Abstract
In its role as a global response regulator, CtrA controls the transcription of a diverse group of genes at different times in the Caulobacter crescentus cell cycle. To understand the differential regulation of CtrA-controlled genes, we compared the expression of two of these genes, the fliQ flagellar gene and the ccrM DNA methyltransferase gene. Despite their similar promoter architecture, these genes are transcribed at different times in the cell cycle. PfliQ is activated earlier than PccrM. Phosphorylated CtrA (CtrA∼P) bound to the CtrA recognition sequence in both promoters but had a 10- to 20-fold greater affinity for PfliQ. This difference in affinity correlates with temporal changes in the cellular levels of CtrA. Disrupting a unique inverted repeat element in PccrM significantly reduced promoter activity but not the timing of transcription initiation, suggesting that the inverted repeat does not play a major role in the temporal control of ccrM expression. Our data indicate that differences in the affinity of CtrA∼P for PfliQ and PccrM regulate, in part, the temporal expression of these genes. However, the timing of fliQ transcription but not of ccrM transcription was altered in cells expressing a stable CtrA derivative, indicating that changes in CtrA∼P levels alone cannot govern the cell cycle transcription of these genes. We propose that changes in the cellular concentration of CtrA∼P and its interaction with accessory proteins influence the temporal expression of fliQ, ccrM, and other key cell cycle genes and ultimately the regulation of the cell cycle.
Temporal regulation of gene expression is central to the progression of the Caulobacter crescentus cell cycle. In each cell cycle (Fig. 1A), a motile swarmer cell releases its flagellum and is transformed into a DNA replication-competent stalked cell, which in turn differentiates into an asymmetric predivisional cell bearing a new flagellum at the pole opposite the stalk. Cell division then yields a flagellated swarmer cell and a nonmotile stalked cell (2, 9, 15). Key events that occur at consecutive stages of the cell cycle include the initiation of DNA replication in the stalked cell, biogenesis of the flagellum, methylation of the newly replicated DNA in predivisional cells, and cell division.
FIG. 1.
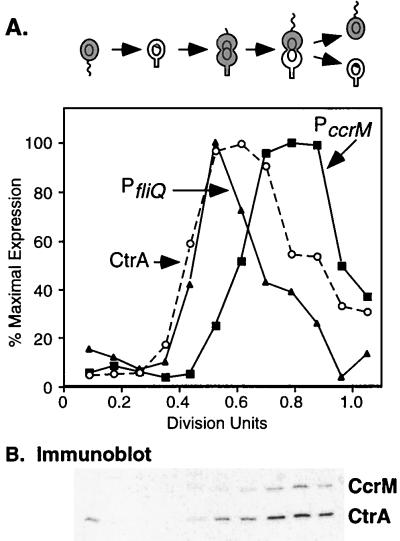
Temporal expression of CtrA and two target promoters in a single synchronized culture. (A) The Caulobacter cell cycle is shown schematically. The gray shading indicates the presence of CtrA (4). The theta structures indicate replicating DNA, and the ring structures within the cells represent nonreplicating DNA. Cultures of LS2531 carrying two transcriptional fusions, PfliQ-neo integrated into the chromosome and PccrM-lacZ on plasmid pCS148, were allowed to progress synchronously through the cell cycle. Every 15 min, samples were pulse-labeled with [35S]methionine and the synthesis of CtrA, neomycin phosphotransferase II, β-galactosidase, and flagellins was assessed by immunoprecipitation as described in Materials and Methods. Labeled proteins were separated by gel electrophoresis and quantitated with a PhosphorImager. CtrA synthesis (○), PfliQ transcription (▴), and PccrM transcription (■) are shown. Flagellin synthesis (data not shown) was assayed as an internal control for cell cycle progression. Cell division occurred at 180 min (1.0 division unit). (B) Immunoblot of CtrA and CcrM in cells from the same synchronized culture. Equal amounts of cellular protein (determined by measuring the A280) were applied to an SDS–12% polyacrylamide gel and probed with antibodies to the C. crescentus CtrA and CcrM proteins.
We have recently identified a regulatory protein that plays a pivotal role in orchestrating all of these cell cycle events. This protein, termed CtrA for cell cycle transcriptional regulator, is a member of the superfamily of response regulators (19). As part of a two-component regulatory system, the CtrA response regulator itself is controlled by phosphorylation. In addition, cell type-specific proteolysis of CtrA is essential for cell cycle progression (4). CtrA, which binds to five sites within the chromosomal origin of replication and inhibits DNA replication initiation, must be cleared from the stalked cell during the transition from swarmer cell to stalked cell (or G1-to-S-phase transition) to allow replication initiation (4, 20). Once DNA replication has begun, CtrA proteolysis stops and CtrA once again accumulates in early predivisional cells and is activated by phosphorylation (4, 19). CtrA is then selectively degraded in the stalked portion of late predivisional cells (4).
CtrA is an essential protein that, in addition to functioning as a negative regulator of DNA replication, controls the transcription of a number of genes. A key feature of the CtrA regulon is that these genes are differentially expressed at distinct times in the cell cycle. In early predivisional cells, CtrA activates the flagellar transcription hierarchy. Later in the cell cycle, CtrA initiates the transcription of ccrM, a gene encoding an essential DNA methyltransferase (19). CtrA also controls its own transcription by a positive feedback loop (5) and represses the transcription of ftsZ, which encodes a tubulin-like protein required for cell division (14).
To understand how CtrA activates or represses the appropriate promoter at the correct time in the cell cycle, we compared two temporally regulated events that are controlled by CtrA: the initiation of the flagellar transcription cascade and, later in the cell cycle, the transcription of the CcrM DNA methyltransferase. To examine these events, we focused on the fliQ operon, one of several class II flagellar operons that encode proteins required for the initial stages of flagellar biogenesis (32), and the ccrM gene, encoding a DNA methyltransferase that converts the newly replicated chromosomes from the hemimethylated to the fully methylated state in late predivisional cells (27). The fliQ and ccrM genes are transcribed sequentially, with fliQ being transcribed earlier than ccrM, yet both are dependent on CtrA for expression (19). The promoters of these genes have a similar architecture: both contain a single CtrA recognition sequence overlapping the −35 promoter element. We have previously shown that purified CtrA binds to this sequence in the fliQ promoter in vitro (19).
