Abstract
The diagnosis of Mendelian disorders requires labor-intensive literature research. Trained clinicians can spend hours looking for the right publication(s) supporting a single gene that best explains a patient’s disease. AMELIE (Automatic Mendelian Literature Evaluation) greatly accelerates this process. AMELIE parses all 29 million PubMed abstracts and downloads and further parses hundreds of thousands of full-text articles in search of information supporting the causality and associated phenotypes of most published genetic variants. AMELIE then prioritizes patient candidate variants for their likelihood of explaining any patient’s given set of phenotypes. Diagnosis of singleton patients (without relatives’ exomes) is the most time-consuming scenario, and AMELIE ranked the causative gene at the very top for 66% of 215 diagnosed singleton Mendelian patients from the Deciphering Developmental Disorders project. Evaluating only the top 11 AMELIE-scored genes of 127 (median) candidate genes per patient resulted in a rapid diagnosis in more than 90% of cases. AMELIE-based evaluation of all cases was 3 to 19 times more efficient than hand-curated database–based approaches. We replicated these results on a retrospective cohort of clinical cases from Stanford Children’s Health and the Manton Center for Orphan Disease Research. An analysis web portal with our most recent update, programmatic interface, and code is available at AMELIE.stanford.edu.
INTRODUCTION
Millions of babies born worldwide each year are affected by severe genetic, often Mendelian disorders (1). Patients with Mendelian diseases have one or two genetic variants in a single gene primarily responsible for their disease phenotypes (2). Roughly 5000 Mendelian diseases, each with a characteristic set of phenotypes, have been mapped to about 3500 genes to date (3). Exome sequencing is often performed to identify candidate causative genes, resulting in a relatively high (currently 30%) diagnostic yield (4). A genetic diagnosis provides a sense of closure to the patient family, aids in patient trajectory prediction and management, allows for better family counseling, and, in the age of gene editing, even provides first hope for a cure. However, identifying the causative mutation(s) in a patient’s exome to arrive at a diagnosis can be very time-consuming, with a typical exome requiring hours of expert analysis (5).
Definitive diagnosis of a known Mendelian disorder is accomplished by matching the patient’s genotype and phenotype to previously described cases from the literature. Manually curated databases (6-10) are used to more efficiently access extracts of the unstructured knowledge in the primary literature. Automatic gene ranking tools (11-18) use these databases to prioritize candidate genes in patients’ genomes for their ability to explain patient phenotypes. An important feature of many gene ranking tools is the use of phenotype match functions on patient phenotypes and gene- or disease-associated phenotypes. Phenotype match functions exploit the structure of a phenotype ontology (9) and known gene-disease-phenotype associations to quantify the inexact match between two sets of phenotypes (11, 12), with recent approaches developed to computationally extract phenotype data from electronic medical notes (19, 20). The goal of all gene ranking tools is to aid a busy clinician in arriving at a definitive diagnosis of any case presented to them in the shortest amount of time by reading up on genes in the order the algorithm has ranked them.
Given the rapidly growing number of rare diseases with a known molecular basis (21) and the difficulty of manually finding a diagnosis for some rare diseases with variable phenotypes, many patients experience long diagnostic odysseys (22). Expert clinician time is expensive and scarce, but machine time is cheap and plentiful. We aimed to accelerate the diagnosis of patients with Mendelian diseases by using information from primary literature to construct gene rankings, thus allowing clinicians to discover the causative gene along with supporting literature in a minimum amount of time.
Here, we introduce AMELIE (Automatic Mendelian Literature Evaluation). AMELIE uses natural language processing (NLP) to automatically construct a homogeneous knowledgebase about Mendelian diseases directly from primary literature. To perform this operation, AMELIE was trained on data from manually curated databases such as Online Mendelian Inheritance in Man (OMIM) (6), Human Gene Mutation Database (HGMD) (8), and ClinVar (10). AMELIE then used a machine learning classifier that integrated knowledge about a patient’s phenotype and genotype with its knowledgebase to rank candidate genes in the patient’s genome for their likelihood of being causative and simultaneously supported its ranking results with annotated citations to the primary literature. We compare this end-to-end machine learning approach to gene ranking methods that rely on manually curated databases using a total of 271 singleton patients from three different sources, including two clinical centers and a research cohort.
RESULTS
Overview of AMELIE
Given a patient’s genome sequencing data and a phenotypic description of the patient, AMELIE aims to both identify the gene causing the patient’s disease (when possible) and supply the clinician with literature supporting the gene’s causal role. To this end, AMELIE creates a ranking of candidate causative genes in the patient’s genome with the aim of ranking the true causative gene at the top. AMELIE constructs its candidate causative gene ranking by comparing information from the primary literature to information about the patient’s genotype and phenotype.
To process information from the full text of primary literature, AMELIE constructs a knowledgebase directly from the primary literature up front using NLP techniques trained on manually curated databases. After knowledgebase construction, AMELIE ranks any patient’s candidate causative genes using a classifier, which compares knowledge from the AMELIE knowledgebase with phenotypic and genotypic information about the patient. AMELIE explains each gene’s ranking to the clinician by citing articles about this gene in the knowledgebase.
Identification and download of relevant Mendelian disease articles based on all of PubMed
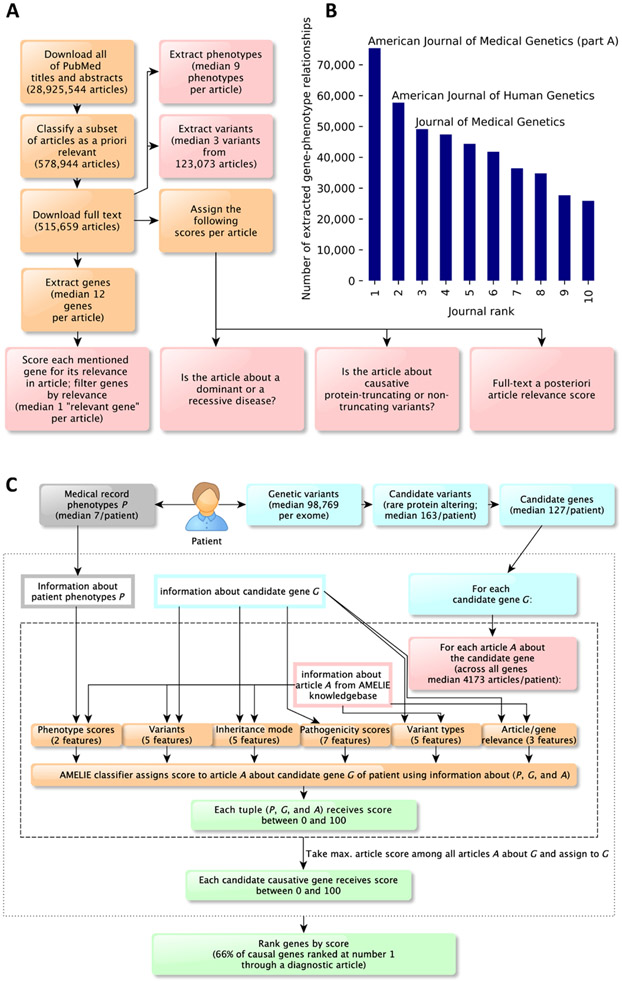
The first step toward building the AMELIE knowledgebase was discovering relevant primary literature. Of 29 million peer-reviewed articles deposited in PubMed, only a fraction is relevant for Mendelian disease diagnosis. We constructed a machine learning classifier that, given titles and abstracts of articles from PubMed, identified potentially relevant articles for the AMELIE knowledgebase.
