different treatments. (C) Quantification of 4T1-GFP cells in the peripheral blood using the flow cytometry (n = 4). (D) Bioluminescence imaging of the pre-metastasis mouse models at preset time points after different treatments. (E) Quantification of bioluminescence intensity of the pre-metastasis mouse models over 15 days after treatment (n = 4). (F) Representative hematoxylin-eosin (H&E)-staining images of the lung sections from different treatment groups (scale bar = 2 mm). Arrowheads indicated tumor areas or nodules. (G) Representative images of Ki67-stained lung slices from different treatment groups (scale bar = 50 μm). Ki67 positive cell nuclei were always stained by diaminobenzidine and appeared as brown, while normal cell nuclei were stained by hematoxylin. (H) The area of tumor metastasis in H&E-staining lung sections from different treatment groups (n = 3). (I) Numbers of the lung metastatic nodule after different treatments (n = 3). (J) Percentage of Ki67-positive cells in lung sections from different treatment groups (n = 4). ∗P < 0.05, and ∗∗P < 0.01.
To further validate whether P-Lipo could inhibit CTC colonization and metastasis formation, the anti-metastasis efficacy of P-Lipo was assessed in a lung metastasis mouse model (Fig. 7D‒J). BLI demonstrated that the growth of 4T1-Luc metastasis had a similar tendency for the PBS, Lipo, and PL-Lipo groups, indicating that Lipo and PL-Lipo had no significant inhibition on CTC colonization and outgrowth (Fig. 7D and E). However, PH-Lipo showed significant suppressive effects on 4T1-Luc metastasis growth in the lung. In addition, H&E staining (Fig. 7F, H, and I) showed that the lungs developed significantly more metastases throughout the entire lobes and bronchi in the PBS and Lipo groups compared with the P-Lipo groups. The PL-Lipo group showed scattered large tumor nodules in the lung, and the PH-Lipo group displayed sporadic tumor nodules in the lung. Ki-67 staining (Fig. 7G and J) revealed significantly lower proliferation of tumor cells in the P-Lipo groups compared with the PBS group or the Lipo group. Considering platelet decoys could not inhibit tumor metastasis without co-incubation with platelets19, these results suggest that P-Lipo could strongly suppress tumor metastasis as nanodecoys with high efficiency.
To test the anti-tumor immune response, the change of immune cells in the peripheral blood after treatment was analyzed via FCM. The gating strategy for FCM analysis was shown in Fig. 8A. As shown in Fig. 8B, there were no significant differences in the percentage of CD8+ and CD4+ T cells in the blood from different groups at different time points after treatment. No significant differences were found in the ratio of CD8+ to CD4+ T cells in the blood from different groups after treatment either. Interestingly, at the early stage of pre-metastasis (4 h and 1 day after CTC inoculation), PH-Lipo treatment significantly increased MDSC levels in the blood compared with PBS treatment. However, on Days 6 and 9 after treatment, PH-Lipo treatment significantly suppressed MDSC levels in the blood to the normal level compared with PBS treatment, indicating PH-Lipo could inhibit the function of MDSC of promoting tumor escape, metastasis and drug resistance80. Compared with PBS treatment, PH-Lipo treatment significantly increased M1 macrophage level (4 h and 3 days after CTC inoculation) and M2 macrophage level (1 and 3 days after CTC inoculation) in the blood at the early stage. These results suggested that P-Lipo treatment could decrease the inflammation at the early stage by upregulating M2 macrophages and suppress tumor cell escape at the latter stage by downregulating MDSC to inhibit CTC colonization and metastasis formation.
Figure 8.
