Abstract
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) infection and associated coronavirus disease 2019 (COVID-19) has severely impacted human well-being. Although vaccination programs have helped in reducing the severity of the disease, drug regimens for clinical management of COVID-19 are not well recognized yet. It is therefore important to identify and characterize the molecular pathways that could be therapeutically targeted to halt SARS-CoV-2 infection and COVID-19 pathogenesis. SARS-CoV-2 hijacks host cell molecular machinery for its entry, replication and egress. Interestingly, SARS-CoV-2 interacts with host cell Calcium (Ca2+) handling proteins and perturbs Ca2+ homeostasis. We here systematically review the literature that demonstrates a critical role of host cell Ca2+ dynamics in regulating SARS-CoV-2 infection and COVID-19 pathogenesis. Further, we discuss recent studies, which have reported that SARS-CoV-2 acts on several organelle-specific Ca2+ transport mechanisms. Moreover, we deliberate upon the possibility of curtailing SARS-CoV-2 infection by targeting host cell Ca2+ handling machinery. Importantly, we delve into the clinical trials that are examining the efficacy of FDA-approved small molecules acting on Ca2+ handling machinery for the management of COVID-19. Although an important role of host cell Ca2+ signaling in driving SARS-CoV-2 infection has emerged, the underlying molecular mechanisms remain poorly understood. In future, it would be important to investigate in detail the signaling cascades that connect perturbed Ca2+ dynamics to SARS-CoV-2 infection.
Keywords: Calcium signaling, SARS-CoV-2, COVID-19, Host cell calcium dynamics, Clinical trials
1. Introduction
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) infection is one of the worst pandemics of the century, affecting millions of people all over the world. The SARS-CoV-2 infection was first reported in Wuhan, China in December 2019 and named coronavirus disease 2019 (COVID-19) [1]. Due to its high infectivity, the virus spread is uncontainable and within three months, it was declared a pandemic by World Health Organization (WHO). The enormity of the pandemic is indicated by the horrifying number of cases and deaths reported worldwide, which summed up to 519 million cases and 6 million deaths by May 2022 (https://covid19.who.int). The majority of SARS-CoV-2 infected patients experience mild symptoms such as sore throat, fever, tiredness and loss of taste and smell, while only 10–15% of patients develop severe symptoms such as shortness of breath, chest pain and decrease oxygen levels [2]. The severity of infection increases with co-morbidities like heart disease and diabetes [3, 4]. COVID-19 related deaths are associated with acute respiratory distress syndrome (ARDS), septic shock and multiple organ failure [2]. Although uncontrolled immune activation and cytokine storm are the main culprits in the SARS-CoV-2 pathogenesis, in many cases treatment with steroids further aggravates the severity of infection [5]. The containment of the SARS-CoV-2 pandemic has become a challenge as the virus smartly escapes the immune response. Therefore, the need of the hour is to develop new drug regimens to treat and/or manage COVID-19 infection.
The initial approach for identifying the drugs against SARS-CoV-2 infection was to screen the major classes of anti-viral drugs such as RNA-dependent RNA polymerase inhibitors, nucleoside analogues and protease blockers [6]. Towards this, Remdesivir, an RNA-dependent RNA polymerase inhibitor was the first FDA approved drug repurposed for the treatment of SARS-CoV-2 infection. The treatment with Remdesivir showed alleviation of symptoms in 68% of treated patients in the first clinical study [7]. In the nucleoside analog category, a prodrug of the nucleoside analog- β-d-N4-hydroxycytidine (NHC), Molnupiravir, developed against the influenza virus, is under investigation (phase 2 clinical trial- https://clinicaltrials.gov/ct2/show/NCT04405570) as an oral treatment for COVID-19 [8]. Molnupiravir stalls viral replication by inhibiting viral RNA synthesis. Another prospective oral treatment is Paxlovid (a combination of drugs PF-07321332 and Ritonavir) which blocks the activity of SARS-CoV-2 Mpro protease and is currently being assessed in Phase 3 clinical trial (https://www.clinicaltrials.gov/ct2/show/NCT04960202) [9]. Additionally, Molnupiravir and Paxlovid were given emergency usage authorization by FDA to treat COVID-19 patients. Another important class of drugs is calcium (Ca2+) channel blockers (CCB) that have been successfully proven to be effective in the case of several viral infections such as influenza A virus, Japanese encephalitis virus (JEV) and hemorrhagic fever arenavirus (NWA) [6,10]. In a recent study, drugs belonging to the Ca2+ channel blockers category such as amlodipine, nifedipine, felodipine, verapamil and diltiazem were demonstrated to inhibit SARS-CoV-2 infection in the epithelial kidney (Vero E6) and epithelial lung (Calu-3) cells [11]. These drugs potentially inhibit viral entry into the host cells by blocking l-type voltage-gated calcium (Cav) channels (LVCC). Certainly, Ca2+ channel blockers are emerging as promising therapeutics against SARS-CoV-2 infection. However, the depth of our understanding of the role of Ca2+ channels involved in regulating SARS-CoV-2 infection and pathogenesis is limited. In this review, we will be focusing on how SARS-CoV-2 hijacks host cell Ca2+ machinery to regulate endocytosis, host mitochondrial energetics, autophagy, exocytosis and immune response for completing a successful infection cycle.
Ca2+ is an important secondary messenger in cells, regulating myriad signaling pathways and cellular functions including apoptosis, autophagy, cellular energetics and transcriptional activation [12], [13], [14], [15]. These functions are tightly regulated by spatiotemporally maintained concentration of Ca2+ across cytosol, ER, mitochondria and other organelles [16], [17], [18], [19]. It is well known from various studies that viral infection undermines host Ca2+homeostasis to facilitate viral entry, replication and egress either by directly interacting with ion channels or by oligomerizing viral proteins to form pores called viroporins for ion transport [10]. A brief overview of host-viral protein interaction is listed in Table 1 . In the majority of the viral infections, the augmented Ca2+ release from ER and impaired SERCA functioning increases Ca2+ concentration in the cytosol and mitochondria [20]. The altered partitioning of Ca2+concentration in different compartments perturbs various cellular and mitochondrial functions. The role of Ca2+ signaling in viral infection is known from various studies, however, our understanding of mechanistic details of how SARS-CoV-2 modulates Ca2+ machinery for driving its infection is by far limited. In the following sections, we will be discussing the indispensable role of Ca2+ dynamics in the SARS-CoV-2 entry, replication and egress.
Table 1.
