Abstract
The gram-positive eubacterium Bacillus subtilis is the organism with the largest number of paralogous type I signal peptidases (SPases) known. These are specified both by chromosomal and plasmid-borne genes. The chromosomally encoded SPases SipS and SipT have a major function in precursor processing, and cells depleted of SipS and SipT stop growing and die. In this study, we show that the SPase SipP, specified by the B. subtilis plasmid pTA1015, can functionally replace SipS and SipT, unlike the three chromosomally encoded SPases with a minor function in protein secretion (i.e., SipU, SipV, and SipW). Unexpectedly, SipP is not specifically required for the processing and secretion of Orf1p, which is specified by a gene that is cotranscribed with sipP. These two genes form a conserved structural module of rolling-circle plasmids from B. subtilis. As previously shown for the chromosomal sipS and sipT genes, the transcription of plasmid-borne copies of sipP is temporally controlled, reaching maximal levels during the post-exponential growth phase when the cells secrete proteins at high levels. However, increased transcription of sipP starts at the end of exponential growth, about 2 h earlier than that of sipS and sipT. These data suggest that SipP fulfills a general role in the secretory precursor processing of pTA1015-containing cells.
Type I signal peptidases (SPases) remove amino-terminal signal peptides from secretory preproteins during or shortly after their translocation across the cytoplasmic membrane, in order to release these proteins from the trans side of this membrane (for reviews, see references 3 and 17). Homologous type I SPases (orthologues) have been identified in archaea, eubacteria, and eukaryotes (3). Notably, various organisms contain multiple (paralogous) type I SPases (3, 21). In eukaryotes, several of these paralogues are localized to membranes of different subcellular compartments, such as the inner membrane of mitochondria, the thylakoid membrane of chloroplasts, and the endoplasmic reticular membrane, consistent with their involvement in different pathways for protein transport. In contrast, the roles of paralogous type I SPases in archaea and eubacteria are less clear, as all of these enzymes are localized in the cytoplasmic membrane.
The greatest number of paralogous type I SPases have been found in the gram-positive eubacterium Bacillus subtilis, which contains five chromosomally encoded type I SPases (20, 21, 26). Two of these, SipS and SipT, are of major importance for protein secretion, and cells depleted of both enzymes stop growing and die. The other three type I SPases, SipU, SipV, and SipW, are of minor importance for protein secretion and cell viability, and they are unable to compensate for the absence of SipS and SipT (21). Despite the fact that the specificities of all five SPases overlap, it seems that at least SipS and SipT have a preference for different precursor proteins (1, 20). Consistent with their important role in secretion, transcription of the sipS and sipT genes is temporally controlled via the DegS-DegU two-component regulatory system, in concert with the transcription of the genes for most secretory proteins (1, 20, 21). Thus, B. subtilis can modulate its capacity for precursor processing, which appears to be a specific mechanism of the cell to prevent SPase limitation under conditions of high-level secretion.
In addition to chromosomally encoded type I SPases, certain strains of B. subtilis (natto) contain a plasmid-encoded type I SPase, designated SipP (10). The two known sipP genes are part of homologous structural modules, which are present on at least two cryptic plasmids, pTA1015 and pTA1040, that replicate via the rolling-circle mechanism. Each sipP gene was found to be preceded by an open reading frame (ORF1), encoding a putative secreted protein (Orf1p) of unknown function (10). In the present study, aimed at a functional analysis of the sipP gene of pTA1015, we demonstrate that SipP is not specifically required for the secretion of Orf1p, as suggested by the presence of genes for a secreted protein and an SPase in one structural module. Instead, SipP may have a more general role in secretion, as it can functionally replace SipS and SipT, unlike the other type I SPases of B. subtilis. Interestingly, the transcription of plasmid-borne copies of sipP is temporally controlled, like that of the chromosomal sipS and sipT genes, but it is most strongly increased in the transition phase between exponential and post-exponential growth, about 2 h before the transcription of sipS and sipT begins to increase.
MATERIALS AND METHODS
Plasmids, bacterial strains, and media.
Table 1 lists the plasmids and bacterial strains used. TY (tryptone-yeast extract) medium contained Bacto Tryptone (1%), Bacto Yeast Extract (0.5%), and NaCl (1%). Minimal medium for B. subtilis was prepared as previously described (21). S7 media 1 and 3, used for labeling of B. subtilis proteins with [35S]methionine (Amersham), were prepared as described by van Dijl et al. (24, 25). When required, media for Escherichia coli were supplemented with ampicillin (50 μg/ml) or erythromycin (100 μg/ml); media for B. subtilis were supplemented with chloramphenicol (5 μg/ml), erythromycin (1 μg/ml), or kanamycin (10 μg/ml).
TABLE 1.
