Abstract
Degenerate oligonucleotides were used to randomize 21 bp of the 53-bp minimal bop promoter in three 7-bp segments, including the putative TATA box and the upstream activator sequence (UAS). The mutagenized bop promoter and the wild-type structural gene and transcriptional terminator were inserted into a shuttle plasmid capable of replication in the halophilic archaeon Halobacterium sp. strain S9. Active promoters were isolated by screening transformants of an orange (Pum− bop) Halobacterium mutant for purple (Pum+ bop+) colonies on agar plates and analyzed for bop mRNA and/or bacteriorhodopsin content. Sequence analysis yielded the consensus sequence 5′-tyT(T/a)Ta-3′, corresponding to the promoter TATA box element 30 to 25 bp 5′ of the transcription start site. A putative UAS, 5′-ACCcnactagTTnG-3′, located 52 to 39 bp 5′ of the transcription start site was found to be conserved in active promoters. This study provides direct evidence for the requirement of the TATA box and UAS for bop promoter activity.
The halophilic archaeon Halobacterium sp. produces the unique membrane protein bacterio-opsin, which complexes with retinal to form bacteriorhodopsin (BR). BR forms a two-dimensional crystalline lattice, called the purple membrane, in the cell membrane of Halobacterium species and acts as a light-driven proton pump (15). The electrochemical proton gradient generated across the membrane is used by the cells for ATP synthesis under low-oxygen, high light intensity conditions. BR synthesis has been shown to be induced by low oxygen tension and high light intensity and supports a period of phototrophic growth (21, 29, 30).
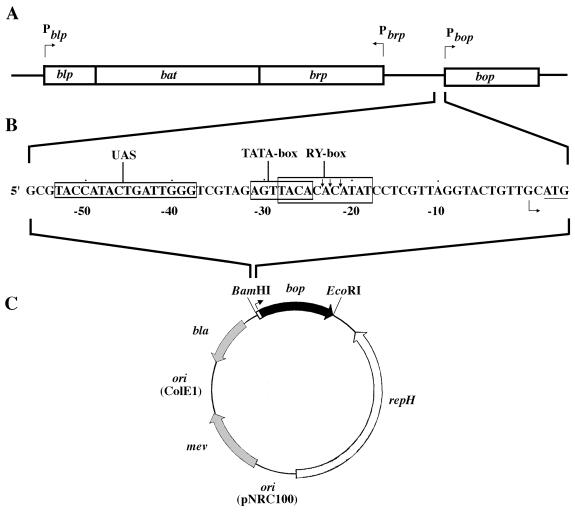
The gene for bacterio-opsin, bop, was one of the first archaeal genes to be cloned (3, 10), and analysis of the transcript showed it to start only two nucleotides upstream of the ATG start codon (6). Expression of the bop gene was detected first in mid- to late-log-phase cultures, with maximum mRNA levels occurring in the stationary phase (17, 34). Insertions in two genes divergently transcribed from bop, brp, and bat resulted in greatly reduced transcription of bop, suggesting their involvement in its regulation (Fig. 1A) (2, 18). The bat gene product showed similarity to the flavin adenine dinucleotide-binding region of nifL (34), which functions in redox sensing in nitrogen-fixing bacteria (9). The function of the brp gene, which encodes a hydrophobic protein, is unknown, and the Bop− phenotype of brp mutants may result from polar effects on bat. A gene downstream of bat, blp, is transcriptionally unlinked but is regulated in a manner similar to that in which bop is regulated (Fig. 1A) (12).
FIG. 1.
Organization of the bop gene region and promoter mutagenesis. (A) Organization of the bop gene region (bop, brp, bat, and blp genes) shown in boxes with the promoters indicated above by arrows. (B) Minimum bop promoter and a few surrounding nucleotides. The sequence is numbered starting with the transcription start point (indicated by an arrow) as +1. The UAS, TATA box, and R-Y box regions are boxed, and the translational start codon is underlined. The locations of osmium tetroxide and S1 nuclease cleavages are indicated by vertical arrows (36). (C) E. coli-Halobacterium shuttle vector (pNB series), which contains the bop gene (black arrow) and promoter (white box). The E. coli replication origin (ori ColE1) and selectable marker (bla [shaded arrow]) are indicated, as are the Halobacterium replication origin (ori pNRC100), repH gene (white arrow), and selectable marker (mev [shaded arrow]).