P-Lipo regulated the immune cell expression in the blood of the pre-metastasis mouse models (n = 3). (A) Representative flow cytometry analysis of CD4+ T cells, CD8+ T cells, MDSC (CD11b+Gr1+), M1 macrophages (F4/80+CD86+), and M2 macrophages (F4/80+CD206+) in the blood (gated on CD45+ cells) of the pre-metastatic mouse models after different treatments. (B) Percentage of CD4+ T cells, CD8+ T cells, CD4+/CD8+ T cell ratio, and myeloid-derived suppressor cells (MDSC), M1 macrophages, M2 macrophages in the blood of the pre-metastasis mouse models after different treatments. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 compared with PBS. ΔP < 0.05 compared with Lipo. (C) White blood cells (WBC), lymphocyte (Lymph), monocyte (Mon), granulocyte (Gran), hemoglobin (HGB), and platelet (PLT) levels in the blood of the pre-metastasis mouse models at 15 days after different treatments. Dotted lines indicated the normal range. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
Furthermore, routine blood examination demonstrated that hematological indexes had obvious alternations at 15 days after different treatments (Fig. 8C). The tumor-bearing mouse who received PBS and Lipo treatment revealed significantly elevated WBC, lymphocytes (Lymph), mononuclear leucocytes (Mon), granulocytes (Gran), and reduced platelets (PLT)81 compared with the Blank (normal) group. Both PL-Lipo and PH-Lipo treatments significantly decreased WBC, Lymph, and Mon to the normal fluctuation range compared with PBS treatment or Lipo treatment but had no damage to major organs (Supporting Information Fig. S13). Moreover, compared with PBS treatment, PH-Lipo treatment could significantly decrease Gan and hemoglobin (HGB) levels, and PL-Lipo treatment could significantly decrease Gan level and increase PLT level. Of note, chronic inflammation may lead to alterations in epithelial cytoarchitecture and enhance genetic mutations along with epigenetic aberrations of epithelial cells82. This alteration in tissue homeostasis will induce a feed-forward chronic inflammatory response, firing up the metastasis of the tumor. Hence, the obtained nanoplateletsomes could suppress the inflammation, and reduce platelet depletion during CTC colonization. In general, P-Lipo treatment could inhibit the inflammation by upregulating M2 macrophages and decreasing WBC, Mon, Lymph, and Gran, suppress tumor cell escape by downregulating MDSC and decreasing the platelet depletion, resulting in decreased CTC survival in the blood and suppressed CTC colonization and metastasis formation. The potential anti-metastasis efficacy could be due to the following reasons. Firstly, P-Lipo takes advantage of both platelet membranes and artificial lipid membranes. It could not only have functional cell surface receptors endowed from natural platelets to capture CTCs, but also have long circulation property similar to PEGylated liposomes, which could provide P-Lipo more chance to capture CTCs or access pre-metastasis. Secondly, compared with natural platelets or platelet decoy, P-Lipo has a large surface-volume ratio and a large quantity. It was estimated that the surface-volume ratio and the number of P-Lipo were 1.92 × 107 and 1.47 × 105 folds those of platelets that were used for P-Lipo preparation, respectively (Supporting Information discussion). P-Lipo would have more opportunities to encase CTCs than natural platelets while not offering a shield against the immune cells like natural platelets. Thus, P-Lipo could compete with natural platelets in vivo, disrupting the platelet-CTC interaction, acting as reversible inhibitors of platelet activation cascades, and breaking the protection of platelets to cancer cells. Finally, the subversion of the host immune responses was prevented by P-Lipo, leading to an outstanding inhibition effect toward the generation of premetastatic tumor cascade.
4. Conclusion
We developed a bioinspired nanoplateletsome, which inherited the function of platelet membrane proteins. It has been demonstrated that these nanoplateletsomes could not only bind with CTCs in vitro and in vivo, but also interfere with CTC arrest by vessel endothelial cells, CTC extravasation through endothelial layers, and EMT of tumor cells as decoys in vitro. Furthermore, in vivo data demonstrated that nanoplateletsomes could decrease CTC survival in the blood, counteract metastasis formation, and slow down the metastasis progress by inhibiting the inflammation and suppressing CTC escape. Therefore, nanoplateletsomes without any drug loading provide a novel strategy for the treatment of tumor metastasis.
