Abstract
Campylobacter rectus is an important periodontal pathogen in humans. A surface-layer (S-layer) protein and a cytotoxic activity have been characterized and are thought to be its major virulence factors. The cytotoxic activity was suggested to be due to a pore-forming protein toxin belonging to the RTX (repeats in the structural toxins) family. In the present work, two closely related genes, csxA and csxB (for C. rectus S-layer and RTX protein) were cloned from C. rectus and characterized. The Csx proteins appear to be bifunctional and possess two structurally different domains. The N-terminal part shows similarity with S-layer protein, especially SapA and SapB of C. fetus and Crs of C. rectus. The C-terminal part comprising most of CsxA and CsxB is a domain with 48 and 59 glycine-rich canonical nonapeptide repeats, respectively, arranged in three blocks. Purified recombinant Csx peptides bind Ca2+. These are characteristic traits of RTX toxin proteins. The S-layer and RTX domains of Csx are separated by a proline-rich stretch of 48 amino acids. All C. rectus isolates studied contained copies of either the csxA or csxB gene or both; csx genes were absent from all other Campylobacter and Helicobacter species examined. Serum of a patient with acute gingivitis showed a strong reaction to recombinant Csx protein on immunoblots.
Campylobacter rectus, formerly Wolinella recta (39), is a gram-negative, anaerobic, motile bacterium which is associated with several forms of human periodontal diseases (9, 34, 45). Little is known about the molecular mechanisms of pathogenicity and host cell interactions; current investigations have concentrated on characterization of a surface-layer (S-layer) protein, Crs, which is assumed to be involved in resistance of C. rectus to phagocytic uptake and to bactericidal activity of serum (31, 41). Recently two groups have simultaneously cloned and sequenced the gene encoding the S-layer protein and named the gene crs (41) and slp respectively (29); in this study, the designation crs will be used. Studies revealed that depending on the strain, the Crs protein varies slightly in size (from 150 to 166 kDa) and amino acid sequence (30, 41). S-layer proteins form regularly arranged structures on the outer surface of various bacteria. They are assumed to play a role in virulence of several pathogens by rendering the bacteria resistant to complement killing and providing structures for adherence to host cells (4). Cultures of C. rectus strains that had lost most of S-layer structures by continuous passage many times on growth medium showed reduced virulence in a mouse abscess model compared to low-passage cultures (10, 20). The primary task of S-layer proteins seems to be to provide the bacterium with a stable shape and surface so as to resist the harsh conditions encountered during the process of infection. S-layer proteins have also been identified in Campylobacter fetus subsp. fetus but not other Campylobacter or Helicobacter species (Table 1).
TABLE 1.
Campylobacter and Helicobacter strains used
| Species | Straina | csx gene(s)b | Gene encoding S-layer protein (reference) |
|---|---|---|---|
| C. rectus | CCUG11640 | csxA | crsc |
| CCUG11643 | csxA | crsc | |
| CCUG27948 | csxA | crsc | |
| CCUG20446BT | csxA and csxB | crsc, crs (41), slp (29) | |
| CCUG11642 | csxA | crsc | |
| CCUG11645 | csxB | crsc | |
| C. curvus | ATCC 35224T | Neg. | NRd |
| C. concisus | ATCC 33237T | Neg. | NR |
| C. fetus subsp. fetus | ATCC 27374T | Neg. | sapA (5) and sapB (8) |
| C. jejuni | NCTC11351T | Neg. | NR |
| C. upsaliensis | LMG8850T | Neg. | NR |
| C. coli | LMG6440T | Neg. | NR |
| C. sputorum biovar sputorum | LMG7795T | Neg. | NR |
| H. pylori | ATCC 43504T | Neg. | None (7) |
| H. cinaedi | ATCC 35683T | Neg. | NR |
| H. pullorum | NCTC12824T | Neg. | NR |
CCUG, Culture Collection, University of Göteborg, Göteborg, Sweden; ATCC, American Type Culture Collection, Rockville, Md.; NCTC, National Collection of Type Cultures and Pathogenic Fungi, London, England; LMG, Laboratorium voor Microbiologie, Universiteit Gent, Ghent, Belgium.
Determined by Southern blotting and partial sequencing. The gene with higher sequence similarity within the determined part (see text) is given. Neg., negative (no hybridization signal).
Determined by PCR with primers BCRSLP-L and BCRSLP-R (Table 2) and subsequent partial sequencing of the PCR product with primer BCRSLP-L.
NR, no reference found.
Cytotoxic activity of gram-negative bacteria such as proposed for C. rectus (16), however, is often due to pore-forming protein toxins belonging to the family of RTX (repeats in the structural toxin) proteins. RTX toxins, which include the α-hemolysin HlyA of Escherichia coli (11), the leukotoxins LktA of Pasteurella haemolytica (26) and AaltA of Actinobacillus actinomycetemcomintans (21), the cytotoxins ApxIA, ApxIIA, and ApxIIIA of Actinobacillus pleuropneumoniae (14), and the adenylate cyclase CyaA of Bordetella pertussis (17), are major virulence factors of these pathogens (42, 43). The RTX proteins are characterized by a domain located generally in the C-terminal part of the protein consisting of a variable number of highly conserved glycine-rich nonapeptide repeats with the consensus sequence (L/I/F)XGGXG(N/D)DX (42). This domain has a high Ca2+-binding capacity and is involved in binding of the toxin to the target cell (3, 27). C. rectus was suggested to contain genes encoding RTX proteins as shown by Kuhnert et al. (22). Furthermore, Gillespie et al. (16) described a cytotoxic fraction of C. rectus which reacted serologically with antiserum directed against leukotoxin of A. actinomycetemcomitans. Surprisingly, the N-terminal amino acid sequence of the main protein of this fraction is identical to that of the Crs protein described by Wang et al. (41). However, neither native nor recombinant Crs showed cytotoxicity against HL-60 cells or human peripheral leukocytes (29). The present study was therefore undertaken to analyze the presence of two new genes in C. rectus coding for bistructural S-layer-RTX proteins named CsxA and CsxB (C. rectus S-layer-RTX protein).
