Key Points
Question
Does combined disease testing provide improved diagnostic yield and clinical utility for patients with a suspected genetic cardiomyopathy or arrhythmia?
Findings
In this cohort study of 4782 patients with a suspected genetic cardiomyopathy or arrhythmia, combined cardiomyopathy and arrhythmia testing revealed clinically relevant variants in 1 in 5 patients, and 66.0% of patients with positive findings had potential clinical management implications. The combined testing approach captured 10.9% of patients who would have been missed if genetic testing had been restricted to a specific suspected disease subtype.
Meaning
This study’s findings suggest that combined cardiomyopathy and arrhythmia genetic testing is able to identify genetic etiologies associated with these diseases that can inform patient management.
Abstract
Importance
Genetic testing can guide management of both cardiomyopathies and arrhythmias, but cost, yield, and uncertain results can be barriers to its use. It is unknown whether combined disease testing can improve diagnostic yield and clinical utility for patients with a suspected genetic cardiomyopathy or arrhythmia.
Objective
To evaluate the diagnostic yield and clinical management implications of combined cardiomyopathy and arrhythmia genetic testing through a no-charge, sponsored program for patients with a suspected genetic cardiomyopathy or arrhythmia.
Design, Setting, and Participants
This cohort study involved a retrospective review of DNA sequencing results for cardiomyopathy- and arrhythmia-associated genes. The study included 4782 patients with a suspected genetic cardiomyopathy or arrhythmia who were referred for genetic testing by 1203 clinicians; all patients participated in a no-charge, sponsored genetic testing program for cases of suspected genetic cardiomyopathy and arrhythmia at a single testing site from July 12, 2019, through July 9, 2020.
Main Outcomes and Measures
Positive gene findings from combined cardiomyopathy and arrhythmia testing were compared with findings from smaller subtype-specific gene panels and clinician-provided diagnoses.
Results
Among 4782 patients (mean [SD] age, 40.5 [21.3] years; 2551 male [53.3%]) who received genetic testing, 39 patients (0.8%) were Ashkenazi Jewish, 113 (2.4%) were Asian, 571 (11.9%) were Black or African American, 375 (7.8%) were Hispanic, 2866 (59.9%) were White, 240 (5.0%) were of multiple races and/or ethnicities, 138 (2.9%) were of other races and/or ethnicities, and 440 (9.2%) were of unknown race and/or ethnicity. A positive result (molecular diagnosis) was confirmed in 954 of 4782 patients (19.9%). Of those, 630 patients with positive results (66.0%) had the potential to inform clinical management associated with adverse clinical outcomes, increased arrhythmia risk, or targeted therapies. Combined cardiomyopathy and arrhythmia gene panel testing identified clinically relevant variants for 1 in 5 patients suspected of having a genetic cardiomyopathy or arrhythmia. If only patients with a high suspicion of genetic cardiomyopathy or arrhythmia had been tested, at least 137 positive results (14.4%) would have been missed. If testing had been restricted to panels associated with the clinician-provided diagnostic indications, 75 of 689 positive results (10.9%) would have been missed; 27 of 75 findings (36.0%) gained through combined testing involved a cardiomyopathy indication with an arrhythmia genetic finding or vice versa. Cascade testing of family members yielded 402 of 958 positive results (42.0%). Overall, 2446 of 4782 patients (51.2%) had only variants of uncertain significance. Patients referred for arrhythmogenic cardiomyopathy had the lowest rate of variants of uncertain significance (81 of 176 patients [46.0%]), and patients referred for catecholaminergic polymorphic ventricular tachycardia had the highest rate (48 of 76 patients [63.2%]).
Conclusions and Relevance
In this study, comprehensive genetic testing for cardiomyopathies and arrhythmias revealed diagnoses that would have been missed by disease-specific testing. In addition, comprehensive testing provided diagnostic and prognostic information that could have potentially changed management and monitoring strategies for patients and their family members. These results suggest that this improved diagnostic yield may outweigh the burden of uncertain results.
This cohort study assesses the diagnostic yield and clinical management implications of combined cardiomyopathy and arrhythmia genetic testing among patients with a suspected genetic cardiomyopathy or arrhythmia.
Introduction
Genetic testing for nonischemic cardiomyopathies and inherited arrhythmias is recommended by cardiovascular societies to establish a genetic diagnosis, guide clinical management, and identify family members at risk.1,2,3,4,5,6,7 However, many individuals with cardiomyopathies or arrhythmias do not receive genetic testing and therefore cannot benefit from gene-specific clinical management.8,9,10 Barriers to testing include limited clinician knowledge of genetics and results interpretation as well as real and perceived concerns regarding cost, insurance coverage, and low diagnostic yields.11,12 Multiple studies13,14,15,16 have described the diagnostic yields of condition-specific genetic tests among patients with clinical diagnoses. However, evidence supporting the shared genetic factors associated with arrhythmia and cardiomyopathy has increased,17,18,19 suggesting that large gene panels encompassing both cardiomyopathies and arrhythmias may increase diagnostic yield. Some evidence supports the use of gene panels restricted to a specific disease subtype, especially hypertrophic cardiomyopathy (HCM), to limit the possibility of detecting variants of uncertain significance, which increases with increasing panel size.3,20 Further exploration of the trade-offs between diagnostic yield and uncertain results is needed.
To assess the utility of inclusive testing, we evaluated real and estimated testing outcomes of patients suspected to have a genetic cardiomyopathy or arrhythmia who were referred for no-charge, sponsored genetic testing for 7 subtypes of cardiomyopathy and arrhythmia (referred to collectively as cardiorhythm) disease. First, we assessed the diagnostic yield of multidisease genetic testing and cascade testing of family members. Second, we examined the proportion of individuals who received positive results with clinical management implications as defined by current literature and treatment guidelines. Third, we explored the possibility of missed positive results if a disease-specific gene panel had been used instead of the multidisease panel.
Methods
Study Population
This cohort study was approved by the independent institutional review board service WCG IRB and met the requirements for waived informed consent because the study analyzed deidentified secondary data. Deidentified clinical data provided by ordering clinicians were reviewed for all individuals referred to a commercial testing laboratory (Invitae Corp; San Francisco, California) via a no-charge, sponsored program from July 12, 2019, through July 9, 2020. This study followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline for cohort studies.