Acknowledgements
This project was supported by the National Natural Science Foundation of China (81773283). We thank the staff members of the Electron Microscopy System and Integrated Laser Microscopy System at the National Facility for Protein Science in Shanghai (NFPS) for providing technical support and assistance in data collection and analysis.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.01.005.
Contributor Information
Jianxin Wang, Email: jxwang@fudan.edu.cn.
Zhiqing Pang, Email: zqpang@fudan.edu.cn.
Author contributions
Longlong Zhang and Yuefei Zhu conducted the experiments and wrote the manuscript with equal contribution. Xunbin Wei and Yongzhuo Huang provided equipment and technical support. Ying Zhu was responsible for Western blot. Yang Li and Jiaxuan Xia participated in immunohistochemistry and flow cytometry. Yiheng Huang took part in the in vivo experiments. Jianxin Wang and Zhiqing Pang designed the research plan, analyzed the data and revised the manuscript. All of the authors have read and approved the final manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Multimedia component 1
References
- 1.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Cristofanilli M., Budd G.T., Ellis M.J., Stopeck A., Matera J., Miller M.C., et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 3.Aceto N., Bardia A., Miyamoto D.T., Donaldson M.C., Wittner B.S., Spencer J.A., et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson R.L., Balasas T., Callaghan J., Coombes R.C., Evans J., Hall J.A., et al. A framework for the development of effective anti-metastatic agents. Nat Rev Clin Oncol. 2019;16:185–204. doi: 10.1038/s41571-018-0134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poudineh M., Sargent E.H., Pantel K., Kelley S.O. Profiling circulating tumour cells and other biomarkers of invasive cancers. Nat Biomed Eng. 2018;2:72–84. doi: 10.1038/s41551-018-0190-5. [DOI] [PubMed] [Google Scholar]
- 6.Tang Y., Qiao G., Xu E., Xuan Y., Liao M., Yin G. Biomarkers for early diagnosis, prognosis, prediction, and recurrence monitoring of non-small cell lung cancer. OncoTargets Ther. 2017;10:4527–4534. doi: 10.2147/OTT.S142149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keller L., Pantel K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat Rev Cancer. 2019;19:553–567. doi: 10.1038/s41568-019-0180-2. [DOI] [PubMed] [Google Scholar]
- 8.Habli Z., AlChamaa W., Saab R., Kadara H., Khraiche M.L. Circulating tumor cell detection technologies and clinical utility: challenges and opportunities. Cancers (Basel) 2020;12:1390–1419. doi: 10.3390/cancers12071930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marquette C.H., Boutros J., Benzaquen J., Ferreira M., Pastre J., Pison C., et al. Circulating tumour cells as a potential biomarker for lung cancer screening: a prospective cohort study. Lancet Respir Med. 2020;8:709–716. doi: 10.1016/S2213-2600(20)30081-3. [DOI] [PubMed] [Google Scholar]
- 10.Castro-Giner F., Aceto N. Tracking cancer progression: from circulating tumor cells to metastasis. Genome Med. 2020;12:31–44. doi: 10.1186/s13073-020-00728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu W., Hu C., Gao H. Advances of nanomedicines in breast cancer metastasis treatment targeting different metastatic stages. Adv Drug Deliv Rev. 2021:113909–113933. doi: 10.1016/j.addr.2021.113909. [DOI] [PubMed] [Google Scholar]
- 12.Schlesinger M. Role of platelets and platelet receptors in cancer metastasis. J Hematol Oncol. 2018;11:125–139. doi: 10.1186/s13045-018-0669-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akpe V., Kim T.H., Brown C.L. Circulating tumour cells: a broad perspective. J R Soc Interface. 2020;17:20200065–20200085. [Google Scholar]
