Abstract
Background
The prognosis of non-small cell lung cancer (NSCLC) with brain metastases (BMs) had been researched in some researches, but the combination of clinical characteristics and serum inflammatory indexes as a noninvasive and more accurate model has not been described.
Methods
We retrospectively screened patients with BMs at the initial diagnosis of NSCLC at Sun Yat-Sen University Cancer Center. LASSO-Cox regression analysis was used to establish a novel prognostic model for predicting OS based on blood biomarkers. The predictive accuracy and discriminative ability of the prognostic model was compared to Adjusted prognostic Analysis (APA), Recursive Partition Analysis (RPA), and Graded Prognostic Assessment (GPA) using concordance index (C-index), time-dependent receiver operating characteristic (td-ROC) curve, Decision Curve Analysis(DCA), net reclassification improvement index (NRI), and integrated discrimination improvement index (IDI).
Results
10-parameter signature's predictive model for the NSCLC patients with BMs was established according to the results of LASSO-Cox regression analysis. The C-index of the prognostic model to predict OS was 0.672 (95% CI = 0.609 ~ 0.736) which was significantly higher than APA,RPA and GPA. The td-ROC curve and DCA of the predictive model also demonstrated good predictive accuracy of OS compared to APA, RPA and GPA. Moreover, NRI and IDI analysis indicated that the prognostic model had improved prediction ability compared with APA, RPA and GPA.
Conclusion
The novel prognostic model demonstrated favorable performance than APA, RPA, and GPA for predicting OS in NSCLC patients with BMs.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-022-02671-2.
Keywords: Brain metastases, Non-small cell lung cancer, Prognostic model, Serum inflammatory indexes, LASSO-Cox regression analysis
Introduction
According to research, almost one third of non-small cell lung cancer (NSCLC) patients will have brain metastases (BMs) and it constitutes the most common source of BMs [1], and the median survival time is only 1 month if without effective treatment [2]. BMs are the most common malignant brain tumors in adults. Over the past decade, the incidence of BMs has increased due to improvements in the diagnosis and systematic treatment of extracranial diseases [3]. However, it is difficult to treat BMs with systemic chemotherapy since the agents have difficulty crossing the blood–brain barrier. Therefore, it is important to estimate the patient's survival prognosis.
Some models that have been reported to predict the prognosis of patients with BMs include Recursive Partition Analysis (RPA) [4, 5], the Score Index for Radiosurgery (SIR) [6], Basic Score for Brain Metastases(BSBM) [7] and Graded Prognostic Assessment (GPA) [8]. Nowadays, the most widely used prognostic model are the RPA and GPA. RPA is based on 4 stratifying factors (Karnofsky Performance Status (KPS), age, extracranial distant metastases and controlled primary tumor), while GPA considers the amount of brain damage as an additional prognostic factor for BMS on the basis of RPA. RPA and GPA models are general prognostic models for BMs which are not directly concerned with lung cancer, unfortunately. In 2017, some researches [9] established a new prognostic model for NSCLC: Adjusted prognostic Analysis (APA) which includes new prognostic indicators smoking history and EGFR genotype. All previously published models have their respective advantages (easy to use and remember) and limitations (qualitative and subjective), and we hope to eliminate the components of other indicators that are difficult to quantify and/or subjective, such as the control of extracranial diseases. Therefore, it is necessary to focus on new markers and explore more reliable prognostic indicators to make up and update for the shortcomings of the existing prognostic models and improve the predictive value of clinical outcomes for NSCLC patients with BMs.
Blood biomarkers can be determined in a simple, quick, and stable manner, with negligible side effects to the patient, which commonly used to detect diagnostic, prognostic, and therapeutic decision-making markers of many cancers [10]. Inflammation is a tissue response to eliminate tissue damage. However, dysregulated inflammation is a recognized cause of cancer [11]. Numerous studies have shown that serum inflammatory indexes are closely related to the prognosis of lung cancer, such as alanine aminotransferase (ALT) [12, 13], Body Mass Index(BMI), serum albumin (ALB) [13, 14], prognostic nutritional index (PNI), systemic immune-inflammation index (SII) [15], lymphocyte count and Neutrophil to lymphocyte ratio(NLR) [16]. Nonetheless, few researchers have utilized a combination of clinical characteristics and serum inflammatory indexes in order to predict a prognosis of NSCLC patients with BMs.
Therefore, this study aims to establish a new prognostic model based on clinical characteristics and inflammation indicators by using LASSO-Cox regression analysis, so as to more accurately reflect the prognostic information of BMs in NSCLC patients comparing with APA, RPA and GPA. And to assess its incremental value in traditional prognostic models and provide a basis for clinicians to formulate reasonable treatment plans.
Materials and methods
Patient population and follow up
We retrospectively screened patients with BMs at the initial diagnosis of NSCLC who received treatment at Sun Yat-Sen University Cancer Center from January 2011 to December 2015. The eligibility criteria included: (1)pathologically confirmed and treatment naive NSCLC; (2) BMs confirmed by brain MRI; (3)available baseline clinical information, laboratory data, and all data collection prior to antitumor treatments; (4) patients did not suffer from any cancer disease prior to NSCLC diagnosis;(5) all deaths were cancer-related.
The survival data of each patient was obtained by reviewing medical records, emailing, and direct telecommunication. The last follow-up was conducted in April 2021. Overall survival (OS) was defined as the time interval from date of diagnosis to the date of patient’s death or last follow-up.
Data collection
Baseline clinical characteristics were collected from the patients’ medical records patient characteristics (gender, age group, smoking history, KPS, BMI and family history), disease characteristics (tumor histology, location, extracranial distant metastases, number of brain metastatic lesions, EGFR mutation status), treatments (thoracic local treatment, EGFR-tyrosine kinase inhibitors (TKIs) treatment, Intracranial metastases local treatment, chemotherapy), previously published models information(RPA,GPA,APA), and the serum inflammatory indexes were collected included white blood cells (WBC), neutrophils (N), lymphocytes (L), platelets (PLT), NLR, platelet/lymphocyte ratio (PLR), derived neutrophil/lymphocyte ratio (dNLR), SII: SII was calculated by the formula: platelet(PLT) × neutrophil (N)/lymphocyte (L) [17], PNI: PNI was calculated by the formula ALB (g/L) + 5 × lymphocyte count × 109/L [18], ALT, aspartate aminotransferase (AST), ALT/AST ratio (LSR), lactate dehydrogenase (LDH), ALB, C-reactive protein (CRP), ALB/CRP ratio (ACR). Advance lung cancer inflammation index(ALI): ALI was calculated by the formula: BMI × Alb/NLR [12]. Patients were randomized intodivided into Derivation cohort (70%) and Validation cohort (30%).