Main inclusion criteria for patients were location in Canada or the US and any level of clinician suspicion of genetic cardiomyopathy or arrhythmia based on personal or family history. Ordering clinicians were also asked to provide 1 or more specific reasons for their clinical suspicion, including (1) suspected or known diagnosis of a cardiomyopathy or arrhythmia, (2) family history of a cardiomyopathy or arrhythmia, (3) family history of unexplained sudden cardiac death, and/or (4) postmortem suspicion of cardiomyopathy or arrhythmia. Ordering clinicians had the option to indicate 1 or more disease subtype. For cardiomyopathies, disease subtypes included HCM, dilated cardiomyopathy (DCM), arrhythmogenic right ventricular cardiomyopathy (as written on the requisition form field; referred to as arrhythmogenic cardiomyopathy in this study), and/or left ventricular noncompaction cardiomyopathy; for arrhythmias, disease subtypes included long QT syndrome, catecholaminergic polymorphic ventricular tachycardia, and/or Brugada syndrome. Clinicians also had the option to provide an index of suspicion (low, moderate, or high) for diagnostic indications and additional clinical information. Additional methods are provided in the eMethods in Supplement 1. An example of the form used by ordering clinicians is available in eFigure 1 in Supplement 1. Cascade family testing was also provided at no charge through the program.
Genetic Testing and Variant Interpretation
Next-generation sequencing gene panels were used to simultaneously test for both sequence and exon-level copy number variants, as previously described.21,22,23 Up to 150 genes associated with cardiomyopathies or arrhythmias were sequenced. The primary panel included 67 genes with established associations with cardiomyopathies and arrhythmias. Four optional add-on panels that included genes with preliminary associations with cardiomyopathies and arrhythmias could be ordered initially or after receipt of initial results without charge (eTable 1 in Supplement 1). Variants were classified as pathogenic or likely pathogenic (P/LP), of uncertain significance, likely benign, or benign using Sherloc,24 a variant interpretation framework that relies on a point-based evidence scoring system built on the joint consensus guidelines from the American College of Medical Genetics and Genomics and the Association for Molecular Pathology.25 Results were categorized as positive, negative, carrier, or uncertain depending on the classification of the variant identified and the inheritance pattern of the associated condition. Additional details are provided in the eMethods in Supplement 1.
Statistical Analysis
Diagnostic yield was calculated as the percentage of patients with positive genetic test results among all patients for whom clinicians had indicated a personal known or suspected cardiomyopathy or arrhythmia. Diagnostic yield was also stratified by referral indication, index of suspicion, age, and vital status. Family history of cardiomyopathy, arrhythmia, or unexplained sudden cardiac death were combined into a single family history variable.
The potential for limited gene testing to miss positive results was assessed among patients with positive results whose clinicians had indicated a specific cardiomyopathy or arrhythmia subtype. Testing results for each patient were reanalyzed as if testing had been restricted to a disease-specific (HCM, DCM, arrhythmogenic cardiomyopathy, left ventricular noncompaction cardiomyopathy, long QT syndrome, catecholaminergic polymorphic ventricular tachycardia, or Brugada syndrome) gene panel (eTable 2 in Supplement 1). Some disease-specific panels had overlapping gene content. Family testing results were excluded from the primary analyses of yield and clinically unexpected results. Family testing results were limited to cascade testing after receipt of a positive result in the index patient.
Genes with clinical management implications were grouped as associated with (1) adverse clinical outcomes in sarcomeric HCM (genes ACTC1, MYL2, MYBPC3, MYH7, MYL3, TNNI3, TNNT2, and TPM1)26 or rapid deterioration (gene LAMP2),27 (2) heightened arrhythmia risk in cardiomyopathy (genes ABCC9, DES, DSP, FLNC, LMNA, PLN, RBM20, RYR2, SCN5A, and TTN),28,29,30,31,32,33 or (3) targeted therapies (genes GAA, GLA, and TTR).34,35
Statistical analyses were performed using R Studio, version 2022.02.3, build 49 (R Foundation for Statistical Computing); the prop.test function, version 3.6.2, was used for group comparisons. Two-tailed P < .05 was considered statistically significant.
Results
Study Population
Over the 12-month study period, 4782 unrelated patients were referred by 1203 clinicians for testing. Of those, 2551 patients (53.3%) were male, and 2231 (46.7%) were female. The mean (SD) age at testing was 40.5 (21.3) years (range, 0-93 years); 1028 patients (21.5%) were 60 years or older. With regard to racial and ethnic ancestry, 39 patients (0.8%) were Ashkenazi Jewish, 113 (2.4%) were Asian, 571 (11.9%) were Black or African American, 375 (7.8%) were Hispanic, 2866 (59.9%) were White, 240 (5.0%) were of multiple races and/or ethnicities, 138 (2.9%) were of other races and/or ethnicities (including American Indian, French Canadian, Mediterranean, and Sephardic Jewish), and 440 (9.2%) were of unknown race and/or ethnicity (eTable 3 in Supplement 1). The proportions of Black or African American and Hispanic patients were greater than those observed in a healthy testing cohort reported by the testing laboratory.36 Overall, 190 cases (4.0%) were post mortem.
Clinicians indicated a suspected or known personal diagnosis of genetic cardiomyopathy or arrhythmia in 4270 of 4782 patients (89.3%) (Figure 1), with cardiomyopathies being the most frequent diagnostic indication (2596 of 4270 patients [60.8%]). For the remaining 512 patients (10.7%), family history and/or death suspected to be associated with cardiorhythm disease were the only reasons provided by clinicians for their suspicion. Overall, 2357 patients (49.3%) were reported to have at least 1 category of family history; 1819 patients (38.0%) had a family history of primary cardiomyopathy or arrhythmia, 1135 (23.7%) had a family history of unexplained sudden cardiac death, and 597 (12.5%) had a family history of both. Clinicians provided multiple reasons for their suspicion for 2002 patients (41.9%).
Figure 1. Clinician-Provided Reasons for Referring Patients.

There were 4782 total patients. The family history category includes patients meeting 1 or more inclusion criterion for a family history of a primary cardiomyopathy or arrhythmia and/or a family history of sudden cardiac death. The multiple cardiomyopathy (CM) and arrhythmia (AR) category includes patients with a combination of suspected or known cardiomyopathies and arrhythmias. ARVC indicates arrhythmogenic right ventricular cardiomyopathy; BrS, Brugada syndrome; CPVT, catecholaminergic polymorphic ventricular tachycardia; DCM, dilated cardiomyopathy; HCM, hypertrophic cardiomyopathy; LVNC, left ventricular noncompaction cardiomyopathy; LQTS, long QT syndrome; and PM, post mortem.