In the present study, we demonstrate that phosphorylated CtrA (CtrA∼P) preferentially binds to its recognition sequence in both the fliQ and the ccrM promoters but has a 10- to 20-fold greater affinity for the fliQ promoter. Because the differences in the timing of the initiation of fliQ transcription and ccrM transcription correlate with changes in cellular levels of total CtrA protein, we propose that the distinct affinities of CtrA∼P for its recognition sequence in these promoters control, at least in part, the sequential transcription of the fliQ and ccrM genes. We evaluated the role of a unique inverted repeat (IR2) in the ccrM promoter and showed that disrupting IR2 without changing the CtrA recognition sequence significantly reduced promoter activity but not the temporal expression of ccrM. A comparison of the timing of fliQ transcription and ccrM transcription in cells expressing a stable derivative of CtrA revealed that CtrA levels alone do not govern the temporal expression of these genes. Transcription of fliQ was prolonged, but the stringent regulation of ccrM transcription was maintained. Thus, in addition to variations in the level of CtrA∼P during the cell cycle, other transcription factors may contribute to the differential expression of these genes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. The synchronizable strain C. crescentus NA1000 (8) was used as the wild-type strain in all experiments. C. crescentus strains were grown at 30°C in PYE medium (18) or M2-glucose minimal medium (M2G) (7) supplemented with kanamycin (5 μg/ml) or tetracycline (1 μg/ml). Plasmids were mobilized from Escherichia coli S17-1 into C. crescentus by bacterial conjugation (6). E. coli strains were cultured at 37°C in Luria-Bertani broth supplemented with ampicillin or kanamycin (50 μg/ml each) or with tetracycline (10 μg/ml).
TABLE 1.
Strains, plasmids, and PCR primers used in this study
| Strain, plasmid, or primer | Relevant genotype or sequence | Reference or source |
|---|---|---|
| C. crescentus strains | ||
| NA1000 | Synchronizable derivative of C. crescentus CB15 | 8 |
| LS2515 | NA1000 with a chromosomal integration of ctrAΔ3M2 as the only copy of ctrA in the cell | 4 |
| LS2531 | NA1000 with a chromosomal integration of the PfliQ::neo reporter | 18a |
| Plasmids | ||
| pAR154 | pCR2.1 + PccrM (−44CGTGGT-to-AACCCC mutant) | This study |
| pAR155 | pCR2.1 + PccrM (−44CGTGG-to-AACCCC mutant) | This study |
| pAR156 | pRKlac290 + PccrM (−44CGTGGT-to-AACCCC mutant) | This study |
| pAR157 | pRKlac290 + PccrM (−44CGTGG-to-AACCCC mutant) | This study |
| pBluescriptII | Ampr cloning vector | Stratagene |
| pCR2.1 | Ampr Kanr vector used for cloning PCR products | Invitrogen |
| pCS148 | pRKlac290 + PccrM (−45 to +19) | 27 |
| pCS155 | pRKlac290 + PccrM (−36AC-to-TG mutant) | 27 |
| pCS156 | pRKlac290 + PccrM (−28CTAA-to-AATT mutant) | 27 |
| pCS179 | pBluescriptII + entire ccrM locus and promoter region | 27 |
| pKJH5 | C terminus of EnvZ in pMAL-c2 (New England Biolabs) | 11 |
| pRKlac290 | lacZ transcriptional fusion vector; pRK290 derivative | 10 |
| pTRC7.4 | pTrcHisA + ctrA coding sequence | 19 |
| pTrcHisA | Vector for expressing His6-containing proteins | Invitrogen |
| pWZ20 | pBluescriptII + entire fliQ locus and promoter | 32 |
| pWZ162 | pRKlac290 + PfliQ (−268 to +378) | 32 |
| pXD51E | ctrAD51E coding sequence inserted in pTRC7.4 | This study |
| PCR primersa | ||
| ccrM725 | (−178)GTCCCTCGCCGATCCATC(−160) | |
| ccrM1047R | (+144)GGGCGGATCCGCGAAGATCAGG(+122) | |
| ccrM1129R | (+226)CGAACGGATCCCAGTGGTCG(+206) | |
| ccrMecoRI | (−104)CTAGACCTTTGAATTCCTTCAACTTTG(−77) | |
| ccrM IR2mut | (−57)GCGCCTGAAAGGCAACCCCTAACGGCCC(−30) | |
| ccrMpstpe | (+159)CAGCTGCAGATTATAGGG(+141) | |
| fliQ399 | (−181)AGGTCCAGGTCGATCTGCT(−162) | |
| fliQ468 | (−112)GTATCCGCATCCGCTAATCA(−92) | |
| fliQ815R | (+235)AGATGAATTCCGCCACGATCT(+214) |
Sequences are oriented 5′ to 3′. Underlined bases are sites at which mutations were introduced to generate restriction sites or promoter mutants.
Culture synchronization, immunoprecipitation, and immunoblot analysis.
Swarmer cells were isolated from late-log-phase cultures by Ludox density centrifugation at 4°C (8), resuspended in M2G at 30°C, and allowed to progress through the cell cycle. The average cell cycle length was 150 min. To assess promoter expression during the cell cycle, C. crescentus NA1000, LS2515, or LS2531 cultures harboring the PccrM-lacZ transcriptional fusion pCS148 or the PfliQ-lacZ transcriptional fusion pWZ162 were synchronized as described above. Samples were taken at 15-min intervals, and 1-ml aliquots were labeled with 10 μCi of [35S]Trans-Label (ICN Pharmaceuticals Inc.) for 5 min. Synthesis of neomycin phosphotransferase II, β-galactosidase, and flagellins was monitored by immunoprecipitating these proteins from equal amounts of labeled cellular proteins as previously described (13). Flagellin synthesis was monitored as an indicator of the quality of synchrony. Proteins were analyzed by gel electrophoresis, and incorporated label was quantitated with a Molecular Dynamics PhosphorImager. Antibodies to NPT II and β-galactosidase were purchased from 5 Prime → 3 Prime, Inc. (Boulder, Colo.).
For immunoblot analysis, cellular proteins were separated on sodium dodecyl sulfate (SDS)–12% polyacrylamide gels and transferred to Immobilon-P membranes (Millipore) by the semidry transfer technique (1). The blots were probed with antibodies to C. crescentus CcrM and CtrA as previously described (4, 25). Bound antibodies were detected by chemiluminescence with peroxidase-conjugated secondary antibodies and a Renaissance chemiluminescence kit (Dupont, NEN Research Products). Prestained SDS-polyacrylamide gel electrophoresis standards were from Bio-Rad.
Protein purification and phosphorylation of His6-CtrA.
The six-histidine (His6)-CtrA and His6-CtrAD51E fusion proteins were overexpressed from pTRC7.4 and pXD51E, respectively, in E. coli BL21 (Novagen). The proteins were purified from the soluble fraction as recommended by Novagen, except that the wash buffer contained 40 mM imidazole and the His6-tagged fusion proteins were eluted with 0.5 M imidazole in binding buffer. Purified proteins were stored in 20 mM Tris (pH 8)–5 mM MgCl2–1 mM dithiothreitol (DTT)–50% glycerol. Maltose-binding protein (MBP)-EnvZ was purified after overexpression from plasmid pKJH5 as described previously (11).
