Abstract
Sequence analysis of Streptomyces lavendulae NRRL 2564 chromosomal DNA adjacent to the mitomycin resistance locus mrd (encoding a previously described mitomycin-binding protein [P. Sheldon, D. A. Johnson, P. R. August, H.-W. Liu, and D. H. Sherman, J. Bacteriol. 179:1796–1804, 1997]) revealed a putative mitomycin C (MC) transport gene (mct) encoding a hydrophobic polypeptide that has significant amino acid sequence similarity with several actinomycete antibiotic export proteins. Disruption of mct by insertional inactivation resulted in an S. lavendulae mutant strain that was considerably more sensitive to MC. Expression of mct in Escherichia coli conferred a fivefold increase in cellular resistance to MC, led to the synthesis of a membrane-associated protein, and correlated with reduced intracellular accumulation of the drug. Coexpression of mct and mrd in E. coli resulted in a 150-fold increase in resistance, as well as reduced intracellular accumulation of MC. Taken together, these data provide evidence that MRD and Mct function as components of a novel drug export system specific to the mitomycins.
Mitomycin C (MC), produced by Streptomyces lavendulae NRRL 2564, is a highly effective antitumor agent used in the treatment of various carcinomas (10). As a prodrug, MC is unreactive until chemical or enzymatic reduction renders the molecule a highly effective alkylating agent (13). The molecular basis of MC bioactivity derives mainly from its propensity to covalently interact with DNA at 5′-CpG sequences, causing lethal intra- and interstrand cross-links as well as monofunctional alkylation (31).
S. lavendulae encounters a daunting challenge in avoiding potentially lethal MC-mediated cross-links, since it has a chromosomal G+C content of over 70%, which translates into at least one million potential drug target sites per cell. Recently, two genetic loci that mediate mitomycin resistance in this organism have been previously reported. One locus (mcr) encodes a protein (MCRA) that catalyzes oxidation of the reduced, bioactivated species of MC via a redox relay mechanism (2, 14). The second locus (mrd) encodes MRD, which functions to sequester the prodrug by a specific mitomycin-binding protein (28). A paradox of our current knowledge regarding mitomycin resistance has been the lack of a clear mechanism for drug transport. Indeed, the observed stoichiometry suggested that it would be ineffective for S. lavendulae to utilize MRD as a solo mechanism for cellular self-protection. Since the majority of MC is found in the culture medium after the drug is presumably excreted from the cell following biosynthesis, the involvement of a specific drug transporter was evident. Export of toxic compounds as a means of resistance is well documented for pathogenic bacteria (22) as well as for antibiotic-producing microorganisms (7, 19).
Here we report the cloning and characterization of a third MC resistance determinant (mct), which encodes a membrane-associated protein involved in excretion of MC from S. lavendulae. Characterization of mct was accomplished by expression and analysis of the gene product in Escherichia coli, along with in vivo mutation of native mct in S. lavendulae. In addition, coexpression of mct and mrd was carried out to investigate potential functional interaction between these resistance determinants. The results establish that MRD maintains a high affinity for MC and may serve as the primary docking site (participating as an accessory component in a drug export system) for subsequent transport by Mct, comparable to the case for several binding-protein-dependent nutrient and cofactor uptake systems (1, 11, 27).
MATERIALS AND METHODS
Bacterial strains, culture conditions, and media.
The strains and plasmids used are described in Table 1. E. coli DH5α used as a host for generation of double-stranded plasmid DNA, was grown at 37°C in Luria-Bertani (LB) medium. E. coli BL21(DE3), used as host for protein expression, was grown at 37°C in NZCYM medium (26). S. lavendulae NRRL 2564 was grown in YEME medium (12) at 30°C for preparation of genomic DNA.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Bacterial strains | ||
| S. lavendulae | ||
| NRRL 2564 | MC+ Mcr | American Type Culture Collection |
| MM105 (mct) | Mcs; aphII insertional disruption of mct | This work |
| E. coli | ||
| S17-1 | RP4 derivative integrated on chromosome | 29 |
| DH5α | F−recA φ80 dlacZΔM15 | Gibco BRL |
| BL21(DE3) | F−ompT hsdS gal dcm (DE3) | Novagen |
| PJS100 (mrd+) | BL21(DE3) containing pDHS7006 | 28 |
| PJS102 (mct+) | BL21(DE3) containing pDHS7023 | This work |
| PJS103 (mrd+ mct+) | BL21(DE3) containing pDHS7024 | This work |
| Plasmids | ||
| pUC119 | High-copy E. coli vector; Apr | 28 |
| pKC1139 | Streptomyces-E. coli conjugal transfer vector; AmroriT | 3 |
| pET17b | Protein expression vector; Apr | Novagen |
| pDHS7006 | pT7SC (4) with 0.7 kb insert containing mrd | 26 |
| pDHS7023 | pET17b with 1.45-kb insert containing mct | This work |
| pDHS7024 | pDHS7023 with 2.1-kb SspI fragment from pDHS7006 | This work |
| pDHS7547 | pNJ1 (32) with 25-kb Sau3A insert from S. lavendulae | This work |
| pDHS7661 | pUC119 with 3.5-kb BamHI subclone from pDHS7547 | This work |
| pDHS7703 | pDHS7661 with aphII gene within the mct sequence | This work |
| pDHS7704 | pKC1139 with 5.4-kb EcoRI-HindIII fragment from pDHS7703 | This work |
Mcr, MC resistant; Mcs, MC sensitive; MC+, MC production.
