Abstract
Short-term glucose starvation prior to chemotherapy has the potential to preferentially weaken cancer cells, making them more likely to succumb to treatment, while protecting normal cells. In this study, we used 3D cell cultures of colorectal cancer and assessed the effects of short-term glucose starvation and chemotherapy compared to treatment of either individually. We evaluated both phenotypic changes and protein expression levels. Our findings indicate that the combined treatment results in more significant phenotypic responses, including decreased cell viability and clonogenicity. These phenotypic responses can be explained by the decreased expression of LDHA and 14-3-3 family proteins, which were found only in the combined treatment groups. This study indicates that short-term glucose starvation has the potential to increase the efficacy of current cancer treatment regimes.
Keywords: Proteomics, Glucose starvation, iTRAQ, Colorectal cancer, Chemotherapy
Graphical Abstract
Introduction
Nutritional intervention is a method to combat disease independently as well as in combination with current cancer treatments [1]. Forms of nutritional intervention include long-term caloric restriction, short-term starvation, intermittent fasting, and time-restricted eating [1–7]. One form of nutrient restriction of particular interest for cancer treatment is short-term starvation. Short-term starvation in the clinic consists of restriction of 75% or more of caloric intake for 72–96 h prior to chemotherapy. Though maintaining long-term nutrition is critical to patient care and to prevent cancer cachexia, short-term fasting before chemotherapy may have a large impact on the efficacy of the treatment. When cells are subject to a short-term fast, healthy cells respond by slowing proliferation and allocating energy for maintenance pathways [8–10]. This normal cell response is in contrast to cancer cells, which have overexpressed oncogenes and other mutations that cause autocrine production of growth factors [2, 8, 11, 12]. This phenomenon is known as differential stress regulation (DSR), coined by Valter Longo [9]. Preliminary pre-clinical results have driven the exploration of nutrient-based intervention clinical trials, but the molecular mechanisms of these interventions remain largely unknown.
In order to analyze nutrient-based interventions, we have previously demonstrated that immortalized colorectal cancer (CRC) cells are a valid, albeit simplified, model of CRC. In particular, 3D cell cultures, known as spheroids, are an ideal in vitro model system to study the effects of nutrient restriction [13, 14]. Spheroids are effective tumor mimics that have pathophysiological gradients of nutrients and oxygen similar to that of an in vivo tumor [15, 16]. After the in vitro tumor mimic is formed, cell culture media can be manipulated and nutrients, including glucose, can be removed to model short-term fasting.
In this study, we investigated the effects of short-term glucose starvation for 96 h on spheroids prior to chemotherapy treatment. We hypothesized that short-term glucose starvation is a method to weaken drug-resistant cells, making them more likely to succumb to treatment. Irinotecan (IR) was the chemotherapeutic used. IR is a first- and second-line chemotherapeutic used for the treatment of CRC, and its metabolism in CRC spheroids has been well characterized [15, 17, 18]. Phenotypic assay results revealed that the addition of short-term glucose restriction to irinotecan treatment decreased cell clonogenicity and viability more significantly than irinotecan treatment or short-term glucose restriction alone.
We employed quantitative proteomics via UPLC-MS/MS to better understand the biochemical mechanisms involved in this increased response to therapeutics. Specifically, we used isobaric tags for relative and absolute quantification (iTRAQ) for quantification. iTRAQ allows identification of peptide fragments simultaneously with protein quantification of the reporter ions in the low mass range [19–22]. This proteomic study revealed that the tandem treatment of short-term glucose starvation and chemotherapy caused the downregulation of proteins that are heavily involved in cancer progression. Of interest was lactate dehydrogenase A (LDHA) and the 14-3-3 family proteins, which were both found to be downregulated only in the tandem treatment condition. This proteomic link may give insight to the molecular mechanisms that cause the decreased cell viability and clonogenicity found in the phenotypic assays.
Methods
Cell Culture Spheroid Growth and Treatment
The human colon carcinoma cell line HCT 116 was obtained from American Type Culture Collection (ATCC) and cultured in RPMI 1640 cell culture medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Thermo Scientific). The provider assured authentication of the cell line by cytogenetic analysis. In addition, the cell line was validated by short tandem repeat (STR) analysis within the last 24 months. To grow spheroids, we cultured HCT 116 cells in agarose-coated 96-well plates, seeding 7000 cells into each well as previously described [16]. RPMI 1640 media was changed every 48 h after 5 days in culture. For spheroids treated with glucose restriction, on day 10 of culture, media was changed to RPMI 1640 containing no glucose (Life Technologies). All spheroids were harvested for analysis on day 14. After 12 days in culture, spheroids to be treated with chemotherapy were treated with 47.8 μM irinotecan (Sigma-Aldrich). Control untreated spheroids had full nutrient media changed every 48 h until harvested also on day 14. Treatment conditions are illustrated in Fig. 1.
Figure 1.
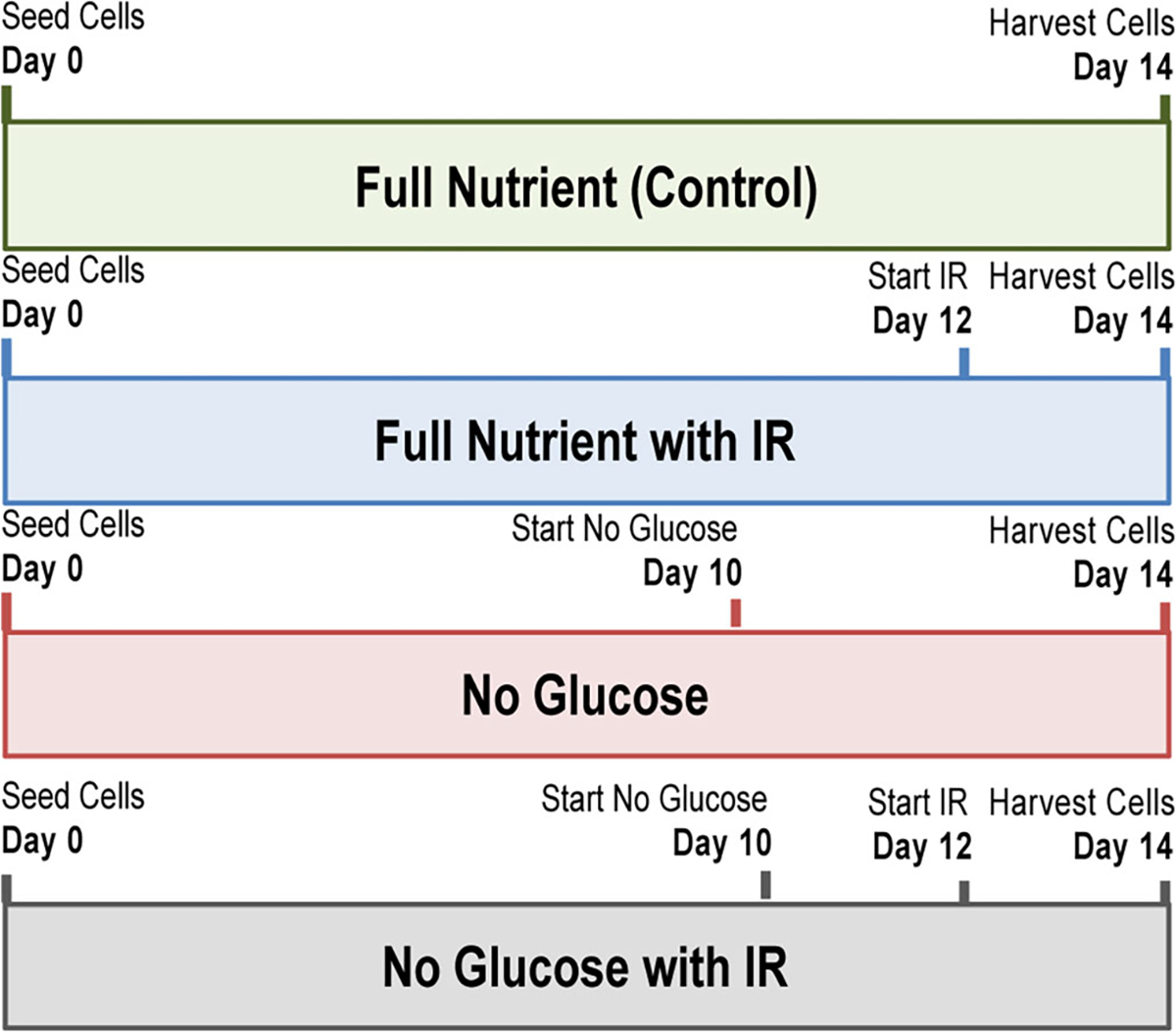
Timeline for each treatment condition used. Day 0 represented when HCT 116 cells were seeded; depending on treatment, glucose restriction was always started at day 10, and IR treatment began at day 12. In all treatment conditions spheroids were harvested for analysis on day 14
Spheroid Gelatin Embedding and Sectioning for MALDI-IMS
On day 13, spheroids were washed with phosphate-buffered saline (PBS) at 37 °C. The spheroids were then placed on a layer of solidified gelatin in a 24-well plate, followed by an additional layer of warm gelatin on top of the spheroids. The gelatin-embedded spheroids were flash frozen at − 80 °C and sliced into 16-μm-thick slices with a cryostat set to − 30 °C. These slices were thaw mounted onto indium tin oxide-coated slides and stored at − 80 °C prior to matrix application.