Plasmids and bacterial strains used
| Plasmid or strain | Relevant properties | Reference |
|---|---|---|
| Plasmids | ||
| pLGW200 | Integration vector for B. subtilis with a promoterless lacZ gene fused to the ribosome-binding site of the spoVG gene; 6.8 kb; Cmr | 28 |
| pLGO201 | pLGW200 derivative with a transcriptional ORF1-lacZ fusion; 7.8 kb | This paper |
| pLGP201 | pLGW200 derivative with a transcriptional sipP-lacZ fusion; 7.9 kb | This paper |
| pTA1015 | Endogenous plasmid from B. subtilis IAM1028; 5.8 kb | 23 |
| pTAB11A | pTA1015 derivative; contains Kmr marker in the ORF2C gene; 7.0 kb | 11 |
| pTAB-OL | pTAB11A with integrated pLGO201 (ORF1-lacZ); Cmr Kmr; 14.6 kb | This paper |
| pTAB-PL | pTAB11A with integrated pLGP201 (sipP-lacZ); Cmr Kmr; 14.7 kb | This paper |
| pGB18 | Vector for the selection of signal sequences; contains SPO2 promoter for transcription of β-lactamase gene fusions; replicates in B. subtilis and E. coli; Emr; 6.1 kb | 15 |
| pOB1 | pGB18 derivative; encodes Orf1p–β-lactamase fusion protein | This paper |
| pKVS263 | Derivative of pKVS242 (6); contains the glgB gene of Bacillus stearothermophilus under control of the sucrose-inducible sacB promoter; lacks the SPO2 promoter; Cmr Emr; 7.9 kb | 7 |
| pJDO | pKVS263 derivative; lacks the glgB gene; contains the ORF1 gene of pTA1015 under control of the sucrose-inducible sacB promoter; 6.3 kb | This paper |
| pJDOH | Like pJDO; encodes a carboxyl-terminally six-histidine-tagged Orf1p | This paper |
| Strains | ||
| E. coli MC1061 | F−araD139 Δ(ara-leu)7696 Δ(lac)X74 galU galK hsdR2 mcrA mcrB1 rspL | 29 |
| B. subtilis | ||
| 8G5 | Derivative of B. subtilis 168, lacking the sipP genes; trpC2 tyr his nic ura rib met ade | 2 |
| 8G5 sipS | Like 8G5; sipS | 1 |
| 8G5 sipT-Cm | Like 8G5; sipT Cmr | 20 |
| 8G5 ΔST(pTAB11A) | Like 8G5; sipS sipT Cmr; contains pTAB11A | This paper |
| 8G5 degU32(Hy) | Like 8G5; degU32(Hy); Kmr | 1 |
| 8G5::pGDE22 | 8G5 carrying pGDE22 (sipS-lacZ) in the chromosome; Cmr | 1 |
| 8G5::pLGV201 | 8G5 carrying pLGV201 (sipV-lacZ) in the chromosome; Cmr | 20 |
DNA and RNA techniques.
Procedures for DNA purification, restriction, ligation, agarose gel electrophoresis, and transformation of E. coli were carried out as described by Sambrook et al. (18). Enzymes were from Boehringer Mannheim. B. subtilis was transformed by growth in minimal medium to an optical density at 600 nm (OD600) of ±1, subsequent addition of plasmid or chromosomal DNA to the culture, and continued incubation for at least 4 hours. PCR was carried out with Vent DNA polymerase (New England Biolabs) as described by van Dijl et al. (27).
To construct an Orf1p–β-lactamase fusion protein, the 5′ sequences of ORF1, specifying the 50 amino-terminal residues of Orf1p, were amplified by PCR with primers orf1-1 (5′-TAT GGA TCC TAT TGA ATT TTG CTA GGA GGG-3′) and orf1-2 (5′-GAT CGT CGA CTC ATA ACT TTT TGT TGA AGA TTG GG-3′), using plasmid pTA1015 as the template. The amplified fragment was cleaved with BamHI and SalI and ligated to the corresponding sites of plasmid pGB18 (15), resulting in plasmid pOB1. The first 57 residues of the Orf1p–β-lactamase fusion protein specified by pOB1 are MVTTIGKSKMWVGIIVVLSLLLVSFSPA/VK/A/DTKDKYYSTTSTQSSTKSY-GDPLEST (putative SPase cleavage sites [/] and the fusion point between Orf1p and β-lactamase [-] are indicated). Putative SPase cleavage sites were predicted with the SignalP algorithm for signal peptides from gram-positive bacteria (14).
To construct plasmids for overproduction of Orf1p or a carboxyl-terminally six-histidine-tagged Orf1p (Orf1p-His), the ORF1 gene was amplified by PCR with primers lbp-1 (5′-TAT TGT CGA CAA GTA TTG AAT GC-3′) and lbp-3 (5′-TCT TTC TAG AGC TCT ATC TAC AAT CGG GAC TCC-3′; no six-histidine tag) or lbp-1 and lbp-4 (5′-TCT TTC TAG AGC TCT AAT GGT GAT GGT GAT GGT GTC TAC AAT CGG GAC TCC-3′; specifying a six-histidine tag). The amplified fragments were cleaved with SalI and XbaI and ligated to the SalI and NheI sites of plasmid pKVS263. The resulting plasmids, in which the tagged ORF1 gene is under control of the sucrose-inducible sacB promoter, were designated pJDO (specifying Orf1p) and pJDOH (specifying Orf1p-His).
To construct pTAB-OL, a DNA fragment comprising the 3′ end of the rep gene and the 5′ end of ORF1 was amplified by PCR with primers bea-3 (5′-GAT CGA ATT CAT AAA GAA CTA AAC CTC GGT G-3′) and bea-4 (5′-GAT CGG ATC CTA TCT ACA ATC GGG ACT CC-3′). Next, the amplified fragment was digested with EcoRI and BamHI and ligated into the corresponding sites of the multiple cloning site upstream of the spoVG-lacZ gene fusion on pLGW200, resulting in pLGO201 (ORF1-lacZ). Finally, the ORF1-lacZ fusion was introduced in pTAB11A by a Campbell-type integration of pLGO201, resulting in pTAB-OL, in which the transcription of lacZ is directed by the promoter(s) of ORF1. Similarly, to construct pTAB-PL (sipP-lacZ), a fragment comprising the 3′ end of ORF1 and the 5′ end of the sipP gene was amplified by PCR with primers bea-3 and bea-6 (5′-GAT CGG ATC CTA TAT CAC AAT AGC CTT GCC CC-3′). Next, the amplified fragment was ligated into pLGW200, resulting in pLGP201 (sipP-lacZ). Finally, the sipP-lacZ gene fusions was introduced in pTAB11A by a Campbell-type integration of pLGP201, resulting in pTAB-PL, in which the transcription of lacZ is directed by the promoter(s) of ORF1 and/or sipP.