DNA preparation and amplification.
S. lavendulae genomic DNA was isolated by the lysozyme-2× Kirby mix method (12). General DNA manipulation was performed as described previously (2). Oligonucleotides for PCR and sequencing were obtained from Gibco BRL (Gaithersburg, MD). PCR amplifications were carried out with a thermal cycler from Hybaid Ltd., (Teddington, United Kingdom).
Cloning and sequencing of mct.
An S. lavendulae NRRL 2564 genomic DNA library was constructed in the cosmid vector pNJ1 (32) as previously described (2). The insert DNA of a cosmid clone containing sequences flanking mrd was digested with BamHI and subcloned into the BamHI site of pUC119. By using exonuclease III (Erase-A-Base kit; Promega, Madison, Wis.), a set of nested deletion clones was generated, and both strands of the insert DNA were sequenced by the dideoxy chain termination method with an ABI Prism kit (PE Applied Biosystems, Warrington, United Kingdom) in coordination with an ABI 373 automated sequencer. Dimethyl sulfoxide (10%) was added to the reaction mixtures to reduce compressions. Sequence data were analyzed by using the GeneWorks/MacVector (Oxford Molecular Group, Mountain View, Calif.) software package. Deduced amino acid sequence data were compared to the available databases by using the BLAST program of the Genetics Computer Group version 9.0 software (Oxford Molecular Group).
Construction and analysis of the mct mutant strain of S. lavendulae.
The mct disruption vector pDHS7704 was constructed as follows. pDHS7661, a subclone containing mct and flanking genomic DNA, was digested with EcoRI, blunt ended, and ligated with the 1.4-kb neomycin resistance gene fragment from pFD666 (ApaLI-HindIII digestion, blunt ended) (1). The 5.4-kb EcoRI-HindIII fragment from the resulting construct (pDHS7703) was subcloned into pKC1139 to create pDHS7704 and was conjugated into S. lavendulae as described by Bierman et al. (3). An mct double-crossover mutant was selected after propagating transconjugants on R5T plates for five generations at 39°C. Kanamycin-resistant and apramycin-sensitive colonies were further tested by Southern blotting to confirm the desired double-crossover genotype. Determinations of MC resistance for wild-type S. lavendulae and the mct mutant were made by growing the strains in YEME medium (24 h at 30°C) and plating 150 μl of this culture on R2YE agar medium (12) containing various concentrations of MC. Growth was scored after 96 h, and the minimum bactericidal concentration (MBC) of drug was determined as the level of MC which inhibited 99.9% of bacterial growth.
Construction of an mct expression plasmid.
For the construction of the E. coli expression plasmid, NdeI and HindIII sites were introduced at the translational start codon and downstream of the translational stop codon of mct, respectively. The primers used for PCR were 5′-GGGAATTCCATATGATGCAGTCCATGTCAC-3′ and 5′-GGGAATTCAAGCTTTCATTCCGCCGGGGTC-3′. The PCR was carried out with 2.5 U of Taq polymerase; 0.4 μg of each primer; 1 μg of pDHS7661 DNA as the template; 10 mM (each) dATP, dGTP, dCTP, and dTTP; 1.5 mM MgCl2; and 10 μl of 10× Promega PCR buffer in a total volume of 100 μl. Amplification was achieved with 30 cycles of denaturation at 94°C for 30 s, annealing at 37°C for 1 min, and extension at 70°C for 2 min. The 1.45-kb PCR product was recovered by 0.8% agarose gel electrophoresis, digested with NdeI-HindIII, and ligated into the T7 expression plasmid pET17b (Novagen, Madison, Wis.), which had been similarly cut with NdeI-HindIII, to give pDHS7023. pDHS7023 was introduced by transformation into E. coli BL21(DE3) to give strain PJS102.