Statistical analysis
Statistical analysis was conducted using R software (version 3.6.1). Continuous variables were represented as mean ± SD and analyzed using t-test or Wilcoxon test. Firstly, we utilized the LASSO-Cox regression algorithm, for which the λ value was determined by tenfold cross validation with the error of the minimum criteria to choose the most useful prognostic factors among all NSCLC-associated serum inflammatory indexes. Then a prognostic model was constructed to predict OS based on the coefficients of significant predictors that were derived from the LASSO-Cox regression. The following formula was used to calculate the risk score:
Subsequently, the predictive accuracy and discriminative ability of the novel prognostic model was compared with RPA,GPA, and APA using the Harrell concordance index (C-index), time-dependent ROC (td-ROC) curves [19], and Decision Curve Analysis(DCA) [20]. The larger C-index and area under the curve (AUC) of td-ROC curves, the better the model was for risk prediction. DCA make curves of different point cuts, pay attention to the relationship between benefits and risks brought by different point cuts in different models, and calculate the improvement after reclassification [21]. Pearson correlation coefficient was used to assess the correlation between the new prognostic model, RPA,GPA and APA. In addition, we developed a nomogram that integrates the prognostic model, RPA,GPA, and APA that may assist in individual survival prediction of NSCLC patients with BMs. Internal validation and calibration of the nomogram were performed using 1000-resample bootstrapping. Finally, we illustrated discrimination by dividing patients into low-risk groups and high-risk groups as per the novel predictive model scores. The Kaplan–Meier method was used to perform OS analysis. The log-rank test was utilized to compare significance of the differences of survival distribution between the groups. Generally, a p value ≤ 0.05 was considered statistically significant across all analyses.
Results
Patient characteristics
A total of 171 consecutive patients (121 in derivation cohort and 50 in validation cohort) with newly diagnosed BMs from NSCLC were enrolled. 94 (55.0%) of these patients were male, and 77 (45.0%) were female. The median age was 57 (inter quartile range [IQR], 51.0–62.0) years. The median follow-up for OS was 21 months. The 1-, 3-, and 5-year OS rates were 66.7%, 39.8%, and 30.4%, respectively. Only 81 patients (66.9%) in the derivation cohort had an identified EGFR genotype whereas more patients in the validation cohort had known EGFR (42, 84.0%) status. Most of other characteristics were similar between the two cohorts. The patients’ baseline characteristics are listed in supplement Table 1.
Table 1.
The C-index of the our prognostic model, APA, RPA and GPA for prediction of OS in the derivation cohort and validation cohort
| Factors | C-index (95% CI) | P value |
|---|---|---|
| For derivation cohort | ||
| Our model | 0.672 (0.609 ~ 0.736) | |
| APA model | 0.597 (0.537 ~ 0.657) | |
| RPA model | 0.517 (0.469 ~ 0.566) | |
| GPA model | 0.514 (0.448 ~ 0.579) | |
| Our model vs APA model | 0.049* | |
| Our model vs RPA model | < 0.001* | |
| Our model vs GPA model | < 0.001* | |
| For validation cohort | ||
| Our model | 0.738 (0.657 ~ 0.819) | |
| APA model | 0.637 (0.550 ~ 0.724) | |
| RPA model | 0.520 (0.456 ~ 0.585) | |
| GPA model | 0.634 (0.548 ~ 0.720) | |
| Our model vs APA model | 0.024* | |
| Our model vs RPA model | < 0.001* | |
| Our model vs GPA model | 0.052 | |
C-index concordance index, CI confidence interval; P values are calculated based on normal approximation using function rcorrp.cens in Hmisc package. *P < 0.05
Construction of prognostic model for OS
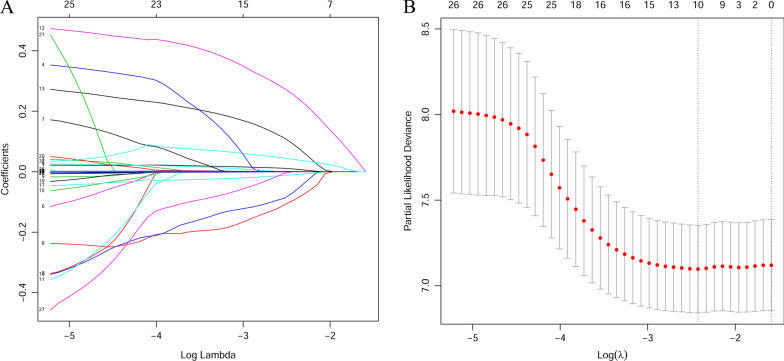
Firstly, LASSO-Cox regression analysis was carried out to extract significant predictors associated with OS in NSCLC patients with BMs. Figure 1A showed the analysis of the trajectory changes of each predictor variable. After, the optimal value for λ was determined through the use of tenfold cross-validation with minimum criteria (Fig. 1B). According to the criteria, the optimal value of λ in this study was 0.088. Its corresponding predictors were considered to be significant prognostic factors for OS, including age, Chemotherapy, TKIs, EGFR, Thoracic local treatment, ALB, ACR, LDH, ALI, and WBC. Finally, a prognostic model was constructed to predict OS based on the coefficients of the ten predictors that were derived from the LASSO-Cox regression. The following formula was used to calculate the risk score: the prognostic model risk score = "age"*0.0093—"Chemotherapy"*0.0950—"TKIs"*0.0690 + "EGFR"*0.2551 + "Thoracic local treatment"*0.0918 − "ALB"*0.0102 − "ACR"*0.0004 + "LDH"*0.0006 − "ALI"*0.0001 + "WBC" *0.0409. In the formula, the values of serum variables represent the respective serum original levels, and the code of clinical characteristic variables are listed in supplement Table 1.
Fig. 1.