RNA for reverse transcription (RT)-PCR was isolated with a RNeasy total RNA kit from Qiagen, dissolved in 100 mM sodium acetate–5 mM Mg2SO4 (pH 5.0) containing DNase I (RNase free; Boehringer Mannheim), and incubated for 30 min at 25°C. Subsequently, the RNA was extracted with phenol-chloroform, precipitated with ethanol, and dissolved in RNAse-free water. First-strand cDNA synthesis with reverse transcriptase and a random primer set was carried out with a first-strand cDNA synthesis kit from Amersham International, and cDNAs were detected by PCR with specific primers.
Plate assay for the secretion of β-lactamase.
The plate assay for the secretion of β-lactamase by B. subtilis cells was carried out as described by van Dijl et al. (26).
β-Galactosidase activity assay.
Overnight cultures were diluted 100-fold in fresh medium, and samples were taken at hourly intervals for OD600 readings and β-galactosidase activity determinations. The assay and the calculation of β-galactosidase units (expressed as units per OD600) were carried out as described by Miller et al. (12).
Protein labeling, immunoprecipitation, SDS-PAGE, and fluorography.
Pulse-chase labeling of B. subtilis, immunoprecipitation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and fluorography were performed as described previously (24, 25).
Western blot analysis.
Western blotting was performed as described by Kyhse-Andersen (9). Spheroplasts of E. coli were prepared as described by van Dijl et al. (24). Samples for SDS-PAGE were prepared as described by van Dijl et al. (25, 26). After separation by SDS-PAGE, proteins were transferred to Immobilon polyvinylidene difluoride membranes (Millipore Corporation). β-Lactamase was visualized with specific antibodies and horseradish peroxidase–anti-rabbit immunoglobulin G conjugates (Amersham International). Orf1p-His was visualized with six-histidine-specific monoclonal antibodies (Amersham International) and horseradish peroxidase–anti-mouse immunoglobulin G conjugates (Amersham International). SipS, SipT, and SipP were visualized as described for β-lactamase, using antibodies directed against SipS, which show a weak cross-reactivity with SipT and SipP.
RESULTS
sipP is cotranscribed with ORF1.
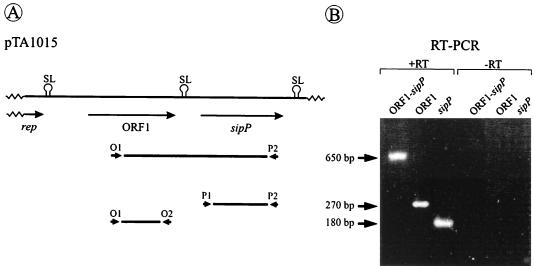
In the sipP module of pTA1015, ORF1 and sipP are separated by a region of 102 bp, containing a potential stem-loop structure (Fig. 1A; see reference 10). As a first approach to investigate a possible functional relationship between these two genes, RT-PCR was performed on total RNA isolated from a B. subtilis strain containing pTA1015. As shown in Fig. 1B, RT-PCR fragments were amplified with ORF1- and sipP-specific primers, showing that both genes are transcribed. Moreover, a 650-bp fragment was amplified with one ORF1- and one sipP-specific primer (Fig. 1B), showing that ORF1 and sipP are cotranscribed. In contrast, no RT-PCR fragment containing sequences of both ORF1 and the upstream-located rep gene for plasmid replication could be amplified (data not shown). Together, these findings show that ORF1 and sipP of pTA1015 form a functional unit, at least with respect to their transcription.
FIG. 1.
Cotranscription of ORF1 and sipP. (A) Schematic presentation of the ORF1-sipP module of pTA1015. The positions of primers used for RT-PCR are indicated. Primers O1 (5′-GAT GGC GCT ACT CTG GG-3′) and O2 (5′-ACT ATC TAC AAT CGG GAC TCC-3′) were used to detect ORF1-specific transcripts; primers P1 (5′-TAG AAA TGA AGA ATG ACC-3′) and P2 (5′-TCG CAT ATT ACT AAA TGG-3′) were used to detect sipP-specific transcripts; primers O1 and P2 were used to detect transcripts of ORF1 and sipP. The positions of potential stem-loop structures (SL) are indicated (10, 11). (B) DNA fragments amplified by RT-PCR with the primer sets O1-O2 (ORF1), P1-P2 (sipP), and O1-P2 (ORF1-sipP) were separated on a 2% agarose gel containing ethidium bromide (1 μg/ml) and visualized by UV illumination (lanes labeled +RT). As a negative control, reactions were also performed in the absence of reverse transcriptase (lanes labeled −RT). The positions of amplified DNA fragments corresponding to ORF1 (180 bp), sipP (270 bp), and ORF1-sipP (650 bp) are indicated.
SipP-independent processing and secretion of the ORF1-encoded protein.
