Abstract
We investigated the potential role of sleep-trait associated genetic loci in conferring a degree of their effect via pancreatic α- and β-cells, given that both sleep disturbances and metabolic disorders, including type 2 diabetes and obesity, involve polygenic contributions and complex interactions. We determined genetic commonalities between sleep and metabolic disorders, conducting linkage disequilibrium genetic correlation analyses with publicly available GWAS summary statistics. Then we investigated possible enrichment of sleep-trait associated SNPs in promoter-interacting open chromatin regions within α- and β-cells, intersecting public GWAS reports with our own ATAC-seq and high-resolution promoter-focused Capture C data generated from both sorted human α-cells and an established human beta-cell line (EndoC-βH1). Finally, we identified putative effector genes physically interacting with sleep-trait associated variants in α- and EndoC-βH1cells running variant-to-gene mapping and establish pathways in which these genes are significantly involved. We observed that insomnia, short and long sleep—but not morningness—were significantly correlated with type 2 diabetes, obesity and other metabolic traits. Both the EndoC-βH1 and α-cells were enriched for insomnia loci (p = .01; p = .0076), short sleep loci (p = .017; p = .022) and morningness loci (p = 2.2 × 10−7; p = .0016), while the α-cells were also enriched for long sleep loci (p = .034). Utilizing our promoter contact data, we identified 63 putative effector genes in EndoC-βH1 and 76 putative effector genes in α-cells, with these genes showing significant enrichment for organonitrogen and organophosphate biosynthesis, phosphatidylinositol and phosphorylation, intracellular transport and signaling, stress responses and cell differentiation. Our data suggest that a subset of sleep-related loci confer their effects via cells in pancreatic islets.
Keywords: sleep, pancreas, alpha cell, beta cell, chromatin conformation capture, epigenetics, metabolism, GWAS
Statement of Significance.
Altered glucose metabolism observed in sleep dysregulation contributes to obesity and type 2 diabetes risk. We investigated if known sleep genetic loci exert any effect via pancreatic alpha and beta-cells. Correlation analyses with GWAS data revealed genetic commonalities between sleep and metabolic disorders. We then intersected GWAS with 3D genomic data to assess enrichment of sleep trait SNPs within promoter-interacting open chromatin regions in the human beta-cell line, EndoC-BH1, and sorted human alpha-cells. EndoC-BH1 and alpha-cells were enriched for chronotype, insomnia and short sleep loci, along with long sleep for just the alpha-cells. Subsequent ‘variant-to-gene mapping’ implicated effector genes for these sleep traits. Our data suggest that a subset of sleep-related genetic loci confer their effects via pancreatic islet cells.
Introduction
Sleep is a phylogenetic conserved state of reversible quiescence essential to many biological functions including metabolism [1]. There is a large body of evidence linking poor sleep to metabolic disorders, such as type 2 diabetes (T2D), obesity, and insulin resistance [2]. One hypothesis behind this link is that poor sleep promotes metabolic dysregulation and immune responses, triggering appetite disruption and the onset of metabolic diseases [3].
In addition to metabolic traits impacting sleep patterns, insomnia is conversely gaining recognition as a key risk factor for T2D; indeed, people with insomnia are 28% more likely to present with T2D than those without insomnia symptoms [4], whereas 50% of individuals with T2D suffer from insomnia [5]. Furthermore, among individuals reporting short sleep (<7 hours per night), there is a high prevalence of higher body mass index (BMI) and obesity [6]. A bidirectional study on possible associations of actigraphic-assessed sleep patterns (total sleep time, sleep efficiency, sleep onset latency, and wake after sleep onset) with BMI and waist circumference (WC) showed that poor sleep was associated with higher BMI/WC and, conversely higher BMI/WC was associated with decreased sleep duration [7]. Moreover, induced sleep deprivation in healthy individuals leads to changes in glucose metabolism, i.e. increased glucose release and insulin resistance, which can be reversed by restoring normal sleep cycles [8]. Sleep restriction also alters the release of hormones such as leptin and ghrelin and stimulates appetite, therefore promoting the intake of high-calorie food and weight gain [2].
There is a strong evidence for a genetic component to the pathogenesis of sleep-related traits. Recent GWAS efforts highlight the complex polygenicity of sleep traits; 248 significant and independent signals have been reported for insomnia [9], 351 for morningness (preference to wake up very early in the morning and being more active in the daytime) [10], 27 for short sleep (< 7 hours per 24 hours) [11] and 8 for long sleep (≥ 9 hours per 24 hours) [11]. Two-sample Mendelian Randomization (MR) analyses use GWAS summary statistic to infer statistically causal relations between risk factors and disease outcomes [12], estimating if genetic risk variants for a trait might predispose to a second trait. Several MR studies have shown that the underlying genetics of insomnia can be a predisposing factor for the onset of T2D [8, 9, 13] or for cardiovascular disease [14]. However, MR results on sleep duration have shown no clear significant causal relationships with T2D, BMI or cardiovascular diseases [11, 15, 16]. MR studies of short sleep and metabolic traits are mixed, some showing that short sleep genetic is a predisposing factor for T2D [13] and cardiovascular disease [8], whereas others show that short sleep is not causal for glycemic-related traits, such as fasting glucose and insulin [16]. Finally, one MR study showed that sleep genetics is linked to human metabolism [17]; indeed, patients with insomnia showed a different global metabolic profile when compared to healthy controls, with elevated amino acid and energy metabolites suggesting that insomnia is associated with metabolic dysregulation [18].
Despite increasing evidence, the physiopathology underpinning the association between poor sleep and metabolic disorders remains unclear. Going beyond the traditional research efforts in brain regions, the pancreatic islet represents an additional tissue worth exploring further. The islets of Langerhans within the pancreas play a major role in glucose homeostasis and metabolism through the secretion of hormones in to the blood stream. α-cells release glucagon while β-cells release insulin, δ-cells produce somatostatin to inhibit the release of both glucagon and insulin, and γ-cells release pancreatic polypeptide to modulate overall pancreatic secretory activity [19].
Given the clinical and genetic associations between sleep and metabolic disorders, and the functional role of pancreatic islets in the pathogenesis of T2D, we investigated a potential role for genetic variants associated with sleep traits in the context of endocrine pancreatic islets. We leveraged both public GWAS reports and our own genomic data. We considered four different sleep traits, namely insomnia, morningness, short, and long sleep. Using published GWAS summary statistics, we calculated the genetic correlation between these sleep phenotypes and metabolic traits, including T2D and obesity. We generated ATAC-seq and promoter-focused Capture C libraries from α-cells isolated from human pancreatic islet samples and from an established model for human β-cells (EndoC-βH1) [20]. We then intersected the GWAS data with these in-house generated genomic datasets to investigate potential enrichment of sleep-trait-associated loci within open chromatin regions of α-cells and of EndoC-βH1 via partitioned linkage disequilibrium score regression analyses (LDSR). We subsequently performed a physical variant to gene mapping effort to implicate putative functional genes at GWAS sleep-trait-associated loci in these two cell types. Finally, we assessed the putative functions of the newly implicated effector genes by running enrichment pathways analyses.