To phosphorylate CtrA, purified MBP-EnvZ (0.5 μM), 0.4 mM ATP, and 10 μCi of [γ-32P]ATP were incubated in phosphorylation buffer (50 mM Tris-HCl [pH 7.5], 50 mM KCl, 20 mM MgCl2, 1 mM DTT) in a total volume of 20 μl. After 5 min at 37°C, a 4-μl aliquot was removed and added to SDS gel loading buffer as described previously (12). Purified His6-CtrA or His6-CtrAD51E (4.5 μM each) was added to the remainder of the reaction mixture, which was then incubated at 37°C. Samples were removed at various times and subjected to electrophoresis on SDS–12% polyacrylamide gels and autoradiography. For experiments that did not require radiolabeled protein, 5 mM ATP was added to the reaction mixture, which was then incubated for 20 min at 37°C.
DNase I protection experiments.
The DNA templates used in the footprinting reactions were generated by PCR as described below and labeled with [γ-32P]ATP by use of T4 DNA polynucleotide kinase. The 3′ end of the labeled template was removed by digestion at introduced BamHI or EcoRI sites. The labeled DNA was then purified with a QIAquik nucleotide removal kit (Qiagen Inc.) and subjected to DNase I footprinting analysis. The 347-bp fliQ promoter fragment (PfliQ −122 to +225) was generated by PCR with fliQ468 and fliQ815R as primers and pWZ20 as the template; the 330-bp wild-type ccrM promoter fragment (PccrM −104 to +226) was generated by PCR with ccrMecoRI and ccrM1129R as primers and pCS179 as the template. For the ccrM mutant promoters 25M and 35M, ccrMecoRI and ccrM1047R were the primers and pCS156 and pCS155 were the templates, respectively. The sequences of the oligonucleotide primers used for PCR are shown in Table 1. Templates for footprinting the ccrM mutant promoters IR2M(a) and IR2M(b) were end-labeled XbaI/AseI fragments of plasmids pAR154 and pAR155, respectively.
Footprinting experiments were performed with unphosphorylated and phosphorylated His6-CtrA (2.5 to 50 μg of protein/ml) essentially as described previously (1) but with the following modifications. Purified His6-CtrA was phosphorylated with MBP-EnvZ as described above and used immediately in DNase I footprinting reactions. The DNase I digestion products were isolated with a QIAquik PCR purification kit (Qiagen), concentrated in a SpeedVac, and separated on 8% polyacrylamide–7 M urea sequencing gels.
Assay of promoter activity.
The IR2M mutant promoters were constructed by PCR with ccrMIR2mut and ccrMpstpe as primers and pCS179 as the template. The promoter fragments (−58 to +159 relative to the transcription start site) were sequenced and then cloned into pRKlac290 upstream of a promoterless lacZ reporter to generate plasmids pAR156 and pAR157. β-Galactosidase activity was measured at 30°C with log-phase cultures as described by Miller (17). Assays were done in duplicate with a minimum of two independent cultures for each promoter construct.
Filter binding assays.
The binding of His6-CtrA to the fliQ and ccrM promoters was determined by measuring the retention of protein-DNA complexes on nitrocellulose filters as previously described (1, 3). DNA templates for the fliQ and ccrM promoters were generated by PCR with the CtrA binding site approximately centered within the fragment. To generate the 416-bp fliQ promoter fragment, the synthetic oligonucleotides fliQ399 and fliQ815R were used as primers and pWZ20 provided the template DNA. For the 337-bp ccrM promoter fragment, ccrM725 and ccrMpstpe were used as primers and pCS179 was used as the template. Table 1 gives the sequences of the oligonucleotide primers used for PCR. After purification with a QIAquik PCR purification kit, the PCR products (5 pmol each) were end labeled with T4 polynucleotide kinase and [γ-32P]ATP, and unincorporated label was removed with a QIAquik nucleotide removal kit according to the manufacturer’s instructions.
In the filter binding assays, 32P-labeled promoter DNA (100 pM) was incubated with increasing amounts of purified His6-CtrA in the presence or absence of MBP-EnvZ and 5 mM ATP in binding buffer (25 mM Tris [pH 7.4], 50 mM KCl, 10 mM MgCl2, 0.1 mM DTT, 0.1 mM EDTA, 0.025% Triton X-100, 100 μg of bovine serum albumin per ml) in a total volume of 100 μl. To minimize variability in the amount of CtrA∼P produced by the phosphorylation reactions, the same batches of purified His6-CtrA and MBP-EnvZ were used in all experiments. After incubation of the DNA template with His6-CtrA or His6-CtrA∼P at 37°C for 20 min, three 30-μl aliquots were filtered through nitrocellulose filters (BA85; Schleicher & Schuell) and washed twice with 250 μl of wash buffer (25 mM Tris [pH 7.4], 0.1 mM DTT, 0.1 mM EDTA). Filters were dried and analyzed by liquid scintillation counting. All data represent the average of 3 to 10 determinations.
RESULTS
Expression of CtrA and two of its target genes during the cell cycle.
A comparison of the temporal expression of CtrA-regulated genes in previous studies suggests that fliQ and other class II flagellar genes are transcribed earlier in the cell cycle than ccrM (26, 27, 32). To confirm this observation, we assessed the transcription of the fliQ and ccrM genes and the synthesis of CtrA protein in the same population of synchronized cells. In this experiment, the fliQ and ccrM promoters were fused to different reporters. A PfliQ-neo transcriptional fusion was integrated at the fliQ locus (strain LS2531), and a PccrM-lacZ transcriptional fusion was present on a low-copy-number plasmid (pCS148). Synchronized LS2531 cells containing pCS148 were pulse-labeled with [35S]methionine at different times in the cell cycle, and the incorporation of label into CtrA and the two reporter proteins was assessed by immunoprecipitation. As shown in Fig. 1A, the induction of CtrA synthesis coincided with fliQ transcription at 0.4 division unit, while the initiation of ccrM transcription was delayed until approximately 0.6 division unit. In addition, fliQ transcription stopped earlier than ccrM transcription. The timing of fliQ and ccrM expression relative to flagellin synthesis (used as an internal control in this experiment) agrees with that previously described in separate experiments with wild-type cultures (27, 32). These data provide definitive evidence that the fliQ and ccrM genes are expressed sequentially during the cell cycle. The pattern of CtrA synthesis mirrors that of ctrA transcription, which also initiates at approximately 0.4 division unit and peaks at 0.6 division unit (19).
The steady-state levels of the CcrM and CtrA proteins in cells isolated during this synchrony experiment are shown in the immunoblot in Fig. 1B. As previously described, CtrA protein is present in swarmer cells at the beginning of the cell cycle, is absent in stalked cells due to rapid proteolysis, and then reappears in predivisional cells at 0.4 division unit (4). In contrast, CcrM protein is present only in late predivisional cells and is first detected at 0.6 division unit, when the cellular levels of CtrA, its transcriptional activator, have increased approximately eightfold (determined by densitometry). CcrM is then rapidly degraded at cell division (30). We have been unsuccessful in raising antibodies to the FliQ integral membrane protein and so were unable measure cellular levels of this protein.
Comparison of the ccrM and fliQ regulatory regions.