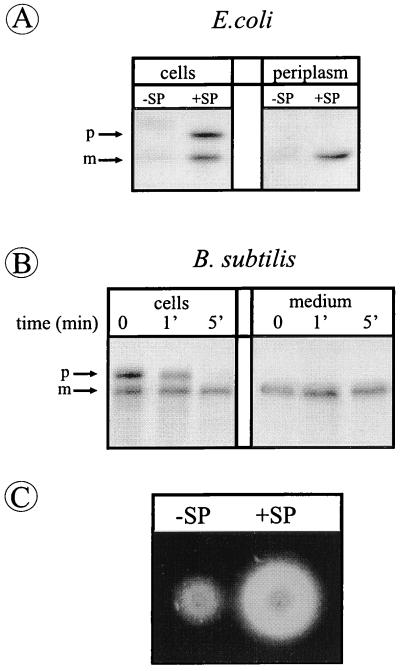
To investigate whether Orf1p contains a cleavable signal peptide as previously predicted (10), the sequence encoding the first 50 amino-terminal residues of this protein was fused to the truncated TEM–β-lactamase gene of pGB18, which lacks its own signal sequence (15). In E. coli, the resulting plasmid pOB1 gave rise to resistance to high levels of ampicillin (>50 μg/ml), showing that the Orf1p–β-lactamase fusion protein was translocated across the cytoplasmic membrane (data not shown). As shown by Western blotting, E. coli cells containing pOB1 (Fig. 2A, +SP) accumulated two forms of β-lactamase with the predicted molecular masses of precursor and mature forms of the Orf1p–β-lactamase fusion protein, respectively. Only the mature form was released into the periplasm (Fig. 2A, +SP), showing that the Orf1p–β-lactamase fusion protein is synthesized with a cleavable signal peptide. Interestingly, this signal peptide was removed in a SipP-independent manner, most likely by the type I SPase (i.e., leader peptidase) of E. coli. Similarly, as shown in pulse-chase labeling experiments, the precursor of the Orf1p–β-lactamase fusion protein was processed to its mature form in B. subtilis 8G5, a strain lacking SipP (Fig. 2B). Subsequently, the mature form was secreted into the growth medium (Fig. 2B and C). Taken together, these results show that Orf1p is synthesized with a signal peptide that is functional in E. coli and B. subtilis and that can be removed from the Orf1p–β-lactamase fusion protein in a SipP-independent manner.
FIG. 2.
SipP-independent processing of the Orf1p-β-lactamase fusion protein. (A) E. coli cells containing pOB1, specifying the Orf1p–β-lactamase fusion protein (+SP), and E. coli cells containing pGB18, which contains a β-lactamase gene without a ribosome-binding site and signal sequence (−SP; negative control) were grown overnight in TY medium. Samples of intact cells (cells) or spheroplast supernatants (periplasm) were used for SDS-PAGE and Western blotting. The positions of precursor (p) and mature (m) forms of the Orf1p–β-lactamase fusion protein are indicated. (B) Processing of Orf1p–β-lactamase in B. subtilis 8G5 and secretion of the corresponding mature form into the medium was analyzed by pulse-chase labeling at 37°C and subsequent immunoprecipitation, SDS-PAGE, and fluorography. Cells were labeled with [35S]methionine for 1 min prior to chase with excess nonradioactive methionine. Samples were withdrawn after the chase at the times indicated, and cells and medium were separated by centrifugation. The positions of precursor (p) and mature (m) forms of the Orf1p–β-lactamase fusion protein are indicated. (C) Secretion of β-lactamase into the growth medium surrounding colonies of B. subtilis 8G5 transformed with pOB1 (+SP) was demonstrated by transfer of cells to fresh plates, overnight growth at 37°C, and a plate assay for β-lactamase activity as described by van Dijl et al. (26). In this assay, β-lactamase activity results in halo formation. Compared to cells containing pOB1, cells containing pGB18 (−SP) formed small halos, due to the secretion of the chromosomally encoded penicillinase PenP (22, 26).
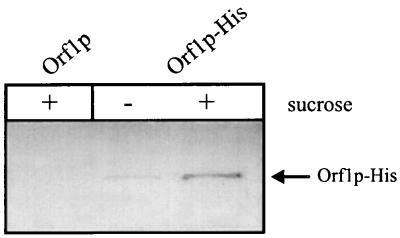
To investigate whether the complete Orf1p is secreted into the growth medium in a SipP-dependent manner, Western blotting experiments were performed with Orf1p-His, which was expressed by using a sucrose-inducible promoter. As shown in Fig. 3, a B. subtilis strain lacking SipP secreted Orf1p-His into the medium upon induction with sucrose. Furthermore, sucrose-induced cells did not accumulate the precursor or mature forms of this protein (data not shown). These findings show that the processing and secretion of Orf1p do not specifically require the presence of SipP, suggesting that the functional relationship between Orf1p and SipP is limited to the cotranscription of the corresponding genes.
FIG. 3.
SipP-independent secretion of Orf1p in B. subtilis. The secretion of Orf1p into the medium of B. subtilis 8G5 was demonstrated by using the sucrose-inducible protein Orf1p-His. Cells of B. subtilis 8G5 containing either pJDO (Orf1p; negative control) or pJDOH (Orf1p-His) were transformed with chromosomal DNA of B. subtilis 8G5 degU32(Hy) for high sacB promoter activity. Upon overnight growth at 37°C in the presence (+) or absence (−) of 2% sucrose, samples were withdrawn. Cells and medium were separated by centrifugation, and the medium fractions were, upon fourfold concentration by trichloroacetic acid precipitation, used for SDS-PAGE and Western blotting. The position of Orf1p-His is indicated. The synthesis of Orf1p-His in the absence of sucrose is due to background activity of the noninduced sacB promoter.