7value for λ was determined using tenfold cross-validation with the minimum criteria(Color should be used for any figures in print)
Assessing performance between the novel prognostic model, APA, RPA and GPA
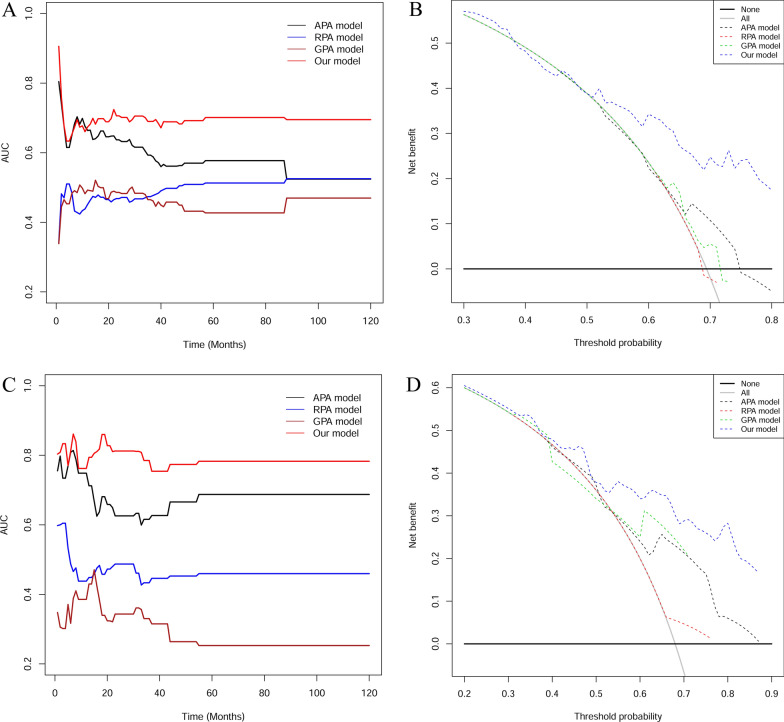
In order to evaluate the predictive values of the novel prognostic model for APA, RPA and GPA of OS, we introduced C-index, td-ROC curve, and DCA to evaluate the predictive accuracy of them. Firstly, we calculated the C-index of the four predictive model in the derivation cohort (Table 1). For OS, the C-index of our model was 0.672 (95% CI = 0.609 ~ 0.736), which was significantly higher than that of the APA [0.597 (95% CI = 0.537 ~ 0.657), p = 0.049], RPA [0.517(95% CI = 0.469 ~ 0.566), p < 0.001], and GPA [0.514 (95% CI = 0.448 ~ 0.579), p < 0.001]. Secondly, we plotted the td-ROC curves and calculated its corresponding AUCs. Results showed that AUCs of our model were higher compared to that of APA, RPA, and GPA with regards to OS at different time points (Fig. 2A). Thirdly, we drew graphs of threshold probabilities and net benefits for different prognostic models, and put them together to form a comparison of DCA for different prognostic models. Results demonstrated that the GPA and RPA curves are very close to the extremes and have little clinical value. APA's benefit is higher than the extreme curve, but it is still much lower than our model's which has a very high benefit over a wide threshold range. This means that compared with the old models, our model has the most practical implications for clinical practice (Fig. 2B). Similarly, we compared C-index (Table 1), AUCs (Fig. 2C) and DCA (Fig. 2D) in validation cohort and the results were consistent with the above.
Fig. 2.
The results of AUCs and DCA in cohorts. The dynamic AUC levels of the four models in derivation cohort (A) and validation cohort (C). DCA for different prognostic models in derivation cohort (B) and validation cohort (D)
Finally, both IDI and NRI calculations were utilized to compare alternative prognostic indices of our model with other models. Positive value represents better accuracy while negative value represents worse accuracy. The results were presented in Table 2. For derivation cohort, IDI analysis indicated that accuracy of our prognostic model was higher compared to that of the APA (0.138, p < 0.001), RPA (0.163, p < 0.001), and GPA (0.156, p < 0.001). NRI analysis indicated that accuracy of our prognostic model was higher compared to that of the APA (0.283, p < 0.001), RPA (0.283, p < 0.001), and GPA (0.388, p < 0.001). The results in validation cohort are similar to the derivation cohort (Table 2).
Table 2.
The IDI and NRI were used to assess reclassification performance and improvement in discrimination of our novel prediction model
| IDIa | P Value | NRIb | P value | |
|---|---|---|---|---|
| For derivation cohort | ||||
| Our model vs APA model | 0.138 | < 0.001* | 0.283 | < 0.001* |
| Our model vs RPA model | 0.163 | < 0.001* | 0.283 | < 0.001* |
| Our model vs GPA model | 0.156 | < 0.001* | 0.388 | < 0.001* |
| For validation cohort | ||||
| Our model vs APA model | 0.139 | 0.158 | 0.272 | 0.178 |
| Our model vs RPA mode | 0.235 | < 0.001* | 0.460 | 0.020* |
| Our model vs GPA model | 0.108 | 0.198 | 0.246 | 0.099 |
IDI integrated discrimination improvement index, NRI net reclassification improvement index. *P < 0.05
a,bPositive velue represents better accuracy, negative velue represents worse accuracy
Construction of a predictive nomogram based on prognostic model, APA, RPA, and GPA
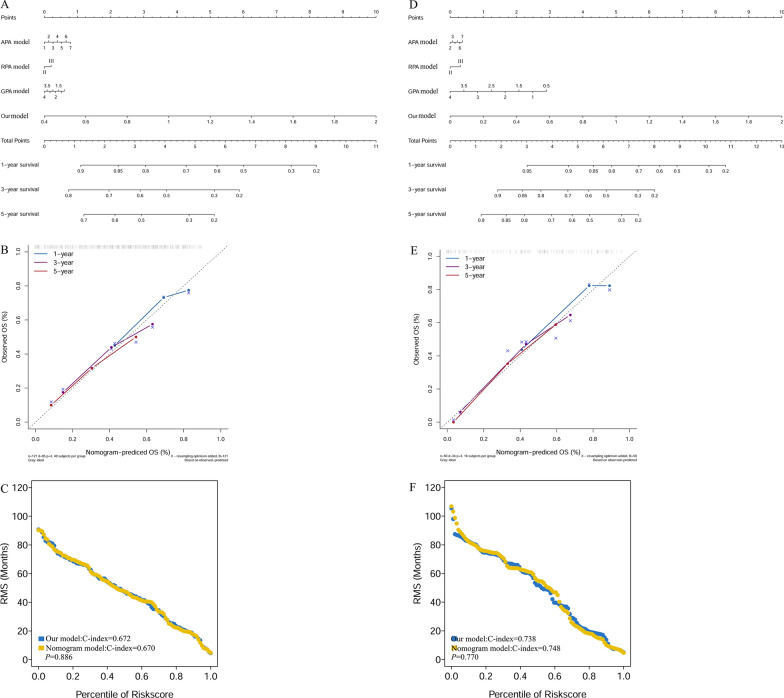
The nomogram incorporated the prognostic model, APA, RPA, and GPA to predict the probability of 1-, 3-, and 5-year OS (Fig. 3A) in the derivation cohort. Each patient was assigned a score for each prognostic variable. All scores were added together to estimate the probability of 1-, 3-, and 5- years OS. The higher the total score, the worse the patient’s prognosis. Besides, the calibration curve indicated good agreement between prediction and observation in 1-, 3-, and 5-year OS (Fig. 3B). Furthermore, we calculated the C-index of all predictive models in the derivation cohort. The results showed that the prognostic model has the same predictive value as nomogram models (p = 0.886, Fig. 3C). The differences of C-index between other models in derivation cohort are detailed in subTable 1. Similarly, we construct nomogram model of the validation cohort (Fig. 3D) and analyzed the calibration curves of nomogram model (Fig. 3E) and compared C-index between nomogram model and our model (Fig. 3F). The results are similar to the derivation cohort. The comparison of C-index among all the five models in validation cohort is shown in supplement Table2.
