Abstract
Remdesivir (RDV, Veklury®) is an FDA-approved prodrug for the treatment of hospitalized patients with COVID-19. Recent in vitro studies have indicated that human carboxylesterase 1 (CES1) is the major metabolic enzyme catalyzing RDV activation. COVID-19 treatment for hospitalized patients typically also involves a number of antibiotics and anti-inflammatory drugs. Further, individuals who are carriers of a CES1 variant (polymorphism in exon 4 codon 143 [G143E]) may experience impairment in their ability to metabolize therapeutic agents which are CES1 substrates. The present study assessed the potential influence of nine therapeutic agents (hydroxychloroquine, ivermectin, erythromycin, clarithromycin, roxithromycin, trimethoprim, ciprofloxacin, vancomycin, and dexamethasone) commonly used in treating COVID-19 and 5 known CES1 inhibitors on the metabolism of RDV. Additionally, we further analyzed the mechanism of inhibition of cannabidiol (CBD), as well as the impact of the G143E polymorphism on RDV metabolism. An in vitro S9 fraction incubation method and in vitro to in vivo pharmacokinetic scaling were utilized. None of the nine therapeutic agents evaluated produced significant inhibition of RDV hydrolysis; CBD was found to inhibit RDV hydrolysis by a mixed type of competitive and noncompetitive partial inhibition mechanism. In vitro to in vivo modeling suggested a possible reduction of RDV clearance and increase of AUC when coadministration with CBD. The same scaling method also suggested a potentially lower clearance and higher AUC in the presence of the G143E variant. In conclusion, a potential CES1-mediated DDI between RDV and the nine assessed medications appears unlikely. However, a potential CES1-mediated DDI between RDV and CBD may be possible with sufficient exposure to the cannabinoid. Patients carrying the CES1 G143E variant may exhibit a slower biotransformation and clearance of RDV. Further clinical studies would be required to evaluate and characterize the clinical significance of a CBD-RDV interaction.
Keywords: Remdesivir, Cannabidiol, CES1, G143E, COVID-19
Abbreviations
- RDV
Remdesivir
- CES1
Human carboxylesterase 1
- G143E
CES1 polymorphism in exon 4 codon 143
- CBD
Cannabidiol
- FDA
The Food and Drug Administration
- DME
Major drug-metabolizing enzyme
- CatA
Cathepsin A
- DDIs
Drug-drug interactions
- THC
Δ9-tetrahydrocannabinol
- CBN
Cannabinol
- IS
Internal standard
- [S]
Substrate concentration
- V
Reaction velocity
- Km
Substrate concentration at half of the maximum reaction velocity
- Vmax
Maximum reaction velocity
- [I]
Inhibitor concentration
- Rv
The ratio of reaction velocity with coadministration of inhibitor divided by reaction velocity in the control condition without inhibitor, expressed as percent
- γ
Exponent in the Hill equation
- IC
Inhibitor concentration producing 50% of maximum inhibition
- Emax
Maximum effect
- E0
Baseline effect
- [I]
The inhibitor concentration
- Ki
Inhibition constant
- α
The mix of non-competitive and competitive inhibition
- β
The ratio of remaining enzyme velocity when bound to the inhibitor
- AUCi
Area under the curve of victim drug with the presence of the inhibitor
- AUCo
Area under the curve of the victim drug in absence of the inhibitor
- CLint, i
Intrinsic clearance by CES1 of the victim drug with the presence of the inhibitor
- CLint, o
Intrinsic clearance by CES1 of the victim drug in absence of the inhibitor
- fm
Fraction of metabolism
- CLi
Clearance of the victim drug in presence of the inhibitor
- CLo
Clearance of the victim drug in absence of the inhibitor
- RAUC
Predicted change in the substrate drug AUC
- RCL
Predicted change in the substrate drug clearance
- MAF
Minor allele frequency
1. Introduction
Remdesivir (RDV, Veklury®) is a SARS-CoV-2 nucleotide analog RNA polymerase inhibitor [1] and the first FDA-approved antiviral to treat COVID-19 [2]. Intravenous remdesivir is approved by the Food and Drug Administration (FDA) for the treatment of mild to moderate COVID-19 in high-risk, nonhospitalized patients and for the treatment of hospitalized patients with COVID-19. Following intravenous administration, the major portion (>90%) of this prodrug is rapidly hydrolyzed to the intermediate metabolite GS-704277 by liver esterases, with a small portion of the drug (7.4%–9.9%) eliminated unchanged [3]. GS-704277 is then further metabolized by intracellular phosphoramides and phosphotransferases to its active form, GS-441524 [4]. Recent in vitro studies have indicated that carboxylesterase 1 (CES1) is the major drug-metabolizing enzyme (DME) catalyzing the initial hydrolyzing step of RDV metabolism with an additional contribution from cathepsin A (CatA) [5] (Fig. 1 ).
Fig. 1.
Metabolic pathway of RDV.
Drug-drug interactions (DDIs), as well as the presence of genetic variants of DMEs, can compromise drug therapy. Importantly, beyond antiviral treatment, patients hospitalized for COVID-19 typically receive a number of concomitant medications including off-label antivirals, antibiotics, anti-inflammatories, and others [6]. Many of these co-administered agents have documented effects on DMEs such as CYP3A [7,8], but few have undergone assessment for their potential influence on CES1, and accordingly, RDV metabolism. The present study utilized previously established in vitro methods [9] to assess the potential influence of nine therapeutic agents frequently utilized in the treatment of COVID-19 and its complications on the initial hydrolysis of RDV. Agents chosen for the evaluation of CES1 inhibitory activity included hydroxychloroquine, ivermectin, erythromycin, clarithromycin, roxithromycin, trimethoprim, ciprofloxacin, vancomycin, and dexamethasone. Additionally, due to the increasing use of medical cannabis and cannabidiol (CBD) supplements, their advocacy for certain COVID-19 associated conditions [10,11] and emerging evidence of CES1 inhibition by the major cannabinoids [12], Δ9-tetrahydrocannabinol (THC), CBD, and cannabinol (CBN) were evaluated (Table 1 ). In addition, since individuals who are carriers of the loss of function CES1 variant G143E have a demonstrated impairment in their ability to metabolize CES1 substrates [[13], [14], [15]], influences of the variant on the hydrolysis of RDV were evaluated as well. An established cell-line and in vitro assay was also utilized to investigate the influence of the variant on the conversion of RDV to GS-704277 [16]. Finally, a mathematical model was applied to evaluate the pharmacokinetic change due to the reduction of CES1 activity. This model was applied to our in vitro data to quantitively evaluate the impact of the coadministration of RDV with the respective inhibitor from the screening result, as well as individuals with the G143E variant.