Methods
Genetic correlation analyses
We performed Linkage Disequilibrium Score Regression analyses (LDSC) to calculate genetic correlations between pairs of traits using LD Score Regression v1.0.0 (https://github.com/bulik/ldsc). The 1000 Genome project phase 3 were used to estimate the LD structure for European populations. We tested insomnia, morningness, short and long sleep against metabolism-related traits (Supplementary Table S1). Quality control of the inputted summary statistics and heritability analysis were automatically performed. We adjusted the p-values for multiple comparisons using the Benjamini–Hochberg false discovery rate (FDR) method and considered two traits to be significantly correlated with an adjusted p-value < .05. The genetic correlations were not biased by sample overlap [21].
Partitioned LD score regression
Partitioned LD score regression estimates heritability from GWAS summary statistics within a subset of regions of the genome after accounting for LD. Partitioned heritability was measured using LD Score Regression v1.0.0 (https://github.com/bulik/ldsc) to identify enrichment of GWAS signals among cis-regulatory elements (cREs) in α-cells and in EndoC-βH1 as previously performed [22]. Briefly, the annotation and heritability estimates for α-cells and for EndoC-βH1 were generated using bed files containing the position of the cRE (OCRs located proximal to a promoter (−1500/+500bp of TSS) + OCRs located within overlapping regions with ±500 bp buffers. hESC-derived hypothalamic-like neurons (HNs) were used as positive control, given the recognized role of the hypothalamus in both circadian rhythms and metabolism [23]. We used the annotation and heritability estimates for HNs had been previously generated by our team [22]. We tested insomnia, morningness, short and long sleep. The enrichment within the α-cells, EndoC-βH1 and HNs were compared to the baseline model for the EUR ancestry, downloaded from https://github.com/bulik/ldsc/wiki/Partitioned-Heritability. The results were visualized as bubble plots using ggplot2, with circle size representing fold enrichment of cREs compared to the base annotation and the color indicating the statistical significance (−log (p-value)). We tested insomnia [9], morningness [10], short and long sleep [11] in this context.
Cell models
Pancreata from deceased organ donors were obtained by Human Pancreas Analysis Program (HPAP) (https://hpap.pmacs.upenn.edu), a Human Islet Research Network consortium. α-Cells were isolated based on the protocol originally published by Dr. Grompe’s Llaboratory [24], and modified as described here (https://hpap.pmacs.upenn.edu/explore/workflow/islet-molecular-phenotyping-studies?protocol=2).
EndoC-βH1(20) were purchased from Univercell Biosolutions and cultured following manufacturer instructions, cells were kept in Matrigel (100 µg/mL)-fibronectin (2 µg/mL)-coated wells and fed with DMEM enriched with 5.6 mM glucose, 2% BSA fraction V (Roche Diagnostics), 50 μM 2-mercaptoethanol, 10 mM nicotinamide (Calbiochem), 5.5 μg/mL transferrin (Sigma–Aldrich), 6.7 ng/mL selenite (Sigma–Aldrich), 100 U/mL penicillin, and 100 μg/mL streptomycin. Passages were performed when confluence was observed. From passage 14 onwards, a 2/3 dilution passage was performed every week.
HNs were previously generated at Columbia University [25].
ATAC-seq
Three technical replicates of ATAC-seq libraries for the EndoC-βH1 cell-line were generated and analyzed using an established protocol [22, 25]. Briefly 50 000 α-cells or EndoC-βH1 were collected, washed in PBS and pelleted for 5 min at 550 rcf at 4°C. The pellet was resuspended in 50 µL of chilled ATAC-seq lysis buffer (10 mM Tris–HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630 (NP-40)) and centrifuged again for 10 min at 550 rcf at 4°C. The pelleted nuclei were tagged using Tn5 Transposase (Illumina, Cat #FC-121–1030) by incubating for 45 min at 37°C. The DNA was purified using the MinElute Kit (Qiagen, Cat #28004) and eluted in 10.5 µL elution buffer. Purified tagged DNA fragments were PCR-amplified using Nextera primers and NEB-Next High-Fidelity PCR Master Mix (New England Labs, Cat #M0541). The generated libraries were cleaned with 1.8X Agencourt AMPureXP beads (BeckmanCoulter, Cat # A63880) and quality was assessed using a bioanalyzer. The libraries were paired-end sequenced using the Illumina Novaseq 6000 platform and 50 bp reads. Open chromatin regions were called using the ENCODE ATAC-seq pipeline (https://github.com/kundajelab/atac_dnase_pipelines), selecting the resulting IDR optimal peaks (with all coordinates referring to hg19). We defined a genomic region as open if it had a 1-bp overlap with an ATAC-seq peak. ATAC-seq libraries for HNs were previously generated and analyzed in our lab [22, 25], whereas processed ATAC-seq data for α-cells were retrieved from the Diabetes Epigenome Atlas (accession number for replicates TSTFF310588, TSTFF447068, TSTFF559678) [26, 27].
Promoter-focused Capture C
We generated α-cells and EndoC-βH1 promoter-focused Capture C libraries as previously described [22, 25, 28]. Each generated library was sonicated using a QSonica Q800R to obtain 350 bp DNA fragments which were purified using AMPureXP beads (BeckmanCoulter, Cat # A63880) and measured via Qubit fluorometer. Fragments quality and sizes were assessed using a 1000 or HS DNA Chip on a Bioanalyzer 2100 (Agilent). DNA ends were repaired and adaptors were ligated using Agilent SureSelectXT Library Prep Kit (Agilent, G9611A). After a bead clean-up step, quality control of the DNA fragments was performed again on the Bioanalyzer 2100. A custom-designed capture probe set (Agilent) [28] was hybridized to the adaptor-ligated DNA fragments using the SureSelectXT capture kit (Agilent, 5190-4901) to generate high-complexity libraries, followed by a final QC on the Bioanalyzer 2100. Each captured library was paired-end sequenced on the Illumina Novaseq 6000 on S2 flow cells (100 bp read length for α-cells, 50 bp read length for EndoC-βH1 cells). Data were analyzed as previously described [22, 25, 28]. Promoter-focused Capture-C libraries from HNs were generated previously and analyzed in our lab [22, 25].
Genetic loci included in variant-to-gene mapping
We used 248, 351, 27 and 8 independent SNPs from published insomnia [9], morningness [10], short- and long-sleep [11] studies. To derive proxy SNPs, we used SNiPA with GRCh37 as human reference assembly; 1000 Genomes phase 1v3 as variant set; European population; Ensembl 87 for genome annotation and an LD threshold of r2 > 0.8. We intersected the genomic coordinates (hg19) of the proxy SNPs with accessible regions defined by ATAC-seq and promoter interacting regions defined by Promoter Focused Capture C with α-cells or EndoC-βH1 separately to pinpoint those sleep-associated open proxies intersecting with the promoters of putative effector genes operating in each of the pancreatic cell settings. We focused on open proxies that contacted open gene promoter regions with r2 > 0.8, as this value represents variants in strong linkage disequilibrium for each reported sentinel SNP.