Construction of an mct-mrd coexpression plasmid.
From plasmid pDHS7006 (28), a 2.1-kb SspI fragment was isolated. The fragment contained the mrd gene under the control of the T7 promoter, including transcriptional terminator sequences (rrnBt1) upstream and downstream of mrd. The fragment was ligated into the MC transporter construct pDHS7023, which had been cut with MscI, to give pDHS7024. pDHS7024 was introduced by transformation into E. coli BL21(DE3) to give strain PJS103.
MC resistance phenotype of E. coli.
To analyze resistance conferred by the expression of the MC transporter in E. coli, strain PJS102 was grown to stationary phase (18 h) in LB broth, and 10 μl of this culture was spread on LB agar medium containing 100 μg of ampicillin per ml, IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1.0 mM, and various concentrations of MC. The cultures were grown overnight at 37°C, and CFU were determined. Similarly, the MC resistance phenotypes of strains PJS100 (mrd+) and PJS103 (mct+ mrd+) were quantified. The MBC of MC was determined as the level of antibiotic which inhibited 99.9% of bacterial growth.
[3H]MC uptake assay of strains PJS102 and PJS103.
[3H]MC was obtained from Kyowa Hakko Kogyo, Ltd. Uptake studies were performed for whole cells of E. coli PJS100, PJS102, PJS103, BL21(DE3)::pT7SC, and BL21(DE3)::pET17b. PJS100, PJS102, and PJS103, as well as vector-only cultures, were cultured (37°C) in 5 ml of NCZYM medium with IPTG added to a final concentration of 1 mM (at approximately 3 h of growth). At 9 h (late exponential phase) cells were harvested by centrifugation and resuspended in 1 ml of NCZYM broth (5× concentration). The concentrated suspension of late-exponential-phase cells was exposed to [3H]MC (59 Ci/mmol) at a final concentration of 0.022 μg/ml (0.0655 nmol). Aliquots (100 μl) were removed at frequent intervals, placed on 1.2-μm-pore-size GF/C filters (Whatman International, Maidstone, United Kingdom), and washed once with 6 ml of 0.85% NaCl poured over the filters under vacuum pressure. Additional aliquots were simultaneously removed for determination of protein content (protein assay kit from Bio-Rad Laboratories, Richmond, Calif.). Radioactivity on the filters was quantified with a Beckman LS7000 scintillation counter. Results were expressed as nanograms of mitomycin per milligram of cell protein.
Nucleotide sequence accession number.
The sequence of the mct gene has been deposited in the GenBank database under accession no. AF120930.
RESULTS
A gene encoding a possible transmembrane protein is physically linked to mrd.
DNA sequence analysis of a cosmid clone containing the mrd locus, a previously characterized MC resistance determinant (28), identified an open reading frame encoding a polypeptide predicted to be highly hydrophobic that shows similarity to a variety of antibiotic export proteins in drug-producing actinomycetes. Significantly, the gene (mct) encoding the putative mitomycin transporter (Mct) protein is located within 9 kb of mrd and is physically linked to the MC biosynthetic gene cluster (18).
Sequence analysis of the mct locus.
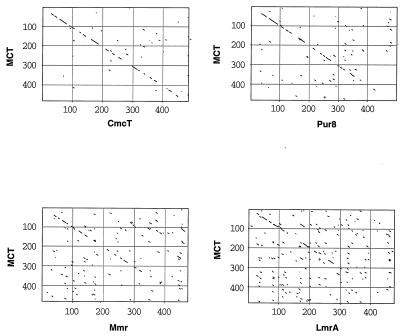
Nucleotide sequence analysis of cosmid clone pDHS7547 revealed an open reading frame predicted to start with the ATG codon at position 132 and end with the TGA codon at nucleotide 1587 (Fig. 1), resulting in a 484-amino-acid polypeptide with a predicted molecular mass of 50,023 Da. Comparison of the deduced amino acid sequence of the mct gene with sequences of proteins in the available databases revealed significant similarity to several integral membrane proteins that confer drug resistance. These include the CmcT protein from the cephamycin producer, Nocardia lactamdurans (6); the Pur8 protein from the puromycin producer, Streptomyces alboniger (30); the Mmr protein from the methylenomycin producer, Streptomyces coelicolor (21); and the LmrA protein from the lincomycin producer, Streptomyces lincolnensis (33). The similarities of the mct gene product and related proteins extend over the entire sequences, with the highest levels of similarity found within the amino-terminal regions (Fig. 2).
FIG. 1.