Overview of molecular outcomes of Host and SARS-CoV-2 protein interactions.
| Viral protein | Interacting host proteins/organelles | Molecular outcome in host cell | Molecular advantage to virus | Techniques used to identify interactions | Reference |
|---|---|---|---|---|---|
| RGD motif of Spike protein | Ca2+ dependent ACE-2 binding | Attachment to host cell | Viral entry | In-silico analysis | [23] |
| NSP4 | IMM proteins | Regulates mitochondrial morphology and CM formation | Viral replication | Affinity purification mass spectrometry (AP-MS) | [36] |
| E-protein | Viroporin non-selective cations conductance like K+ and Na+, Ca2+ and Mg2+ located predominantly in ER | Secretion of Viral-like particles | Release of viral particles |
[40] [41] |
|
| ORF3a | Interacts with TRPML3 in lysosomes. Non-selective cation channel with permeability in order of Ca2+> K+ > Na+ located predominantly on lysosomes. | Regulates autophagy, pro-apoptotic pathways and NLRP3 inflammasome activation | Exocytosis of viral particles |
[42] [46] [44] [43] |
|
| mPTP proteins such as ANT and ATP synthase | Opening of mPTP | Unknown | Co-immunoprecipitation | [37] | |
| Mitochondrial complexes | Decreases mitochondrial OCR | Unknown | Co-immunoprecipitation | [37] | |
| ORF9c | OMM proteins | Regulates MAVS | Immune evasion | Affinity purification mass spectrometry (AP-MS) | [36] |
| mPTP proteins such as ANT and ATP synthase | Opening of mPTP | Unknown | Co-immunoprecipitation | [37] | |
| NSP6, ORF9b, and ORF10 | mPTP proteins such as ANT and ATP synthase | Opening of mPTP | Unknown | Co-immunoprecipitation | [37] |
| M-protein | CCDC58 | Reduction in mitochondrial CRC | Unknown | Overexpression and co-immunoprecipitation | [37] |
| Mitochondrial complexes | Decreases mitochondrial OCR | Unknown | Co-immunoprecipitation | [37] |
2. SARS-CoV-2 utilizes host Ca2+ signaling for viral entry
SARS-CoV-2 spike protein (S-protein) binds to the human angiotensin-converting enzyme 2 (ACE2) receptor for attachment to the host cells [21]. The S-protein is made up of two subunits, S1 and S2. The S1 subunit has the receptor-binding domain (RBD), which binds to the ACE2 receptor and the S2 subunit helps in the fusion with the host cell membrane. The cleavage of S1/S2 is mediated by the host cell proteases such as transmembrane serine protease 2 (TMPRSS2) or cathepsin to facilitate virus-host fusion. The three-dimension crystal structure for the binding of SARS-CoV-2 S-protein with the human ACE2 receptor is resolved [22]. The data based on physical interactions suggest that SARS-CoV-2 binds to ACE2 receptors with more affinity than SARS-CoV-1. S-protein receptor binding domain of SARS-CoV-1 is 253 amino acids and that of SARS-CoV-2 is 254 amino acids. Two sequences showed a mismatch of 49 amino acids and 1 deletion/insertion resulting in 80% sequence similarity between SARS-CoV-1 and SARS-CoV-2 S-protein. A recent bioinformatics-based analysis of the S-protein of SARS-CoV-1 and SARS-CoV-2 showed that SARS-CoV-2 has a unique RGD motif (Arginine-Glycine-Aspartate tripeptide) in RBD that may bind to the integrins present on lung epithelium [23]. In this study, the authors carried out motif scanning and prediction using MyHits Motif Scan at SIB server and showed the presence of RGD motif in SARS-CoV-2 RBD but not in SARS-CoV-1 [23]. Docking studies of viral S-protein RGD motif onto the integrin α5β1 and αvβ6 (most abundantly expressed in lung epithelium) suggests that spike protein binds to the integrins via RGD motif. S-protein RGD motif interaction is facilitated by the nearby sequence containing a Ca2+ binding site [23]. Parallelly, integrins also have Ca2+ binding domains that facilitate cell adhesion. This analysis suggests that the interaction of the RGD motif with integrin is dependent on the presence of Ca2+. According to another study, Ca2+ plays an important role in attachment of SARS-CoV-2 S2 fragment with the lipid bilayer [24]. MD simulation based studies demonstrated that the extent of fusion of S2 fragment to the bilayer and penetration in the membrane is significantly dependent on the presence of extracellular Ca2+ ions [24]. This simulation-based study provides a tentative role of extracellular Ca2+ in fusion of S2 peptide with the membrane.
Independent of the above in-silico analysis, a recent study shows that low pH and Ca2+induce a conformational change in S-protein and thereby facilitate membrane fusion [25]. In this study, the fluorescently labelled in vitro liposome membranes were reconstructed with either SARS-CoV-2 S-protein or host ACE2 receptors along with a self-quenching concentration of lipophilic dye DiO. The fusion dynamics was measured by dequenching of the fluorescent probe under varying Ca2+and pH conditions. The study demonstrated that dequenching only occurs in presence of 0.5 µM Ca2+ at low pH 4.6 [25]. The authors observed dynamic sensitivity of S-protein for Ca2+ as an increase in the Ca2+ concentration from 100 µM to 500 µM effectively enhances the fusion efficiency and any further increase in Ca2+inactivates the receptor for fusion. Low pH and Ca2+concentrations used in this assay mimic the physiological conditions of the endo/lysosomal compartment, indicating the viral entry through this route. The authors further examined the viral fusogenicity in HEK 293T cells expressing ACE2 and reported that the D614G spike variant is found to be dependent on Ca2+ for fusion [25]. Moreover, chelating intracellular Ca2+ of infected cells with BAPTA-AM abrogated the membrane fusion of the virus. While the limitation of this study is that it is conducted in liposome-based pseudo virion membranes, the major highlight of the study is the correlation of Ca2+ sensitivity across various evolved spike variants, showing the highest Ca2+ dependence of Delta variant as compared to Alpha and Beta variants. Taken together, above studies suggest that Ca2+is a critical determinant of the virus binding to the host cell and the subsequent fusion process.
After the attachment and fusion of the virus to the host cell, the entry of enveloped viruses is mediated by the clathrin-dependent and independent endocytosis pathway. Viruses are taken up by cells in the vesicles through the membrane transport system. Viruses mature as they pass from the early to the late endosome. These late endosomes further fuse with low pH organelle lysosomes. Somewhere during this process, the viral envelope fuses with the endo-lysosomal membrane and forms a pore to release its genomic material into the cytosol for viral replication [26]. An important regulator of early to late endosome dynamics is Fab1/PIKfye kinase that mediates the conversion of phosphatidylinositol-3-phosphate (PI(3)P) to phosphatidylinositol-3,5-biphosphate (PI(3,5)P2), which is late endosomal marker [26, 27]. In SARS-CoV-2 infection, the understanding of fusion pore formation during the endocytosis process is not very clear. However, a recent study by Ou et al. showed that the inhibition of PIKfyve, the main enzyme synthesizing PI(3,5)P2 in the early endosome, using the chemical inhibitor apilimod, significantly reduces the entry of SARS-CoV-2 pseudo-virions in HEK293/hACE2 cells in a dose-dependent manner [28]. Apilimod is currently in phase 2 clinical trial for evaluating its efficacy in curtailing the progression of COVID-19 (https://clinicaltrials.gov/ct2/show/NCT04446377). Two main downstream effectors of PI(3,5)P2 in lysosomes are two-pore channel subtype 2 (TPC2) and transient receptor potential mucolipin 1 (TRPML1). Inhibition of TPC2 with tetrandrine decreases the entry of SARS-CoV-2 S pseudo-virions but no effect was observed with TRPML1 inhibitor [28]. These studies suggest that PIKfyve and TPC2 are crucial targets to stall SARS-CoV-2 entry. TPC antagonist tetrandrine is under phase 4 clinical trial for controlling pulmonary fibrosis associated with SARS-CoV-2 infection (https://clinicaltrials.gov/ct2/show/study/NCT04308317). These studies certainly underline the critical role of Ca2+ dynamics in SARS-CoV-2 endocytosis, however, further investigation is required for understanding the mechanistic details.