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmid cloning vectors.
C. rectus strains (Table 1) were grown at 37°C in an atmosphere of 10% CO2 and 1% O2 in H2 either on blood agar plates (Trypticase soy agar supplemented with 0.1% CaCl2 and 5% sheep blood) supplemented with sodium formate (Merck, Darmstadt, Germany) and disodium fumarate (Fluka, Buchs, Switzerland) (each at 0.6 g/liter) or in mff broth described by Gillespie and Holt (15) (PPLO broth without CV [20 g/liter] [Difco Laboratories, Detroit, Mich.] supplemented with 25 mM sodium formate, 50 mM sodium fumarate, and 5 mg of hemin [Sigma-Aldrich, St. Louis, Mo.]). The other Campylobacter spp. and the Helicobacter spp. were routinely cultured at 37°C on blood agar plates under microaerophilic conditions (7% CO2, 7% H2, 80% N2, 6% O2).
E. coli K-12 strain XL1-Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)]) (6) and BL21(DE3) [E. coli B F′ dcm ompT hsdS(rB− mB−) gal λ(DE3)] (37) were used for cloning and gene expression, respectively. Plasmid pBluescriptIISK− (Stratagene, La Jolla, Calif.) was used as the cloning vector. The T7 promoter-based expression vector pETHIS-1 (36a), which is derived from cloning vector pET14b (Novagen, Madison, Wis.) and allows addition of polyhistidine tails at both N- and C-terminal ends of cloned proteins, was used for expression of cloned genes. E. coli strains were grown at 37°C in Luria-Bertani broth supplemented when necessary with ampicillin (50 μg/ml) for selection and maintenance of recombinant plasmids. For blue-white selection with pBluescriptIISK−, 125 μM X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) were added.
Gene cloning and DNA sequencing.
DNA was extracted by the method of Pitcher et al. (33). Plasmid libraries were constructed by cloning restricted total DNA of the C. rectus type strain into pBluescriptIISK−, using conventional techniques (2). Recombinant plasmids were screened by a colony blot assay (2) using digoxigenin (DIG)-labelled DNA probes detected either with CDP-Star (Boehringer Mannheim, Mannheim, Germany) or with NBT (4-nitroblue tetrazolium chloride)-BCIP (5-bromo-4-chloro-3-indolylphosphate) (Boehringer Mannheim) according to the manufacturer’s protocol.
Subclones were generated either with a double-stranded nested deletion kit (Pharmacia LKB, Biotechnology AB, Uppsala, Sweden) or by cutting with restriction enzymes and religating with T4 DNA ligase (Boehringer Mannheim). Sequencing reactions were performed with a Taq Dye Deoxy Terminator cycle sequencing kit (Applied Biosystems/Perkin-Elmer Cetus, Norwalk, Conn.) with either T3 and T7 matching primers flanking the inserts or custom-synthesized internal primers such that all sequences ultimately were read from both directions. Reaction products were analyzed on an ABI Prism 310 genetic analyzer (Applied Biosystems/Perkin-Elmer Cetus).
For direct DNA sequencing from genomic DNA, extraction of DNA was performed with the QIAamp tissue kit (Qiagen GmbH, Hilden, Germany). Seven micrograms of DNA was preincubated with 2 μg of RNase at 37°C for 10 min followed by addition of 16 μl of sequencing premix (ABI Prism Bigdye ABI Primer Cycle Sequencing Ready Reaction kit; Perkin-Elmer), 21 pmol of primer, and an appropriate amount of water to make up total volume of 40 μl. After 3 min at 95°C, 99 cycles of 30 s at 95°C, 20 s at 55°C, and 4 min at 65°C with primer CRS1HisKin (Table 2) and 99 cycles of 30 s at 95°C and 5 min at 65°C with primer CRS1HisKin2 were performed. Reaction products were analyzed on an ABI Prism 310 genetic analyzer.
TABLE 2.
Oligonucleotide primers
| Name | Sequencea | Positionb | Annealing temp (°C) |
|---|---|---|---|
| CRSAP1C-L | ggactagtgATCACCTCGCATAAAGAT | 2560–2577 | 55 |
| CRSAP1C-R | cgcggatccCATACCTGCAAGACCGAC | 4293–4276 | 55 |
| CRSAP1N-Lc | ggactagtgATGTCCCTAACCCAGTCC | 583–600 | 55 |
| CRSAP1N-R | cgcggatccCAAGCGCGGTTTTGATTGTT | 2775–2757 | 55 |
| BCR1B391c | AGCGTATCGTTTAGCTCGCC | 3914–3895 | 55 |
| CRS1HisKin | TTTTGTATGACGAGCGCAGCTG | 5368–5389 | 55 |
| CRS1HisKin2 | AGATGGTTTTAGAGGCGGTTTTTATG | 5551–5576 | 65 |
| BCRSLP-Ld | GGCGCCTCTCAAGATTTGTT | 57 | |
| BCRSLP-Re | CCGCCGTCGATATTTGTTCT | 57 |
Lowercase letters indicate nucleotides added to create restriction enzyme recognition sites (underlined) for cloning.
Based on nucleotide sequence AF035192 (csxB).
Common primers for csxA and csxB.
Based on nucleotide sequence AF010143 (crs) positions 552 to 575.
Based on nucleotide sequence AF010143 (crs) positions 1681 to 1662.
Sequence data analyses.
