Abstract
The pelA gene from the N2-fixing plant-associated bacterium Azospirillum irakense, encoding a pectate lyase, was isolated by heterologous expression in Escherichia coli. Nucleotide sequence analysis of the region containing pelA indicated an open reading frame of 1,296 bp, coding for a preprotein of 432 amino acids with a typical amino-terminal signal peptide of 24 amino acids. N-terminal amino acid sequencing confirmed the processing of the protein in E. coli at the signal peptidase cleavage site predicted by nucleotide sequence analysis. Analysis of the amino acid sequence of PelA revealed no homology to other known pectinases, indicating that PelA belongs to a new pectate lyase family. PelA macerates potato tuber tissue, has an alkaline pH optimum, and requires Ca2+ for its activity. Of several divalent cations tested, none could substitute for Ca2+. Methyl-esterified pectin (with a degree of esterification up to 93%) and polygalacturonate can be used as substrates. Characterization of the degradation products formed upon incubation with polygalacturonate indicated that PelA is an endo-pectate lyase generating unsaturated digalacturonide as the major end product. Regulation of pelA expression was studied by means of a translational pelA-gusA fusion. Transcription of this fusion is low under all growth conditions tested and is dependent on the growth phase. In addition, pelA expression was found to be induced by pectin. An A. irakense pelA::Tn5 mutant still displayed pectate lyase activity, suggesting the presence of multiple pectate lyase genes in A. irakense.
The capacity to degrade pectin, a major constituent of the primary plant cell wall and middle lamella, is a feature of many plant-associated bacteria, especially plant pathogens. Pectin is a heteropolysaccharide with a backbone consisting of partially methyl- and acetyl-esterified galacturonic acid. Pectin is depolymerized by a combination of enzymatic activities: pectin methyl esterase, pectin acetyl esterase, pectin lyase, pectate lyase, and polygalacturonase. Lyases cleave internal glycosidic bonds via β-elimination, producing oligomers with 4,5-unsaturated residues at the nonreducing end. In contrast, polygalacturonase hydrolyzes the polymer, yielding saturated products. The preference for highly methylated pectin and the absence of activity on polygalacturonate (PGA) differentiate pectin lyase and pectate lyase. Pectin methyl esterase and pectin acetyl esterase facilitate degradation of pectin by pectate lyase by generating PGA through demethylation and deacetylation of pectin, respectively.
In the past decade, the genetics of bacterial pectinase biosynthesis has been extensively studied in phytopathogens, especially in the soft-rotting Erwinia species Erwinia carotovora and Erwinia chrysanthemi. Both species were found to produce a set of pectin-depolymerizing activities such as pectate lyases, polygalacturonases, pectin methyl esterases, and a pectin acetyl esterase (3, 17, 41, 47, 48). Among these, the pectate lyases are the major pectinases and play a key role in the development of the soft rot disease. In E. chrysanthemi 3937, nine extracellular pectate lyases have been identified so far, i.e., one exo-Pel (PelX) (6) and eight endo-Pels (PelA, PelB, PelC, PelD, PelE, PelL, PelZ, and PelI) (17, 41, 47). Erwinia pel genes are transcribed from independent cistrons, and their differential expression is believed to reflect their distinct roles during plant pathogenesis. Besides Erwinia, pel genes have been cloned from other phytopathogens such as Xanthomonas and soft rot-causing Pseudomonas and Bacillus species (24, 33, 37). Little information is available on the genes that encode pectolytic activity in nonphytopathogenic bacteria. A Yersinia pseudotuberculosis pel gene (pelY) was cloned and found to be homologous to genes encoding periplasmic Pel enzymes in E. carotovora strains (29).
The genus Azospirillum comprises free-living nitrogen fixing soil bacteria that have been isolated from the rhizosphere of a wide range of plant species, including commercially important crops such as maize, rice, wheat, and sorghum (38). Plant growth promotion by Azospirillum has been attributed to the potential of these bacteria to produce the phytohormone indole-3-acetic acid. Five species have been identified within the genus: A. brasilense, A. lipoferum, A. amazonense, A. halopraeferans, and A. irakense. Some isolates of A. irakense were obtained from surface-sterilized field-grown rice roots (20), indicating their capacity to penetrate plant roots and suggesting the involvement of plant cell wall-degrading enzymes in this infection process. Here we report on the molecular characterization of a Pel (PelA) and the cloning of the corresponding structural gene from Azospirillum irakense KBC1. A. irakense PelA defines a new class of Pel enzymes since it displays no homology to other known bacterial, plant or fungal pectinases.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Azospirillum irakense | ||
| KBC1 (LMG10653) | Wild type | 20 |
| FAJ0602 | LMG10653pelA::Tn5 | This study |
| Azospirillum brasilense Sp245 | Wild type | 2 |
| Azospirillum lipoferum SpBr17 | Wild type | 52 |
| Azospirillum halopraeferans Au4 | Wild type | 43 |
| Escherichia coli | ||
| HB101 | F−hsdS20(r−B m−B) recA13 ara-14 proA2 rpsL20 supE44 | 46 |
| S17-1::Tn5 | S17.1 (49) carrying chromosomal Tn5, Kmr | Laboratory collection |
| ABG4.1 | MG1655 pro::Tn10 dCam, Cmr | Laboratory collection |
| Plasmids | ||
| pLAFR1 | IncP broad-host-range cosmid, Tcr | 10 |
| pLAFR3 | IncP broad-host-range cosmid, Tcr | 50 |
| pRK2013 | ColE1 replicon, Tra+, Kmr | 8 |
| pSUP202 | Suicide vector for A. irakense, oriT from RP4, Cmr Tcr Apr | 49 |
| pUC19 | Plasmid vector, Apr | 56 |
| pUC18 | Plasmid vector, Apr | 56 |
| pKW118 | pUC8 derivative containing the gusA gene followed by the trpA terminator, Apr | 55 |
| pFAJ0601 | pLAFR1 carrying the 25-kb Pel+ DNA region of A. irakense LMG10653, Tcr | This study |
| pFAJ0610 | pLAFR1 carrying the 9.2-kb Pel+EcoRI fragment from pFAJ0601, Tcr | This study |
| pFAJ0611 | pUC19 carrying the 9.2-kb Pel+EcoRI fragment from pFAJ0601, Apr | This study |
| pFAJ0612 | pUC19 carrying the 3.2-kb Pel+HindIII-EcoRI fragment from pFAJ0601, Apr | This study |
| pFAJ0613 | pUC18 with the 0.9-kb NaeI fragment containing the regulatory region and 360 bp of the coding sequence of pelA inserted in SmaI, Apr | This study |
| pFAJ0614 | pSUP202 carrying the 3.15-kb Pel+PstI-EcoRI fragment from pFAJ0601, Tcr | This study |
| pFAJ0615 | pFAJ0614, pelA::Tn5, Tcr Kmr | This study |
| pFAJ0617 | pLAFR3 containing the pelA-gusA fusion on a 2.8-kb EcoRI-HindIII fragment, Tcr | This study |
Media and culture conditions.
Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) medium (46). Azospirillum strains were grown at 30°C in minimal medium AB (MMAB) containing 0.5% malate as the carbon source (54) or in LB* medium (LB medium supplemented with 2.5 mM CaCl2 and 2.5 mM MgSO4). Growth on PGA or pectin was evaluated in MMAB in which malate was replaced by 0.5% PGA or 0.5% citrus pectin (10% methylesterified; Sigma Chemical Co.), respectively. Acetylene reduction activity was determined in N-free MMAB as described previously (31). Ampicillin, chloramphenicol, tetracycline, and kanamycin were used at 100, 25, 10, and 50 μg/ml, respectively.