Fig. 3.
Construction of predictive nomogram and comparisons.The nomogram and calibration plots for estimating OS at 1, 3, and 5 years in derivation cohort (A, B), and validation cohort (D, E). The differences of C-index between nomogram model and our model in the derivation cohort (C) and validation cohort (F)
The correlation between the novel prognostic models and other models
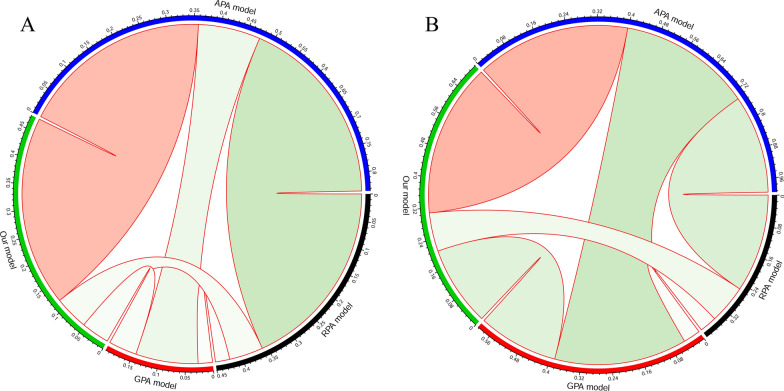
We evaluated the correlation between the prognostic model and other models (Fig. 4). In this plot, the green represented negative correlation and red represented positive correlation. The size of a circle and color intensity were directly proportional to the correlation coefficients. We utilized Pearson's correlation coefficient to determine a significant linear correlation between the variables. The results demonstrated that our prognostic model was positively correlated with APA (correlation coefficient = 0.357, p < 0.001) while with other models were negatively correlated in derivation cohort. There were same correlation trends and differences in validation cohort showed in Table 3.
Fig. 4.
The correlations between the prognostic model, APA, RPA, and GPA. The red represented positive correlation and the green represented negative correlation. Significant linear dependence between variables was identified using Pearson's correlation coefficient (PCC)
Table 3.
The correlation between our model and other models
| Models | Correlation coefficients a | P value |
|---|---|---|
| For training cohort | ||
| Our model vs APA model | 0.357 | < 0.001* |
| Our model vs RPA model | − 0.064 | 0.486 |
| Our model vs GPA model | − 0.053 | 0.564 |
| For external validation cohort | ||
| Our model vs APA model | 0.398 | 0.004* |
| Our model vs RPA model | − 0.100 | 0.490 |
| Our model vs GPA model | − 0.208 | 0.148 |
aPearson's correlation coefficient. *P < 0.05
Survival analyses according to the prognostic model, APA, RPA, and GPA
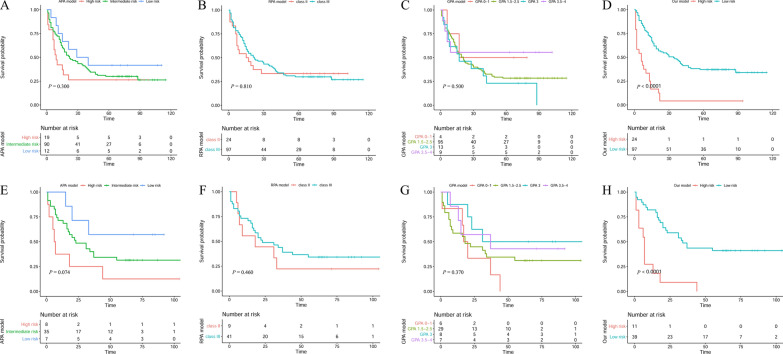
We classified patients into low-risk patients and high-risk patients based on the prognostic model risk score and made the Kaplan–Meier curve for our prognostic model. The APA, RPA, and GPA were grouped according to their respective classification methods. In the derivation cohort, the results indicated that patients with higher risk scores (risk score > 1.33) had a significantly lower OS (Fig. 5D; p < 0.001) rate compared to the low-risk (risk score ≤ 1.33) counterparts. So did the validation cohort (Fig. 5H, p < 0.001). However, patients could not be effectively distinguished between different risk groups in derivation cohort (Fig. 5A–C) and in validation cohort (Fig. 5E–G) based on APA, RPA, and GPA. The results indicated that our prognostic model had improved performance in distinguishing the prognosis of patients in NSCLC with BMs than others.
Fig. 5.
Kaplan–Meier analysis in different models. APA, RPA, GPA, and our prognostic model in derivation cohort (A–D) and in validation cohort (E–H)
Discussion
In this study, we analyzed the clinical characteristics and serum inflammatory indexes of individuals based on survival analysis to establish a novel prognostic model to predict OS in NSCLC patients with BMs by using LASSO-Cox regression algorithm. Compared to traditional prognostic model APA, RPA, and GPA, our prognostic model had improved prediction accuracy and ability to distinguish between the groups. Our prognostic model successfully classified those patients into high-risk and low-risk subgroups, which were significantly different with regards to OS.
Algorithm LASSO-Cox regression analysis has been applied across many studies as a statistical method because of adjusting the model’s over fitting to avoid extreme predictions and significantly improving the prediction accuracy [22–24]. In this research, we utilized the new algorithm LASSO-Cox regression analysis, we identified a ten individual prognostic factors (some were reported and others were new) and incorporated them into a novel predictive model to predict OS in NSCLC patients with BMs. Age is a factor common to all four models, while chemotherapy, TKIs, and EGFR mutation status are specific to APA. This may be explained by the specificity of APA focusing on predicting patients in NSCLC with BMs, while other models are common to all kinds of tumors with BMs. In our model, we identified six new individual prognostic factors (thoracic local treatment, ALB, ACR, LDH, ALI, and WBC) which had not been considered in all those previously published prognosis models. Although surgery is the main treatment for early lung cancer, it had been reported that surgery for the primary tumor was associated with a superior patient outcome in NSCLC with BMs [25].