Most patients (3360 of 4270 [78.7%]) with an indication of a personal cardiomyopathy or arrhythmia had at least 1 subtype; among those, the most common subtypes were HCM (1393 patients [41.5%]), DCM (965 patients [28.7%]), and long QT syndrome (554 patients [16.5%]). In addition, 1996 of 3360 patients (59.4%) with at least 1 subtype listed had an associated index of suspicion, with 1226 patients (61.4%) having a high index of suspicion, 683 (34.2%) having a moderate index of suspicion, and 87 (4.4%) having a low index of suspicion.
Variants in Study Cohort
Overall, 1227 P/LP variants were identified in the cohort (eFigure 2 and eTable 4 in Supplement 1 and spreadsheet of variants observed in the comprehensive testing cohort in Supplement 2), including 42 copy number variants (3.4% of all P/LP variants). Seven of the 42 copy number variants (16.7%) encompassed only a single exon. Most copy number variants (34 [81.0%]) were in genes associated with autosomal dominant inheritance, whereas 6 copy number variants (14.3%) were in the X-linked DMD gene (4 male patients and 2 female patients). Two copy number variants (4.8%) were found in autosomal recessive genes but were monoallelic. Across all 1227 P/LP sequence variants and copy number variants, variants were most frequent in the MYBPC3 gene (205 patients [16.7%]) followed by the TTN gene (143 patients [11.7%]) and the MYH7 gene (111 patients [9.0%]). Across all 4782 patients, 1132 (23.7%) had at least 1 P/LP variant, and 89 (1.9%) had more than 1 P/LP variant; 2446 patients (51.2%) had 1 or more variant of uncertain significance in the absence of any P/LP variant.
Diagnostic Findings
Among all 4782 patients, 954 (19.9%) had a positive result (molecular diagnosis) (Figure 2). Of those, 25 patients (2.6%) had positive results in more than 1 gene. Notably, 8 patients (0.8%) had positive results associated with both cardiomyopathy and arrhythmia. Patients with a high index of suspicion were more likely to have a positive result than patients with a low index of suspicion (371 of 1441 patients [25.7%] vs 11 of 122 patients [9.0%]; P < .001) (eTable 4 in Supplement 1). However, those 11 patients with a low index of suspicion had a positive result. If testing had followed recent American Heart Association guidance and excluded patients with low and moderate indices of suspicion,6,37 at least 137 of 954 patients (14.4%) with positive results would have been missed.
Figure 2. Flowchart of Testing Results for Original Cohort and Family Members.
Family-specific follow-up testing could include up to 150 genes associated with cardiomyopathies and arrhythmias or be limited to a specific gene or variant.
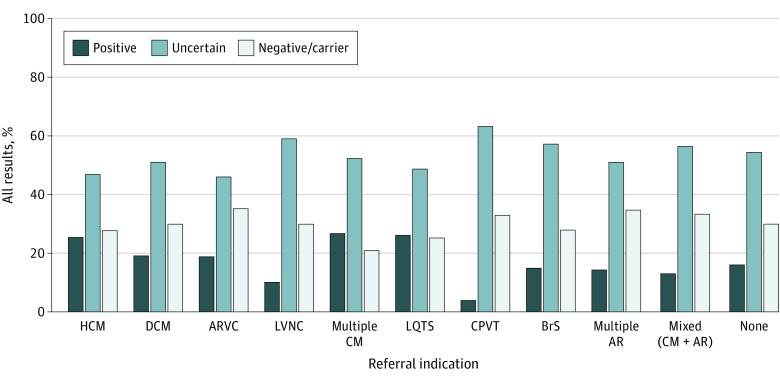
The diagnostic yield for those with an indication of a personal cardiomyopathy or arrhythmia was 867 of 4270 patients (20.3%). Diagnostic yield varied across conditions and age groups, with patients aged 19 to 39 years referred for suspicion of HCM having the highest yield (95 of 235 patients [40.4%]) (eTable 5 in Supplement 1). Among the 2596 patients with a cardiomyopathy indication, 574 (22.1%) had a positive result, 1273 (49.0%) had an uncertain (1 or more variants of uncertain significance only) result, and 749 (28.9%) had a negative or carrier result (eTable 4 in Supplement 1). Among patients with a single cardiomyopathy subtype indicated, HCM was associated with the highest yield (336 of 1321 patients [25.4%]), and left ventricular noncompaction cardiomyopathy was associated with the lowest yield (16 of 144 patients [11.1%]) (Figure 3). For patients with multiple cardiomyopathy subtypes indicated, the diagnostic yield was 23 of 86 patients (26.7%). Among 686 patients with an arrhythmia-specific diagnostic indication, 145 (21.1%) had a positive result, 355 (51.7%) had an uncertain result, and 186 (27.1%) had a negative or carrier result. Within the arrhythmia-specific diagnostic indications, long QT syndrome was associated with the highest yield (120 of 460 patients [26.1%]), and catecholaminergic polymorphic ventricular tachycardia was associated with the lowest yield (3 of 76 patients [3.9%]) (Figure 3). For those with multiple arrhythmia subtypes indicated, the yield was 7 of 49 patients (14.3%). Patients referred for arrhythmogenic cardiomyopathy had the lowest rate of variants of uncertain significance (81 of 176 patients [46.0%]), whereas patients referred for catecholaminergic polymorphic ventricular tachycardia had the highest rate of variants of uncertain significance (48 of 76 patients [63.2%]).
Figure 3. Test Results Stratified by Diagnostic Indication.
Negative or carrier category indicates results in which no molecular diagnosis was identified, including observation of only benign or likely benign variant(s), pseudodeficiency allele(s), or a pathogenic or likely pathogenic variant in an autosomal recessive gene or an X-linked recessive gene in female patients. AR indicates arrhythmia; ARVC, arrhythmogenic right ventricular cardiomyopathy; BrS, Brugada syndrome; CM, cardiomyopathy; CPVT, catecholaminergic polymorphic ventricular tachycardia; DCM, dilated cardiomyopathy; HCM, hypertrophic cardiomyopathy; LQTS, long QT syndrome; and LVNC, left ventricular noncompaction cardiomyopathy.
