Abstract
The role of the C terminus of Escherichia coli DNA helicase II (UvrD), a region outside the conserved helicase motifs, was investigated by using three mutants: UvrDΔ107C (deletion of the last 107 C-terminal amino acids), UvrDΔ102C, and UvrDΔ40C. This region, which lacks sequence similarity with other helicases, may function to tailor UvrD for its specific in vivo roles. Genetic complementation assays demonstrated that mutant proteins UvrDΔ107C and UvrDΔ102C failed to substitute for the wild-type protein in methyl-directed mismatch repair and nucleotide excision repair. UvrDΔ40C protein fully complemented the loss of helicase II in both repair pathways. UvrDΔ102C and UvrDΔ40C were purified to apparent homogeneity and characterized biochemically. UvrDΔ102C was unable to bind single-stranded DNA and exhibited a greatly reduced single-stranded DNA-stimulated ATPase activity in comparison to the wild-type protein (kcat = 0.01% of the wild-type level). UvrDΔ40C was slightly defective for DNA binding and was essentially indistinguishable from wild-type UvrD when single-stranded DNA-stimulated ATP hydrolysis and helicase activities were measured. These results suggest a role for a region near the C terminus of helicase II in binding to single-stranded DNA.
DNA helicase-catalyzed unwinding of duplex DNA provides the single-stranded DNA (ssDNA) intermediates required for the reactions of DNA metabolism, including DNA replication, recombination, transcription, bacterial conjugation, and DNA repair (25, 26, 31, 32). Helicase enzymes use energy derived from nucleoside 5′-triphosphate (NTP) hydrolysis to catalyze the disruption of hydrogen bonds between the two strands in duplex DNA. The mechanism by which ATP hydrolysis is coupled with strand separation is not well understood. However, several models have been proposed and are currently being tested (27, 28, 48). An important element of these models is the requirement of multiple DNA binding sites for active translocation along DNA. For monomeric helicases, this requirement suggests the presence of multiple DNA binding sites on each individual protomer (36). In the case of oligomeric helicases (generally dimers or hexamers), individual ligand binding sites are on each monomer and oligomerization provides multiple DNA binding sites (27).
DNA helicases have been characterized in a variety of organisms, including bacteria, bacteriophages, viruses, and eukaryotes (for reviews see references 31 and 32). Eleven distinct helicases have been identified in Escherichia coli (30–32). The existence of multiple helicases within a single cell could reflect functional redundancy, either direct substitution of one helicase for another or an overlap in the biochemical pathways in which the proteins are involved. There is no evidence for the direct substitution of one helicase for another in a specific pathway, and at least in E. coli, each helicase appears to have a distinct biological role (31).
Alignments of the primary structures of helicases have been used to categorize helicases into several superfamilies (8–10, 17). Within superfamily I and II helicases, seven regions of sequence similarity, the so-called helicase motifs, have been identified. Sequence similarity among helicases is greatest within the motifs and tends to diverge outside these regions (8–10, 17). The conserved motifs are believed to have functional significance in the biochemistry of helicase-catalyzed unwinding. Indeed, this has been demonstrated for several different helicases in structure-function studies (2, 7, 11, 14, 15, 29, 41, 42, 50, 51). From these studies it has become clear that the helicase-associated motifs are required for the biological and biochemical functions of these proteins. The sequence conservation suggests that regions of a helicase outside the defined motifs may be responsible for dictating the biological specificity of a DNA helicase. Unique regions in each protein may direct participation in a particular biochemical pathway by guiding interactions with a specific DNA substrate or other proteins, or by directing its oligomerization for those proteins that form dimers or hexamers.
The biological specificity of DNA helicases has been well documented with the closely related DNA helicase II and Rep protein. DNA helicase II is required in methyl-directed mismatch repair (12, 13, 21, 37) and UvrABC-mediated excision repair (4, 18, 39, 44). Rep protein, which exhibits 40% identity with helicase II along its entire length and 90% identity within the helicase-associated motifs (9), cannot substitute for helicase II in either pathway. Conversely, Rep protein has a role in φX174 DNA replication (6), and helicase II cannot substitute for Rep protein in this capacity. Presumably this high degree of specialization is due to specific protein-protein or protein-DNA interactions.
The amino acid sequences of Rep protein and helicase II diverge most dramatically in the C-terminal region of each protein, outside motif VI (9). The functional importance of the C terminus is evident for many proteins with helicase activity including the gpα helicase-primase from bacteriophage P4 and the Rad25 helicase from yeast (40, 52). In addition, several mutations of the Werner’s syndrome protein have been identified as C-terminal truncations outside the conserved helicase motifs (49).
A region near the C-terminal end of E. coli helicase II, and outside conserved motif VI, was defined as required for biological and biochemical function in assays using three mutants: UvrDΔ107C (deletion of 107 C-terminal amino acids), UvrDΔ102C, and UvrDΔ40C. The mutant proteins UvrDΔ107C and UvrDΔ102C failed to substitute for the wild-type protein in methyl-directed mismatch repair and nucleotide excision repair, while UvrDΔ40C protein fully restored activity in both genetic assays. The defect in UvrDΔ102C was confirmed in vitro by its failure to bind ssDNA and a dramatically reduced ssDNA-stimulated ATPase activity. UvrDΔ40C was essentially indistinguishable from wild-type UvrD in biochemical activity assays. The results suggest a role for a region near the C terminus, residues 618 to 680, of helicase II in DNA binding.
MATERIALS AND METHODS
Bacterial strains.
E. coli BL21(DE3) (F− ompT[lon] hsdSB(rB−mB−) gal dcm λDE3) was from Novagen, Inc. E. coli GE1752 (metE::cam) was obtained from G. Weinstock. BL21(DE3) ΔuvrD and GE1752 ΔuvrD were constructed previously in this laboratory (7).
DNA and nucleotides.
pET11d and pLysS were from Novagen, Inc. pMal-C2 and pTYB4 were from New England Biolabs, Inc. (NEB). M13mp7 ssDNA was prepared as described elsewhere (24). Unlabeled nucleotides were from U.S. Biochemical Corp. Radioactively labeled nucleotides were from Amersham Corp. pET11d-UvrD was constructed previously in this laboratory (7).
Mutagenesis.
Plasmids capable of expressing UvrDΔ107C, UvrDΔ102C, maltose-binding protein (MBP)–UvrDΔ102C, and intein-UvrDΔ102C were constructed by PCR with Vent polymerase and appropriate primers. pET11d-UvrD was used as a template for amplification. UvrDΔ107C and UvrDΔ102C were amplified by using a primer (5′-GGAATTGTGAGCGGATAACAATTCCCC-3′) that annealed upstream of the uvrD coding sequence and primer Δ107C (5′CGGAGATCTCATTAGGTCAGCGTCAGTTTCTGC-3′) or primer Δ102C (5′-GCCCAGATCTAAGCTTATTAGCGGGTTTCCGCGTAGG-3′). Primer Δ107C altered codon 614 of helicase II from TAC (tyrosine) to TAA (stop) and codon 615 GCG (alanine) to TGA (stop). Primer Δ102C changed codon 619 from CGT (arginine) to TAA (stop) and codon 620 from CTG (leucine) to TAA (stop). Following amplification, the DNA was cleaved with restriction enzymes NcoI and BglII. These restriction sites were engineered into the primers. Gel-purified DNA fragments were subcloned into the pET11d vector that had been digested with NcoI and BamHI.
Intein-UvrDΔ102C was constructed using the upstream primer used for UvrDΔ102C and primer inteinΔ102C (5′TTTTCCATAAGTACTGCGGGTTTCCGCGTAGGTC-3′). Primer inteinΔ102C altered codon 619 from CGT (arginine) to AGT (serine) in order to create a ScaI restriction enzyme cleavage site. After amplification, the purified fragment was cleaved with NcoI and ScaI. This fragment was subcloned into pTYB4 that had been digested with NcoI and SmaI. UvrDΔ102C purified from this construct contained an additional glycine on the C terminus.