Recently, deletion analysis of the bop promoter showed that a 53-bp region upstream of the transcription start site is sufficient for wild-type transcriptional activity (Fig. 1B) (12, 36). Sequence analysis of this minimal promoter region revealed weak homology to the archaeal TATA box element located approximately 25 bp 5′ of the start site. Further upstream, sequence homology was noted between the bop promoter and the blp gene promoter (12). This region was referred to as the upstream activator sequence (UAS) and predicted to be involved in bop gene regulation (12). In addition to the TATA box and UAS, an 11-bp alternating purine-pyrimidine sequence overlapping the TATA box and centered 23 bp 5′ of the transcription start site was also identified. The region was hypothesized to undergo a change in DNA structure under conditions of low oxygen tension to a non-B-DNA form, which could explain the observed inhibition of bop transcription in the presence of the DNA gyrase inhibitor novobiocin (34). Direct evidence for a supercoiling-dependent structural alteration in this region was provided by showing sensitivity to osmium tetroxide and S1 nuclease (36).
In order to study the sequence requirements for transcription in more detail, we conducted saturation mutagenesis of key sequences within the minimal bop promoter. In this report, we provide evidence for the requirement of the TATA box and UAS for bop promoter activity.
MATERIALS AND METHODS
Halobacterium strains and culturing.
Halobacterium sp. strain S9, a purple-membrane (Pum) constitutive strain, and strain SD23, a Pum− derivative of S9 with an ISH1 insertion at the 5′ end of bop, have already been described (34). Culturing of these Halobacterium strains was done at 37°C in a CM+ medium containing 4.5 M NaCl and trace metals as previously described (7). Culturing for studying effects of treatment with novobiocin (Sigma, St. Louis, Mo.) was conducted as described by Yang et al. (36). Briefly, novobiocin was added to a final concentration of 0.05 μg/ml to cultures grown to an optical density at 600 nm (OD600) of 0.2 (early log phase), the cultures were allowed to grow to stationary phase (OD600 of >1.8) in the presence of the drug, at which point the cells were harvested for purple-membrane or RNA preparations (see below).
Mutagenesis.
Mutagenesis of the promoter was accomplished by PCR amplification of the cloned bop gene in pMS1 (10) with a mutagenic primer having degeneracies in seven positions corresponding to the region to be mutated, NB10 for the first seven nucleotides of the UAS, starting at position −52 (5′ TCGCGGATCCTAAATTCCGTCACGAGCGTNNNNNNNTGATTGGGTCGTAGAGTTA-3′); NB11 for the subsequent seven nucleotides of the UAS, from position −46 to position −40 (5′-TCGCGGATCCGTCACGAGCGTACCATACNNNNNNNGTCGTAGAGTTACACACATATCCTC-3′); NB6 for the TATA box (5′TCGCGGATCCGCGTACCATACTGATTGGGTCGTAGNNNNNNNCACATATCCTCGTTAGG-3′); and downstream oligonucleotide NB3 (5′-GGGAATTCTACAAGACCGAGTGG-3′). The synthetic oligonucleotides were purchased from Genosys Biotechnologies, Inc., Woodlands, Tex. The PCR amplifications were done by using Taq polymerase and standard conditions on a GeneAmp PCR system 2400 thermal cycler (Perkin-Elmer, Foster City, Calif.).
Construction and screening of UAS and TATA box libraries.
The products obtained from the PCR amplifications with one of the three mutagenic primers (NB10, NB11, or NB6) and NB3 were fractionated on a 0.8% agarose gel, gel purified, and digested with a fivefold excess (each) of EcoRI and BamHI (New England Biolabs, Beverly, Mass.). The digested inserts were repurified after fractionation on a 0.8% agarose gel and used for cloning. The vector was prepared by removing a 1.2-kb EcoRI/BamHI stuffer fragment from Halobacterium-Escherichia coli shuttle plasmid pNB148, a pNG168 derivative (Fig. 1C) (8), and subsequently gel purified. The insert DNAs were ligated to this vector and electroporated into Electromax E. coli DH10B cells (Gibco-BRL, Rockville, Md.). A fraction of the transformation was plated on Luria-Bertani agar plates containing ampicillin (100 μg/ml) to determine the efficiency of cloning, and the remainder was amplified in a 1-liter culture. Each library contained 25,000 to 30,000 members. The libraries were amplified in E. coli DH10B, and plasmid DNA was prepared on a large scale from a 1-liter culture by the alkaline lysis method. Plasmid DNA was purified on a CsCl density gradient and analyzed spectrophotometrically. All of the standard recombinant DNA procedures used have been previously described (27).