DNA manipulations and nucleotide sequencing.
DNA was isolated and manipulated by using standard techniques (46). Subclones for nucleotide sequencing were constructed in pUC18. Nucleotide sequencing was accomplished on an automated sequencer (A.L.F.; Pharmacia Biotech) by using the Autoread sequencing kit (Pharmacia Biotech) and fluorescein-labeled universal and synthetic oligonucleotide primers. Sequence compilation and analyses were carried out with the aid of the PC/Gene software package (IntelliGenetics Inc.). The program BLAST 2.0 (1) was used to search for related sequences. For Southern hybridizations, probe DNA was labeled with digoxigenin-dUTP by using the random-primed labeling kit from Boehringer Mannheim. Detection was performed with a chemiluminescence detection kit (Boehringer Mannheim). Low-stringency hybridization was carried out at 56°C. Megaplasmid DNA was extracted from A. irakense and separated on an agarose gel as described previously (19).
Construction of a genomic library of A. irakense KBC1.
Total genomic DNA of A. irakense KBC1 was partially digested with EcoRI and ligated into the cosmid pLAFR1, restricted with EcoRI. The ligation mixture was packaged in phage heads with an in vitro packaging system (Boehringer Mannheim). This mixture was used to infect E. coli HB101 as recommended by the manufacturer of the kit.
Detection tests and pectic enzyme assays on cultures.
Pectinolytic activity was assayed after 7 days of growth on solid medium containing 1% citrus pectin (10% methylesterified) by overlaying the plates with 2% hexadecyltrimethylammonium bromide (CTAB) (42). The appearance of halos around the colonies indicates the degradation of pectin. For enzyme assays performed on culture supernatants and cell extracts, the thiobarbituric acid (TBA) test (22) was used. Cells were grown in complex LB* medium and centrifuged at 6,000 × g. Pelleted cells were resuspended in physiological solution (0.8% NaCl) and lysed in Fast RNA tubes with the Fast Prep FP120 device (Bio 101-Savant) as described by the manufacturer. For Pel activity, an appropriate volume of culture supernatant or cell lysate was incubated at 30°C in a reaction mixture containing 50 mM Tris-HCl (pH 8.5), 0.5 mM CaCl2, and 0.2% PGA. Two volumes of 0.5 N HCl and 4 volumes of 0.01 M TBA, were then added and the reaction mixture was heated at 100°C for 1 h and centrifuged. For pectin lyase activity, 0.2% pectin (93% methylesterified) was used as substrate and 3 mM EDTA was added to inhibit any contaminating Pel activity. The reaction mixture for polygalacturonase activity contained 50 mM Tris-HCl (pH 8.5), 0.2% PGA, and 3 mM EDTA. Enzyme activities were calculated from the increase in the absorbance of the supernatants at 550 nm (A550) (Pel and pectin lyase) or 515 nm (A515) (polygalacturonase). One unit of activity was defined as an increase of 1 A550 or 1 A515 unit in 1 h at 30°C.
Construction of the translational pelA-gusA fusion, pFAJ0617.
The NaeI fragment containing 540 bp of the upstream region and 360 bp of the coding sequence of the A. irakense pelA gene was cloned into the SmaI site of pUC18, yielding pFAJ0613. A 1.9-kb BamHI-HindIII fragment carrying the promoterless gusA gene from pKW118 (55) was then ligated into BamHI-HindIII-digested pFAJ0613. Nucleotide sequence analysis with an internal gusA primer (5′-GATTTCACGGGTTGGGGTTTCT-3′) confirmed that the pelA-gusA fusion was in frame. The EcoRI-HindIII fragment of approximately 2.8 kb carrying the entire pelA-gusA fusion was then inserted in the corresponding sites of the broad-host-range plasmid pLAFR3, yielding pFAJ0617. Finally, pFAJ0617 was transferred to A. irakense KBC1 and A. brasilense Sp245 by triparental mating as described previously with pRK2013 as a helper plasmid (54).
Site-directed Tn5 mutagenesis of pFAJ0614.
E. coli S17-1::Tn5 was transformed with pFAJ0614. Tn5-containing derivatives of pFAJ0614 were isolated by biparental conjugation to the acceptor E. coli ABG4.1 (donor/acceptor ratio, 1:1) and selected on plates containing tetracycline, kanamycin, and chloramphenicol. Transconjugants no longer expressing pectinolytic activity were screened by the TBA assay.
Purification and amino acid sequencing of PelA.
E. coli DH5α(pFAJ0612) was grown at 37°C in LB broth to stationary growth phase. The culture was centrifuged at 6,000 × g, and the pellet was resuspended to a final concentration of 20% (wt/vol) in phosphate-buffered saline containing 250 U of DNase per ml. Cells in this suspension were lysed in FastRNA tubes with a FastPrep FP120 device. This lysate was further cleared by ultracentrifugation (Beckman SW50.1 rotor; 120,000 × g for 2.5 h at 4°C) and filtration through a Millipore Millex 0.22-μm-pore-size filter. The filtrate was then concentrated on Microcon10 microconcentrators (Amicon) and fractionated on a gel chromatography column (Superdex 200 Prep Grade, packed in HR16/50; Pharmacia) with phosphate-buffered saline (flow rate, 1 ml/min). The Pel activity in the different fractions was assayed by monitoring the increase in A232 upon incubation with PGA as described below. Fractions exhibiting Pel activity were concentrated on Microcon10 microconcentrators and further fractionated by cation-exchange chromatography (Mono S HR5/5; Pharmacia). Proteins were eluted with a linear gradient of NaCl in 50 mM sodium acetate buffer (pH 5) (from 0 to 0.5 M NaCl in 20 min; flow rate, 0.5 ml/min). Pel-active fractions were pooled, 5 volumes of HIC buffer [1.7 M (NH4)2SO4, 20 mM Tris-HCl (pH 7.4)] was added, and the mixture was loaded onto a hydrophobic interaction column (phenyl-Sepharose HP, packed in XK16 [Pharmacia]). Elution was carried out with a decreasing linear gradient of (NH4)2SO4 in the same buffer (flow rate, 1 ml/min). Fractions exhibiting Pel activity were concentrated, desalted on Microcon10 microconcentrators, and stored at −20°C.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was carried out in a vertical Laemmli system with a 12% polyacrylamide separating gel. A PhastGel IEF3-9 (Pharmacia) gel was used for isoelectric focusing. N-terminal amino acid sequence analysis was accomplished by Edman degradation with a 477A protein sequencer (Applied Biosystems/Perkin-Elmer, Foster City, Calif.). Internal sequences were obtained by digesting the protein for 18 h with endoproteinase Asp-N (sequencing grade; Boehringer Mannheim). The resulting peptides were separated by C8 reverse-phase high-pressure liquid chromatography on an Aquapore RP-300 column (50 by 1 mm; Brownlee/Perkin-Elmer) and eluted in an acetonitrile gradient prior to Edman degradation.
Pel assays with purified PelA.