Table 1.
In vivo concentration ranges of the study compounds.
| Name of compound | Molecular weight | Concentration use in this study | In vivo Cmax | Dose | Subjects | N | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| g/mol | μg/mL | μM | μg/mL | μM | |||||
| Aripiprazole | 448.39 | 10.00 | 22.30 | 0.14 | 0.31 | P.O. 10 mg/day tablets | Healthy subjects | 24 | [17] |
| Ziprasidone | 412.94 | 10.00 | 24.22 | 0.09 | 0.22 | P.O. 40 mg tablets | Healthy subjects | 10 | [18] |
| CBN | 310.43 | 10.00 | 32.21 | 0.01 | 0.02 | Case report | – | – | [19] |
| THC | 314.45 | 10.00 | 31.80 | 1.10 | 3.50 | Case report | – | – | [19] |
| CBD | 314.47 | 10.00 | 31.80 | 1.39 | 4.40 | P.O. 1500 mg CBD twice daily | Healthy subjects | 9 | [20] |
| Dexamethasone | 392.46 | 10.00 | 25.48 | 0.26 | 0.66 | P.O. 20 mg tablet under fasting conditions | Healthy subjects | 34 | [21] |
| Trimethoprim | 290.32 | 10.00 | 34.44 | 2.70 | 9.30 | P.O. 400:2000 mg trimethoprim: sulfamethoxazole | Healthy subjects | 24 | [22] |
| Ivermectin | 875.10 | 10.00 | 11.43 | 0.08 | 0.09 | P.O. 12 mg solution | Healthy subjects | 11 | [23] |
| Azithromycin | 749.00 | 10.00 | 13.35 | 0.40 | 0.53 | P.O. 500 mg | Healthy subjects | 12 | [24,25] |
| Clarithromycin | 747.95 | 10.00 | 13.37 | 2.85 | 3.81 | P.O. 500 mg | Healthy subjects | 17 | [26] |
| Ciprofloxacin | 331.35 | 10.00 | 30.18 | 1.10 | 3.32 | P.O. 200 mg | Healthy subjects | 12 | [27] |
| Erythromycin | 733.93 | 10.00 | 13.63 | 3.49 | 4.75 | P.O. 800 mg every 12 h for 3 doses | Healthy subjects | 23 | [28] |
| Roxithromycin | 837.05 | 10.00 | 11.95 | 9.70 | 11.59 | P.O. 300 mg | Healthy subjects | 20 | [29] |
| Vancomycin | 1449.30 | 10.00 | 6.90 | 10.00 | 6.90 | I.V. 10 mg/kg | Patients | 6 | [30] |
| Hydroxychloroquine | 335.87 | 10.00 | 29.77 | 3.30 | 9.83 | I.V. 310 mg over 0.5 h | Healthy subjects | 4 | [31] |
2. Methods and materials
2.1. Materials
Remdesivir (RDV), GS704277, THC, CBD, CBN, hydroxychloroquine were purchased from Cayman Chemical (Ann Arbor, MI). Azithromycin was purchased from Pfizer Inc. (New York NY). Erythromycin, clarithromycin was purchased from Abbott Laboratories Inc. (Chicago, IL). Dexamethasone, trimethoprim, ivermectin ciprofloxacin, roxithromycin, vancomycin, aripiprazole, ziprasidone, and phenacetin were purchased from Sigma-Aldrich Inc. (St. Louis, MO). Phosphate buffered saline (PBS) was purchased from Thermo Fisher Scientific Inc. (Waltham, MA). All other chemicals and reagents were of the highest analytical grade and were commercially available.
2.2. Preparation of cell S9 fractions containing wild-type CES1
Wild-type and G143E S9 fractions were prepared as previously described [13]. In brief, human embryonic kidney cell line (Flp-In-293, Invitrogen, Carlsbad, CA) stably expressing WT and CES1 G143E [13] were cultured in Dulbecco's modified Eagle medium with 10% FBS and 100 mg/ml hygromycin. Upon reaching approximately 90% confluency, cells were then harvested in PBS and sonicated for 10 s. The mixture was then centrifuged at 9000 g for 30 min to isolate the S9 fraction which was then stored at −70 °C until use. The total protein concentrations in S9 fractions were determined using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific Inc.).
2.3. General in vitro assay conditions
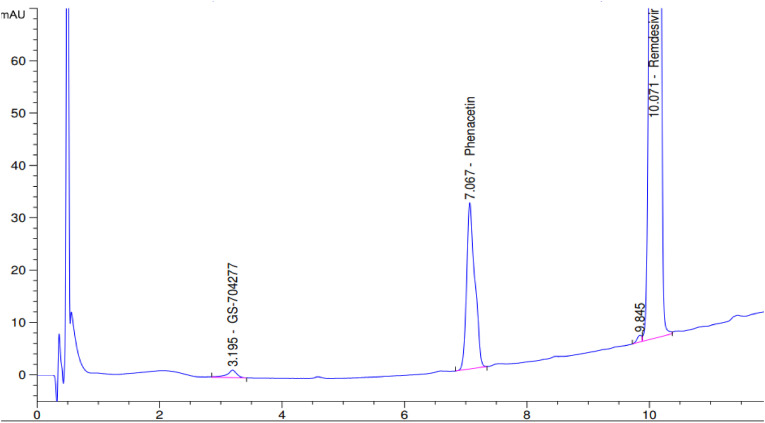
Stock solutions of RDV and the agents assessed for inhibition were made in DMSO at a concentration of 20 mM. Prior to experiments, RDV and prospective inhibitors were prediluted with PBS into an intermediate concentration and further diluted into 96-well plates in a series of concentrations (7.8, 15.6, 31.2, 62.5, 125, 250 μM). The S9 fractions were diluted in PBS and maintained on ice during the experimental workup. In addition to the substrate (RDV) and prospective inhibitors, the mixture contains 40 μg/ml of S9 fraction for a total volume of 100 μl. The reaction was initiated by the addition of the S9 fraction and terminated by the addition of 30 μl of 100 μM phenacetin (internal standard; IS) in acetonitrile. Ninety-six well plates were kept in a water bath maintained at 37 °C. The incubation time was determined in the linear phase of the metabolite formation vs the incubation time curve determined in our preliminary experiments and was chosen to be 45 min. After the reaction was terminated, samples were then centrifuged at 3486 g for 10 min to separate the protein, and the supernatant was collected and analyzed for RDV, GS704277 and IS. The analysis was achieved utilizing an 1100 series Agilent HPLC with Diode Array Detector (Agilent Technologies, Palo Alto, USA). Agilent Openlab software was used for data acquisition and analysis (Santa Clara, CA). A gradient method was utilized based on a previously published method with minor modifications [32]. Briefly, the mobile phase was initiated with 98% 5 mM (pH = 4) KH2PO4 buffer balanced with methanol and kept for 0.2 min, then the gradient changed to 10% buffer at 10 min. The ratio was then held for 1 min, and switched back to the 98% buffer for 0.5 min, balanced for another 3.5 min for a total run time of 15 min. A Synergi™ 4 μm Max-RP 80 Å, 100 × 4.6 mm LC column (Phenomenex, Torrance, CA, USA) was utilized for analysis. The UV absorbance was set at 240 nm. The concentration of GS-704277 was determined by the peak area ratio of the GS704277 and the IS phenacetin, and was compared with a standard curve. Representative chromatographic peaks are shown in Fig. 2 .