Machine learning classifiers take as input a numerical vector describing the input, called the “feature vector.” Here, we used a so-called term frequency–inverse document frequency transformation to convert input text into a feature vector. We implemented the title/abstract document classifier as a logistic regression classifier. Logistic regression transforms its output using the logistic sigmoid function to return a probability value that is then mapped into binary (positive/negative) decision-making (23).
Machine learning classifiers learn to classify an input as positive (relevant) or negative (irrelevant) by being exposed to a large number of labeled positive and negative examples (the training set). OMIM (6) is an online database of Mendelian diseases, genes, and associated phenotypes. HGMD (8) is a database of disease-causing mutations in the human genome. The training set for the title/abstract relevance classifier consisted of titles and abstracts of 56,479 Mendelian disease–related articles cited in OMIM and HGMD as positive training examples and 67,774 random titles and abstracts of PubMed articles (largely unrelated to Mendelian disease) as negative training examples.
Precision and recall are two standard measures of evaluating classifier performance. Precision measures the fraction of all inputs classified positive that are truly relevant. Recall measures the fraction of truly positive inputs that are classified positive. Fivefold cross-validation (splitting all available labeled training data to include 80% in a training set and evaluating on the remaining 20%, five times in round-robin fashion) returned an average precision of 98% and an average recall of 96%.
All 28,925,544 titles and abstracts available in PubMed on 30 September 2018 were downloaded and processed by the document classifier. The classifier identified 578,944 articles as possibly relevant on the basis of their PubMed title and abstract, of which we downloaded 515,659 (89%) full-text articles directly from dozens of different publishers.
Building a structured database of information about Mendelian diseases from full text
From the full text of an article, multiple types of information were extracted. Gene mentions in full text were identified using lists of gene and protein names and synonyms from the HUGO Gene Nomenclature Committee (24), UniProt (25), and the automatically curated PubTator (26), a National Center for Biotechnology Information service combining gene mentions found by multiple previously published automatic gene recognition methods. AMELIE recognized about 93% of disease-causing gene names. However, through a combination of unfortunate gene synonyms (such as “FOR,” “TYPE,” “ANOVA,” or “CO2”), as well as genes mentioned only in titles of cited references, or interaction partners of causative genes, a median of 12 distinct gene candidates was found in each article (table S1).
To discover which gene(s) were the subject of the PubMed article, each distinct gene candidate extracted from an article received a “relevant gene score” between 0 and 1 indicating the likelihood of the gene being important in the context of the article. Training data for the relevant gene classifier were obtained from OMIM and HGMD. A total of 304,471 downloaded full-text articles contained at least one gene with a relevance score of 0.1 or higher. These articles, along with their above-threshold scoring genes, formed the AMELIE knowledgebase. Articles in the AMELIE knowledgebase contained a median of 1 gene with a relevant gene score between 0.1 and 1 (table S1). Furthermore, genetic variants (for example, “p.Met88Ile” or “c.251A>G”) were identified in the full text of each article and converted to genomic coordinates (chromosome, position, reference, and alternative allele) using the AVADA (Automatic Variant Evidence Database) variant extraction method (27). A median of three distinct genetic variants was extracted from 123,073 full-text articles in the AMELIE knowledgebase.
Phenotype mentions were recognized in full-text articles using a list of phenotype names compiled from Human Phenotype Ontology (HPO) (9). By linking all genes with a relevant gene score of at least 0.5 in an article with all phenotypes mentioned in the same article, we arrived at a total of 872,080 gene-phenotype relationships covering 11,537 genes (fig. S1).
Five scores between 0 and 1 were assigned to the full text of each article. A “full-text document relevance” score assessed the likely relevance of the article for the diagnosis of Mendelian diseases. A “protein-truncating” and a “nontruncating” score each gave an assessment of whether the article was about a disease caused by protein-truncating (splice site, frameshift, and stopgain) or nontruncating (other) variants. A “dominant” and a “recessive” score each gave an assessment of the discussed inheritance mode(s) in the article.
Precision and recall of full-text article information (relevant genes, extracted phenotypes, and full-text article scores) varied between 74 and 96%. All the data described in this section were entered into the AMELIE knowledgebase, keyed on the article that they were extracted from (Fig. 1A). The top journals from which the most gene-phenotype relationships were extracted are shown in Fig. 1B and table S2. We estimated that the number of newly described gene-phenotype relationships has increased by an average of 10.5% every 2 years since the year 2000 (fig. S2).
Fig. 1. AMELIE knowledgebase creation and subsequent patient causal gene ranking classifier.
(A) AMELIE knowledgebase creation. AMELIE applies multiple machine learning classifiers to all (current) 29 million PubMed abstracts to parse, predict relevance, download full text, and lastly extract Mendelian gene-phenotype relationships and related attributes automatically. (B) Number of gene-phenotype relationships extracted from the 10 journals that AMELIE extracted most gene-phenotype relationships from. (C) The AMELIE classifier combines 27 features to rank all articles in the AMELIE knowledgebase for their ability to explain any input patient.
The AMELIE classifier assigns patient genes a likelihood of being causative
Given a patient with a suspected Mendelian disease, AMELIE aims to speed up discovery of the causative gene by ranking patient genes for their ability to describe a set of patient phenotypes. AMELIE performs standard filtering of the patient variant list (21, 28) to keep only “candidate causative variants” that are rare in the unaffected population and are predicted to change a protein-coding region (missense, frameshift, nonframeshift indel, core splice site, stoploss, and stopgain variants). Core splice sites were defined to consist of the 2 base pairs at either end of each intron. Genes containing candidate causative variants were called candidate causative genes (or “candidate genes”). AMELIE ranked about 97% of known disease-causing mutations, excluding only those in deeper intronic and non–protein-coding intergenic regions.
We defined an article in the AMELIE knowledgebase to be about a candidate causative gene if the candidate causative gene had a relevant gene score of at least 0.1 in the article to maximize recall while maintaining a median of 1 relevant gene per article. We constructed a machine learning classifier called the “AMELIE classifier” that assigns a score between 0 and 100 to triples (P, G, and A), consisting of a set of patient phenotypes P, a candidate causative gene G, and an article A about the candidate gene. Given a patient with phenotypes P and a candidate gene G, the AMELIE score indicates whether the article A is likely helpful for diagnosing the patient because it links mutations in G to the patient’s phenotypes P. Higher AMELIE scores indicate articles more likely relevant to diagnosis. The AMELIE classifier was implemented as a logistic regression classifier and returns a score between 0 and 100 called the “AMELIE score.” The AMELIE score is used to both rank patient candidate genes and explain rankings by citing primary literature, as described below.