Another defining feature of SARS-CoV-2 and MERS-CoV is the formation of pneumocytes syncytia in the infected patients [29]. This predominantly occurs due to the presence of a multi-basic amino acid sequence at the interface of S1/S2 that can be cleaved by ubiquitously expressed serine protease furin, resulting in multi-nucleated giant cells. Expression of SARS-CoV-2 S-protein in Vero cells induces syncytia formation. Along with the syncytia formation, transient Ca2+ oscillations were also observed in the S-protein expressing cells [29]. Braga et al. screened over 3000 small molecule inhibitors to target syncytia formation and identified 83 drugs that inhibit S-protein mediated cell fusion. While these 83 drugs belong to different pharmacological classes, the majority of them share a common property of regulating intracellular Ca2+ levels [29]. The authors identified that the most effective drug in reducing syncytia formation and Ca2+ oscillations is Niclosamide, which is a potent inhibitor of Ca2+-activated chloride channel (TMEM16F). TMEM16F also functions as a Ca2+-dependent phospholipid scramblase that mediates cell surface exposure of phosphatidylserine. The downregulation of TMEM16F in Spike-expressing cells showed blunted syncytia formation similar to anti-ACE2 siRNA [29]. As previously reported, TMEM16F drives externalization of phosphatidylserine residues, which can mediate cell fusion. This study mechanistically delineates that the SARS-CoV-2 S-protein increases Ca2+ oscillations in the cells thereby enhancing the activity of plasma membrane TMEM16 channel [29]. Recently, Sim et al. corroborated a critical role of TMEM16F in SARS-CoV-2 fusion with host cell plasma membrane and subsequent entry into host cells [30]. The authors demonstrated that SARS-CoV-2 S-protein induces rise in cytosolic Ca2+ which in turn leads to TMEM16F mediated plasma membrane exposure of phosphatidylserine in ACE2 expressing cells. Importantly, the authors carried out a high throughput screening of 56,000 small molecules for identifying the TMEM16F inhibitors and identified three independent structural classes of drugs that could inhibit TMEM16F. Notably, a highly specific and potent inhibitor of TMEM16F (A6–001) was identified, which abrogated both SARS-CoV-2 stimulated phosphatidylserine exposure and SARS-CoV-2 replication in a variety of host cells [30]. Taken together, these studies establish a crucial role for TMEM16F in SARS-CoV-2 entry into host cells.
Recently, Luu et al. demonstrated that Pannexin 1 (Panx1), a plasma membrane channel, plays a critical role in COVID-19 lung pathogenesis [31]. Panx1, along with purinergic receptors, regulates intracellular Ca2+ signaling [32]. Panx1 mediates release of nucleotides, lipids, and small RNA into the extracellular space. The release of ATP upon opening of the Panx1 channel enables increase in extracellular ATP, which in turn facilitates Ca2+ and K + fluxes via purinergic receptors. The authors demonstrated that SARS-CoV-2 infection leads to opening of Panx1 channels. Panx1 remains closed under healthy conditions, while during viral infection this channel opens transiently and releases ATP, prostaglandins and cytokines to activate immune response [31]. Further, using publicly available datasets the authors reported that the expression of Panx1 mRNA is higher in nasal and bronchial epithelial cells isolated from SARS-CoV-2 patients in comparison to healthy controls. Moreover, it was demonstrated that Panx1 protein expression is greater in the lung tissues retrieved from lethal COVID-19 cases as compared to lung tumor biopsies used as control in this analysis [31]. The increased Panx1 expression was associated with enhanced accumulation of ATP, PGE2 and IL-1β in bronchiolar alveolar lavage of SARS-CoV-2 patients. Importantly, Probenecid (a Panx1 blocker) prevented SARS-CoV-2 replication [31]. Therefore, Panx1 shows a dual role in SARS-CoV2 infection i.e. facilitation of viral replication and induction of immune response. Collectively, the literature suggests that Panx1 is a critical regulator of COVID-19 pathogenesis.
3. SARS-CoV-2 hijacks host mitochondrial signaling for viral replication
SARS-CoV-2 is a positive sense, single-stranded RNA virus encoding a RNA-dependent RNA polymerase for replication. Once the viral genome is released in the cytosol, transcription of viral genes takes place in cytosol on the convoluted membranes (CMs). Similar to other viruses, SARS-CoV-2 utilizes host cellular metabolites like nucleotides, amino acids and fatty acids for its replication [33]. In response to viral infection, the host cell releases mitochondrial DNA (mtDNA) and activates mitochondrial antiviral signaling (MAVS) proteins for interferon stimulation and initiation of the innate immune response. Thus, the hijacking of mitochondria by the virus is not only pertinent for controlling host metabolism but also for regulating mitochondria-associated danger signals and immune activation. The majority of viruses target mitochondria by orchestrating morphological changes in mitochondria [34]. SARS-CoV infection results in elongation of mitochondria that is associated with convoluted membrane (CM) formation required for viral replication [34]. Mitochondrial elongation and viral replication also disrupt mitochondrial associated membranes (MAMs), which are ER-mitochondrial contact sites thereby altering downstream processes like autophagy [35]. Some viruses induce mitophagy to prevent the release of mtDNA and the initiation of MAVS. Viral infection also prevents activation of apoptosis during the early viral cycle by modulating the permeability of voltage-dependent anion channel (VDAC) and activating Ca2+ and ROS dependent survival pathways [34].
SARS-CoV-2 utilizes similar strategies to modulate mitochondrial function, metabolism, autophagy, immune activation and apoptosis for its replication [33]. To induce the above changes in host cell function, SARS-CoV-2 proteins interact with the host protein and localize to different organelles. Recently, Gordon et al. expressed 26 out of 29 viral proteins in a human cell line (HEK293T/17) and identified 332 high confidence physical interactions of the human proteins with SARS-CoV-2 proteins using affinity purification mass spectrometry (AP-MS) [36]. Interestingly, a variety of SARS-CoV-2 proteins are shown to interact with mitochondrial proteins. SARS-CoV-2 ORF9c interacts with electron transport chain (ETC) complexes and has a possible role in the regulation of oxidative phosphorylation [36]. ORF9c also interacts with outer mitochondrial membrane (OMM) protein probably resulting in negative regulation of MAVS [36]. SARS-CoV-2 Nsp4 protein that resides on ER potentially interacts with inner mitochondrial membrane (IMM) proteins thereby regulating mitochondrial morphology and CM formation. Similarly, M-protein is shown to interact with a number of mitochondrial metabolism proteins. Further studies are required to validate the protein-protein interaction networks emerging from this study and to characterize the implications of these interactions on host cell functions.