DNAs from the amplified libraries were transformed into Pum− Halobacterium sp. strain SD23 by the polyethylene glycol-EDTA transformation method (4). Halobacterium transformants were selected on CM+ plates containing 16-μg/ml mevinolin. A total of >25,000 CFU was considered a good representation of the library (each library can have a maximum of 47 or 16,384 different sequences). The purple-colony (Pum+) phenotype was used as a screen to select candidates for further analysis. The presence of plasmids was confirmed by PCR analysis of total DNA from the Halobacterium transformants, prepared by previously described methods (19), with NB3 and T7 primers, and by recovering the plasmids after transformation into E. coli DH5α. Plasmid DNA extracted from single isolated E. coli colonies was retransformed into Halobacterium sp. strain SD23 to confirm the observed phenotype.
BR assays.
Halobacterium cultures grown to an OD600 of approximately 1.8 were used for purple-membrane preparation by the method described by Oesterhelt (22). Cells from 50 ml of each culture were harvested by centrifugation at 7,000 rpm for 20 min. The cell paste was resuspended in 4 ml of a basal salts solution containing 20-μg/ml DNase I. The cells were gradually lysed by overnight dialysis (Spectrum Laboratories, Laguna Hills, Calif.) against distilled water, and the lysate was then analyzed spectrophotometrically at 568 nm to quantitate BR content. The absorption values were normalized to an OD280 of 1.5 and corrected by subtracting the background (normalized reading for host Halobacterium sp. strain SD23).
Primer extension analysis.
Crude RNA was prepared from 3- or 6-ml Halobacterium cultures by either the hot-phenol method (36) or use of the RNeasy kit (Qiagen, Valencia, Calif.). Primer extension was performed on approximately 10 μg of RNA by using an end-labeled bop2 primer (5′CCTGCGATACCCCCT-3′) and a primer extension kit (Promega Corporation, Madison, Wis.) in accordance with the manufacturer’s instructions. End labeling was done by using [γ-32P]ATP (Amersham Life Science, Arlington Heights, Ill., and NEN Life Science Products, Inc., Boston, Mass.) in a T4 polynucleotide kinase (New England Biolabs)-catalyzed reaction. The cDNA product was analyzed by electrophoresis under denaturing conditions on a 6% polyacrylamide–8.3 M urea gel. Band intensities were quantified by densitometric analysis of the autoradiograms with a Bio-Rad densitometer with Molecular Analyst software (Bio-Rad Laboratories, Hercules, Calif.) or by PhosphorImager analysis of the gel with a Storm 860 scanner connected to the PhosphorImager SI system with ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.).
Sequence analysis.
Promoter plasmids prepared by the alkaline lysis method from cultures of E. coli DH5α transformants were used as templates for sequencing. T7 and bop2 oligonucleotides were used to sequence both strands by the dideoxy cycle-sequencing method (25, 28) using either radioisotope-based chemistry (Genozyme Cycle-Sequencing Kit [Genomyx Corp., Foster City, Calif.] or Sequenase version 2.0 Sequencing kit [Amersham Life Science, Cleveland, Ohio]) on a Genomyx LR sequencer or fluorescent-dye terminator chemistry (Thermosequenase dye terminator cycle sequencing premix kit; Amersham Life Science) on an ABI373A sequencer (Perkin-Elmer). Sequences were analyzed by using the GCG software package (Genetics Computer Group Inc., Madison, Wis.) running on an SGI O2 workstation (Silicon Graphics Inc., Mountain View, Calif.).
RESULTS
Saturation mutagenesis of the TATA box and UAS.
The bop gene was mutagenized by PCR amplification using one of three oligonucleotides, each with degeneracies at seven positions within the promoter sequence, and a nonmutagenic oligonucleotide located downstream of the transcriptional terminator. The mutagenic regions corresponded to two segments of the UAS and the TATA box (Fig. 1B). The amplified bop gene fragments were cloned into a Halobacterium-E. coli shuttle vector (Fig. 1C) and transformed into E. coli. At least 25,000 transformants were obtained for each library. Since the total number of different sequences that could be obtained with seven degeneracies was 16,384, we calculate that 78% of the possible sequences were represented. The libraries were amplified in E. coli DH10B and transformed into Halobacterium sp. strain SD23 (a bop mutant derivative of strain S9). Transformant colonies were visually inspected for purple-membrane content and scored as Pum+ or Pum−. Phenotypes were confirmed by recovery of plasmids and retransformation of SD23. Pum+ and Pum− representatives were analyzed for bop gene expression by assaying mRNA levels using primer extension analysis and/or for BR content by spectrophotometric analysis.
TATA box mutagenesis.