The AMELIE classifier uses a set of 27 real-valued features, falling into six feature groups (Fig. 1C). The six feature groups comprise (i) five features containing information about disease inheritance mode extracted from the article and patient variant zygosity, (ii) five features containing information about AVADA-extracted variants from the article and overlap of these variants with patient variants, (iii) two features containing information about patient phenotypes based on the Phrank (11) phenotypic match score of phenotypes in article A with the patient phenotypes P, (iv) five features containing information about article and patient variant types, (v) three features containing information about article relevance and relevance of the candidate gene in the article, and (vi) seven features containing a priori information about the patient’s candidate causative variants in G such as in silico pathogenicity scores (29) and gene-level mutation intolerance scores (30, 31).
To train the AMELIE classifier, we constructed a set of 681 simulated patients using data from OMIM (6), ClinVar (10), and the 1000 Genomes Project (32). Each simulated patient s was assigned a disease from OMIM, with phenotypes noisily sampled from the phenotypes associated with the disease. The genome of each simulated patient was based on genome sequencing data from the 1000 Genomes Project. An appropriate disease-causing variant from ClinVar was added to each simulated patient’s genome. Each simulated patient was assigned a diagnostic article As describing the genetic cause of the patient’s disease. In total, the simulated patients covered a total of 681 OMIM diseases (1 per patient) and a total of 1090 distinct phenotypic abnormalities (table S3). The sampled phenotypes for each disease covered an average of 21% of the phenotypes manually associated with the disease by HPO.
The AMELIE classifier was trained to recognize the diagnostic article As out of all articles about genes with candidate causative variants in a patient s. Of a total of 681 training “patients” constructed using data in OMIM and ClinVar, the single positively labeled article was recognized and downloaded during AMELIE knowledgebase construction in 664 cases (98%), creating 664 positive training examples. The negative training set for the AMELIE classifier consisted of triples (Ps, G, and A) for each simulated patient s, where G was a noncausative candidate gene in patient s and A was an article about G. For training efficiency, we used only 664,000 random negative training examples out of all available negative training examples.
The AMELIE classifier assigns each candidate gene G an AMELIE score, defined as the best AMELIE classifier score for any paper A about gene G, as it relates to patient P (Fig. 1C). Candidate causative genes were ranked in descending order of their associated score.
Evaluating AMELIE on a retrospective patient test set
We evaluated AMELIE on a set of 215 real singleton patients with an established diagnosis from the Deciphering Developmental Disorders (DDD) project (33). The DDD dataset included HPO phenotypes (a median of 7 per patient), exome data in variant call format, and the causative gene for each patient (1 per patient). AMELIE’s goal was to rank the established causative gene at or near the top of its ranked list of candidate genes for each patient. Filtering for candidate causative variants resulted in a median of 163 variants in 127 candidate genes per patient Fig. 1C). We used the set of 215 patients obtained from the DDD study to evaluate AMELIE against Exomiser (14), Phenolyzer (15), Phen-Gen (16), eXtasy (17), and PubCaseFinder (18). The output of all methods, consisting of a list of ranked genes, was subset to the (median) 127 candidate genes that AMELIE used for each patient based on the filtering criteria previously described (Fig. 2A). This ensured the fair evaluation of all gene ranking methods against the same set of genes.
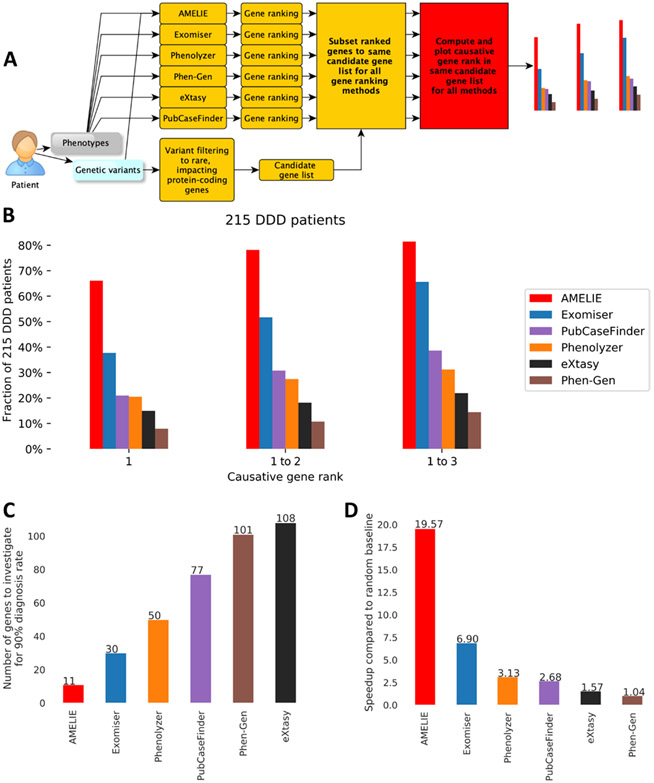
Fig. 2. AMELIE patient causative gene ranking outperforms methods based on manually curated databases.
(A) Evaluation scheme. The output gene ranking of all algorithms was subset to the same list of candidate genes AMELIE uses its gene ranking to ensure a fair comparison. (B) Fraction of (n = 215) DDD cases ranked as 1, 1 to 2, or 1 to 3 by six different tools. (C) The number of top-ranked genes needed to achieve a 90% diagnosis rate across (n = 215) DDD cases by various gene ranking tools. By evaluating up to AMELIE’s 11th top-ranked gene, a 90% diagnosis yield on the DDD cases was achieved. The next best tool, Exomiser, achieved a 90% diagnosis yield by evaluating up to Exomiser’s 30th gene. (D) The speedup in terms of number of genes to investigate when perusing the ranked gene lists provided by each tool from top to bottom until the causative gene was found compared to the expected value of a random baseline gene ordering for (n = 215) DDD cases.
AMELIE analyzed a median of 4173 articles per patient and ranked the causative gene at the very top in 142 (66%) of 215 cases and in the top 10 in 193 cases (89.7%). Other methods ranked the causative gene at the top between 38% of cases (Exomiser) and 8% of cases (Phen-Gen) (Fig. 2B). AMELIE performed significantly better than all compared methods (all P values ≤1.68 × 10−9; one-sided Wilcoxon signed-rank test; table S4). Of 117 distinct top-ranked articles supporting the DDD patients where AMELIE ranked the test set causative gene at number 1, only 36 (31%) were cited in OMIM as determined by a systematic Google search of omim.org (table S5).
Because of the large number of patients expected to be sequenced for Mendelian diagnosis (34), one may want to set guidelines for rapid versus in-depth exome or genome analysis. In our test set of 215 patients, AMELIE offered a diagnosis for 90% of diagnosable cases when evaluating only up to the top 11 AMELIE-ranked genes per case or 9% of a median of 127 candidate causative genes. If using any of the other methods, the clinician would have to investigate between a median of 30 genes (when using Exomiser to rank patient candidate causative genes) and 108 genes per patient to arrive at the diagnosis in 90% of diagnosable cases (Fig. 2C).