Recently, it was demonstrated that SARS-CoV-2 can maneuver host cell Ca2+ signaling by targeting its protein to host intracellular organelles [37]. In this study, transcriptome analysis was performed on peripheral blood mononuclear cells (PBMCs) isolated from healthy control, mildly symptomatic and severely-ill COVID-19 patients admitted to the intensive care unit (ICU). The data shows an association of the severity of the COVID-19 disease with transcriptomic changes in three prominent cellular machineries, namely mitochondrial complexes, Ca2+ channel subunits and autophagy-related translational precursors. Ramachandran et al. further studied the effect of ectopic expression of numerous SARS-CoV-2 proteins, such as M-protein, NSP6, NSP10, ORF3a and ORF9a on host cellular functions. They showed that the viral membrane protein (M-protein) localizes particularly to mitochondria and potentially alters mitochondrial morphology. M-protein also interacts with mitochondrial protein CCDC58. The authors observed that CCDC58 expression was significantly upregulated in PBMC of COVID-19 patients. Previously, it was reported that the CCDC58 is involved in regulating the mitochondrial Ca2+ retention capacity of the cells [38]. The effect of increased CCDC58 expression and its interaction with SARS-CoV-2 M-protein on mitochondrial Ca2+ retention capacity (CRC) was studied in CCDC58 silenced HEK cells. The ectopic expression of SARS-CoV-2 M-protein in CCDC58 knockdown cells decreases mitochondrial CRC, but does not affect the activity of mitochondrial calcium uniporter (MCU). The study further highlights the interaction of other SARS-CoV-2 proteins such as NSP6, ORF3a, ORF9b, ORF9c and ORF10 with mPTP proteins such as adenine nucleotide transporter (ANT) and ATP synthase in the COS7 cell line [37]. Based on the co-immunoprecipitation and immunofluorescence studies, the authors reported that the SARS-CoV-2 proteins target mitochondria and interact with mPTP leading to pore opening and mitochondrial dysfunction. In continuation, the ectopic expression of SARS-CoV-2 M-protein or ORF3a showed a remarkable decrease in basal respiration rate while the ectopic expression of SARS-CoV-2 NSP6 and NSP7 elicited partial suppression of maximal oxygen consumption rate (OCR). Interaction of these SARS-CoV-2 proteins with the host mPTP proteins and respiratory complexes can modulate mitochondrial metabolism and cellular energetics for its survival.
Ramachandran and colleagues further studied the effect of SARS-CoV-2 infection on cardiomyocytes as severely-ill SARS-CoV-2 patients show elevated cardiac injury markers such as troponin-C, interleukin-6 (IL-6) and lactate dehydrogenase (LDH) [37]. In human-induced pluripotent stem cells derived cardiomyocytes (hiPSC—CMs), SARS-CoV-2 infection collapses cytoskeleton, ER morphology and increases autophagic flux. Transcriptome analysis of SARS-CoV-2 infection in hiPSC—CMs showed a significant alteration in the gene expression of ion channels. While an elevated expression of Ca2+channels such as l-Type calcium channel (LTCC) subunits, transient receptor potential channel (TRP), inositol 1,4,5-triphosphate (IP3R) and MCU was observed in SARS-CoV-2 infected cardiomyocytes, the levels of Ca2+ release-activated Ca2+ channel (CRAC) Orai3 and mitochondrial magnesium channel MRS2 were downregulated. The authors further showed that the alteration in the expression of ion channels severely affects cardiomyocyte rhythmicity and function. Taken together, this study reveals that the SARS-CoV-2 infection transcriptionally alters the expression of Ca2+ channels and regulates Ca2+ homeostasis. Further, SARS-CoV-2 targets its protein to mitochondrial compartments to alter cellular processes such as the mitochondrial morphology and Ca2+ retention capacity. Importantly, this hijacking of host mitochondrial Ca2+ signaling by SARS-CoV-2 modulates cellular functions like respiration and apoptosis for enabling its pathogenesis.
4. SARS-CoV-2 regulates host Ca2+ permeability for viral egress
Viral assembly and release from the host cell are the crucial stage of the virus life cycle for the spread of infection. To carry out this, viruses target host cell ion permeability to disrupt the ionic gradients across ER and plasma membrane by utilizing their own proteins [39]. These proteins are termed as viroporins. Viroporins have a transmembrane hydrophilic pore, that possesses selectivity for charged molecules and ions along with the concentration gradients [39]. Viroporins are mostly present on the ER membrane but may also be present on the plasma membrane. So far, three viroporins are reported in SARS-CoV family viruses i.e. envelope protein (E-protein), open reading frame (ORF) 3a and ORF8 protein [39]. E-protein is a non-selective cation channel. Studies on SARS-CoV-2 E-protein reconstituted in lipid bilayer membrane shows that E-protein is highly permeable to monovalent cations like K + and Na+ ions and has low permeability to divalent ions like Ca2+ and Mg2+ [40]. E-protein is an important player in the maturation and localization of SARS-CoV-2 S-protein in ER Golgi intermediate component (ERGIC) by slowing down the cell secretory pathway [41]. In HEK293 cells, co-expression of SARS-CoV M and N structural proteins in absence of E-protein showed sufficient budding of virus-like particles (VLPs). However, these cells were inefficient in the secretion of VLPs. Thus, E-protein may have a potential role in regulating secretory pathway and viral exocytosis [41].
SARS-CoV-2 ORF3a contains short N-terminal domain present in the ER/Golgi lumen, three transmembrane domains and a longer C-terminal domain situated in the cytosol [39]. ORF3a localizes mostly on Golgi but is also expressed on ER and lysosomes. Reconstruction studies of SARS-CoV-2 ORF3a in proteo-liposome membrane discs suggest that ORF3a is a non-selective cation channel with a permeability order of Ca2+ > K+ > Na+ [42]. Due to its high selectivity towards Ca2+, ORF3a is previously shown to play a role in the regulation of autophagy, pro-apoptotic pathways and NLRP3 inflammasome activation in SARS-CoV infection [43], [44], [45]. Recent studies of SARS-CoV-2 ORF3a suggest that it may potentially regulate exocytosis for viral egress [46]. Overexpression of SARS-CoV-2 ORF3a on lysosomes of HeLa and COS7 cells enhances the expression of lysosomal associated membrane protein 1 (LAMP1) in the plasma membrane fraction, indicating the interaction of lysosomes to the plasma membrane. This increase in the expression of LAMP1 in membrane fraction is not observed upon SARS-CoV ORF3a overexpression [46] suggesting that it's a highly specific phenomenon associated with SARS-CoV-2. The authors further showed that ORF3a increases BLOC related complex – Arf like protein 8b (BORC-ARL8b) expression on the lysosome to promote its movement towards the plasma membrane. ORF3a also interacts with SNARE complex protein and promotes its localization on the plasma membrane to mediate exocytosis [46]. Interestingly, this study demonstrates that the enhanced fusion of lysosomes and plasma membrane upon ectopic expression of ORF3a is mediated by an increase in cytosolic Ca2+levels. Co-localization experiments with ORF3a revealed involvement of TRPML channels in lysosomal exocytosis. TRPML3, a widely distributed (PM, ER, lysosomes) Ca2+channel was shown to be facilitating this process by mobilizing intracellular Ca2+ stores. Depletion of TRPML3 reduced PM-localized LAMP1 levels and suppressed lysosomal exocytosis [46]. Therefore, the interplay between ORF3a and TRPML3 increases the cytosolic Ca2+ levels and drives viral exocytosis. Collectively, SARS-CoV-2 regulates host cell ionic permeability by expressing viroporin on different organelles and hijacking host cell Ca2+ homeostasis for promoting its egress.