Enrichment pathways analyses
Enrichment pathway analyses were performed using the “Compute Overlaps” tool provided by Gene Set Enrichment Analysis (GSEA) website (https://www.gsea-msigdb.org/gsea/index.jsp). Only genes with RPKM >1 in α-cells; and TPM >1 in EndoC-βH1 were considered. The Compute Overlaps tool first calculated the overlap of our selected genes with Molecular Signatures Database (MSigDB) annotated gene sets. Then it determined the statistical significance of the overlaps using a hypergeometric test. The p-values were corrected by the FDR method. We considered pathways with at least two overlapping genes and with an adjusted p < .05 as significant.
Data and resource availability
The datasets generated during and/or analyzed during the current study are not publicly available due to their utilization in two other parallel manuscripts in preparation but are available from the corresponding author upon reasonable request and will be made publicly available once all three studies are published.
Results
Genetic correlation analyses
We ran linkage disequilibrium score regression analyses (LDSC) to estimate the genetic correlation between insomnia, morningness, short and long sleep with T2D, BMI, obesity class 1 (BMI ≥30 kg/m2), obesity class 2 (BMI ≥35 kg/m2), obesity class 3 (BMI ≥40 kg/m2), extreme BMI, and extreme waist to hip ratio, overweight, fasting glucose, fasting insulin, fasting proinsulin, Homeostatic Model Assessment for Insulin Resistance (HOMA-IR) and ß-cell function (HOMA-b), glycated hemoglobin (HbA1c), waist circumference, waist circumference adjusted to BMI, hip circumference, hip circumference adjusted BMI and waist to hip ratio.
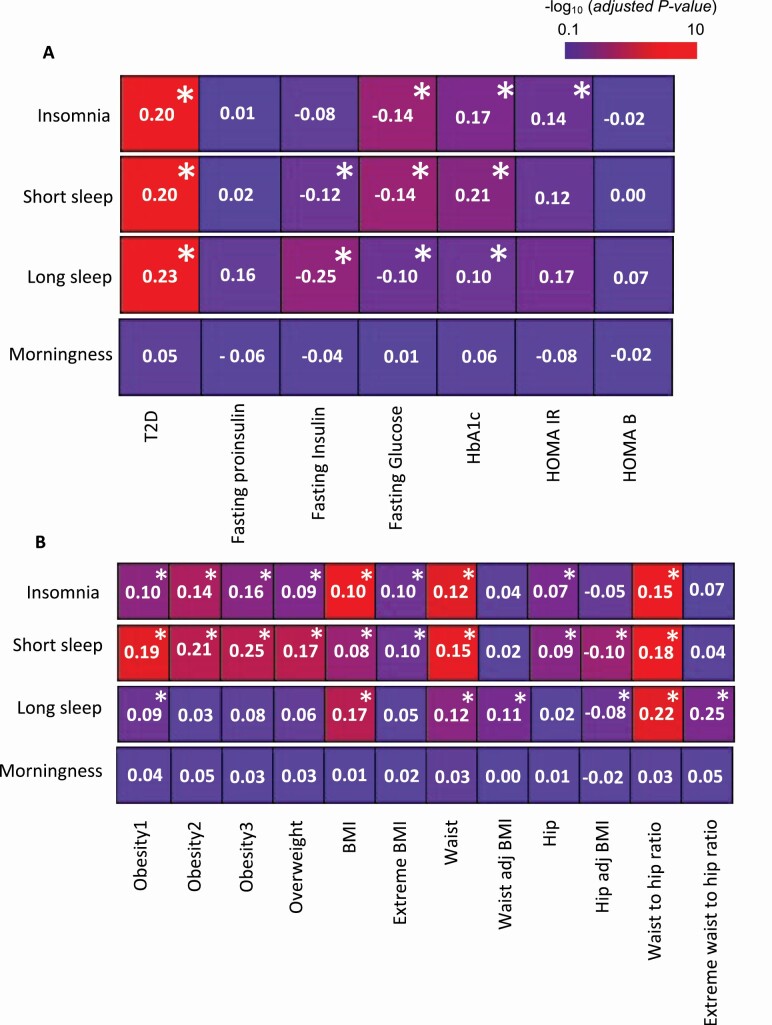
Insomnia, short and long sleep were significantly correlated with T2D (genetic correlation coefficient (rg) 0.20 and adjusted p =1.7 × 10−10, rg=0.2; padj = 1.2 × 10−9, rg = 0.22; padj = 2.6 × 10−8, respectively). As well as other metabolic traits commonly associated with T2D: fasting glucose (insomnia rg = −0.14; padj = 5.6 × 10−5, short sleep rg = −0.14; padj = 1.6 × 10−4, long sleep rg = −0.1; padj = 3.3 × 10−2) and Hba1c (insomnia rg = 0.17; padj = 2.6 × 10−3, short sleep rg = 0.21; padj = 1.2 × 10−3, long sleep rg = 0.10; padj = 4.1 × 10−2). Insomnia was also significantly correlated with HOMA-IR (rg = 0.14; padj = 2.4 × 10−2), while short and long sleep showed a significant correlation with fasting insulin (short sleep rg = −0.12; padj = 4.3 × 10−2, long sleep rg = −0.25; padj = 3.4 × 10−4) [Figure 1A, Supplementary Table S2].
Figure 1.
Genetic correlation estimates for sleep traits with T2D-related (A) or obesity- related (B) traits. Numbers indicate the genetic correlation coefficients (rg). Colors indicate the statistical significance (−log10 (adjusted p-value)) of the correlations. Asterisks (*) indicate adjusted p-value < .05.
Insomnia and short sleep showed similar correlations with nine different obesity-related traits, obesity class 1, class 2 and class 3, overweight, BMI and extreme BMI, waist and hip circumference and their ratio. Short sleep was also significantly correlated with hip-circumference adjusted for BMI [Figure 1B, Supplementary Table S3]. Long sleep was significantly correlated with seven obesity-related traits: obesity class 1, BMI, waist circumference, waist circumference adjusted for BMI, hip circumference adjusted for BMI, waist-to-hip ratio and extreme waist to hip circumference ratio adjusted for BMI [Figure 1B, Supplementary Table S3]. Finally, and in contrast, after correction for multiple comparisons, morningness revealed no significant correlation with any metabolic traits [Figure 1, Supplementary Table S2 and S3].
As such, the genetics of all sleep traits, with the notable exception of morningness, were significantly correlated with multiple metabolic traits. These results suggest that a portion of the genetic variation is shared between sleep duration and T2D associated metabolic traits. We then elected to investigate deeper these shared genetics potentially functioning in a pancreatic context by running partitioned LD score regression (LDSR) on 3D genomic datasets we generated on α−cells and EndoC-βH1 cells.