Matrix Application for MALDI-IMS
The MALDI matrix 2,5-dihydroxybenzoic acid (DHB) (Sigma-Aldrich, St. Louis, MO, USA) was prepared in 60:40 ACN/H2O with 0.2% TFA (EMD, Billerica, MA, USA) to a final concentration of 30 mg/mL. Before use, the matrix solution was filtered (0.22 μm filter) and applied to samples using a TM-Sprayer (HTX Technologies, Carrboro, NC, USA) with heated nitrogen sheath flow gas. The solvent pump flow rate was set to 0.1 mL/min and the temperature was set to 70 °C. Matrix was applied five times in a crisscross pattern; samples were dried in a desiccator for 1 h prior to MALDI-MSI.
Mass Spectrometry Imaging
MALDI-IMS analysis was performed using an ultrafleXtreme MALDI-TOF-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). Mass spectra were acquired in reflectron positive ion mode with 600 laser shots per spot with a mass range set to 300–1000 m/z. The experiment was performed using a laser spot size of 35 μm. A sampling frequency of 2 kHz was used and the laser intensity was set at 69%. Images were processed with flexImaging 4.1software (Bruker Daltonics, Bremen, Germany) to generate ion maps with a semiquantitative red color scale bar.
Clonogenic Assay
Sixty spheroids per condition were grown for 10 days in full nutrient medium; no-glucose treatment started at day 10 and/or IR began at day 12. All spheroids were harvested on day 14. When ready for harvest, the spheroids were homogenized using a mechanical pipet and filtered through a 40-μm cell strainer (Corning Incorporated) to obtain a single cell suspension. The number of live cells was determined using Trypan Blue (Thermo Scientific) and a hemocytometer according to the manufacturer’s protocol. Once cell density was determined, 2000 live cells were seeded in triplicate to an adherent 15-mm dish and incubated at 37 °C in 5% CO2 for the 5.25 days, which corresponds to six cell divisions for HCT 116 cells. First, media was removed and the cells were washed with 5 mL PBS. Next, the cells were fixed by adding 5 mL neutral buffered formalin solution (10%) to the 15-mm dish for 15 min. For staining, 5 mL of 0.01% crystal violet in dH2O (Sigma-Aldrich) was added to the 15-mm dish for 120 min. After, any excess solution was rinsed with dH2O. For analysis, digital images were obtained and colonies were counted by hand and verified with ImageJ (Fiji Version 1.44a).
CellTiter-Blue® Cell Viability Assay
Spheroids were grown for 14 days and treated accordingly. For cell viability, the spheroids were each individually transferred to a new 96-well plate containing only 100 μL of PBS. For staining, 20 μL of the CellTiter-Blue® (Promega) solution was added to each well and incubated for 1 h at 37 °C. For analysis, a Molecular Devices Spectramax M5 plate reader was used according to the manufacturer’s protocol for the cell viability assay.
Proteomics Sample Preparation
On day 14, spheroids (25 per biological replicate) were mechanically lysed in buffer containing 5% sodium dodecyl sulfate (SDS) in 100 mM tetraethylammonium bicarbonate (TAEB), ½ protease inhibitor cocktail tablet (Complete Mini, Roche), 1 mM sodium fluoride, 1 mM sodium orthovanadate, and 1 mM sodium pyrophosphate. The samples were then sonicated at 15% amplitude for four rounds of 1 min on, 1 min off. The samples were then heated for 10 min at 90 °C. The samples were centrifuged at 13,000×g for 10 min and the supernatant was collected. Total protein was quantified using the Pierce BCA Protein Assay Kit (Thermo) according to the manufacturer’s protocol. For each sample, 100 μg of protein was reduced with 5 mM dithiothreitol (DTT) for 1 h at 37 °C, alkylated with 14 mM iodoacetamide in the dark for 30 min at RT, and quenched with 5 mM DTT for 20 min at RT. After these additions, the SDS concentration in the solutions was increased to 5% using 40% SDS.
For digestion, suspension trapping (STrap) was used [23]. STraps were purchased from Protifi, and the manufacturer’s protocol was followed for digestion using trypsin overnight at 37 °C. Samples were dried using a speed vacuum and labeled with isobaric tags for relative and absolute quantification (iTRAQ) 8-plex reagents according to the manufacturer’s protocols (AB Sciex). After each individual sample was labeled, all eight samples were combined, dried, and desalted with a 100-mg C18 Sep-Paks (Waters).
Strong Cation Exchange (SCX) Offline Fractionation
Mobile phases were prepared for strong cation exchange (SCX) liquid chromatography fractionation using a Waters Alliance HPLC System (Waters) with a SCX guard column (2.1 mm i.d. × 50 mm length, 5 μM particles, Agilent Technologies). Buffer A: 10 mM KH2PO4 in 20% ACN, pH 2.85, and buffer B: 750 mM KCl in buffer A, pH 2.85. Prior to fractionation, the sample was suspended in 120 μL buffer A. The sample was then fractioned as previously described [13]. Fractions were collected every minute of the linear gradient for a total of 31 fractions. Due to low concentration of sample eluting off the column in the first 5 min, the first five of these fractions were combined for analysis. Following SCX fractionation, the samples were dried down and resuspended in 60 μL of 0.1% formic acid (FA) in H2O. Twenty microliters of each sample was desalted with a 5-μg C18 ZipTip (ZTC18S096, Millipore) and again dried down. Peptides were then resuspended in 5 μL of 0.1% FA in H2O for analysis by ultra-performance liquid chromatography (UPLC)-ESI-MS/MS.