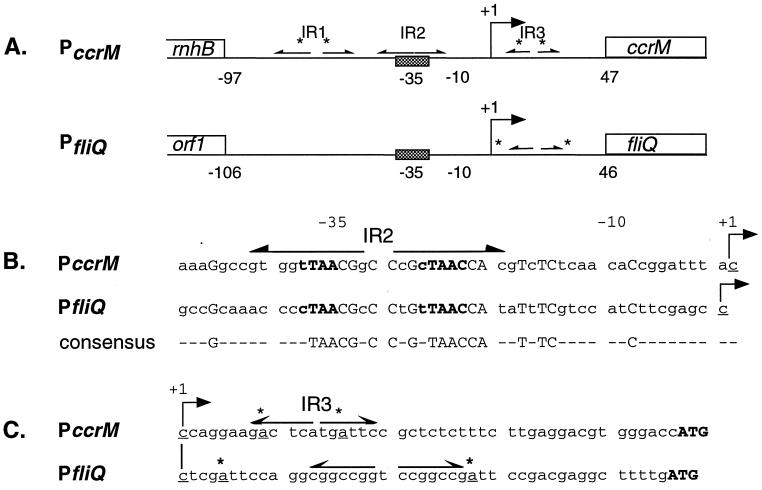
The ccrM and fliQ promoters have a similar architecture. As shown in Fig. 2A, both contain a single CtrA binding site overlapping the −35 region, with the CtrA recognition sequence closely matching the published consensus sequence (TTAA-n7-TTAAC) (19, 27, 32). Mutations in the CtrA recognition sequence dramatically decrease the transcription of both fliQ and ccrM (27, 32), indicating that CtrA activates the transcription of these target genes. In the ccrM promoter, the CtrA recognition sequence is embedded in a 25-bp inverted repeat (IR2) centered at −32. The regions from −20 to −45 in both promoters are similar: 14 of the 25 nucleotides in IR2 are identical to bases in the analogous region of the fliQ promoter, with the least homology in the 5′ region (Fig. 2B). The ccrM promoter is the only identified CtrA target promoter that contains the CtrA binding site in an inverted repeat.
FIG. 2.
Comparison of the ccrM and fliQ promoters. (A) Diagram of the ccrM and fliQ promoter regions. The bent arrows indicate the transcription start sites, the cross-hatched boxes denote the CtrA recognition sequence, inverted repeats are shown by divergent arrows, and GAnTC methylation sites are indicated by asterisks. The three inverted repeats in the ccrM promoter are labeled IR1, IR2, and IR3. (B) Sequences of the promoter regions from −50 to the transcription start sites. The consensus CtrA recognition sequence is in boldface type, the IR2 in the ccrM promoter is indicated by divergent arrows, and the start sites are indicated by bent arrows. Identical bases in both promoters are capitalized and are also shown below in the consensus sequence. (C) Sequences of the promoter regions from the transcription start sites to the putative translation start sites. Symbols are as described above.
In both promoters, inverted repeat elements flanked by CcrM methylation sites (GAnTC) are located immediately downstream of the start sites (Fig. 2C). In fact, a pair of CcrM methylation sites generates the 10-bp inverted repeat (IR3) in the ccrM leader region. When both methylation sites in IR3 are mutated, PccrM activity is elevated and transcription is maintained in predivisional and swarmer cells (27), suggesting that the methylation of this promoter element functions to inhibit transcription. The role of the inverted repeat in the fliQ leader region has not been defined. A striking difference between the promoters is the presence of three inverted repeats in the ccrM regulatory region. Deleting IR1 does not affect the level or timing of ccrM transcription, but both IR2 and IR3 are required for promoter activity (27).
CtrA∼P binds to the CtrA recognition sequence in the ccrM and fliQ promoters.
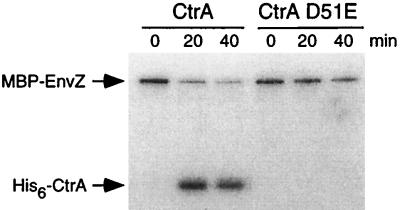
In the two-component system paradigm, phosphorylation activates the response regulator, enhancing its binding to promoter DNA and allowing the control of transcription of its target genes. We used an in vitro method to phosphorylate purified CtrA in order to assess the binding of CtrA∼P to its target promoters. Purified MBP-EnvZ, a fusion of E. coli MBP and the C terminus of the E. coli histidine kinase EnvZ (11), was used as the phosphate donor. In the presence of [32P]ATP, MBP-EnvZ underwent autophosphorylation, and the phosphate was transferred to purified His6-CtrA (Fig. 3). The His6-CtrAD51E mutant protein, in which the aspartate 51 phosphorylation site has been mutated to glutamate, was not phosphorylated in the in vitro reaction. These data indicate that the phosphate was transferred to the conserved aspartate 51 residue of His6-CtrA. Using pulse-chase experiments (data not shown), we determined that His6-CtrA∼P was stable in vitro in the presence of MBP-EnvZ (half-life, 50 min).
FIG. 3.
CtrA is phosphorylated at conserved aspartate 51 in vitro. Purified His6-CtrA and the His6-CtrAD51E mutant protein were incubated at 37°C with the MBP-EnvZ fusion protein and [γ-32P]ATP as described in Materials and Methods. Samples were removed at the indicated times and analyzed by SDS–12% polyacrylamide gel electrophoresis and autoradiography. MBP-EnvZ phosphorylated His6-CtrA but not the His6-CtrAD51E mutant protein, in which the conserved aspartate 51 has been mutated to glutamate.