MBP-UvrDΔ102C was constructed by amplifying the uvrD sequence on pET11d-UvrD by using a primer (5′-TGTGTCTAGAATGGACGTTTCTTACCTGC-3′) that annealed at the initiation site of the uvrD gene and primer Δ102C. This was cloned into pMal-C2, using XbaI and HindIII restriction sites found both on the DNA insert and in the vector. pET11d-UvrDΔ40C was constructed as described previously (36). The stop codon generated to terminate UvrDΔ40C was the result of a +1 frameshift. As a result, amino acids 678, 679, and 680 were changed from histidine, alanine, and lysine in the wild-type protein to threonine, proline, and arginine in UvrDΔ40C.
All mutant clones were characterized by restriction digestion and mutant proteins were verified by small-scale inductions resolved on polyacrylamide gels run in the presence of sodium dodecyl sulfate (SDS) (to compare sizes of induced proteins with wild-type protein). The uvrDΔ40C mutation was confirmed by DNA sequencing using a Sequenase kit (U.S. Biochemical). The double termination codons used to construct the uvrDΔ102C mutation were confirmed as the only mutations in the uvrD gene by sequencing the entire gene with an Applied Biosystems 373A DNA sequencer.
Enzymes.
Restriction endonucleases, DNA polymerase I (large fragment), phage T4 polynucleotide kinase, and Vent DNA polymerase were from NEB and used as recommended by the supplier. Phage T4 DNA ligase was from Boehringer Mannheim and used as advised.
Helicase II and helicase II mutants were overexpressed, prior to purification, by growing a 10-liter culture of BL21(DE3)/pLysS containing pET11d-UvrD or a 2-liter culture of BL21(DE3)ΔuvrD/pLysS containing pET11d-UvrDΔ102C, pMal-UvrDΔ102C, or pET11d-UvrDΔ40C in LB-glucose (0.4%) medium to mid-log phase. BL21(DE3)ΔuvrD/pLysS containing pTYB4-UvrDΔ102C was grown in 2× YT instead of LB-glucose medium. Cells were induced for protein expression by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to 0.5 mM. Cells containing either pET11d-UvrD or pET11d-UvrDΔ40C were incubated for an additional 4 h at 37°C. Cells containing pET11d-UvrDΔ102C, pMal-UvrDΔ102C, or pTYB4-UvrΔ102C were incubated at 25°C. Wild-type helicase II protein was purified as described elsewhere (43). UvrDΔ40C was purified by the same procedure, with one modification. UvrDΔ40C was loaded onto an ssDNA-cellulose column (5.8 mg of ssDNA/g of cellulose) at 0.1 M NaCl in buffer A (20 mM Tris-HCl [pH 8.3 at 25°C], 20% [vol/vol] glycerol, 1 mM EDTA, 0.5 mM EGTA, 15 mM 2-mercaptoethanol) instead of buffer A plus 0.2 M NaCl. The column was washed with buffer A plus 0.2 M NaCl and eluted with buffer A plus 1 M NaCl.
The procedure for purification of UvrDΔ102C was modified considerably due to the DNA binding defect inherent in this protein. Cells overexpressing UvrDΔ102C were lysed, and the protocol for purification of the wild-type protein was followed through the polymin-P precipitation step. The pellet from this precipitation (which contained UvrDΔ102C) was resuspended in 1/2 volume of fraction 1 (soluble fraction of the whole cell lysate) with buffer A containing 1 M NaCl to extract UvrDΔ102C. After a clearing spin, the supernatant was precipitated with ammonium sulfate added to 44% saturation. The pellet was resuspended in 0.59 volume of fraction 1 with buffer A. This resuspension was incubated with DEAE-cellulose resin (26-ml bed volume), equilibrated to 0.1 M NaCl in buffer A. After a 60-min incubation, a column (2.7-cm inside diameter by 4.5 cm) was poured and washed with buffer A plus 0.15 M NaCl. Protein was eluted with a linear gradient from 0.2 M to 0.6 M NaCl in buffer A. Fractions containing UvrDΔ102C, which eluted at 0.22 M NaCl, were pooled and dialyzed against buffer A plus 1 M ammonium sulfate. The dialysate was loaded onto a 10-ml phenyl-Sepharose CL-6B (Pharmacia) column equilibrated with buffer A and 1 M ammonium sulfate. The column was washed to baseline with 1 M ammonium sulfate, followed by 0.4 and 0.15 M ammonium sulfate. A linear gradient from 0.1 to 0 M ammonium sulfate, followed by extensive washing with buffer A, was required to elute protein off this column. Pooled fractions were applied to a 2-ml ssDNA-cellulose column (5.8 mg of ssDNA/g of cellulose) (Amersham) equilibrated with buffer A plus 0.1 M NaCl. The column was washed with buffer A plus 0.1 M NaCl, and protein that failed to bind was collected and concentrated by ammonium sulfate precipitation (55% saturation). The pellet was suspended in a minimal volume of buffer A containing 0.5 M NaCl and loaded on a 60-ml Sephacryl S-200 sizing column (Pharmacia). Peak fractions were pooled and concentrated by ammonium sulfate precipitation (55% saturation) and resuspended in 1 ml of helicase II storage buffer (20 mM Tris-HCl [pH 7.5 at 25°C], 0.2 M NaCl, 50% [vol/vol] glycerol, 1 mM EDTA, 0.5 mM EGTA, 25 mM 2-mercaptoethanol) (43).
UvrDΔ102C was also purified as an MBP fusion. Cells expressing MBP-UvrDΔ102C were lysed by the procedure described for the wild-type protein. The conductivity of the whole-cell lysate was adjusted with buffer A to a conductivity equivalent to that of buffer A plus 0.2 M NaCl. MBP-UvrDΔ102C was purified by binding to an amylose column (NEB) followed by elution with 10 mM maltose as described by the manufacturer. Protein from pooled fractions was concentrated by ammonium sulfate precipitation (55% saturation). The precipitated protein was suspended in helicase II storage buffer and dialyzed against storage buffer to remove excess salt.
Additional UvrDΔ102C was purified by using the IMPACT system from NEB. After UvrDΔ102C was cloned into pTYB4, the protein was expressed as an intein fusion. Cells containing the intein-UvrDΔ102C fusion were lysed by sonication in lysis buffer (3 ml/g of cells). Lysis buffer was 50 mM Tris-HCl (pH 8.3 at 25°C), 10% (wt/vol) sucrose, 0.2 M NaCl, 5 mM EDTA, 0.5 mM EGTA, and 0.1% Triton X-100. Lysate was loaded onto a chitin resin column (25 ml) that had been equilibrated in buffer A plus 0.2 M NaCl plus 0.1% Triton X-100. UvrDΔ102C (without the intein) was eluted from the column by incubation with 30 mM dithiothreitol according to the instructions of the manufacturer. Fractions were pooled and protein was concentrated by ammonium sulfate precipitation (55% saturation). The precipitated protein was suspended in CD buffer (20 mM sodium phosphate [pH 8.0], 0.2 M sodium sulfate, 20% glycerol, 0.1 mM EDTA, 1 mM dithiothreitol) and dialyzed to remove excess salt.
The concentration of helicase II was determined spectrophotometrically, using the published extinction coefficient of 1.29 ml mg−1cm−1 at 280 nm (43). Concentrations of UvrDΔ102C and UvrDΔ40C were determined based on a Bradford protein assay (Bio-Rad) and a standard curve generated by using wild-type helicase II protein. The concentration of MBP-UvrDΔ102C was based on Bradford protein assay using bovine serum albumin as a standard. The concentration of UvrDΔ102C purified from the intein fusion was determined based on absorbance at 280 nm and the extinction coefficient calculated with SEDNTERP (22).
Genetic assays.