The Pel activity of the purified PelA protein was determined at 30°C by monitoring the formation of unsaturated products from PGA at 232 nm. Unless stated otherwise, the reaction buffer contained 50 mM Tris-HCl (pH 8.5), 0.1 mM CaCl2, and 0.2% PGA. One unit of activity is defined as the amount of enzyme required to produce 1 μmol of unsaturated uronide per min at 37°C. The specific activity is expressed as units per milligram of protein. For determination of the optimum pH, 100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer (pH 6 to 7) and 100 mM Tris-HCl (pH 7.5 to 10) buffer were used. The effect of CaCl2 was assayed by adding CaCl2 up to 2 mM. The effect of other divalent cations (Mg2+, Mn2+, Ba2+, Cu2+, Zn2+, and Co2+) was evaluated by using the corresponding chloride salts at 0.1 mM. The influence of esterification of the substrate was tested by replacing PGA in the reaction mixture by 10, 28, 64, and 93% methylated citrus pectin. The Vmax and Km values were determined in the standard reaction mixture by using PGA at 0.1 to 1 g/liter.
Characterization of PGA degradation products by HPAEC.
PelA ranging from 0 to 0.5 U/ml was incubated with 0.1% PGA for 10 min at 30°C in a 0.1 M glycine–NaOH (pH 9.4) buffer containing 1 mM CaCl2. After the incubation, PelA was inactivated by boiling for 5 min. The degradation products in the reaction mixtures were separated by high-performance anion-exchange chromatography (HPAEC) with a BioLC system (Dionex, Sunnyvale, Calif.) equipped with a Dionex Carbopack PA-100 column (250 by 4 mm) as described previously (9).
Maceration of potato tuber tissue.
Plant tissue maceration was assessed on small potato cubes (9 mm3) placed in 1 ml of 0.1 M Tris-HCl (pH 8.5)–0.5 mM CaCl2 by inoculating 1 ml of a bacterial suspension (107 to 109 cells ml−1) or by adding 0.01 to 2 U of purified A. irakense PelA or 0.01 to 0.1 U of purified E. chrysanthemi PelD per ml. Noninoculated samples were used as negative control. The samples were incubated at 37°C and examined for tissue softening at regular times by using a spatula.
Construction of the A. irakense pelA mutant FAJ0602.
The 3.15-kb PstI-EcoRI fragment, carrying the entire pelA gene, was subcloned into pSUP202, resulting in pFAJ0614. Site-directed Tn5 mutagenesis on pFAJ0614 by using E. coli S17-1::Tn5 yielded pFAJ0615, carrying a Tn5 insertion in the pelA gene (approximately 1.65 kb from the PstI site). pFAJ0615 was introduced in A. irakense by triparental mating with HB101(pRK2013) as a helper. Transconjugants were selected on MMAB containing kanamycin. Double recombinants were screened for sensitivity to tetracycline and verified by Southern hybridization. One of the recombinants showing a correct genetic configuration was named FAJ0602.
Nucleotide sequence accession number.
The nucleotide sequence of the A. irakense pelA gene will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AF121904.
RESULTS
Pectinolytic activity in Azospirillum species.
The capacity of different Azospirillum species to hydrolyze pectin was tested on LB* agar containing citrus pectin by overlaying the plates with CTAB. A clear halo around the colonies, indicating bacterial degradation of pectin, was observed for A. irakense. However, in this assay, none of the other Azospirillum species tested (A. brasilense, A. lipoferum, and A. halopraeferans) exhibited visible pectinolytic activity.
A. irakense KBC1 grew and fixed N2 in media containing pectin or PGA as the sole carbon source, indicating the presence of a catabolic pathway for further breakdown of the oligogalacturonides generated by the pectinolytic activity. Acetylene reduction activities with PGA or pectin as the sole carbon source were determined to be 54 and 95%, respectively, of those observed with malate.
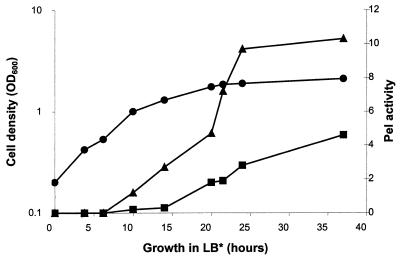
To further characterize the pectinase(s) produced by A. irakense, cultures of strain KBC1 were grown to different optical densities in LB* medium and examined for Pel, pectin lyase, and polygalacturonase activities in the TBA assay. Only Pel activity could be observed. Pel production was induced in the second half of exponential growth and reached a maximal level during stationary growth (Fig. 1). In both the exponential and stationary growth phase cultures, more than 60% of the activity was detected in the supernatant, suggesting that the majority of the enzyme is secreted.
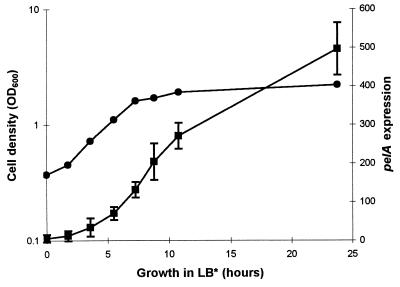
FIG. 1.
Pel activity in cell extracts and culture supernatants of A. irakense KBC1 during growth in LB* medium. Samples were taken periodically to determine the Pel activity in the cell extracts (squares) and culture supernatants (triangles). Pel activity was measured by the TBA assay as described in Materials and Methods and expressed in (units per milliliter of culture/OD600) × 1,000. Circles: optical density at 600 nm (OD600). Data points are the means of duplicate determinations from a representative experiment.
Cloning of the A. irakense pelA gene.
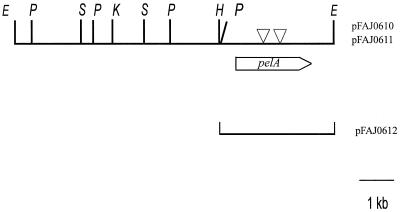
To isolate the gene(s) encoding pectinolytic activity in A. irakense, a genomic library of A. irakense KBC1 was constructed in cosmid pLAFR1. Approximately 3,000 recombinant E. coli clones were subsequently screened for pectinolytic activity by the CTAB overlay assay. After a 7-day incubation, eight transfectants exhibiting pectinase activity were isolated. DNA restriction analysis of the cosmids purified from these clones revealed one common EcoRI fragment of 9.2 kb. Subcloning experiments and site-directed Tn5 mutagenesis mapped the putative A. irakense pectinase gene on a 3.2-kb HindIII-EcoRI fragment (Fig. 2).
FIG. 2.
Physical map of the 9.2-kb EcoRI fragment carrying the A. irakense pelA gene. Triangles indicate the position of Tn5 insertions that abolish pectinolytic activity in E. coli. The position of the pelA gene, deduced from the nucleotide sequence, is indicated by an arrow. E, EcoRI; P, PstI; S, SalI; K, KpnI; H, HindIII.
To determine the nature of the pectinolytic activity associated with the 3.2-kb HindIII-EcoRI fragment, cell extracts of DH5α(pFAJ0612) were screened for Pel, pectin lyase, and polygalacturonase activities in the TBA assay. Only Pel activity could be detected (0.1 U per ml of cell culture).
Purification and physical properties of PelA.
The Pel enzyme (PelA) encoded by the pFAJ0612 insert was purified to apparent homogeneity from the E. coli transformant by a combination of gel chromatography, cation-exchange chromatography, and hydrophobic interaction high-pressure liquid chromatography. After each purification step, Pel-active samples were screened by measuring the increase in A232 upon incubation with PGA. The preparation contained a single polypeptide with an apparent molecular mass of 44.4 kDa. The isoelectric point as determined by isoelectric focusing was 6.2 (data not shown). The specific activity of purified PelA in the standard reaction buffer was 59 U/mg.
The amino acid sequence of A. irakense PelA shows no homology to any known Pel protein.