- 14.Kapeleris J., Kulasinghe A., Warkiani M.E., Vela I., Kenny L., O'Byrne K., et al. The prognostic role of circulating tumor cells (CTCs) in lung cancer. Front Oncol. 2018;8:311–323. doi: 10.3389/fonc.2018.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gay L.J., Felding-Habermann B. Contribution of platelets to tumour metastasis. Nat Rev Cancer. 2011;11:123–134. doi: 10.1038/nrc3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaertner F., Massberg S. Patrolling the vascular borders: platelets in immunity to infection and cancer. Nat Rev Immunol. 2019;19:747–760. doi: 10.1038/s41577-019-0202-z. [DOI] [PubMed] [Google Scholar]
- 17.Li N. Platelets in cancer metastasis: to help the “villain” to do evil. Int J Cancer. 2016;138:2078–2087. doi: 10.1002/ijc.29847. [DOI] [PubMed] [Google Scholar]
- 18.Kanikarla-Marie P., Lam M., Menter D.G., Kopetz S. Platelets, circulating tumor cells, and the circulome. Cancer Metastasis Rev. 2017;36:235–248. doi: 10.1007/s10555-017-9681-1. [DOI] [PubMed] [Google Scholar]
- 19.Papa A.-L., Jiang A., Korin N., Chen M.B., Langan E.T., Waterhouse A., et al. Platelet decoys inhibit thrombosis and prevent metastatic tumor formation in preclinical models. Sci Transl Med. 2019;11:5898–5910. doi: 10.1126/scitranslmed.aau5898. [DOI] [PubMed] [Google Scholar]
- 20.Pasto A., Giordano F., Evangelopoulos M., Amadori A., Tasciotti E. Cell membrane protein functionalization of nanoparticles as a new tumor-targeting strategy. Clin Transl Med. 2019;8:8–16. doi: 10.1186/s40169-019-0224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan H., Shao D., Lao Y.H., Li M., Hu H., Leong K.W. Engineering cell membrane-based nanotherapeutics to target inflammation. Adv Sci. 2019;6:1900605–1900627. doi: 10.1002/advs.201900605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhen X., Cheng P., Pu K. Recent advances in cell membrane-camouflaged nanoparticles for cancer phototherapy. Small. 2019;15:1804105–1804124. doi: 10.1002/smll.201804105. [DOI] [PubMed] [Google Scholar]
- 23.Thanuja M.Y., Anupama C., Ranganath S.H. Bioengineered cellular and cell membrane-derived vehicles for actively targeted drug delivery: so near and yet so far. Adv Drug Deliv Rev. 2018;132:57–80. doi: 10.1016/j.addr.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Jin K., Luo Z., Zhang B., Pang Z. Biomimetic nanoparticles for inflammation targeting. Acta Pharm Sin B. 2018;8:23–33. doi: 10.1016/j.apsb.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X., Zhong X., Li C. Challenges in cell membrane-camouflaged drug delivery systems: development strategies and future prospects. Chin Chem Lett. 2021;32:2347–2358. [Google Scholar]
- 26.Le Q.V., Lee J., Lee H., Shim G., Oh Y.K. Cell membrane-derived vesicles for delivery of therapeutic agents. Acta Pharm Sin B. 2021;11:2096–2113. doi: 10.1016/j.apsb.2021.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han X., Shen S., Fan Q., Chen G., Archibong E., Dotti G., et al. Red blood cell-derived nanoerythrosome for antigen delivery with enhanced cancer immunotherapy. Sci Adv. 2019;5:6870–6879. doi: 10.1126/sciadv.aaw6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y., Li R., Li H., Zhang S., Dai W., Wu Q., et al. Erythroliposomes: integrated hybrid nanovesicles composed of erythrocyte membranes and artificial lipid membranes for pore-forming toxin clearance. ACS Nano. 2019;13:4148–4159. doi: 10.1021/acsnano.8b08964. [DOI] [PubMed] [Google Scholar]