All genetic assays were performed with BL21(DE3)/pLysS or BL21(DE3)ΔuvrD/pLysS. UV irradiation survival assays were as described previously (1). To determine the spontaneous mutation rate, 11 independent isolates of each indicated cell strain were grown in LB-glucose (0.4%) medium under antibiotic selection at 37°C. LB-glucose medium was necessary to alleviate problems associated with plasmid loss in these experiments. Serial dilutions of each saturated culture were made in M9 minimal medium salts. Cell titer was determined by plating appropriate dilutions on LB-agar or LB-agar plus ampicillin (200 μg/ml). Dilutions were plated on LB-agar plus rifampin (100 μg/ml) to ascertain the number of spontaneously arising rifampin-resistant colonies. Plates were incubated at 37°C for at least 24 h. Colonies were counted, and the spontaneous mutation rate was calculated for each strain by the method of the median as described elsewhere (23).
DNA binding assays.
DNA binding was evaluated by measuring the retention of a [32P]DNA ligand on nitrocellulose filters as described previously (15, 34). Reaction mixtures (20 μl) contained 25 mM Tris-HCl (pH 7.5), 3 mM MgCl2, 20 mM NaCl, 3 mM adenosine 5′-O-(thiotriphosphate) (ATPγS), 5 mM 2-mercaptoethanol, 50 μg of bovine serum albumin per ml, and a 32P-labeled 90-base oligonucleotide at approximately 0.9 μM DNA phosphate (0.01 μM molecules) (6.74 × 108 cpm μmol−1). Reactions were initiated by adding the indicated amount of protein, and reaction mixtures were incubated at 37°C for 10 min. After incubation, 1 ml of prewarmed (37°C) reaction buffer was added. The entire mixture was filtered on a HAWP (Millipore) filter at a flow rate of about 4 ml/min. Filters were washed two times with 1 ml of reaction buffer and dried for liquid scintillation counting. Background values were typically less than 1% of the total radioactivity.
The binding of (dT)10 to UvrD and UvrDΔ40C was measured in the presence or absence of 1 mM β,γ-imidoadenosine 5′-triphosphate (AMP-PNP) by measuring the decrease in the intrinsic fluorescence of the proteins upon titration with DNA. Reaction mixtures (2 ml) contained 25 mM Tris-HCl (pH 7.5), 10% glycerol, 3 mM MgCl2, 20 mM NaCl, and 5 mM 2-mercaptoethanol. UvrD was at a concentration of 38 nM protein, and UvrDΔ40C was at 76 nM. The excitation wavelength was 290 nm, and the emission wavelength was 340 nm. Titrations of (dT)10 were performed at 25°C. Measurements were made with an SLM-Aminco 8100 spectrofluorometer. Experimental data were corrected for filter effects and normalized according to the procedures described by Hall et al. (16). Since it is possible that multiple helicase II enzymes bind on a (dT)10 molecule, and the mechanism of binding of helicase II is not completely understood, we may not be measuring true Kd for binding of helicase II to ssDNA. Therefore, we chose to define KD as the effective macroscopic DNA binding constant reflecting binding of all helicase II molecules to this substrate, which may be influenced by effects such as cooperativity. This value is valid for qualitative comparison of DNA binding affinities. Relative macroscopic KD values were calculated as described elsewhere (16).
ATPase assays.
Measurements of DNA-stimulated ATP hydrolysis were completed as described elsewhere (33). Reaction mixtures for evaluating the kcat and Km contained 25 mM Tris-HCl (pH 7.5), 3 mM MgCl2, 20 mM NaCl, 5 mM 2-mercaptoethanol, and M13mp7 ssDNA (30 μM nucleotide phosphate). In kcat assays, 30- or 60-μl (for duplicate trials) reaction mixtures were prewarmed at 37°C and initiated by the addition of protein. Dilutions of protein were made in storage buffer. [3H]ATP was at a concentration of 0.8 mM. Samples (5 μl) were removed at 2-min intervals from reaction mixtures containing UvrD or UvrDΔ40C. Samples (5 μl) were removed at 5, 10, 20, 40, and 60 min from reaction mixtures containing UvrDΔ102C. Reaction tubes without protein were incubated in the same manner and used as controls. Each sample was quenched with 5 μl of stop solution (33 mM EDTA, 7 mM ADP, 7 mM ATP). Quenched reactions were processed as described elsewhere (33). The UvrDΔ102C-catalyzed ATP hydrolysis rate remained in the linear range throughout the entire time course (60 min) and thus reflects an initial rate. Thirty-minute incubations produced the same calculated kcat values (data not shown).
Experiments to determine the Km were performed with [α-32P]ATP at concentrations ranging from 0 to 500 μM. Reactions (20 μl) were initiated by addition of ATP. Protein concentrations were 40, 51, or 103 nM for UvrDΔ102C and 4 nM for UvrD and UvrDΔ40C for reactions with ssDNA. UvrD and UvrDΔ40C were at 100 nM in those reaction mixtures in the absence of ssDNA. Reaction mixtures containing UvrD and UvrDΔ40C were incubated for 10 min, and those containing UvrDΔ102C were incubated for 60 min at 37°C. Duplicate samples (8 μl) were removed at each concentration of [α-32P]ATP and quenched with 5 μl of stop solution. Reaction mixtures without enzyme (storage buffer alone), at each ATP concentration, served as controls. Quenched reactions were processed as described elsewhere (33). Results were visualized with a PhosphorImager and quantified by using ImageQuant software (Molecular Dynamics). Km values were calculated by fitting the Michaelis-Menten equation to the data, using Sigma Plot (Jandel Scientific).
Helicase assays.
The DNA unwinding activities of UvrD, UvrDΔ40C, and MBP-UvrDΔ102C were compared by using a 238-bp blunt duplex substrate. The DNA unwinding activities of UvrD and MBP-UvrDΔ102C were also evaluated with a 92-bp partial duplex. The 92-bp partial duplex was prepared as described elsewhere (2, 35). Reaction mixtures contained approximately 1.3 μM nucleotide phosphate.
The 238-bp blunt duplex was prepared by digesting pLitmus28 with BglII and PvuII. The 238-bp fragment was purified from a 1% NuSieve agarose (FMC BioProducts) gel, using Gene Clean (Bio101). Once the fragment was isolated, the 5′ overhanging end generated from the BglII digestion was filled in with DNA polymerase I (large fragment), [α-32P]dCTP, and a 50 μM concentration of each of the other dNTPs. After a 20-min incubation at 25°C, the reaction was chased with 3 mM unlabeled dCTP. This substrate was purified by using a Sephacryl S-200 sizing column. Helicase reaction mixtures contained 0.17 μM nucleotide phosphate.
Helicase reaction conditions were as above for the kcat ATPase assay with the exception that M13mp7 ssDNA was omitted and the ATP concentration was 3 mM. Protein was added to prewarmed reaction mixtures to initiate the unwinding reaction; reaction mixtures were incubated at 37°C for 10 min. For experiments using the blunt duplex, comparing UvrD and MBP-UvrDΔ102C, the UvrD concentration ranged from 0 to 503 nM and the MBP-UvrDΔ102C concentration ranged from 0 to 362 nM. In those experiments comparing UvrD with UvrDΔ40C, the UvrD concentration ranged from 0 to 251 nM and UvrDΔ40C ranged from 0 to 368 nM. To terminate the reactions, 10 μl of stop solution (37.5% glycerol, 50 mM EDTA, 0.5% each xylene cyanol and bromophenol blue, 0.3% SDS) were added to each tube. Products were resolved on an 8% nondenaturing polyacrylamide gel, imaged with a PhosphorImager, and quantified with ImageQuant software (Molecular Dynamics).
Proteolytic digestion.