UPLC-ESI-MS/MS Analysis
For ultra-performance liquid chromatography, a nano-ACQUITY UltraPerformance LC® (UPLC®) system (Waters) with mobile phases buffer A (0.1% FA in H2O) and buffer B (0.1% FA in ACN) was used. For each sample analyzed in technical triplicate, 2 μL was automatically loaded onto a C18 reversed-phase column (Waters, 100 μm × 100 mm, 1.7 μm particle, part No. 186003546, column temperature 40 °C) via a method that has been previously described [13]. For the MS/MS portion, the sample was electrosprayed (ESI) into a Q-Exactive mass spectrometer (Thermo Fisher Scientific). The mass spectrometer was operated in data dependent acquisition (DDA) mode using a top 12 method with automated switching of MS to MS/MS. The electrospray voltage was set to 2.1 kV, and the ion transfer tube temperature was set to 280 °C. Full MS scans were acquired in the Orbitrap mass analyzer 350–2000 m/z range with resolution of 70,000, ACG target 1.00 E+06 ions, and fill time of 250 ms. The tandem mass spectra were acquired from 100 to 1500 m/z with a resolution of 17,500, ACG target of 5.00 E+05 ions, and fill time of 120 ms. The isolation window was set as 2.0 m/z for fragmentation and the normalized collision energy was 31.0%. Peptide match and exclude isotopes were turned on. Dynamic exclusion was set as 40 s. All injections were run in triplicate.
Data Analysis
Raw data files were searched with MaxQuant version 1.5.6.0 using the Andromeda search engine against the UniProt human and decoy databases. The enzyme for digestion was chosen to be trypsin with a maximum of two missed cleavages. Additional settings included first search peptide tolerance of 20 ppm and a main search peptide tolerance of 4.5 ppm. The protein FDR was set at 0.01. Deamidation (NQ), oxidation (M), protein N-terminal acetylation, and iTRAQ 8-plex tags on tyrosine residues were set as dynamic modifications. Static modifications included carbamidomethylation (C) and iTRAQ 8-plex tags (peptide N-term and K). iTRAQ 8-plex was set for quantification, and the instrument was designated as an Orbitrap MS and MS/MS. All functional classification and pathway analysis was completed using Ingenuity Pathway Analysis (IPA).
Statistical Analysis
Phenotypic experiments were performed in biological triplicate, and significant data was determined by comparing means of treated and control using an unpaired Student’s t test, with statistically significant cutoff at *P < 0.01. A Pearson correlation coefficient was determined to measure the linear correlation between two variables.
Results and Discussion
Treatment of Short-Term Glucose Starvation and IR, and Visualization of IR in HCT 116 Spheroids
These experiments used HCT 116 three-dimensional cell cultures, or spheroids. First developed by the Sutherland lab [24], and further characterized by our lab, spheroids are an in vitro model system that mimics a solid tumor [14–16]. Similar to a solid tumor found in vivo, spheroids have decreased amounts of available oxygen and nutrients in the center of the cell mass, leading to different cellular regions. In the outer region of a spheroid, there are proliferative cells, and on the inner core of the spheroids, there are apoptotic and necrotic cells. To study short-term glucose starvation and its effects on the proteome of CRC, we first grew HCT 116 spheroids for 10 days in full nutrient media. This growth period allowed for the spheroid maturation and development of regions of proliferative and apoptotic cells that replicate an in vivo CRC tumor. After 10 days in culture, we switched the cell culture media to RPMI 1640 containing no glucose and began the short-term glucose starvation. On day 12 in culture, spheroids treated with both glucose starvation and chemotherapy were subject to a media change with media containing IR but devoid of glucose. Spheroids were then harvested after 14 days in culture. The four individual treatment regimens are illustrated in Fig. 1.
Prior to phenotypic assays and quantitative proteomic analysis, we confirmed IR penetration in both full-nutrient and short-term glucose starvation condition spheroids using MALDI-mass spectrometry imaging (MSI) (Fig. 2). After 48 h of IR treatment, using an IC50 previously determined [13], penetration of IR through the center of the spheroid was observed in both the full-nutrient and glucose-starved spheroids. Spheroids have radial symmetry and the sample in Fig. 3 is an equatorial slice. Thus, penetration into the equatorial slice confirms that there is penetration of IR in all regions of the spheroid. Therefore, it is evident that IR fully penetrated the spheroid after 48 h of treatment and there is not a bias in distribution when glucose is removed from the cell culture media, which could affect the proteomic changes observed.
Figure 2.

MALDI-IMS images of IR penetration in no glucose and full nutrient spheroids. Images are of IR (m/z 587) in spheroids and are normalized to total ion current with intensity scale on the right. IR is distributed throughout the equatorial slice of a spheroid in both conditions
Figure 3.
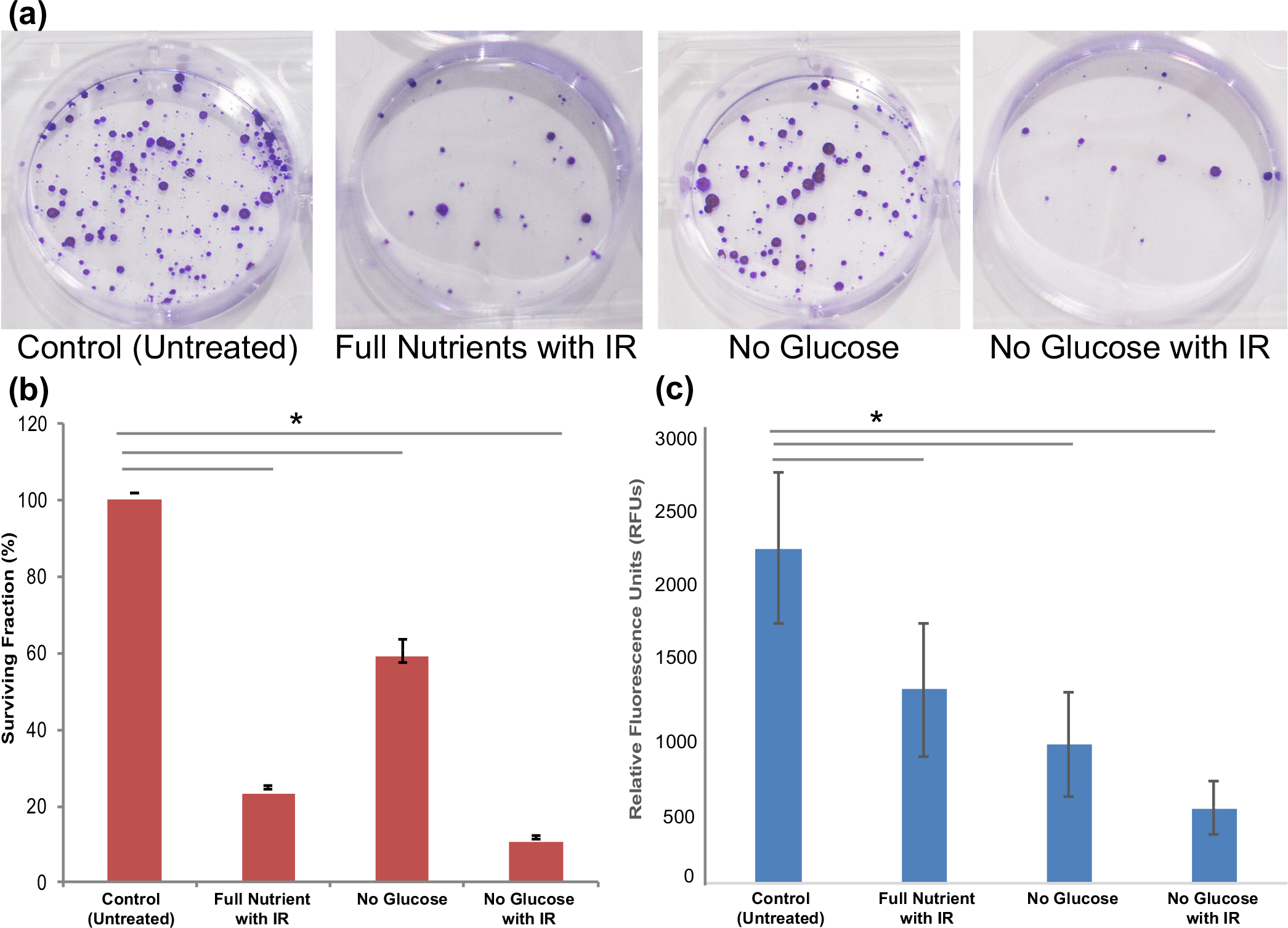
Phenotypic assays to assess clonogenicity and cell viability of treatment on HCT (a) Images of clonogenicity assay in a six-well plate after fiv e doublings of HCT 116 cells. (b) Surviving fraction (%) from clonogenicity assay compared to the full nutrient control, to determine ability of cells to proliferate after treatment. (c) Cell viability assay results reporting cell viability of spheroids after treatment in relative fluorescence units (RFUs) using the Promega cell titer blue assay
Phenotypic Assays Reveal Decreased Clonogenicity and Cell Viability When Treated with Short-Term Glucose Starvation and IR
Short-term starvation altered the efficacy of IR treatment in HCT 116 spheroids. The first phenotypic assay used was a clonogenic assay (Fig. 3A, B) which measures the ability of cells to survive a specific condition or treatment. The cells were treated according to the scheme shown in Fig. 1. The spheroids were dissociated into individual cells, and the same number of live cells was plated (n = 3) for each condition in triplicate. After six doublings, the cells were stained and colonies were counted. The spheroids treated with IR showed a significant decrease in their ability to proliferate after treatment, known as clonogenicity, with a 23% surviving fraction. The spheroids treated with only short-term glucose starvation also had a significantly decreased clonogenicity to 59%. The spheroids treated with both short-term glucose starvation and IR had the greatest impact on clonogenicity with only a 10% surviving fraction. This assay revealed that cells were not able to proliferate after any type of treatment of IR or short-term glucose starvation, but the treatment with both had an additive effect on their ability to proliferate.