Sequence alignment and editing were done with the software Sequencher (Gene Codes Corporation, Ann Arbor, Mich.). Sequence comparisons were done as described by Altschul et al. (1), alignments were done with the Wisconsin Package (Genetics Computer Group, Inc. [GCG], Madison, Wis.), and PEST scores were calculated by the method of Rechsteiner and Rogers (36). RNA secondary structures were calculated by using the PCGENE software (Oxford Molecular Ltd., Oxford, England) or the GCG package. The theoretical isoelectric pHs (pIs) and molecular masses of proteins were calculated with the GCG software.
Southern blot analysis.
DNA fragments used as probes were purified twice over an agarose gel, using a JETsorb gel extraction kit (Genomed GmbH, Bad Oeynhausen, Germany) and DIG labelled overnight with a High Prime DIG labelling kit (Boehringer Mannheim). Southern blotting was done by alkaline transfer onto positively charged nylon membranes (Boehringer Mannheim) with an LKB 2016 VacuGene vacuum blotting pump (Pharmacia LKB). Gels were depurinated for 10 min in 0.25 M HCl, and subsequent transfer was performed with 0.4 M NaOH. After blotting, filters were baked for 30 min at 80°C under vacuum. After at least 1 h of prehybridization, hybridization was carried out in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–1% blocking reagent (Boehringer Mannheim)–0.1% N-lauroylsarcosine sodium salt–0.02% sodium dodecyl sulfate (SDS) at 68°C overnight. Filters were washed under nonstringent conditions twice for 5 min each with 50 ml of 0.2× SSC–0.1% SDS per 100 cm2 at room temperature (RT), followed by medium-stringency washing twice for 15 min each with 50 ml of 0.2× SSC–0.1% SDS per 100 cm2 at RT. The filters were then processed with phosphatase-labelled anti-DIG antibody according to the producer’s protocol. Signals were produced with chemiluminescent substrate (CDP-Star; Boehringer Mannheim) or with the chromogene substrate NBT-BCIP. Luminescence was detected on X-ray films.
Expression of His-tailed fusion proteins and mouse immunization.
Sequences of the primers used to amplify segments of the csxB gene are given in Table 2. Amplification was done on a DNA thermal cycler (GeneAmp 9600; Perkin-Elmer Cetus), using an Expand long template PCR kit (Boehringer Mannheim) an annealing temperature of 55°C, 2-min expansion time, and genomic DNA of C. rectus CCUG20446BT (type strain) as the template. The PCR products were purified by using a QIAquick PCR purification kit (Qiagen). Plasmid pJFFCsxBN-His (Fig. 1), encoding the polyhistidine-tailed N-terminal part of CsxB (CsxBN-His; amino acids [aa] 1 to 723) was obtained by cloning the PCR product amplified with primers CRSAP1N-L and CRSAP1N-R into the BamHI-SpeI restriction sites of pETHIS-1. Plasmid pJFFCsxBC-His (Fig. 1), encoding the polyhistidine-tailed C-terminal part of CsxB (CsxBC-His; aa 670 to 1238), was obtained with the same procedure, using primers CRSAP1C-L and CRSAP1C-R. The clones were sequenced for confirmation of the gene fusions with the vector and transformed into BL21(DE3) cells. One of each positive clone was inoculated in 50 ml of LB-ampicillin at 37°C to an optical density at 650 nm of 0.3 and induced with 10 mM IPTG for another 3 h. The cells were sedimented by centrifugation at 4,000 rpm, resuspended in 5 ml of PN buffer (50 mM NaH2PO4, 300 mM NaCl; pH 8), sonicated with a microtip for 4 min at 50% and 1-s interval in a Branson Sonifier 250 (Branson Ultrasonics, Danbury, Conn.), and then centrifuged at 15,000 rpm for 20 min. The supernatant consisting of the cytosolic fraction was kept, and the pelleted cell debris was resuspended in 5 ml of PN buffer (cell debris). Analysis of the sonicated fractions on SDS–10% acrylamide gels showed the induced protein to be in the cytosolic fraction. The cytosolic fraction was loaded onto a prewashed 1.25-ml-bed-volume Ni-nitrilotriacetic acid-agarose column (Qiagen) and washed once more with 10 ml of buffer B (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-Cl [pH 8]). Elution of the protein was performed with a 40-ml buffer B-to-buffer F (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-Cl [pH 3]) gradient with a flow rate of 0.25 ml/min and collection of fractions of 1 ml with a HiLoad system (Pharmacia LKB). The fractions were analyzed on SDS–10% acrylamide gels. Those containing the purified fusion protein were pooled and dialyzed overnight against PN buffer.
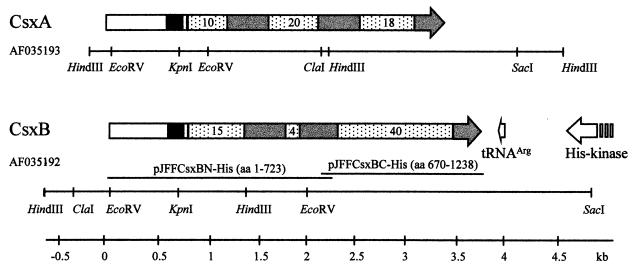
FIG. 1.
Physical and genetic map of the gene clones containing the genes csxA and csxB. Boxes with arrowheads indicate ORFs. Black boxes indicate the Pro-rich region. Dotted boxes represent the regions with the Gly-rich nonapeptide repeats; numerals indicate the number of repeats. Grey boxes represent the regions found interspaced and at the ends of the nonapeptide repeat regions. White boxes at the N terminus represent the S-layer-homologous region. The cloned segments of csxB in pJFFCsxBN-His and pJFFCsxBC-His used for His-tailed protein expression are indicated. Note that the N-terminal part (white) and the Pro-rich region (black) are identical for CsxA and CsxB.