If the clinician used AMELIE to determine the order in which they evaluate their entire candidate gene list, one gene after the other, on the DDD set of 215 patients, they would evaluate a total of 735 gene-patient matches to arrive at the causative gene for all 215 patients. If the clinician went through the list of candidate genes in random order, they would evaluate an expected total sum of 14,383 gene-patient matches to arrive at the causative gene for all patients. By this metric, AMELIE improved diagnosis time by a factor of 19.6× over a random baseline. The next best tool, Exomiser, would require the clinician to read about 2085 genes until arriving at the causative gene for all patients, an improvement of 6.9× faster over a random baseline. The performance of other methods ranged from a speedup of 3.13× to 1.04× (Fig. 2D). The speedup provided by AMELIE was thus more than twice that provided by the next best tool, Exomiser.
Replication of AMELIE performance on 56 clinical cases from two sites
To test for the result replication across data sources, we evaluated AMELIE using 56 singleton clinical cases seen by the Medical Genetics Service at Stanford Children’s Health and the Manton Center for Orphan Disease Research at Boston Children’s Hospital. Patient genotype and phenotype data were obtained from Stanford and the Manton Center Gene Discovery Core.
We performed a comparison of gene ranking performance using AMELIE against other methods as above for the DDD patients. AMELIE ranked the causative gene at the very top in 33 (59%) of 56 cases and in the top 10 in 50 cases (89%). Again, AMELIE significantly outperformed all other methods (all P values ≤6.65 × 10−3; one-sided Wilcoxon signed-rank test; fig. S3A and table S6). AMELIE offered a diagnosis for 90% of patients in the test set of 56 Stanford and Manton patients if evaluating the top 15 candidate genes per patient (9% of a median of 172.5), replicating its performance on the DDD set (fig. S3B).
To arrive at the causative gene for each patient in the clinical test set from Stanford and Manton when using AMELIE, a clinician would need to evaluate 300 genes compared to a baseline of 6106 genes if evaluating genes in random order. Similar to the DDD patient test set, AMELIE resulted in a speedup of 20× compared to the baseline, 2× to 20× faster than other methods (fig. S3C). Because the other methods do not use simulated patients for training, gene ranking results using other methods were obtained by running each respective method once on the simulated patient set. Fivefold cross-validation on the 681 simulated patients showed that AMELIE generated significantly better causative gene rankings compared to the other methods (all P values ≤5.24 × 10−10; fig. S4 and table S7).
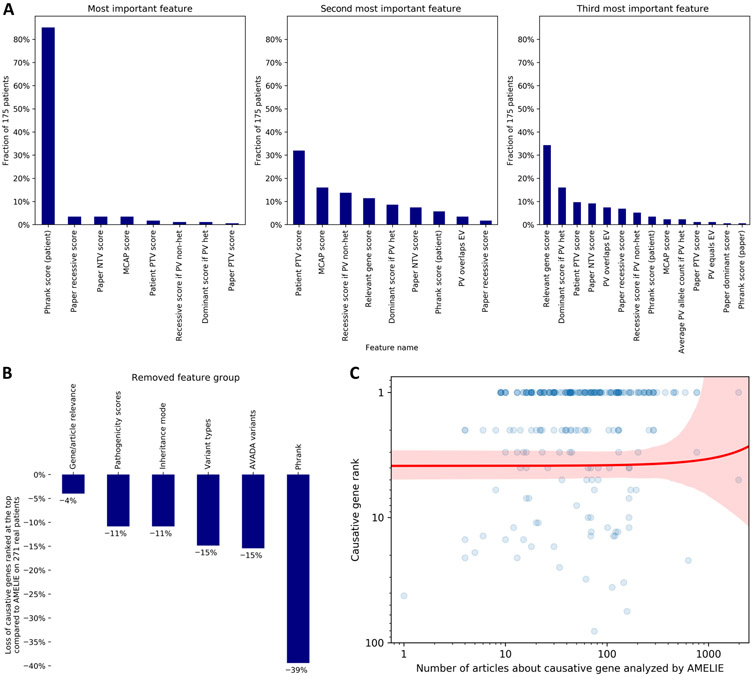
We ran multiple tests with modified AMELIE knowledgebases and AMELIE classifiers to dissect the relative contribution of different AMELIE components to its causative gene ranking performance. For all 175 test cohort patients with the causative gene ranked at the top, we investigated which machine learning features of the AMELIE classifier contributed most to the high score of the causative gene. Overwhelmingly, for 149 (85%) of 175 real test patients, the feature contributing most to the high score was a high phenotypic match between the patient and the article. However, 14 of a total 27 AMELIE classifier features (52%) occurred at least once within the three features contributing most to the top rank of a patient’s causative gene (Fig. 3A and table S8).
Fig. 3. Investigating AMELIE’s gene ranking performance.
(A) For each of the 175 patients with AMELIE causative gene rank 1 among all (n = 271) real DDD, Stanford, and Manton patients, the 27 features to the AMELIE classifier were ranked by their contribution to the top-ranked article’s high score. The panels (left to right) show the fraction of patients for which certain features were ranked most, second most, or third most contributing. PTV, protein-truncating variant; NTV, non–protein-truncating variant; MCAP, Mendelian clinically applicable pathogenicity score, an in silico pathogenicity score; PV, patient variant; het, heterozygous; EV, full-text article–extracted variant. (B) Retraining the AMELIE classifier with fivefold cross-validation, each time omitting one of AMELIE’s six feature groups, shows the degree to which feature groups aided performance across all (n = 271) DDD, Stanford, and Manton patients. (C) Each blue dot represents one of (n = 271) real DDD, Stanford, or Manton patients in this log-log plot. The red line is a linear regression line between number of articles about causative gene (x axis) and causative gene rank (y axis), with red denoting the 95% confidence interval.
To measure how much AMELIE relied on certain feature groups, we retrained the AMELIE classifier six times, each time dropping one of its six feature groups. With dropped-out features, the number of causative genes ranked at the top across the test set of 271 real patients shrank between 4 and 39% (Fig. 3B and table S9). AMELIE did not better rank causative genes when phenotype recognition was augmented by data from Unified Medical Language System (35), Medical Subject Headings (36), and Systematized Nomenclature of Medicine–Clinical Terms (37), three databases containing additional phenotype names and synonyms. However, AMELIE ranked 32% more causative genes at the top when using full-text data rather than data gathered only from titles and abstracts.
AMELIE’s performance is not correlated with number of articles about a causative gene
We investigated whether the number of articles about the causative gene in the AMELIE knowledgebase is correlated with the causative gene rank by performing linear regression between the causative gene rank and number of articles analyzed for the causative gene. The regression revealed no significant relationship (P = 0.85 that the slope of regression is equal to 0 according to a Wald test with t distribution of the test statistic; Fig. 3C), suggesting that AMELIE performs well independent of the number of papers it has analyzed about a causative gene. For the 22 patients (8% of a total of 271 real test patients) with less than 10 papers analyzed for the causative gene, AMELIE ranked causative genes at the top for 10 (45%) cases. In contrast, Exomiser ranked the causative gene at the top in six (27%) of these cases.