Nucleotide sequence analysis of the 3.2-kb HindIII-EcoRI fragment revealed an open reading frame of 1,296 bp. The G+C content of this open reading frame is 61.8%, which is somewhat lower than the overall G+C content of A. irakense DNA (64 to 67%) (20). The G+C content in the third position of the codons is 71.2%. A putative Shine-Dalgarno sequence (GAGGAA) is located 12 bases upstream of the potential GTG initiation codon. Database searching failed to disclose homology of the deduced amino acid sequence to known peptides. A signal peptidase I cleavage site located between amino acids 24 and 25 was identified by the program SignalP (36) and confirmed by N-terminal amino acid sequencing of the mature PelA protein. The signal sequence resembles the amino-terminal signal peptides of gram-negative bacteria (45); i.e., it contains a positively charged amino terminus, a central hydrophobic core of 12 residues, and two alanine residues at positions −1 and −3 relative to the processing site. Amino acid sequencing of internal peptides generated by Asp-N digestion of PelA confirmed the open reading frame deduced from the nucleotide sequence. The mature PelA protein contains 408 amino acids and has a calculated molecular mass of 44.5 kDa and a pI of 5.74. This molecular mass is in good agreement with that determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the purified PelA. The calculated pI, however, is slightly lower than the value determined by isoelectric focusing of the secreted enzyme.
Enzymatic properties of PelA.
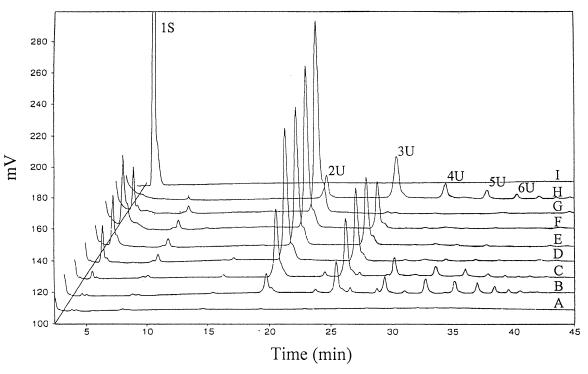
To determine whether PelA was an exo- or endo-enzyme, the reaction products formed during depolymerization of PGA were characterized by HPAEC. For this, a dilution series of PelA was incubated with 0.1% PGA for 10 min at 30°C. The HPAEC profiles of the degradation products obtained in the reaction mixtures are presented in Fig. 3. Clearly, the formation of multiple unsaturated oligogalacturonates (ranging in a degree of polymerization from 2 to 9) was observed during depolymerization, suggesting that PelA is an endo-Pel. High PelA concentrations or longer incubation times resulted in the specific accumulation of unsaturated digalacturonate.
FIG. 3.
HPAEC profiles of oligogalacturonides generated by PelA. The reactions were performed in a 0.1 M glycine buffer (pH 9.4) containing 1 mM CaCl2, 0.1% PGA, and various PelA concentrations: 0 μg/ml (A), 1 μg/ml (B), 2 μg/ml (C), 4 μg/ml (D and G), 6 μg/ml (E), and 8 μg/ml (F). Incubation was carried out at 30°C for 10 min (A to F) or at 23°C for 48 h (G). The reactions were stopped by boiling for 5 min. A mixture of unsaturated oligogalacturonides is also shown (H). Numbers indicate the degree of polymerization; U, unsaturated; I, galacturonic acid (1S).
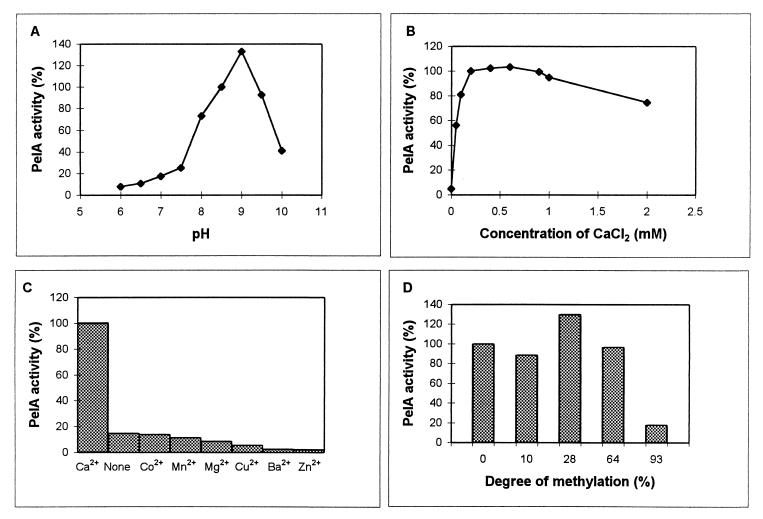
Similarly to the situation for endo-Pels from phytopathogenic bacteria, PelA was found to have an alkaline pH optimum (about pH 9) and to require Ca2+ for its activity. A narrow pH zone of optimum activity was observed (Fig. 4A). At neutral pH, the activity was less than 15% of the maximal activity. PelA activity was completely inhibited by the addition of 3 mM EDTA. Optimal activity was observed throughout a wide range of CaCl2 concentrations (from 0.2 to 1 mM) (Fig. 4B). No other divalent cation (Mg2+, Mn2+, Ba2+, Cu2+, Zn2+, or Co2+) could substitute for Ca2+ (Fig. 4C). In contrast, most of these cations strongly inhibited PelA activity. Zn2+ and Ba2+ had the strongest inhibitory effect. With PGA as a substrate, the Km and Vmax values for PelA were determined to be 0.076 g liter−1 and 23 μmol min−1 mg−1, respectively. PelA was shown to be more active toward partially esterified pectins than toward PGA. Optimal activity was found on pectin, with a degree of methylation of up to 28% (Fig. 4D). More than 74% of the maximum activity was retained with a substrate methylation of up to 64%. Optimal PelA activity was found at 40°C for incubations of 20 min. Above the optimal temperature, the activity dropped sharply, with only 17 and 6% of the activity being retained at 50 and 55°C, respectively.
FIG. 4.
Enzymatic properties of PelA. Enzyme activity was assayed as described in Materials and Methods. (A) Effect of pH on PelA activity. The influence of pH was tested by using PIPES (pH 6 to 7) or Tris-HCl (pH 7.5 to 10) buffers at 100 mM. The activities are expressed relative to that obtained at pH 8.5, which was arbitrarily defined as 100. (B) Effect of CaCl2 concentration on PelA activity. The activity obtained in the presence of 0.2 mM CaCl2 was arbitrarily defined as 100. (C) Effect of divalent cations on PelA activity. The influence of each divalent cation was tested by using the corresponding chloride salt (0.1 mM) instead of CaCl2. The activity in the presence of CaCl2 was arbitrarily defined as 100. (D) Effect of the degree of pectin methylation on PelA activity. The influence of pectin methylesterification was tested by replacing PGA in the standard reaction mixture by pectins with different degrees of methylation. The activity obtained in the presence of PGA was arbitrarily defined as 100.
The maceration capacity of PelA was compared with that of E. chrysanthemi PelD by incubating serial dilutions of purified enzymes with potato tuber cubes. E. chrysanthemi PelD, at 0.1 U ml−1, showed evidence of maceration after only 1 h of incubation at 37°C. A. irakense PelA was much less active than E. chrysanthemi PelD, since tissue softening was observed only after 4 to 5 h of incubation with 1.6 U ml−1.
Analysis of pelA expression.