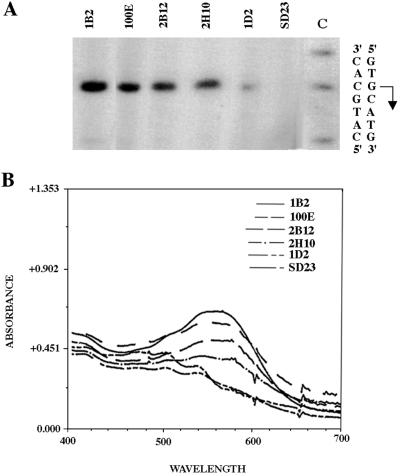
The putative TATA box region of the bop promoter (31 to 25 bp 5′ of the transcription start point) was mutagenized as described above. Approximately 2 to 3% of the transformants were phenotypically Pum+, suggesting that two or three nucleotides in this region are critical for bop gene expression. The levels of bop mRNA and BR protein produced from the wild-type promoter and four mutated Pum+ promoters (1B2, 2B12, 2H10, and 1D2) were compared by primer extension analysis and spectrophotometric assays (Fig. 2). There was close correspondence between the Pum phenotypes, bop mRNA levels, and BR levels. Moreover, the transcription start points were unchanged, irrespective of the level of transcription observed.
FIG. 2.
Analysis of TATA box mutants at the transcriptional (A) and translational (B) levels. (A) Primer extension analysis of bop mRNA in four TATA box mutants using crude RNA with the bop2 primer. Strain designations are above the lanes. The controls used in this experiment were Pum− (bop) host strain SD23, strain 100E (SD23 containing a plasmid with an unmutated promoter), and TATA box mutants (1B2, 2B12, 2H10, and 1D2). A sequencing reaction (lane C) performed with the bop2 primer on a pMS1 template (plasmid containing the cloned bop gene) is shown, as is the double-stranded sequence across the transcription start point (the start site and direction are indicated by the arrow). (B) Spectra (absorbance versus wavelength from 400 to 700 nm) of purple membrane preparations for the four TATA box mutant strains and the two control strains (same as in panel A). Relative BR content was quantified by comparison of absorbance at 568 nm.
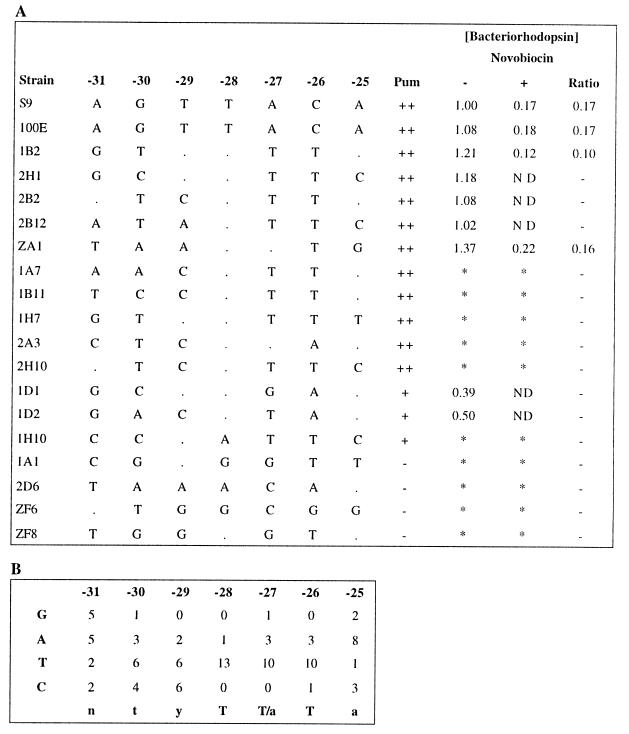
We selected 13 Pum+ and 4 Pum− transformants for further characterization. The promoter regions were sequenced and aligned, and a consensus sequence was derived from the most active promoters [5′-tyT(T/a)Ta-3′, −30 to −25 bp 5′ of the transcription start point] (Fig. 3). Significantly, two positions of the consensus (two T nucleotides at positions −28 and −26) were highly conserved (present in 9 of the 10 most active promoters). A third T nucleotide (at position −27) was nearly as highly conserved, although an A is also tolerated. Those promoters containing a TTT or TAT sequence in the region between −28 and −26 (i.e., 1B2, 2H1, 2B2, 2B12, and ZA1) produced higher levels of transcription than the wild type, which contained a TAC sequence in this region. Inactive promoters (1A1, ZF6, ZF8, and 2D6), in contrast, contained two or more differences within the most highly conserved region in the consensus. All of the functional promoters were found to be inhibited or inactive in the presence of novobiocin, indicating that they are sensitive to DNA supercoiling, like the wild type.
FIG. 3.