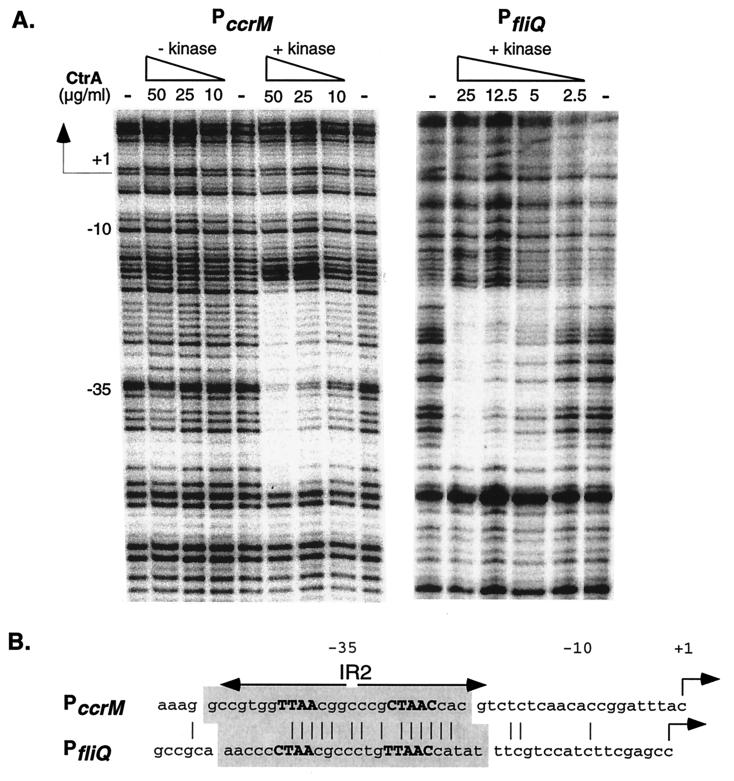
DNase I footprinting was used to compare the binding of His6-CtrA∼P prepared in the in vitro reaction to the ccrM and fliQ promoters. Template DNA was generated by PCR as described in Materials and Methods and labeled at the 5′ end. The PccrM template was a 330-bp fragment corresponding to positions −104 to +226 with respect to the transcription start site (27), and the PfliQ template was a 347-bp fragment corresponding to positions −122 to +225 (32). As shown in Fig. 4A, CtrA∼P specifically protected a single 26-bp region from −21 to −46 relative to the transcription start site in the ccrM promoter. However at the same protein concentrations, unphosphorylated CtrA did not bind to this promoter, indicating that CtrA∼P has a greater affinity for the CtrA binding site in the ccrM promoter. As expected, CtrA∼P protected the CtrA recognition sequence in the fliQ promoter. The single 26-bp protected region corresponded to the region protected by higher concentrations of unphosphorylated CtrA (19). Because CtrA was phosphorylated by use of MBP-EnvZ and both proteins were present in the DNase I protection reactions, we determined that MBP-EnvZ alone does not bind to either promoter (data not shown). Figure 4B compares the sequences protected from DNase I digestion in the ccrM and fliQ promoters. In both promoters, the protected regions overlap the −35 promoter element and are the same size. In the ccrM promoter, the region protected by CtrA∼P coincides with IR2.
FIG. 4.
DNase I footprinting of the ccrM and fliQ promoters by phosphorylated CtrA. (A) DNase I protection of PccrM and PfliQ with purified His6-CtrA (− kinase) and His6-CtrA∼P (+ kinase). His6-CtrA concentrations (2.5 to 50 μg/ml) in each footprinting reaction are indicated above the lanes. His6-CtrA was omitted from the reactions in the lanes marked with a minus sign. The transcription start site (+1) and the −10 and −35 sites are shown. (B) Comparison of the regions protected by CtrA∼P in the ccrM and fliQ promoters. The protected sequences are shaded; the CtrA recognition sequence in each promoter is in boldface capitalized letters; identical bases are shown by vertical lines. Other symbols are as described in the legend to Fig. 2.
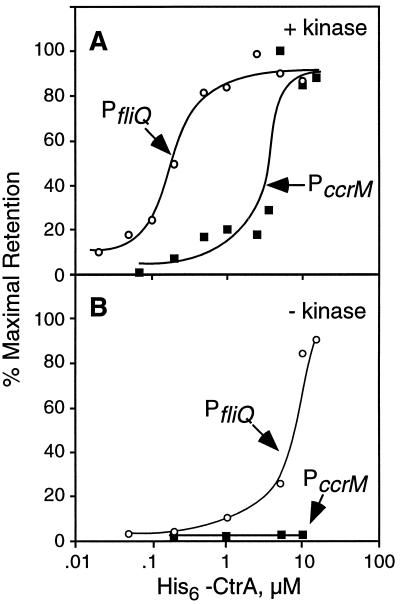
CtrA∼P has a greater affinity for the fliQ promoter than for the ccrM promoter.
The DNase I footprinting analysis shown in Fig. 4 suggests that CtrA∼P has a greater affinity for the fliQ promoter than for the ccrM promoter. To confirm this observation, we compared the affinity of CtrA∼P for both promoters by using a filter binding assay (3). Although we cannot directly calculate the actual CtrA∼P concentrations in the in vitro phosphorylation reactions, we used the same preparations of His6-CtrA and MBP-EnvZ in multiple assays to reveal relative binding affinities. As shown in Fig. 5A, CtrA∼P had a 10- to 20-fold greater affinity for PfliQ than for PccrM (half-maximal binding occurred at 0.2 and 3 to 4 μM CtrA for the fliQ and ccrM promoters, respectively). The sigmoidal shape of these curves suggests cooperative binding of CtrA∼P to both promoters. To verify this notion, we performed a fit of the Hill equation (23) to the data by using the program Statistica (Statsoft, Inc., Tulsa, Okla.). The Hill coefficient, an indicator of cooperativity, was 2.4 for the fliQ promoter (r2, 0.96), indicating that CtrA∼P binds as a dimer to this promoter. We were unable to calculate a statistically significant value for the Hill coefficient for the ccrM promoter due to the variability in the data. The greater-than-10-fold difference in the apparent binding constants is consistent with our in vivo results indicating that ccrM transcription initiates after fliQ transcription, at a time when CtrA protein levels have increased at least eightfold (Fig. 2B). Together, our results suggest that changes in the cellular concentration of CtrA∼P account, at least in part, for the differences in the timing of fliQ and ccrM transcription initiation.
FIG. 5.
Affinity of CtrA∼P and unphosphorylated CtrA for the fliQ and ccrM promoters. The binding of CtrA and CtrA∼P to the fliQ (○) and ccrM (■) promoters was determined by measuring the retention of protein-DNA complexes on nitrocellulose filters as described in Materials and Methods. (A) His6-CtrA was first phosphorylated with the MBP-EnvZ kinase and then incubated with labeled fliQ or ccrM promoter fragments for 20 min at 37°C as described in Materials and Methods. (B) The kinase was omitted from the reactions. The relative amounts of the protein-DNA complexes retained on the filters are plotted as a function of His6-CtrA concentration. Each point represents an average of 3 to 10 determinations. Standard error was typically less than 15%.
The filter binding assay also confirmed the observation that phosphorylation significantly increased the affinity of CtrA for both promoters. Phosphorylation enhanced the affinity of CtrA for the fliQ promoter approximately 50-fold (compare the half-maximal binding in Fig. 5A and B). Consequently, the contribution of unphosphorylated CtrA present in the in vitro reaction to the apparent binding of CtrA∼P to the fliQ promoter is negligible (about 2%). Unphosphorylated CtrA did not bind to the ccrM promoter at the concentrations tested (Fig. 5B).
Role of promoter architecture in regulating ccrM transcription.