To study the expression of A. irakense pelA, a plasmid-encoded translational fusion between pelA and the reporter gene gusA was constructed (pFAJ0617). Analysis of GusA activity during growth of KBC1(pFAJ0617) in complex LB* medium revealed that pelA is expressed at very low levels. Also, pelA expression was shown to depend on the growth phase of the cells, with the highest level of induction being observed during the stationary growth phase (Fig. 5). These expression results are in agreement with the previously observed cell density-dependent pattern of Pel activity.
FIG. 5.
Temporal expression of pelA in A. irakense KBC1. A culture of KBC1(pFAJ0617) was grown in LB* medium. Samples were taken throughout growth, and the optical density at 600 nm (OD600) (circles) and the expression of the pelA-gusA fusion, pFAJ0617 (squares) were assessed. Quantitative analysis of β-glucuronidase activity was carried out in microtiter plates with 4-methylumbelliferyl-β-d-glucuronide as the substrate (18) and expressed as picomoles of product formed per minute per milligram protein. The data points are the means of duplicates from one representative experiment, which was confirmed twice in additional independent experiments.
pelA expression in stationary-phase cultures of A. irakense in MMAB was twofold lower than in LB* medium (Table 2). Both in MMAB and in LB* medium, the presence of pectin significantly stimulated pelA transcription, although this induction was stronger in LB* medium. Additionally, pelA showed a higher level of expression in the presence of the easily metabolizable carbon sources, malate and glucose, than in the presence of starch.
TABLE 2.
Expression of pelA-gusA (pFAJ0617) in A. irakense, A. brasilense, and E. coli
| Strain and growth mediuma | β-Glucuronidase activityb |
|---|---|
| A. irakense KBC1 | |
| LB* medium | 420 ± 91 |
| LB* medium + pectin | 975 ± 37 |
| MMAB (malate) | 118 ± 23 |
| MMAB (malate) + pectin | 183 ± 8 |
| MMAB (glucose) | 131 ± 19 |
| MMAB (starch) | 75 ± 1 |
| A. brasilense Sp245 | |
| LB* medium | 1,249 ± 119 |
| E. coli HB101 | |
| LB* medium | 1,886 ± 148 |
Cells were grown to the stationary growth phase in the different culture media. Pectin (64% methylated) was added at 4 g/liter. The carbon sources in MMAB were present at 4 g/liter.
Quantitative analysis of β-glucuronidase activity was carried out in microtiter plates with 4-methylumbelliferyl-β-d-glucuronide as the substrate (18). Glucuronidase activity is expressed as picomoles of 4-methylumbelliferone produced per minute per milligram of protein. The results are given as mean ± standard deviation of duplicate determinations.
The low level of pelA expression suggests a tight transcriptional control of the gene in A. irakense. The presence of such a strict regulatory system in A. irakense was further evidenced by the fact that we observed a threefold-higher pelA expression level in the closely related A. brasilense strain Sp245 and a four- to fivefold-higher expression level in E. coli HB101 (Table 2). Concomitant with the enhanced pelA transcription in this strain, Pel activity in A. brasilense Sp245 containing pFAJ0610 was 5-fold higher than in wild-type A. irakense and 3.5-fold higher than in A. irakense containing pFAJ0610 (data not shown).
PelA is chromosomally located in A. irakense and absent from A. brasilense, A. lipoferum, and A. halopraeferans.
A. irakense has previously been shown to contain a megaplasmid of 135 MDa (14). In the best studied Azospirillum species, A. brasilense, many traits related to the plant interaction are encoded by a 90-MDa megaplasmid (7). To evaluate whether pelA is chromosomally or plasmid located, Southern hybridizations were performed on plasmid profiles of A. irakense KBC1 with the 3.2-kb HindIII-EcoRI fragment carrying the entire pelA gene as probe. Hybridization could be detected only with chromosomal DNA (data not shown). With the same probe, no signal was detected on genomic digests prepared from A. brasilense, A. lipoferum, and A. halopraeferans, even when hybridized at low stringency.
PelA is not the only pectate lyase produced by A. irakense KBC1.
To determine the role of PelA in pectate metabolism, an A. irakense pelA mutant (FAJ0602) was constructed. Plasmid pFAJ0615, carrying a Tn5 insertion into the pelA gene, was used for marker exchange. No Pel activity could be detected in cell extracts of E. coli ABG4.1(pFAJ0615). Although reduced compared to the activity in the wild type, Pel activity was still detected in culture supernatants of FAJ0602, indicating the existence of multiple Pel enzymes in A. irakense KBC1 (Fig. 6). Introduction of pFAJ0610, carrying the entire pelA gene, into FAJ0602 restored the Pel activity to the wild-type level (Fig. 6).
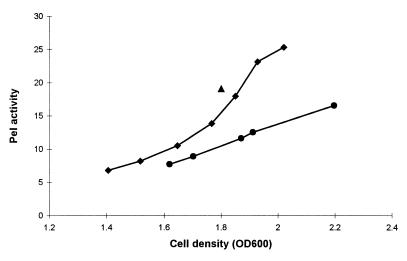
FIG. 6.
Pel activity in culture supernatants of A. irakense KBC1 (⧫), FAJ0602 (●), and FAJ0602(pFAJ0610) (▴). Cells were grown in LB* medium. Pel activity in the culture supernatant was measured by the TBA assay as described in Materials and Methods. For KBC1 and FAJ0602, samples were taken in the late exponential and stationary growth phases. For FAJ0602(pFAJ0610), activity was determined only at the stationary growth phase (optical density at 600 nm [OD600], 1.8). Pel activity is expressed in units per milliliter of culture supernatant × 1,000. Data points are the means of duplicate determinations from one representative experiment, which was confirmed twice in additional independent experiments.
DISCUSSION
The nitrogen-fixing soil bacterium A. irakense was demonstrated to possess pectinolytic activity. Degradation of PGA yielded products with an absorbance peak at 550 nm upon incubation with TBA, characteristic for unsaturated oligogalacturonides generated by a sugar lyase. By means of heterologous expression in E. coli, the A. irakense pelA gene, coding for a 44-kDa acidic endo-Pel, was isolated and further characterized at the genetic and biochemical levels.
The A. irakense PelA protein defines a new family of Pel enzymes, since it displays no homology to other known bacterial, fungal, or plant pectinases according to sequence comparison programs. During the past 5 years, sequence information for many bacterial pel genes has become available. In addition to the previously described five major endo-Pel enzymes (PelA, PelB, PelC, PelD, and PelE) (21), three secondary endo-Pel enzymes, PelL, PelZ, and PelI (28, 41, 47), have recently been cloned from E. chrysanthemi. Additionally, a number of pel genes have been sequenced from non-Erwinia phytopathogens, including X. campestris pv. malvacearum (24), and soft rot-producing fluorescent Pseudomonas strains (P. viridiflava [24], P. fluorescens [24], and P. marginalis [37]). On the basis of sequence alignments, distinct families of Pel enzymes can be distinguished (47). A first family corresponds to several well-studied Pel enzymes, including the major extracellular Pel enzymes of E. chrysanthemi. This family is further divided into two subfamilies, of which the first subfamily contains, in distinct clusters, PelA, PelD, and PelE of E. chrysanthemi and the Pel proteins originating from non-Erwinia phytopathogens such as Xanthomonas and soft rot-causing Pseudomonas and Bacillus strains. E. chrysanthemi PelB and PelC and several extracellular E. carotovora Pel enzymes constitute the second subfamily. Structural analysis of E. chrysanthemi PelC (57), E. chrysanthemi PelE (25, 58), and B. subtilis PelK (39) revealed that these proteins fold into a unique motif of parallel β-strands coiled in a large helix stabilized by a stack of asparagine residues. A second family corresponds to the periplasmic Pel enzymes of E. carotovora subsp. carotovora, i.e., PelB and Pel153 (16, 53), and includes PelY of the nonphytopathogenic Y. pseudotuberculosis (29). The recently identified secondary Pel enzymes of E. chrysanthemi constitute three additional families: a third family containing PelL (28) and the exo-pectate lyase PelX (6) of E. chrysanthemi, a fourth family containing PelI of E. chrysanthemi (47) and Pel3 of E. carotovora (26), and a fifth family corresponding to PelZ of E. chrysanthemi (41). The PelA protein of A. irakense, identified in the present paper, defines a sixth new family.