- 29.Molinaro R., Pasto A., Corbo C., Taraballi F., Giordano F., Martinez J.O., et al. Macrophage-derived nanovesicles exert intrinsic anti-inflammatory properties and prolong survival in sepsis through a direct interaction with macrophages. Nanoscale. 2019;11:13576–13586. doi: 10.1039/c9nr04253a. [DOI] [PubMed] [Google Scholar]
- 30.Deng G., Sun Z., Li S., Peng X., Li W., Zhou L., et al. Cell-membrane immunotherapy based on natural killer cell membrane coated nanoparticles for the effective inhibition of primary and abscopal tumor growth. ACS Nano. 2018;12:12096–12108. doi: 10.1021/acsnano.8b05292. [DOI] [PubMed] [Google Scholar]
- 31.Wang H., Wu J., Williams G.R., Fan Q., Niu S., Wu J., et al. Platelet-membrane-biomimetic nanoparticles for targeted antitumor drug delivery. J Nanobiotechnol. 2019;17:60–75. doi: 10.1186/s12951-019-0494-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang M., Ye J.J., Xia Y., Wang Z.Y., Li C.X., Wang X.S., et al. Platelet-mimicking biotaxis targeting vasculature-disrupted tumors for cascade amplification of hypoxia-sensitive therapy. ACS Nano. 2019;13:14230–41240. doi: 10.1021/acsnano.9b07330. [DOI] [PubMed] [Google Scholar]
- 33.Ding K., Zheng C., Sun L., Liu X., Yin Y., Wang L. NIR light-induced tumor phototherapy using ICG delivery system based on platelet-membrane-camouflaged hollow bismuth selenide nanoparticles. Chin Chem Lett. 2020;31:1168–7112. [Google Scholar]
- 34.Kumar S., Han J.A., Michael I.J., Ki D., Sunkara V., Park J., et al. Human platelet membrane functionalized microchips with plasmonic codes for cancer detection. Adv Funct Mater. 2019:1902669–1902675. [Google Scholar]
- 35.Teng Y., Ren Y., Sayed M., Hu X., Lei C., Kumar A., et al. Plant-derived exosomal microRNAs Shape the gut microbiota. Cell Host Microbe. 2018;24:637–652. doi: 10.1016/j.chom.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J., Ai Y., Wang L., Bu P., Sharkey C.C., Wu Q., et al. Targeted drug delivery to circulating tumor cells via platelet membrane-functionalized particles. Biomaterials. 2016;76:52–65. doi: 10.1016/j.biomaterials.2015.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Q., Sun W., Qian C., Wang C., Bomba H.N., Gu Z. Anticancer platelet-mimicking nanovehicles. Adv Mater. 2015;27:7043–7050. doi: 10.1002/adma.201503323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu H., Jin F., Liu D., Shu G., Wang X., Qi J., et al. ROS-responsive nano-drug delivery system combining mitochondria-targeting ceria nanoparticles with atorvastatin for acute kidney injury. Theranostics. 2020;10:2342–2357. doi: 10.7150/thno.40395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao L., Meng Q.-F., Huang Q., Wang Z., Yu G.-T., Li A., et al. Platelet–leukocyte hybrid membrane-coated immunomagnetic beads for highly efficient and highly specific isolation of circulating tumor cells. Adv Funct Mater. 2018;28:1803531–1803540. [Google Scholar]
- 40.Luk B. UC San Diego; 2016. Cell membrane-cloaked nanoparticles for targeted therapeutics.https://escholarship.org/uc/item/1894m1cb Available from: [Google Scholar]
- 41.Hu C.M., Fang R.H., Wang K.C., Luk B.T., Thamphiwatana S., Dehaini D., et al. Nanoparticle biointerfacing by platelet membrane cloaking. Nature. 2015;526:118–121. doi: 10.1038/nature15373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato Y.T., Umezaki K., Sawada S., Mukai S.A., Sasaki Y., Harada N., et al. Engineering hybrid exosomes by membrane fusion with liposomes. Sci Rep. 2016;6:21933–21944. doi: 10.1038/srep21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molinaro R., Corbo C., Martinez J.O., Taraballi F., Evangelopoulos M., Minardi S., et al. Biomimetic proteolipid vesicles for targeting inflamed tissues. Nat Mater. 2016;15:1037–1046. doi: 10.1038/nmat4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H. Thin-film hydration followed by extrusion method for liposome preparation. Methods Mol Biol. 2017;1522:17–22. doi: 10.1007/978-1-4939-6591-5_2. [DOI] [PubMed] [Google Scholar]