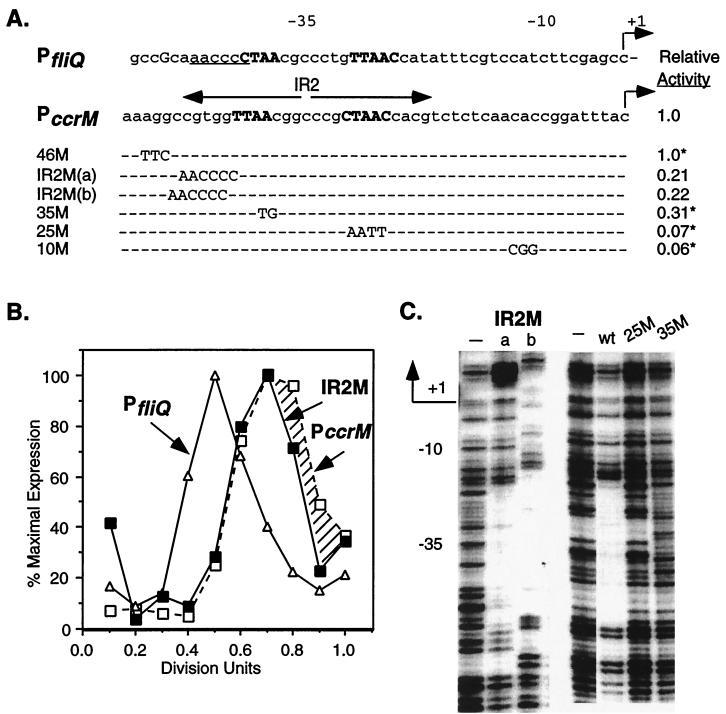
To assess the role of the IR2 element in the temporal control of ccrM expression, we mutated the IR2 region in PccrM and examined the effects of the mutations on transcriptional activity, temporal expression, and CtrA∼P binding (Fig. 6). First, the 5′ end of IR2 was disrupted without changing the CtrA consensus sequence by mutating bases −39 to −44. Two IR2M mutant promoters were constructed: IR2M(a), in which the first 6 nucleotides in IR2 were replaced with bases present at these positions in the fliQ promoter; and IR2M(b), which is identical to IR2M(a) except that the mutant sequence was inserted 1 bp upstream and retained the complete TTAA arm of the CtrA recognition sequence. As shown in Fig. 6A, both IR2M mutations reduced promoter activity by approximately 80%. The effects of mutations in PccrM from a previous study (27) are shown for comparison. A 3-bp change in the region directly upstream of IR2 (46M) had no effect on promoter activity. A 2-bp substitution at −35 (35M) reduced activity by 70%, while a 4-bp change in the CtrA recognition sequence (25M) and a 3-bp substitution at −10 (10M) reduced activity by greater than 90% (27). Comparable mutations in the −10, −25, and −35 regions of PfliQ also reduced promoter activity by greater than 90% (32).
FIG. 6.
Role of the IR2 in regulating PccrM expression. (A) Effects of mutations on ccrM promoter activity. The wild-type fliQ and ccrM promoters are shown; nucleotides in the CtrA recognition sequence are capitalized and boldfaced. Mutant bases in PccrM are shown beneath the wild-type bases being replaced. Promoters with changes in the 5′ region of the inverted repeat [IR2M(a) and IR2M(b)] were constructed by replacing the first 6 nucleotides in IR2 with the bases at these positions in PfliQ (underlined in the wild-type sequence). The IR2M mutations were cloned into pRKlac290 to create transcriptional fusions, and the mutants were assayed for β-galactosidase activity. The activity of control plasmid pCS145, defined here as 1.0, was 3,200 Miller units. Shown for comparison are PccrM mutants and their relative β-galactosidase activities (asterisks) from a previous study (27). (B) Temporal expression of wild-type PfliQ (▵), IR2M (■), and wild-type PccrM (□) in synchronous LS2531 cultures. See the legend to Fig. 1 for details. The hatched region indicates the differences in the expression of the wild-type and mutant ccrM promoters. (C) DNase I protection of ccrM mutant promoters and the wild-type ccrM promoter (wt) by His6-CtrA∼P (50 μg/ml). Data for the IR2M(a) and IR2M(b) mutant promoters and promoters with changes at conserved bases −25 to −28 (25M) and bases −35 and −36 (35M) are shown. The regions protected by CtrA∼P in the wild-type and IR2M promoters are of the same size. The protected regions in the IR2M(a) and IR2M(b) promoters are at different positions because the 5′ end of the IR2M(b) template contained an additional 3 nucleotides. Control reactions without His6-CtrA∼P are indicated by minus signs.
To determine if disrupting IR2 affects the temporal regulation of PccrM, we assessed the timing of the expression of the IR2M mutant promoters. LS2531 cells containing the IR2M-lacZ transcriptional fusions pAR156 and pAR157 were synchronized, and the expression of the ccrM, fliQ, and IR2M promoters was monitored throughout the cell cycle. As shown in Fig. 6B, the transcription of the IR2M(b) and wild-type ccrM promoters was induced at the same time in the cell cycle. Both exhibited the wild-type PccrM time of transcription initiation, but the activity of the mutant promoter decreased earlier in the cell cycle. Similar results were obtained for the IR2M(a) promoter (data not shown). Although the first 6 bases of the inverted repeat are important for promoter activity, they are not required for the temporal control of transcription initiation from PccrM.
DNase I footprinting assays were used to determine if CtrA∼P binds to the ccrM mutant promoters in vitro. The same concentration of CtrA∼P protected both IR2M promoters and wild-type PccrM but not the 25M and 35M promoters, with mutations in each arm of the CtrA recognition sequence (Fig. 6C). The 25M and 35M mutations decreased promoter activity and disrupted CtrA∼P footprinting, providing further evidence that CtrA acts as a transcriptional activator at PccrM. Although the nucleotides in IR2 upstream of the CtrA recognition sequence are important for promoter function, they do not appear to be essential for CtrA∼P binding or for the timing of transcription initiation.
Temporal regulation of fliQ and ccrM transcription in cells expressing a stable CtrA derivative.
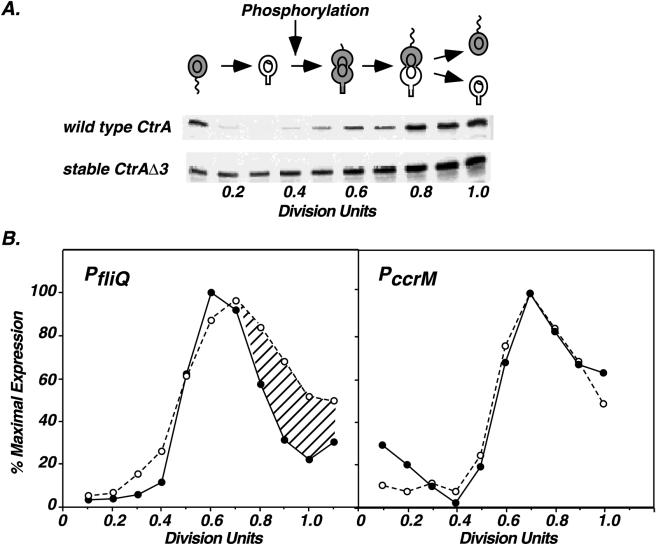
To determine if changes in the cellular level of CtrA affect the timing of ccrM and fliQ transcription, we compared the in vivo expression of these genes in wild-type cells and in cells expressing CtrAΔ3, a biologically active, stable CtrA derivative that lacks the C-terminal degradation signal (4). In these cells, the stable ctrAΔ3M2 allele is expressed from the ctrA promoter and replaces the chromosomal copy of ctrA. The immunoblot in Fig. 7A compares the steady-state levels of wild-type CtrA and the stable CtrAΔ3 derivative during the cell cycle. As previously described, the wild-type protein is present in swarmer cells and predivisional cells but absent in stalked cells. In contrast, the stable CtrAΔ3 derivative is present throughout the cell cycle (4). Phosphorylation of the wild-type CtrA protein occurs shortly after its synthesis at 0.4 division unit. The timing of CtrA phosphorylation is tightly regulated: if CtrA is forced to be present throughout the cell cycle, it is still only phosphorylated at 0.4 division unit in early predivisional cells. Although CtrAΔ3 is present in stalked cells, it is not phosphorylated (4).