Increased transcription of sipP in the transition phase between exponential and postexponential growth.
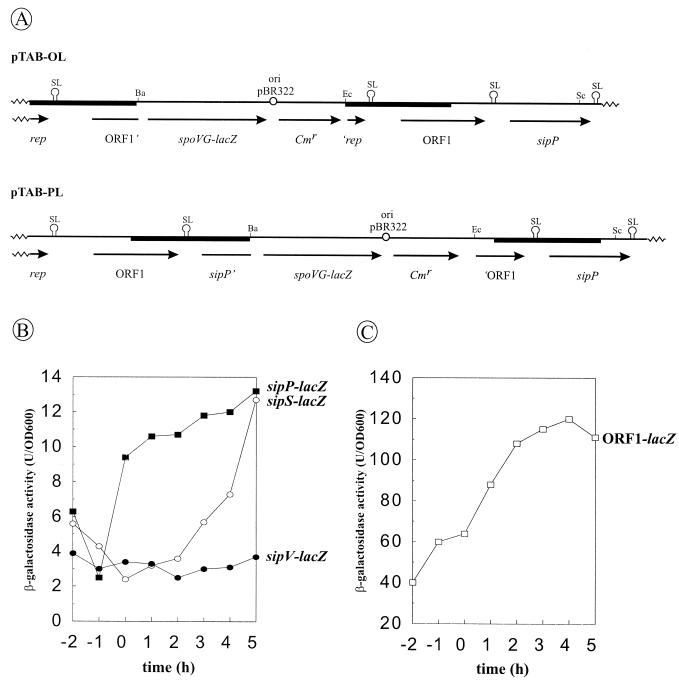
Previous results showed that the transcription of the sipS and sipT genes is temporally controlled, in concert with most genes for secretory proteins, whereas the sipU, sipV, and sipW genes are constitutively transcribed (1, 20, 21). To analyze the transcription of plasmid-borne copies of ORF1 and sipP, transcriptional ORF1-lacZ and sipP-lacZ fusions were introduced in plasmid pTAB11A, a derivative of pTA1015 containing a selectable kanamycin resistance (Kmr) marker. The resulting plasmids were named pTAB-OL (ORF1-lacZ) and pTAB-PL (sipP-lacZ), respectively (the ORF1-sipP regions of both plasmids are schematically shown in Fig. 4A). Next, B. subtilis 8G5 cells transformed with pTAB-OL or pTAB-PL were grown in TY medium, and samples withdrawn at hourly intervals were assayed for β-galactosidase activity. B. subtilis 8G5::pGDE22 (sipS-lacZ; temporally controlled expression) and 8G5::pLGV201 (sipV-lacZ; constitutive expression) were used as control strains to compare the expression levels of the plasmid-borne sipP gene with those of the chromosomal sipS and sipV genes. The results show that the transcription of sipP is temporally controlled, being low in the exponential growth phase and strongly increased in the transition phase (time zero), between exponential and post-exponential growth; in contrast, sipS transcription starts to increase about 2 h after the transition phase, reaching levels comparable to those of sipP transcription about 3 h later (Fig. 4B). The time course of ORF1 transcription (Fig. 4C) appears to be comparable to that of sipP, but the ORF1 transcription levels are about 10-fold higher during all growth phases and the increase of ORF1 transcription in the transition phase is less pronounced. The latter findings suggest that the stem-loop structure between ORF1 and sipP has a regulating effect on sipP transcription, terminating at least 90% of the transcriptional activity from the ORF1 promoter. Taken together, these results show that the transcription of plasmid-borne copies of sipP is well coordinated with that of the plasmid-borne ORF1 and the chromosomal genes for secreted degradative enzymes (5, 13), even better than has been found for sipS and sipT of B. subtilis (1, 20, 21).
FIG. 4.
Temporally controlled transcription of ORF1 and sipP. (A) Schematic presentation of the ORF1-sipP regions of pTAB-OL and pTAB-PL, containing transcriptional ORF1-lacZ and sipP-lacZ gene fusions, respectively. The ORF1- and sipP-lacZ gene fusions were constructed with plasmid pLGW200 (28), an integration plasmid for B. subtilis containing a promoterless spoVG-lacZ gene fusion (see Materials and Methods for details). In pTAB-OL, the transcription of lacZ is directed by the promoter(s) of ORF1. In pTAB-PL, the transcription of lacZ is directed by the promoter(s) of ORF1 and/or sipP. DNA fragments amplified by PCR are indicated with black bars. Only restriction sites relevant for the constructions are shown (Ba, BamHI; Ec, EcoRI; Sc, SacI). ‘rep, 5′ truncated rep gene; ORF1’, 3′ truncated ORF1 gene; sipP’, 3′ truncated sipP gene; ‘ORF1, 5′ truncated ORF1 gene; ori pBR322, replication functions of pBR322; SL, potential stem-loop structures. (B) Time course of the transcription of sip-lacZ gene fusions were determined in cells growing in TY medium at 37°C. β-Galactosidase activities (in units per OD600) were determined for B. subtilis 8G5(pTAB-PL) (■) (sipP-lacZ), B. subtilis 8G5::pGDE22 (○) (sipS-lacZ) (1), and B. subtilis 8G5::pLGV201 (●) (20) (sipV-lacZ). Time zero indicates the transition point between the exponential and post-exponential growth phases. (C) The time course of transcription of the ORF1-lacZ gene fusion was determined as for panel B, using B. subtilis 8G5(pTAB-OL) (□).