Nucleotide sequence of mct. The deduced amino acid sequence of mct is indicated under the nucleotide sequence with the one-letter designations. A conserved motif characteristic of 14 TMS proteins is boxed, while the invariant β-turn motif is denoted with a hatched underline. The putative drug extrusion consensus is marked with a hatched box. A potential ribosome binding site is marked with a solid underline.
FIG. 2.
Dot matrix alignment of the deduced amino acid sequence of mct with those of other actinomycete antibiotic efflux proteins. Comparable parameters were utilized in generating the alignments (see Materials and Methods).
Within the N-terminal regions of several antibiotic efflux proteins, including Mmr and LmrA, several highly conserved structural motifs have been established. The β-turn motif (VxGxLxDxxGRKxxxL), found within the highly conserved cytoplasmic loop sequence separating transmembrane segments (TMS) 2 and 3 of most eucaryotic and procaryotic transport proteins (24, 33), is clearly evident in Mct at positions 79 to 95 (Fig. 1). A motif (LDxTVxNVALP) found at the end of transmembrane domain 1, specific to the 14-TMS family (24), is present in Mct at positions 41 to 51 (Fig. 1). Additionally, several other invariant motifs are apparent in the Mct sequence, including a domain (gxxxGgxxGG) encompassing amino acids 164 to 173 of TMS 5 that resembles a putative drug extrusion motif (gxxxGPxxGG) found within several drug transport proteins (23).
Transport proteins that mediate resistance to antibiotics and antiseptics by active efflux are highly related, usually containing 12 or 14 transmembrane regions. Notably, the actinomycete drug transport proteins that have homology with Mct appear to contain 14 membrane-spanning regions and fall within the major facilitator drug/H+ antiport family of drug resistance translocases. By utilizing the membrane structure and topology program MEMSAT (University College, London, United Kingdom) and hydropathy analyses based on the algorithm of Kyte and Doolittle (15), a prediction of 14 membrane-spanning domains was made for the deduced amino acid sequence of Mct (data not shown). Taken together, the sequence analyses suggest that Mct is a new member of the major facilitator drug/H+ antiport family of drug resistance integral membrane proteins.
Inactivation of mct results in greater sensitivity to MC.
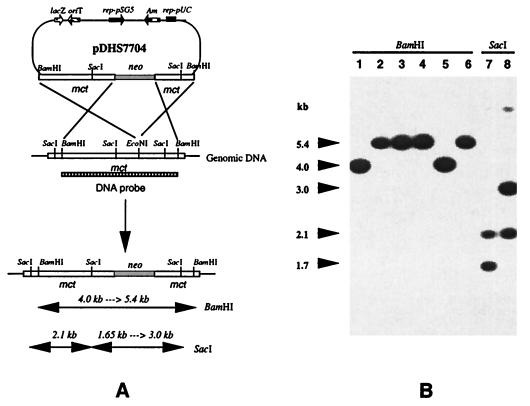
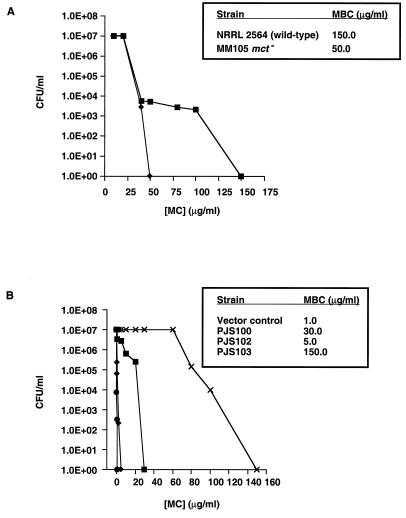
To establish a physiological role for Mct in S. lavendulae, the corresponding gene (mct) was inactivated by insertion of the aphII gene from transposon Tn5 to give pDHS7704. After conjugal transfer of pDHS7704 from E. coli to S. lavendulae and growth of the transconjugants under selective conditions, targeted replacement of native mct was achieved by double-crossover homologous recombination (Fig. 3A). Gene disruption was confirmed by Southern blot hybridization of total DNA from the S. lavendulae wild type and mutant with a DNA probe that included the mct locus. Analytical digests of the genomic DNA resulted in detection of the predicted band shifts in the mutant and wild-type strains (Fig. 3B). The S. lavendulae mct mutant strain (MM105) exhibited approximately a threefold increase in sensitivity to MC (Fig. 4A). In medium lacking MC, the growth kinetics of strain MM105 were comparable to those of the wild-type S. lavendulae strain (data not shown).
FIG. 3.