The AMELIE knowledgebase and AMELIE classifier work together to arrive at high causative gene ranks
We investigated the relative contribution of the AMELIE classifier and the AMELIE knowledgebase to AMELIE’s overall gene ranking performance. We retrained the AMELIE classifier using data from DisGeNET (38), a text mining–based database containing gene-phenotype relationships, disease-causing variants, and links to primary literature from PubMed. Using DisGeNET data resulted in significantly worse causative gene rankings compared to the AMELIE knowledgebase (P ≤ 4.76 × 10−23; table S10). We then replaced the AMELIE classifier (Fig. 1C) with the Phrank (11) phenotypic match score to estimate the impact of the AMELIE classifier on overall AMELIE performance. Gene ranking by the Phrank phenotypic match score resulted in ranking 94 (35%) of 271 real patients’ causative genes at the top, significantly worse compared to the AMELIE classifier, which ranked 175 causative genes at the top (P = 1.33 × 10−11, one-sided Wilcoxon signed-rank test). We conclude that the AMELIE knowledgebase and the AMELIE classifier work together to achieve AMELIE’s high causative gene ranking performance.
Interactive and programmatic access to AMELIE-based literature analysis
AMELIE can be used through its web portal at https://AMELIE.stanford.edu for patient analysis. The portal offers both an interactive interface (fig. S5) and an application programming interface that enables integrating AMELIE into any computer-assisted clinical workflow. The AMELIE knowledgebase will be updated every year. A pilot of AMELIE has been running at this web address since August 2017, as a service to the community, using an AMELIE knowledgebase automatically curated from articles published until June 2016 and has since served many thousands of queries from more than 40 countries.
DISCUSSION
We present AMELIE, a method for ranking candidate causative genes and supporting articles from the primary literature in patients with suspected Mendelian disorders. We show that AMELIE ranks the causative gene first (among a median of 127 genes) in two of three of patients and within the top 11 genes in over 90% of 215 real patient cases. These results were closely replicated on a cohort of 56 clinical patients from Stanford Children’s Health and the Manton Center for Orphan Disease Research.
Mendelian disease diagnosis is a complex problem and clinicians or researchers can spend many hours evaluating a single case. With 5000 diagnosable Mendelian diseases caused by roughly 3500 different genes that manifest in different subsets of more than 13,000 documented phenotypes, manual patient diagnosis from the primary literature is highly labor intensive. Manually curated databases such as OMIM, Orphanet, and HGMD take a step toward alleviating clinician burden by attempting to summarize the current literature. However, manual curation is growing even more challenging because the literature about Mendelian diseases is increasing at an accelerating rate. On the basis of AMELIE analysis, the number of gene-phenotype relationships in Mendelian literature has been increasing by an average of 10.5% every 2 years since the year 2000. Because AMELIE is an automatic curation approach requiring only an initial critical mass of human-curated data to train on, it is not constrained by the bottleneck of on-going human curation. For example, of 117 top-ranked articles supporting the DDD patients where AMELIE ranked the test set causative gene at number 1, only 36 (31%) were cited in OMIM. OMIM, a manually curated database, does not, of course, promise to capture all papers pertaining to any given disease gene but an automated effort like AMELIE can.
Compared to existing gene ranking approaches, AMELIE replaces the notion of a fixed disease description (that is, a single set of phenotypes) with the notion of an article and the phenotypes described in it. This approach has multiple advantages. First, it is often fastest to convince a clinician about a diagnosis given an article directly describing the disease, which often includes disease information such as patient images and related literature. In addition, with considerable phenotypic variability in Mendelian diseases (39), matching patients to specific reports in the literature is conceptually more helpful for definitive diagnosis than matching to a disease, which is effectively a compendium of previously described patient phenotypes.
Because of its dependence on literature and exome sequencing data, AMELIE is subject to a number of limitations. Biomedical literature is not guaranteed to contain the full set of phenotypes known to be associated with a disease, and AMELIE makes no claim about capturing this full set. Rather, AMELIE focuses on causal gene ranking using its knowledgebase, and as we show, it already does it to great practical utility. Certain articles about Mendelian diseases may mention a very small number of phenotypes (or none at all) and just mention disease and causative genes. Although this situation does not appear to be very common in practice (as seen by the good performance of AMELIE), the problem could be alleviated by automatically parsing disease names from such articles and associating diseases with manually curated phenotype information from resources such as HPO. NLP approaches could also be used to read additional texts, such as electronic medical notes (19, 20). Furthermore, AMELIE requires, as input, a list of HPO terms to describe patient phenotypes, although these may be provided by tools such as ClinPhen (19) that automatically extract HPO phenotypes directly from free-text clinical notes. Last, AMELIE is hampered by access to literature. Although AMELIE successfully obtained 80% of full-text articles that it deemed relevant on the basis of title and abstract, better publisher programmatic access to full-text literature for the purposes of text mining may lead to even better gene ranking results.
Understanding the impact of hundreds of thousands of variants in thousands of different genes against a body of knowledge of millions of peer reviewed papers that is ever expanding is a challenging task. Because a diagnosis shapes the future management of a patient, there must always be a human expert approving every diagnosis. However, the sheer number of patients that can benefit from a molecular diagnosis and our intention to sequence millions of them in the next few years absolutely necessitate automating, as much as possible, the diagnostic process to potentiate rapid, affordable, reproducible, and accessible clinical genome-wide diagnosis. Hence, along with complementary medical record parsing tools (19, 20), AMELIE provides a step toward integrating personal genomics into standard clinical practice.
MATERIALS AND METHODS
Study design
We implemented an NLP and machine learning system dubbed “AMELIE” to automatically identify candidate causative genes in patients with Mendelian (monogenic) diseases based on information in primary literature. The system consists of two components: a knowledgebase constructed directly from primary literature and a classifier that ranks candidate causative genes for a patient with a Mendelian disease.
To construct the AMELIE knowledgebase, we trained logistic regression classifiers (23) largely on OMIM (6) and HGMD (8) data to identify potentially relevant PubMed abstracts. Similar classifiers were used to determine full-text relevance and identify disease-causing genes, phenotypes, disease inheritance modes, disease-causing variants, and disease-causing variant types from abstract and article text. The AMELIE classifier was implemented as a logistic regression classifier (23). We constructed a set of 681 simulated patients with a single disease-causing variant using data from the 1000 Genomes Project (32), OMIM (6), HPO (9), and ClinVar. The AMELIE classifier was trained to recognize the simulated patients’ disease-causing genes (positive training examples) against a background of non–disease-causing genes (negative training examples).
We evaluated AMELIE against other knowledgebases and gene ranking tools using a set of 215 previously diagnosed patients from the DDD project (33). The DDD study has U.K. Research Ethics Committee (REC) approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12, granted by the Republic of Ireland REC). Each patient was associated with a candidate gene list generated using variant frequency filtering techniques, by restricting variant frequency to ≤0.5% minor allele frequency in a large control cohort (30). Using the DDD patient data, we compared AMELIE against five other gene ranking tools [Exomiser (14), Phenolyzer (15), Phen-Gen (16), eXtasy (17), and PubCaseFinder (18)]. We replicated the results on the DDD cohort by combining 35 patients from Stanford Children’s Health and 21 patients from the Manton Center for Orphan Disease Research into a further set of 56 test patients. Informed consent was obtained from all participants. Further details about the AMELIE algorithm are provided in Supplementary Materials and Methods.