Clinical Management Implications
Overall, combined cardiomyopathy and arrhythmia gene panel testing identified clinically relevant variants for 1 in 5 patients suspected of having a genetic cardiomyopathy or arrhythmia. Genetic testing results would have supported more intensive monitoring or changes in medication or device use for 630 of 954 patients (66.0%) with a positive result (Table 1). Genetic information from those 954 patients estimated a more severe clinical course for 283 individuals (29.7%); 279 patients (29.2%) had results indicating sarcomeric HCM (associated with more adverse outcomes than nonsarcomeric HCM), and 4 patients (0.4%) had results in the LAMP2 gene (associated with quickly progressing disease). Positive results were associated with heightened arrhythmia risk in 300 patients (31.4%). In addition, 57 positive results (6.0%) were in a gene with a targeted molecular therapy (genes GAA, GLA, or TTR). Among all patients tested, 1 in 8 (13.2%) had a positive result that could potentially inform prognosis or clinical management.
Table 1. Possible Prognostic or Management Changes for Patients With Positive Test Results.
| Category | Genes | Potential clinical implications | Patients with positive test results, No. (%) (n = 954) |
|---|---|---|---|
| Adverse clinical outcomes among patients with HCM | ACTC1, MYL2, MYBPC3, MYH7, MYL3, TNNI3, TNNT2, TPM1 | Sarcomeric HCM with earlier adverse cardiovascular outcomes vs nonsarcomeric HCM, which necessitates earlier monitoring and medical and/or device intervention to address greater incidence of AF, VT, and/or HF26 | 279 (29.2)a |
| LAMP2 | Rapid deterioration with earlier consideration for heart transplant27 | 4 (0.4) | |
| Heightened arrhythmia risk | ABCC9, DES, DSP, FLNC, LMNA, PLN, RBM20, RYR2, SCN5A, TTN | More intense cardiac monitoring and/or device intervention to address AF, VT, and/or heart block28,29,30,31,32,33 | 300 (31.4) |
| Genes with targeted therapies | GAA, GLA, TTR | Treatment with enzyme replacement therapy (GAA and GLA) or molecular inhibitors or stabilizers (TTR)34,35 | 57 (6.0) |
| Any gene-specific management | Any gene listed above | Prognostic or treatment implications as described above | 630 (66.0)b |
Abbreviations: AF, atrial fibrillation; HCM, hypertrophic cardiomyopathy; HF, heart failure; VT, ventricular tachycardia.
Only patients with a diagnostic indication of HCM (alone or in combination with other indications) were included.
Patients with multiple gene-specific management implications (n = 10) were only counted once in this overall sum.
Genetic Diagnoses Gained With Combined Disease Testing
Among all 689 patients with a single disease subtype indicated, combined testing produced 75 more genetic diagnoses (10.9%) compared with estimated results of disease-specific testing offered by the commercial laboratory (Table 2). Most of this increased yield produced a genetic result within the same broad category of the diagnostic indication (eg, a specific cardiomyopathy was indicated, but genetic testing identified a different cardiomyopathy). These within-category differences comprised 33 of 75 gained diagnoses (44.0%), and all but 1 diagnosis were cardiomyopathies (eTable 6 in Supplement 1). Cardiomyopathy indications with arrhythmia-consistent genetic test results comprised 12 of 75 gained diagnoses (16.0%), and arrhythmia indications with cardiomyopathy-consistent genetic test results comprised 15 of 75 gained diagnoses (20.0%). Among 689 patients with positive results and a specific diagnostic indication, 27 of 75 (36.0%; 3.9% of all 689 patients) had an arrhythmia diagnosis but a positive test result for cardiomyopathy or vice versa.
Table 2. Comparison of Diagnostic Yield by Disease-Specific Gene Panel or Broad Panel Testing Among Patients With Single Disease-Specific Diagnostic Indications.
| Referral indicationa | Total patients with referral indication, No. | Patients, No./total No. (%) | P valued | ||
|---|---|---|---|---|---|
| With positive results | With positive results for genes not on disease-specific panel | ||||
| Broad panel diagnostic yieldb | Disease-specific diagnostic yield (estimated)c | ||||
| Total patients, No. | 3147 | 689 | 614 | 75 | .02 |
| HCM | 1321 | 336/1321 (25.4) | 303/1321 (22.9) | 33/336 (9.8) | .13 |
| DCM | 869 | 166/869 (19.1) | 161/869 (18.5) | 5/166 (3.0) | .76 |
| ACM | 176 | 33/176 (18.8) | 28/176 (15.9) | 5/33 (15.2) | .48 |
| LVNC | 144 | 16/144 (11.1) | 2/144 (1.4) | 14/16 (87.5) | .001 |
| LQTS | 460 | 120/460 (26.1) | 106/460 (23.0) | 14/120 (11.7) | .28 |
| CPVT | 76 | 3/76 (3.9) | 2/76 (2.6) | 1/3 (33.3) | .65 |
| Brugada syndrome | 101 | 15/101 (14.9) | 12/101 (11.9) | 3/15 (20.0) | .54 |
Abbreviations: ACM, arrhythmogenic cardiomyopathy; CPVT, catecholaminergic polymorphic ventricular tachycardia; DCM, dilated cardiomyopathy; HCM, hypertrophic cardiomyopathy; LQTS, long QT syndrome; LVNC, left ventricular noncompaction cardiomyopathy.
Based on clinician complete test requisition form. Individuals with zero or multiple suspected or known diagnostic indications were excluded.
Positive results were defined as 1 pathogenic or likely pathogenic (P/LP) variant in a gene associated with an autosomal dominant or X-linked disease, 2 P/LP variants in the same gene associated with an autosomal recessive disease, or 1 P/LP variant and 1 variant of uncertain significance, each in a different copy of the same gene (ie, in trans) in a gene associated with autosomal recessive disease.
Results for each indication-specific panel are based on an estimation of actual testing results in the broad panel. The genes included in the broad panel are shown in eTable 1 in Supplement 1, and those included in each disease-specific panel are shown in eTable 2 in Supplement 1.
P values from significance testing for difference between broad testing yield and disease-specific testing yield. The significance threshold was P < .05.