5. SARS-CoV-2 modulates Ca2+signaling for mounting immune response
The SARS-CoV-2 related deaths are typically characterized by dysfunctional immune responses and severe pulmonary injury. They are a consequence of cytokine storm and subsequent acute respiratory distress syndrome [40]. The uncontrolled activation of immune response attacking various tissues is due to excessive secretion of pro-inflammatory cytokines such as IL-1β, IL-6, IL-10, TNF-α etc. and chemokines like CXCL10, CXCL9, CCL2, CCL3, CCL5 etc. After the inflammatory outburst, the immune system in severe COVID-19 patients may be programed to an immunosuppressive condition, leading to the collapse of the whole immune system. Upon infection, SARS-CoV-2 induces an influx of Ca2+ in host epithelial and immune cells [47]. As evident from other inflammatory conditions, increased intracellular Ca2+ levels through CRAC is a critical regulator of the immune response. It activates phosphatase calcineurin that induces one of the following three signaling pathways: NFAT pathway, NF-κB pathway and c-Jun NH2-terminal kinase (JNK) pathway [48], [49], [50]. CRAC channels are a sub-class of store-operated calcium entry (SOCE), which as the name suggests are activated by depletion of Ca2+ from ER stores. Two main components of SOCE that regulate the entry of extracellular Ca2+ are plasma membrane-localized pore unit (Orai1) and ER-localized sensor protein (STIM1). The role of SOCE was studied in SARS-CoV-2 infection in HEK-293-ACE-2 cells by silencing Orai1 and STIM1 [51]. Both Orai1 and STIM1 knockout cells show complete abrogation of SOCE. Further, measurement of cytosolic Ca2+ in presence of 2 mM and 20 mM extracellular Ca2+ without depleting ER stores showed a reduction in baseline Ca2+ levels only in Orai1 silenced cells suggesting an additional role of Orai1 in regulating cytosolic Ca2+ homeostasis. Interestingly, infection of SARS-CoV-2 upon either Orai1 or STIM1 silencing showed the opposite phenotype. While Orai1 knockout cells showed high infectivity, increased viral genome copy number and enhanced cell lysis post-infection, STIM1 knockout cells showed resistance to infection [51]. The resistance to SARS-CoV-2 infection upon STIM1 silencing is attained by increased levels of baseline interferon-β (IFN-β) levels, which on the other hand, is reduced in Orai1 knockout cells. Transcriptomic analysis of Orai1 knockout cells showed differential regulation of immune-related genes like MEF2C, FOS, JUN and ATF2. The authors suggested that the inefficient mounting of tonic IFN-I response in Orai1 knockout cells is probably due to changes in Ca2+ dynamics resulting in insufficient activation of the c-JUN pathway. Further, the expression of several antiviral signaling cascades were decreased in Orai1 knockout cells. [51]. Taken together, this study demonstrates an intriguing role of Orai1 in maintaining the basal Ca2+ concentration required for mounting an immune response against SARS-CoV-2 infection.
While the above study shows that Orai1 plays an important role in mounting an inflammatory response against SARS-CoV-2 to prevent the spread of infection, under several other inflammatory conditions such as acute pancreatitis (AP), Orai1 mediated sustained Ca2+ influx leads to hyper-inflammatory pathogenesis [52]. In acute pancreatitis, unknown factors cause the release of Ca2+ from ER, leading to the depletion of ER Ca2+. This is followed by activation and translocation of STIM1 to ER-plasma membrane junctions for gating Orai1. This results in a sustained overload of Ca2+ in the cytosol. Breaking this vicious loop of Ca2+ influx by targeting Orai1 is proven to be effective in the case of AP. Considering that SARS-CoV-2 pathogenesis is associated with uncontrolled cytokine storm and inflammation similar to AP, CalciMedica tested a small molecule “Auxora” for COVID-19 management. Auxora is a potent selective inhibitor of the Orai1 channel. Auxora was tested in three clinical trials. The first clinical trial was to study Auxora (CARDEA) in patients with severe COVID-19 pneumonia (https://clinicaltrials.gov/ct2/show/NCT04345614). The drug is in Phase 2 clinical trial. 284 participants were enrolled to determine the efficacy, safety and pharmacokinetic profile of Auxora in severe COVID-19 patients. The inclusion of patients was done based on the baseline ratio of arterial oxygen partial pressure (PaO2 in mmHg) to fractional inspired oxygen (FiO2 expressed as a fraction), PaO2/FiO2 ≤ 200. The study reported that the recovery time was reduced to 7 days from 10 days for patients who received Auxora vs placebo [53]. The all-cause mortality rate with Auxora vs placebo on day 30 was 7.6% and 17.6% while on day 60 was 13.8% and 20.6% respectively [52]. The second clinical trial was a single-blind dose-ranging pharmacodynamic study of Auxora (CM4620- injectable emulsion) in patients with critical COVID-19 Pneumonia (https://clinicaltrials.gov/ct2/show/NCT04661540). The study is in Phase 2 clinical trial with 36 patients enrolled. The third clinical trial study of Auxora is for the extended treatment of high-risk patients with critical COVID-19 pneumonia (CARDEA-Plus) (https://www.clinicaltrials.gov/ct2/show/NCT05171920). Although this particular trial was withdrawn due to the declining COVID-19 cases, the data clearly suggest a promising effect of Auxora in treatment and management COVID-19 [53].
6. Future perspectives
The initial understanding of SARS-CoV-2 mainly comes from the clinical studies revealing the pathological changes in blood and tissues of the COVID-19 patients. Among these pathological markers, one of the striking features observed in SARS-CoV-2 patients is hypocalcemia [54]. Hypocalcemia is often associated with the disease severity in COVID-19 patients. It is debatable whether hypocalcemia is induced by the host to inhibit SARS-CoV-2 entry into the cells or a viral strategy to maneuver the host defense response. A larger perception is that hypocalcemia is a host triggered response that inhibits Ca2+ entry in the cells to disrupt the viral cycle [55]. While at the systemic level host decreases Ca2+ in the plasma mimicking hypocalcemia, at the cellular level SARS-CoV-2 increases the intracellular flux of Ca2+, most likely by augmenting Ca2+ release from ER. Although the mechanistic details pertaining to the regulation of Ca2+ signaling by SARS-CoV-2 are by far limited, based on inhibitor screenings, we propose that Ca2+ signaling is regulated at multiple levels in SARS-CoV-2 infection [6]. Inhibitors targeting H1 histamine receptor, M1, M2, and M5 muscarinic receptor and 5-HT2 serotonin receptors showed decrease syncytia formation [29]. These receptors regulate Ca2+ signaling through Gqα receptors to activate PLCß, which in turn hydrolyses PIP2 to generate Ins(1,4,5)P3 (IP3). IP3 acts as a second messenger that activates Ca2+ release into the cytoplasm from the ER stores upon binding to IP3R. Other molecules shown to be effective in prevention of syncytia formation belongs to the tricyclic antidepressants and the l-type Ca2+ channel blockers that inhibit the voltage-dependent l-type calcium channels [29]. Non-canonical channels such TMEM16F and Panx1 are also shown to modulate Ca2+ signaling upon SARS-CoV-2 infection [29], [30], [31], [32]. The tussle between the host and the virus for modulating Ca2+ signaling is critical in mounting infection and pathogenesis. Indeed, a number of small molecules that regulate cellular Ca2+ dynamics have shown promising results in stalling SARS-CoV-2 infection and/or curtailing COVID-19 pathogenesis in clinical trials (https://clinicaltrials.gov/ct2/show/NCT04345614, https://clinicaltrials.gov/ct2/show/NCT04661540 https://clinicaltrials.gov/ct2/show/NCT04446377, https://clinicaltrials.gov/ct2/show/study/NCT04308317). These molecules target several independent Ca2+ handling proteins such as CRAC channel (Orai1), PIKfyve and TPC2. Based on the early results from these trials, Ca2+ signaling pathways appear to be a promising target for clinical management of COVID-19. Please refer to Fig. 1 for the details of role of Ca2+ signaling in SARS-CoV-2 infection and the site of action of the small molecules, which are under clinical trials.