Statistical analysis
To test performance differences between any two different gene ranking methods, we used the one-sided Wilcoxon signed-rank test throughout the manuscript. P < 0.05 was considered significant. No adjustments to alpha level or multiple testing correction methods were applied. The Wilcoxon signed-rank test is a nonparametric test and does not assume any particular distribution of data. We used this test to compare two matched samples: in our case, two lists of causative gene ranks on the same set of patients generated by two different methods. To test for significance of the slope of the regression line in Fig. 3C, we used the Wald test with t distribution of the test statistic.
Supplementary Material
Fig. S1. Number of phenotypes associated with genes through articles in the AMELIE knowledgebase.
Fig. S2. The accelerated accumulation of curatable facts in Mendelian genomics.
Fig. S3. Replication of AMELIE’s causative gene ranking performance on 56 clinical patients from Stanford and Manton.
Fig. S4. Cross-validation of AMELIE’s causative gene ranking performance on 681 simulated patients.
Fig. S5. Essence of the AMELIE interface at https://AMELIE.stanford.edu.
Table S1. Full-text gene extraction statistics.
Table S2. Extraction statistics from the 100 most used journals.
Table S3. Simulated patient details.
Table S4. DDD patient details.
Table S5. Searching for top-ranked AMELIE articles in OMIM.
Table S6. Stanford and Manton clinical patient details.
Table S7. Simulated patient gene ranking results.
Table S8. Most important features for patients with top-ranked causative genes.
Table S9. AMELIE classifier feature ablation results.
Table S10. DisGeNET gene ranking results.
Table S11. Regular expression patterns used to parse variant type from OMIM allelic variant entries.
Table S12. Phenotypes extracted from full-text articles by AMELIE, indicating whether the phenotype was extracted correctly or not.
Table S13. Assignment of features to feature groups.
Acknowledgments:
We thank K. Wang for assistance with ANNOVAR and E. Weiler for support and guidance. We thank P. McDonagh and T. Defay, as well as the members of the Bejerano Lab, for technical advice and helpful discussions. We thank E. Kravets, J. Buckingham, and K. MacMillen for assistance with obtaining patient data. We thank P. B. Agrawal, C. A. Brownstein, M. Danowski, C. A. Genetti, J. A. Madden, N. Nori, H. Paterson, K. Schmitz-Abe, and T. Yu from the Manton Center for their work. We thank all data sources used in AMELIE, including HPO, Ensembl, HGNC, UniProt, OMIM, ClinVar, HGMD, PubTator, ExAC, pLI, RVIS, M-CAP, and the GWAS catalog. We thank the European Genome-Phenome Archive (40) (EGA) and the Deciphering Developmental Disorders (33) (DDD) project. We thank P. B. Agrawal, C. A. Brownstein, M. Danowski, C. A. Genetti, J. A. Madden, N. Nori, H. Paterson, K. Schmitz-Abe, and T Yu from the Manton Center for Orphan Disease Research for providing patient data and advice. The research team acknowledges the support of the National Institute for Health Research through the Comprehensive Clinical Research Network. C.R. is affiliated with SambaNova Systems and Apple.
Funding:
All computational work was funded, in part, by a Bio-X SIGF fellowship to J.B., DARPA (C.R. and G.B.), the Stanford Pediatrics Department (J.A.B. and G.B.), a Packard Foundation Fellowship (G.B.), a Microsoft Faculty Fellowship (G.B.), NHGRI U41HG002371-15 (M.H.), and the Stanford Data Science Initiative (G.B. and C.R.). Manton Center sequence analysis and diagnosis was supported by NIH 1U54HD090255 IDDRC Molecular Genetics Core grant. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The DDD study presents an independent research commissioned by the Health Innovation Challenge Fund (grant no. HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute (grant no. WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health.
Footnotes
Competing interests: D.N.C. and P.D.S. acknowledge the receipt of financial support from Qiagen Inc. through a License Agreement with Cardiff University. C.R. is affiliated with SambaNova Systems and Apple Inc. The other authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. Data from public databases can be downloaded using the provided links and materials. Deidentified DDD data were obtained through EGA study number EGAS00001000775. Data for Stanford and Manton Center patients require patient consent and are available upon reasonable request, in agreement with the Stanford Children’s Health and Manton Center for Orphan Disease Research clinical coordinators and Institutional Review Boards. The AMELIE portal and code are available at https://AMELIE.stanford.edu and https://zenodo.org/deposit/3707012.
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Posey JE, O’Donnell-Luria AH, Chong JX, Harel T, Jhangiani SN, Akdemir ZHC, Buyske S, Pehlivan D, Carvalho CMB, Baxter S, Sobreira N, Liu P, Wu N, Rosenfeld JA, Kumar S, Avramopoulos D, White JJ, Doheny KF, Witmer PD, Boehm C, Sutton VR, Muzny DM, Boerwinkle E, Günel M, Nickerson DA, Mane S, MacArthur DG, Gibbs RA, Hamosh A, Lifton RP, Matise TC, Rehm HL, Gerstein M, Bamshad MJ, Valle D, Lupski JR; Centers for Mendelian Genomics, Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet. Med 21,798–812 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ, Exome sequencing identifies the cause of a Mendelian disorder. Nat. Genet 42, 30–35 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.OMIM Gene Map Statistics (2020); https://omim.org/statistics/geneMap.