To further characterize this phenomenon, we analyzed cell viability after treatment using the CellTiter-Blue® (Promega) and observed that tandem treatment of short-term glucose starvation and IR had the largest decrease in cell viability (Fig. 3C). Specifically, the spheroids treated with only IR had a 42% decrease in cell viability compared to the control. The spheroids treated with only short-term glucose starvation had a 58% decrease in cell viability compared to the control. Finally, the spheroids treated with tandem short-term glucose starvation and IR had a 77% decrease in cell viability compared to the control. These results are noteworthy because short-term glucose starvation by itself caused more cell death than treatment of IR alone. When cultured after treatment, cells overcame short-term glucose starvation and proliferated more than with IR treatment alone. The overall conclusion from these phenotypic assays is that adding short-term glucose starvation to IR treatment intensified cell death and decreased the ability to proliferate after treatment.
Comparing Proteomic Changes Between Treatment Conditions
It is important to understand the molecular mechanisms underwriting these phenotypic results. To further discern these mechanisms, we performed quantitative proteomic analysis. We used eight iTRAQ tags to analyze our control, IR-treated, short-term glucose fasted, and combined short-term fasted and IR-treated conditions (Fig. 4A). Each treatment was performed and analyzed in biological duplicate. For mass spectrometric analysis, we injected all 29 fractions in triplicate and searched the data using MaxQuant. To determine which proteins were differentially regulated, we used ProteoSign [25], an open-source web-based platform for statistical quantitative proteomic analysis. To determine differentially regulated proteins, ProteoSign examines proteins that have two or more unique peptide sequences identified and then utilizes Linear Models for Microarray Data (LIMMA) to determine if a protein is statistically up- or down-regulated compared to the control. The log2 iTRAQ reporter ion intensity ratios for the treatment conditions relative to control are visualized with volcano plots (Fig. 4B). To determine which proteins were differentially regulated by treatment, we determined log2 fold changes cutoffs, as well as a p value cutoff. To determine the log2 fold change cutoff, we plotted histograms of the fold change ratios from control replicates to determine inherent biological and experimental variation. The fold changes were distributed around zero on the log2 scale with a standard deviation of 0.15 (data not shown). The fold change threshold for differential regulation was then determined to be three times the standard deviation of the controls (log2 fold change ≤− 0.45 or ≥ 0.45). Additionally, we used calculated p values for each protein as determined by Proteosign and chose a p value < 0.05 as the cutoff for differentially regulated proteins. Using these criteria, it was determined that 65 proteins were differentially regulated when spheroids were treated with only IR. When spheroids were treated with only short-term glucose starvation, there were 130 proteins differentially regulated, and when spheroids were treated with combined short-term glucose starvation and IR, there were 133 proteins differentially regulated (Fig. 5).
Figure 4.
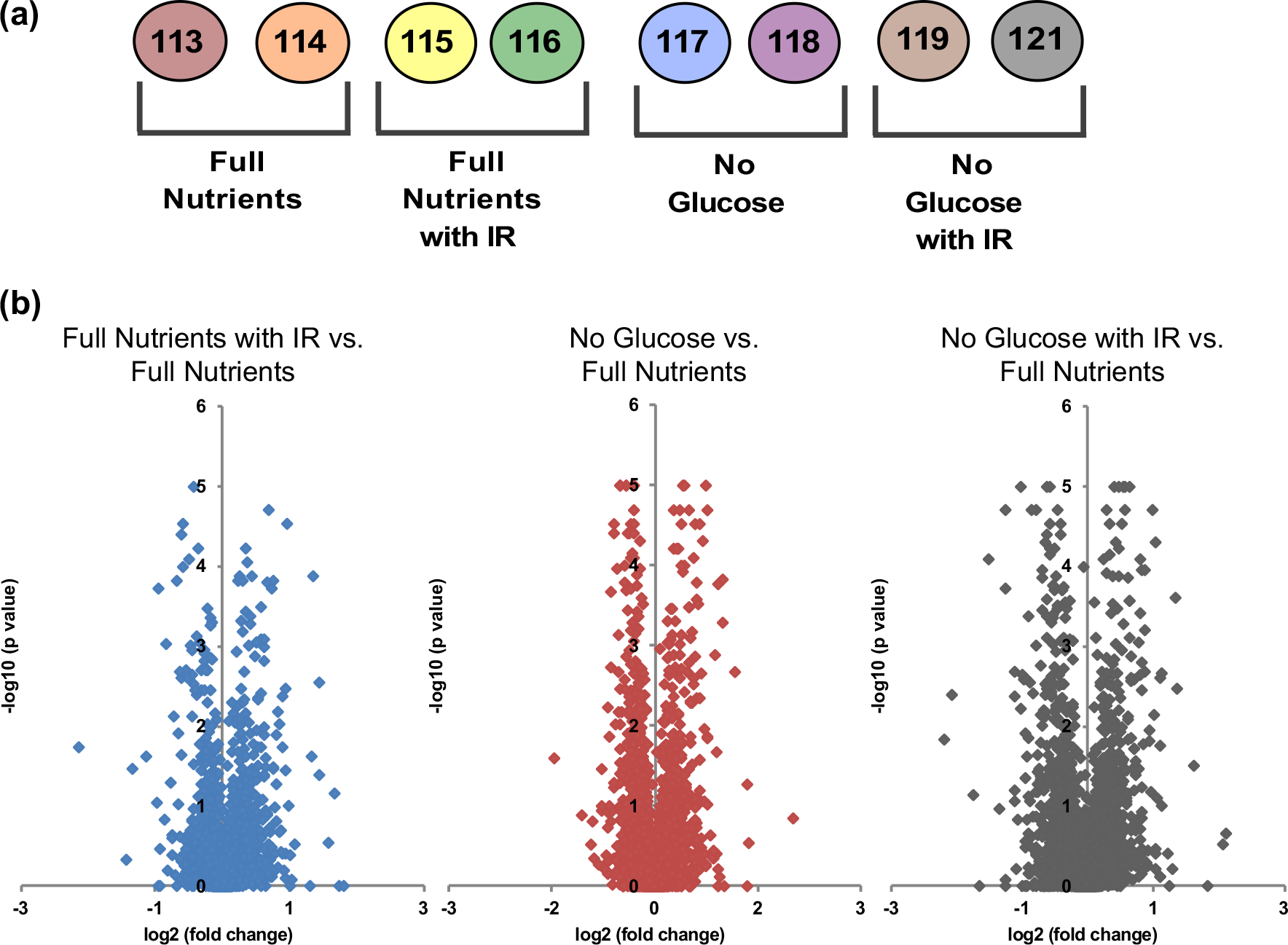
(a) Setup of iTRAQ experiment, labeled each of the four biological conditions in biological duplicate utilizing all eight iTRAQ reporter ion tags. (b) Volcano plots plotting log2 (fold change) of iTRAQ reporter ion ratio on the x-axis, and − log10(p value) determined by ProteoSign on the y-axis for each treatment condition compared to the full-nutrient control. Data points, which were +/− 0.50 log2(fold change) and > 1 − log10(p value), were considered statistically significantly up- or downregulated and used for further analysis
Figure 5.