Fig. 2.
Chromatograph of GS-704277, the IS phenacetin, and RDV. The standard solution was made with 6.25 μM GS704277 and 625 μM RDV in water mixed with 3:1 (v/v) 100 μM phenacetin in acetonitrile as the internal standard.
2.4. Determination of in vitro inhibition prospective to the RDV metabolism
The screening of the candidate CES1 inhibitor drugs was performed by co-incubating RDV with 10 μg/mL of each drug and comparing it with a control in which no prospective inhibitor was added. The conversion of 10 μg/ml to the respective molar concentration units are provided in Table 1.
The IC50 of the potential inhibitors from the screen results was measured by coincubation of varying concentrations of inhibitors incubated with 40 μM RDV and compared with a control.
To determine if the potential inhibitor produced time-dependent inhibition, the S9 fraction and inhibitor were co-incubated for 30 min before the addition of RDV, and the result was compared with a control group in which the S9 fraction was exposed to substrate and inhibitor immediately.
To investigate the kinetic inhibition mechanism and calculated inhibition constant (Ki), various concentrations of RDV (7.8, 15.6, 31.2, 62.5, 125 μM) co-incubated with varying concentrations of CBD (0, 0.78, 1.56, 3.13, 6.25, 12.5 μM). All incubations were performed in duplicate.
2.5. Statistical analysis
Models were fitted to the data points by nonlinear regression using GraphPad PRISM 9 (San Diego, CA). Basic models were used as previously described [33]. The fundamental kinetic model utilized was the Michaelis-Menten model [34]:
| (1) |
Data points are: [S]: substrate concentration, and V: reaction velocity. Iterated variables are: Km: substrate concentration at half of the maximum reaction velocity, and Vmax: maximum reaction velocity.
The kinetic model for determination of the IC50 was the Hill Equation with modification (Eq. (2)) [35]. The derivation of the equation to calculate IC50 (Eq. (3)) has been described previously [36], which was derived by setting the left side of Eq. (2) to be 50. The substrate concentration is fixed and chosen to be close to the substrate Km.
| (2) |
| (3) |
Data points are: [I]: inhibitor concentration, and Rv: the ratio of reaction velocity with coadministration of inhibitor divided by reaction velocity in the control condition without inhibitor, expressed as percent. Iterated variables are: γ: exponent in the Hill equation, IC: inhibitor concentration producing 50% of maximum inhibition, Emax: maximum effect, E0: Baseline effect. The true IC50 is calculated from Equation (3) based on the iterated variables from nonlinear regression.
A mixed-type partial inhibition model [33] was used to determine Ki, a, and b (Equation (4)).
| (4) |
Data points are: [S], substrate concentration depending on the experiments, and [I], the inhibitor concentration. Iterated variables are: Ki: inhibition constant, α: the mix of non-competitive and competitive inhibition, β: the ratio of remaining enzyme velocity when bound to the inhibitor.
The risk of coadministration of RDV and CBD was evaluated with an FDA-suggested model for DDI [37,38] with the modification to the mix-type partial model (Equation (5)) [39]. Without a suggested dose of using CBD in COVID-19 treatment available, we chose the maximum plasma concentration of approved oral CBD administration from a previously reported PK study [20] to estimate a “worst-case scenario” of high systemic exposure. Other parameters incorporated into the model were either from this in vitro enzyme kinetic experiment or from previously published literature, the details of which are listed in Table 1.
| (5) |
Iterated variables: AUCi: Area under the curve of victim drug with the presence of the inhibitor; AUCo: Area under the curve of the victim drug in absence of the inhibitor; CLint, i: Intrinsic clearance by CES1 of the victim drug with the presence of the inhibitor; CLint, o: Intrinsic clearance by CES1 of the victim drug in absence of the inhibitor; fm: fraction metabolism; CLi: Clearance of the victim drug in presence of the inhibitor; CLo: clearance of the victim drug in absence of the inhibitor. Predicted change in the substrate drug AUC (RAUC) was calculated by AUCi/AUCo, predicted change in the substrate drug clearance (RCL) was calculated by CLi/CLo Other variables are the same as previous equations.
3. Results
3.1. CES1 catalyzed RDV hydrolysis and Michaelis–Menten kinetics
The result of in vitro incubations of RDV is shown in Fig. 3 r2 ≥ 0.95 indicates a good fit of the Michaelis-Menten model confirming our assumptions of the mechanism of CES1 catalyzation is Michaelis-Menten kinetics. Vmax = 2.34 nmol/min/mg protein and Km = 14.19 μM is close to the result from the other group [16].
Fig. 3.
S9 fraction incubation of RDV. Model: Eq. (2). r2 = 0.95. Each data point was average of the duplication. Values are shown with best fit value ± SE.
3.2. CBD exhibited the highest degree of inhibition of RDV among all evaluated compounds
The 2nd generation antipsychotic compounds aripiprazole and ziprasidone, as well as the major cannabinoids CBD, CBN, and THC have been identified as potent in vitro CES1 inhibitors in our previous research [12,40]. Of these agents, all produced a ≥50% inhibition of RDV hydrolysis. For ziprasidone and CBN, RDV metabolite formation was completely inhibited at the selected concentration (10 μg/mL). Dexamethasone, trimethoprim, ivermectin, azithromycin, clarithromycin, ciprofloxacin, erythromycin, roxithromycin, vancomycin, hydroxychloroquine, CBD were selected as co-administratered drugs to treat COVID-19 [6]. Among all the evaluated compounds, CBD produced the highest degree of inhibition (residual enzyme activity = ∼16.5%) (Fig. 4 ).