The purified and dialyzed recombinant protein CsxBN-His was mixed 1:1 with complete Freund’s adjuvant (Difco Laboratories) and 100 μl of the emulsion containing 0.5 μg of recombinant protein and then injected into a female mouse. The mouse was booster immunized with the same amount of protein emulsified with Freund’s incomplete adjuvant 20 days later. On day 33 after the first immunization, the mice were bled, and serum was collected and stored at −20°C.
Acid extraction of C. rectus S-layer protein.
Cells from 48-h cultures of C. rectus CCUG20446BT of three blood agar plates were suspended in 10 ml of 0.9% NaCl and centrifuged for 6 min at 20,000 rpm (SS-34 rotor, 4°C). The cell pellet was resuspended in 10 ml of 0.1 M glycine and 500 μl of 1 M HCl were added, bringing the pH to 2.2. After being shaken on ice for 30 min, the suspension was centrifuged for 1 h at 25,000 rpm (Ti-50 rotor, 4°C); the supernatant was neutralized by addition of 1 ml of 1 M Tris and incubated on ice for another 30 min. The supernatant was then centrifuged under the same conditions as before. The final supernatant containing the S-layer protein was stored at −20°C until further use.
Analyses of proteins in the supernatant of C. rectus cells grown in broth.
A 20-h culture of C. rectus type strain at an optical density at 600 nm of 0.25 grown in 100 ml of mff broth was centrifuged for 20 min at 4,000 rpm and 4°C; 2 ml of the supernatant was added to 18 ml of 4°C ethanol and centrifuged for another 30 min at 20,000 rpm. The ethanol precipitant was resuspended in 100 μl of SDS loading buffer, and 20 μl thereof was analyzed in parallel with 3 μl of the cell pellet resuspended in 20 μl of SDS loading buffer on an SDS–10% polyacrylamide gel.
SDS-PAGE and immunoblot analyses.
Proteins were separated by SDS–10% polyacrylamide gel electrophoresis (SDS-PAGE) as described by Laemmli (23) and transferred to a nitrocellulose membrane (Bio-Rad Laboratories, Hercules, Calif.). Western blots were used either for the Ca2+ binding assay or for immunoblotting. For immunoblotting, Western blots were blocked with 1% milk buffer for 30 min and then incubated with the antiserum (1:1,000) in milk buffer overnight at 4°C. After a thorough wash with water, the appropriate phosphatase-labelled conjugate (Ac to human [catalog no. 15-10-06] or goat anti-mouse immunoglobulins G plus M [heavy plus light chain] [catalog no. 15-18-09]; Kirkegaard & Perry, Gaithersburg, Md.) diluted 1:2,000 in milk buffer was added, and the reaction was visualized 90 min later by incubation with BCIP-NBT as the substrate.
Ca2+ binding assay.
The binding of Ca2+ to proteins was detected by 45Ca autoradiography. Western blots were soaked and washed in calcium binding buffer (60 mM KCl, 5 mM MgCl2, 10 mM imidazole hydrochloride [pH 7.2]) for 10 min followed by 10 min of incubation in binding buffer supplemented with 1.0 μCi of 45CaCl2 per ml with a specific activity of 0.02 mCi/μg (Amersham, Little Chalfont, Buckinghamshire, England). The membranes were then rinsed with deionized water for 5 min, dried at room temperature, and exposed to autoradiography.
Nucleotide sequence accession numbers.
The sequences derived from clones were submitted to EMBL, and the assigned accession numbers are AF035193 for csxA and AF035192 for csxB.
RESULTS
Cloning and sequence analyses of two RTX genes, csxA and csxB.
Analysis of different Campylobacter spp. with the broad-range RTX gene probes developed by Kuhnert et al. (22) revealed a signal for a potential RTX gene for C. rectus with probe RTXCYAA, derived from the Bordetella pertussis cyaA toxin gene. This probe was then used to screen a gene library of C. rectus CCUG20446BT made by cloning HindIII-digested genomic DNA fragments in cloning vector pBluescript SK−. Two clones containing two different partial RTX genes were found. To obtain the full genes, two additional gene libraries made from KpnI/SacI-digested DNA and from ClaI/HindIII-digested DNA were made and screened with gene probes derived from the initial clones. This strategy allowed us to obtain composite clones of 4,649 and 5,473 bp containing two highly similar genes which were designated csxA and csxB (Fig. 1). Nucleotide sequence analysis done on both strands of the two cloned genes revealed 83% identity (BESTFIT parameters, gap weight = 50 and length weight = 1) with 22 gaps.
Analysis of the sequenced DNA showed a low G+C content of 42.2%, which is below the average G+C content for C. rectus of 45.4% (39). The csxA open reading frame (ORF) is located from positions 136 to 3507, with an ATG initiation codon and TGA stop codon. It encodes a protein with 1,123 amino acid residues and a calculated molecular mass of 118 kDa. The ORF for csxB is 3,717 bp long, starting with ATG at position 583 and ending with TAA at position 4299. It encodes a protein with 1,238 amino acid residues and a calculated molecular mass of 130.8 kDa. For both ORFs, a putative ribosomal binding site (RBS), AGGAG, was found 5 bp upstream of the ATG. Upstream of the RBS of csxA, a putative promoter −10 box (TATAAT) but no canonical −35 box was found. The csxA gene is flanked by two hairpins, one of them 21 bp upstream of the ATG consisting of a 13-bp stem and a 5-bp loop with a ΔG of −24.4 kcal/mol and the other positioned 17 bp downstream of the stop codon with a 11-bp stem, 6-bp loop, and ΔG of −21.8 kcal/mol. These structures could serve as rho-independent transcription termination signals. The csxB gene is preceded by a characteristic promoter with −35 (TAAACT) and −10 (TATAAT) sequences identical to those found upstream of crs (41). Between the promoter and RBS of csxB, there are two 11-bp direct repeats (AAATATATTAT). The csxB gene is followed by a 19-bp palindrome (TTTTTAATTTAATTAAAAA) 19 bp downstream of the stop codon.