In 2013, Jafri SH et al. proposed a new prognostic index ALI for patients with advanced lung cancer [12]. ALI is combined with BMI, plasma ALB, and NLR [16, 26, 27]. Previous studies have shown that ALI has certain prognostic value for patients with esophageal cancer, lung cancer and malignant lymphoma [28]. BMI and serum ALB are not only represent the nutritional status of patients to some extent, but also closely related to tumor progression [13]. Besides, malnutrition is positively correlated with the decrease of quality of life score and the severity of patients' symptoms [14]. Carcinogenesis is a complex, stepwise process that involves the acquisition of genetic mutations and epigenetic changes [29], the environmental and hereditary factors, stochastic effects [30] and inflammation [31, 32]. Rodents animal studies have revealed that chronic inflammation significantly enhances lung carcinogenesis, and inhibition of inflammation suppresses cancer progression and reduces the tumor volume [33]. Besides, the role of inflammation in increasing the risk of lung tumorigenesis driven primarily by oncogenic KRAS has been researched and the results showed that inflammatory responses may increase KRAS mutation rate and create a vicious cycle of chronic inflammation and KRAS mutation [34]. Fortunately, many studies have clarified the molecular mechanism and roles of chronic inflammation in lung cancer [35] and various immune cells, cytokines and signaling pathways participate in inflammation mediated lung carcinogenesis [33]. Acute lung inflammation is dominated by neutrophils, whereas chronic reactions mainly involve macrophages and lymphocytes [35], so it is easy to understand the predictive value of WBC in cancers. In 2021, Berghoff [36] et al. also reported inflammatory markers were associated with OS in patients with newly diagnosed brain metastases. Compare to that study, we focused on the prognosis of patients with BMS in NSCLC only, making the study more targeted for clinical application. More importantly, our study is a quantitative prognostic risk model jointly constructed by multiple indicators, which is convenient for doctors to conduct personalized assessment and treatment of patients.
Increasing uptake of glucose and preferential conversing of glucose to lactate are a generic feature in types of cancer although the precise role of the Warburg effect [37] is not fully understood. LDH is the enzyme responsible for conversion of pyruvate to lactate at the endpoint of glycolysis. LDH regulates the rapid growth of tumor cells and makes the disease progress [38–40]. Studies have shown that elevated serum LDH concentration can lead to poor prognosis of lung cancer patients with different pathological types [41–46] and be useful in monitoring of treatment in advanced NSCLC [47]. The preoperative LDH concentration and postoperative LDH concentration change trend were independent prognostic factors for patients in lung large-cell neuroendocrine carcinoma (L-LCNEC) [48]. Based on the evidence above, LDH as a predictor in our model is valid and credible.
Compared to previous models, the novel model had several advantages. Including more potential prognostic factors in which serum inflammatory factors are mentioned for the first time is the most striking feature in our study. This method significantly improves the accuracy compared with traditional COX regression analysis. However, there were still some limitations in this study. Selection bias may be unavoidable in all retrospective analysis, and especially in this single cancer center with a small sample size relatively. So it is necessary to carry out multi-center and large-scale studies in the future to further verify the generalizability of our prognostic model established in this study. Although these predictors in our model were easy to obtain, it was undeniable that they were all non-specific predictors for NSCLC with BMs. NSCLC-related immunohistochemical markers [49] may be incorporated into prognostic models to improve specificity, such as PD-1 [50–53], EML4-ALK [54–58], and VEGF [59–61]. In addition, we established the model with initial diagnosis data, so we could not know the prognosis of the patient after each treatment. We can also collaborate clinically in the future, focusing on the establishment of prognostic models related to treatment duration.
Conclusions
In summary, we established a novel prognostic model successfully based on clinical characteristics and serum inflammatory factors which outperformed APA, RPA, and GPA in predicting OS in NSCLC patients with BMs. This prognostic model may act as a potential tool for clinicians to provide consultation, personalized treatment and follow-up for NSCLC patients with BMs due to the low cost, easy operation, precision, and stability. However, the wide practical application of this model required more clinical data and multi-center verification to verify the accuracy of our model in predicting prognosis of NSCLC patients with BMs.
Supplementary Information
Additional file 1: Table S1. Clinical characteristics of derivation and validation cohorts.
Acknowledgements
Not applicable.
Abbreviations
- NSCLC
Non-small cell lung cancer;BMs: brain metastases;
- C-index
Concordance index
- td-ROC
Time-dependent receiver operating characteristic curve
- KPS
Karnofsky Performance Status
- DCA
Decision Curve Analysis
- NRI
Net reclassification improvement index
- IDI
Integrated discrimination improvement index
- BMI
Body Mass Index
- EGFR
Epidermal Growth Factor Receptor
- TKIs
EGFR-tyrosine kinase inhibitors
- APA
Adjusted prognostic Analysis
- RPA
Recursive Partition Analysis
- GPA
Graded Prognostic Assessment
- WBC
White blood cells
- NLR
Neutrophil/lymphocyte ratio
- PLR
Platelet/lymphocyte ratio
- dNRI
Derived neutrophil/lymphocyte ratio
- SII
Systemic immune-inflammation index
- PNI
Prognostic nutritional index
- ALB
Albumin
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- LSR
ALT/AST ratio
- LDH
Lactate dehydrogenase
- CRP
C-reactive protein
- ACR
ALB/CRP ratio
- ALI
Advance lung cancer inflammation index
Author contributions
SC, XL and WG: conceptualization and visualization; YL, XW, and LT: investigation and data curation; SC: funding acquisition; SC and XL: methodology, software and formal analysis; SY: supervision; XL and SY: writing- original draft; SY and WG: writing-review & editing.
Funding
This work was funded by the Guangdong Esophageal Cancer Institute(Guangdong Esophageal Cancer Institute Science and Technology Program, No.Q202001).
Availability of data and materials
The datasets analysed during the current study are available in the Research Data Deposit public platform (www.researchdata.org.cn) provided by RDD Management Committee in the Sun Yat-sen University Cancer Center.
Declarations
Ethics approval and consent to participate
This study was granted approval by the Clinical Research Ethics Committee of the Sun Y at-sen University Cancer Center. All patients were provided with written informed consent at the first visit. This work complies with the principles laid down in the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors report no competing interests in this work.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaohui Li, Wenshen Gu, and Yijun Liu contributed equally to this work
Contributor Information
Shumei Yan, Email: yanshm@sysucc.org.cn.
Shulin Chen, Email: chenshl@sysucc.org.cn.