Fig. 4.
In vitro coincubation of the COVID-19 therapeutic agents. The coadministration drug's final concentration is 10 μg/mL. Among all compounds assessed, CBD produced the strongest inhibition of RDV hydrolysis at 10 μg/mL.
3.3. Clarithromycin weakly inhibits RDV hydrolysis
Since the clarithromycin results indicated 36% residual enzyme activity, to validate this result, follow-up experiments were conducted and confirmed that clarithromycin inhibits RDV hydrolysis albeit, weakly (Fig. 5 ). An IC50 could not be calculated since the Emax was less than 0.5.
Fig. 5.
Clarithromycin weakly inhibits RDV hydrolysis. Eq. (2) was used for regression. r2 = 0.94.
3.4. CBD inhibits RDV hydrolysis by reversible inhibition with an IC50 = 10.22 μM
Further preincubation results showed that at a substrate concentration of 40 μM, CBD inhibited RDV hydrolysis with an IC50 = 10.22 μM (Fig. 6 ). The preincubation experiment performed on the preincubation group and control group, paired by the same CBD concentration indicated a non-significant difference (P = 0.2087; paired t-test). This result suggests CBD inhibited RDV metabolism by a reversible inhibition mechanism (Fig. 6).
Fig. 6.
IC50 of CBD-associated RDV hydrolysis and determination of the inhibition type. Panel A: IC50 of CBD vs RDV. Eq. (2) was used for regression. r2 = 0.94. Panel B: determination of the inhibition type. A paired t-test was utilized to compare the preincubation group and control group. Control group r2 = 0.94; Pre-incubation group r2 = 0.95. Panel C: paired t-test of control group vs preincubation group, P = 0.21 (nonsignificant). Each data point was the average of the duplicate determinations. Values are shown with best fit value ± SE.
3.5. CBD inhibited RDV hydrolysis via a mixed type-partial mechanism
To further investigate the mechanism of inhibition, we coincubated a series of concentrations of RDV with various concentrations of CBD. The results indicated that CBD inhibits RDV metabolism via a mixed type of competitive and noncompetitive partial inhibition. The inhibition type was closer to being characterized as a competitive inhibition (α = 1.05), and the inhibition constant Ki = 0.89 μM. When binding with CBD, CES1 reduced the catalyzing efficiency to 24% (β = 0.24) (Fig. 7 ).
Fig. 7.
Mechanism of CBD inhibiting RDV hydrolysis. Model: Eq. (4). r2 = 0.85. Each data point represented the average of duplication. Values are shown with best fit value ± SE.
3.6. The CES1 G143E variant has 14% intrinsic clearance of that of wild-type
One extra concentration (500 μM) was added to the G143E group to evaluate maximum velocity. The in vitro evaluation shows the intrinsic clearance (Vmax/Km) in the presence of the CES1 G143E variant is 14% of wild type CES1 on RDV metabolism (Fig. 8 ).
Fig. 8.
S9 fraction incubation for RDV hydrolysis with CES1 G143E. r2 for the wild-type condition is > 0.99, r2 for G143E is 0.97. Each data point was the mean of the duplicate time points. Values are shown with best fit value ± SE.
3.7. In vitro to in vivo scaling of clearance and AUC difference for RDV coadministration with CBD and CES1 G143E carrier and noncarrier
The result of in vitro to in vivo scaling was listed in Table 2 . We predicted a potential decrease in clearance and an increase in AUC of RDV in coadministration with CBD over administration of RDV alone. We also predicted a potential decrease in clearance and an increase in AUC of RDV in G143E carrier subjects over non-carrier subjects.
Table 2.
In vitro to clinical scaling of CL and AUC change of co-administration of CBD and G143E polymorphism.
| Coadministration with CBD | |||||
|---|---|---|---|---|---|
| Input Parameter | fm | α | β | Ki (μM) | [I] (μM) |
| Best fit value | 0.91 | 1.05 | 0.24 | 0.89 | 4.40 |
| SE | – | 1.23 | 0.04 | 0.79 | – |
| Reference | [3] | Experimental | Experimental | Experimental | [20] |
| Scaling Result | CLint,I/CLint,o | RCL | RAUC | ||
| 0.36 | 0.42 | 2.40 | |||
| Genetic polymorphism | |||||
| Input Parameter | fm | Vmaxwt (nmol/min/mg protein) | Kmwt (μM) | VmaxG143E (nmol/min/mg protein) | KmG143E (μM) |
| Best fit value | 0.91 | 2.35 | 15.10 | 2.08 | 93.94 |
| SE | – | 0.03 | 0.80 | 0.17 | 20.92 |
| Reference | [3] | Experimental | Experimental | Experimental | Experimental |
| Scaling Result | CLint,I (mL/min/mg protein) | CLint,o (mL/min/mg protein) | CLint,I/CLint,o | RCL | RAUC |
| 0.02 | 0.16 | 0.14 | 0.22 | 4.56 |
4. Discussion
Intravenous RDV has been routinely utilized in the treatment of mild to moderate COVID-19 in high-risk, nonhospitalized patients and in hospitalized patients with COVID-19. An assessment of nine therapeutic agents frequently prescribed and often coadministered with RDV failed to produce significant inhibition of RDV hydrolysis. Clarithromycin shows a weak inhibition on RDV hydrolysis in our IC50 study (IC50 unable to be calculated). The lower residual enzyme activity in the screening results could be due to the simultaneous degradation of the substrate during the incubation. Nevertheless, considering the Cmax attained in healthy subjects (2.85 μg/mL, 3.81 μM, Table 1) receiving clinically relevant doses, it would appear unlikely to produce a significant inhibitory effect [26]. However, consistent with previous in vitro studies employing other substrates, CBD produced potent inhibition of CES1-catalyzed hydrolysis [12]. The in vitro catalyzation velocity reduction in the presence of the CES1 G143E variant was also identical to our previous report [41].
The likelihood of CBD-RDV concurrent use and an attendant DDI liability is a possibility given the long elimination half-life of CBD and the interest in and promotion of CBD to treat COVID-19 and related conditions. For example, following in vitro study results suggesting the potential activity of CBD in reducing coronavirus replication and virus-induced cytokine release [10,42], CBD was recently approved by the FDA for a clinical trial to treat lung inflammation induced by COVID-19 [43] (Clinical trial identifier: NCT04686539, NCT04731116). In addition to our present evaluation, recent research from another group using microsomal incubations demonstrated that CBD inhibited RDV hydrolysis [44]. The potential DDI between CBD and the COVID-19 therapeutic agent RDV should be appreciated by clinicians using the antiviral. As our results (Table 1) suggested, when coadministered with CBD, the intrinsic clearance of RDV could likely be reduced – by as much as 36% (compared with using the same dose of RDV only). Additionally, the total clearance of RDV will be reduced to 42% of the value without CBD, and the AUC will be increased to 2.4-fold, compared with a condition of no coadministration with CBD.