SipP can functionally replace the major SPases SipS and SipT.
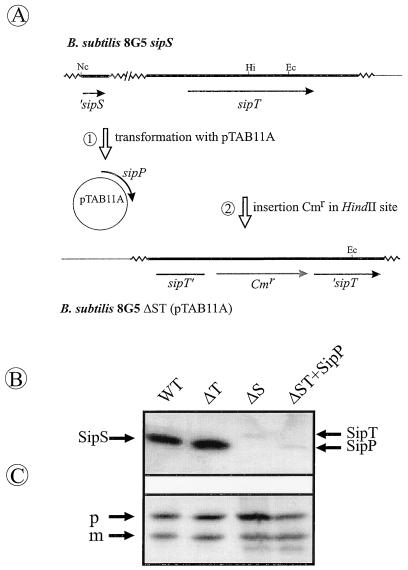
B. subtilis cells depleted of SipS and SipT are not viable, and SipU, SipV, and SipW cannot functionally replace these two major SPases (21), even if the sipU, sipV, or sipW gene is placed on multicopy plasmids (22). To investigate whether SipP can compensate for the absence of SipS and SipT, we made use of plasmid pTAB11A, which carries the sipP gene of pTA1015 (10). First, pTAB11A was used to transform B. subtilis 8G5 sipS, which lacks the sipS gene (1); subsequently, the sipT gene of the resulting strain was disrupted with a chloramphenicol resistance (Cmr) marker (schematically shown in Fig. 5A). As verified by Southern blotting, the resulting strain, 8G5 ΔST(pTAB11A), lacked the sipS and sipT genes (data not shown). Furthermore, as shown by Western blotting, the latter strain lacked the SipS and SipT proteins but produced SipP, showing that SipP can functionally replace SipS and SipT (Fig. 5B; weak SipT- and SipP-specific signals were detected in strains with intact sipT or sipP genes due to cross-reactivity of the anti-SipS antibodies with SipT and SipP). Similarly, the chromosomal sipS and sipT genes could be replaced by plasmid-borne copies of either sipS or sipT but not by an empty vector lacking a sip gene (data not shown). Finally, Western blotting experiments showed that the replacement of SipS and SipT by SipP did not significantly affect the accumulation of the Orf1p–β-lactamase fusion protein in cells of B. subtilis (Fig. 5C). In summary, these observations demonstrate that SipP of pTA1015 is a functional equivalent of the major chromosomally encoded SPases SipS and SipT.
FIG. 5.
Functional replacement of sipS and sipT by sipP. (A) Schematic presentation of the construction of B. subtilis ΔST(pTAB11A) in two steps. First, B. subtilis 8G5 sipS (1) was transformed with pTAB11A expressing the sipP gene. Second, the sipT gene of the resulting strain was disrupted with a Cmr marker by transformation with chromosomal DNA of B. subtilis sipT-Cm containing a Cmr marker inserted in the sipT gene. The Cmr marker was integrated into the sipT gene of the transformed strain via homologous recombination (20). Only restriction sites relevant for the constructions are shown (Hi, HindII; Ec, EcoRI; Nc, NcoI). ‘sipS, 5′ truncated sipS gene (1); sipT’, 5′ end of the disrupted sipT gene; ‘sipT, 3′ end of the disrupted sipT gene; ori pBR322, replication functions of pBR322; SL, potential stem-loop structures. (B) Detection of SipS, SipT, and SipP. B. subtilis 8G5 (wild type [WT]), 8G5 sipT-Cm (ΔT), 8G5 sipS (ΔS), and ΔST(pTAB11A) (ΔST+SipP) were transformed with pOB1, which specifies the Orf1p–β-lactamase fusion protein. Next, pOB1-containing cells were grown in TY medium until 5 h after the transition phase between exponential and post-exponential growth. Cells were collected by centrifugation and used for SDS-PAGE and Western blotting to detect SipS, SipT, and SipP with antibodies directed against SipS. These antibodies show weak cross-reactivity with SipT and SipP. The positions of SipS, SipT, and SipP are indicated. (C) The presence of the pre-Orf1p–β-lactamase fusion protein in samples of B. subtilis 8G5 (WT), 8G5 sipT-Cm (ΔT), 8G5 sipS (ΔS), or ΔST(pTAB11A) (ΔST+SipP), all of which contained pOB1, was visualized with specific antibodies as for panel B. Positions of precursor (p) and mature (m) forms are indicated.
DISCUSSION
About 47% of the B. subtilis genes belong to paralogous gene families (8). As very little is known about why these paralogous genes have evolved in B. subtilis (and other organisms), we have initiated a functional analysis, starting with the sip gene family, which consists of five chromosomal (21) and at least two plasmid-borne genes (10). We have recently reported that under laboratory conditions, the presence of one chromosomally encoded SPase (i.e., SipS or SipT) is sufficient for protein secretion, growth, and viability, suggesting that the secretory precursor processing machinery of B. subtilis is functionally redundant (21). The presence of multiple SPases is, however, required for optimal processing of various precursors (1, 26). These findings suggest that the sip gene family has evolved both to provide the cell with backup SPases and to prevent potential bottlenecks for protein secretion, in particular under conditions of high-level synthesis of secretory proteins. In addition, the presence of multiple sip genes allows the cell to use different mechanisms to regulate their expression, as illustrated by the concerted, temporally regulated transcription of sipS, sipT, and the genes for secreted degradative enzymes (1, 20, 21). Finally, it seems that at least the chromosomally encoded SPases have acquired a preference for different precursor proteins (1, 19–21). Under natural conditions, these properties of the sip gene family may be important for the fitness of B. subtilis, which can secrete large amounts of proteins into the medium as an adaptive response to changes in the environment. If our explanations for the presence of paralogous sip genes in B. subtilis are correct, there remains the intriguing question of why other genes that are essential for protein secretion and viability, such as secA and secY, were not multiplied during the evolution of this organism.