Creation of the mct disruption mutant. (A) The chromosomal mct gene was disrupted by inserting a neomycin resistance marker (shaded) within the coding region. Following double-crossover recombination, specific restriction bands were predicted to be shifted in the mct mutant genomic DNA compared to the wild-type strain. (B) Southern blot analysis of the mct mutant. As expected, when probed with the 4.0-kb BamHI insert from pDHS7661, the 4.0-kb BamHI hybridization band in wild-type S. lavendulae was shifted to 5.4 kb in mct knockouts, while a 1.65-kb SacI hybridization band was shifted to 3.0 kb in size. Lanes 1 and 5, wild-type genomic DNA digested with BamHI; lanes 2, 3, 4, and 6, genomic DNAs from four double-crossover mutant strains digested with BamHI; lane 7, wild-type genomic DNA digested with SstI; lane 8, genomic DNA from double-crossover clone 6 digested with SacI.
FIG. 4.
Resistance of S. lavendulae and E. coli strains to MC. (A) Resistance of S. lavendulae NRRL 2564 (wild type) (■) and MM105 (mct mutant) (⧫) to various concentrations of MC. (B) Resistance of E. coli strains PJS100 (mrd+) (■), PJS102 (mct+) (⧫), PJS103 (mrd+ mct+) (×), and the BL21(DE3)::pET17b vector control strain (●) to various concentrations of MC. The insets show the MBC (see Materials and Methods) of MC for each strain.
Expression of mct in E. coli.
To investigate further the function of mct, heterologous expression of the gene in E. coli was pursued. mct was amplified by PCR and cloned into the protein expression vector pET17b to give pDHS7023. pDHS7023 was then introduced into E. coli BL21(DE3) to give strain PJS102. After disruption of the cells by sonication, Mct was found to be associated mainly with the membrane fraction of the cell lysate (data not shown), as expected for an integral membrane protein. To determine if strain PJS102 was resistant to MC, cultures were grown up and plated on agar medium containing various concentrations of MC. Significantly, IPTG-induced cultures of PJS102 exhibited resistance to MC at drug concentrations fivefold greater than those for E. coli BL21(DE3) containing vector alone (Fig. 4B).
Coexpression of mct and mrd in E. coli.
To address the notion that the MRD and Mct proteins participate as components of a binding-protein-dependent drug export system, the mct and mrd genes were coexpressed in E. coli. From plasmid pDHS7006 (mrd expression construct) (28), a DNA fragment containing the mrd gene under the control of the T7 promoter was ligated into pDHS7023 to give pDHS7024. pDHS7024 was then introduced into E. coli BL21(DE3) to give strain PJS103. To determine if strain PJS103 was resistant to MC, cultures were grown up and plated on agar medium containing various concentrations of MC. Significantly, IPTG-induced cultures of PJS103 exhibited resistance to MC at drug concentrations 150-fold greater than those for E. coli BL21(DE3) containing vector alone (150 versus 1.0 mg of MC per ml) (Fig. 4B). In addition to PJS103 maintaining levels of resistance greater than that of the vector control strain, coexpression of mct and mrd confers resistance to MC at drug concentrations 5- and 30-fold greater than those for PJS100 (containing the mrd gene alone) (28) or strain PJS102 (containing the mct gene alone), respectively. Strain PJS103 also displayed high-level resistance to mitomycin B (data not shown), a mitomycin analog also produced by S. lavendulae.
MC uptake by E. coli cells expressing mct, mrd, or mct and mrd.
Since the deduced amino acid sequence of the mct gene was similar to those of antibiotic export proteins, reduced accumulation of MC in Mct-expressing cells would be expected. An assay, modeled after experiments used to study tetracycline efflux-mediated resistance in E. coli (16), was designed to study the uptake of [3H]MC by the susceptible vector control and resistant mct-, mrd-, and mct- and mrd-expressing E. coli strains (see Materials and Methods).
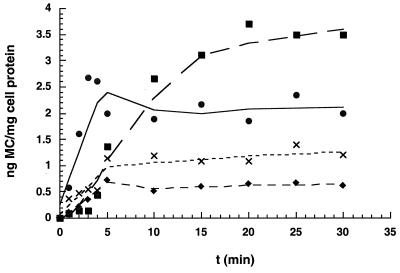
MC accumulation by the susceptible vector control strain [BL21(DE3)::pET17b] was found to reach a maximum level at 5 min and thereafter was maintained at constant concentrations. In contrast, the quantity of MC accumulation in the resistant, mct-expressing strain (PJS102) was only 25% of that in the susceptible control at 5 min and thereafter remained at reduced concentrations (Fig. 5). Reduced accumulation of the drug in PJS102 suggests that mct encodes a protein that facilitates MC export from the cell. To determine if the coexpression of mct and mrd in E. coli also resulted in reduced accumulation of MC, strain PJS103 was analyzed by using the [3H]MC uptake assay. Analyses of drug uptake by cultures of strain PJS100 (28) were also performed to determine drug accumulation levels in this MC-resistant E. coli strain.