Analyses of sequences downstream of csxB.
The 19-bp palindrome downstream of csxB is followed by a sequence with high similarity to the Helicobacter pylori tRNAArg(GCG) gene sequence (GenBank/EMBL accession no. AE000551; bp 73 to 149) in the opposite direction to csxB (Fig. 1). Folding analysis by the program of Zuker et al. (46) gave an energy of −19.3 kcal/mol (13). The typical prokaryotic 3′ CCA terminus (12, 40) and the signatures A20, C35, G36, and A74 (38) further confirm this sequence to be a tRNAArg gene with GCG as the anticodon.
The tRNAArg gene is preceded by a sequence with high similarity to histidine protein kinase (His-kinase) genes (TIGR database no. HP1364; 29% identity, 45% similarity) (Fig. 1). The conserved residues for the N, D/F, and G boxes (32) among members of the His-kinase superfamily are also present within this sequence. To determine regions of this gene which are not present on the plasmid clones, genomic sequencing was performed with the primers CRS1HisKin and CRS1HisKin2. This allowed the determination of a further 300 bp of the putative His-kinase with the characteristic H-box consensus (FIRDTTHEINTPLSVILM) for His-kinases.
Specificity of csx genes for C. rectus.
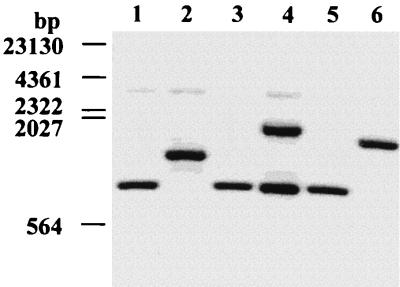
Southern blot analysis of EcoRV-digested genomic DNA of different Campylobacter and Helicobacter species (Table 1) with a probe common for csxA and csxB made from the 263-bp KpnI-EcoRV fragment of csxA (Fig. 1) revealed two bands representing csxA and csxB for C. rectus CCUG20446BT and only one band for the other C. rectus strains analyzed (Fig. 2). Other Campylobacter and Helicobacter species (Table 1) showed no hybridization signals. Further determination of the csx genes in C. rectus strains which showed only one gene on Southern blots was performed by PCR amplification with primers CRSAP1N-L and BCR1B931, which are common for both csx genes, and subsequent partial sequencing of the PCR product with primer BCR1B931, which differentiated csxA from csxB. These analyses revealed that five of the six strains, i.e., CCUG20446BT, CCUG11640, CCUG11642, CCUG11643, and CCUG27948, harbored a copy of gene csxA. In these strains, csxA showed some variability which result in 3 to 6% different amino acid positions of CsxA compared to that of the type strain. One strain analyzed, CCUG11645, did not harbor csxA but contained only a copy of csxB.
FIG. 2.
Southern hybridization analysis of total genomic DNA from C. rectus with a csxA probe. Genomic C. rectus DNA was cut with EcoRV and probed with the DIG-labelled 263-bp EcoRV-KpnI fragment from csxA. Samples of 1 μg of total genomic DNA of strains CCUG11640 (lane 1), CCUG11643 (lane 2), CCUG27948 (lane 3), CCUG20446BT (lane 4), CCUG11642 (lane 5), and CCUG11645 (lane 6) were separated by gel electrophoresis and transferred to a positively charged nitrocellulose membrane. Positions of DIG-labelled lambda HindIII fragments and their sizes are indicated on the left.
To determine whether the C. rectus strains analyzed in this study for the presence of csxA and csxB coharbor the crs gene encoding the S-layer protein, we have developed a crs-specific PCR using primers BCRSLP-L and BCRSLP-R (Table 2). PCR analysis revealed the presence of the crs gene in all six C. rectus strains analyzed as shown by the amplification of the specific 1,130-bp fragment and subsequent partial DNA sequence determination of the PCR products using oligonucleotide primer BCRSLP-L.
Structural analysis of the CsxA and CsxB sequences.
The amino acid sequences for CsxA and CsxB were deduced from the genes, using the universal genetic code. As expected from the high nucleotide similarity, CsxA and CsxB showed a very similar amino acid sequence. Both proteins have high contents of Gly (16.6 and 15.3%) and Asp (15.1 and 14.0%) residues and a calculated pI of 4.1.
Both (CsxA and CsxB) show two main domains with different structural features (Fig. 1). The first 100 aa, which are identical between CsxA and CsxB, show structures typical for S-layer proteins and exhibit high similarity with the S-layer protein Crs of C. rectus (41) (58% identical and 78% similar amino acid residues), SapA of C. fetus (5) (36% identity and 74% similarity), and SapB of C. fetus (8) (34% identity and 76% similarity).
The S-layer domain of each of CsxA and CsxB is followed by a proline-rich segment from aa 222 to 268 containing 23 Pro residues out of 47 aa. This region also shows a very high PEST score of +25, with a molar fraction of the amino acids P, E, D, S, and T of 0.68 and a hydrophobicity index of 24. The remaining part of each protein contains the typical RTX structures. CsxA contains three blocks of 10, 20, and 18 glycine-rich nonapeptide repeats interspaced and ended by a segment of 140 aa which is present twice between the blocks and partially at the C-terminal end of CsxA (Fig. 1). A very similar modular structure is observed in the slightly larger protein CsxB. The 310 N-terminal aa including the S-layer structure and the proline-rich segment are identical in CsxA. In the RTX part, CsxB contains three blocks of 15, 4, and 40 glycine-rich repeats which are interspaced and flanked C terminally by segments homologous to those found in CsxA, the segments showing 38 to 79% identity (45 to 82% similarity) within and between the Csx proteins. More detailed investigations in the Gly-rich nonapeptide repeat sequences of CsxA and CsxB reveal, respectively, 13 and 22 of the total of 48 and 59 repeats to fit the consensus (L/I/F)XGGXG(N/D)D.