Genome-wide ATAC-seq and promoter focused Capture C
Three ATAC-seq libraries derived from EndoC-βH1 cells were generated, sequenced and subsequently open chromatin regions were called following the ENCODE pipeline (see Methods Section), yielding 224 968 open chromatin peaks. Processed publicly available ATAC-seq data for α-cells were retrieved from the Diabetes Epigenome Atlas [26]. Peaks were filtered to those that were present in at least two libraries, which yielded 125 729 IDR peaks. Capture C libraries from α-cells and EndoC-βH1 were generated and analyzed following established protocols [22, 25, 28]. The Capture C libraries yielded high coverage (an average of ~1.4 billion reads per each library), with an average of 37.9% valid reads pairs and 75.8% capture efficiency. Using the CHiCAGO pipeline [29], we called 488 480 promoter interactions in EndoC-BH1s and 245 310 promoter interactions in α- cells. We leveraged the α- cells and EndoC-βH1 ATAC-seq and Capture C libraries to call significant interactions between gene promoters and open chromatin regions and identified short- and long-distance interactions performing analyses at 1- or 4-fragment resolution, and then merged the results.
Partitioned LD score regression
To assess for enrichment of sleep-trait associated SNPs within cells relevant to metabolic disorders, we ran partitioned LD score regression (LDSR). This approach intersected the ATAC-seq and promoter-focused Capture C dataset described above with insomnia [9], morningness [10], short and long sleep [11] GWAS summary statistics. Given the established role of the hypothalamus as master regulator of both circadian rhythms and metabolism, we ran partitioned LDSR intersecting the same GWAS data with 3D genomic data previously generated from hESC-derived hypothalamic-like neurons (HNs) [22, 25] as a positive control.
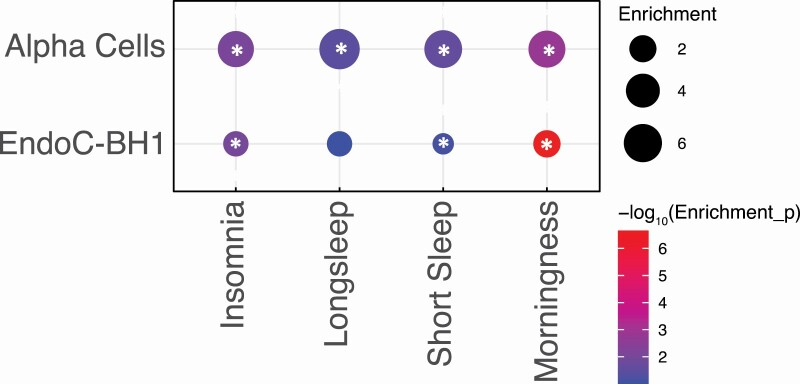
Promoter-interacting open chromatin regions of α-cells revealed significant enrichment for all sleep traits, with a ~5-fold enrichment for insomnia loci (p = 7.61 × 10−3), a ~6-fold enrichment for short sleep loci (p = 2.15 × 10−2), a ~7-fold enrichment for long sleep loci (p = 3.37 × 10−2) and a ~5-fold enrichment for morningness loci (p = 1.58 × 10−3) [Figure 2, Supplementary Table S4]. EndoC-βH1 open chromatin regions were significantly enriched for insomnia loci (~2-fold; p = 1.03 × 10−2), for short sleep loci (~2-fold; p = 1.72 × 10−2) and for morningness loci (~2-fold; p = 2.23 × 10−7) [Figure 2, Supplementary Table S4]. HNs showed significant enrichments for all the sleep trait-associated variants: insomnia loci (~3-fold; p = 1.40 × 10−3), short sleep loci (~3-fold; p = 4.17 × 10−5), long sleep loci (~3-fold; p = 3.06 × 10−3) and morningness loci (~3-fold; p = 6.88 × 10−6) [Supplementary Table S4].
Figure 2.
Heritability enrichment of sleep traits loci within α-cells or EndoC-βH1 cells. Circles sizes represent fold enrichment of sleep traits associated open variants within α-cells or EndoC-βH1 cells promoter-interacting open chromatin regions. Colors indicate the statistical significance (−log10(p-value)) of the enrichments. Asterisks (*) indicate p-value < .05.
The enrichment of sleep-traits associated variants within α-cells and EndoC-βH1 were comparable both in terms of fold increases and statistical significance to the enrichment observed within hESC-derived hypothalamic-like neurons. These results implicate a subset of sleep-trait associated variants operating within α-cells and EndoC-βH1 cells. To identify genes putatively contacted/regulated by sleep-trait relevant variants, we next identified genes with promoters in contact with proxy SNPs of genome-wide significant sleep-trait loci located in open chromatin regions of EndoC-BH1 and α-cells.
Variant to gene mapping
We sought to implicate putative functional effector genes by intersecting sleep-traits associated SNPs with ATAC-seq and promoter-focused Capture C datasets. GWAS studies reported 248, 351, 27, and 8 statistically significant independent sentinel SNPs for insomnia (9), morningness (10), short and long sleep (11). respectively. We identified proxy SNPs with an r2 > 0.8 for insomnia (7836), morningness (9536), short sleep (927), and long sleep (2187). To focus on putatively functional variants residing within α-cells or EndoC-βH1 open chromatin regions, we overlapped these proxy SNPs with ATAC-seq peaks. In α-cells, we identified a total of 1551 open proxies (603 for insomnia, 862 for morningness, 50 for short sleep, and 36 for long sleep). In EndoC-βH1 we found 1789 open proxies (752 for insomnia, 964 for morningness, 56 for short sleep, and 17 for long sleep).
Implicated genes
EndoC-βH1.
In the EndoC-βH1 setting, the above-described open proxies contacted the open promoters of 63 protein-coding-genes (27 genes implicated by insomnia associated SNPs, 29 genes implicated by morningness associated SNPs, 6 genes implicated by short-sleep associated SNPs, and 1 gene implicated by long-sleep associated SNPs) [Supplementary Table S5]. Of these genes, 32 had previously reported circadian rhythm expression in murine β -cells [30] and 13 have already been implicated functionally in diabetes, glucose homeostasis, insulin secretion and/or adipogenesis [Table 1]. Mutations in EIF2AK3 result in the Wolcott–Rallison syndrome characterized by a permanent neonatal diabetes [31]. In vivo and in vitro ablation of DOC2A in β-cells led to a marked impairment of insulin secretion [32]. Overexpression of KLF7 in β-cells inhibits glucose-stimulated insulin secretion (GSIS) [33]. RFX3 encodes a transcription factor which regulates and maintains β-cells differentiation and insulin secretion [34]. BMP5 signaling is necessary for pancreas morphogenesis and promotes insulin-positive cells differentiation [35]. FOXP1 and FOXP4 gene products are both necessary for the development of α-cells [36]. IP6K1 encodes a hexakiphosphate kinase necessary for the generation of the diphosphoinositol pentakisphosphate (IP7), which is critical for β-cells exocytosis and basal insulin secretion [37]. Inhibition of the phosphodiesterase PDE1C increases GSIS [38]. SOX6 overexpression decreases GSIS in β-cells [39]. The GNAO1 gene product is known to regulate insulin secretion [40]. The PAM2 gene product is a monooxygenase involved in insulin secretion from β-cells [41]. SIM1 heterozygosity in mice leads to early-onset obesity, with hyperinsulinemia and hyperleptinemia [42].
Table 1.