Limited digestion of UvrD and UvrDΔ102C was performed with α-chymotrypsin (Sigma). Reaction mixtures (25 μl) contained 1.5 μM UvrD or 1.6 μM UvrDΔ102C (purified from an intein fusion). UvrD-containing reaction mixtures were incubated with 4.3 ng of chymotrypsin, and UvrDΔ102C-containing reaction mixtures were incubated with 5.6 ng. Tubes were prewarmed for 30 s before reactions were initiated. Reactions were initiated by the addition of chymotrypsin, the mixtures were incubated for 4 min at 37°C, and the reactions were stopped by addition of 25 μl of gel loading buffer (250 mM Tris-HCl [pH 6.8], 3.4% SDS, 1.1 M 2-mercaptoethanol, 20% glycerol, 0.01% bromophenol blue) and boiling. Products were resolved on a 9.6% polyacrylamide gel (32:1 cross-linking ratio) run in the presence of 0.1% SDS. Proteins were visualized with Coomassie brilliant blue R-250 (Sigma).
Cleavage products were also analyzed in an experiment probing conformation changes in the presence of ATP. In this assay, 0.6 μM UvrD and 0.63 μM UvrDΔ102C (purified from an intein fusion) were incubated on ice for 5 min in the presence or absence of 3.2 mM ATP with 3.2 mM MgCl2. Reactions were carried out as described above and terminated by addition of gel loading buffer. Samples were resolved on a 9.6% polyacrylamide gel, and proteins were visualized by Western blotting using affinity-purified helicase II antibody. The blot was developed with 5-bromo-4-chloro-3-indolylphosphate and p-nitroblue tetrazolium chloride.
Circular dichroism.
Samples were prepared for circular dichroism measurements by extensive dialysis into CD buffer (see above). Protein samples were filtered (0.2 μm) to remove any aggregated material. Measurements were made with a Jasco J600 spectropolarimeter. Wavelength scans of UvrD (4.6 μM), UvrDΔ40C (6.9 μM), and UvrDΔ102C (6.5 μM) were from 200 to 260 nm at 20°C. The cuvette used was a 0.1-cm quartz cell. Scans of protein material were normalized to scans of the buffer without protein.
RESULTS
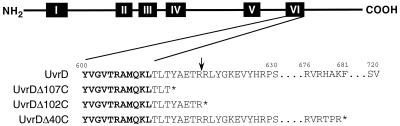
The significance of the C terminus of helicase II (UvrD), a region outside the conserved helicase-associated motifs, was investigated by constructing and evaluating the biological and biochemical function of three truncation mutants: UvrDΔ40C, UvrDΔ102C, and UvrDΔ107C. These mutants were constructed as described in Materials and Methods and are shown in Fig. 1. A protein essentially identical to UvrDΔ102C was the subject of a previous study (5). UvrDΔ40C was constructed because this truncation was significantly further away from the end of motif VI than the other two mutant proteins.
FIG. 1.
Structures and nomenclature of helicase II (UvrD) C-terminal deletion mutants. Approximate positions of six conserved helicase motifs are shown for UvrD (motif Ia is not shown). The sequence of motif VI, as described by Gorbalenya et al. (10) is in boldface. The arrow points to a trypsin cleavage site previously described by Chao and Lohman (5). An asterisk denotes the end of each polypeptide. Amino acid residue numbers are indicated above the sequence for wild-type UvrD.
The ability of each truncation mutant to complement a deletion of the uvrD gene in genetic assays was evaluated. In addition, UvrDΔ40C, UvrDΔ102C, and MBP-UvrDΔ102C were purified and biochemically characterized. The MBP-UvrDΔ102C and the intein-UvrDΔ102C construct were made to facilitate the purification of UvrDΔ102C (see Materials and Methods). Previous studies have shown that an MBP fusion to wild-type UvrD results in a protein with wild-type activity (data not shown). UvrDΔ102C purified from the intein fusion had a Km for ATP hydrolysis similar to that of UvrDΔ102C purified from pET11d (data not shown).
Genetic characterization of UvrDΔ40C and UvrDΔ102C.
E. coli DNA helicase II has an essential role in methyl-directed mismatch repair and UvrABC-directed nucleotide excision repair (4, 18, 21). The ability of each truncation mutant to function in these pathways was evaluated by genetic complementation tests using strains lacking the wild-type uvrD gene. Both complementation assays required the use of bacterial strains containing the λDE3 prophage that encodes T7 RNA polymerase. In each case, the mutant gene was introduced on an expression plasmid that utilized phage T7 transcription-translation initiation signals. For reasons that are not understood, and unlike other mutants and wild-type UvrD, expression of UvrDΔ102C and UvrDΔ107C was not detectable in the absence of the prophage (data not shown). Thus, we assume that expression of these plasmid-encoded truncation mutants is driven by T7 RNA polymerase. It should be noted that induction of the expression of T7 RNA polymerase by the addition of IPTG was not required to observe expression of these mutant uvrD alleles. The level of expression of UvrD, UvrDΔ102C and UvrDΔ40C in BL21(DE3)/pLysS cells was somewhat greater (less than 10-fold) than chromosomal levels of UvrD expression, as judged by Western blot analysis of cell lysates (data not shown).
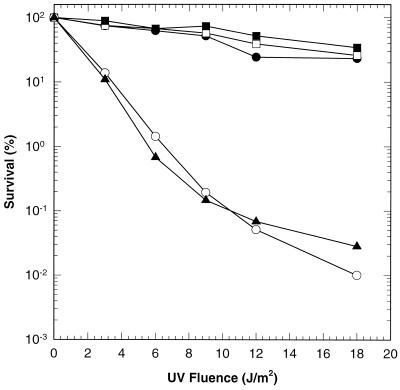
Due to the absence of helicase II, BL21(DE3)ΔuvrD/pLysS was more sensitive to UV irradiation than BL21(DE3)/pLysS cells (Fig. 2). Transformation of the uvrD deletion strain with either pET11d-UvrD or pET11d-UvrDΔ40C restored wild-type UV resistance to these cells. On the other hand, UvrDΔ102C failed to complement the loss of UvrD. UvrDΔ107C also failed to function in excision repair, as judged by qualitative excision repair assays (data not shown).
FIG. 2.
UV survival of BL21(DE3)/pLysS cells and derivatives. E. coli BL21(DE3)/pLysS (uvrD+) (●), BL21(DE3)ΔuvrD/pLysS (ΔuvrD) (○), BL21(DE3)ΔuvrD/pLysS/pET11d-UvrD (uvrD+) (■), BL21(DE3)ΔuvrD/pLysS/pET11d-UvrDΔ40C (uvrDΔ40C) (□), and BL21(DE3)ΔuvrD/pLysS/pET11d-UvrDΔ102C (uvrDΔ102C) (▴) were exposed to increasing doses of UV light as indicated; 100% survival was defined as the number of colonies present in an unirradiated sample. Data are averages of three individual experiments.
To assess function in methyl-directed mismatch repair, the rate of spontaneous mutation was measured by quantifying the number of spontaneously arising rifampin-resistant colonies. Rifampin resistance arises due to mutations at the rpoB locus. Previous studies have shown that wild-type helicase II, expressed from a plasmid, fully complements the loss of chromosomally encoded helicase II (1–3, 7, 15). The mutation rate of BL21(DE3)ΔuvrD/pLysS harboring plasmid pET11d was 45.3-fold that of the same strain with plasmid pET11d-UvrD. This finding is consistent with the absence of helicase II in the former strain (Table 1). UvrDΔ40C complemented the deletion of helicase II, while UvrDΔ102C did not. Preliminary data suggested that UvrDΔ107C was also defective in this repair pathway (data not shown).
TABLE 1.
Spontaneous mutation rates
| Strain | Relevant genotype | Spontaneous mutation rate (no. of mutations [10−9]/cell division)a | Relative mutation rateb |
|---|---|---|---|
| BL21(DE3)ΔuvrD/pLysS/pET11d-UvrD | uvrD+ | 5.67 | 1 |
| BL21(DE3)ΔuvrD/pLysS/pET11d | ΔuvrD | 257 | 45.3 |
| BL21(DE3)ΔuvrD/pLysS/pET11d-UvrDΔ102C | uvrDΔ102C | 296 | 52.2 |
| BL21(DE3)ΔuvrD/pLysS/pET11d-UvrDΔ40C | uvrDΔ40C | 13.9 | 2.5 |
Determined as described in Materials and Methods.