There has been considerable interest in the potential role of pharmacogenomics in outcomes associated with COVID-19 therapeutics [45,46]. The CES1 G143E variant was reported to have a clinical impact on several drugs that are CES1 substrates [47]. As a relatively new identified variant of a less well-studied DME with a lower MAF, the G143E polymorphism is not included on most of the commercial gene chips utilized in clinical genotyping efforts. For these potential reasons and others, the potential clinical implications (if any) of G143E carrier status in RDV-treated patients remains unknown. To our knowledge, our experiment is the first evaluation of the impact of the G143E variant in RDV hydrolysis through an in vitro approach. Notwithstanding the significant limitations of in vitro assessments and extrapolation of results to patients, our findings suggest that carriers of the G143E polymorphism might experience a significant reduction in the intrinsic clearance of RDV compared to non-carriers. According to FDA criteria for in vitro evaluation of DDIs, RAUC greater than 1.02 may be considered to potentially produce a significant DDI (https://www.fda.gov/regulatory-information/search-fda-guidance-documents/invitro-drug-interaction-studies-cytochrome-p450-enzyme-and-transporter-mediated-drug-interactions). Our results suggested the intrinsic clearance of RDV could potentially be reduced to as much as 14% of the non-carriers. The total clearance of RDV was estimated to be reduced to 22% of the non-carriers, and the AUC was predicted to be increased to 4.6-fold compared with the non-carriers.
For RDV, which demonstrates significant antiviral efficacy against SARS-CoV-2 in vitro [48], it has lacked a robust clinical performance with variable outcomes and multiple associated toxicities [49]. Common side effects of RDV include nausea, vomiting, diarrhea, and elevation of hepatic enzymes ALT and AST [50]. More serious and potentially fatal side effects including bradycardia [51] and renal failure [52] have been reported. Currently, there is no clinical data regarding the potential for a DDI with RDV due to the CES1 inhibition, but the potential for a DDI mediated by CES1 inhibition has been noted in the literature [53]. The full toxicology of RDV remains incompletely understood and continues to be investigated. Recent in vitro research suggested that GS-704277 may be the critical mediator leading to nephrotoxicity [54]. Since the metabolism of RDV is sequential, the role of CES1 inhibition and changes in the dynamic plasma concentration change of GS-704277 and GS-441524 remains a matter of speculation, as well as any relationship to RDV efficacy and toxicity.
The results of the present in vitro evaluations assessing the potential CES1-mediated DDI potential between RDV and at least nine medications that have been commonly coadministered with the antiviral appear to be unlikely. However, there are a number of limitations to our research. First, our in vitro experiment was only designed to assess the hydrolysis and inhibition of hydrolysis mediated by CES1. Previous reports have indicated that beyond CES1, the enzyme CatA was also able to catalyze RDV hydrolysis [55]. It has been shown that the expression of CES1 and CatA is different across organ types [56], thus, the major contributing hydrolytic enzyme may differ across tissues [55]. Secondly, clinical studies have indicated that there is a significant systemic exposure to the CBD metabolites 6-OH-CBD, 7-OH-CBD, and 7-COOH-CBD after the administration of CBD [20]. The potential influences of these metabolites were not evaluated in this study. Additionally, the CES1/CatA expression pattern in the CES1-overexpressing HEK293 cells utilized in our assay may be substantially different from the organs of interest (e.g. liver vs lung) [13]. These potential differences may influence the accuracy of the in vitro to in vivo scaling model. Also, though the majority of RDV is biotransformed by hydrolysis, RDV is also a known substrate of several CYP enzymes including CYP2C8, CYP2D6, and CYP3A4 [57,58]. Several of the antibiotics screened in this study are also recognized as CYP inhibitors [8,59]. Notably, erythromycin, clarithromycin, and roxithromycin are known to be time-dependent inhibitors of CYP3A [60,61]. Although no appreciable effect on CES1-mediated RDV hydrolysis was demonstrated in our study, a potential DDI otherwise mediated by the CYP system was not evaluated. Finally, the influence of the assessed compounds on enzymes catalyzing the biotransformation of GS-704277 to GS-441524, and the further downstream biotransformation were not evaluated in these studies. As we mentioned in the introduction section, the further cascade activations involve multiple intracellular enzymes including a number of phosphoramides and phosphotransferases [55]. The quantitative distribution of the intermedia metabolic enzymes catalyzing RDV biotransformation in each step has not been studied clearly. With an in vitro enzyme or microsomal incubation system, it is hard to further metabolize GS-704277 to the final active form, thus a study in hepatocytes may need to be performed to estimate how CES1 inhibition could affect the formation of the final active metabolite GS-443902. Nonetheless, the results of these in vitro-in vivo predictions are of potential clinical concern. However, a clinical study designed to assess this suspected DDI would be required to confirm the prediction.
5. Conclusion
The results of the present in vitro evaluations assessing the potential CES1-mediated DDI potential between RDV and nine medications (hydroxychloroquine, ivermectin, erythromycin, clarithromycin, roxithromycin, trimethoprim, ciprofloxacin, vancomycin, and dexamethasone) commonly coadministered with the antiviral find no evidence for a likely interaction. On the other hand, the identified CES1 inhibitor aripiprazole and ziprasidone, as well as the major cannabinoids CBD, CBN, and THC, inhibited RDV metabolism by CES1. Cannabidiol inhibits RDV metabolism by a mixed type of competitive and noncompetitive partial inhibition mechanism, with a = 1.05, Ki = 0.89 μM. When binding with CBD, CES1 reduces the catalyzing capacity to 24% of the nonbinding enzyme. The intrinsic clearance (Vmax/Km) of the G143E carrier is 14% compared with the wild-type. In vitro to in vivo mathematical scaling predicted a potential 42% reduction in total clearance of RDV and potential 2.4-fold increase in RDV AUC when coadministered CBD. Predictive scaling also suggested a potentially 22% lower total clearance, and potential 4.6-fold higher AUC among G143E carriers. Our research was only designed to evaluate CES1-mediated RDV metabolism but biotransformation catalyzed by other enzymes were not evaluated. Further research needs to be performed to evaluate polymorphism any potential changes in RDV efficacy and potential toxicity secondary to CES1 inhibition. The coadministration of RDV with CBD or its use in patients carrying the G143E variant should be approached with caution due to the potential of impaired activation and clearance of RDV.