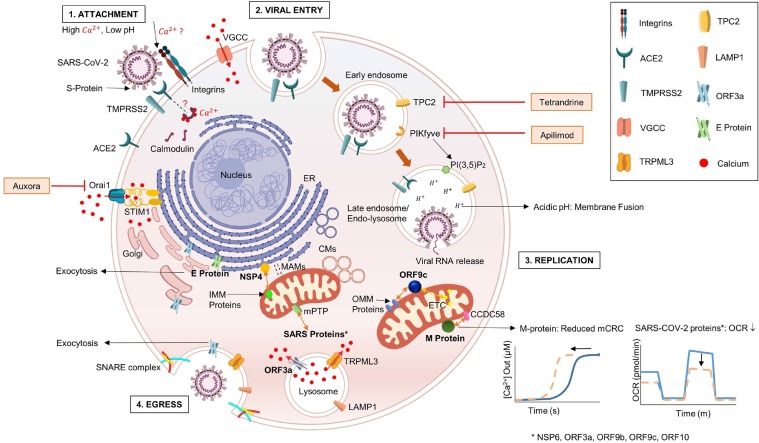
Fig. 1.
Crosstalk between SARS-CoV-2 proteins and host cell Ca2+dynamics. Host cell Ca2+signaling regulates all four stages of viral life cell i.e., 1. Attachment to the host cell; 2. Entry; 3. Replication and 4. Egress. SARS-CoV-2 entry into the host cell is mediated by the interaction of viral S-protein with the host ACE2 surface receptors. The process is facilitated by cell surface serine protease-TMPRSS2, which cleaves S1/S2 subunits of the S-protein. Ca2+ plays an important role in manipulating viral entry. S-protein-integrin interaction is possibly maneuvered by Ca2+. Calmodulin, a downstream target protein of Ca2+ can regulate infection by altering ACE2 stability. Following entry, early endosomal conversion to late endosomes is mediated by PIKfyve enzyme which synthesizes PI(3,5)P2. Apilimod, a drug targeting PIKfyve enzyme is in phase 2 clinical trial. Viral replication and protein synthesis begins following viral fusion with the endo-lysosome and release of viral RNA into the host. Tetrandrine targets Ca2+efflux channel-TPC2 localized on the endo-lysosomal system to inhibit viral entry. A small molecule, Auxora inhibits Orai1 to limit the sustained Ca2+influx into the cytosol via SOCE. It can tackle the uncontrolled cytokine storm and inflammation involved in SARS-CoV-2 pathogenesis. Crosstalk between several viral and host proteins modulate host cellular processes. ORF9c interacts with ETC complexes and OMM proteins to regulate oxidative phosphorylation and MAVS, respectively. Viral M-protein interacts with mitochondrial protein CCDC58 leading to reduced mitochondrial CRC. Several SARS proteins such as NSP6, ORF3a, ORF9b, ORF9c and ORF10 interact with mPTP causing pore opening, OCR reduction and mitochondrial dysfunction. Viral NSP4 interacts with IMM proteins to regulate mitochondrial morphology and convoluted membranes formation. Interaction between host TRPML3 and viral ORF3a plays a crucial role in lysosomal exocytosis by manipulating cytosolic Ca2+levels and interacting with key proteins involved in the exocytic pathway, thereby promoting egress of mature virions.
Interestingly, all these inhibitors decrease the flux of Ca2+ in the cytosol resulting in reduced infection, however, the underlying mechanism remains poorly understood. One of the possible mechanisms is through regulation of the presence of ACE2 receptor on the plasma membrane. The mechanism that stabilizes the ACE2 receptor on the plasma membrane was studied back in 2008 by two independent groups [56,57]. These studies identified the calmodulin-binding domain within the cytoplasmic domain of ACE2 and showed that the interaction of ACE2 with calmodulin prevents the shedding of its ectodomain. The first study predicted the 10 amino acid sequence “TGIRDRKKKN”, conserved in mice, rats and humans, that contains calmodulin-binding domain [57]. The authors demonstrated that the immunoprecipitation of HEK-ACE2 cells lysate with calmodulin antibody showed a 120 KDa band, which is the expected size of ACE2 [57]. The other study utilized a 16 amino acid synthetic peptide mimicking the cytoplasmic region of ACE2 and performed an electrophoretic mobility shift assay (EMSA) with calmodulin (CaM) [56]. Both the groups showed that calmodulin inhibitors, stimulate ACE2 ectodomain shedding. These studies corroborate the binding of CaM with ACE2 to stabilize its expression on the plasma membrane. As calmodulin activation is dependent on intracellular Ca2+ levels, the stability of ACE2 on the host plasma membrane is mediated by Ca2+ dynamics [58]. It should be noted that the stability of ACE2 on the host plasma membrane is not studied in SARS-CoV-2 infection. Here, based on the above discussed literature [56], [57], [58], [59], we hypothesize that the increased cytosolic Ca2+ upon SARS-CoV-2 infection would activate calmodulin, which in turn binds to the cytosolic domain of the ACE2 receptor. Upon inhibition of the cytosolic Ca2+ influx, calmodulin remains in its inactive state. Thereby, the binding of calmodulin with ACE2 is weaker and stability of the ACE2 receptor on the host plasma membrane is poor. As a result, the ectodomain of ACE2 is shed off leading to less binding with viral S-protein and decreased internalization of the virus. Going forward, it would be interesting to test this hypothesis experimentally.
The subtle changes in the compartmentalization of intracellular Ca2+ are strategically mediated by the localization of SARS-CoV-2 proteins to host lysosomes, Golgi and ER, which leads to the release of Ca2+ in the cytosol [46]. Interestingly, SARS-CoV-2 M-protein interacts with mPTP and regulates mitochondrial permeability, CRC and respiration [37]. Several studies have shown that SARS-CoV-2 induces perturbations in Ca2+ homeostasis by employing its viroporins for hijacking key cellular processes like autophagy, apoptosis and immune response [34,44,45]. As evident from numerous studies discussed above, Ca2+ signaling is important for viral attachment, entry and egress. These studies have laid down the map of SARS-CoV-2 protein interactions with the host proteins (Please refer Table 1 for the details of such interactions). However, the mechanistic details of these interactions and their outcome on the pathogenesis are yet to be understood. Further studies are required to validate high-throughput genomics and proteomics data for detailed characterization of the role of SARS-CoV-2 proteins in modulating host cell Ca2+dynamics and its consequence on COVID-19 severity.
Declaration of Competing Interest
Authors report no conflicts of interest.
Acknowledgments
This work was supported by the DBT/Wellcome Trust India Alliance Fellowship (IA/I/19/2/504651) awarded to Rajender K Motiani. Authors also acknowledge RCB core funding. KA acknowledges her Junior Research Fellowship from DBT, India.