Variant-to-gene mapping implicated sleep traits putative effector genes in EndoC-βH1 or α-cells. Genes implicated leveraging publicly available insomnia, morningness, short and long sleep summary statistic and ATAC-seq and promoter-focused Capture C libraries generated in EndoC-βH1 or α-cells, and their reported known function
| Cell setting | Gene | PubMed search (reference) |
|---|---|---|
| EndoC-ßH1 | EIF2AK3 | Wolcott–Rallison syndrome characterized by a permanent neonatal diabetes [31] |
| DOC2A | Marked impairment of insulin secretion [32] | |
| KLF7 | Glucose-stimulated insulin secretion inhibition [33] | |
| RFX3 | β-Cells differentiation and insulin secretion [34] | |
| BMP5 | Insulin-positive cells differentiation [35] | |
| FOXP1 | Development of α-cells [36] | |
| FOXP4 | Development of α-cells [36] | |
| IP6K1 | Cell exocytosis and basal insulin secretion [37] | |
| PDE1C | Increased glucose-stimulated insulin secretion [38] | |
| SOX6 | Decreased glucose-stimulated insulin secretion [39] | |
| GNAO1 | Regulation of insulin secretion [40] | |
| PAM2 | Insulin secretion [41] | |
| SIM1 | Early-onset obesity, with hyperinsulinemia and hyperleptinemia in mice [42] | |
| α-cells | MT2A | KO ameliorates glucose tolerance increasing insulin release in mice [43] |
| PLCXD3 | Reduced expression in diabetic islets of T2D subjects, inhibition of insulin secretion and regulation of glucose sensing [45] | |
| Variants are associated with increased risk of metabolic syndrome [46] | ||
| HDAC3 | Proposed as a novel diabetes drug target [47] | |
| HMGA1 | One variant associated with increased risk of T2D [48] | |
| PTPM1 | Increased ATP production and insulin secretion in β-cell line [49] | |
| PACSIN3 | Increased glucose uptake via GLUT1 in adipocytes [50] | |
| NDUFS3 | Aberrant mitochondrial hyperactivity in both type 1 and 2 diabetes [51] | |
| CUGBP1 | Inhibition of glucose- and GLP1-induced insulin secretion in mice [52] |
α-Cells.
In α-cells, 76 protein-coding-genes were implicated by sleep-associated open proxies (36 genes implicated by insomnia loci, 35 genes implicated by morningness loci, and 5 genes implicated by short-sleep loci) [Supplementary Table S6]. Of these genes, 41 are circadially expressed in α-cells of mice [30] and 8 have already been associated with T2D or implicated functionally in insulin and glucose metabolism or [Table 1]. MT2A expression is reduced by stimulation with glucose in humans and murine islets and its ablation in mice increases insulin release [43]. MT2A expression is higher in adipose tissue of T2D subjects [44]. PLCXD3 expression is reduced in human diabetic islets, and when silenced in INS-1 cells inhibits insulin secretion and alters glucose sensing [45]. Two PLCXD3 variants are associated with increased risk of metabolic syndrome [46]. HDAC3 has been proposed as a novel diabetes drug target [47]. One variant of HMGA1 has been associated with an increased risk of T2D development [48] and its gene product is critical for insulin gene expression and β-cell function. Suppression of PTPMT1 in INS-1 cell line enhances insulin secretion [49]. In adipocytes, PACSIN3 overexpression enhances glucose uptake [50]. NDUFS3 encodes for a component of the mitochondrial complex I which shows an aberrant hyperactivity in both type 1 and 2 diabetes [51]. CUGBP1 is expressed in the islets of diabetic mice, where its overexpression reduces insulin secretion [52].
Common genetic architecture
α-Cells and EndoC-βH1 cells shared 14 common open insomnia proxy SNPs in LD with the same sentinel SNPs (r2 > 0.8). Six of these common proxies implicated the same genes in the two different cell settings. Three of these genes are protein-coding; MEIS1, PITPNM2, and SNAPC5. The MEIS1 and SNAPC5 observations were very close to the promoter (~18 bp), suggesting that they are directly affecting gene expression by altering binding of the basal transcriptional machinery.
Comparably, there were 10 shared morningness proxies in accessible regions in both α-cells and EndoC-βH1. Six of the common proxies contacted the same gene, three of which encode for proteins; RNF10, COQ5, and USP34. Finally, in α-cells and EndoC-βH1 four common short sleep proxies implicated two common coding-protein genes, namely PPIP5K2, GIN1.
Pathways analyses.
We assessed the expression of the sleep-implicated genes using RNA-seq generated in α-cells and EndoC-BH1. We then ran pathway enrichment analyses through the “Compute Overlaps” tool provided by Gene Set Enrichment Analysis (GSEA) website (https://www.gsea-msigdb.org/gsea/index.jsp) to verify if the implicated genes were enriched for metabolic relevant terms.
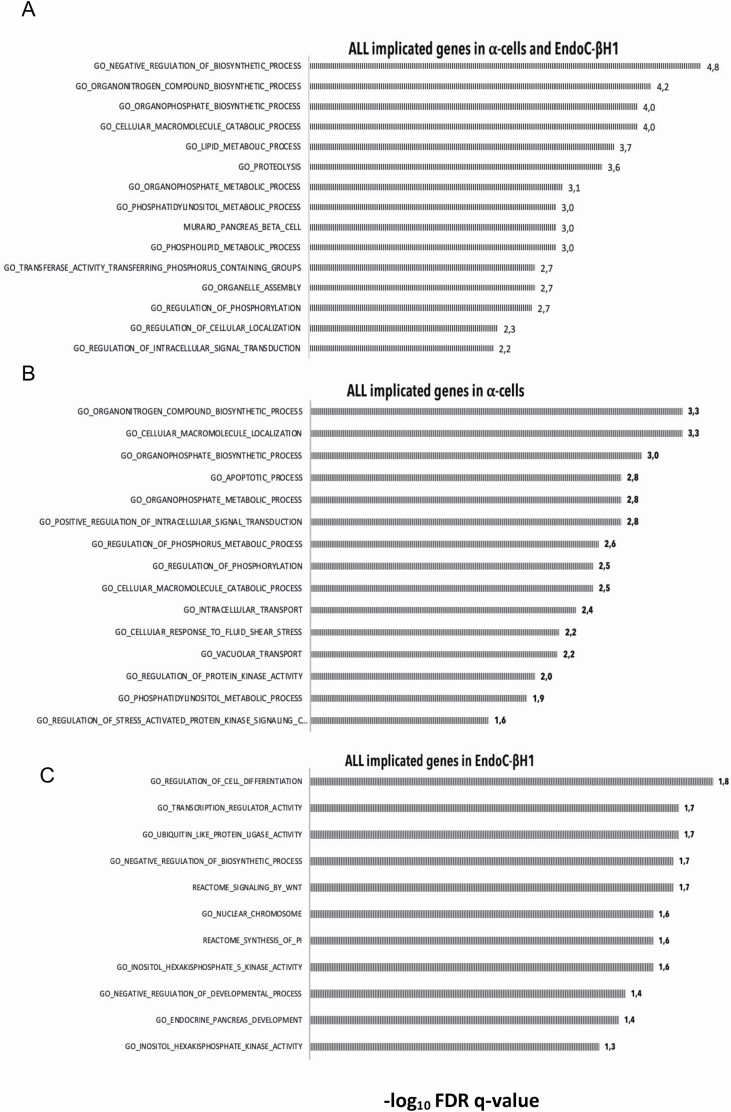
First, we pooled all the genes from both cellular settings. We observed significant enrichment (padj < .05) for metabolic and catabolic process; for organonitrogen and organophosphate biosynthesis; for phosphatidylinositol and phosphorylation (e.g., phospholipid, phosphatidylinositol, phosphorylation and glycerophospholipid process); for response to endogenous stimulus, for organelle assembly and for intracellular signal transduction [Figure 3A; Supplementary Table S7].