Author contributions
Study design: Q.Z., J.S.M.
Experimental design: Q.Z., J.S.M., P.W.M.
Analysis of data: Q.Z.
Preparation of the manuscript: Q.Z., J.S.M., P.W.M.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was partially supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development [R01 HD093612].
Data availability
Data will be made available on request.
References
- 1.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9(2) doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin H.X.J., Cho S., Meyyur Aravamudan V., Sanda H.Y., Palraj R., Molton J.S., Venkatachalam I. Remdesivir in Coronavirus Disease 2019 (COVID-19) treatment: a review of evidence. Infection. 2021;49:401–410. doi: 10.1007/s15010-020-01557-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Humeniuk R., Mathias A., Cao H., Osinusi A., Shen G., Chng E., Ling J., Vu A., German P. Safety, tolerability, and pharmacokinetics of remdesivir, an antiviral for treatment of COVID-19, in healthy subjects. Clin. Transl. Sci. 2020;13:896–906. doi: 10.1111/cts.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humeniuk R., Mathias A., Kirby B.J., Lutz J.D., Cao H., Osinusi A., Babusis D., Porter D., Wei X., Ling J., Reddy Y.S., German P. Pharmacokinetic, pharmacodynamic, and drug-interaction profile of remdesivir, a SARS-CoV-2 replication inhibitor. Clin. Pharmacokinet. 2021;60:569–583. doi: 10.1007/s40262-021-00984-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pashaei Y. Analytical methods for the determination of remdesivir as a promising antiviral candidate drug for the COVID-19 pandemic. Drug Discov. Ther. 2021;14:273–281. doi: 10.5582/ddt.2020.03097. [DOI] [PubMed] [Google Scholar]
- 6.Badary O.A. Pharmacogenomics and COVID-19: clinical implications of human genome interactions with repurposed drugs. Pharmacogenomics J. 2021;21:275–284. doi: 10.1038/s41397-021-00209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yousefifard M., Zali A., Zarghi A., Madani Neishaboori A., Hosseini M., Safari S. Non-steroidal anti-inflammatory drugs in management of COVID-19; A systematic review on current evidence. Int. J. Clin. Pract. 2020;74 doi: 10.1111/ijcp.13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayhew B.S., Jones D.R., Hall S.D. An in vitro model for predicting in vivo inhibition of cytochrome P450 3A4 by metabolic intermediate complex formation. Drug Metab. Dispos. 2000;28:1031–1037. [PubMed] [Google Scholar]
- 9.Rhoades J.A., Peterson Y.K., Zhu H.-J., Appel D.I., Peloquin C.A., Markowitz J.S. Prediction and in vitro evaluation of selected protease inhibitor antiviral drugs as inhibitors of carboxylesterase 1: a potential source of drug-drug interactions. Pharm. Res. (N. Y.) 2012;29:972–982. doi: 10.1007/s11095-011-0637-9. [DOI] [PubMed] [Google Scholar]
- 10.Khalsa J.H., Bunt G., Maggirwar S.B., Kottilil S. COVID-19 and cannabidiol (CBD) J. Addiction Med. 2021;15:355–356. doi: 10.1097/ADM.0000000000000771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malinowska B., Baranowska-Kuczko M., Kicman A., Schlicker E. Opportunities, challenges and pitfalls of using cannabidiol as an adjuvant drug in COVID-19. Int. J. Mol. Sci. 2021;22(4) doi: 10.3390/ijms22041986. 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian Y., Wang X., Markowitz J.S. In vitro inhibition of carboxylesterase 1 by major cannabinoids and selected metabolites. Drug Metab. Dispos. 2019;47:465–472. doi: 10.1124/dmd.118.086074. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H.-J., Patrick K.S., Yuan H.-J., Wang J.-S., Donovan J.L., DeVane C.L., Malcolm R., Johnson J.A., Youngblood G.L., Sweet D.H., Langaee T.Y., Markowitz J.S. Two CES1 gene mutations lead to dysfunctional carboxylesterase 1 activity in man: clinical significance and molecular basis. Am. J. Hum. Genet. 2008;82:1241–1248. doi: 10.1016/j.ajhg.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis J.P., Horenstein R.B., Ryan K., O'Connell J.R., Gibson Q., Mitchell B.D., Tanner K., Chai S., Bliden K.P., Tantry U.S., Peer C.J., Figg W.D., Spencer S.D., Pacanowski M.A., Gurbel P.A., Shuldiner A.R. The functional G143E variant of carboxylesterase 1 is associated with increased clopidogrel active metabolite levels and greater clopidogrel response. Pharmacogenetics Genom. 2013;23:1–8. doi: 10.1097/FPC.0b013e32835aa8a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Z.-Y., Edginton A.N., Laizure S.C., Parker R.B. Physiologically based pharmacokinetic modeling of impaired carboxylesterase-1 activity: effects on oseltamivir disposition. Clin. Pharmacokinet. 2014;53:825–836. doi: 10.1007/s40262-014-0160-3. [DOI] [PubMed] [Google Scholar]
- 16.Zhang F., Li H.-X., Zhang T.-T., Xiong Y., Wang H.-N., Lu Z.-H., Xiong L., He Y.-Q., Ge G.-B. Human carboxylesterase 1A plays a predominant role in the hydrolytic activation of remdesivir in humans. Chem. Biol. Interact. 2021;351 doi: 10.1016/j.cbi.2021.109744. 109744. [DOI] [PubMed] [Google Scholar]