- 4.Iglesias A, Anyane-Yeboa K, Wynn J, Wilson A, Truitt Cho M, Guzman E, Sisson R, Egan C, Chung WK, The usefulness of whole-exome sequencing in routine clinical practice. Genet. Med 16, 922–931 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Dewey FE, Grove ME, Pan C, Goldstein BA, Bernstein JA, Chaib H, Merker JD, Goldfeder RL, Enns GM, David SP, Pakdaman N, Ormond KE, Caleshu C, Kingham K, Klein TE, Whirl-Carrillo M, Sakamoto K, Wheeler MT, Butte AJ, Ford JM, Boxer L, Ioannidis JPA, Yeung AC, Altman RB, Assimes TL, Snyder M, Ashley EA, Quertermous T, Clinical interpretation and implications of whole-genome sequencing. JAMA 311, 1035–1045 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amberger JS, Bocchini CA, Scott AF, Hamosh A, OMIM.org: Leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 47, D1038–D1043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pavan S, Rommel K, Mateo Marquina ME, Höhn S, Lanneau V, Rath A, Clinical practice guidelines for rare diseases: The Orphanet Database. PLOS ONE 12, e0170365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, Cooper DN, The Human Gene Mutation Database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet 136, 665–677 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Köhler S, Carmody L, Vasilevsky N, Jacobsen JOB, Danis D, Gourdine J-P, Gargano M, Harris NL, Matentzoglu N, McMurry JA, Osumi-Sutherland D, Cipriani V, Balhoff JP, Conlin T, Blau H, Baynam G, Palmer R, Gratian D, Dawkins H, Segal M, Jansen AC, Muaz A, Chang WH, Bergerson J, Laulederkind SJF, Yüksel Z, Beltran S, Freeman AF, Sergouniotis PI, Durkin D, Storm AL, Hanauer M, Brudno M, Bello SM, Sincan M, Rageth K, Wheeler MT, Oegema R, Lourghi H, Rocca MGD, Thompson R, Castellanos F, Priest J, Cunningham-Rundles C, Hegde A, Lovering RC, Hajek C, Olry A, Notarangelo L, Similuk M, Zhang XA, Gómez-Andrés D, Lochmüller H, Dollfus H, Rosenzweig S, Marwaha S, Rath A, Sullivan K, Smith C, Milner JD, Leroux D, Boerkoel CF, Klion A, Carter MC, Groza T, Smedley D, Haendel MA, Mungall C, Robinson PN, Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res. 47, D1018–D1027 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, Karapetyan K, Katz K, Liu C, Maddipatla Z, Malheiro A, McDaniel K, Ovetsky M, Riley G, Zhou G, Holmes JB, Kattman BL, Maglott DR, ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jagadeesh KA, Birgmeier J, Guturu H, Deisseroth CA, Wenger AM, Bernstein JA, Bejerano G, Phrank measures phenotype sets similarity to greatly improve Mendelian diagnostic disease prioritization. Genet. Med 21, 464–470 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Köhler S, Schulz MH, Krawitz P, Bauer S, Dölken S, Ott CE, Mundlos C, Horn D, Mundlos S, Robinson PN, Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am. J. Hum. Genet 85, 457–464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singleton MV, Guthery SL, Voelkerding KV, Chen K, Kennedy B, Margraf RL, Durtschi J, Eilbeck K, Reese MG, Jorde LB, Huff CD, Yandell M, Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am. J. Hum. Gen 94, 599–610 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smedley D, Jacobsen JOB, Jäger M, Köhler S, Holtgrewe M, Schubach M, Siragusa E, Zemojtel T, Buske OJ, Washington NL, Bone WP, Haendel MA, Robinson PN, Next-generation diagnostics and disease-gene discovery with the Exomiser. Nat. Protoc 10, 2004–2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H, Robinson PN, Wang K, Phenolyzer: Phenotype-based prioritization of candidate genes for human diseases. Nat. Methods 12, 841–843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Javed A, Agrawal S, Ng PC, Phen-Gen: Combining phenotype and genotype to analyze rare disorders. Nat. Methods 11, 935–937 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Sifrim A, Popovic D, Tranchevent L-C, Ardeshirdavani A, Sakai R, Konings P, Vermeesch JR, Aerts J, De Moor B, Moreau Y, eXtasy: Variant prioritization by genomic data fusion. Nat. Methods 10, 1083–1084 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara T, Yamamoto Y, Kim J-D, Buske O, Takagi T, PubCaseFinder: A case-report-based, phenotype-driven differential-diagnosis system for rare diseases. Am. J. Hum. Genet 103, 389–399 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deisseroth CA, Birgmeier J, Bodle EE, Kohler JN, Matalon DR, Nazarenko Y, Genetti CA, Brownstein CA, Schmitz-Abe K, Schoch K, Cope H, Signer R, Martinez-Agosto JA, Shashi V, Beggs AH, Wheeler MT, Bernstein JA, Bejerano G, ClinPhen extracts and prioritizes patient phenotypes directly from medical records to expedite genetic disease diagnosis. Genet. Med 21, 1585–1593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Son JH, Xie G, Yuan C, Ena L, Li Z, Goldstein A, Huang L, Wang L, Shen F, Liu H, Mehl K, Groopman EE, Marasa M, Kiryluk K, Gharavi AG, Chung WK, Hripcsak G, Friedman C, Weng C, Wang K, Deep phenotyping on electronic health records facilitates genetic diagnosis by clinical exomes. Am. J. Hum. Genet 103, 58–73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wenger AM, Guturu H, Bernstein JA, Bejerano G, Systematic reanalysis of clinical exome data yields additional diagnoses: Implications for providers. Genet. Med 19, 209–214 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Carmichael N, Tsipis J, Windmueller G, Mandel L, Estrella E, “Is it going to hurt?”: The impact of the diagnostic odyssey on children and their families. J. Genet. Couns 24, 325–335 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Hastie T, Tibshirani R, Friedman J, The Elements of Statistical Learning (Springer, 2009); www.springer.com/us/book/9780387848570. [Google Scholar]
- 24.Yates B, Braschi B, Gray KA, Seal RL, Tweedie S, Bruford EA, Genenames.org: The HGNC and VGNC resources in 2017. Nucleic Acids Res. 45, D619–D625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bateman A, Martin MJ, O’Donovan C, Magrane M, Alpi E, Antunes R, Bely B, Bingley M, Bonilla C, Britto R, Bursteinas B, Bye-A-Jee H, Cowley A, Silva AD, Giorgi MD, Dogan T, Fazzini F, Castro LG, Figueira L, Garmiri P, Georghiou G, Gonzalez D, Hatton-Ellis E, Li W, Liu W, Lopez R, Luo J, Lussi Y, MacDougall A, Nightingale A, Palka B, Pichler K, Poggioli D, Pundir S, Pureza L, Qi G, Renaux A, Rosanoff S, Saidi R, Sawford T, Shypitsyna A, Speretta E, Turner E, Tyagi N, Volynkin V, Wardell T, Warner K, Watkins X, Zaru R, Zellner H, Xenarios I, Bougueleret L, Bridge A, Poux S, Redaschi N, Aimo L, Argoud-Puy G, Auchincloss A, Axelsen K, Bansal P, Baratin D, Blatter M-C, Boeckmann B, Bolleman J, Boutet E, Breuza L, Casal-Casas C, de Castro E, Coudert E, Cuche B, Doche M, Dornevil D, Duvaud S, Estreicher A, Famiglietti L, Feuermann M, Gasteiger E, Gehant S, Gerritsen V, Gos A, Gruaz-Gumowski N, Hinz U, Hulo C, Jungo F, Keller G, Lara V, Lemercier P, Lieberherr D, Lombardot T, Martin X, Masson P, Morgat A, Neto T, Nouspikel N, Paesano S, Pedruzzi I, Pilbout S, Pozzato M, Pruess M, Rivoire C, Roechert B, Schneider M, Sigrist C, Sonesson K, Staehli S, Stutz A, Sundaram S, Tognolli M, Verbregue L, Veuthey A-L, Wu CH, Arighi CN, Arminski L, Chen C, Chen Y, Garavelli JS, Huang H, Laiho K, McGarvey P, Natale DA, Ross K, Vinayaka CR, Wang Q, Wang Y, Yeh L-S, Zhang J, UniProt: The universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei C-H, Kao H-Y, Lu Z, PubTator: A web-based text mining tool for assisting biocuration. Nucleic Acids Res. 41, W518–W522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birgmeier J, Deisseroth CA, Hayward LE, Galhardo LMT, Tierno AP, Jagadeesh KA, Stenson PD, Cooper DN, Bernstein JA, Haeussler M, Bejerano G, AVADA: Toward automated pathogenic variant evidence retrieval directly from the full-text literature. Genet. Med 22, 362–370 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jagadeesh KA, Wu DJ, Birgmeier JA, Boneh D, Bejerano G, Deriving genomic diagnoses without revealing patient genomes. Science 357, 692–695 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Jagadeesh KA, Wenger AM, Berger MJ, Guturu H, Stenson PD, Cooper DN, Bernstein JA, Bejerano G, M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet 48, 1581–1586 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won H-H, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG; Exome Aggregation Consortium, Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB, Genic intolerance to functional variation and the interpretation of personal genomes. PLOS Genet. 9, e1003709 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR, A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deciphering Developmental Disorders Study, Large-scale discovery of novel genetic causes of developmental disorders. Nature 519, 223–228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birney E, Vamathevan J, Goodhand P, Genomics in healthcare: GA4GH looks to 2022. bioRxiv, 203554 (2017). [Google Scholar]
- 35.Bodenreider O, The Unified Medical Language System (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 32, D267–D270 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogers FB, Medical subject headings. Bull. Med. Libr. Assoc 51, 114–116 (1963). [PMC free article] [PubMed] [Google Scholar]
- 37.SNOMED CT, www.nlm.nih.gov/healthit/snomedct/.