Figure 3.
Pathways enrichment in α-cells and/or EndoC-βH1 cells. Enriched pathways for α-cells and EndoC-βH cells (A), only α-cells (B), or only EndoC-βH1 cells (C) implicated genes. Selection of significant enriched pathways (adjusted p-value < .05) are reported. Bars indicate the statistical significance (−log10 (adjusted p-value)) of the enrichments.
We then analyzed the genes implicated within the two pancreatic cell types separately.
In α-cells, we observed enrichment of organonitrogen and organophosphate biosynthetic process; phosphatidylinositol and phosphorylation associated terms; intracellular signal transduction and transport; for kinase activity and for stress-response [Figure 3b; Supplementary Table S8]. In EndoC-βH1, we found enrichment for cell differentiation and apoptosis, transcription regulation, Wnt signaling, endocrine pancreas development and phosphatidylinositol-associated terms (inositol-hexakisphosphate-5-kinase-activity, synthesis-of-pi) [Figure 3c; Supplementary Table S9].
Overall, the pathway enrichments showed that sleep-trait loci were contacting putative effector genes involved in cell metabolism and catabolism, organonitrogen and organophosphate biosynthesis, stress responses, cell trafficking and signaling, phosphatidylinositol terms, and phosphorylation.
Conclusions
Poor sleep and metabolic disorders are tightly associated, but the pathophysiology of this association remains unclear. Genetic correlation analyses have revealed that insomnia, short and long sleep are significantly correlated with T2D, obesity and several other metabolic-related traits.
Our results are in agreement with other published genetic correlation analyses except for T2D [11]. The different result might be due to the fact that we used a newer and larger T2D GWAS, highlighting the importance of statistical power when performing such analyses. Insomnia and short sleep yielded very similar correlations, suggesting that loss of sleep—rather than longer sleep and morning preference—are genetically intertwined with metabolic dysfunction [Figure 1; Supplementary Table S2 and S3]. Indeed, MR studies have shown that genetic variants underlying insomnia/short sleep is a predisposing factor for the onset of T2D and cardiovascular disease [8, 9, 13, 14], while long sleep-associated risk variants are causal for lower BMI in children [15] but are unlikely to be causal for cardiovascular diseases [53]. In line with other published studies [10], we did not observe significant correlation between morningness and any metabolic trait [Figure 1; Supplementary Table S2 and S3]. This result might be partially explained by the fact that in the current literature eveningness, rather than morningness, is related to metabolic dysfunctions and T2D [54]. Also, MR studies have shown that being a “morning person” predisposes to an increased intake of healthy foods [55], higher subjective well-being and decreased liability of depression [10]; therefore, suggesting health benefits of adopting a more morning diurnal preference.
Partitioned LDSR analyses revealed that loci for insomnia, morningness, short sleep were significantly enriched in open chromatin regions of both α-cells and EndoC-βH1, while long sleep loci were significantly enriched only in α-cells [Figure 2, Supplementary Table S4]. The fold increase as well as the significance of the enrichments in both cell settings were comparable with enrichments observed in hypothalamic neurons [Supplementary Table S4]. These results suggest that sleep genetic variants have a regulatory function in the context of the pancreatic islets, enhancing/inhibiting the expression of genes via cis interactions.
Our variant-to-gene mapping analyses identified interactions with putative effector genes, implicating a total of 76 genes in α-cells and 63 gene in EndoC-βH1 [Supplementary Table S5 and S6]. Twenty-one of all the implicated genes have known functions in insulin secretion and resistance, glucose homeostasis, diabetes and/or obesity [Table 1], corroborating the notion that sleep-traits associated SNPs alter the expression of genes also important for glucose metabolism in the pancreatic cells.
Other implicated genes have not yet been functionally associated with metabolic disorders, but they are involved in pathways related to insulin and glucagon secretion or glucose uptake. CDIPT and PITPNM2 encode for proteins involved in the syntheses of phosphatidylinositol (PI) the precursor of all the phosphoinositides (PPIs). PPIs are essential in insulin signaling since they are involved in glucose uptake, insulin release and resistance, and in glycogen and lipid storage. The PITPNM2 transcript is also involved in exocytosis [56]. NDUFS3 encodes for a component of the mitochondrial complex 1; and mitochondrial oxidoreductase activity is essential for normal insulin release in β-cells [57] and for glucagon release in α-cells [58].
These results suggest that sleep-trait associated variants alter the expression of genes involved in insulin or glucagon secretion in the pancreatic cells, impacting glucose homeostasis and favoring or exacerbating metabolic disorders.
Pathway analyses conducted on genes implicated by sleep-traits associated variants in α- and EndoC-βH1 cells showed significant enrichments for organonitrogen biosynthesis and stress responses which are functions normally regulated by mitochondria. Mitochondrial dysfunction, ER stress and oxygen reactive species accumulation can indeed impair insulin/glucagon release [57, 58]. The implicated genes were also enriched for cell signaling and intracellular trafficking, which are mechanisms required for glucagon/insulin secretion [59]. Genes were enriched also for organophosphate biosynthesis, phosphatidylinositol (PI)-associated terms and phosphorylation; with PI and their phosphorylation status being key elements in glucose homeostasis. Notably, genes implicated in EndoC-βH1 were specifically enriched for cell differentiation and endocrine pancreas development. Taken together the pathways analyses support the hypothesis that sleep-trait associated variants influence the expression of genes relevant to metabolic disorders.
Our data implicate pancreatic α- and β-cells as effectors cells for insomnia, morningness, long and short associated variants. We propose that sleep-traits associated variants alter insulin/glucagon secretion or ß-cells differentiation/development, thus impairing glucose homeostasis and predisposing to metabolic dysfunction. Our results therefore warrant functional follow-up to validate the interaction between the sleep-traits associated SNPs and their implicated genes, along with the effect of the novel implicated genes on insulin or glucagon release in both in vitro and in vivo settings.