Family Testing
Among the 954 patients with a positive result, 306 (32.1%) had family members referred and tested. A total of 958 family members received testing (Figure 2), with 1 to 17 family members (mean [SD], 3.1 [2.8] family members) tested per index patient. Of those, 402 family members (42.0%) received a positive result, and 445 (46.5%) received a negative result. If the index patient had received testing with the laboratory’s disease-specific gene panel instead of the combined disease panel, 18 of 402 positive cascade test results (4.5%; approximately 1 in 23 family members) would have been missed.
Postmortem Testing
Among 4782 patients tested, 190 (4.0%) were decedents whose age at death ranged from 2 days to 85 years (median, 26 years). For 73 of 190 patients (38.4%), being deceased was the only inclusion criterion provided beyond the general program requirement of a suspected genetic cardiomyopathy or arrhythmia. A total of 102 decedents (53.7%) also had a diagnostic indication of a cardiomyopathy or arrhythmia, which was most frequently unspecified (48 individuals [47.1%]); 16 individuals (15.7%) had HCM and 11 individuals (10.8%) had DCM as diagnostic indications. Among the 190 decedents, a positive result was found for 18 individuals (9.5%), an uncertain result for 104 individuals (54.7%), and a negative or carrier result for 68 individuals (35.8%) (eFigure 3 in Supplement 1). After receipt of a positive result for the decedent, at least 1 family member of 11 decedents (61.1% of the 18 decedents with a positive result) received cascade testing, with a mean (range) of 4.0 (2.0-17.0) family members tested per decedent with a positive result. In total, 44 family members were tested; of those, 18 family members (40.9%) had a positive result.
Discussion
This cohort study evaluated combined cardiomyopathy and arrhythmia genetic testing without cost as a barrier among 4782 patients with a suspected cardiorhythm disease. The overall diagnostic yield (19.9%) was lower than that of a different cohort38 who received combined cardiomyopathy and arrhythmia testing to evaluate 105 genes (which found an overall diagnostic yield of approximately 30%). Our study’s permissive inclusion criteria, which required only a low level of clinician suspicion for a genetic association, may account for the overall lower diagnostic yield. In contrast to recent American Heart Association guidance recommending that testing be limited to patients with a high likelihood of disease,6,37 our results support a broader approach given that at least 14.4% of patients with a positive genetic result had been referred with a low or moderate suspicion of inherited cardiomyopathy or arrhythmia.
Although the index of clinical suspicion was able to estimate disease in some cases, 9.0% of tests with low clinician suspicion had a positive finding. This result suggests the importance of increasing access to comprehensive genetic testing because relevant genetic findings may be present even when phenotype data are inconclusive or unavailable. Age should also not exclude patients from genetic testing because this testing can reveal pathogenic variants in patients older than 60 years.39 Moreover, cardiomyopathies and arrhythmias may have overlapping clinical presentations, and variants in the same gene may produce more than 1 phenotype. For example, patients with positive findings in the SCN5A gene had a mix of referral indications, including specific arrhythmia subtypes, specific cardiomyopathy subtypes, and unknown subtypes. Without comprehensive disease testing, some of these patients would likely have missed opportunities for comprehensive care that addressed their full medical needs. Testing that encompasses multiple genetic etiologies may also be especially helpful for patients with left ventricular noncompaction cardiomyopathy given that the condition is not well defined and has few definitive genes.40,41,42 Combined disease testing may more efficiently identify a patient’s disease-causing variant.
Notably, this study found a 10.9% gain in genetic diagnoses that would have been missed if testing had been limited to genes associated with a single cardiomyopathy or arrhythmia subtype. Among the 689 patients with positive results and a specific diagnostic indication, 27 (3.9%) had an arrhythmia diagnosis but a positive test result for cardiomyopathy or vice versa. In addition, gained positive results were amplified by cascade testing; approximately 1 in 23 family members with positive results would have been missed by disease-restricted panels. Although testing in this study was performed by a single commercial laboratory, it is likely that any combined cardiomyopathy and arrhythmia panel (offered by many commercial laboratories) would have identified these patients. These findings aligned with other evidence against limited gene testing,43,44,45,46,47 including findings that 5% of patients with DCM and arrhythmias harbor variants in both ion channel and cardiomyopathy genes,44 10% of patients with early-onset atrial fibrillation harbor variants in cardiomyopathy genes,47 and 22% of patients with idiopathic cardiac arrest harbor variants in cardiorhythm genes.45
Most of the patients with positive results (66.0%) in this study had findings that could potentially inform patient care or prognosis. Findings from a previous study26 suggested that sarcomeric HCM was associated with worse outcomes than nonsarcomeric HCM, with increased risk of heart failure, earlier atrial fibrillation, and major ventricular arrhythmias. Positive genetic findings in arrhythmogenic cardiomyopathy genes have implications for monitoring and management of arrhythmia-associated complications.28,29,30,31,32,33 Disease-causing TTR variants have specific therapies.35 Moreover, all positive results may be actionable if there are living first-degree relatives because clinical screening of close relatives of index patients with positive results is recommended.3,4,5,6,7 Overall, 42.0% of family members who received testing had a positive result, and 46.5% had a negative result that allowed them to avoid intensive longitudinal screening. Although cascade testing of family members was used in 32.1% of positive cases overall, it was used in 61.1% of positive postmortem cases. This finding highlights the importance of including decedents in genetic testing, despite complexities in covering the cost of this testing.
There is an inherent trade-off in multidisease testing; although broad testing can identify more patients with genetic disease than disease-specific testing, it also detects more variants of uncertain significance. Within the full cohort, 51.2% of patients had 1 or more variant of uncertain significance in the absence of any P/LP variant, highlighting the need to resolve variants of uncertain significance. In some cases, clinical assessment and assessment of variants of uncertain significance in family members can promote definitive classification by clarifying the variant’s association with disease. Rates of variants of uncertain significance may be higher among non-White patients in clinical testing, and broader participation should help to reduce the disparity in these rates.48 In addition, academic and commercial groups are pursuing various methods for resolving variants of uncertain significance, including biochemical and cellular assays and machine learning algorithms.49,50
It is important to provide genetic counseling and clinician support services, both before and after testing, to manage expectations and increase understanding of variants of uncertain significance, which are considered medically nonactionable.6 Expert centers, where available, may be consulted to potentially aid in family testing and other means to promote resolution of variants of uncertain significance.51 Although there are concerns that genetic testing may increase health care costs, research is needed to reconcile the balance of those costs with the potential improved outcomes among patients found to harbor an actionable variant.