- 45.Pang Z., Lu W., Gao H., Hu K., Chen J., Zhang C., et al. Preparation and brain delivery property of biodegradable polymersomes conjugated with OX26. J Control Release. 2008;128:120–127. doi: 10.1016/j.jconrel.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 46.Song Y., Zhang N., Li Q., Chen J., Wang Q., Yang H., et al. Biomimetic liposomes hybrid with platelet membranes for targeted therapy of atherosclerosis. Chem Eng J. 2021;408:127296–127312. [Google Scholar]
- 47.Wang X., Huang H., Zhang L., Bai Y., Chen H. PCM and TAT co-modified liposome with improved myocardium delivery: in vitro and in vivo evaluations. Drug Deliv. 2017;24:339–345. doi: 10.1080/10717544.2016.1253121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banerjee R., Purkayastha P. Revival of the nearly extinct fluorescence of coumarin 6 in water and complete transfer of energy to rhodamine 123. Soft Matter. 2017;13:5506–5508. doi: 10.1039/c7sm01198a. [DOI] [PubMed] [Google Scholar]
- 49.Borsig L., Wong R., Feramisco J., Nadeau D.R., Varki N.M., Varki A. Heparin and cancer revisited: mechanistic connections involving platelets, P-selectin, carcinoma mucins, and tumor metastasis. Proc Natl Acad Sci U S A. 2001;98:3352–3357. doi: 10.1073/pnas.061615598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walther A., Czabanka M., Gebhard M.M., Martin E. Glycoprotein IIB/IIIA-inhibition and microcirculatory alterations during experimental endotoxemia—an intravital microscopic study in the rat. Microcirculation. 2004;11:79–88. doi: 10.1080/10739680490266216. [DOI] [PubMed] [Google Scholar]
- 51.Rashid O.M., Nagahashi M., Ramachandran S., Dumur C.I., Schaum J.C., Yamada A., et al. Is tail vein injection a relevant breast cancer lung metastasis model? J Thorac Dis. 2013;5:385–392. doi: 10.3978/j.issn.2072-1439.2013.06.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang T., Zhu Q., Wei D., Feng J., Yao J., Jiang T., et al. Nanoparticles coated with neutrophil membranes can effectively treat cancer metastasis. ACS Nano. 2017;11:1397–1411. doi: 10.1021/acsnano.6b06477. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y., Zhu X., Chen X., Chen Q., Zhou W., Guo Q., et al. Activated platelets-targeting micelles with controlled drug release for effective treatment of primary and metastatic triple negative breast cancer. Adv Funct Mater. 2019;29:1806620–1806635. [Google Scholar]
- 54.Nedosekin D.A., Verkhusha V.V., Melerzanov A.V., Zharov V.P., Galanzha E.I. In vivo photoswitchable flow cytometry for direct tracking of single circulating tumor cells. Chem Biol. 2014;21:792–801. doi: 10.1016/j.chembiol.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Justus C.R., Leffler N., Ruiz-Echevarria M., Yang L.V. In vitro cell migration and invasion assays. J Vis Exp. 2014:51046–51053. doi: 10.3791/51046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parodi A., Quattrocchi N., van de Ven A.L., Chiappini C., Evangelopoulos M., Martinez J.O., et al. Synthetic nanoparticles functionalized with biomimetic leukocyte membranes possess cell-like functions. Nat Nanotechnol. 2013;8:61–68. doi: 10.1038/nnano.