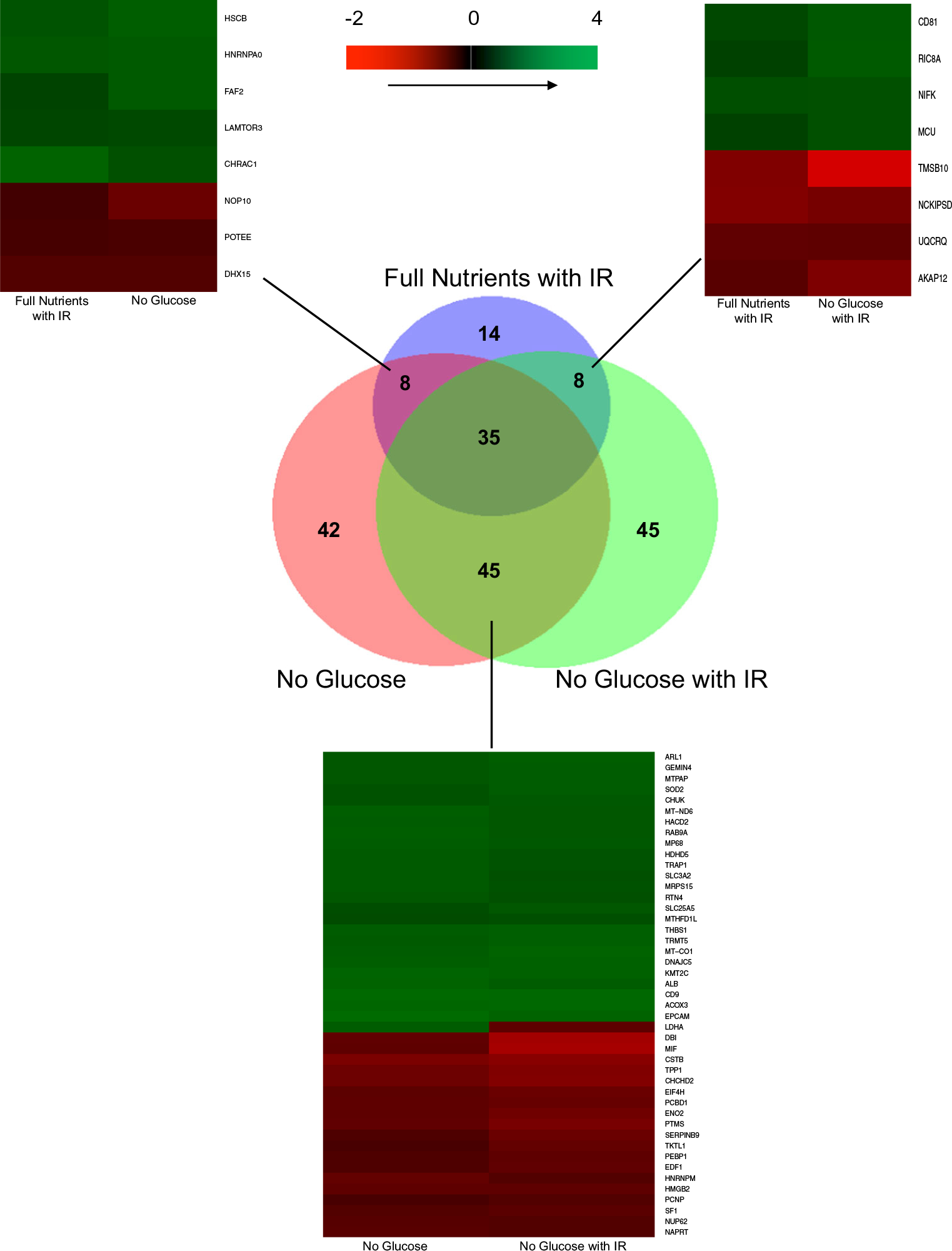
A proportional Venn diagram was made comparing the statistically significant up- or downregulated proteins for each treatment condition compared to the full-nutrient control. Corresponding heat maps for proteins that overlapped between two conditions were created to compare regulation between conditions
We next identified proteins that were found to be differentially regulated in multiple conditions, as shown in Fig. 5. The IR treatment condition and the glucose-starved treatment condition had eight proteins that overlapped. The proteins were regulated in the same direction, with five proteins downregulated and three proteins upregulated. Additionally, when comparing the IR treatment and glucose starved with IR treatment, there were eight proteins that were found to be differentially regulated in both treatments. These eight proteins were also similarly regulated, with four upregulated and four downregulated.
Finally, there were 45 proteins that overlapped between the glucose-starved condition and glucose-starved with IR. Of the 45 proteins, only one was oppositely regulated. Lactate dehydrogenase A (LDHA) was upregulated when glucose starved with a log2 fold change value of 0.707, but in the glucose-starved with IR treatment, LDHA was downregulated with a log2 fold change value of − 0.679. It is intriguing to note that LDHA is regulated in the opposite direction between these treatments. LDHA is a key regulator of anaerobic glycolysis and is responsible for the conversion of pyruvate to lactate. Pyruvate is the terminal product formed from glucose during glycolysis [26]. Thus, it is noteworthy that the enzyme that converts pyruvate to lactate, LDHA, was upregulated when glucose is taken out of cell culture media. It may be that pyruvate is formed not only from glucose; another known source of pyruvate is from the oxidation of lactate and the transamination of alanine [26]. LDHA is essential for the Warburg Effect often observed in cancer cells, which allows the metabolic switch of cancer cells to upregulate glycolysis when oxygen supply is limited [27]. LDHA is ubiquitously overexpressed in cancer cells [27, 28]. When spheroids were treated with glucose starvation, we found LDHA to be upregulated, which was unexpected, but possibly a mechanism by the cancer cells to produce as much energy from glycolysis as possible. When treated with glucose starvation and IR, LDHA was downregulated in the spheroids. This difference showed that the additional treatment caused a significant change in cellular metabolism. LDHA is a target for cancer treatment, and its downregulation has been shown to decrease tumor progression and metastasis [27]. Our results showed that the combined effects of treating spheroids with both short-term glucose starvation and IR caused a decrease of an important metabolism regulator, and this decrease could mitigate the metastatic potential of the cancer cell.