- 17.Koller D., Almenara S., Mejía G., Saiz-Rodríguez M., Zubiaur P., Román M., Ochoa D., Navares-Gómez M., Santos-Molina E., Pintos-Sánchez E., Abad-Santos F. Metabolic effects of aripiprazole and olanzapine multiple-dose treatment in a randomised crossover clinical trial in healthy volunteers: association with pharmacogenetics. Adv. Ther. 2021;38:1035–1054. doi: 10.1007/s12325-020-01566-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilner K.D., Hansen R.A., Folger C.J., Geoffroy P. The pharmacokinetics of ziprasidone in healthy volunteers treated with cimetidine or antacid. Br. J. Clin. Pharmacol. 2000;49(Suppl 1):57S–60S. doi: 10.1046/j.1365-2125.2000.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gronewold A., Skopp G. A preliminary investigation on the distribution of cannabinoids in man. Forensic Sci. Int. 2011;210:e7–e11. doi: 10.1016/j.forsciint.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Taylor L., Gidal B., Blakey G., Tayo B., Morrison G. A phase I, randomized, double-blind, placebo-controlled, single ascending dose, multiple dose, and food effect trial of the safety, tolerability and pharmacokinetics of highly purified cannabidiol in healthy subjects. CNS Drugs. 2018;32:1053–1067. doi: 10.1007/s40263-018-0578-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bashir Q., Acosta M. Comparative safety, bioavailability, and pharmacokinetics of oral dexamethasone, 4-mg and 20-mg tablets, in healthy volunteers under fasting and fed conditions: a randomized open-label, 3-way crossover study. Clin. Lymphoma, Myeloma & Leukemia. 2020;20:768–773. doi: 10.1016/j.clml.2020.06.022. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan S.A., Weinfeld R.E., Abruzzo C.W., McFaden K., Jack M.L., Weissman L. Pharmacokinetic profile of trimethoprim-sulfamethoxazole in man. J. Infect. Dis. 1973;128(Suppl):547–555. doi: 10.1093/infdis/128.supplement_3.s547. [DOI] [PubMed] [Google Scholar]
- 23.González Canga A., Sahagún Prieto A.M., Diez Liébana M.J., Fernández Martínez N., Sierra Vega M., García Vieitez J.J. The pharmacokinetics and interactions of ivermectin in humans--a mini-review. AAPS J. 2008;10:42–46. doi: 10.1208/s12248-007-9000-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lalak N.J., Morris D.L. Azithromycin clinical pharmacokinetics. Clin. Pharmacokinet. 1993;25:370–374. doi: 10.2165/00003088-199325050-00003. [DOI] [PubMed] [Google Scholar]
- 25.Foulds G., Shepard R.M., Johnson R.B. The pharmacokinetics of azithromycin in human serum and tissues. J. Antimicrob. Chemother. 1990;25(Suppl A):73–82. doi: 10.1093/jac/25.suppl_a.73. [DOI] [PubMed] [Google Scholar]
- 26.Rodvold K.A. Clinical pharmacokinetics of clarithromycin. Clin. Pharmacokinet. 1999;37:385–398. doi: 10.2165/00003088-199937050-00003. [DOI] [PubMed] [Google Scholar]
- 27.Bergan T., Dalhoff A., Rohwedder R. Pharmacokinetics of ciprofloxacin. Infection. 1988;16(Suppl 1):S3–S13. doi: 10.1007/BF01650500. [DOI] [PubMed] [Google Scholar]
- 28.Chun A.H.C., Seitz J.A. Pharmacokinetics and biological availability of erythromycin. Infection. 1977;5:14–22. doi: 10.1007/BF01639131. [DOI] [Google Scholar]
- 29.Lassman H.B., Puri S.K., Ho I., Sabo R., Mezzino M.J. Pharmacokinetics of roxithromycin (RU 965) J. Clin. Pharmacol. 1988;28:141–152. doi: 10.1002/j.1552-4604.1988.tb05738.x. [DOI] [PubMed] [Google Scholar]
- 30.Bunke C.M., Aronoff G.R., Brier M.E., Sloan R.S., Luft F.C. Vancomycin kinetics during continuous ambulatory peritoneal dialysis. Clin. Pharmacol. Ther. 1983;34:631–637. doi: 10.1038/clpt.1983.225. [DOI] [PubMed] [Google Scholar]
- 31.Nicol M.R., Joshi A., Rizk M.L., Sabato P.E., Savic R.M., Wesche D., Zheng J.H., Cook J. Pharmacokinetics and pharmacological properties of chloroquine and hydroxychloroquine in the context of COVID-19 infection. Clin. Pharmacol. Ther. 2020;108:1135–1149. doi: 10.1002/cpt.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamdy M.M.A., Abdel Moneim M.M., Kamal M.F. Accelerated stability study of the ester prodrug remdesivir: recently FDA-approved Covid-19 antiviral using reversed-phase-HPLC with fluorimetric and diode array detection. Biomed. Chromatogr. 2021;35 doi: 10.1002/bmc.5212. [DOI] [PubMed] [Google Scholar]
- 33.Segel I.H., Mixed-Type Inhibition . first ed. Wiley; Canada: 2014. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems; pp. 170–202. [Google Scholar]
- 34.Michaelis L., Menten M.L., Johnson K.A., Goody R.S. The original Michaelis constant: translation of the 1913 Michaelis-Menten paper. Biochemistry. 2011;50:8264–8269. doi: 10.1021/bi201284u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volpe D.A., Hamed S.S., Zhang L.K. Use of different parameters and equations for calculation of IC₅₀ values in efflux assays: potential sources of variability in IC₅₀ determination. AAPS J. 2014;16:172–180. doi: 10.1208/s12248-013-9554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenblatt D.J., Zhao Y., Venkatakrishnan K., Duan S.X., Harmatz J.S., Parent S.J., Court M.H., von Moltke L.L. Mechanism of cytochrome P450-3A inhibition by ketoconazole. J. Pharm. Pharmacol. 2011;63:214–221. doi: 10.1111/j.2042-7158.2010.01202.x. [DOI] [PubMed] [Google Scholar]
- 37.Volak L.P., Greenblatt D.J., von Moltke L.L. In: Drug–Drug Interactions in Pharmaceutical Development. Li A.P., editor. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2007. In vitro approaches to anticipating clinical drug interactions; pp. 31–74. [DOI] [Google Scholar]
- 38.Rowland M., Matin S.B. Kinetics of drug-drug interactions. J. Pharmacokinet. Biopharm. 1973;1:553–567. doi: 10.1007/BF01059791. [DOI] [Google Scholar]
- 39.Zhang Q., Duan S.X., Harmatz J.S., Wei Z., Singleton C.A., Greenblatt D.J. Mechanism of dasabuvir inhibition of acetaminophen glucuronidation. J. Pharm. Pharmacol. 2021:131–138. doi: 10.1093/jpp/rgab144. [DOI] [PubMed] [Google Scholar]
- 40.Zhu H.-J., Appel D.I., Peterson Y.K., Wang Z., Markowitz J.S. Identification of selected therapeutic agents as inhibitors of carboxylesterase 1: potential sources of metabolic drug interactions. Toxicology. 2010;270:59–65. doi: 10.1016/j.tox.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 41.Zhu H.-J., Markowitz J.S. Carboxylesterase 1 (CES1) genetic polymorphisms and oseltamivir activation. Eur. J. Clin. Pharmacol. 2013;69:733–734. doi: 10.1007/s00228-012-1350-2. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen L.C., Yang D., Nicolaescu V., Best T.J., Ohtsuki T., Chen S.-N., Friesen J.B., Drayman N., Mohamed A., Dann C., Silva D., Gula H., Jones K.A., Millis J.M., Dickinson B.C., Tay S., Oakes S.A., Pauli G.F., Meltzer D.O., Randall G., Rosner M.R. Cannabidiol inhibits SARS-CoV-2 replication and promotes the host innate immune response. BioRxiv [Preprint] 2021 doi: 10.1101/2021.03.10.432967. [DOI] [Google Scholar]