Calculated by dividing the mutation rate of each strain by the mutation rate of BL21(DE3)ΔuvrD/pLysS/pET11d-UvrD.
From the data presented in Fig. 2 and Table 1, it is evident that UvrDΔ102C cannot function in either mismatch repair or excision repair whereas UvrDΔ40C is active in both pathways. Thus, the C-terminal 40 amino acid residues are not critical for the function of helicase II in these two pathways. However, residues between positions 618 and 680 are essential. These results might be explained by the loss of a region of the protein required for specific protein-protein interactions or, alternatively, by the loss of biochemical activity. To distinguish between these possibilities, UvrDΔ102C and UvrDΔ40C were purified and evaluated in biochemical assays.
Biochemical characterization of UvrDΔ102C and UvrDΔ40C.
UvrDΔ40C was purified by using a previously published helicase II purification procedure (43), with one exception. The protein failed to bind an ssDNA-cellulose column equilibrated at 0.2 M NaCl. Purification of this protein required the loading of the ssDNA-cellulose at a lower salt concentration (0.1 M NaCl).
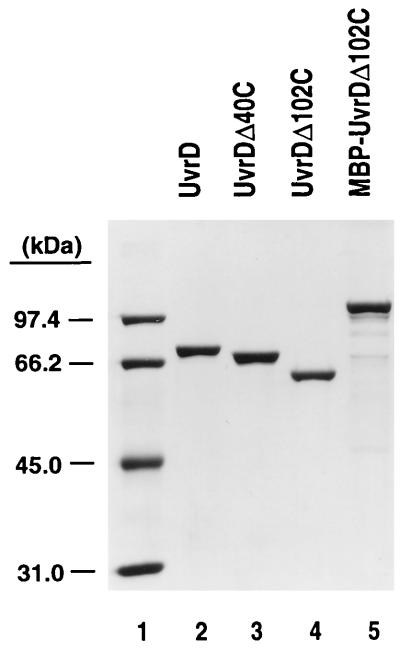
The protocol used to purify UvrDΔ102C deviated significantly from the standard procedure. UvrDΔ102C precipitated in the presence of polymin-P and required extraction with a high concentration of NaCl (1 M NaCl). UvrDΔ102C and UvrDΔ107C consistently failed to bind either heparin-agarose or ssDNA-cellulose. Purification of UvrDΔ102C was accomplished using DEAE-cellulose, phenyl Sepharose CL-6B, a flowthrough step with ssDNA-cellulose, and a Sephacryl S-200 sizing column as detailed in Materials and Methods. MBP-UvrDΔ102C and UvrDΔ102C purified from the intein fusion were purified as described in Materials and Methods. UvrD, UvrDΔ40C, and UvrDΔ102C were purified to apparent homogeneity, as ascertained by the appearance of a single species on SDS-polyacrylamide gels (Fig. 3). MBP-UvrDΔ102C was purified to near homogeneity, though some breakdown products of this fusion protein were visible. UvrDΔ102C was purified from the intein fusion to near homogeneity, with some breakdown products visible (see Fig. 5A, lane 1). These were confirmed as breakdown products by Western blotting.
FIG. 3.
SDS-polyacrylamide gel analysis of purified proteins. Purified proteins were resolved on a 9.6% polyacrylamide gel in the presence of SDS and stained with Coomassie blue. Lane 2, 1.2 μg of UvrD; lane 3, 1.1 μg of UvrDΔ40C; lane 4, 1 μg of UvrDΔ102C; lane 5, 1.2 μg of MBP-UvrDΔ102C. Molecular weight standards (lane 1) were rabbit muscle phosphorylase b (97.4 kDa), bovine serum albumin (66.2 kDa), hen egg white ovalbumin (45.0 kDa), and bovine carbonic anhydrase (31.0 kDa).
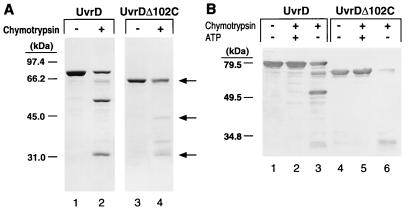
FIG. 5.
Limited proteolysis of UvrD and UvrDΔ102C with α-chymotrypsin. (A) UvrD (1.5 μM) and UvrDΔ102C (1.6 μM) were subjected to limited proteolysis as described in Materials and Methods (lanes 2 and 4, respectively). Lanes 1 and 3 contain UvrD and UvrDΔ102C in the absence of chymotrypsin. This gel was stained with Coomassie blue. Molecular weight standards were the same as in Fig. 3. Arrows indicate full-length UvrDΔ102C and the 42- and 29-kDa proteolytic fragments. (B) UvrD (lanes 1 to 3; 0.6 μM) and UvrDΔ102C (lanes 4 to 6; 0.6 μM) were incubated in the absence (lanes 1 and 4) or presence (lanes 2, 3, 5, and 6) of 10 ng of chymotrypsin. Reaction mixtures were preincubated in the presence (lanes 2 and 5) or absence (lanes 3 and 6) of 3.2 mM ATP and MgCl2. Previous studies have shown that chymotrypsin is not inhibited by the presence of either MgCl2 or ATP (2, 16). The products of digestion were resolved on a 9.6% polyacrylamide gel in the presence of SDS. Products were visualized by Western blotting with antibodies to wild-type helicase II. Molecular weight standards were prestained broad-range markers (Bio-Rad).
(i) DNA binding.
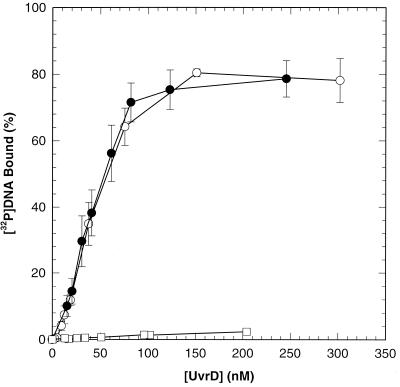
The behavior of UvrDΔ40C and UvrDΔ102C on ssDNA-cellulose columns during purification suggested possible defects in the interaction of these proteins with DNA. The binding of UvrDΔ102C and UvrDΔ40C to ssDNA was directly evaluated by using a nitrocellulose filter binding assay and a [32P]DNA ligand (90-mer oligonucleotide). DNA concentration was held constant while protein concentration was varied as indicated. Results of DNA binding experiments performed in the presence of a poorly hydrolyzed ATP analog, ATPγS, are shown in Fig. 4. UvrDΔ40C behaved like wild-type helicase II under these conditions. UvrDΔ102C, however, was markedly defective for binding to ssDNA.
FIG. 4.
DNA binding using UvrD, UvrDΔ40C, and UvrDΔ102C. Nitrocellulose filter binding experiments with UvrD (○), UvrDΔ40C (●), and UvrDΔ102C (□) were performed as described in Materials and Methods, using the indicated concentration of each purified protein. Data represent averages of several trials. Error bars show the standard error about the mean.
We took advantage of the fact that the intrinsic fluorescence of helicase II is quenched upon binding DNA to more rigorously determine the KD for binding ssDNA by UvrD and UvrDΔ40C. These experiments were performed in the presence and the absence of a poorly hydrolyzable ATP analog, AMP-PNP. In the presence of 1 mM AMP-PNP, the apparent KD values for UvrD and UvrDΔ40C were 11 ± 1 and 24 ± 13 nM, respectively. These values were consistent with the results obtained in the filter binding assay. The KD values in the absence of AMP-PNP were 276 ± 18 and 345 ± 16 nM for UvrD and UvrDΔ40C, respectively. In both cases, UvrDΔ40C appears to have a slightly higher KD value than UvrD. The purification results also suggest a minor DNA binding defect due to the loss of the 40 C-terminal amino acids.
(ii) ATPase and helicase activities.