Tabulation of strain designations, promoter sequences, phenotypes, and BR contents of TATA box mutants (A) and analysis of promoter sequences (B). In panel A, the strain designations are shown in the first column, and the sequence of nucleotides −31 to −25 (identity to the wild type base is denoted by a dot), the Pum phenotype (−, negative; +, positive; ++, overproducer), and relative BR content are shown to the right. ND, not detectable; ∗, not done. For panel B, the TATA box consensus sequence was determined by tallying individual nucleotides observed at each position in Pum+ strains. The consensus sequence is indicated at the bottom.
UAS mutagenesis.
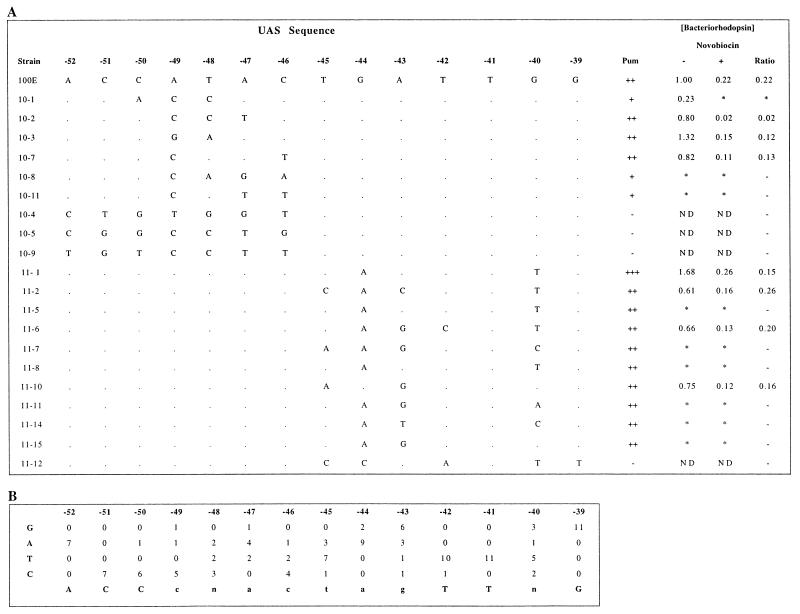
Because of the larger size of the UAS region, it was mutagenized in two separate 7-bp segments, from −52 to −46 (UAS10) and from −45 to −39 (UAS11) from the transcription start site. The importance of this entire region for bop gene expression was shown by the finding of about 1% Pum+ transformants, which indicated that the nucleotide sequence is somewhat less mutable than the TATA box region for active promoter activity. We selected 6 Pum+ and 3 Pum− transformants from the UAS10 region and 10 Pum+ and 1 Pum− transformants from the UAS11 region for further characterization. The BR contents in eight Pum+ and four Pum− mutants were analyzed spectrophotometrically (Fig. 4), and the mRNA levels in the Pum+ mutants were measured by primer extension analysis (Fig. 5). The results confirmed the observed phenotypes. The transcription start site was found to be unchanged, irrespective of the promoter strength (Fig. 5). The consensus sequence obtained from alignment of the mutated UASs was 5′-ACCcnactagTTnG-3′, very similar (three differences in 12 bp) to the wild-type sequence, 5′-ACCATACTGATTGG-3′. Significantly, four positions (−52, −51, −41, and −39) were completely invariant within the UASs of active promoters. One mutant promoter (11-1), with improved similarity to the consensus UAS, produced 68% more BR protein than the wild type, while another (10-3) with comparable similarity produced 33% more BR than the wild type. By comparison, the inactive promoters contained multiple differences in the most-conserved nucleotides of the consensus sequence. All of the active promoters were also inhibited by novobiocin, indicating that they are sensitive to DNA supercoiling.
FIG. 4.
Tabulation of strain designations, promoter sequences, phenotypes, and BR contents of UAS mutants (A) and analysis of promoter sequences (B). In panel A, the strain designations are shown in the first column, and sequence of nucleotides −52 to −39 (identity to the wild type base is denoted by a dot), the Pum phenotype (for definitions, see the legend to Fig. 3), and relative BR content are shown to the right. ND, not detectable; ∗, not done. For panel B, the UAS element consensus sequence was determined by tallying individual nucleotides observed at each position in Pum+ strains. The consensus sequence is indicated at the bottom.
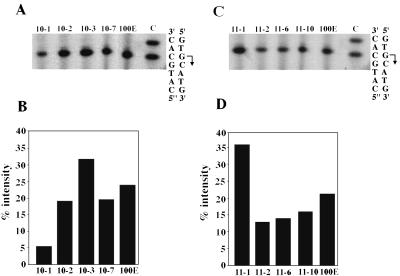
FIG. 5.