Data Availability
No data was used for the research described in the article.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mokhtari T., Hassani F., Ghaffari N., Ebrahimi B., Yarahmadi A. COVID-19 and multiorgan failure : a narrative review on potential mechanisms. J. Mol. Histol. 2020 doi: 10.1007/s10735-020-09915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.AL-KINDI S., ZIDAR D.A. COVID-lateral damage: cardiovascular manifestations of SARS-CoV-2 infection. Transl. Res. 2022;241:25–40. doi: 10.1016/j.trsl.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellido V., Pérez A. Covid-19 and diabetes. J. Clin. Med. 2021;10:5341. doi: 10.3390/jcm10225341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaur S., Bansal R., Kollimuttathuillam S., Gowda A.M., Singh B., Mehta D., Maroules M. The looming storm: blood and cytokines in COVID-19. Blood Rev. 2021;46 doi: 10.1016/j.blre.2020.100743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berlansky S., Sallinger M., Grabmayr H., Fahrner M., Frischauf I., Humer C., Bernhard A. Calcium signals during SARS-CoV-2 infection: assessing the potential of emerging therapies. Cells. 2022;11:253. doi: 10.3390/cells11020253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.-.X., Nicastri E., Oda R., Yo K., Quiros-Roldan E., Studemeister A., Redinski J., Ahmed S., Bernett J., Chelliah D., Chen D., Chihara S., Cohen S.H., Cunningham J., D'Arminio Monforte A., Ismail S., Kato H., Lapadula G., L'Her E., Maeno T., Majumder S., Massari M., Mora-Rillo M., Mutoh Y., Nguyen D., Verweij E., Zoufaly A., Osinusi A.O., DeZure A., Zhao Y., Zhong L., Chokkalingam A., Elboudwarej E., Telep L., Timbs L., Henne I., Sellers S., Cao H., Tan S.K., Winterbourne L., Desai P., Mera R., Gaggar A., Myers R.P., Brainard D.M., Childs R., Flanigan T. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/nejmoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon C.J., Tchesnokov E.P., Schinazi R.F., Götte M. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y., Fang C., Zhang Q., Zhang R., Zhao X., Duan Y., Wang H., Zhu Y., Feng L., Zhao J., Shao M., Yang X., Zhang L., Peng C., Yang K., Ma D., Rao Z., Yang H. Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell. 2021 doi: 10.1007/s13238-021-00883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saurav S., Tanwar J., Ahuja K., Motiani R.K. Dysregulation of host cell calcium signaling during viral infections: emerging paradigm with high clinical relevance. Mol. Aspects Med. 2021;81 doi: 10.1016/j.mam.2021.101004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Straus M.R., Bidon M.K., Tang T., Jaimes J.A., Whittaker G.R., Daniel S. Inhibitors of L-type calcium channels show therapeutic potential for treating SARS-CoV-2 infections by preventing virus entry and spread. ACS Infect. Dis. 2021;7:2807–2815. doi: 10.1021/acsinfecdis.1c00023. [DOI] [PubMed] [Google Scholar]
- 12.Bagur R., Hajnóczky G. Intracellular Ca(2+) sensing: its role in calcium homeostasis and signaling. Mol. Cell. 2017;66:780–788. doi: 10.1016/j.molcel.2017.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanwar J., Motiani R.K. Role of SOCE architects STIM and Orai proteins in cell death. Cell Calcium. 2018;69:19–27. doi: 10.1016/j.ceca.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Clapham D.E. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 15.Tanwar J., Trebak M., Motiani R.K. Cardiovascular and hemostatic disorders: role of STIM and orai proteins in vascular disorders. Adv. Exp. Med. Biol. 2017;993:425–452. doi: 10.1007/978-3-319-57732-6_22. [DOI] [PubMed] [Google Scholar]
- 16.Sharma N., Arora S., Saurav S., Motiani R.K. Pathophysiological significance of calcium signaling at mitochondria-associated endoplasmic reticulum membranes (MAMs) Curr. Opin. Physiol. 2020;17:234–242. doi: 10.1016/j.cophys.2020.08.012. [DOI] [Google Scholar]
- 17.Tanwar J., Singh J.B., Motiani R.K. Molecular machinery regulating mitochondrial calcium levels: the nuts and bolts of mitochondrial calcium dynamics. Mitochondrion. 2020;57:9–22. doi: 10.1016/j.mito.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parys J.B., Guse A.H. Full focus on calcium. Sci. Signal. 2019;12:1–4. doi: 10.1126/scisignal.aaz0961. [DOI] [PubMed] [Google Scholar]
- 19.Patel S., Muallem S. Acidic Ca2+ stores come to the fore. Cell Calcium. 2011;50:109–112. doi: 10.1016/j.ceca.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Bai D., Fang L., Xia S., Ke W., Wang J., Wu X., Fang P., Xiao S. Porcine deltacoronavirus (PDCoV) modulates calcium influx to favor viral replication. Virology. 2020;539:38–48. doi: 10.1016/j.virol.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson C.B., Farzan M., Chen B. Mechanisms of SARS-CoV-2 entry into cells. Mol. Cell Biol. 2022;23:20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 23.Dakal T.C. SARS-CoV-2 attachment to host cells is possibly mediated via RGD-integrin interaction in a calcium-dependent manner and suggests pulmonary EDTA chelation therapy as a novel treatment for COVID 19. Immunobiology. 2021;226 doi: 10.1016/j.imbio.2020.152021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khelashvili G., Plante A., Doktorova M., Weinstein H. Ca2+-dependent mechanism of membrane insertion and destabilization by the SARS-CoV-2 fusion peptide. Biophys. J. 2021;120:1105–1119. doi: 10.1016/j.bpj.2021.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh P., Mukherji S., Basak S., Hoffmann M., Das D.K. Dynamic Ca2+ sensitivity stimulates the evolved SARS-CoV-2 spike strain-mediated membrane fusion for enhanced entry. Cell Rep. 2022 doi: 10.1016/j.celrep.2022.110694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Z., Qin P., Huang Y. Lysosomal ion channels involved in cellular entry and uncoating of enveloped viruses: implications for therapeutic strategies against SARS-CoV-2. Cell Calcium. 2021;94 doi: 10.1016/j.ceca.2021.102360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin N., Lang M.J. Phosphatidylinositol 3,5-bisphosphate: regulation of cellular events in space and time. Inf. Theory Quantum Phys. 2016;44:177–184. doi: 10.1007/978-3-642-57162-6_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braga L., Ali H., Chiavacci E., Neves G., Penn R., Jimenez-guardeño J.M., Ortega-prieto A.M., Cannatà A., Rizzari G., Collesi C., Schneider E. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 Spike- induced syncytia. Nature. 2021;594:88–93. doi: 10.1038/s41586-021-03491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sim J.-.R., Shin D.H., Park P.-.G., Park S.-.H., Bae J.-.Y., Lee Y., Kang D.-Y., Kim Y.J., Aum S., Noh S.H., Hwang S.J., Cha H.-.R., Kim C.B., Ko S.H., Park S., Jeon D., Cho S., Lee G.E., Kim J., Moon Y.-.H., Kim J.-.O., Nam J.-.S., Kim C.-.H., Moon S., Chung Y.W., Park M.-.S., Ryu J.-.H., Namkung W., Lee J.M., Lee M.G. Amelioration of SARS-CoV2 infection by ANO6 phospholipid scramblase infection. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luu R., Valdebenito S., Scemes E., Cibelli A., Spray D.C., Rovegno M., Tichauer J., Cottignies-Calamarte A., Rosenberg A., Capron C., Belouzard S., Dubuisson J., Annane D., de la Grandmaison G.L., Cramer-Bordé E., Bomsel M., Eugenin E. Pannexin-1 channel opening is critical for COVID-19 pathogenesis. IScience. 