To better define the enzymatic properties of UvrDΔ102C and UvrDΔ40C, both proteins were assayed for ATPase and helicase activities. The kcat values for ssDNA-stimulated ATP hydrolysis catalyzed by UvrD and UvrDΔ40C were essentially the same. UvrDΔ102C was significantly deficient in DNA-stimulated ATP hydrolysis (Table 2). The kcat for this protein was 0.01% that of the wild-type protein.
TABLE 2.
DNA-stimulated ATP hydrolysis by UvrD, UvrDΔ40C, and UvrDΔ102Ca
| Enzyme | kcatb (s−1) | Kmb (μM) |
|---|---|---|
| UvrD | 147 ± 21 | 62 ± 11 |
| UvrDΔ40C | 157 ± 14 | 50 ± 9 |
| UvrDΔ102C | 0.013 ± 0.004 | 95 ± 19 |
The hydrolysis of ATP was measured as described in Materials and Methods.
Reaction mixtures contained M13mp7 ssDNA (30 μM nucleotide phosphate).
The Km for ATP in the hydrolysis reaction was also determined for the wild-type protein and both truncation mutants. The experimental data for all three proteins were well described by a hyperbolic saturation curve as a function of increasing ATP concentration (data not shown). This type of curve reflects an ability of these proteins to interact with nucleotides. Km values, determined by fitting the Michaelis-Menten equation, were not significantly different for UvrD, UvrDΔ40C, and UvrDΔ102C (Table 2). Km values were also determined for UvrD and UvrDΔ40C in the absence of ssDNA as 53 ± 10 and 153 ± 31 μM, respectively. We conclude that both UvrDΔ40C and UvrDΔ102C are capable of interacting with ATP with wild-type affinity.
Measurements of the unwinding reaction catalyzed by each protein were made using a 92-bp partial duplex and a blunt duplex substrate. UvrDΔ102C was defective in both binding ssDNA and ssDNA-stimulated ATP hydrolysis. ATP hydrolysis is thought to provide the energy for the helicase reaction, and it has been shown to be required for DNA unwinding (7). The defect in DNA binding would result in the failure of UvrDΔ102C to interact with the DNA substrate. Thus, it was expected that UvrDΔ102C would fail to catalyze an unwinding reaction.
Wild-type UvrD (251 nM) catalyzed unwinding of greater than 50% of the 238-bp blunt duplex DNA substrate in the 10-min reaction. However, 362 nM MBP-UvrDΔ102C was unable to unwind a detectable fraction of this substrate (data not shown) in the reaction time. MBP-UvrDΔ102C also failed to unwind a 92-bp partial duplex substrate at concentrations up to 72 nM (data not shown). UvrD and UvrDΔ40C catalyzed the unwinding of both substrates tested with equivalent efficiency (data not shown) (36).
Protein conformation studies.
The ability of UvrDΔ102C to interact with ATP indicates that loss of the 102 C-terminal amino acids has not caused the disruption of the global fold of the protein. The possibility that UvrDΔ102C may be defective as a helicase due to an alteration of the overall or local conformation or due to an inability of this protein to make some of the necessary conformational changes believed to be associated with ATP hydrolysis was further addressed by the use of circular dichroism measurements and limited proteolytic cleavage.
Circular dichroism measurements were done with UvrD, UvrDΔ40C and UvrDΔ102C. Wavelength scans produced a pattern consistent with a significant amount of α-helical content in these proteins. Each of these proteins exhibited peak signals at 220 and 209 nm. Therefore, UvrDΔ102C has a wavelength spectrum consistent with both the wild-type protein and UvrDΔ40C, indicating that it has similar secondary structure content (data not shown).
It has been shown that when wild-type helicase II is subjected to limited digestion with chymotrypsin, two predominant proteolytic species that migrate at 53 and 29 kDa on SDS-polyacrylamide gels are formed (5) (Fig. 5A, lane 2). This property of the protein was used to probe the global conformation of UvrDΔ102C. Since UvrDΔ102C is a C-terminal truncation, the predicted sizes of the corresponding fragments upon limited digestion are 29 and 42 kDa. These species were formed, suggesting that UvrDΔ102C was correctly folded (Fig. 5A, lane 4). An additional proteolytic cleavage fragment appears in lane 4. It should be noted that this proteolytic fragment is also visible, although at much reduced levels, when wild-type UvrD is digested with chymotrypsin. The loss of the C-terminal portion of the protein could make an additional region of the protein more accessible to protease.
The ability of UvrDΔ102C to change conformational states in the presence of nucleotide was also examined with limited proteolysis. Both UvrD and UvrDΔ102C were incubated with chymotrypsin in the presence and absence of ATP. Wild-type UvrD was protected from cleavage in the presence of ATP, as previously reported (5, 16). UvrDΔ102C was also protected from digestion in the presence of ATP (Fig. 5B). This finding is consistent with the fact that UvrDΔ102C exhibits a wild-type Km for ATP and provides further evidence that this truncation has not disturbed the overall stability of the protein. Moreover, this observation supports the notion that this protein can make the appropriate conformational changes associated with the interaction with ATP.
DISCUSSION
The results presented here indicate that a region near the C-terminal end of DNA helicase II is necessary for in vivo function of the protein in both the excision repair and methyl-directed mismatch repair pathways. In vivo defects correlate with in vitro defects including a markedly impaired interaction with DNA and nearly undetectable ATPase and helicase activities. This result was, perhaps, unexpected since this region of the protein is located outside of the conserved helicase motifs. Amino acids associated with these motifs are assumed, and in many cases known, to be essential for the biochemical activities of this class of enzymes. However, the results presented here indicate that regions outside of the identified motifs also play significant roles in the biochemical mechanism of DNA unwinding.
It was previously reported that the C terminus was dispensable for the unwinding and ATPase activities of helicase II (5). In those experiments helicase II was digested with trypsin to produce a 72-kDa polypeptide lacking the C-terminal end of the protein (see Fig. 1 for the location of the trypsin cleavage site). The results reported here with UvrDΔ102C reveal a very significant defect in the ATPase/helicase reaction catalyzed by this protein. A possible explanation for this discrepancy centers on the fact that the 72-kDa polypeptide was not purified in the previous study (5). The 10-kDa C-terminal polypeptide may have remained associated with the larger fragment, masking the defect caused by the truncation. It is also possible that under the conditions chosen for the trypsin reaction, UvrD had oligomerized further aiding in the association of the C-terminal peptide fragment with the remainder of the protein. There is precedence for the proteolytic fragments of UvrD remaining associated with each other. Chao and Lohman (5) report that chymotrypsin cleavage products (29 and 53 kDa) copurify under nondenaturing conditions, suggesting a physical association between these cleavage products. Moreover, these associated chymotrypsin cleavage products were able to bind ssDNA despite the fact that the cleavage site is located in the middle of motif III, which has been implicated in DNA binding (2, 20). Alternatively, there is the potential for contamination by full-length helicase II in the proteolytically digested preparations. The genetically altered UvrD truncations used in these studies were not subject to any of these potential problems.
Nitrocellulose filter binding experiments clearly demonstrate the failure of UvrDΔ102C to bind ssDNA. These results were consistent with the failure of this protein to bind ssDNA-cellulose during purification. This defect is sufficient to explain the genetic results—UvrDΔ102C is unable to function in excision repair or methyl-directed mismatch repair because it cannot bind the necessary DNA substrates. UvrDΔ40C appeared slightly less efficient in DNA binding, suggesting that this truncation was beginning to affect a region of the protein that was important for DNA binding. However, this very slight DNA binding defect did not impair in vivo function of this enzyme. It should be noted here that UvrDΔ40C has been shown to fail to dimerize and that dimerization is not essential for the in vitro and in vivo activities of helicase II (36). In other helicases, for example, gpα and eIF-4B, regions at or near the C terminus have been shown to have a role in DNA and RNA binding (38, 52).