Supplementary Material
Acknowledgments
This manuscript used data acquired from the Human Pancreas Analysis Program (HPAP-RRID:SCR_016202) Database (https://hpap.pmacs.upenn.edu), a Human Islet Research Network (RRID:SCR_014393) consortium (UC4-DK-112217, U01-DK-123594, UC4-DK-112232, and U01-DK-123716). This work was in part supported by National Institutes of Health (NIH) UM1 DK126194, NIH R01 HL143790. C.L. designed the study, collected and analyzed data, wrote and edited manuscript. M.C.P. collected/analyzed data, wrote/edited manuscript J.A.P collected and analyzed data, reviewed and edited manuscript. C.S. collected and analyzed data, reviewed and edited manuscript. M.E.J. collected and analyzed data, reviewed and edited manuscript. A.C. collected and analyzed data, reviewed and edited manuscript. K.B. collected and analyzed data, reviewed and edited manuscript. E.M. collected and analyzed data, reviewed and edited manuscript. K.O. collected and analyzed data, reviewed and edited manuscript. M.L.G. collected and analyzed data, reviewed and edited manuscript. A.D.W. reviewed and edited manuscript. K.H.K. reviewed and edited manuscript. S.F.A.G. supervised the study, reviewed and edited manuscript. Lasconi and Grant are the guarantors of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Contributor Information
Chiara Lasconi, Center for Spatial and Functional Genomics, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA.
Matthew C Pahl, Center for Spatial and Functional Genomics, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA.
James A Pippin, Center for Spatial and Functional Genomics, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA.
Chun Su, Center for Spatial and Functional Genomics, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA.
Matthew E Johnson, Center for Spatial and Functional Genomics, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA.
Alessandra Chesi, Center for Spatial and Functional Genomics, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA; Department of Pathology and Laboratory Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Keith Boehm, Center for Spatial and Functional Genomics, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA.
Elisabetta Manduchi, Institute for Biomedical Informatics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
Kristy Ou, Department of Genetics, University of Pennsylvania, Philadelphia, PA 19104, USA; Institute for Diabetes, Obesity and Metabolism, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Maria L Golson, Department of Genetics, University of Pennsylvania, Philadelphia, PA 19104, USA; Institute for Diabetes, Obesity and Metabolism, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Andrew D Wells, Center for Spatial and Functional Genomics, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA; Department of Pathology and Laboratory Medicine, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Klaus H Kaestner, Department of Genetics, University of Pennsylvania, Philadelphia, PA 19104, USA; Institute for Diabetes, Obesity and Metabolism, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Struan F A Grant, Center for Spatial and Functional Genomics, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA; Department of Genetics, University of Pennsylvania, Philadelphia, PA 19104, USA; Institute for Diabetes, Obesity and Metabolism, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Pediatrics, The University of Pennsylvania Perelman School of Medicine, Philadelphia, PA 19104, USA; Division of Human Genetics, Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA.
Disclosure Statement
None declared.
References
- 1. Anafi RC, et al. Exploring phylogeny to find the function of sleep. Nat Rev Neurosci. 2019;20(2):109–116. [DOI] [PubMed] [Google Scholar]
- 2. Reutrakul S, et al. Sleep influences on obesity, insulin resistance, and risk of type 2 diabetes. Metab Clin Exp. 2018;84:56–66. [DOI] [PubMed] [Google Scholar]
- 3. Grandner MA, et al. Sleep duration and diabetes risk: population trends and potential mechanisms. Curr Diab Rep. 2016;16(11):106. doi: 10.1007/s11892-016-0805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. LeBlanc ES, et al. Insomnia is associated with an increased risk of type 2 diabetes in the clinical setting. BMJ Open Diabetes Res Care 2018;6(1):e000604. doi: 10.1136/bmjdrc-2018-000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hein M, et al. Prevalence and risk factors of type 2 diabetes in insomnia sufferers: a study on 1311 individuals referred for sleep examinations. Sleep Med. 2018;46:37–45. doi: 10.1016/j.sleep.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 6. St-Onge M-P. Sleep-obesity relation: underlying mechanisms and consequences for treatment. Obes Rev Off J Int Assoc Study Obes. 2017;18(Suppl 1):34–39. [DOI] [PubMed] [Google Scholar]
- 7. Koolhaas CM, et al. Objectively measured sleep and body mass index: a prospective bidirectional study in middle-aged and older adults. Sleep Med. 2019;57:43–50. [DOI] [PubMed] [Google Scholar]
- 8. Gao X, et al. Investigating causal relations between sleep-related traits and risk of type 2 diabetes mellitus: a mendelian randomization study. Front Genet [Internet]. 2020;11:1–9. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7770175/[cited 2021 Mar 31] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jansen PR, et al. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat Genet. 2019;51(3):394–403. [DOI] [PubMed] [Google Scholar]
- 10. Jones SE, et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun. 2019;10(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dashti HS, et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat Commun. 2019;10(1):1100. doi: 10.1038/s41467-019-08917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davies NM, et al. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yuan S, et al. An atlas on risk factors for type 2 diabetes: a wide-angled Mendelian randomisation study. Diabetologia. 2020;63(11):2359–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yuan S, et al. Genetic liability to insomnia in relation to cardiovascular diseases: a Mendelian randomisation study. Eur J Epidemiol. 2021;36(4):393–400. doi: 10.1007/s10654-021-00737-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang J, et al. Sleep duration and adiposity in children and adults: Observational and Mendelian randomization studies. Obes Silver Spring Md. 2019;27(6): 1013–1022. [DOI] [PubMed] [Google Scholar]
- 16. Bos MM, et al. The association between habitual sleep duration and sleep quality with glycemic traits: Assessment by cross-sectional and Mendelian randomization analyses. J Clin Med. [Internet]. 2019;8(5):1–13. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6572144/ [cited 2021 May 6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bos MM, et al. Investigating the relationships between unfavourable habitual sleep and metabolomic traits: evidence from multi-cohort multivariable regression and Mendelian randomization analyses. BMC Med. 2021;19(1):69. doi: 10.1186/s12916-021-01939-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gehrman P, et al. Altered diurnal states in insomnia reflect peripheral hyperarousal and metabolic desynchrony: a preliminary study. Sleep. 2018;41(5):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Röder PV, et al. Pancreatic regulation of glucose homeostasis. Exp Mol Med. 2016;48:e219. doi: 10.1038/emm.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsonkova VG, et al. The EndoC-βH1 cell line is a valid model of human beta cells and applicable for screenings to identify novel drug target candidates. Mol Metab. 2018 Feb;8:144–157. doi: 10.1016/j.molmet.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lasconi C, et al. Variant-to-gene-mapping analyses reveal a role for the hypothalamus in genetic susceptibility to inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2021;11(3):667–682. doi: 10.1016/j.jcmgh.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Challet E. The circadian regulation of food intake. Nat Rev Endocrinol. 2019 Jul;15(7):393–405. doi: 10.1038/s41574-019-0210-x. [DOI] [PubMed] [Google Scholar]