Ca2+ binding.
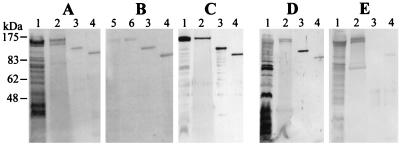
Autoradiography of the 45Ca2+ blots indicates that recombinant CsxBN-His, representing the N-terminal part of CsxB, which contains two blocks of 15 and 4 glycine-rich nonapeptide repeats, and CsxBC-His, representing the C-terminal part of CsxB, which contains one block of 40 glycine-rich nonapeptide repeats both (CsxBN-His and CsxBC-His), strongly bound Ca2+ (Fig. 3B). Analysis of the proteins obtained by acid extraction, which is supposed to extract preferentially proteins with S-layer characteristics, revealed a distinct band in the 120-kDa range on 45Ca2+ blots (Fig. 3B). This band is also seen in total cell protein but is strongly enriched in the acid glycine extract.
FIG. 3.
Ca2+ binding and antigenicity of csx. SDS-PAGE (Coomassie blue-stained gel) (A), 45Ca2+ binding (B), and immunoblot (C to E) analysis of C. rectus CCUG20446BT grown in broth. Total cells (lanes 1), culture supernatant (lanes 2), Ni-nitrilotriacetic acid-purified recombinant CsxBN-His (lanes 3), and CsxBC-His (lanes 4) were used. Total cells of C. rectus grown on blood agar plates (lane 5) and acid extraction thereof (lane 6) fractions are represented on the 45Ca2+ binding blot (B). The following sera were reacted on the immunoblots: polyclonal monospecific mouse serum raised against the N terminus of CsxB (anti-CsxBN-His) (C), serum from a human patient with acute gingivitis from whom C. rectus was isolated (D), and serum from a human with no known C. rectus infection (E).
Antigenic properties of Csx.
Mouse serum directed against the recombinant peptide CsxBN-His, representing the N-terminal part of CsxB, strongly reacted with proteins in the 120- to 130-kDa size range of total cells and of culture supernatants of all C. rectus strains analyzed (Table 1) except strain CCUG11645, lacking CsxA as shown above. The 120- to 130-kDa protein reacting with anti-CsxBN-His was also found in the acid-extracted protein fraction of C. rectus CCUG20446BT. The serum also reacted with the recombinant CsxBN-His and CsxBC-His due to domains shared between these peptides (Fig. 1). An immunoblot with serum from a patient suffering from acute gingivitis and from whom C. rectus had been isolated showed a very strong reaction to recombinant CsxBN-His and a clear but weaker reaction against CsxBC-His (Fig. 3D). This serum also reacted against several other antigens of total cells of C. rectus CCUG20446BT; the reaction in the 100- to 130-kDa range of the culture supernatant was weak. Serum from a patient with no history of gingivitis and who never had dental problems showed very weak reactions with recombinant CsxBN-His and CsxBC-His peptides (Fig. 3E). Such weak reactions were also observed in nine sera from randomly chosen blood donors.
DISCUSSION
By applying RTX-specific broad-range gene probes (22), we were able to detect and characterize two new RTX genes in C. rectus. The strong relatedness found within the amino acid sequences of the two proteins, CsxA and CsxB, encoded by these genes indicate that they have common function, whereas the variability found upstream of the genes suggests that they are regulated differently. The proteins show a two-domain structure. The N-terminal S-layer domain and the C-terminal part with RTX domains are separated by a sequence with a high PEST score, giving the protein a two-domain structure. High PEST scores indicate possible proteolytic signals (36) and imply a potential cleavage of the two domains.
The N-terminal parts of CsxA and CsxB show significant similarity to S-layer proteins. In addition, they have the low cysteine content, low pI, and high molecular weight which are typical for S-layer proteins. The N-terminal parts of CsxA and CsxB may therefore be involved in binding of lipopolysaccharide as was demonstrated for S-layer proteins from C. fetus (44). CsxA and CsxB are both distinguished by their large number of glycine-rich nonapeptide repeats which are arranged in three blocks. We attribute the potential to bind Ca2+ by these proteins to these RTX structures. RTX structures are involved together with Ca2+ binding in receptor recognition on the target cell and have been shown for certain toxins to determine the host cell specificity (24, 25). Thus, the RTX domains of Csx are suggested to be involved in target binding. No potential function could be attributed to the domain which is located between and C terminally to these RTX sequences. Thus far, we cannot suggest any possible toxic function for CsxA or CsxB. It remains, however, to be clarified whether the cytotoxic activity as measured by Gillespie et al. (16) might be due to CsxA or CsxB protein rather than to the S-layer protein Crs (29).
The genes csxA and csxB seem to be located on monocistronic operons, as indicated by the flanking putative transcription termination signals. The presence of a tRNAArg(GCG) gene, which represents a low-abundance Arg codon in Helicobacter and Campylobacter species, immediately downstream of csxB is noteworthy in view of the frequent association of tRNA genes with pathogenicity islands (18). For Corynebacterium diphtheriae, a tRNAArg gene was reported to be the chromosomal integration site for toxinogenic bacteriophages (35).
Immediately next to the tRNAArg gene we found a gene encoding a His-kinase. His-kinases commonly function as transmitter domains within the sensors of two-component signal transduction systems. The two-component system is the major mechanism of signal transduction in bacteria (19) and is often involved in the regulation of expression of virulence genes in pathogenic bacteria by sensing environmental signals (28).