The 35 proteins were found to be differentially regulated in all three conditions (online resource ESM2) and mapped to known pathways using ingenuity pathway analysis (IPA) (data not shown). All of the 35 proteins were regulated in the same direction across the three treatment conditions. The pathways that were enriched, regardless of the treatment of IR, glucose starvation, or glucose starvation with IR, were the sirtuin signaling pathway (p value = 8.32E−04) and granzyme A signaling (p value = 3.87E−04). Granzyme A is an enzyme that activates a programmed cell death that resembles apoptosis [29]. Thus, our results indicate that chemotherapy treatment alone, short-term glucose starvation, or the combination of both, enrich a killer cell pathway that protects against tumorigenesis.
Gene Ontology Analysis for Each Individual Condition
For each individual treatment condition, the respective differentially regulated proteins were mapped to metabolic pathways using IPA. IPA identified the top enriched pathways based on the significance of protein expression. The top five pathways enriched for each condition are listed in Table 1. The IR-treated and glucose-starved conditions had similar enriched pathways overall, even though they had fewer overlapping proteins (Fig. 5). The top canonical pathways enriched in both of these treatments included granzyme A signaling, sirtuin signaling, and oxidative phosphorylation. Sirtuins are involved in stress response [30] and can be expected to be differentially regulated due to chemotherapy or short-term glucose starvation, which are both stressors for a cell. Additionally, oxidative phosphorylation was enriched in both chemotherapy-treated and glucose-starved spheroids. This change is likely due to the increased fatty acid oxidation creating substrates for oxidative phosphorylation that occurs both when glucose is restricted and when there is DNA damage caused by IR [2, 31].
Table 1. Gene Ontology Pathway Analysis.
The top five canonical pathways enriched from the differentially regulated proteins of each treatment condition determined using IPA
| Full nutrients with IR | No glucose | No glucose with IR | |||
|---|---|---|---|---|---|
|
| |||||
| Canonical pathway | p value | Canonical pathway | p value | Canonical pathway | p value |
| Granzyme A signaling | 1.50E–03 | Sirtuin Signaling Pathway | 8.81E-12 | IGF-1 Signaling | 1.39E-06 |
| Pyrimidine deoxyribonucleotides de novo biosynthesis I | 1.99E–03 | Mitochondrial Dysfunction | 4.58E-08 | p70S6K Signaling | 4.11E-06 |
| Oxidative phosphorylation | 3.84E–03 | Oxidative Phosphorylation | 3.52E-05 | ERK5 signaling | 6.36E-06 |
| Sirtuin signaling pathway | 4.04E-03 | Granzyme A Signaling | 1.84E-04 | Myc Mediated Apoptosis Signaling | 8.04E-06 |
| p53 Signaling | 9.99E–03 | Acute phase response signaling | 2.76E-03 | ERL/MAPK Signaling | 3.08E-05 |
A completely independent set of pathways were enriched when spheroids were treated with combined glucose starvation and IR. The top five pathways are listed in Table 1. These five pathways are all intimately involved with one another and many, including MAPK, ERK5, IGF-1, are signaling pathways that are exploited for progression of many cancers [32–34]. Furthermore, when looking at the proteins that match these enriched pathways, our data showed the identification of several members of the 14-3-3 family of proteins, which is a conserved family of proteins in eukaryotic cells involved in mitogenic cell signaling [35]. The 14-3-3 proteins are often found overexpressed in various cancers and can have a role in not only tumor progression but also chemosensitivity [36, 37]. Specifically, we found that YWHAB, YWHAQ, and YWHAG all were downregulated when spheroids were treated with short-term glucose starvation and IR. The following log2 fold changes for each protein were as follows: YWHAB − 0.647, YWHAQ − 0.667, and YWHAG − 0.631. These proteins were also unique to this treatment condition and were not found to be differentially regulated by the other treatments. This result indicated that again there is a unique effect of treating spheroids with both short-term glucose starvation and IR that causes a decrease in 14-3-3 family proteins, leading to decreased mitogenic signaling that is vital for cancer progression. In addition to the signaling pathways, the p70s6k signaling pathway was enriched, also known as the mammalian target of rapamycin (mTOR). mTOR is the master energy regulator in the cell and is traditionally turned on in response to stress, particular nutrient restriction [38]. Thus, we hypothesized that mTOR signaling would be intensified during short-term glucose starvation. However, this pathway was only enriched during the combined treatment of short-term glucose starvation and IR.
Identification of Upstream Regulators for Each Treatment
Utilizing the upstream regulator tool in IPA, we identified molecules that are likely to be activated or inactivated upstream of the proteins that were quantified based on an activation z-score (Fig. 6). The upstream regulator tool is able to identify the cascade of upstream regulators based on the proteins identified in the data set. The upstream regulators were first identified using an overlap p value of the proteins identified in the dataset and the known targets of those proteins. An activation z-score was then used to predict activation state of the upstream regulators based on a model that assigns random regulation directions [39]. This tool is predictive and can be used to determine which biological functions are implicated due to short-term glucose starvation and chemotherapy treatment.