An obvious concern when one is evaluating the biochemical activity of a truncated protein is disturbance of the conformation of the protein. Several lines of evidence suggest that UvrDΔ102C is properly folded. First, the protein was successfully overexpressed and was soluble in whole-cell lysates. Misfolded proteins are often quickly degraded or insoluble in the cell. Second, based on previous limited proteolysis experiments (5), UvrDΔ102C represents an independently folding domain. Data from circular dichroism measurements demonstrate that this protein has significant secondary structure. Third, additional proteolysis experiments indicate that this protein is folded correctly, as evidenced by the presence of the appropriate chymotrypsin cleavage site (Fig. 5). Finally, UvrDΔ102C interacts with nucleotide in the same fashion as wild-type helicase II. The Km values for UvrD and UvrDΔ102C were essentially identical. Moreover, UvrDΔ102C and UvrD are both protected from digestion with chymotrypsin in the presence of ATP (Fig. 5).
Even though UvrDΔ102C had a Km for ATP similar to that of UvrD, it had a significantly lower kcat value for ATP hydrolysis than UvrD. Not surprisingly, this reaction was not stimulated by the addition of ssDNA. It should be noted that the kcat value was lower than that measured for DNA-independent ATP hydrolysis catalyzed by wild-type UvrD (0.4 s−1). The reason for this difference is not clear. If the region between amino acids 618 and 680 was simply required for ssDNA binding, then one might expect UvrDΔ102C to have a DNA-independent hydrolysis activity equivalent to that of wild-type UvrD. However, the mechanism of DNA-independent ATP hydrolysis is not well characterized. Evidence suggests that ATP may bind in the ssDNA binding region, albeit with low affinity. Substrate inhibition of ssDNA-stimulated ATP hydrolysis at high ATP concentrations has been observed for the wild-type enzyme. Kinetic studies suggest this is the result of competition for the DNA binding site (42a). Therefore, in the case of the wild-type enzyme, ATP could bind in the DNA binding pocket, when present at sufficiently high concentration, and act to stimulate ssDNA-independent hydrolysis by acting as an allosteric effector. Due to the absence of this region in UvrDΔ102C, this allosteric stimulation by ATP would presumably not occur with UvrDΔ102C.
Other explanations are also possible. This region may be directly involved in the ATP hydrolysis reaction. The ssDNA binding regions are located near the ATP binding pocket in the structure of Rep protein (20). Another possibility is that the C-terminal amino acids serve as a bridge of communication between domains of helicase II. The individual domains of helicase II might have independent roles in the cycle of ATP binding, hydrolysis, and product release. Coordination of the activities of these domains could be required for DNA-independent ATP hydrolysis. Finally, the C terminus may function as a switch that regulates the cycling of conformational states in order to couple ATP hydrolysis to the disruption of hydrogen bonds. Although the protein is able to change conformation in response to ATP binding, perhaps without the C-terminal region the protein cannot continuously cycle, preventing efficient hydrolysis of ATP.
The data presented here clearly implicate residues 618 to 680 as part of helicase II required for a stable interaction with ssDNA. It is known, from site-directed mutagenesis studies (2, 16) and from crystal structures of the related Rep and PcrA proteins (20, 47), that other regions of the protein also have roles in DNA binding. In the Rep cocrystal with bound dT(pT)15 several amino acids that contact bound ssDNA were identified. Some of these residues lie in motifs Ia, III, and V, between motifs IV and V, and between motifs V and VI. In this structural model, the DNA molecule was located in a large cleft that runs between the subdomains of the protein (20). The DNA contact regions identified in the Rep crystal structure were consistent with several of the putative DNA binding regions proposed for PcrA, based on its structural similarity with RecA (47). Consistent with the idea of multiple DNA binding regions, the C terminus of helicase II may serve as an additional portion of the protein with a role in DNA binding. The C terminus is absent in the solved structures of PcrA and Rep protein (it was too disordered to be resolved). This flexible region (residues 618 to 680) could be seen as overlying the central pocket of the protein, where ssDNA is thought to bind, serving as a type of clamp (20, 47). This clamp could stabilize the interaction between helicase II and ssDNA either by physically holding the DNA in place or by locking the protein in a DNA-bound conformational state. The multiple regions of the protein involved in DNA binding may or may not be independently regulated. There is not enough known to differentiate between these possibilities. Ultimately, the role of the C terminus will only be fully understood once a structure of helicase II is determined with resolution of this C-terminal region.
All of the data presented establish that while the helicase motifs may be associated with all helicases, and are required for unwinding activity, these motifs alone do not define a functional helicase domain for UvrD. Moreover, there are several known examples of putative helicases that contain all the helicase-associated motifs but fail to catalyze a helicase reaction in vitro. These include the E. coli and eukaryotic transcription repair coupling factor (45, 46) and the yeast Rad5 protein (19). Thus, precisely what determines a functional helicase is not yet known with certainty. While helicases contain conserved motifs that are critical for the biochemical mechanism of unwinding, these motifs are not the only regions of these proteins required for enzyme activity. Thus, regions lying outside of the motifs may tailor these proteins to specific biochemical roles.
ACKNOWLEDGMENTS
We thank Dan Bean, Jim George, and Mark Hall for critical reading of the manuscript. We also thank Susan Whitfield for help with preparation of figures. We are grateful to Roopa Thapor for help with circular dichroism measurements.
This work was supported by NIH grant GM33476 to S.W.M.
Footnotes
Corresponding author. Mailing address: Department of Biology, CB 3280, Coker Hall, University of North Carolina, Chapel Hill, NC 27599-3280. Phone: (919) 962-0005. Fax: (919) 962-1625. E-mail: smatson@bio.unc.edu.
REFERENCES
- 1.Brosh R M, Jr, Matson S W. Mutations in motif II of Escherichia coli DNA helicase II render the enzyme nonfunctional in both mismatch repair and excision repair with differential effects on the unwinding reaction. J Bacteriol. 1995;177:5612–5621. doi: 10.1128/jb.177.19.5612-5621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosh R M, Jr, Matson S W. A partially functional DNA helicase II mutant defective in forming stable binary complexes with ATP and DNA. A role for helicase motif III. J Biol Chem. 1996;271:25360–25368. doi: 10.1074/jbc.271.41.25360. [DOI] [PubMed] [Google Scholar]
- 3.Brosh R M, Jr, Matson S W. A point mutation in Escherichia coli DNA helicase II renders the enzyme nonfunctional in two DNA repair pathways. Evidence for initiation of unwinding from a nick in vivo. J Biol Chem. 1997;272:572–579. doi: 10.1074/jbc.272.1.572. [DOI] [PubMed] [Google Scholar]
- 4.Caron P R, Kushner S R, Grossman L. Involvement of helicase II (uvrD gene product) and DNA polymerase I in excision mediated by the uvrABC protein complex. Proc Natl Acad Sci USA. 1985;82:4925–4929. doi: 10.1073/pnas.82.15.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao K, Lohman T M. DNA and nucleotide-induced conformational changes in the Escherichia coli Rep and helicase II (UvrD) proteins. J Biol Chem. 1990;265:1067–1076. [PubMed] [Google Scholar]
- 6.Denhardt D T, Iwaya M, Larison L L. The rep mutation. II. Its effect on Escherichia coli and on the replication of bacteriophage phi X174. Virology. 1972;49:486–496. doi: 10.1016/0042-6822(72)90500-4. [DOI] [PubMed] [Google Scholar]