Limitations
This study has limitations. Because clinical data were not confirmed with medical records, and ordering clinicians were only required to document a patient’s eligibility for the program, some clinical information may have been omitted. The Electronic Medical Records and Genomics (eMERGE) Network52 and other efforts to better harness electronic records in genomic research could address this gap. Furthermore, family testing data were only available from the study laboratory; therefore, the amount of cascade testing performed may have been underestimated. Although more individuals of non-White ancestry were represented in the present cohort compared with similar studies,53,54,55 non-White participants remained underrepresented. This issue is of special concern for Black or African American patients, who receive fewer testing referrals than White patients and may gain less utility (or even false-positive results because of variant misclassifications) from testing when it is ordered.56,57,58 In addition, ancestry was determined by clinician report. Although the no-charge program was established to increase access to testing, the inclusion of patients who would not agree to receive typical for-cost testing may have introduced bias.
Conclusions
In this cohort study, comprehensive genetic testing for cardiomyopathies and arrhythmias revealed diagnoses that would have been missed by disease-specific testing or testing restricted to patients with high likelihood of disease. Furthermore, comprehensive testing provided diagnostic and prognostic information that could have potentially changed management and monitoring strategies for patients and their relatives. Although the chance of finding variants of uncertain significance increases as more genes are tested, the benefits of identifying more cases of genetic disease outweigh the challenges of higher detection of variants of uncertain significance.
eMethods. Next-Generation Sequencing, Patient Testing Outcomes, and Inheritance of Conditions Associated With Genetic Testing Results
eTable 1. Genes Analyzed in Comprehensive Cardiomyopathy and Arrhythmia Genetic Testing
eTable 2. Genes Addressed by Each Disease-Specific Panel Projected for Patients in the Comprehensive Cardiomyopathy and Arrhythmia Genetic Testing Cohort
eTable 3. Patient Characteristics
eTable 4. Diagnostic Yields by Diagnostic Indication Stratified by the Index of Clinical Suspicion
eTable 5. Test Results Stratified by Diagnostic Indication and Age Group
eTable 6. Gained Diagnoses in Combined Disease Testing Compared With Limited Gene Panel Testing
eFigure 1. Testing Requisition Form for the No Charge Program
eFigure 2. Prevalence of Pathogenic and Likely Pathogenic Variants by Gene and Diagnostic Indication
eFigure 3. Flowchart of Results for Postmortem Patients and Their Family Members
eReferences
Variants Observed in the Cardiomyopathy and Arrhythmia Comprehensive Testing Cohort
References
- 1.Ackerman MJ, Priori SG, Willems S, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Heart Rhythm. 2011;8(8):1308-1339. doi: 10.1016/j.hrthm.2011.05.020 [DOI] [PubMed] [Google Scholar]
- 2.Priori SG, Wilde AA, Horie M, et al. HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes. Heart Rhythm. 2013;10(2);1932-1963. doi: 10.1016/j.hrthm.2013.05.014 [DOI] [PubMed] [Google Scholar]
- 3.Hershberger RE, Givertz MM, Ho CY, et al. Genetic evaluation of cardiomyopathy—a Heart Failure Society of America practice guideline. J Card Fail. 2018;24(5):281-302. doi: 10.1016/j.cardfail.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Khatib SM, Stevenson WG, Ackerman MJ, et al. 2017 AHA/ACC/HRS guideline for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. Circulation. 2018;138(13):e272-e391. doi: 10.1161/CIR.0000000000000549 [DOI] [PubMed] [Google Scholar]
- 5.Towbin JA, McKenna WJ, Abrams DJ, et al. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardiomyopathy: executive summary. Heart Rhythm. 2019;16(11):e373-e407. doi: 10.1016/j.hrthm.2019.09.019 [DOI] [PubMed] [Google Scholar]
- 6.Musunuru K, Hershberger RE, Day SM, et al. ; American Heart Association Council on Genomic and Precision Medicine; Council on Arteriosclerosis, Thrombosis and Vascular Biology; Council on Cardiovascular and Stroke Nursing; and Council on Clinical Cardiology . Genetic testing for inherited cardiovascular diseases: a scientific statement from the American Heart Association. Circ Genom Precis Med. 2020;13(4):e000067. doi: 10.1161/HCG.0000000000000067 [DOI] [PubMed] [Google Scholar]
- 7.Ommen SR, Mital S, Burke MA, et al. 2020 AHA/ACC guideline for the diagnosis and treatment of patients with hypertrophic cardiomyopathy: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J Am Coll Cardiol. 2020;76(25):e159-e240. doi: 10.1016/j.jacc.2020.08.045 [DOI] [PubMed] [Google Scholar]
- 8.Morales A, Hershberger RE. The rationale and timing of molecular genetic testing for dilated cardiomyopathy. Can J Cardiol. 2015;31(11):1309-1312. doi: 10.1016/j.cjca.2015.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maron MS, Hellawell JL, Lucove JC, Farzaneh-Far R, Olivotto I. Occurrence of clinically diagnosed hypertrophic cardiomyopathy in the United States. Am J Cardiol. 2016;117(10):1651-1654. doi: 10.1016/j.amjcard.2016.02.044 [DOI] [PubMed] [Google Scholar]
- 10.Schwartz PJ, Ackerman MJ, Antzelevitch C, et al. Inherited cardiac arrhythmias. Nat Rev Dis Primers. 2020;6(1):58. doi: 10.1038/s41572-020-0188-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mital S, Musunuru K, Garg V, et al. ; American Heart Association Council on Functional Genomics and Translational Biology; Council on Cardiovascular Disease in the Young; Council on Cardiovascular and Stroke Nursing; Stroke Council; Council on Lifestyle and Cardiometabolic Health; and Council on Quality of Care and Outcomes Research . Enhancing literacy in cardiovascular genetics: a scientific statement from the American Heart Association. Circ Cardiovasc Genet. 2016;9(5):448-467. doi: 10.1161/HCG.0000000000000031 [DOI] [PubMed] [Google Scholar]