- 43.Crippa J.A.S., Pacheco J.C., Zuardi A.W., Guimarães F.S., Campos A.C., Osório F. de L., Loureiro S.R., Dos Santos R.G., Souza J.D.S., Ushirohira J.M., Ferreira R.R., Mancini Costa K.C., Scomparin D.S., Scarante F.F., Pires-Dos-Santos I., Mechoulam R., Kapczinski F., Fonseca B.A.L., Esposito D.L.A., Passos A.D.C., Hallak J.E.C. Cannabidiol for COVID-19 patients with mild to moderate symptoms (candidate study): a randomized, double-blind, placebo-controlled clinical trial. Cannabis Cannabinoid. Res. 2021 doi: 10.1089/can.2021.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saraswat A., Vartak R., Patki M., Patel K. Cannabidiol inhibits in vitro human liver microsomal metabolism of remdesivir: a promising adjuvant for COVID-19 treatment. Cannabis Cannabinoid. Res. 2021 doi: 10.1089/can.2021.0109. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 45.Stanković B., Kotur N., Gašić V., Klaassen K., Ristivojević B., Stojiljković M., Pavlović S., Zukić B. Pharmacogenomics landscape of COVID-19 therapy response in Serbian population and comparison with worldwide populations. J. Med. Biochem. 2020;39:488–499. doi: 10.5937/jomb0-26725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi T., Luzum J.A., Nicol M.R., Jacobson P.A. Pharmacogenomics of COVID-19 therapies. NPJ Genom. Med. 2020;5:35. doi: 10.1038/s41525-020-00143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X., Her L., Xiao J., Shi J., Wu A.H., Bleske B.E., Zhu H.-J. Impact of carboxylesterase 1 genetic polymorphism on trandolapril activation in human liver and the pharmacokinetics and pharmacodynamics in healthy volunteers. Clin. Transl. Sci. 2021;14:1380–1389. doi: 10.1111/cts.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jorgensen S.C.J., Kebriaei R., Dresser L.D. Remdesivir: review of pharmacology, pre-clinical data, and emerging clinical experience for COVID-19. Pharmacotherapy. 2020;40:659–671. doi: 10.1002/phar.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan V.C., Muller F.L. Why remdesivir failed: preclinical assumptions overestimate the clinical efficacy of remdesivir for COVID-19 and ebola. Antimicrob. Agents Chemother. 2021;65 doi: 10.1128/AAC.01117-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elsawah H.K., Elsokary M.A., Abdallah M.S., ElShafie A.H. Efficacy and safety of remdesivir in hospitalized Covid-19 patients: systematic review and meta-analysis including network meta-analysis. Rev. Med. Virol. 2021;31 doi: 10.1002/rmv.2187. [DOI] [PubMed] [Google Scholar]
- 51.Touafchia A., Bagheri H., Carrié D., Durrieu G., Sommet A., Chouchana L., Montastruc F. Serious bradycardia and remdesivir for coronavirus 2019 (COVID-19): a new safety concerns. Clin. Microbiol. Infect. 2021 doi: 10.1016/j.cmi.2021.02.013. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gérard A.O., Laurain A., Fresse A., Parassol N., Muzzone M., Rocher F., Esnault V.L.M., Drici M.-D. Remdesivir and acute renal failure: a potential safety signal from disproportionality analysis of the WHO safety database. Clin. Pharmacol. Ther. 2021;109:1021–1024. doi: 10.1002/cpt.2145. [DOI] [PubMed] [Google Scholar]
- 53.Rezaee H., Pourkarim F., Pourtaghi-Anvarian S., Entezari-Maleki T., Asvadi-Kermani T., Nouri-Vaskeh M. Drug-drug interactions with candidate medications used for COVID-19 treatment: an overview. Pharmacol. Res. Perspect. 2021;9 doi: 10.1002/prp2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan V.C., Muller F.L. Captisol and GS-704277, but not GS-441524, are credible mediators of remdesivir's nephrotoxicity. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.01920-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li R., Liclican A., Xu Y., Pitts J., Niu C., Zhang J., Kim C., Zhao X., Soohoo D., Babusis D., Yue Q., Ma B., Murray B.P., Subramanian R., Xie X., Zou J., Bilello J.P., Li L., Schultz B.E., Sakowicz R., Feng J.Y. Key metabolic enzymes involved in remdesivir activation in human lung cells. Antimicrob. Agents Chemother. 2021;65 doi: 10.1128/AAC.00602-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Her L., Zhu H.-J. Carboxylesterase 1 and precision pharmacotherapy: pharmacogenetics and nongenetic regulators. Drug Metab. Dispos. 2020;48:230–244. doi: 10.1124/dmd.119.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deb S., Arrighi S. Potential effects of COVID-19 on cytochrome P450-mediated drug metabolism and disposition in infected patients. Eur. J. Drug Metab. Pharmacokinet. 2021;46:185–203. doi: 10.1007/s13318-020-00668-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang K. What do we know about remdesivir drug interactions? Clin. Transl. Sci. 2020;13:842–844. doi: 10.1111/cts.12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Izzo A.A., Ernst E. Interactions between herbal medicines and prescribed drugs: an updated systematic review. Drugs. 2009;69:1777–1798. doi: 10.2165/11317010-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 60.Zhou S., Yung Chan S., Cher Goh B., Chan E., Duan W., Huang M., McLeod H.L. Mechanism-based inhibition of cytochrome P450 3A4 by therapeutic drugs. Clin. Pharmacokinet. 2005;44:279–304. doi: 10.2165/00003088-200544030-00005. [DOI] [PubMed] [Google Scholar]
- 61.Yamazaki H., Shimada T. Comparative studies of in vitro inhibition of cytochrome P450 3A4-dependent testosterone 6beta-hydroxylation by roxithromycin and its metabolites, troleandomycin, and erythromycin. Drug Metab. Dispos. 1998;26:1053–1057. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.