FIG. 5.
MC uptake analysis of strains PJS100 (mrd+) (■), PJS102 (mct+) (⧫), PJS103 (mrd+ mct+) (×), and the BL21(DE3)::pET17b vector control strain (●).
The results show a clear difference in MC accumulation between the MC-sensitive and -resistant strains. Compared to that in E. coli cells bearing vector alone, MC accumulation in PJS103 was only 35% at 5 min and thereafter remained at reduced concentrations. The accumulation of the drug in strain PJS103 was found to parallel that in strain PJS102, albeit at slightly higher levels (about 23% greater) of drug over the course of the experiment. Interestingly, strain PJS100, although resistant to significant concentrations of MC, accumulated the drug to levels 42% higher than those in the drug-sensitive vector control strain at 30 min (Fig. 5).
DISCUSSION
Most antibiotics inhibit bacterial growth by binding to proteins or other macromolecular components involved in essential metabolic processes of the cell (7). For instance, DNA alkylation by MC results in disruption of chromosomal replication, leading to cell death (13). In many antibiotic-producing streptomycetes, a macromolecular target site(s) is likewise susceptible to endogenous cytotoxic compounds (that is certainly the case in S. lavendulae). Thus, pumping the antibiotic out of the cell at a rate equal to its production and/or reuptake would prevent drug access to intracellular target sites. Based on the levels of drug found in most antibiotic fermentation broths (concentrations of intracellular drug being low), it is apparent that drug-producing organisms often depend on efficient antibiotic transport mechanisms. Indeed, a growing number of membrane systems implicated in transport of (and therefore resistance to) a variety of antibiotics have been discovered in drug-producing streptomycetes (19, 23).
In general, genes coding for drug export proteins are physically linked to the corresponding biosynthetic genes within the genome of the antibiotic-producing microorganism. Presumably, the tight linkage of antibiotic export and biosynthetic genes ensures coordinate gene regulation. Interestingly, the presence of back-to-back and overlapping divergent promoters of antibiotic export and regulatory genes has been observed within the tetracenomycin (9) and actinorhodin (5) biosynthetic gene clusters. Conforming to this example, S. lavendulae possesses a gene coding for an integral membrane drug export protein within the MC biosynthetic gene cluster (18, 18a). Analysis of the deduced amino acid sequence of Mct revealed several similarities with actinomycete proteins predicted to function as drug exporters. By virtue of homology to tetracycline resistance proteins, which have been shown to use proton motive force to energize transport (17), the actinomycete drug resistance translocases cited in this study are predicted to power excretion by a proton-dependent electrochemical gradient. Previous work has suggested that highly conserved sequences within the amino-terminal regions of these proteins play a role in proton translocation (25), while the less well conserved C-terminal regions may be involved in drug binding (reference 23) and references therein) or recognition of a protein-drug complex.
Disruption of mct in S. lavendulae resulted in a threefold increase in sensitivity to exogenously added MC, providing evidence that Mct maintains a role in providing drug resistance in S. lavendulae. Although the effect is significant, alternative mechanisms of cellular self-protection clearly continue to operate. These evidently include MCRA, the novel redox-relay protein that reoxidizes activated MC in S. lavendulae (2, 14). It is also likely that unidentified xenobiotic transporters provide an alternative mode of drug transport in the absence of Mct, albeit with lower efficiency.
In order to probe the ability of Mct to transport drug in the presence and absence of the MC-binding protein, accumulation of [3H]MC in E. coli was analyzed. Expression of mct in E. coli resulted in MC-resistant cultures that accumulated lower levels of the drug than strains bearing the vector control (Fig. 5). Interestingly, strain PJS102 (expressing mct only) accumulates less drug intracellularly than strain PJS103 (expressing mrd and mct) (Fig. 5). Increased drug accumulation in strain PJS100 may lend support to the model of equimolar binding between MRD and MC (28). Significantly, higher levels of drug accumulation in strain PJS100 may be the result of intracellular sequestration of MC by MRD. Accordingly, the presence of MRD could also account for the slightly greater levels of MC accumulation in strain PJS103 (mct+ mrd+) than in strain PJS102 (expressing mct alone). Comparable to the case for binding-protein-dependent import systems (20), the binding of MC by MRD may be rate limiting in the drug excretion process.