References
- 1.Kvale PA, Simoff M, Prakash UB. Lung cancer. Palliative Care CHEST. 2003;123(1 Suppl):284S–311S. doi: 10.1378/chest.123.1_suppl.284s. [DOI] [PubMed] [Google Scholar]
- 2.Pesce GA, Klingbiel D, Ribi K, Zouhair A, von Moos R, Schlaeppi M, Caspar CB, Fischer N, Anchisi S, Peters S, Cathomas R, Bernhard J, Kotrubczik NM, D'Addario G, Pilop C, Weber DC, Bodis S, Pless M, Mayer M, Stupp R. Outcome, quality of life and cognitive function of patients with brain metastases from non-small cell lung cancer treated with whole brain radiotherapy combined with gefitinib or temozolomide. A randomised phase II trial of the Swiss Group for Clinical Cancer Research (SAKK 70/03) Eur J Cancer. 2012;48(3):377–384. doi: 10.1016/j.ejca.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 3.Ricciardi S, de Marinis F. Multimodality management of non-small cell lung cancer patients with brain metastases. Curr Opin Oncol. 2010;22(2):86–93. doi: 10.1097/CCO.0b013e3283350106. [DOI] [PubMed] [Google Scholar]
- 4.Gaspar LE, Scott C, Murray K, Curran W. Validation of the RTOG recursive partitioning analysis (RPA) classification for brain metastases. Int J Radiat Oncol Biol Phys. 2000;47(4):1001–1006. doi: 10.1016/S0360-3016(00)00547-2. [DOI] [PubMed] [Google Scholar]
- 5.Gaspar L, Scott C, Rotman M, Asbell S, Phillips T, Wasserman T, McKenna WG, Byhardt R. Recursive partitioning analysis (RPA) of prognostic factors in three Radiation Therapy Oncology Group (RTOG) brain metastases trials. Int J Radiat Oncol Biol Phys. 1997;37(4):745–751. doi: 10.1016/S0360-3016(96)00619-0. [DOI] [PubMed] [Google Scholar]
- 6.Weltman E, Salvajoli JV, Brandt RA, de Morais HR, Prisco FE, Cruz JC, de Oliveira BS, Wajsbrot DB. Radiosurgery for brain metastases: a score index for predicting prognosis. Int J Radiat Oncol Biol Phys. 2000;46(5):1155–1161. doi: 10.1016/S0360-3016(99)00549-0. [DOI] [PubMed] [Google Scholar]
- 7.Lorenzoni J, Devriendt D, Massager N, David P, Ruiz S, Vanderlinden B, Van Houtte P, Brotchi J, Levivier M. Radiosurgery for treatment of brain metastases: estimation of patient eligibility using three stratification systems. Int J Radiat Oncol Biol Phys. 2004;60(1):218–224. doi: 10.1016/j.ijrobp.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 8.Sperduto PW, Berkey B, Gaspar LE, Mehta M, Curran W. A new prognostic index and comparison to three other indices for patients with brain metastases: an analysis of 1,960 patients in the RTOG database. Int J Radiat Oncol Biol Phys. 2008;70(2):510–514. doi: 10.1016/j.ijrobp.2007.06.074. [DOI] [PubMed] [Google Scholar]
- 9.Dinglin XX, Ma SX, Wang F, Li DL, Liang JZ, Chen XR, Liu Q, Zeng YD, Chen LK. Establishment of an adjusted prognosis analysis model for initially diagnosed non-small-cell lung cancer with brain metastases from sun Yat-Sen University Cancer Center. Clin Lung Cancer. 2017;18(3):e179–e186. doi: 10.1016/j.cllc.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Vandenbrouck Y, Christiany D, Combes F, Loux V, Brun V. Bioinformatics tools and workflow to select blood biomarkers for early cancer diagnosis: an application to pancreatic cancer. Proteomics. 2019;19(21–22):e1800489. doi: 10.1002/pmic.201800489. [DOI] [PubMed] [Google Scholar]
- 11.Dong J. Signaling pathways implicated in carbon nanotube-induced lung inflammation. Front Immunol. 2020;11:552613. doi: 10.3389/fimmu.2020.552613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jafri SH, Shi R, Mills G. Advance lung cancer inflammation index (ALI) at diagnosis is a prognostic marker in patients with metastatic non-small cell lung cancer (NSCLC): a retrospective review. BMC Cancer. 2013;13:158. doi: 10.1186/1471-2407-13-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakagawa T, Toyazaki T, Chiba N, Ueda Y, Gotoh M. Prognostic value of body mass index and change in body weight in postoperative outcomes of lung cancer surgery. Interact Cardiovasc Thorac Surg. 2016;23(4):560–566. doi: 10.1093/icvts/ivw175. [DOI] [PubMed] [Google Scholar]
- 14.Polanski J, Jankowska-Polanska B, Uchmanowicz I, Chabowski M, Janczak D, Mazur G, Rosinczuk J. Malnutrition and quality of life in patients with non-small-cell lung cancer. Adv Exp Med Biol. 2017;1021:15–26. doi: 10.1007/5584_2017_23. [DOI] [PubMed] [Google Scholar]
- 15.Li W, Qu Y, Wen F, Yu R, He X, Jia H, Liu H, Yu H. Prognostic nutritional index and systemic immune-inflammation index are prognostic biomarkers for non-small-cell lung cancer brain metastases. Biomark Med. 2021;15(13):1071–1084. doi: 10.2217/bmm-2020-0786. [DOI] [PubMed] [Google Scholar]
- 16.Berardi R, Rinaldi S, Santoni M, Newsom-Davis T, Tiberi M, Morgese F, Caramanti M, Savini A, Ferrini C, Torniai M, Fiordoliva I, Bower M, Cascinu S. Prognostic models to predict survival in patients with advanced non-small cell lung cancer treated with first-line chemo- or targeted therapy. Oncotarget. 2016;7(18):26916–26924. doi: 10.18632/oncotarget.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yilmaz A, Mirili C, Bilici M, Tekin SB. A novel predictor in patients with gastrointestinal stromal tumors: systemic immune-inflammation index (SII) J Buon. 2019;24(5):2127–2135. [PubMed] [Google Scholar]
- 18.He X, Li JP, Liu XH, Zhang JP, Zeng QY, Chen H, Chen SL. Prognostic value of C-reactive protein/albumin ratio in predicting overall survival of Chinese cervical cancer patients overall survival: comparison among various inflammation based factors. J Cancer. 2018;9(10):1877–1884. doi: 10.7150/jca.23320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamarudin AN, Cox T, Kolamunnage-Dona R. Time-dependent ROC curve analysis in medical research: current methods and applications. BMC Med Res Methodol. 2017;17(1):53. doi: 10.1186/s12874-017-0332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26(6):565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzgerald M, Saville BR, Lewis RJ. Decision curve analysis. JAMA. 2015;313(4):409–410. doi: 10.1001/jama.2015.37. [DOI] [PubMed] [Google Scholar]