Our present studies suggest that the presence of a plasmid-borne sipP gene from B. subtilis (natto) provides this organism with yet another system to prevent SPase limitation. First, using strains derived from B. subtilis 168 which, in contrast to B. subtilis (natto), are highly amenable to genetic analyses, we show that SipP is a functional equivalent of the two major SPases, SipS, and SipT. For the latter two SPases, we have previously shown that their limitation is highly detrimental to the cell (21). Second, we show that the expression of plasmid-borne copies of sipP is temporally regulated in concert with the expression of at least one other plasmid-specific gene for a secretory protein, ORF1. In fact, as the transcription of sipP starts to increase most strongly in the transition phase between exponential and post-exponential growth, about 2 h before the transcription of sipS and sipT starts to increase, it seems that the transcription of sipP is coordinated even better with the transcription of genes for secreted enzymes than that of sipS and sipT. The observation that SipP is not specifically required for processing of Orf1p is in line with a more general role of SipP in the processing of chromosomally encoded precursor proteins. Finally, the presence of the sipP gene on pTA1015 is likely to be advantageous because this plasmid can spread by conjugative mobilization throughout a Bacillus population (11). Even though these considerations are based on experiments with B. subtilis 168-derived strains, we are convinced that they are also relevant for B. subtilis (natto), in view of the close genetic relationship between these two different isolates of the same organism (16).
The observation that the cell needs a functional copy of either sipS or sipT (21) suggests that the enzymes specified by these genes share the ability to process at least one essential protein that is exported from the cytoplasm, either to the cell wall or to the growth medium. In this respect, we conclude that SipS, SipT, and SipP must have similar substrate specificities, as SipP can functionally replace SipS and SipT. The substrate specificities of SipS, SipT, and SipP seem to differ, at least partly, from those of SipU, SipV, and SipW, which cannot complement the absence of SipS and SipT (21). As illustrated by the observation that the α-amylase AmyQ of Bacillus amyloliquefaciens is a preferred substrate of SipT but not of SipS (20), even the substrate specificities of SipS and SipT are not identical. At present, we do not know whether the substrate specificity of SipP is more similar to that of SipS or SipT, and whether SipP has a preference for Orf1p. Our observation that SipP is not specifically required for the processing of Orf1p does not exclude the latter possibility. It will be a major challenge for future research to identify the factors that determine the similarities and differences in the substrate specificities of the type I SPases of B. subtilis. This should lead to an increased understanding of SPase function in general, and the development of algorithms to predict which of the approximately 180 putative secreted proteins of B. subtilis (22) is processed by which SPase(s) in particular.
The question of why the sipP modules of pTA1015 and pTA1040 contain the ORF1 gene remains to be answered. The identification of this module on endogenous plasmids of B. subtilis (natto) strains suggests that ORF1 has a role during the production of natto, a traditional Japanese food product based on fermented soy beans. However, the production of Orf1p in B. subtilis 168 strains did not result in the production of extracellular polymers, such as polyglutamate and levan (11), showing that Orf1p by itself is not sufficient for the production of natto.
Finally, the temporally controlled transcription of genes on naturally occurring rolling-circle plasmids of a gram-positive bacterium has thus far been reported only for certain rep genes, their expression being high in the exponential growth phase and low in the post-exponential growth phase (4). The temporally controlled transcription of plasmid-encoded genes, such as ORF1-sipP, in concert with the transcription of chromosomally encoded genes, as documented in the present report is unprecedented. To explain the observed differences in the regulation of the sipP and sipS genes, which may simply be due to the fact that sipP is located on a multicopy plasmid, whereas sipS is located on the chromosome, it will be important to identify the factors that control the transcription of ORF1-sipP. This will also be important to increase our understanding of the ways in which eubacteria, such as B. subtilis, which are challenged by highly fluctuating environmental conditions exploit plasmids to increase their fitness.
ACKNOWLEDGMENTS
We thank J. A. K. W. Kiel for providing plasmid pKVS263, A. Bolhuis, M. L. van Roosmalen, J. D. H. Jongbloed, and other members of the European Bacillus Secretion Group for useful discussions, and C. Driessen for technical support.
H.T. and W.J.J.M. were supported by Gist-brocades B.V. (Delft, The Netherlands). In addition, H.T. was supported by Genencor International (Rijswijk, The Netherlands). S.B. and J.M.D. were supported by Biotechnology Grants Bio2-CT93-0254, Bio4-CT95-0278, and Bio4-CT96-0097 from the European Union.