The absence of homology between the primary sequences of Pel enzymes of distinct families suggests that they have evolved from different lineages. The fact that such catalytically similar enzymes have evolved independently may reflect different functions in nature. Strikingly, homology was observed between E. chrysanthemi PelI and four Pel proteins of a phytopathogenic fungus, Nectria haemotococca (Fusarium solani) (11, 12, 13, 47), indicating that Pel proteins belonging to the same family are found in distantly related organisms.
The fact that Pel activity is not completely abolished in the A. irakense pelA mutant FAJ0602 suggests the production of multiple Pel isoenzymes by A. irakense. This situation of pel gene redundancy is similar to that for the Erwinia species. On the other hand, most other pectinolytic bacteria, including P. viridiflava (23), B. subtilis (32), X. campestris pv. vesicatoria (5), and P. syringae pv. lachrymans (4), produce only a single Pel enzyme. A screening of the A. irakense genomic library for clones able to depolymerize pectin, however, yielded only clones carrying pelA, suggesting no or poor expression of the other Pel isoenzyme(s) in E. coli. Also, the additional pel gene(s) has no homology to pelA, since only hybridizing bands corresponding to pelA were detected in Southern hybridizations with the entire pelA gene as a probe (data not shown).
Thus far, detailed information on the enzymatic properties of Pel enzymes is restricted to the isoenzymes of Erwinia species. Cloning of the distinct structural genes made it possible to purify each of the Pel enzymes without contamination of the other isoenzymes and to characterize the associated catalytic activity (28, 41, 47, 51). Similarly, the A. irakense PelA protein was purified and studied at the biochemical level. Like other Pel enzymes, A. irakense PelA requires Ca2+ for its activity and has an highly alkaline pH optimum. Three sequence patterns, onserved among the major extracellular Pel enzymes of E. chrysanthemi and some Pel enzymes from non-Erwinia phytopathogens and suspected of being involved in Ca2+-binding and catalytic activity (15), could not be detected in the PelA protein of A. irakense. Unlike the E. chrysanthemi Pel enzymes (28, 47, 51), high activity of purified A. irakense PelA was observed throughout a wide range of Ca2+ concentrations (from 0.2 to 2 mM). Other cations cannot substitute for Ca2+, and some were even found to inhibit PelA activity. Similarly to what has been observed for the five major E. chrysanthemi Pel enzymes (51), Zn2+ was the strongest inhibitor of PelA activity.
Evidence that PelA is an endo-Pel enzymes comes from the fact that multiple unsaturated oligogalacturonides are formed during PGA depolymerization by purified PelA. Complete digestion results in the accumulation of unsaturated dimers as major reaction product. A. irakense PelA is active on PGA as well as on pectins with a degree of methylesterification up to 93% but it presented the highest activity on pectins with a lower degree of methylation (28%). In plants, pectins with different degrees of methylation are present. Pectin methylation varies among plant species, among tissues, and among domains within a single cell wall. In E. chrysanthemi, variation in the activity between Pel enzymes as a function of the degree of pectin methylation has been observed (51). Therefore, given the large variation in pectic substances and the presence of a large array of Pel isoenzymes, a possible specialization (class of Pel per type of pectin) can be suggested.
More than 60% of the Pel activity is detected in the supernatant of A. irakense mid- and late-exponential-phase cultures as well as stationary-phase cultures, indicating transport of the degrading enzymes to the extracellular medium. A clear reduction in extracellular Pel activity compared to that in the parental strain was observed for the pelA mutant strain FAJ0602, suggesting secretion of the PelA protein.
It has been suggested that differences in the rate of synthesis and secretion of Pel enzymes determine whether a plant-microbe interaction becomes pathogenic (17). The Pel activity detected in A. irakense culture supernatant is low compared with that detected in Erwinia cultures. Purified PelA, however, exhibits a specific activity which is higher than that reported for E. chrysanthemi PelA (51). Maceration of potato tuber tissue was observed with purified A. irakense PelA but not with intact A. irakense cells. These observations strongly point to a restricted pelA gene expression and/or PelA secretion in A. irakense. Indeed, expression studies with a pelA-gusA fusion indicated a low level of pelA transcription under all growth conditions tested.
Similarly to what has been observed for E. chrysanthemi Pel enzymes, PelA synthesis in A. irakense was found to be stimulated by the presence of pectin. In E. chrysanthemi, degradation products of pectin, such as 2-keto-3-deoxygluconate, are the actual intracellular pel gene inducers rather than pectin itself (17). For most E. chrysanthemi pel genes, inducibility by pectin degradation intermediates is mediated mainly by the KdgR product (44). In the absence of pectic inducers, KdgR represses expression by binding to a specific operator sequence, termed the KdgR box, upstream of these genes (34). However, no sequence resembling the Erwinia KdgR box could be found in the promoter region of the A. irakense pelA gene.
In addition to the induction by pectin, pelA transcription was found to be dependent on other physiological conditions such as the growth phase and the presence of several carbon sources. The observed growth phase-dependent pelA inducibility might involve a positively acting quorum-sensing system similarly to what has been demonstrated for the expression of pectinase genes in E. chrysanthemi and E. carotovora (35, 40).
Ultrastructural studies have provided evidence that plant cell wall-degrading enzymes (pectinolytic and cellulolytic) are involved in the various steps of the root hair infection process in the Rhizobium-legume symbiosis (27, 30). A. irakense strains were found associated with rice roots and were also isolated from the root interior (20). Interestingly, we recently observed cellulolytic activity (i.e., β-glucosidase and cellobiohydrolase activity) in A. irakense in addition to its pectinolytic activity (unpublished data). As proposed for the Rhizobium-legume interaction, one might anticipate that degradation of the plant cell wall by bacterial pectinases and cellulases at a root infection site would be required for penetration of A. irakense into the host plant. However, if cell wall-degrading enzymes are involved, their production and activity must be tightly controlled to account for a slow, localized penetration without destruction of the plant tissues surrounding the infection site. Alternatively, pectin degradation might have a strictly catabolic function related to bacterial nutrition. A. irakense KBC1 was shown to grow and to fix nitrogen on PGA or pectin as the sole carbon source, indicating the presence of catabolic enzymes for further breakdown of the oligogalacturonides that result from the Pel activity. The ability of A. irakense to catabolize pectic polysaccharides and to use them as carbon and energy sources for growth might therefore be advantageous for its survival in the rhizosphere and in the soil.
ACKNOWLEDGMENTS
M.A.B. is a recipient of a predoctoral fellowship of the Onderzoeksraad K.U. Leuven. P.P. and A.V.B. are recipients of postdoctoral fellowships of the Fund for Scientific Research of Flanders (Belgium). This work was financially supported by GOA93/98 of the Bijzonder Onderzoeksfonds K.U. Leuven.