Effect of UAS mutagenesis on transcription start site location (A and C) and promoter strength (B and D). For panels A and B, transcription start sites were mapped by comparing the mobilities of primer extension products to that of a sequencing ladder generated with the bop2 primer and the pMS1 template (plasmid with a cloned bop gene). Lane C of the sequencing reaction is shown. The strain designations are shown over the corresponding lanes. For panels C and D, promoter strength was measured by quantifying the bop message by PhosphorImager analysis of the primer extension gels. The strain designations (corresponding to A and C, respectively) are shown below the bars plotted against relative intensity.
DISCUSSION
We have conducted saturation mutagenesis of the TATA box and UAS region in the bop promoter. Degenerate oligonucleotides were used to produce over 25,000 different promoter mutants, which were screened by using the purple (Pum+) or orange (Pum−) phenotype. A wide range of phenotypes, from purple-membrane overproducers to completely purple-membrane-deficient strains, were characterized at both the transcriptional and translational levels. The results were found to be consistent at the phenotypic, mRNA, and protein levels, confirming that the observed effects resulted from promoter mutations. The mutations had no effect on the transcription start site, ruling out the possible activation of alternate promoters. Taken together, the findings constitute a detailed mutagenic analysis of two putative bop promoter elements with clear demonstration of their requirement for wild-type promoter activity.
The bop promoter has been of considerable interest because of its complex regulation (responsive to oxygen, light, and DNA supercoiling). Moreover, unlike most archaeal (and eukaryotic) promoters which have a distinctive TATA box (also called BoxA) centered at 25 bp 5′ of the transcription start site, which is recognized by the TATA-binding protein (TBP) transcription factor (1a, 11, 14, 16, 32), the bop promoter has weak homology to the TATA sequence located several nucleotides further upstream. Our mutagenic analysis has definitively established that the sequence in the TATA box region is involved in bop promoter activity. Interestingly, the wild-type bop promoter sequence is different from the consensus TATA box sequence, even in the most highly conserved region, and the 3′ four nucleotides of the consensus are more similar in sequence and position to the consensus sequences derived from alignment and mutagenesis of other archaeal promoters (5, 13, 23, 31).
The Halobacterium genome project has shown the presence of multiple TBP-encoding genes (20). Four tbp genes are present on a 191-kb minichromosome named pNRC100. This finding suggests that alternate TBPs are probably involved in the recognition of different subsets of genes in the Halobacterium genome. If so, it is possible that an alternate TBP is involved in the recognition of the bop promoter in the wild type. A similar mechanism may be used to regulate transcription of heat shock promoters in the related halophile Haloferax volcanii (33). It is also possible that different TBP factors are involved in promoting the transcription of some mutated and wild-type bop promoters. However, the same transcription start site is used in all cases.
The UAS has been hypothesized to function in bop gene regulation (12, 36). Deletion analysis showed that the UAS is required for bop transcription, and the sequence requirement in this region has been confirmed by the mutagenic analysis described in this report. The consensus sequence of the UAS derived from mutagenesis has several interesting properties, including a sequence slightly different from that of the wild type, which likely explains the less-than-maximum promoter activity observed in the wild type. A greater degree of conservation is observed near the 5′ and 3′ ends than in the middle. It is likely that the UAS is a site of action of a global regulator of gene expression, which is suggested by its occurrence at many genomic sites (1). An interesting possibility is that the bop regulatory gene products, BRP and/or BAT (or a protein interacting with these proteins), bind to the UASs near the bop, blp, and other genes and modulate transcription from these promoters in response to oxygen and/or light.
One of the most intriguing aspects of bop promoter function is its property of supercoiling sensitivity. We previously observed that like that of some other supercoiling sensitive genes in bacteria (24), the DNA gyrase inhibitor reduces bop transcription by a factor of 5 to 10 at concentrations subinhibitory for growth (35). Novobiocin prevents the increased supercoiling observed at late logarithmic phase which accompanies bop gene induction. At the high negative supercoiling density found under inducing conditions, the 11-bp alternating purine-pyrimidine sequence (the R-Y box) adopts a non-B-DNA structure. We hypothesized that the supercoiling-stimulated non-B-DNA structure is necessary for full induction of the bop gene via transcriptional activation. Mutagenesis of the UAS and TATA box showed that neither region is responsible for the sensitivity of the promoter to supercoiling. Preliminary data suggest that the region 3′ of the TATA box (middle of the alternating purine-pyrimidine R-Y box region) mediates the response to a change in supercoiling (1).