- 38.Piñero J, Queralt-Rosinach N, Bravo À, Deu-Pons J, Bauer-Mehren A, Baron M, Sanz F, diseases and their genes. Database 2015, bav028 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornett KMD, Menezes MP, Bray P, Halaki M, Shy RR, Yum SW, Estilow T, Moroni I, Foscan M, Pagliano E, Pareyson D, Laurá M, Bhandari T, Muntoni F, Reilly MM, Finkel RS, Sowden J, Eichinger KJ, Herrmann DN, Shy ME, Burns J; Inherited Neuropathies Consortium, Phenotypic variability of childhood Charcot-Marie- Tooth disease. JAMA Neurol. 73, 645–651 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lappalainen I, Almeida-King J, Kumanduri V, Senf A, Spalding JD, ur-Rehman S, Saunders G, Kandasamy J, Caccamo M, Leinonen R, Vaughan B, Laurent T, Rowland F, Marin-Garcia P, Barker J, Jokinen P, Torres AC, de Argila JR, Llobet OM, Medina I, Puy MS, Alberich M, de la Torre S, Navarro A, Paschall J, Flicek P, The European Genome-phenome Archive of human data consented for biomedical research. Nat. Genet 47, 692–695 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É, Scikit-learn: Machine learning in Python. J. Mach. Learn. Res 12, 2825–2830 (2011). [Google Scholar]
- 42.Haeussler M, Download, convert and process the full text of scientific articles: Maximilianh/pubMunch3 (2018); https://github.com/maximilianh/pubMunch3. [Google Scholar]
- 43.Wang K, Li M, Hakonarson H, ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tange O, Gnu parallel—The command-line power tool. The USENIXMag. 36, 42–47 (2011). [Google Scholar]
- 45.Li H, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio] (2013). [Google Scholar]
- 46.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ, A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zemojtel T, Köhler S, Mackenroth L, Jäger M, Hecht J, Krawitz P, Graul-Neumann L, Doelken S, Ehmke N, Spielmann M, Øien NC, Schweiger MR, Krüger U, Frommer G, Fischer B, Kornak U, Flöttmann R, Ardeshirdavani A, Moreau Y, Lewis SE, Haendel M, Smedley D, Horn D, Mundlos S, Robinson PN, Effective diagnosis of genetic disease by computational phenotype analysis of the disease-associated genome. Sci. Transl.Med 6, 123 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson PN, Köhler S, Oellrich A; Sanger Mouse Genetics Project, Wang K, Mungall CJ, Lewis SE, Washington N, Bauer S, Seelow D, Krawitz P, Gilissen C, Haendel M, Smedley D, Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res. 24, 340–348 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singhal A, Simmons M, Lu Z, Text mining genotype-phenotype relationships from biomedical literature for database curation and precision medicine. PLOS Comput. Biol 12, e1005017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doughty E, Kertesz-Farkas A, Bodenreider O, Thompson G, Adadey A, Peterson T, Kann MG, Toward an automatic method for extracting cancer- and other disease-related point mutations from the biomedical literature. Bioinformatics 27, 408–415 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xing W, Qi J, Yuan X, Li L, Zhang X, Fu Y, Xiong S, Hu L, Peng J, A gene–phenotype relationship extraction pipeline from the biomedical literature using a representation learning approach. Bioinformatics 34, i386–i394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coulet A, Shah N, Garten Y, Musen M, Altman RB, Using text to build semantic networks for pharmacogenomics. J. Biomed. Inform 43, 1009–1019 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei C-H, Kao H-Y, Lu Z, GNormPlus: An integrative approach for tagging genes, gene families, and protein domains. Biomed. Res. Int 2015, 918710 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campos D, Matos S, Lewin I, Oliveira JL, Rebholz-Schuhmann D, Harmonization of gene/protein annotations: Towards a gold standard MEDLINE. Bioinformatics 28, 1253–1261 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Chun H-W, Tsuruoka Y, Kim J-D, Shiba R, Nagata N, Hishiki T, Tsujii J, Extraction of gene-disease relations from Medline using domain dictionaries and machine learning. Pac. Symp. Biocomput 2006, 4–15 (2006). [PubMed] [Google Scholar]
- 56.Özgür A, Vu T, Erkan G, Radev DR, Identifying gene-disease associations using centrality on a literature mined gene-interaction network. Bioinformatics 24, i277–i285 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rindflesch TC, Tanabe L, Weinstein JN, Hunter L, EDGAR: Extraction of drugs, genes and relations from the biomedical literature. Pac. Symp. Biocomput 2000, 517–528 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng D, Knox C, Young N, Stothard P, Damaraju S, Wishart DS, PolySearch: A web-based text mining system for extracting relationships between human diseases, genes, mutations, drugs and metabolites. Nucleic Acids Res. 36, W399–W405 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Collier N, Groza T, Smedley D, Robinson PN, Oellrich A, Rebholz-Schuhmann D, PhenoMiner: From text to a database of phenotypes associated with OMIM diseases. Database 2015, bav104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J, Kim J-J, Lee H, An analysis of disease-gene relationship from Medline abstracts by DigSee. Sci. Rep 7, 40154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Number of phenotypes associated with genes through articles in the AMELIE knowledgebase.
Fig. S2. The accelerated accumulation of curatable facts in Mendelian genomics.
Fig. S3. Replication of AMELIE’s causative gene ranking performance on 56 clinical patients from Stanford and Manton.
Fig. S4. Cross-validation of AMELIE’s causative gene ranking performance on 681 simulated patients.
Fig. S5. Essence of the AMELIE interface at https://AMELIE.stanford.edu.
Table S1. Full-text gene extraction statistics.
Table S2. Extraction statistics from the 100 most used journals.
Table S3. Simulated patient details.
Table S4. DDD patient details.
Table S5. Searching for top-ranked AMELIE articles in OMIM.
Table S6. Stanford and Manton clinical patient details.
Table S7. Simulated patient gene ranking results.
Table S8. Most important features for patients with top-ranked causative genes.
Table S9. AMELIE classifier feature ablation results.
Table S10. DisGeNET gene ranking results.
Table S11. Regular expression patterns used to parse variant type from OMIM allelic variant entries.
Table S12. Phenotypes extracted from full-text articles by AMELIE, indicating whether the phenotype was extracted correctly or not.
Table S13. Assignment of features to feature groups.