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu W., Wu B., Fu L., Chen J., Wang Z., Huang F., et al. Comparison of three different methods for the detection of circulating tumor cells in mice with lung metastasis. Oncol Rep. 2017;37:3219–3226. doi: 10.3892/or.2017.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warren J.S.A., Feustel P.J., Lamar J.M. Combined use of tail vein metastasis assays and real-time in vivo imaging to quantify breast cancer metastatic colonization and burden in the lungs. J Vis Exp. 2019:60687–60701. doi: 10.3791/60687. [DOI] [PubMed] [Google Scholar]
- 59.van den Bijgaart R.J., Kong N., Maynard C., Plaks V. Ex vivo live imaging of lung metastasis and their microenvironment. J Vis Exp. 2016:53741–53754. doi: 10.3791/53741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Menitove J.E., Frenzke M., Aster R.H. Use of prostacyclin to inhibit activation of platelets during preparation of platelet concentrates. Transfusion. 1984;24:528–531. doi: 10.1046/j.1537-2995.1984.24685066818.x. [DOI] [PubMed] [Google Scholar]
- 61.Hawker R.J., Turner V.S., Mitchell S.G. Use of prostaglandin E1 during preparation of platelet concentrates. Transfus Med. 1996;6:249–254. doi: 10.1111/j.1365-3148.1996.tb00076.x. [DOI] [PubMed] [Google Scholar]
- 62.Ben-Akiva E., Meyer R.A., Yu H., Smith J.T., Pardoll D.M., Green J.J. Biomimetic anisotropic polymeric nanoparticles coated with red blood cell membranes for enhanced circulation and toxin removal. Sci Adv. 2020;6:9035–9043. doi: 10.1126/sciadv.aay9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fox C.B., Mulligan S.K., Sung J., Dowling Q.M., Fung H.W., Vedvick T.S., et al. Cryogenic transmission electron microscopy of recombinant tuberculosis vaccine antigen with anionic liposomes reveals formation of flattened liposomes. Int J Nanomed. 2014;9:1367–1377. doi: 10.2147/IJN.S56582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zelena A., Isbaner S., Ruhlandt D., Chizhik A., Cassini C., Klymchenko A.S., et al. Time-resolved MIET measurements of blood platelet spreading and adhesion. Nanoscale. 2020;12:21306–21315. doi: 10.1039/d0nr05611a. [DOI] [PubMed] [Google Scholar]
- 65.Piao J.G., Wang L., Gao F., You Y.Z., Xiong Y., Yang L. Erythrocyte membrane is an alternative coating to polyethylene glycol for prolonging the circulation lifetime of gold nanocages for photothermal therapy. ACS Nano. 2014;8:10414–10425. doi: 10.1021/nn503779d. [DOI] [PubMed] [Google Scholar]
- 66.Molinaro R., Evangelopoulos M., Hoffman J.R., Corbo C., Taraballi F., Martinez J.O., et al. Design and development of biomimetic nanovesicles using a microfluidic approach. Adv Mater. 2018;30:1702749–1702757. doi: 10.1002/adma.201702749. [DOI] [PubMed] [Google Scholar]
- 67.Qureshi A.H., Chaoji V., Maiguel D., Faridi M.H., Barth C.J., Salem S.M., et al. Proteomic and phospho-proteomic profile of human platelets in basal, resting state: insights into integrin signaling. PLoS One. 2009;4:7627–7643. doi: 10.1371/journal.pone.0007627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kassassir H., Karolczak K., Siewiera K.M., Wojkowska D.W., Braun M., Watala C.W. Time-dependent interactions of blood platelets and cancer cells, accompanied by extramedullary hematopoiesis, lead to increased platelet activation and reactivity in a mouse orthotopic model of breast cancer— implications for pulmonary and liver metastasis. Aging. 2020;12:5091–5120. doi: 10.18632/aging.102933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stefanini L., Bergmeier W. RAP GTPases and platelet integrin signaling. Platelets. 