The results obtained thus far have established the importance of the TATA box and the UAS in bop gene expression and suggested the involvement of transcription factors such as a TBP and other regulatory proteins in the activation and modulation of transcription. However, more detailed understanding of the mechanisms of promoter recognition, activation, and regulation requires further genetic and biochemical analysis, including development of an in vitro transcription system using both purified proteins and DNA topoisomers.
ACKNOWLEDGMENTS
We thank Stacy Ciufo for help with sequence analysis and Fazeela Morshed for help with halobacterial transformations.
This work was supported by grant MCB-9604443 from the National Science Foundation to S.D.
ADDENDUM IN PROOF
Further mutagenesis has shown the requirement of a G at position −38, which is likely to be part of the functional UAS.
REFERENCES
- 1.Baliga, N. S., and S. DasSarma. Unpublished data.
- 1a.Baumann P, Qureshi S A, Jackson S P. Transcription: new insights from studies on archaea. Trends Genet. 1995;11:279–283. doi: 10.1016/s0168-9525(00)89075-7. [DOI] [PubMed] [Google Scholar]
- 2.Betlach M, Friedman J, Boyer H W, Pfeifer F. Characterization of a halobacterial gene affecting bacterio-opsin gene expression. Nucleic Acids Res. 1984;12:7949–7959. doi: 10.1093/nar/12.20.7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang S, Majumdar H A, Dunn R, Makabe O, RajBhandary U L, Khorana H G, Ohtsuka E, Tanaka T, Taniyama Y, Ikehara M. Bacteriorhodopsin: partial sequence of mRNA provides amino acid sequence in the precursor region. Proc Natl Acad Sci USA. 1981;78:3398–3402. doi: 10.1073/pnas.78.6.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cline S, Doolittle W F. Efficient transfection of the archaebacterium Halobacterium halobium. J Bacteriol. 1987;169:1341–1344. doi: 10.1128/jb.169.3.1341-1344.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danner S, Soppa J. Characterization of the distal promoter element of halobacteria in vivo using saturation mutagenesis and selection. Mol Microbiol. 1996;19:1265–1276. doi: 10.1111/j.1365-2958.1996.tb02471.x. [DOI] [PubMed] [Google Scholar]
- 6.DasSarma S, RajBhandary U L, Khorana H G. Bacterio-opsin mRNA in wild-type and bacterio-opsin deficient Halobacterium halobium strains. Proc Natl Acad Sci USA. 1984;81:125–129. doi: 10.1073/pnas.81.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DasSarma S, Fleischmann E M, Rodriguez-Valera F. Media for halophiles. In: DasSarma S, et al., editors. Archaea: a laboratory manual—halophiles. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 225–230. [Google Scholar]
- 8.DasSarma S. Natural plasmids and plasmid vectors for halophiles. In: DasSarma S, et al., editors. Archaea: a laboratory manual—halophiles. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 241–250. [Google Scholar]
- 9.Dixon R. The oxygen-responsive NIFL-NIFA complex: a novel two-component regulatory system controlling nitrogenase synthesis in γ-proteobacteria Arch. Microbiol. 1998;169:371–380. doi: 10.1007/s002030050585. [DOI] [PubMed] [Google Scholar]
- 10.Dunn R, McCoy J, Simsek M, Majumdar A, Chang S H, Rajbhandary U L, Khorana H G. The bacteriorhodopsin gene. Proc Natl Acad Sci USA. 1981;78:6744–6748. doi: 10.1073/pnas.78.11.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gohl H P, Gröndahl B, Thomm M. Promoter recognition in archaea is mediated by transcription factors: identification of transcription factor aTFB from Methanococcus thermolithotrophicus as archaeal TATA-binding protein. Nucleic Acids Res. 1995;20:5423–5428. doi: 10.1093/nar/23.19.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gropp F, Gropp R, Betlach M C. Effects of upstream deletions on light- and oxygen-regulated bacterio-opsin gene expression in Halobacterium halobium. Mol Microbiol. 1995;16:357–364. doi: 10.1111/j.1365-2958.1995.tb02307.x. [DOI] [PubMed] [Google Scholar]