- 22.Tian MX, Liu WR, Wang H, Zhou YF, Jin L, Jiang XF, Tao CY, Tang Z, Zhou PY, Fang Y, Qu WF, Ding ZB, Peng YF, Dai Z, Qiu SJ, Zhou J, Lau WY, Fan J, Shi YH. Tissue-infiltrating lymphocytes signature predicts survival in patients with early/intermediate stage hepatocellular carcinoma. BMC Med. 2019;17(1):106. doi: 10.1186/s12916-019-1341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu WH, Wu J, Wang J, Wan FN, Wang HK, Cao DL, Qu YY, Zhang HL, Ye DW. Screening and identification of potential prognostic biomarkers in adrenocortical carcinoma. Front Genet. 2019;10:821. doi: 10.3389/fgene.2019.00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z, Liang C, Tian J, Liang C. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology. 2016;281(3):947–957. doi: 10.1148/radiol.2016152234. [DOI] [PubMed] [Google Scholar]
- 25.Enders F, Geisenberger C, Jungk C, Bermejo JL, Warta R, von Deimling A, Herold-Mende C, Unterberg A. Prognostic factors and long-term survival in surgically treated brain metastases from non-small cell lung cancer. Clin Neurol Neurosurg. 2016;142:72–80. doi: 10.1016/j.clineuro.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 26.Sierzega M, Lenart M, Rutkowska M, Surman M, Mytar B, Matyja A, Siedlar M, Kulig J. Preoperative neutrophil-lymphocyte and lymphocyte-monocyte ratios reflect immune cell population rearrangement in resectable pancreatic cancer. Ann Surg Oncol. 2017;24(3):808–815. doi: 10.1245/s10434-016-5634-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salim DK, Mutlu H, Eryilmaz MK, Salim O, Musri FY, Tural D, Gunduz S, Coskun HS. Neutrophil to lymphocyte ratio is an independent prognostic factor in patients with recurrent or metastatic head and neck squamous cell cancer. Mol Clin Oncol. 2015;3(4):839–842. doi: 10.3892/mco.2015.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park YH, Yi HG, Lee MH, Kim CS, Lim JH. Prognostic value of the pretreatment advanced lung cancer inflammation index (ALI) in diffuse large B cell lymphoma patients treated with R-CHOP chemotherapy. Acta Haematol. 2017;137(2):76–85. doi: 10.1159/000452991. [DOI] [PubMed] [Google Scholar]
- 29.Gomperts BN, Spira A, Massion PP, Walser TC, Wistuba II, Minna JD, Dubinett SM. Evolving concepts in lung carcinogenesis. Semin Respir Crit Care Med. 2011;32(1):32–43. doi: 10.1055/s-0031-1272867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347(6217):78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakano Y, Yokohira M, Hashimoto N, Yamakawa K, Kishi S, Ninomiya F, Kanie S, Saoo K, Imaida K. Rat strain differences in levels and effects of chronic inflammation due to intratracheal instillation of quartz on lung tumorigenesis induced by DHPN. Exp Toxicol Pathol. 2014;66(8):391–401. doi: 10.1016/j.etp.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Zaynagetdinov R, Sherrill TP, Polosukhin VV, Han W, Ausborn JA, McLoed AG, McMahon FB, Gleaves LA, Degryse AL, Stathopoulos GT, Yull FE, Blackwell TS. A critical role for macrophages in promotion of urethane-induced lung carcinogenesis. J Immunol. 2011;187(11):5703–5711. doi: 10.4049/jimmunol.1100558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakano-Narusawa Y, Yokohira M, Yamakawa K, Ye J, Tanimoto M, Wu L, Mukai Y, Imaida K, Matsuda Y. Relationship between lung carcinogenesis and chronic inflammation in rodents. Cancers (Basel) 2021;13(12):2910. doi: 10.3390/cancers13122910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keohavong P, Peter DY. Pulmonary inflammation and KRAS mutation in lung cancer. Adv Exp Med Biol. 2021;1303:71–87. doi: 10.1007/978-3-030-63046-1_5. [DOI] [PubMed] [Google Scholar]
- 35.Moldoveanu B, Otmishi P, Jani P, Walker J, Sarmiento X, Guardiola J, Saad M, Yu J. Inflammatory mechanisms in the lung. J Inflamm Res. 2009;2:1–11. [PMC free article] [PubMed] [Google Scholar]
- 36.Starzer AM, Steindl A, Mair MJ, Deischinger C, Simonovska A, Widhalm G, Gatterbauer B, Dieckmann K, Heller G, Preusser M, Berghoff AS. Systemic inflammation scores correlate with survival prognosis in patients with newly diagnosed brain metastases. Br J Cancer. 2021;124(7):1294–1300. doi: 10.1038/s41416-020-01254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–270. doi: 10.1126/science.124.3215.269. [DOI] [PubMed] [Google Scholar]
- 38.Seagroves TN, Ryan HE, Lu H, Wouters BG, Knapp M, Thibault P, Laderoute K, Johnson RS. Transcription factor HIF-1 is a necessary mediator of the pasteur effect in mammalian cells. Mol Cell Biol. 2001;21(10):3436–3444. doi: 10.1128/MCB.21.10.3436-3444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5(5):343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 40.Ostergaard L, Tietze A, Nielsen T, Drasbek KR, Mouridsen K, Jespersen SN, Horsman MR. The relationship between tumor blood flow, angiogenesis, tumor hypoxia, and aerobic glycolysis. Cancer Res. 2013;73(18):5618–5624. doi: 10.1158/0008-5472.CAN-13-0964. [DOI] [PubMed] [Google Scholar]
- 41.Galvano A, Peri M, Guarini AA, Castiglia M, Grassadonia A, De Tursi M, Irtelli L, Rizzo S, Bertani A, Gristina V, Barraco N, Russo A, Natoli C, Bazan V. Analysis of systemic inflammatory biomarkers in neuroendocrine carcinomas of the lung: prognostic and predictive significance of NLR, LDH, ALI, and LIPI score. Ther Adv Med Oncol. 2020;12:431409878. doi: 10.1177/1758835920942378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Guo M, Fan J, Lv Z, Huang Q, Han J, Wu F, Hu G, Xu J, Jin Y. Prognostic significance of serum LDH in small cell lung cancer: a systematic review with meta-analysis. Cancer Biomark. 2016;16(3):415–423. doi: 10.3233/CBM-160580. [DOI] [PubMed] [Google Scholar]
- 43.Chen C, Zhu YH, Huang JA. Clinical evaluation of potential usefulness of serum lactate dehydrogenase level in follow-up of small cell lung cancer. J Cancer Res Ther. 2018;14(Supplement):S336–S340. doi: 10.4103/0973-1482.168994. [DOI] [PubMed] [Google Scholar]
- 44.Fiegl M, Pircher A, Waldthaler C, Gamerith G, Kocher F, Pall G, Nevinny M, Schmid T, Sterlacci W, Jamnig H, Zangerl G, Zabernigg A, Oberaigner W, Hilbe W. Small steps of improvement in small-cell lung cancer (SCLC) within two decades: a comprehensive analysis of 484 patients. Lung Cancer. 2014;84(2):168–174. doi: 10.1016/j.lungcan.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Jiang R, Li K. Prognostic significance of pretreatment laboratory parameters in combined small-cell lung cancer. Cell Biochem Biophys. 2014;69(3):633–640. doi: 10.1007/s12013-014-9845-3. [DOI] [PubMed] [Google Scholar]