Taken together, these results suggest that cellular protection afforded by Mct is a function of drug transport from the cytoplasm. Interestingly, coexpression of mrd and mct in E. coli led to cultures that are dramatically more resistant to exogenously added drug. While normally required for the transport systems with which they are associated, in many instances binding proteins are not integral to the process of solute translocation (11). Similarly, the presence of MRD is not required for MC translocation but dramatically enhances drug tolerance. Hence, the binding protein (MRD) may be considered an accessory component, a rather specific adaptation required for optimal drug resistance. The drug resistance phenotypes of E. coli strains expressing mct alone and in combination with mrd, along with the MC uptake analysis of these strains, provide evidence that MRD and Mct are components of a novel drug transport system. Such a resistance mechanism, sequestering the intact drug for efficient excretion to the environment, represents a unique cellular strategy for self-preservation by the MC-producing organism.
ACKNOWLEDGMENTS
P.J.S. was supported in part by NIGMS Biotechnology Training grant GM08347 and by a grant from the Biological Process Technology Institute, University of Minnesota (to D.H.S.).
We thank Tetsuo Oka of Kyowa Hakko Kogyo for generous gifts of mitomycins and [3H]MC. A gift of MC from R. Schwindinger of Bristol-Myers Squibb Pharmaceutical Research Institute is gratefully acknowledged.
REFERENCES
- 1.Ames G. Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu Rev Biochem. 1986;55:397–425. doi: 10.1146/annurev.bi.55.070186.002145. [DOI] [PubMed] [Google Scholar]
- 2.August P R, Flickinger M C, Sherman D H. Cloning and analysis of a locus (mcr) involved in mitomycin C resistance in Streptomyces lavendulae. J Bacteriol. 1994;176:4448–4454. doi: 10.1128/jb.176.14.4448-4454.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bierman M, Logan R, O’Brien K, Seno E, Rao R N, Schoner B. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 4.Brown W C, Campbell J L. A new cloning vector and expression strategy for genes encoding proteins toxic to Escherichia coli. Gene. 1993;127:99–103. doi: 10.1016/0378-1119(93)90622-a. [DOI] [PubMed] [Google Scholar]
- 5.Caballero J, Malpartida F, Hopwood D A. Transcriptional organization and regulation of an antibiotic export complex in the producing Streptomyces culture. Mol Gen Genet. 1991;228:372–380. doi: 10.1007/BF00260629. [DOI] [PubMed] [Google Scholar]
- 6.Coque J, Liras P, Martin J. Genes for a β-lactamase, a penicillin-binding protein and a transmembrane protein are clustered with the cephamycin biosynthetic genes in Nocardia lactamdurans. EMBO J. 1993;12:631–639. doi: 10.1002/j.1460-2075.1993.tb05696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cundliffe E. Self-protection mechanisms in antibiotic producers. Ciba Found Symp. 1992;171:199–208. doi: 10.1002/9780470514344.ch12. [DOI] [PubMed] [Google Scholar]
- 8.Denis F, Brzezinski R. A versatile shuttle cosmid vector for use in Escherichia coli and actinomycetes. Gene. 1992;111:115–118. doi: 10.1016/0378-1119(92)90611-r. [DOI] [PubMed] [Google Scholar]
- 9.Guilfoile P, Hutchinson C R. The Streptomyces glaucescens TcmR protein represses transcription of the divergently oriented tcmR and tcmA genes by binding to an intergenic operator region. J Bacteriol. 1992;174:3659–3666. doi: 10.1128/jb.174.11.3659-3666.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson C I. Recent advances in the usage of mitomycin. Oncology. 1993;50(Suppl. 1):1–84. doi: 10.1159/000227240. [DOI] [PubMed] [Google Scholar]
- 11.Higgins C, Hyde S, Mimmack M, Gileadi U, Gill D, Gallagher M. Binding protein-dependent transport systems. J Bioenerg Biomembr. 1990;22:571–592. doi: 10.1007/BF00762962. [DOI] [PubMed] [Google Scholar]
- 12.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces. A laboratory manual. Norwich, England: John Innes Foundation; 1985. [Google Scholar]
- 13.Iyer V N, Szybalski W. Mitomycin or porfiromycin: chemical mechanism of activation and cross-linking of DNA. Science. 1964;145:55–56. doi: 10.1126/science.145.3627.55. [DOI] [PubMed] [Google Scholar]