2019;30:41–47. doi: 10.1080/09537104.2018.1476681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bennett J.S. Structure and function of the platelet integrin alphaIIbbeta3. J Clin Invest. 2005;115:3363–3369. doi: 10.1172/JCI26989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bianchi E., Norfo R., Pennucci V., Zini R., Manfredini R. Genomic landscape of megakaryopoiesis and platelet function defects. Blood. 2016;127:1249–1259. doi: 10.1182/blood-2015-07-607952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qi C., Li B., Guo S., Wei B., Shao C., Li J., et al. P-Selectin-mediated adhesion between platelets and tumor cells promotes intestinal tumorigenesis in Apcmin/+ mice. Int J Biol Sci. 2015;11:679–687. doi: 10.7150/ijbs.11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mellor P., Harvey J.R., Murphy K.J., Pye D., O'Boyle G., Lennard T.W., et al. Modulatory effects of heparin and short-length oligosaccharides of heparin on the metastasis and growth of LMD MDA-MB 231 breast cancer cells in vivo. Br J Cancer. 2007;97:761–768. doi: 10.1038/sj.bjc.6603928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stevenson J.L., Varki A., Borsig L. Heparin attenuates metastasis mainly due to inhibition of P- and L-selectin, but non-anticoagulant heparins can have additional effects. Thromb Res. 2007;120:107–111. doi: 10.1016/S0049-3848(07)70138-X. [DOI] [PubMed] [Google Scholar]
- 75.Manne B.K., Getz T.M., Hughes C.E., Alshehri O., Dangelmaier C., Naik U.P., et al. Fucoidan is a novel platelet agonist for the C-type lectin-like receptor 2 (CLEC-2) J Biol Chem. 2013;288:7717–7726. doi: 10.1074/jbc.M112.424473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuo C.W., Chueh D.Y., Chen P. Real-time in vivo imaging of subpopulations of circulating tumor cells using antibody conjugated quantum dots. J Nanobiotechnol. 2019;17:26–36. doi: 10.1186/s12951-019-0453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heeke S., Mograbi B., Alix-Panabieres C., Hofman P. Never travel alone: the crosstalk of circulating tumor cells and the blood microenvironment. Cells. 2019;8:714–725. doi: 10.3390/cells8070714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takemoto A., Okitaka M., Takagi S., Takami M., Sato S., Nishio M., et al. A critical role of platelet TGF-beta release in podoplanin-mediated tumour invasion and metastasis. Sci Rep. 2017;7:42186–42197. doi: 10.1038/srep42186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang K.K., Yoon C., Yi B.C., Tap W.D., Simon M.C., Yoon S.S. Platelet-derived growth factor receptor-alpha and -beta promote cancer stem cell phenotypes in sarcomas. Oncogenesis. 2018;7:47–59. doi: 10.1038/s41389-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Malek E., de Lima M., Letterio J.J., Kim B.G., Finke J.H., Driscoll J.J., et al. Myeloid-derived suppressor cells: the green light for myeloma immune escape. Blood Rev. 2016;30:341–348. doi: 10.1016/j.blre.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Næser E., Møller H., Fredberg U., Frystyk J., Vedsted P. Routine blood tests and probability of cancer in patients referred with non-specific serious symptoms: a cohort study. BMC Cancer. 2017;17:817–827. doi: 10.1186/s12885-017-3845-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu Y.R., Hotten D.F., Malakhau Y., Volker E., Ghio A.J., Noble P.W., et al. Flow cytometric analysis of myeloid cells in human blood, bronchoalveolar lavage, and lung tissues. Am J Respir Cell Mol Biol. 2016;54:13–24. doi: 10.1165/rcmb.2015-0146OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1