- 7.George J W, Robert J, Brosh M, Matson S W. A dominant negative allele of the Escherichia coli uvrD gene encoding DNA helicase II: a biochemical and genetic characterization. J Mol Biol. 1994;235:424–435. doi: 10.1006/jmbi.1994.1003. [DOI] [PubMed] [Google Scholar]
- 8.Gorbalenya A E, Koonin E V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 9.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 1988;235:16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4729. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graves-Woodward K L, Gottlieb J, Challberg M D, Weller S K. Biochemical analyses of mutations in the HSV-1 helicase-primase that alter ATP hydrolysis, DNA unwinding, and coupling between hydrolysis and unwinding. J Biol Chem. 1997;272:4623–4630. doi: 10.1074/jbc.272.7.4623. [DOI] [PubMed] [Google Scholar]
- 12.Grilley M, Griffith J, Modrich P. Bidirectional excision in methyl-directed mismatch repair. J Biol Chem. 1993;268:11830–11837. [PubMed] [Google Scholar]
- 13.Grilley M, Holmes J, Yashar B, Modrich P. Mechanisms of DNA-mismatch correction. Mutat Res. 1990;236:253–267. doi: 10.1016/0921-8777(90)90009-t. . (Review.) [DOI] [PubMed] [Google Scholar]
- 14.Gross C H, Shuman S. The QRxGRxGRxxxG motif of the vaccinia virus DExH box RNA helicase NPH-II is required for ATP hydrolysis and RNA unwinding but not for RNA binding. J Virol. 1996;70:1706–1713. doi: 10.1128/jvi.70.3.1706-1713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall M C, Matson S W. Mutation of a highly conserved arginine in motif IV of Escherichia coli DNA helicase II results in an ATP-binding defect. J Biol Chem. 1997;272:18614–18620. doi: 10.1074/jbc.272.30.18614. [DOI] [PubMed] [Google Scholar]
- 16.Hall M C, Ozsoy A Z, Matson S W. Site-directed mutations in motif VI of Escherichia coli DNA helicase II result in multiple biochemical defects: evidence for the involvement of motif VI in the coupling of ATPase and DNA binding activities via conformational changes. J Mol Biol. 1998;277:257–271. doi: 10.1006/jmbi.1997.1614. [DOI] [PubMed] [Google Scholar]
- 17.Hodgman T C. A conserved NTP-motif in putative helicases. Nature. 1988;333:22–23. doi: 10.1038/333022a0. [DOI] [PubMed] [Google Scholar]
- 18.Husain I, Van Houten B, Thomas D C, Abdel-Monem M, Sancar A. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc Natl Acad Sci USA. 1985;82:6774–6778. doi: 10.1073/pnas.82.20.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson R E, Prakash S, Prakash L. Yeast DNA repair protein RAD5 that promotes instability of simple repetitive sequences is a DNA-dependent ATPase. J Biol Chem. 1994;269:28259–28262. [PubMed] [Google Scholar]
- 20.Korolev S, Hseigh J, Gauss G H, Lohman T M, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 21.Lahue R S, Au K G, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 22.Laue T M, Shah B D, Ridgeway T M, Pelletier S L. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe A, Horton J C, editors. Analytical ultracentrifugation in biochemistry and polymer science. Cambridge, England: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 23.Lea D E, Coulson C A. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 24.Lechner R L, Richardson C C. A preformed, topologically stable replication fork. J Biol Chem. 1983;258:11185–11196. [PubMed] [Google Scholar]
- 25.Lohman T M. Escherichia coli DNA helicases: mechanisms of DNA unwinding. Mol Microbiol. 1992;6:5–14. doi: 10.1111/j.1365-2958.1992.tb00831.x. [DOI] [PubMed] [Google Scholar]
- 26.Lohman T M. Helicase-catalyzed DNA unwinding. J Biol Chem. 1993;268:2269–2272. [PubMed] [Google Scholar]
- 27.Lohman T M, Bjornson K P. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 28.Makhov A M, Boehmer P E, Lehman I R, Griffith J D. Visualization of the unwinding of long DNA chains by the herpes simplex virus type 1 UL9 protein and ICP8. J Mol Biol. 1996;258:789–799. doi: 10.1006/jmbi.1996.0287. [DOI] [PubMed] [Google Scholar]
- 29.Martinez R, Shao L, Weller S K. The conserved helicase motifs of the herpes simplex virus type 1 origin-binding protein UL9 are important for function. J Virol. 1992;66:6735–6746. doi: 10.1128/jvi.66.11.6735-6746.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matson S W. DNA helicases of Escherichia coli. Prog Nucleic Acid Res Mol Biol. 1991;40:289–326. doi: 10.1016/s0079-6603(08)60845-4. [DOI] [PubMed] [Google Scholar]
- 31.Matson S W, Bean D W, George J W. DNA helicases: enzymes with essential roles in all aspects of DNA. Bioessays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 32.Matson S W, Kaiser-Rogers K A. DNA helicases. Annu Rev Biochem. 1990;59:289–329. doi: 10.1146/annurev.bi.59.070190.001445. [DOI] [PubMed] [Google Scholar]
- 33.Matson S W, Richardson C C. DNA-dependent nucleoside 5′-triphosphatase activity of the gene 4 protein of bacteriophage T7. J Biol Chem. 1983;258:14009–14016. [PubMed] [Google Scholar]
- 34.Matson S W, Richardson C C. Nucleotide-dependent binding of the gene 4 protein of bacteriophage T7 to single-stranded DNA. J Biol Chem. 1985;260:2281–2287. [PubMed] [Google Scholar]
- 35.Matson S W, Tabor S, Richardson C C. The gene 4 protein of bacteriophage T7: characterization of helicase activity. J Biol Chem. 1983;258:14017–14024. [PubMed] [Google Scholar]
- 36.Mechanic, L. E., M. C. Hall, and S. W. Matson. E. coli DNA helicase II is active as a monomer. J. Biol. Chem., in press. [DOI] [PubMed]
- 37.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 38.Naranda T, Strong W B, Menaya J, Fabbri B J, Hershey J W B. Two structural domains of initiation factor eIF-4B are involved in binding to RNA. J Biol Chem. 1994;269:14465–14472. [PubMed] [Google Scholar]
- 39.Orren D K, Selby C P, Hearst J E, Sancar A. Post-incision steps of nucleotide excision repair in Escherichia coli. Disassembly of the UvrBC-DNA complex by helicase II and DNA polymerase I. J Biol Chem. 1992;267:780–788. [PubMed] [Google Scholar]
- 40.Park E, Guzder S N, Koken M H, Jaspers-Dekker I, Weeda G, Hoeijmakers J H, Prakash S, Prakash L. RAD25 (SSL2), the yeast homolog of the human xeroderma pigmentosum group B DNA repair gene, is essential for viability. Proc Natl Acad Sci USA. 1992;89:11416–11420. doi: 10.1073/pnas.89.23.11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pause A, Methot N, Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol Cell Biol. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Porter, D. Glaxo-Wellcome, Inc. Unpublished observations.
- 43.Runyon G T, Wong I, Lohman T M. Overexpression, purification, DNA binding, and dimerization of the Escherichia coli uvrD gene product (helicase II) Biochemistry. 1993;32:602–612. doi: 10.1021/bi00053a028. [DOI] [PubMed] [Google Scholar]
- 44.Sancar A. Mechanisms of DNA excision repair. Science. 1994;266:1954–1956. doi: 10.1126/science.7801120. . (Review.) [DOI] [PubMed] [Google Scholar]
- 45.Selby C P, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 46.Selby C P, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem. 1997;272:1885–1890. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- 47.Subramanya H S, Bird L E, Brannigan J A, Wigley D B. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 48.Wong I, Lohman T M. Allosteric effects of nucleotide cofactors on Escherichia coli Rep helicase-DNA binding. Science. 1992;256:350–355. doi: 10.1126/science.256.5055.350. [DOI] [PubMed] [Google Scholar]
- 49.Yu C-E, Oshima J, Fu Y-H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin G M, Mulligan J, Schellenberg G D. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 50.Zavitz K H, Marians K J. ATPase-deficient mutants of the Escherichia coli DNA replication protein PriA are capable of catalyzing the assembly of active primosomes. J Biol Chem. 1992;267:6933–6940. [PubMed] [Google Scholar]
- 51.Zhu L, Weller S K. The six conserved helicase motifs of the UL5 gene product, a component of the herpes simplex virus type 1 helicase-primase, are essential for its function. J Virol. 1992;66:469–479. doi: 10.1128/jvi.66.1.469-479.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ziegelin G, Linderoth N A, Calendar R, Lanka E. Domain structure of phage P4 a protein deduced by mutational analysis. J Bacteriol. 1995;177:4333–4341. doi: 10.1128/jb.177.15.4333-4341.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]