- 14.Johnson D A, August P R, Shackleton C, Liu H W, Sherman D H. Microbial resistance to mitomycins involves a redox relay mechanism. J Am Chem Soc. 1997;119:2576–2577. [Google Scholar]
- 15.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 16.Levy S, McMurry L. Plasmid-mediated tetracycline resistance involves alternative transport systems for tetracycline. Nature. 1978;276:90–92. doi: 10.1038/276090a0. [DOI] [PubMed] [Google Scholar]
- 17.Littlejohn T, Paulsen I, Gillespie M, Tennent J, Midgley M, Jones I, Purewal A, Skurray R. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol Lett. 1992;95:259–266. doi: 10.1016/0378-1097(92)90439-u. [DOI] [PubMed] [Google Scholar]
- 18.Mao Y, Varoglu M, Sherman D H. Genetic localization and molecular characterization of two key genes (mitAB) required for biosynthesis of the antitumor antibiotic mitomycin C. J Bacteriol. 1999;181:2199–2208. doi: 10.1128/jb.181.7.2199-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Mao, Y., M. Varoglu, and D. H. Sherman. Molecular characterization and analysis of the biosynthetic gene cluster for the antitumor antibiotic mitomycin C from Streptomyces Tavendulae NRRL 2564. Chem. Biol., in press. [DOI] [PubMed]
- 19.Mendez C, Salas J A. ABC transporters in antibiotic-producing actinomycetes. FEMS Microb Lett. 1998;158:1–8. doi: 10.1111/j.1574-6968.1998.tb12792.x. [DOI] [PubMed] [Google Scholar]
- 20.Miller J, Olson J, Plfugrath J, Quiocho F. Rates of ligand binding to periplasmic proteins involved in bacterial transport and chemotaxis. J Biol Chem. 1983;258:13665–13672. [PubMed] [Google Scholar]
- 21.Neal R J, Chater K F. Nucleotide sequence analysis reveals similarities between proteins determining methylenomycin A resistance in Streptomyces and tetracycline resistance in eubacteria. Gene. 1987;58:229–241. doi: 10.1016/0378-1119(87)90378-7. [DOI] [PubMed] [Google Scholar]
- 22.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 23.Paulsen I, Brown M, Skurray R. Proton-dependent multidrug efflux pumps. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulsen I, Skurray R. Topology, structure and evolution of two families of proteins involved in antibiotic and antiseptic resistance in eukaryotes and prokaryotes—an analysis. Gene. 1993;124:1–11. doi: 10.1016/0378-1119(93)90755-r. [DOI] [PubMed] [Google Scholar]
- 25.Rouch D, Cram D, DiBerardino D, Littlejohn T, Skurray R. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline and sugar-transport proteins. Mol Microbiol. 1990;4:2051–2062. doi: 10.1111/j.1365-2958.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch T, Maniatis E F. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Schlosser A, Schrempf H. A lipid-anchored binding protein is a component of an ATP-dependent cellobiose/-triose transport system from the cellulose degrader Streptomyces reticuli. Eur J Biochem. 1996;242:332–338. doi: 10.1111/j.1432-1033.1996.0332r.x. [DOI] [PubMed] [Google Scholar]
- 28.Sheldon P J, Johnson D A, August P R, Liu H-W, Sherman D H. Characterization of a mitomycin-binding drug resistance mechanism from the producing organism, Streptomyces lavendulae. J Bacteriol. 1997;179:1796–1804. doi: 10.1128/jb.179.5.1796-1804.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 30.Tercero J, Lacalle R, Jimenez A. The pur8 gene from the pur cluster of Streptomyces alboniger encodes a highly hydrophobic polypeptide which confers resistance to puromycin. Eur J Biochem. 1993;218:963–971. doi: 10.1111/j.1432-1033.1993.tb18454.x. [DOI] [PubMed] [Google Scholar]
- 31.Tomasz M. Mitomycin C: small, fast and deadly (but very selective) Chem Biol. 1995;2:575–579. doi: 10.1016/1074-5521(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 32.Tuan J, Weber J M, Staver M, Leung J, Donadio S, Katz L. Cloning of genes involved in erythromycin biosynthesis from Saccharopolyspora erythraea using a novel actinomycete-Escherichia coli cosmid. Gene. 1990;90:21–29. doi: 10.1016/0378-1119(90)90435-t. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H Z, Schmidt H, Piepersberg W. Molecular cloning and characterization of two lincomycin-resistance genes, lmrA and lmrB, from Streptomyces lincolnensis 78-11. Mol Microbiol. 1992;6:2147–2157. doi: 10.1111/j.1365-2958.1992.tb01388.x. [DOI] [PubMed] [Google Scholar]