- 12.Spoonamore KG, Johnson NM. Who pays? coverage challenges for cardiovascular genetic testing in U.S. patients. Front Cardiovasc Med. 2016;3:14. doi: 10.3389/fcvm.2016.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behere SP, Weindling SN. Inherited arrhythmias: the cardiac channelopathies. Ann Pediatr Cardiol. 2015;8(3):210-220. doi: 10.4103/0974-2069.164695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cirino AL, Harris S, Lakdawala NK, et al. Role of genetic testing in inherited cardiovascular disease: a review. JAMA Cardiol. 2017;2(10):1153-1160. doi: 10.1001/jamacardio.2017.2352 [DOI] [PubMed] [Google Scholar]
- 15.Mazzarotto F, Olivotto I, Boschi B, et al. Contemporary insights into the genetics of hypertrophic cardiomyopathy: toward a new era in clinical testing? J Am Heart Assoc. 2020;9(8):e015473. doi: 10.1161/JAHA.119.015473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gal DB, Morales A, Rojahn S, et al. Comprehensive genetic testing for pediatric hypertrophic cardiomyopathy reveals clinical management opportunities and syndromic conditions. Pediatr Cardiol. 2022;43(3):616-623. doi: 10.1007/s00246-021-02764-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi SH, Weng LC, Roselli C, et al. ; DiscovEHR Study and the NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium . Association between titin loss-of-function variants and early-onset atrial fibrillation. JAMA. 2018;320(22):2354-2364. doi: 10.1001/jama.2018.18179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haggerty CM, Damrauer SM, Levin MG, et al. Genomics-first evaluation of heart disease associated with titin-truncating variants. Circulation. 2019;140(1):42-54. doi: 10.1161/CIRCULATIONAHA.119.039573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah S, Henry A, Roselli C, et al. ; Regeneron Genetics Center . Genome-wide association and mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun. 2020;11(1):163. doi: 10.1038/s41467-019-13690-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alfares AA, Kelly MA, McDermott G, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med. 2015;17(11):880-888. doi: 10.1038/gim.2014.205 [DOI] [PubMed] [Google Scholar]
- 21.Lincoln SE, Kobayashi Y, Anderson MJ, et al. A systematic comparison of traditional and multigene panel testing for hereditary breast and ovarian cancer genes in more than 1000 patients. J Mol Diagn. 2015;17(5):533-544. doi: 10.1016/j.jmoldx.2015.04.009 [DOI] [PubMed] [Google Scholar]
- 22.Truty R, Paul J, Kennemer M, et al. Prevalence and properties of intragenic copy-number variation in mendelian disease genes. Genet Med. 2019;21(1):114-123. doi: 10.1038/s41436-018-0033-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lincoln SE, Truty R, Lin CF, et al. A rigorous interlaboratory examination of the need to confirm next-generation sequencing–detected variants with an orthogonal method in clinical genetic testing. J Mol Diagn. 2019;21(2):318-329. doi: 10.1016/j.jmoldx.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nykamp K, Anderson M, Powers M, et al. ; Invitae Clinical Genomics Group . Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med. 2017;19(10):1105-1117. doi: 10.1038/gim.2017.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho CY, Day SM, Ashley EA, et al. Genotype and lifetime burden of disease in hypertrophic cardiomyopathy: insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe). Circulation. 2018;138(14):1387-1398. doi: 10.1161/CIRCULATIONAHA.117.033200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maron BJ, Roberts WC, Arad M, et al. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. JAMA. 2009;301(12):1253-1259. doi: 10.1001/jama.2009.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Spaendonck-Zwarts KY, van Hessem L, Jongbloed JDH, et al. Desmin-related myopathy. Clin Genet. 2011;80(4):354-366. doi: 10.1111/j.1399-0004.2010.01512.x [DOI] [PubMed] [Google Scholar]
- 29.Akhtar M, Elliott PM. Risk stratification for sudden cardiac death in non-ischaemic dilated cardiomyopathy. Curr Cardiol Rep. 2019;21(12):155. doi: 10.1007/s11886-019-1236-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akhtar MM, Lorenzini M, Cicerchia M, et al. Clinical phenotypes and prognosis of dilated cardiomyopathy caused by truncating variants in the TTN gene. Circ Heart Fail. 2020;13(10):e006832. doi: 10.1161/CIRCHEARTFAILURE.119.006832 [DOI] [PubMed] [Google Scholar]
- 31.Sammani A, Kayvanpour E, Bosman LP, et al. Predicting sustained ventricular arrhythmias in dilated cardiomyopathy: a meta-analysis and systematic review. ESC Heart Fail. 2020;7(4):1430-1441. doi: 10.1002/ehf2.12689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith ED, Lakdawala NK, Papoutsidakis N, et al. Desmoplakin cardiomyopathy, a fibrotic and inflammatory form of cardiomyopathy distinct from typical dilated or arrhythmogenic right ventricular cardiomyopathy. Circulation. 2020;141(23):1872-1884. doi: 10.1161/CIRCULATIONAHA.119.044934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akhtar MM, Lorenzini M, Pavlou M, et al. ; European Genetic Cardiomyopathies Initiative Investigators . Association of left ventricular systolic dysfunction among carriers of truncating variants in filamin C with frequent ventricular arrhythmia and end-stage heart failure. JAMA Cardiol. 2021;6(8):891-901. doi: 10.1001/jamacardio.2021.1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun A. Lysosomal storage disease overview. Ann Transl Med. 2018;6(24):476. doi: 10.21037/atm.2018.11.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siddiqi OK, Ruberg FL. Cardiac amyloidosis: an update on pathophysiology, diagnosis, and treatment. Trends Cardiovasc Med. 2018;28(1):10-21. doi: 10.1016/j.tcm.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sturm AC, Truty R, Callis TE, et al. Limited-variant screening vs comprehensive genetic testing for familial hypercholesterolemia diagnosis. JAMA Cardiol. 2021;6(8):902-909. doi: 10.1001/jamacardio.2021.1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landstrom AP, Kim JJ, Gelb BD, et al. ; American Heart Association Council on Genomic and Precision Medicine; Council on Lifelong Congenital Heart Disease and Heart Health in the Young; Council on Arteriosclerosis, Thrombosis and Vascular Biology; and Council on Lifestyle and Cardiometabolic Health. Genetic testing for heritable cardiovascular diseases in pediatric patients: a scientific statement from the American Heart Association. Circ Genom Precis Med. 2021;14(5):e000086. doi: 10.1161/HCG.0000000000000086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janin A, Januel L, Cazeneuve C, Delinière A, Chevalier P, Millat G. Molecular diagnosis of inherited cardiac diseases in the era of next-generation sequencing: a single center’s experience over 5 years. Mol Diagn Ther. 2021;25(3):373-385. doi: 10.1007/s40291-021-00530-w [DOI] [PubMed] [Google Scholar]