Figure 6.
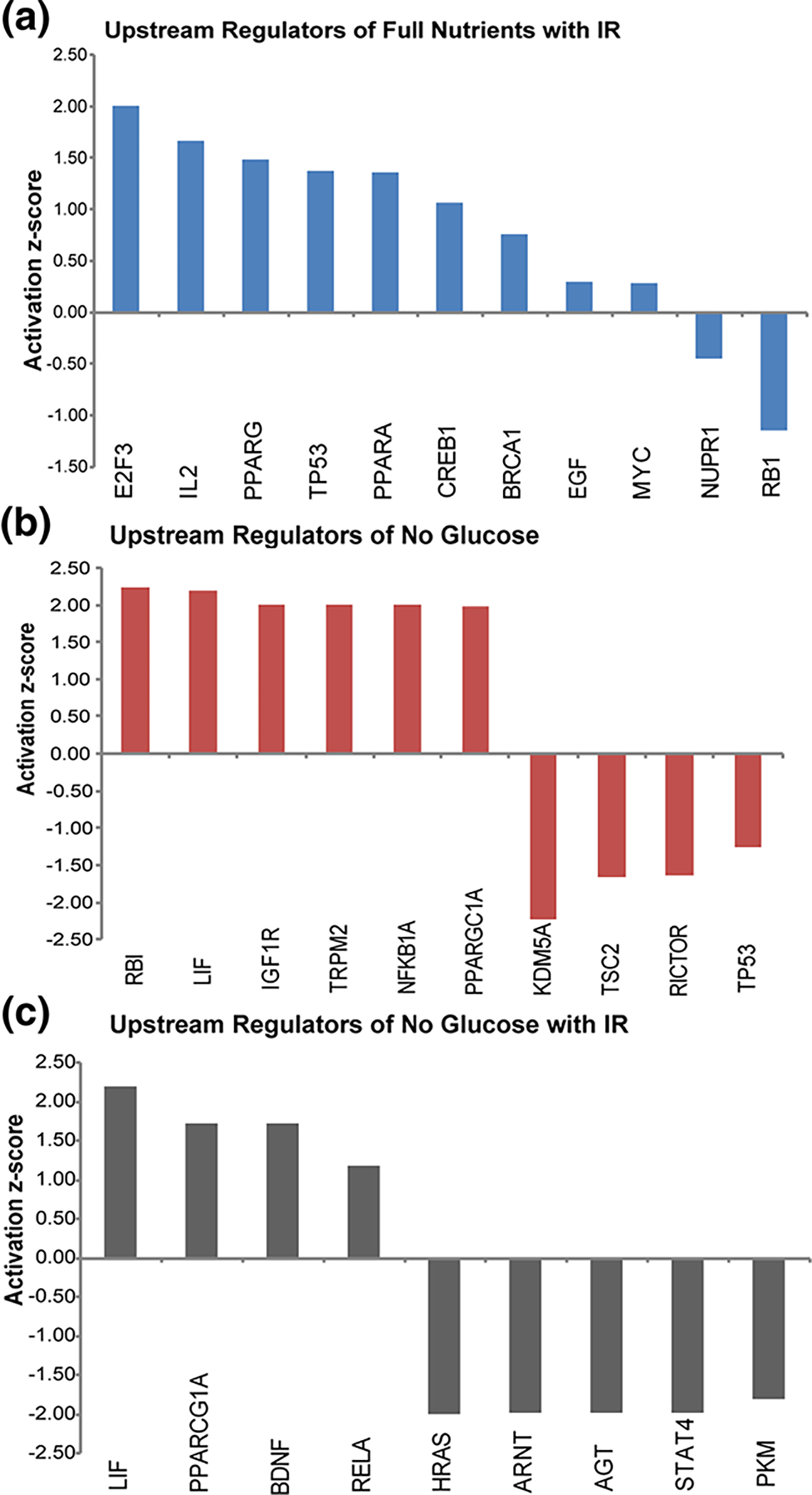
Using the upstream regulator function on IPA, upstream transcription factors of proteins found to be differentially regulated in each treatment condition were determined to be putatively activated or inactivated. (a, b, c) The transcription factors activation z-score is plotted on the y-axis for each transcription factor for all treatment conditions
The upstream regulators identified varied for each treatment condition. No single upstream regulator was found to be significantly activated or inactivated across all three treatment conditions. Most of the upstream regulators found were transcription factors that are implicated in cancer. Of note, p53 was found to be putatively activated when spheroids were treated with only IR, but found to be putatively deactivated when spheroids were treated with glucose starvation. Though p53 is one of the most highly mutated genes in cancer, the HCT 116 cell line has wild type p53 [40]. With abundant literature on p53, it is known that upon DNA damage, p53 expression is induced. [41] This fact explains p53’s putative activation upon IR treatment. In contrast, our results show that p53 was putatively inactivated in cancer cells treated with short-term glucose starvation. This result has been consistent through multiple studies, as inactivation of p53 has also been found when glucose is restricted and autophagy is inhibited with the small molecule chloroquine [13]. The role of p53 in cancer under stress conditions is complex. Under normal conditions, p53 plays a role in promoting apoptosis, but under nutrient stress conditions, p53 can promote cell survival [41, 42]. Upstream regulators identified by the tandem treatment of short-term glucose starvation and IR include many essential tumor oncogenes (Fig. 6). Signal transducers and activators of transcription 4 (STAT4) are critical for cell proliferation and differentiation and are found to be overexpressed in colorectal cancer [43]. Experiments knocking down STAT4 in colorectal cancer have shown to suppress cell proliferation and invasion [44]. STAT4 was found to be putatively inactivated when spheroids were treated with tandem short-term glucose starvation and IR.
Conclusions
In the current literature, there is a lack of proteomic data on the effect of food components and diet as related to cancer. It has been suggested that the factors hindering nutritional proteomics are limited accessibility to proteomic technologies and insufficient preanalytical sample handling and sample processing training [45]. Our lab has made significant progress toward eliminating some of these limitations by establishing a 3D cell culture platform to study the global proteomic changes of food component restrictions and completing the study of combined treatment of short-term glucose starvation and chemotherapy in colorectal cancer. These studies showed that the tandem treatment of short-term glucose starvation and chemotherapy have synergistic effects on the phenotype and proteome of cancer. One of the limitations of this study is that 3D cell cultures, though more complex than 2D cell cultures, still do not recapitulate all the complexity of an organism. Thus, though we find these studies exciting, further studies in animal models must be completed before this treatment regimen is translated into the clinic.
Our results showed that compared to short-term glucose starvation or chemotherapy treatment alone, the tandem treatment caused decreased clonogenicity and decreased cell viability. The phenotypic results indicated that the tandem treatment is not only able to kill more cancer cells in a spheroid model but the treatment also renders the cells that do survive less capable of proliferating after treatment. From the proteomic data, we compared the differentially regulated proteins found in common between the conditions. We compared the 45 proteins that were in common between the treatment of only short-term glucose starvation and tandem short-term glucose starvation and IR. Only one protein, LDHA, was differentially regulated between the two conditions. LDHA was found to be upregulated in glucose-starved treatment and downregulated in glucose-starved with IR treatment. LDHA is overexpressed in colorectal cancer, and we found it intriguing that this protein downregulated only with the tandem treatment. Additionally, the quantitative proteomic results revealed that a subset of proteins, the 14-3-3 family proteins, were downregulated by the tandem treatment of short-term glucose starvation and chemotherapy. The 14-3-3 family proteins are implicated in cancer, responsible for cell growth, division, and migration. The downregulation of both the LDHA and the 14-3-3 family proteins, only in the tandem treatment, may be a promising molecular link to the phenotype of decreased cell viability and clonogenicity.
Supplementary Material
Acknowledgements
MMS was supported by National Institutes of Health Training Grant –Chemistry Biochemistry Biology Interface Program (T32GM075762). ABH was supported by the National Institutes of Health (R01GM110406), and the National Science Foundation (CAREER Award, CHE-1351595). The UltrafleXtreme instrument (MALDI-TOF-TOF) was acquired through National Science Foundation award #1625944. We gratefully acknowledge the assistance of the Notre Dame Mass Spectrometry and Proteomics Facility (MSPF).
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s13361-018-2013-z) contains supplementary material, which is available to authorized users.