- 13.Hain J, Reiter W-D, HÅdepohl U, Zillig W. Elements of an archaeal promoter defined by mutational analysis. Nucleic Acids Res. 1992;20:5423–5428. doi: 10.1093/nar/20.20.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kosa P F, Ghosh G, DeDecker B S, Sigler P B. The 2.1-Å crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box. Proc Natl Acad Sci USA. 1995;94:6042–6047. doi: 10.1073/pnas.94.12.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krebs M P, Khorana H G. Mechanism of light-dependent proton translocation by bacteriorhodopsin. J Bacteriol. 1993;175:1555–1560. doi: 10.1128/jb.175.6.1555-1560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langer D, Hain J, Thuriaux P, Zillig W. Transcription in archaea: similarity to that in eucarya. Proc Natl Acad Sci USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leong D, Boyer H, Betlach M. Transcription of genes involved in bacterio-opsin gene expression in mutants of a halophilic archaebacterium. J Bacteriol. 1988;170:4910–4915. doi: 10.1128/jb.170.10.4910-4915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leong D, Pfeifer F, Boyer H, Betlach M. Characterization of a second gene involved in bacterio-opsin gene expression in a halophilic archaebacterium. J Bacteriol. 1988;170:4903–4909. doi: 10.1128/jb.170.10.4903-4909.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng W-L, Yang C-F, Halladay J T, Arora P, DasSarma S. Isolation of genomic and plasmid DNAs from Halobacterium halobium. In: DasSarma S, et al., editors. Archaea: a laboratory manual—halophiles. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 179–184. [Google Scholar]
- 20.Ng W-L, Ciufo S A, Smith T M, Bumgardner R E, Baskin D, Faust J, Hall B, Loretz C, Seto J, Hood L, DasSarma S. Snapshot of a dynamic replicon in a halophilic archaeon: megaplasmid or minichromosome? Genome Res. 1998;8:1131–1141. doi: 10.1101/gr.8.11.1131. [DOI] [PubMed] [Google Scholar]
- 21.Oesterhelt D, Stoeckenius W. Functions of a new photoreceptor membrane. Proc Natl Acad Sci USA. 1973;70:2853–2857. doi: 10.1073/pnas.70.10.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oesterhelt D. Isolation of purple membranes. In: DasSarma S, et al., editors. Archaea: a laboratory manual—halophiles. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 55–57. [Google Scholar]
- 23.Palmer J R, Daniels C J. In vivo definition of an archaeal promoter. J Bacteriol. 1995;177:1844–1849. doi: 10.1128/jb.177.7.1844-1849.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruss G J, Drlica K. DNA supercoiling and prokaryotic transcription. Cell. 1989;56:521–523. doi: 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- 25.Reeve M A, Fuller C W. A novel thermostable polymerase for DNA sequencing. Nature. 1995;376:796–797. doi: 10.1038/376796a0. [DOI] [PubMed] [Google Scholar]
- 26.Reiter W-D, Hüdepohl U, Zillig W. Mutational analysis of an archaebacterial promoter: essential role of a TATA box for transcription efficiency and start site selection in vitro. Proc Natl Acad Sci USA. 1990;87:9509–9513. doi: 10.1073/pnas.87.24.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shand F R, Betlach M C. Expression of the bop gene cluster of Halobacterium halobium is induced by low oxygen tension and by light. J Bacteriol. 1991;173:4692–4699. doi: 10.1128/jb.173.15.4692-4699.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sumper M, Reitmeier H, Oesterhelt D. Biosynthesis of the purple membrane of halobacteria. Angew Chem Int Ed Engl. 1976;16:187–194. doi: 10.1002/anie.197601871. [DOI] [PubMed] [Google Scholar]
- 31.Thomm M, Wich G. An archaebacterial promoter element for stable RNA genes with homology to the TATA box of higher eukaryotes. Nucleic Acids Res. 1988;16:151–163. doi: 10.1093/nar/16.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomm M. Archaeal transcription factors and their role in transcription initiation. FEMS Microbiol Rev. 1996;18:159–171. doi: 10.1111/j.1574-6976.1996.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 33.Thompson D K, Daniels C J. Heat shock inducibility of an archaeal TATA-like promoter is controlled by adjacent sequence elements. Mol Microbiol. 1998;27:541–551. doi: 10.1046/j.1365-2958.1998.00698.x. [DOI] [PubMed] [Google Scholar]
- 34.Yang C-F, DasSarma S. Transcriptional induction of purple membrane and gas vesicle synthesis in the archaebacterium Halobacterium halobium is blocked by a DNA gyrase inhibitor. J Bacteriol. 1990;172:4118–4121. doi: 10.1128/jb.172.7.4118-4121.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang C-F. Ph.D. thesis. Amherst: University of Massachusetts; 1994. [Google Scholar]
- 36.Yang C-F, Kim J-M, Molinari E, DasSarma S. Genetic and topological analyses of the bop promoter of Halobacterium halobium: stimulation by DNA supercoiling and non-B-DNA structure. J Bacteriol. 1996;178:840–845. doi: 10.1128/jb.178.3.840-845.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]