2021:24. doi: 10.1016/j.isci.2021.103478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nadeali Z., Mohammad-rezaei F., Aria H., Nikpour P. Possible role of pannexin 1 channels and purinergic receptors in the pathogenesis and mechanism of action of SARS-CoV-2 and therapeutic potential of targeting them in COVID-19. Life Sci. 2022;297 doi: 10.1016/j.lfs.2022.120482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh K.K., Chaubey G., Chen J.Y., Suravajhala P. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am. J. Physiol. Cell Physiol. 2020;319:258–267. doi: 10.1152/ajpcell.00224.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gatti P., Ilamathi H.S., Todkar K., Germain M. Mitochondria targeted viral replication and survival strategies — prospective on SARS-CoV-2. Front. Pharmacol. 2020:11. doi: 10.3389/fphar.2020.578599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong N.A., Saier M.H. The sars-coronavirus infection cycle: a survey of viral membrane proteins, their functional interactions and pathogenesis. Int. J. Mol. Sci. 2021;22:1–63. doi: 10.3390/ijms22031308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A. A SARS-CoV-2 protein interaction map reveals targets for drug- repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9.A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramachandran K., Maity S., Brian W., El-aziz T.M.A., Allen C., Sun Y., Venkatesan M., Madaris T.R., Stockand J.D., Singh B.B., Srikantan S., Reeves W.B., Madesh M. SARS-CoV-2 infection enhances mitochondrial PTP. IScience. 2022;25 doi: 10.1016/j.isci.2021.103722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shanmughapriya S., Rajan S., Hoffman N.E., Higgins A.M., Tomar D., Nemani N., Hines K.J., Smith D.J., Eguchi A., Vallem S., Shaikh F., Cheung M., Leonard N.J., Stolakis R.S., Wolfers M.P., Ibetti J., Chuprun J.K., Jog N.R., Houser S.R., Koch W.J., Elrod J.W., Madesh M. SPG7 is an essential and conserved component of the mitochondrial permeability transition pore. Mol. Cell. 2015;60:47–62. doi: 10.1016/j.molcel.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Breitinger U., Farag N.S., Sticht H., Breitinger H.G. Viroporins: structure, function, and their role in the life cycle of SARS-CoV-2. Int. J. Biochem. Cell Biol. 2022;145 doi: 10.1016/j.biocel.2022.106185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia B., Shen X., He Y., Pan X., Liu F., Wang Y., Yang F., Fang S., Wu Y., Duan Z. SARS-CoV-2 envelope protein causes acute respiratory distress syndrome (ARDS) -like pathological damages and constitutes an antiviral target. Cell Res. 2021;31:847–860. doi: 10.1038/s41422-021-00519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boson B., Legros V., Zhou B., Siret E., Mathieu C., Cosset F., Lavillette D., Denolly S. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 2021;296 doi: 10.1074/jbc.RA120.016175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kern D.M., Sorum B., Hoel C.M., Sridharan S., Remis J.P., Toso D.B., Kotecha A., Bautista D.M., Brohawn S.G. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat. Struct. Mol. Biol. 2021;28:573–582. doi: 10.1038/s41594-021-00619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu H., Akinyemi I.A., Chitre S.A., Loeb J.C., Lednicky J.A., McIntosh M.T., Bhaduri-McIntosh S. SARS-CoV-2 viroporin encoded by ORF3a triggers the NLRP3 inflammatory pathway. Virology. 2022;568:13–22. doi: 10.1016/j.virol.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su W.Q., Yu X.J., Zhou C.M. SARS-CoV-2 ORF3a induces incomplete autophagy via the unfolded protein response. Viruses. 2021;13:2467. doi: 10.3390/v13122467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., Han Y., Zhang X., Zhou W. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen D., Zheng Q., Sun L., Ji M., Li Y., Deng H., Zhang H. ORF3a of SARS-CoV-2 promotes lysosomal exocytosis-mediated viral egress. Dev. Cell. 2021;56:3250–3263. doi: 10.1016/j.devcel.2021.10.006. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danta C.C. SARS-CoV-2, hypoxia, and calcium signaling: the consequences and therapeutic options. ACS Pharmacol. Transl. Sci. 2021;4:400–402. doi: 10.1021/acsptsci.0c00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang C.C., Wang J.M., Kikkawa U., Mukai H., Shen M.R., Morita I., Chen B.K., Chang W.C. Calcineurin-mediated dephosphorylation of c-Jun Ser-243 is required for c-Jun protein stability and cell transformation. Oncogene. 2008;27:2422–2429. doi: 10.1038/sj.onc.1210888. [DOI] [PubMed] [Google Scholar]
- 49.Sanna B., Bueno O.F., Dai Y.-.S., Wilkins B.J., Molkentin J.D. Direct and indirect interactions between calcineurin-NFAT and MEK1-extracellular signal-regulated kinase 1/2 signaling pathways regulate cardiac gene expression and cellular growth. Mol. Cell. Biol. 2005;25:865–878. doi: 10.1128/mcb.25.3.865-878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alzuherri H., Chang K.C. Calcineurin activates NF-κB in skeletal muscle C2C12 cells. Cell. Signal. 2003;15:471–478. doi: 10.1016/S0898-6568(02)00120-1. [DOI] [PubMed] [Google Scholar]
- 51.Wu B., Ramaiah A., Garcia G., Hasiakos S., Arumugaswami V., Srikanth S., Alerts E. ORAI1 Limits SARS-CoV-2 Infection by Regulating Tonic Type I IFN Signaling. J. Immunol. 2022;208:74–84. doi: 10.4049/jimmunol.2100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerasimenko O.V., Gerasimenko J.V. CRAC channel inhibitors in pancreatic pathologies. J. Physiol. 2022;600:1597–1598. doi: 10.1113/JP282826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruen C., Al-Saadi M., Michelson E.A., Tanios M., Mendoza-Ayala R., Miller J., Zhang J., Stauderman K., Hebbar S., Hou P.C. Auxora vs. placebo for the treatment of patients with severe COVID-19 pneumonia: a randomized-controlled clinical trial. Crit. Care. 2022;26:1–11. doi: 10.1186/s13054-022-03964-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di Filippo L., Formenti A.M., Rovere-Querini P., Carlucci M., Conte C., Ciceri F., Zangrillo A., Giustina A. Hypocalcemia is highly prevalent and predicts hospitalization in patients with COVID-19. Endocrine. 2020;68:475–478. doi: 10.1007/s12020-020-02383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crespi B., Alcock J. Conflicts over calcium and the treatment of COVID-19. Evol. Med. Public Heal. 2021:149–156. doi: 10.1093/emph/eoaa046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Z.W. Lai, R.A. Lew, M.A. Yarski, F.T. Mu, R.K. Andrews, A.I. Smith, The identification of a calmodulin-binding domain within the cytoplasmic tail of angiotensin-converting enzyme-2, Endocrinology. 150 (2009) 2376–2381. https://doi.org/10.1210/en.2008-1274. [DOI] [PMC free article] [PubMed]
- 57.Lambert D.W., Clarke N.E., Hooper N.M., Turner A.J. Calmodulin interacts with angiotensin-converting enzyme-2 (ACE2) and inhibits shedding of its ectodomain. FEBS Lett. 2008;582:385–390. doi: 10.1016/j.febslet.2007.11.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.García-Escobar A., Vera-Vera S., Jurado-Román A., Jiménez-Valero S., Galeote G., Moreno R. Calcium signaling pathway is involved in the shedding of ACE2 catalytic ectodomain: new insights for clinical and therapeutic applications of ACE2 for COVID-19. Biomolecules. 2022;12:76. doi: 10.3390/biom12010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ragia G., Manolopoulos V.G. Inhibition of SARS-CoV-2 entry through the ACE2/TMPRSS2 pathway: a promising approach for uncovering early COVID-19 drug therapies. Eur. J. Clin. Pharmacol. 2020;76:1623–1630. doi: 10.1007/s00228-020-02963-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.