FIG. 7.
Temporal expression of ccrM and fliQ in cells expressing wild-type CtrA and stable CtrAΔ3. (A) CtrA immunoblots of cell extracts from synchronous cultures of wild-type cells (NA1000) and LS2515 cells expressing stable CtrAΔ3. Extracts from equal amounts of cells were applied to each lane. The point in the cell cycle when CtrA is first phosphorylated (0.4 division unit) is indicated. See the legend to Fig. 1 for an explanation of symbols. (B) Comparison of fliQ and ccrM transcription in synchronous wild-type (●) and LS2515 (○) cultures. Temporal expression of PfliQ-lacZ (from pWZ162) and PccrM-lacZ (from pCS148) was determined by quantitating labeled β-galactosidase as described in the legend to Fig. 1. Flagellin synthesis was used as an internal control for comparing the timing of fliQ and ccrM transcription in NA1000 and LS2515 cultures. Differences in PfliQ expression between the wild-type and LS2515 cultures are indicated by hatching. The transcription of ccrM and fliQ in LS2515 cultures represents an average of two and three experiments, respectively. Variations among experiments were <7% for PccrM activity and <13% for PfliQ activity.
As shown in Fig. 7B, the timing of fliQ transcription was altered in cells expressing the stable protein. Transcription was prolonged and remained elevated in late predivisional cells. Using immunofluorescence microscopy, we have previously shown that wild-type CtrA is found only in the swarmer compartment of late predivisional cells, while the stable derivative is present at both poles (4). The presence of CtrAΔ3 in the stalked compartment of late predivisional cells may signal the inappropriate transcription of fliQ. In contrast, the timing of ccrM transcription was identical in cells expressing either wild-type CtrA or the stable CtrAΔ3 derivative. Thus, altering the temporal and spatial distribution of total CtrA protein affected the timing of fliQ transcription but not ccrM transcription, which appears to be more stringently controlled. These data imply that changes in CtrA∼P levels alone cannot govern the temporal expression of these genes. In addition, fliQ and ccrM are not transcribed in swarmer cells, and the transcription of these genes decreases in predivisional cells, although CtrA is present and active. Furthermore, neither gene is transcribed efficiently in stalked cells when CtrA is forced to be present, suggesting that unphosphorylated CtrA cannot activate their transcription.
DISCUSSION
The response regulator family of two-component signal transduction systems controls the expression of a wide variety of genes. One of these, the CtrA transcriptional activator, plays a key role in regulating diverse events that occur at different times in the Caulobacter cell cycle, including the initiation of DNA replication, the initiation of cell division, flagellar biogenesis, and the synthesis of the CcrM DNA methyltransferase (14, 19, 20). In order to understand how CtrA controls the timing of these events, we studied factors that contribute to the regulation of two genes, fliQ and ccrM, that are dependent on CtrA and that are transcribed sequentially. Differences in the affinity of CtrA∼P for PfliQ and PccrM accompanied by changes in the cellular concentration of CtrA during the cell cycle contribute to the observed temporal regulation of fliQ and ccrM transcription. Similar differences in the affinity of the BvgA response regulator for promoter DNA regulate the expression of early and late virulence genes in Bordetella pertussis (33). In addition, changes in the intracellular concentration of BvgA correlate with the timing of the transcription of these virulence genes (22).
Phosphorylation increased the affinity of CtrA for its recognition sequence in promoter DNA in vitro, as is the case for OmpR and other response regulators (16, 19, 21). Moreover, CtrA∼P exhibited distinct affinities for two promoters that are expressed sequentially in predivisional cells, the fliQ and ccrM promoters. Changes in the CtrA∼P concentration during the cell cycle may therefore play an important role in the temporal regulation of its target promoters. Like those of other response regulators, CtrA∼P levels are likely to be modulated by opposing kinase and phosphatase activities. In addition, controlled proteolysis of CtrA results in variations in the CtrA concentration during the cell cycle (4).
Although CtrA∼P has a 10- to 20-fold greater affinity for the fliQ promoter than for the ccrM promoter, both promoters contain a single CtrA recognition sequence, and CtrA binds at the same location in each promoter. These differences in affinity appear to be related to subtle changes in promoter architecture. Even though the consensus CtrA recognition sequence is found in both promoters, this motif is embedded in an inverted repeat in the ccrM promoter. Altering the IR2 in the ccrM promoter without changing the CtrA recognition sequence did not change its time of transcription initiation but significantly reduced promoter activity. These data suggest that the inverted repeat structure may provide a binding site for an accessory factor that is required for activating ccrM transcription.
Other evidence supports the contention that changes in the cellular CtrA∼P concentration alone cannot control the sequential activation of fliQ and ccrM transcription. In experiments with the ctrAΔ3M2 allele, which encodes the stable CtrAΔ3 derivative, the precise timing of ccrM expression was unchanged despite constant CtrA levels throughout the cell cycle. In contrast, fliQ transcription was prolonged under the same conditions. CtrAΔ3 not only is biologically active but also is phosphorylated at the same time in the cell cycle as the wild-type protein (0.4 division unit) (4). If variations in the affinity of CtrA∼P for promoter DNA alone control the timing of gene expression, one would expect that ccrM would be transcribed earlier in cells expressing the stable CtrAΔ3 derivative.
There is precedence for the control of gene expression by complexes of regulatory proteins in other prokaryotes. In E. coli, the RcsC-RcsB two-component regulatory system that activates the synthesis of capsule polysaccharides uses RcsA as an accessory factor. RcsA is an unstable protein that appears to interact directly with the RcsB response regulator to activate transcription (28). The interaction of two other transcriptional regulators, cyclic AMP receptor protein (CRP) and the CytR repressor, controls the expression of genes in the CytR regulon which encode enzymes and transport proteins required for nucleoside catabolism and recycling. Repression of CytR-regulated promoters requires direct protein-protein contact between CRP and CytR, while the binding of CRP alone activates transcription (24). A novel regulator, KipI, that inhibits the autophosphorylation of kinase A has recently been identified (29). Kinase A is one of the key proteins in the phosphorelay that activates Spo0A and initiates the sporulation pathway in Bacillus subtilis. Such interactions among regulatory proteins provide a mechanism that allows a small number of transcription factors, such as CRP, Spo0A, and CtrA, to control global regulatory programs.