Southern blot analysis, PCR amplifications, and partial DNA sequence analysis of the PCR amplification products revealed that the csxA gene was present in five of six C. rectus strains tested. The csxB gene was present in only two strains, the type strain CCUG20446BT (together with csxA) and strain CCUG11645. Interestingly, the latter strain did not react on immunoblots with anti-CsxBN-His, whereas all other strains did. This result might be interpreted as signifying that anti-CsxBN-His recognizes protein CsxA in all strains where it would be expressed from gene csxA, while csxB would not be expressed under the given culture conditions, thus leading to an apparent Csx− phenotype in strain CCUG11645. It also should be noted that the very N-terminal parts of CsxA and CsxB show similarity to the analogous part of the S-layer protein Crs (41), whose gene is present in all C. rectus strains analyzed (Table 1) and therefore could generate to some extent serological cross-reactions.
The strong immunological reaction of a serum from a patient with a confirmed C. rectus infection with purified recombinant CsxBN-His peptide suggests the N-terminal half of Csx to be highly immunogenic upon infection of C. rectus, while the C-terminal part seems to be less antigenic. While in this patient tissue invasion by C. rectus apparently led to a strong immune response, the weak reaction to C. rectus proteins observed in normal sera suggests that exposure to this agent is limited in the general population.
In summary, we have shown that a putative RTX homologue detected in C. rectus by screening with broad-range RTX gene probes (22) represents a new class of putative bifunctional proteins containing both RTX and S-layer domains. This study also confirms the value of using probes for the general detection of toxin families.
ACKNOWLEDGMENTS
We thank Jürg Meyer, Department of Preventive Dentistry, University of Basel, Basel, Switzerland, for the serum of a patient with acute gingivitis, Yvonne Schlatter for technical support, and Isabelle Brodard for culturing Campylobacter strains.
This work was supported by the Priority Program Biotechnology of the Swiss National Science Foundation (grant 5002-045027).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1990. [Google Scholar]
- 3.Bauer M E, Welch R A. Association of RTX toxins with erythrocytes. Infect Immun. 1996;64:4665–4672. doi: 10.1128/iai.64.11.4665-4672.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beveridge T J, Pouwels P H, Sara M, Kotiranta A, Lounatmaa K, Kari K, Kerosuo E, Haapasalo M, Egelseer E M, Schocher I, Sleytr U B, Morelli L, Callegari M L, Nomellini J F, Bingle W H, Smit J, Leibovitz E, Lemaire M, Miras I, Salamitou S, Beguin P, Ohayon H, Gounon P, Matuschek M, Sahm K, Bahl H, GrogonoThomas R, Dworkin J, Blaser M J, Woodland R M, Newell D G, Kessel M, Koval S F. Functions of S-layers. FEMS Microbiol Rev. 1997;20:99–149. doi: 10.1111/j.1574-6976.1997.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 5.Blaser M J, Gotschlich E C. Surface array protein of Campylobacter fetus. Cloning and gene structure. J Biol Chem. 1990;265:14529–14535. . (Erratum, 265:19372.) [PubMed] [Google Scholar]
- 6.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high frequency efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–378. [Google Scholar]
- 7.Dunn B E, Perez-Perez G I, Blaser M J. Two-dimensional gel electrophoresis and immunoblotting of Campylobacter pylori proteins. Infect Immun. 1989;57:1825–1833. doi: 10.1128/iai.57.6.1825-1833.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dworkin J, Blaser M J. Generation of Campylobacter fetus S-layer protein diversity utilizes a single promoter on an invertible DNA segment. Mol Microbiol. 1996;19:1241–1253. doi: 10.1111/j.1365-2958.1996.tb02469.x. [DOI] [PubMed] [Google Scholar]
- 9.Dzink J L, Socransky S S, Haffajee A D. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J Clin Periodontol. 1988;15:316–323. doi: 10.1111/j.1600-051x.1988.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 10.Ebersole J L, Kesavalu L, Schneider S L, Machen R L, Holt S C. Comparative virulence of periodontopathogens in a mouse abscess model. Oral Dis. 1995;1:115–128. doi: 10.1111/j.1601-0825.1995.tb00174.x. [DOI] [PubMed] [Google Scholar]
- 11.Felmlee T, Pellett S, Lee E Y, Welch R A. Escherichia coli hemolysin is released extracellularly without cleavage of a signal peptide. J Bacteriol. 1985;163:88–93. doi: 10.1128/jb.163.1.88-93.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fournier M J, Ozeki H. Structure and organization of the transfer ribonucleic acid genes of Escherichia coli K-12. Microbiol Rev. 1985;49:379–397. doi: 10.1128/mr.49.4.379-397.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freier S M, Kierzek R, Jaeger J A, Sugimoto N, Caruthers M H, Neilson T, Turner D H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frey J. Virulence in Actinobacillus pleuropneumoniae and RTX toxins. Trends Microbiol. 1995;3:257–261. doi: 10.1016/s0966-842x(00)88939-8. [DOI] [PubMed] [Google Scholar]
- 15.Gillespie J, Holt S C. Growth studies of Wolinella recta, a gram-negative periodontopathogen. Oral Microbiol Immunol. 1987;2:105–111. doi: 10.1111/j.1399-302x.1987.tb00271.x. [DOI] [PubMed] [Google Scholar]
- 16.Gillespie M J, Smutko J, Haraszthy G G, Zambon J J. Isolation and partial characterization of the Campylobacter rectus cytotoxin. Microb Pathog. 1993;14:203–215. doi: 10.1006/mpat.1993.1020. [DOI] [PubMed] [Google Scholar]