We gratefully acknowledge Nicole Hugouvieux-Cotte-Pattat for providing E. chrysanthemi PelD and René De Mot for critical reading of the manuscript.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST, a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldani V L D, Alvarez M A, Baldani J L, Döbereiner J. Establishment of inoculated Azospirillum spp. in the rhizosphere and in roots of field grown wheat and sorghum. Plant Soil. 1986;90:35–46. [Google Scholar]
- 3.Barras F, van Gijsegem F, Chatterjee A K. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu Rev Phytopathol. 1994;32:201–234. [Google Scholar]
- 4.Bauer D W, Collmer A. Molecular cloning, characterization and mutagenesis of a pel gene from Pseudomonas syringae pv. lachrymans encoding a member of the Erwinia chrysanthemi PelADE family of pectate lyases. Mol Plant-Microbe Interact. 1997;10:369–379. doi: 10.1094/MPMI.1997.10.3.369. [DOI] [PubMed] [Google Scholar]
- 5.Beaulieu C, Minsavage G V, Canteros B L, Stall R E. Biochemical and genetic analysis of a pectate lyase gene from Xanthomonas campestris pv. vesicatoria. Mol Plant-Microbe Interact. 1991;4:446–451. doi: 10.1094/mpmi-4-628. [DOI] [PubMed] [Google Scholar]
- 6.Brooks A D, He S Y, Gold S, Keen N T, Collmer A, Hutcheson S W. Molecular cloning of the structural gene for exopolygalacturonate lyase from Erwinia chrysanthemi EC16 and characterization of the enzyme product. J Bacteriol. 1990;172:6950–6958. doi: 10.1128/jb.172.12.6950-6958.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croes C, Van Bastelaere E, Declercq E, Eyers M, Vanderleyden J, Michiels K. Identification and mapping of loci involved in motility, adsorption to wheat roots, colony morphology and growth in minimal medium on the Azospirillum brasilense Sp7 90 MDa plasmid. Plasmid. 1991;26:83–93. doi: 10.1016/0147-619x(91)90048-2. [DOI] [PubMed] [Google Scholar]
- 8.De Vos G F, Walker G C, Signer E R. Genetic manipulations in Rhizobium meliloti utilizing two new transposon Tn5 derivatives. Mol Gen Genet. 1986;204:485–491. doi: 10.1007/BF00331029. [DOI] [PubMed] [Google Scholar]
- 9.Fraaije B A, Bosveld M, Van den Bulk R W, Rombouts F M. Analysis of conductance responses during depolymerization of pectate by soft rot Erwinia spp. and other pectolytic bacteria isolated from potato tubers. J Appl Microbiol. 1997;83:17–24. doi: 10.1046/j.1365-2672.1997.d01-396.x. [DOI] [PubMed] [Google Scholar]
- 10.Friedman A M, Long S R, Brown S E, Buikema S E, Ausubel F M. Construction of a broad host range cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Candelas L, Kolattukudy P E. Isolation and analysis of a novel inducible pectate lyase gene from the phytopathogenic fungus Fusarium solani f. sp. pisi (Nectria haematococca, mating population VI) J Bacteriol. 1992;174:6343–6349. doi: 10.1128/jb.174.20.6343-6349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo W L, Gonzalez-Candelas L, Kolattukudy P E. Cloning of a new pectate lyase gene pelC from Fusarium solani f. sp. pisi (Nectria haematococca, mating group VI) and characterization of the gene product expressed in Pichia pastoris. Arch Biochem Biophys. 1995;232:352–360. doi: 10.1006/abbi.1995.9954. [DOI] [PubMed] [Google Scholar]
- 13.Guo W L, Gonzalez-Candelas L, Kolattukudy P E. Cloning of a novel constitutively expressed pectate lyase gene pelB from Fusarium solani f. sp. pisi and characterization of the gene product expressed in Pichia pastoris. J Bacteriol. 1995;177:7070–7077. doi: 10.1128/jb.177.24.7070-7077.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haurat J, Faure D, Bally R, Normand P. Molecular relationship of an atypical Azospirillum strain 4T to other Azospirillum species. Res Microbiol. 1994;145:633–640. doi: 10.1016/0923-2508(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 15.Henrissat B, Heffron S E, Yoder M D, Lietzke S E, Jurnak F. Functional implications of structure-based sequence alignment of proteins in the extracellular pectate lyase superfamily. Plant Physiol. 1995;107:963–976. doi: 10.1104/pp.107.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinton J C D, Sidebotham J M, Gill D R, Salmond G P C. Extracellular and periplasmic isoenzymes of pectate lyase from Erwinia carotovora subspecies carotovora belong to different gene families. Mol Microbiol. 1989;3:1785–1795. doi: 10.1111/j.1365-2958.1989.tb00164.x. [DOI] [PubMed] [Google Scholar]
- 17.Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. Regulation of pectinolysis in Erwinia chrysanthemi. Annu Rev Microbiol. 1996;50:213–257. doi: 10.1146/annurev.micro.50.1.213. [DOI] [PubMed] [Google Scholar]
- 18.Jefferson R A. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- 19.Kado C, Liu S. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khammas K M, Ageron E, Grimont P A D, Kaiser P. Azospirillum irakense sp. nov., a nitrogen-fixing bacterium associated with rice roots and rhizosphere soil. Res Microbiol. 1989;140:679–693. doi: 10.1016/0923-2508(89)90199-x. [DOI] [PubMed] [Google Scholar]
- 21.Kotoujansky A. Molecular genetics of pathogenesis by soft-rot Erwinias. Annu Rev Phytopathol. 1987;25:405–430. [Google Scholar]
- 22.Lei S, Lin H, Heffernan L, Wilcox G. Evidence that polygalacturonase is a virulence determinant in Erwinia carotovora. J Bacteriol. 1985;164:831–835. doi: 10.1128/jb.164.2.831-835.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao C, Hung H, Chatterjee A K. An extracellular pectate lyase is the pathogenicity factor of the soft-rotting bacterium Pseudomonas viridiflava. Mol Plant-Microbe Interact. 1988;1:199–206. [Google Scholar]
- 24.Liao C, Gaffney T D, Bradley S P, Wong L C. Cloning of a pectate lyase gene from Xanthomonas campestris pv. malvacearum and comparison of its sequence relationship with pel genes of soft-rot Erwinia and Pseudomonas. Mol Plant-Microbe Interact. 1996;9:14–21. doi: 10.1094/mpmi-9-0014. [DOI] [PubMed] [Google Scholar]
- 25.Lietzke S E, Yoder M D, Keen N T, Jurnak F. The three dimensional structure of pectate lyase E, a plant virulence factor from Erwinia chrysanthemi. Plant Physiol. 1994;106:849–862. doi: 10.1104/pp.106.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Chatterjee A, Chatterjee A K. Nucleotide sequence and expression of a novel pectate lyase gene (pel-3) and a closely linked endo-polygalacturonase gene (peh-1) of Erwinia carotovora subsp. carotovora 71. Appl Environ Microbiol. 1994;60:2545–2552. doi: 10.1128/aem.60.7.2545-2552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ljunggren H, Fahraeus G. The role of polygalacturonase in root-hair invasion by nodule bacteria. J Gen Microbiol. 1961;26:521–528. doi: 10.1099/00221287-26-3-521. [DOI] [PubMed] [Google Scholar]