We speculate that interactions of CtrA with other proteins influence the binding specificity of CtrA for different promoters at different stages of the cell cycle. Because the initiation of transcription of class II flagellar genes coincides with the initiation of CtrA synthesis and phosphorylation in early predivisional cells, we propose that the transcription of these genes is activated by low levels of CtrA∼P alone. This concept is supported by our demonstration that the fliQ promoter contains a high-affinity binding site for CtrA∼P and by the activation of in vitro transcription of other class II flagellar genes by CtrA∼P (31). The initiation of ccrM transcription later in the cell cycle requires higher concentrations of CtrA∼P and a putative accessory protein. Because mutations in the IR3 element extend ccrM expression to swarmer cells (27), we also propose that a repressor might bind to the inverted repeat element downstream of the ccrM start site to terminate transcription. Experiments to identify these additional regulators are in progress. In summary, accessory proteins may contribute to the specificity of CtrA∼P for different promoters at different stages of the cell cycle. Changes in the cellular concentration of CtrA∼P and its interaction with accessory proteins could influence the temporal expression of key cell cycle genes and ultimately the regulation of the cell cycle.
ACKNOWLEDGMENTS
We thank Rachel Wright and other members of the Shapiro laboratory for helpful discussions and for reading the manuscript. We are grateful to Adam Arkin for calculating the Hill coefficients; John Wang for constructing plasmid pXD51E, used for overexpressing His6-CtrAD51E; and Michele Igo and Ke-Jung Huang (University of California, Davis) for plasmid pKJH5, used for overexpressing the MBP-EnvZ fusion protein.
This work was supported by National Institutes of Health grants GM 32506/5120MZ and GM 51426.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 2.Brun Y, Marczynski G, Shapiro L. The expression of asymmetry during Caulobacter cell differentiation. Annu Rev Biochem. 1994;63:419–450. doi: 10.1146/annurev.bi.63.070194.002223. [DOI] [PubMed] [Google Scholar]
- 3.Dombroski A J. Sigma factors: purification and DNA binding. Methods Enzymol. 1996;273:134–144. doi: 10.1016/s0076-6879(96)73013-6. [DOI] [PubMed] [Google Scholar]
- 4.Domian I J, Quon K C, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 5.Domian, I. J., A. M. Reisenauer, and L. Shapiro. Unpublished results.
- 6.Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 7.Ely B, Johnson R C. Generalized transduction in Caulobacter crescentus. Genetics. 1977;87:391–399. doi: 10.1093/genetics/87.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gober J W, Marques M V. Regulation of cellular differentiation in Caulobacter crescentus. Microbiol Rev. 1995;59:31–47. doi: 10.1128/mr.59.1.31-47.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gober J W, Shapiro L. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol Biol Cell. 1992;3:913–926. doi: 10.1091/mbc.3.8.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang K-J, Igo M M. Identification of the bases in the ompF regulatory region which interact with the transcription factor OmpR. J Mol Biol. 1996;262:615–628. doi: 10.1006/jmbi.1996.0540. [DOI] [PubMed] [Google Scholar]
- 12.Igo M M, Ninfa A J, Stock J B, Silhavy T J. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989;3:1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- 13.Jenal U, White J, Shapiro L. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J Mol Biol. 1994;243:227–244. doi: 10.1006/jmbi.1994.1650. [DOI] [PubMed] [Google Scholar]
- 14.Kelly A J, Sackett M J, Din N, Quardokus E, Brun Y V. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lane T, Benson A, Hecht G B, Burton J B, Newton A. Switches and signal transduction networks in the Caulobacter crescentus cell cycle. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 296–301. [Google Scholar]
- 16.McCleary W R. The activation of PhoB by acetylphosphate. Mol Microbiol. 1996;20:1155–1163. doi: 10.1111/j.1365-2958.1996.tb02636.x. [DOI] [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 18.O’Regan M, Gloeckler R, Bernard S, Ledoux C, Ohsawa I, Lemoine Y. Nucleotide sequence of the bioH gene of Escherichia coli. Nucleic Acids Res. 1989;17:8004. doi: 10.1093/nar/17.19.8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Quon, K. Unpublished data.
- 19.Quon K, Marczynski G T, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 20.Quon K C, Yang B, Domian I J, Shapiro L, Marczynski G T. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rampersaud A, Harlocker S L, Inouye M. The OmpR protein of Escherichia coli binds to sites in the ompF promoter region in a hierarchical manner determined by its degree of phosphorylation. J Biol Chem. 1994;269:12559–12566. [PubMed] [Google Scholar]
- 22.Scarlato V, Arico B, Prugnola A, Rappuoli R. Sequential activation and environmental regulation of the virulence genes in Bordetella pertussis. EMBO J. 1991;10:3971–3975. doi: 10.1002/j.1460-2075.1991.tb04967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segel I H. Biochemical calculations. 2nd ed. New York, N.Y: John Wiley & Sons, Inc.; 1976. [Google Scholar]
- 24.Sogaard-Andersen L, Valentin-Hansen P. Protein-protein interactions in gene regulation: the cAMP-CRP complex sets the specificity of a second DNA-binding protein, the CytR repressor. Cell. 1993;75:557–566. doi: 10.1016/0092-8674(93)90389-8. [DOI] [PubMed] [Google Scholar]
- 25.Stephens C, Reisenauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci USA. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens C M, Shapiro L. An unusual promoter controls cell-cycle regulation and dependence on DNA replication of the Caulobacter fliLM early flagellar operon. Mol Microbiol. 1993;9:1169–1179. doi: 10.1111/j.1365-2958.1993.tb01246.x. [DOI] [PubMed] [Google Scholar]
- 27.Stephens C M, Zweiger G, Shapiro L. Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J Bacteriol. 1995;177:1662–1669. doi: 10.1128/jb.177.7.1662-1669.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stout V, Torres-Cabassa A, Maurizi M R, Gutnick D, Gottesman S. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J Bacteriol. 1991;173:1738–1747. doi: 10.1128/jb.173.5.1738-1747.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Grau R, Perego M, Hoch J A. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev. 1997;11:2569–2579. doi: 10.1101/gad.11.19.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright R, Stephens C, Zweiger G, Shapiro L, Alley M R K A. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 1996;10:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]
- 31.Wu J, Ohta N, Newton A. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc Natl Acad Sci USA. 1998;95:1443–1448. doi: 10.1073/pnas.95.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhuang W Y, Shapiro L. Caulobacter FliQ and FliR membrane proteins, required for flagellar biogenesis and cell division, belong to a family of virulence factor export proteins. J Bacteriol. 1995;177:343–356. doi: 10.1128/jb.177.2.343-356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zu T, Manetti R, Rappuoli R, Scarlato V. Differential binding of BvgA to two classes of virulence genes of Bordetella pertussis directs promoter selectivity by RNA polymerase. Mol Microbiol. 1996;21:557–565. doi: 10.1111/j.1365-2958.1996.tb02564.x. [DOI] [PubMed] [Google Scholar]