- 17.Glaser P, Ladant D, Sezer O, Pichot F, Ullmann A, Danchin A. The calmodulin-sensitive adenylate cyclase of Bordetella pertussis: cloning and expression in Escherichia coli. Mol Microbiol. 1988;2:19–30. [PubMed] [Google Scholar]
- 18.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 19.Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. [Google Scholar]
- 20.Kesavalu L, Holt S C, Crawley R R, Borinski R, Ebersole J L. Virulence of Wolinella recta in a murine abscess model. Infect Immun. 1991;59:2806–2817. doi: 10.1128/iai.59.8.2806-2817.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolodrubetz D, Dailey T, Ebersole J, Kraig E. Cloning and expression of the leukotoxin gene from Actinobacillus actinomycetemcomitans. Infect Immun. 1989;57:1465–1469. doi: 10.1128/iai.57.5.1465-1469.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhnert P, Heyberger-Meyer B, Burnens A P, Nicolet J, Frey J. Detection of RTX toxin genes in gram-negative bacteria with a set of specific probes. Appl Environ Microbiol. 1997;63:2258–2265. doi: 10.1128/aem.63.6.2258-2265.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lally E T, Golub E E, Kieba I R. Identification and immunological characterization of the domain of Actinobacillus actinomycetemcomitans leukotoxin that determines its specificity for human target cells. J Biol Chem. 1994;269:31289–31295. [PubMed] [Google Scholar]
- 25.Lally E T, Kieba I R, Sato A, Green C L, Rosenbloom J, Korostoff J, Wang J F, Shenker B J, Ortlepp S, Robinson M K, Billings P C. RTX toxins recognize a beta 2 integrin on the surface of human target cells. J Biol Chem. 1997;272:30463–30469. doi: 10.1074/jbc.272.48.30463. [DOI] [PubMed] [Google Scholar]
- 26.Lo R Y, Strathdee C A, Shewen P E. Nucleotide sequence of the leukotoxin genes of Pasteurella haemolytica A1. Infect Immun. 1987;55:1987–1996. doi: 10.1128/iai.55.9.1987-1996.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig A, Jarchau T, Benz R, Goebel W. The repeat domain of Escherichia coli haemolysin (HlyA) is responsible for its Ca2+-dependent binding to erythrocytes. Mol Gen Genet. 1988;214:553–561. doi: 10.1007/BF00330494. [DOI] [PubMed] [Google Scholar]
- 28.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto M, Maeda H, Kitanaka M, Kokeguchi S, Takashiba S, Murayama Y. The S-layer protein from Campylobacter rectus: sequence determination and function of the recombinant protein. FEMS Microbiol Lett. 1998;166:275–281. doi: 10.1111/j.1574-6968.1998.tb13901.x. [DOI] [PubMed] [Google Scholar]
- 30.Nitta H, Holt S C, Ebersole J L. Purification and characterization of Campylobacter rectus surface layer proteins. Infect Immun. 1997;65:478–483. doi: 10.1128/iai.65.2.478-483.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okuda K, Kigure T, Yamada S, Kaneko T, Ishihara K, Miura T, Kato T, Takazoe I. Role for the S-layer of Campylobacter rectus ATCC33238 in complement mediated killing and phagocytic killing by leukocytes from guinea pig and human peripheral blood. Oral Dis. 1997;3:113–120. doi: 10.1111/j.1601-0825.1997.tb00022.x. [DOI] [PubMed] [Google Scholar]
- 32.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 33.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 34.Rams T E, Feik D, Slots J. Campylobacter rectus in human periodontitis. Oral Microbiol Immunol. 1993;8:230–235. doi: 10.1111/j.1399-302x.1993.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 35.Ratti G, Covacci A, Rappuoli R. A tRNA2Arg gene of Corynebacterium diphtheriae is the chromosomal integration site for toxinogenic bacteriophages. Mol Microbiol. 1997;25:1179–1181. doi: 10.1046/j.1365-2958.1997.5191887.x. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 36.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 36a.Segers, R., and J. Frey. Unpublished data.
- 37.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K, Himeno H, Asahara H, Hasegawa T, Shimizu M. In vitro study of E. coli tRNA(Arg) and tRNA(Lys) identity elements. Nucleic Acids Res. 1992;20:2335–2339. doi: 10.1093/nar/20.9.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandamme P, Falsen E, Rossau R, Hoste B, Segers P, Tytgat R, De Ley J. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int J Syst Bacteriol. 1991;41:88–103. doi: 10.1099/00207713-41-1-88. [DOI] [PubMed] [Google Scholar]
- 40.Vold B S. Structure and organization of genes for transfer ribonucleic acid in Bacillus subtilis. Microbiol Rev. 1985;49:71–80. doi: 10.1128/mr.49.1.71-80.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang B, Kraig E, Kolodrubetz D. A new member of the S-layer protein family: characterization of the crs gene from Campylobacter rectus. Infect Immun. 1998;66:1521–1526. doi: 10.1128/iai.66.4.1521-1526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch R A. Pore-forming cytolysins of gram-negative bacteria. Mol Microbiol. 1991;5:521–528. doi: 10.1111/j.1365-2958.1991.tb00723.x. [DOI] [PubMed] [Google Scholar]
- 43.Welch R A, Bauer M E, Kent A D, Leeds J A, Moayeri M, Regassa L B, Swenson D L. Battling against host phagocytes: the wherefore of the RTX family of toxins? Infect Agents Dis. 1995;4:254–272. [PubMed] [Google Scholar]
- 44.Yang L Y, Pei Z H, Fujimoto S, Blaser M J. Reattachment of surface array proteins to Campylobacter fetus cells. J Bacteriol. 1992;174:1258–1267. doi: 10.1128/jb.174.4.1258-1267.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zambon J J, Reynolds H, Fisher J G, Shlossman M, Dunford R, Genco R. Microbiological and immunological studies of adult periodontitis in patients with noninsulin-dependent diabetes mellitus. J Periodontol. 1988;59:23–31. doi: 10.1902/jop.1988.59.1.23. [DOI] [PubMed] [Google Scholar]
- 46.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]