- 39.Cannatà A, Merlo M, Dal Ferro M, et al. Association of titin variations with late-onset dilated cardiomyopathy. JAMA Cardiol. 2022;7(4):371-377. doi: 10.1001/jamacardio.2021.5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross SB, Jones K, Blanch B, et al. A systematic review and meta-analysis of the prevalence of left ventricular non-compaction in adults. Eur Heart J. 2020;41(14):1428-1436. doi: 10.1093/eurheartj/ehz317 [DOI] [PubMed] [Google Scholar]
- 41.Ross SB, Singer ES, Driscoll E, et al. Genetic architecture of left ventricular noncompaction in adults. Hum Genome Var. 2020;7:33. doi: 10.1038/s41439-020-00120-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rojanasopondist P, Nesheiwat L, Piombo S, Porter GA Jr, Ren M, Phoon CKL. Genetic basis of left ventricular noncompaction. Circ Genom Precis Med. 2022;15(3):e003517. doi: 10.1161/CIRCGEN.121.003517 [DOI] [PubMed] [Google Scholar]
- 43.Hoss S, Habib M, Silver J, et al. Genetic testing for diagnosis of hypertrophic cardiomyopathy mimics: yield and clinical significance. Circ Genom Precis Med. 2020;13(2):e002748. doi: 10.1161/CIRCGEN.119.002748 [DOI] [PubMed] [Google Scholar]
- 44.Li Z, Chen P, Li C, et al. Genetic arrhythmias complicating patients with dilated cardiomyopathy. Heart Rhythm. 2020;17(2):305-312. doi: 10.1016/j.hrthm.2019.09.012 [DOI] [PubMed] [Google Scholar]
- 45.Isbister JC, Nowak N, Butters A, et al. “Concealed cardiomyopathy” as a cause of previously unexplained sudden cardiac arrest. Int J Cardiol. 2021;324:96-101. doi: 10.1016/j.ijcard.2020.09.031 [DOI] [PubMed] [Google Scholar]
- 46.Webster G, Puckelwartz MJ, Pesce LL, et al. Genomic autopsy of sudden deaths in young individuals. JAMA Cardiol. 2021;6(11):1247-1256. doi: 10.1001/jamacardio.2021.2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoneda ZT, Anderson KC, Quintana JA, et al. Early-onset atrial fibrillation and the prevalence of rare variants in cardiomyopathy and arrhythmia genes. JAMA Cardiol. 2021;6(12):1371-1379. doi: 10.1001/jamacardio.2021.3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landry LG, Rehm HL. Association of racial/ethnic categories with the ability of genetic tests to detect a cause of cardiomyopathy. JAMA Cardiol. 2018;3(4):341-345. doi: 10.1001/jamacardio.2017.5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo H, Liu L, Nishiga M, Cong L, Wu JC. Deciphering pathogenicity of variants of uncertain significance with CRISPR-edited iPSCs. Trends Genet. 2021;37(12):1109-1123. doi: 10.1016/j.tig.2021.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stein A, Fowler DM, Hartmann-Petersen R, Lindorff-Larsen K. Biophysical and mechanistic models for disease-causing protein variants. Trends Biochem Sci. 2019;44(7):575-588. doi: 10.1016/j.tibs.2019.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahmad F, McNally EM, Ackerman MJ, et al. Establishment of specialized clinical cardiovascular genetics programs: recognizing the need and meeting standards: a scientific statement from the American Heart Association. Circ Genom Precis Med. 2019;12(6):e000054. doi: 10.1161/HCG.0000000000000054 [DOI] [PubMed] [Google Scholar]
- 52.Gottesman O, Kuivaniemi H, Tromp G, et al. ; eMERGE Network . The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15(10):761-771. doi: 10.1038/gim.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rampersaud E, Siegfried JD, Norton N, Li D, Martin E, Hershberger RE. Rare variant mutations identified in pediatric patients with dilated cardiomyopathy. Prog Pediatr Cardiol. 2011;31(1):39-47. doi: 10.1016/j.ppedcard.2010.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kapplinger JD, Landstrom AP, Salisbury BA, et al. Distinguishing arrhythmogenic right ventricular cardiomyopathy/dysplasia-associated mutations from background genetic noise. J Am Coll Cardiol. 2011;57(23):2317-2327. doi: 10.1016/j.jacc.2010.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kapplinger JD, Landstrom AP, Bos JM, Salisbury BA, Callis TE, Ackerman MJ. Distinguishing hypertrophic cardiomyopathy–associated mutations from background genetic noise. J Cardiovasc Transl Res. 2014;7(3):347-361. doi: 10.1007/s12265-014-9542-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manrai AK, Funke BH, Rehm HL, et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. 2016;375(7):655-665. doi: 10.1056/NEJMsa1507092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cragun D, Weidner A, Lewis C, et al. Racial disparities in BRCA testing and cancer risk management across a population-based sample of young breast cancer survivors. Cancer. 2017;123(13):2497-2505. doi: 10.1002/cncr.30621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muller C, Lee SM, Barge W, et al. Low referral rate for genetic testing in racially and ethnically diverse patients despite universal colorectal cancer screening. Clin Gastroenterol Hepatol. 2018;16(12):1911-1918. doi: 10.1016/j.cgh.2018.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Next-Generation Sequencing, Patient Testing Outcomes, and Inheritance of Conditions Associated With Genetic Testing Results
eTable 1. Genes Analyzed in Comprehensive Cardiomyopathy and Arrhythmia Genetic Testing
eTable 2. Genes Addressed by Each Disease-Specific Panel Projected for Patients in the Comprehensive Cardiomyopathy and Arrhythmia Genetic Testing Cohort
eTable 3. Patient Characteristics
eTable 4. Diagnostic Yields by Diagnostic Indication Stratified by the Index of Clinical Suspicion
eTable 5. Test Results Stratified by Diagnostic Indication and Age Group
eTable 6. Gained Diagnoses in Combined Disease Testing Compared With Limited Gene Panel Testing
eFigure 1. Testing Requisition Form for the No Charge Program
eFigure 2. Prevalence of Pathogenic and Likely Pathogenic Variants by Gene and Diagnostic Indication
eFigure 3. Flowchart of Results for Postmortem Patients and Their Family Members
eReferences
Variants Observed in the Cardiomyopathy and Arrhythmia Comprehensive Testing Cohort