References
- 1.Donaldson MS: Nutrition and cancer: a review of the evidence for an anti-cancer diet. Nutrition. 3, 19 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee C, Longo VD: Fasting vs dietary restriction in cellular protection and cancer treatment: from model organisms to patients. Oncogene. 30, 3305–3316 (2011) [DOI] [PubMed] [Google Scholar]
- 3.Longo VD, Mattson MP: Fasting: molecular mechanisms and clinical applications. Cell Metab. 19, 181–192 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen BG, Bhatia SK, Buatti JM, Brandt KE, Lindholm KE, Button AM, Szweda LI, Smith BJ, Spitz DR, Fath MA: Ketogenic diets enhance oxidative stress and radio-chemo-therapy responses in lung cancer xenografts. Clin. Cancer Res. 19, 3905–3913 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattison JA, Colman RJ, Beasley TM, Allison DB, Kemnitz JW, Roth GS, Ingram DK, Weindruch R, De Cabo R, Anderson RM: Caloric restriction improves health and survival of rhesus monkeys. Nat. Commun. 8, 1–12 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colman RJ, Anderson RM, Johnson SC, Kastman EK, Simmons HA, Kemnitz JW, Weindruch R: Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 325, 201–204 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothschild J, Hoddy KK, Jambazian P, Varady KA: Time-restricted feeding and risk of metabolic disease: a review of human and animal studies. Nutr. Rev. 72, 308–318 (2014) [DOI] [PubMed] [Google Scholar]
- 8.Raffaghello L, Safdie F, Bianchi G, Dorff T, Longo VD: Fasting and differential chemotherapy protection in patients. Cell Cycle. 9, 4474–4476 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raffaghello L, Lee C, Safdie FM, Wei M, Madia F, Bianchi G, Longo VD: Starvation-dependent differential stress resistance protects normal but not cancer cells against high-dose chemotherapy. Proc. Natl. Acad. Sci. 105, 8215–8220 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C, Raffaghello L, Brandhorst S, Safdie FM, Bianchi G, Martin-Montalvo A, Pistoia V, Wei M, Hwang S, Merlino A, Emionite L, de Cabo R, Longo VD: Fasting cycles retard growth of tumors and sensitize a range of cancer cell types to chemotherapy. Sci. Transl. Med. 4, 124–127 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Niekerk G: Enhanced therapeutic efficacy in Cancer patients by short-term fasting: the autophagy connection. Front. Oncologia. 6, 1–7 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorff TB, Groshen S, Garcia A, Shah M, Tsao-Wei D, Pham H, Cheng C-W, Brandhorst S, Cohen P, Wei M, Longo V, Quinn DI: Safety and feasibility of fasting in combination with platinum-based chemotherapy. BMC Cancer. 16, (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroll MM, LaBonia GJ, Ludwig KR, Hummon AB: Glucose restriction combined with autophagy inhibition and chemotherapy in HCT 116 spheroids decreases cell clonogenicity and viability regulated by tumor suppressor genes. J. Proteome Res. 16, 3009–3018 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroll MM, Liu X, Herzog SK, Skube SB, Hummon AB: Nutrient restriction of glucose or serum results in similar proteomic expression changes in 3D Colon Cancer cell cultures. Nutr. Res. 36, 1068–1080 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Weaver EM, Hummon AB: Evaluation of therapeutics in three-dimensional cell culture systems by MALDI imaging mass spectrometry. Anal. Chem. 85, 6295–6302 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Hummon AB: Imaging mass spectrometry of three-dimensional cell culture systems. Anal. Chem. 83, 8794–8801 (2011) [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Hummon AB: Quantitative determination of irinotecan and the metabolite SN-38 by nanoflow liquid chromatography-tandem mass spectrometry in different regions of multicellular tumor spheroids. J. Am. Soc. Mass Spectrom. 26, 577–586 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaBonia GJ, Lockwood SY, Heller AA, Spence DM, Hummon AB: Drug penetration and metabolism in 3D cell cultures treated in a 3D printed fluidic device: assessment of irinotecan via MALDI imaging mass spectrometry. Proteomics. 16, 1814–1821 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiese S, Reidegeld KA, Meyer HE, Warscheid B: Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 7, 340–350 (2007) [DOI] [PubMed] [Google Scholar]
- 20.Karp N.a., Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS: Addressing accuracy and precision issues in iTRAQ quantitation. Mol. Cell. Proteomics. 9, 1885–1897 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu WW, Wang G, Baek SJ, Shen RF: Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J. Proteome Res. 5, 651–658 (2006) [DOI] [PubMed] [Google Scholar]
- 22.Burkhart JM, Vaudel M, Zahedi RP, Martens L, Sickmann A: iTRAQ protein quantification: a quality-controlled workflow. Proteomics. 11, 1125–1134 (2011) [DOI] [PubMed] [Google Scholar]
- 23.Zougman A, Selby PJ, Banks RE: Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. Proteomics. 14, 1006–1000 (2014) [DOI] [PubMed] [Google Scholar]
- 24.Sutherland RM: Microregions: the spheroid model. Science. 240, 177–240 (1988) [DOI] [PubMed] [Google Scholar]
- 25.Efstathiou G, Antonakis AN, Pavlopoulos GA, Theodosiou T, Divanach P, Trudgian DC, Thomas B, Papanikolaou N, Aivaliotis M, Acuto O, Iliopoulos I: ProteoSign: an end-user online differential proteomics statistical analysis platform. Nucleic Acids Res. 45, W300–W306 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray LR, Tompkins SC, Taylor EB: Regulation of pyruvate metabolism and human disease. Cell. Mol. Life Sci. 71, 2577–2604 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miao P, Sheng S, Sun X, Liu J, Huang G: Lactate dehydrogenase a in cancer: a promising target for diagnosis and therapy. IUBMB Life. 65, 904–910 (2013) [DOI] [PubMed] [Google Scholar]
- 28.Xie H, Hanai J, Ren J-G, Kats L, Burgess K, Bhargava P, Signoretti S, Billiard J, Duffy KJ, Grant A, Wang X, Lorkiewicz PK, Schatzman S, Bousamra II M, Lane AN, Higashi RM, Fan TWM, Pandolfi PP, Sukhatme VP, Seth P: Targeting lactate dehydrogenase-a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 19, 795–809 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lieberman J: Granzyme A activates another way to die. Immunol. Rev. 235, 93–104 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bosch-Presegué L, Vaquero A: The dual role of Sirtuins in Cancer. Genes Cancer. 2, 648–662 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brace LE, Vose SC, Stanya K, Gathungu RM, Marur VR, Longchamp A, Treviño-Villarreal H, Mejia P, Vargas D, Inouye K, Bronson RT, Lee C-H, Neilan E, Kristal BS, Mitchell JR: Increased oxidative phosphorylation in response to acute and chronic DNA damage. Aging Mech. Dis. 2(16022), (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laviola L, Natalicchio A, Giorgino F: The IGF-I signaling pathway. Curr. Pharm. Des. 13, 663–669 (2007) [DOI] [PubMed] [Google Scholar]
- 33.Simões AES, Rodrigues CMP, Borralho PM: The MEK5/ERK5 signalling pathway in cancer: a promising novel therapeutic target. Drug Discov. Today. 21, 1654–1663 (2016) [DOI] [PubMed] [Google Scholar]
- 34.Dhillon AS, Hagan S, Rath O, Kolch W: MAP kinase signalling pathways in cancer. Oncogene. 26, 3279–3290 (2007) [DOI] [PubMed] [Google Scholar]
- 35.Wilker E, Yaffe MB: 14-3-3 proteins—a focus on cancer and human disease. J. Mol. Cell. Cardiol. 37, 633–642 (2018) [DOI] [PubMed] [Google Scholar]
- 36.Wu Y-J, Jan Y-J, Ko B-S, Liang S-M, Liou J-Y: Involvement of 14-3-3 proteins in regulating tumor progression of hepatocellular carcinoma. Cancers (Basel). 7, 1022–1036 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neal CL, Yu D: 14-3-3ζ as a prognostic marker and therapeutic target for cancer. Expert Opin. Ther. Targets. 14, 1343–1354 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panieri E, Toietta G, Mele M, Labate V, Ranieri SC, Fusco S, Tesori V, Antonini A, Maulucci G, De Spirito M, Galeotti T, Pani G: Nutrient withdrawal rescues growth factor-deprived cells from mTOR-dependent damage. Aging. 2, 487–503 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krämer A, Green J, Pollard Jack J, Tugendreich S: Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 30, 523–530 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Bodmer WF: Analysis of P53 mutations and their expression in 56 colorectal cancer cell lines. Proc. Natl. Acad. Sci. U. S. A. 103, 976 LP–976981 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozaki T, Nakagawara A: Role of p53 in cell death and human cancers. Cancers (Basel). 3, 994–1013 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maddocks ODK, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH: Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 493, 542–546 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slattery ML, Lundgreen A, Kadlubar SA, Bondurant KL, Wolff RK: JAK/STAT/SOCS-signaling pathway and colon and rectal cancer. Mol. Carcinog. 52, 155–166 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He R, Chen H, Feng Z, Dang Y, Gan T, Chen G, Yang L: High level of STAT4 expression is associated with the deterioration of breast cancer. Int. J. Clin. Exp. Med. 9, 11612–11618 (2016) [Google Scholar]
- 45.Romagnolo DF, Milner JA: Opportunities and challenges for nutrional proteomics in cancer prevention. Jounal Nutr. 142, 225–229 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