- 24. Dorrell C, et al. Isolation of mouse pancreatic alpha, beta, duct and acinar populations with cell surface markers. Mol Cell Endocrinol. 2011;339(1–2):144–50. 10.1016/j.mce.2011.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pahl MC, et al. Cis-regulatory architecture of human ESC-derived hypothalamic neuron differentiation aids in variant-to-gene mapping of relevant complex traits. Nat Commun. 2021;12(1):6749. doi: 10.1038/s41467-021-27001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. CMDGA – Common Metabolic Diseases Genome Atlas consortium [Internet]. [cited 2022 Apr 24]. Available from: https://cmdga.org/
- 27. Ackermann AM, et al. Integration of ATAC-seq and RNA-seq identifies human alpha cell and beta cell signature genes. Mol Metab. 2016;5(3):233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chesi A, et al. Genome-scale Capture C promoter interactions implicate effector genes at GWAS loci for bone mineral density. Nat Commun. 2019;10(1):1260. doi: 10.1038/s41467-019-09302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cairns J, et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 2016;17(1):127. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Petrenko V, et al. Pancreatic α- and β-cellular clocks have distinct molecular properties and impact on islet hormone secretion and gene expression. Genes Dev. 2017;31(4):383–398. doi: 10.1101/gad.290379.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Julier C, et al. Wolcott–Rallison syndrome. Orphanet J Rare Dis. 2010;5:29. 10.1186/1750-1172-5-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li J, et al. DOC2 isoforms play dual roles in insulin secretion and insulin-stimulated glucose uptake. Diabetologia. 2014;57(10):2173–2182. [DOI] [PubMed] [Google Scholar]
- 33. Kawamura Y, et al. Overexpression of Kruppel-like factor 7 regulates adipocytokine gene expressions in human adipocytes and inhibits glucose-induced insulin secretion in pancreatic beta-cell line. Mol Endocrinol Baltim Md. 2006;20(4):844–856. [DOI] [PubMed] [Google Scholar]
- 34. Ait-Lounis A, et al. The transcription factor Rfx3 regulates beta-cell differentiation, function, and glucokinase expression. Diabetes. 2010;59(7):1674–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang F-X, et al. Bone morphogenetic proteins promote development of fetal pancreas epithelial colonies containing insulin-positive cells. J Cell Sci. 2002;115(Pt 4):753–60. [DOI] [PubMed] [Google Scholar]
- 36. Spaeth JM, et al. The FOXP1, FOXP2 and FOXP4 transcription factors are required for islet alpha cell proliferation and function in mice. Diabetologia. 2015;58(8):1836–1844. doi: 10.1007/s00125-015-3635-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rajasekaran SS, et al. Inositol hexakisphosphate kinase 1 is a metabolic sensor in pancreatic β-cells. Cell Signal. 2018;46:120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Han P, et al. The calcium/calmodulin-dependent phosphodiesterase PDE1C down-regulates glucose-induced insulin secretion. J Biol Chem. 1999;274(32):22337–44. 10.1074/jbc.274.32.22337 [DOI] [PubMed] [Google Scholar]
- 39. Iguchi H, et al. SOX6 attenuates glucose-stimulated insulin secretion by repressing PDX1 transcriptional activity and is down-regulated in hyperinsulinemic obese mice. J Biol Chem. 2005;280(45):37669–37680. doi: 10.1074/jbc.M505392200. [DOI] [PubMed] [Google Scholar]
- 40. Leiss V, et al. Insulin secretion stimulated by L-arginine and its metabolite L-ornithine depends on Gα(i2). Am J Physiol Endocrinol Metab. 2014;307(9):E800–E812. doi: 10.1152/ajpendo.00337.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thomsen SK, et al. Type 2 diabetes risk alleles in PAM impact insulin release from human pancreatic β-cells. Nat Genet. 2018;50(8):1122–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Michaud JL, et al. Sim1 haploinsufficiency causes hyperphagia, obesity and reduction of the paraventricular nucleus of the hypothalamus. Hum Mol Genet. 2001;10(14):1465–1473. doi: 10.1093/hmg/10.14.1465. [DOI] [PubMed] [Google Scholar]
- 43. Bensellam M, et al. Metallothionein 1 negatively regulates glucose-stimulated insulin secretion and is differentially expressed in conditions of beta cell compensation and failure in mice and humans. Diabetologia. 2019;62(12):2273–2286. doi: 10.1007/s00125-019-05008-3. [DOI] [PubMed] [Google Scholar]
- 44. Haynes V, et al. Metallothionein 2a gene expression is increased in subcutaneous adipose tissue of type 2 diabetic patients. Mol Genet Metab. 2013;108(1):90–94. [DOI] [PubMed] [Google Scholar]
- 45. Aljaibeji H, et al. Reduced expression of PLCXD3 associates with disruption of glucose sensing and insulin signaling in pancreatic β-cells. Front Endocrinol. 2019;10:735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aljaibeji H, et al. Genetic variants of the PLCXD3 gene are associated with risk of metabolic syndrome in the Emirati population. Genes. 2020;11(6):665. 10.3390/genes11060665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meier BC, et al. Inhibition of HDAC3 as a strategy for developing novel diabetes therapeutics. Epigenomics. 2014;6(2):209–214. [DOI] [PubMed] [Google Scholar]
- 48. Bianco A, et al. The association between HMGA1 rs146052672 variant and Type 2 diabetes: a transethnic meta-analysis. PLoS One. 2015;10(8):e0136077. doi: 10.1371/journal.pone.0136077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pagliarini DJ, et al. Involvement of a mitochondrial phosphatase in the regulation of ATP production and insulin secretion in pancreatic beta cells. Mol Cell. 2005;19(2):197–207. doi: 10.1016/j.molcel.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 50. Roach W, et al. PACSIN3 overexpression increases adipocyte glucose transport through GLUT1. Biochem Biophys Res Commun. 2007;355(3):745–750. doi: 10.1016/j.bbrc.2007.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu J, et al. Pancreatic mitochondrial complex I exhibits aberrant hyperactivity in diabetes. Biochem Biophys Rep. 2017;11:119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhai K, et al. RNA-binding protein CUGBP1 regulates insulin secretion via activation of phosphodiesterase 3B in mice. Diabetologia. 2016;59(9):1959–1967. [DOI] [PubMed] [Google Scholar]
- 53. Ai S, et al. Causal associations of short and long sleep durations with 12 cardiovascular diseases: linear and nonlinear Mendelian randomization analyses in UK Biobank. Eur Heart J. 2021;42(34):3349–3357. [DOI] [PubMed] [Google Scholar]
- 54. Hashemipour S, et al. Association of evening chronotype with poor control of type 2 diabetes: roles of sleep duration and insomnia level. Int J Endocrinol Metab. 2020;18(3):e99701. doi: 10.5812/ijem.99701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dashti HS, Chen A, Daghlas I, Saxena R. Morning diurnal preference and food intake: a Mendelian randomization study. Am J Clin Nutr. 2020;112(5):1348–1357. 10.1093/ajcn/nqaa219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cockcroft S. The diverse functions of phosphatidylinositol transfer proteins. Curr Top Microbiol Immunol. 2012;362:185–208. doi: 10.1007/978-94-007-5025-8_9. [DOI] [PubMed] [Google Scholar]
- 57. Martin SD, et al. The role of mitochondria in the aetiology of insulin resistance and type 2 diabetes. Biochim Biophys Acta. 2014;1840(4):1303–1312. [DOI] [PubMed] [Google Scholar]
- 58. Grubelnik V, et al. Modelling of dysregulated glucagon secretion in type 2 diabetes by considering mitochondrial alterations in pancreatic α-cells. R Soc Open Sci. 2020;7(1):191171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Seino S, et al. Pancreatic beta-cell signaling: toward better understanding of diabetes and its treatment. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86(6):563–577. doi: 10.2183/pjab.86.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.