REFERENCES
- 1.Bolhuis A, Sorokin A, Azevedo V, Ehrlich S D, Braun P G, de Jong A, Venema G, Bron S, van Dijl J M. Bacillus subtilis can modulate its capacity and specificity for protein secretion by temporally controlled expression of the sipS gene for signal peptidase I. Mol Microbiol. 1996;22:605–618. doi: 10.1046/j.1365-2958.1996.d01-4676.x. [DOI] [PubMed] [Google Scholar]
- 2.Bron S, Venema G. Ultraviolet inactivation and excision repair in Bacillus subtilis. Construction and characterization of a transformable eightfold auxotrophic strain and two ultraviolet-sensitive derivatives. Mutat Res. 1972;15:1–10. doi: 10.1016/0027-5107(72)90086-3. [DOI] [PubMed] [Google Scholar]
- 3.Dalbey R E, Lively M O, Bron S, van Dijl J M. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espinosa M, del Solar G, Rojo F, Alonso J C. Plasmid rolling circle replication and its control. FEMS Microbiol Lett. 1995;130:111–120. doi: 10.1111/j.1574-6968.1995.tb07707.x. [DOI] [PubMed] [Google Scholar]
- 5.Ferrari E, Jarnagin A S, Schmidt B F. Commercial production of extracellular enzymes. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 917–937. [Google Scholar]
- 6.Kiel J A K W, Boels J M, Beldman G, Venema G. Molecular cloning and nucleotide sequence of the glycogen branching enzyme gene (glgB) from Bacillus stearothermophilus and expression in Escherichia coli and Bacillus subtilis. Mol Gen Genet. 1991;230:136–144. doi: 10.1007/BF00290661. [DOI] [PubMed] [Google Scholar]
- 7.Kiel, J. A. K. W. Unpublished data.
- 8.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 9.Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- 10.Meijer W J J, de Jong A, Wisman G B A, Tjalsma H, Venema G, Bron S, van Dijl J M. The endogenous Bacillus subtilis (natto) plasmids pTA1015 and pTA1040 contain signal peptidase-encoding genes: identification of a new structural module on cryptic plasmids. Mol Microbiol. 1995;17:621–631. doi: 10.1111/j.1365-2958.1995.mmi_17040621.x. [DOI] [PubMed] [Google Scholar]
- 11.Meijer W J J, Wisman G B A, Terpstra P, Thorsted P B, Thomas C M, Holsappel S, Venema G, Bron S. Rolling-circle plasmids from Bacillus subtilis: complete nucleotide sequences and analyses of genes of pTA1015, pTA1040, pTA1050 and pTA1060, and comparisons with related plasmids from Gram-positive bacteria. FEMS Microbiol Rev. 1998;21:337–368. doi: 10.1111/j.1574-6976.1998.tb00357.x. [DOI] [PubMed] [Google Scholar]
- 12.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 13.Msadek T, Kunst F, Rapoport G. Two-component regulatory systems. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 729–745. [Google Scholar]
- 14.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Martinez G, Kok J, Venema G, van Dijl J M, Smith H, Bron S. Protein export elements from Lactococcus lactis. Mol Gen Genet. 1992;234:401–411. doi: 10.1007/BF00538699. [DOI] [PubMed] [Google Scholar]
- 16.Priest F G. Systematics and ecology of Bacillus. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 3–16. [Google Scholar]
- 17.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Stöver, A. G., and A. Driks. Personal communication.
- 20.Tjalsma H, Noback M A, Bron S, Venema G, Yamane K, van Dijl J M. Bacillus subtilis contains four closely related type I signal peptidases with overlapping substrate specificities: constitutive and temporally controlled expression of different sip genes. J Biol Chem. 1997;272:25983–25992. doi: 10.1074/jbc.272.41.25983. [DOI] [PubMed] [Google Scholar]
- 21.Tjalsma H, Bolhuis A, van Roosmalen M L, Wiegert T, Schumann W, Broekhuizen C P, Quax W, Venema G, Bron S, van Dijl J M. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 1998;12:2318–2331. doi: 10.1101/gad.12.15.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tjalsma, H., and J. M. van Dijl. Unpublished results.
- 23.Uozumi T, Ozaki A, Beppu T, Arima K. New cryptic plasmids of Bacillus subtilis and restriction analysis of other plasmids found by general screening. J Bacteriol. 1980;142:315–318. doi: 10.1128/jb.142.1.315-318.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Dijl J M, de Jong A, Smith H, Bron S, Venema G. Signal peptidase I overproduction results in increased efficiencies of export and maturation of hybrid secretory proteins in Escherichia coli. Mol Gen Genet. 1991;227:40–48. doi: 10.1007/BF00260704. [DOI] [PubMed] [Google Scholar]
- 25.van Dijl J M, de Jong A, Smith H, Bron S, Venema G. Non-functional expression of Escherichia coli signal peptidase I in Bacillus subtilis. J Gen Microbiol. 1991;137:2073–2083. doi: 10.1099/00221287-137-9-2073. [DOI] [PubMed] [Google Scholar]
- 26.van Dijl J M, de Jong A, Vehmaanperä J, Venema G, Bron S. Signal peptidase I of Bacillus subtilis: patterns of conserved amino acids in prokaryotic and eukaryotic type I signal peptidases. EMBO J. 1992;11:2819–2828. doi: 10.1002/j.1460-2075.1992.tb05349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Dijl J M, de Jong A, Venema G, Bron S. Identification of the potential active site of the signal peptidase SipS of Bacillus subtilis. J Biol Chem. 1995;270:3611–3618. doi: 10.1074/jbc.270.8.3611. [DOI] [PubMed] [Google Scholar]
- 28.van Sinderen D, Withoff S, Boels H, Venema G. Isolation and characterisation of comL, a transcriptional unit involved in competence development of Bacillus subtilis. Mol Gen Genet. 1990;224:396–404. doi: 10.1007/BF00262434. [DOI] [PubMed] [Google Scholar]
- 29.Wertman K F, Wyman A R, Botstein D. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene. 1986;49:253–262. doi: 10.1016/0378-1119(86)90286-6. [DOI] [PubMed] [Google Scholar]