- 28.Lojkowska E, Masclaux C, Boccara M, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Characterization of the pelL gene encoding a novel pectate lyase of Erwinia chrysanthemi 3937. Mol Microbiol. 1995;16:1183–1195. doi: 10.1111/j.1365-2958.1995.tb02341.x. [DOI] [PubMed] [Google Scholar]
- 29.Manulis S, Kobayashi D Y, Keen N T. Molecular cloning and sequencing of a pectate lyase gene from Yersinia pseudotuberculosis. J Bacteriol. 1988;170:1825–1830. doi: 10.1128/jb.170.4.1825-1830.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mateos P F, Jimenez-Zurdo J L, Chen J, Squartini A S, Haack S K, Martinez-Molina E, Hubbell D H, Dazzo F. Cell-associated pectinolytic and cellulolytic enzymes in Rhizobium leguminosarum biovar trifolii. Appl Environ Microbiol. 1992;58:1816–1822. doi: 10.1128/aem.58.6.1816-1822.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milcamps A, Van Dommelen A, Stigter J, Vanderleyden J, de Bruijn F J. The Azospirillum brasilense rpoN gene is involved in nitrogen fixation, nitrate assimilation, ammonium uptake and flagellar biosynthesis. Can J Microbiol. 1996;42:467–478. doi: 10.1139/m96-064. [DOI] [PubMed] [Google Scholar]
- 32.Nasser W, Chalet F, Robert-Baudouy J. Purification and characterization of extracellular pectate lyase from Bacillus subtilis. Biochimie. 1990;72:689–695. doi: 10.1016/0300-9084(90)90053-j. [DOI] [PubMed] [Google Scholar]
- 33.Nasser W, Awadé A C, Reverchon S, Robert-Baudouy J. Pectate lyase from Bacillus subtilis: molecular characterization of the gene, and properties of the cloned enzyme. FEBS Lett. 1993;335:319–326. doi: 10.1016/0014-5793(93)80410-v. [DOI] [PubMed] [Google Scholar]
- 34.Nasser W, Reverchon S, Condemine G, Robert-Baudouy J. Specific interactions of Erwinia chrysanthemi KdgR repressor with different operators of genes involved in pectinolysis. J Mol Biol. 1994;236:427–440. doi: 10.1006/jmbi.1994.1155. [DOI] [PubMed] [Google Scholar]
- 35.Nasser W, Bouillant M L, Salmond G, Reverchon S. Characterization of the Erwinia chrysanthemi expI-expR locus directing the synthesis of two N-acyl-homoserine lactone signal molecules. Mol Microbiol. 1998;29:1391–1405. doi: 10.1046/j.1365-2958.1998.01022.x. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen H, Engelbrecht J, Brunak S, Von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 37.Nikaidou N, Kamio Y, Izaki K. Molecular cloning and nucleotide sequence of the pectate lyase gene from Pseudomonas marginalis N6301. Biosci Biotechnol Biochem. 1993;57:957–960. doi: 10.1271/bbb.57.957. [DOI] [PubMed] [Google Scholar]
- 38.Okon Y, Vanderleyden J. Root-associated Azospirillum species can stimulate plants. ASM News. 1997;63:366–370. [Google Scholar]
- 39.Pickersgill R, Jenkins J, Harris G, Nasser W, Robert-Baudouy J. The structure of Bacillus subtilis pectate lyase in complex with calcium. Nat Struct Biol. 1994;1:717–723. doi: 10.1038/nsb1094-717. [DOI] [PubMed] [Google Scholar]
- 40.Pirhonen M, Flego D, Heikinheimo R, Palva E T. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in plant pathogen Erwinia carotovora. EMBO J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pissavin C, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Regulation of pelZ, a gene of the pelB-pelC cluster encoding a new pectate lyase of Erwinia chrysanthemi 3937. J Bacteriol. 1996;178:7187–7196. doi: 10.1128/jb.178.24.7187-7196.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plazinski J, Rolfe B G. Analysis of pectinolytic activity of Rhizobium and Azospirillum strains isolated from Trifolium repens. J Plant Physiol. 1985;120:181–187. [Google Scholar]
- 43.Reinhold B, Hurek T, Fendrik I, Pot B, Gillis M, Kersters K, Thielmans S, De Ley J. Azospirillum halopraeferans sp. nov., a nitrogen-fixing organism associated with roots of Kallar grass (Leptochloa fusca (L.) Kunth.) Int J Syst Bacteriol. 1987;37:43–51. [Google Scholar]
- 44.Reverchon S, Nasser W, Robert-Baudouy J. Characterization of kdgR, a gene of Erwinia chrysanthemi that regulates pectin degradation. Mol Microbiol. 1991;5:2203–2216. doi: 10.1111/j.1365-2958.1991.tb02150.x. [DOI] [PubMed] [Google Scholar]
- 45.Rush S L, Kendall D A. Protein transport via amino-terminal targeting sequences: common themes in diverse systems. Mol Membr Biol. 1995;12:295–307. doi: 10.3109/09687689509072431. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 47.Shevchik V E, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Pectate lyase PelI of Erwinia chrysanthemi 3937 belongs to a new family. J Bacteriol. 1997;179:7321–7330. doi: 10.1128/jb.179.23.7321-7330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shevchik V E, Hugouvieux-Cotte-Pattat N. Identification of a bacterial pectin acetyl esterase in Erwinia chrysanthemi 3937. Mol Microbiol. 1997;24:1285–1301. doi: 10.1046/j.1365-2958.1997.4331800.x. [DOI] [PubMed] [Google Scholar]
- 49.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 50.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tardy F, Nasser W, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: enzyme characteristics and potential inhibitors. J Bacteriol. 1997;179:2503–2511. doi: 10.1128/jb.179.8.2503-2511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tarrand J J, Krieg N R, Döbereiner J. A taxonomic study of the Spirillum lipoferum group, with description of a new genus, Azospirillum gen. nov., and two species Azospirillum lipoferum (Beijerinck) sp. nov. and Azospirillum brasilense sp. nov. Can J Microbiol. 1978;24:967–980. doi: 10.1139/m78-160. [DOI] [PubMed] [Google Scholar]
- 53.Trollinger D, Berry S, Belser W, Keen N T. Cloning and characterization of a pectate lyase gene from Erwinia carotovora EC153. Mol Plant-Microbe Interact. 1989;2:17–25. doi: 10.1094/mpmi-2-017. [DOI] [PubMed] [Google Scholar]
- 54.Vanstockem M, Michiels K, Vanderleyden J, Van Gool A. Transposon mutagenesis of Azospirillum brasilense and Azospirillum lipoferum, physical analysis of Tn5 and Tn5-mob insertion mutants. Appl Environ Microbiol. 1987;53:410–415. doi: 10.1128/aem.53.2.410-415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson K J, Sessitsch A, Corbo J C, Giller K E, Akkermans A D L, Jefferson R A. β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other Gram-negative bacteria. Microbiology. 1995;141:1691–1705. doi: 10.1099/13500872-141-7-1691. [DOI] [PubMed] [Google Scholar]
- 56.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 57.Yoder M D, Keen N T, Jurnak F. New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science. 1993;260:1503–1507. doi: 10.1126/science.8502994. [DOI] [PubMed] [Google Scholar]
- 58.Yoder M D, Lietzke S E, Jurnak F. Unusual structural features in the parallel β-helix in pectate lyases. Structure. 1993;1:241–251. doi: 10.1016/0969-2126(93)90013-7. [DOI] [PubMed] [Google Scholar]