- 46.Lee DS, Park KR, Kim SJ, Chung MJ, Lee YH, Chang JH, Kang JH, Hong SH, Kim MS, Kim YS. Serum lactate dehydrogenase levels at presentation in stage IV non-small cell lung cancer: predictive value of metastases and relation to survival outcomes. Tumour Biol. 2016;37(1):619–625. doi: 10.1007/s13277-015-3776-5. [DOI] [PubMed] [Google Scholar]
- 47.de Jong C, Deneer V, Kelder JC, Ruven H, Egberts T, Herder G. Association between serum biomarkers CEA and LDH and response in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Thorac Cancer. 2020;11(7):1790–1800. doi: 10.1111/1759-7714.13449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, Shan D, Dong Y, Yang X, Yu Z. Correlation analysis of serum LDH concentration before and after operation and prognosis of large cell neuroendocrine lung cancer patients. Zhongguo Fei Ai Za Zhi. 2021;24(5):351–356. doi: 10.3779/j.issn.1009-3419.2021.103.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osmani L, Askin F, Gabrielson E, Li QK. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52(Pt 1):103–109. doi: 10.1016/j.semcancer.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lantuejoul S, Sound-Tsao M, Cooper WA, Girard N, Hirsch FR, Roden AC, Lopez-Rios F, Jain D, Chou TY, Motoi N, Kerr KM, Yatabe Y, Brambilla E, Longshore J, Papotti M, Sholl LM, Thunnissen E, Rekhtman N, Borczuk A, Bubendorf L, Minami Y, Beasley MB, Botling J, Chen G, Chung JH, Dacic S, Hwang D, Lin D, Moreira A, Nicholson AG, Noguchi M, Pelosi G, Poleri C, Travis W, Yoshida A, Daigneault JB, Wistuba II, Mino-Kenudson M. PD-L1 testing for lung cancer in 2019: perspective from the IASLC Pathology Committee. J Thorac Oncol. 2020;15(4):499–519. doi: 10.1016/j.jtho.2019.12.107. [DOI] [PubMed] [Google Scholar]
- 51.Arpin D, Charpentier MC, Bernardi M, Monnet I, Boni A, Watkin E, Goubin-Versini I, Lamy R, Geriniere L, Geier M, Forest F, Gervais R, Madrosyk A, Guisier F, Serrand C, Locher C, Decroisette C, Fournel P, Auliac JB, Jeanfaivre T, Letreut J, Doubre H, Francois G, Piton N, Chouaid C, Damotte D. PD-L1-expression patterns in large-cell neuroendocrine carcinoma of the lung: potential implications for use of immunotherapy in these patients: the GFPC 03–2017 "EPNEC" study. Ther Adv Med Oncol. 2020;12:431414284. doi: 10.1177/1758835920937972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torous VF, Rangachari D, Gallant BP, Shea M, Costa DB, VanderLaan PA. PD-L1 testing using the clone 22C3 pharmDx kit for selection of patients with non-small cell lung cancer to receive immune checkpoint inhibitor therapy: are cytology cell blocks a viable option? J Am Soc Cytopathol. 2018;7(3):133–141. doi: 10.1016/j.jasc.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gniadek TJ, Li QK, Tully E, Chatterjee S, Nimmagadda S, Gabrielson E. Heterogeneous expression of PD-L1 in pulmonary squamous cell carcinoma and adenocarcinoma: implications for assessment by small biopsy. Mod Pathol. 2017;30(4):530–538. doi: 10.1038/modpathol.2016.213. [DOI] [PubMed] [Google Scholar]
- 54.Zhang SS, Nagasaka M, Zhu VW, Ou SI. Going beneath the tip of the iceberg. Identifying and understanding EML4-ALK variants and TP53 mutations to optimize treatment of ALK fusion positive (ALK+) NSCLC. Lung Cancer 2021; 158: 126–136. [DOI] [PubMed]
- 55.Rossi E, Aieta M, Tartarone A, Pezzuto A, Facchinetti A, Santini D, Ulivi P, Ludovini V, Possidente L, Fiduccia P, Minicuci N, Zamarchi R. A fully automated assay to detect the expression of pan-cytokeratins and of EML4-ALK fusion protein in circulating tumour cells (CTCs) predicts outcome of non-small cell lung cancer (NSCLC) patients. Transl Lung Cancer Res. 2021;10(1):80–92. doi: 10.21037/tlcr-20-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reclusa P, Laes JF, Malapelle U, Valentino A, Rocco D, Gil-Bazo I, Rolfo C. EML4-ALK translocation identification in RNA exosomal cargo (ExoALK) in NSCLC patients: a novel role for liquid biopsy. Transl Cancer Res. 2019;8(Suppl 1):S76–S78. doi: 10.21037/tcr.2018.11.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christopoulos P, Kirchner M, Endris V, Stenzinger A, Thomas M. EML4-ALK V3, treatment resistance, and survival: refining the diagnosis of ALK(+) NSCLC. J Thorac Dis. 2018;10(Suppl 17):S1989–S1991. doi: 10.21037/jtd.2018.05.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ulivi P, Puccetti M, Capelli L, Chiadini E, Bravaccini S, Calistri D, Zoli W, Amadori D, Candoli P. Molecular determinations of EGFR and EML4-ALK on a single slide of NSCLC tissue. J Clin Pathol. 2013;66(8):708–710. doi: 10.1136/jclinpath-2013-201502. [DOI] [PubMed] [Google Scholar]
- 59.Fu BH, Fu ZZ, Meng W, Gu T, Sun XD, Zhang Z. Platelet VEGF and serum TGF-beta1 levels predict chemotherapy response in non-small cell lung cancer patients. Tumour Biol. 2015;36(8):6477–6483. doi: 10.1007/s13277-015-3338-x. [DOI] [PubMed] [Google Scholar]
- 60.Schwaederle M, Lazar V, Validire P, Hansson J, Lacroix L, Soria JC, Pawitan Y, Kurzrock R. VEGF-A expression correlates with TP53 mutations in non-small cell lung cancer: implications for antiangiogenesis therapy. Cancer Res. 2015;75(7):1187–1190. doi: 10.1158/0008-5472.CAN-14-2305. [DOI] [PubMed] [Google Scholar]
- 61.Ma W, Xu M, Liu Y, Liu H, Huang J, Zhu Y, Ji LJ, Qi X. Safety profile of combined therapy inhibiting EFGR and VEGF pathways in patients with advanced non-small-cell lung cancer: a meta-analysis of 15 phase II/III randomized trials. Int J Cancer. 2015;137(2):409–419. doi: 10.1002/ijc.29377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Clinical characteristics of derivation and validation cohorts.
Data Availability Statement
The datasets analysed during the current study are available in the Research Data Deposit public platform (www.researchdata.org.cn) provided by RDD Management Committee in the Sun Yat-sen University Cancer Center.