Abstract
Endometrial health is affected by molecular processes that underlie estrogen responses. We assessed estrogen regulation of endometrial function by integrating the estrogen receptor α (ESR1) cistromes and transcriptomes of endometrial biopsies taken from the proliferative and mid-secretory phases of the menstrual cycle together with hormonally stimulated endometrial epithelial organoids. The cycle stage–specific ESR1 binding sites were determined by chromatin immunoprecipitation and next-generation sequencing and then integrated with changes in gene expression from RNA sequencing data to infer candidate ESR1 targets in normal endometrium. Genes with ESR1 binding in whole endometrium were enriched for chromatin modification and regulation of cell proliferation. The distribution of ESR1 binding sites in organoids was more distal from gene promoters when compared to primary endometrium and was more similar to the proliferative than the mid-secretory phase ESR1 cistrome. Inferred organoid estrogen/ESR1 candidate target genes affected formation of cellular protrusions and chromatin modification. Comparison of signaling effected by candidate ESR1 target genes in endometrium vs organoids reveals enrichment of both overlapping and distinct responses. Our analysis of the ESR1 cistromes and transcriptomes from endometrium and organoids provides important resources for understanding how estrogen affects endometrial health and function.
Keywords: estrogen receptor, chromatin, endometrium
Uterine function depends on the precise coordination of ovarian steroid hormone signaling. The actions of estrogen and progesterone, via their cognate receptors, estrogen receptor α (ESR1) and progesterone receptor (PGR), regulate the ability of the uterus to support embryo implantation and fetal growth. Alterations in estrogen action can result in diseases of the uterus such as endometriosis and endometrial cancer (1, 2). Understanding how these hormones regulate uterine biology is critical to understanding their roles in uterine function and dysfunction.
The uterus contains multiple types of cells arranged into layered compartments with an interior lumen that is lined by epithelial cells (3). Glandular structures with specialized epithelial cells emanate from the lumen into the uterine stromal tissue and secrete cytokines to facilitate the establishment and maintenance of pregnancy. Underlying the single-cell epithelial lumen and glands are stromal fibroblasts, blood vessel endothelium, and bone marrow–derived immune cells. The stromal fibroblasts undergo a marked structural and functional transformation, beginning before implantation in the mid-secretory phase, into decidual cells, in a process termed decidualization. The endometrium includes the stromal and epithelial compartments and when pregnancy does not occur, the corpus luteum fails and ovarian hormones decrease, causing the endometrium to be shed by menstruation (4). The hormone-dependent endometrial stages are classified based on histological examination as the proliferative (preovulatory, estrogen dominant) and secretory (postovulatory, progesterone dominant) phases. The mid-secretory phase is the time when the endometrium is receptive to embryo implantation (5).
During the proliferative phase of the menstrual cycle, estrogen induces uterine cell growth and endometrial tissue thickening, and acts to increase the progesterone sensitivity of the endometrial cells (5). Situations or conditions that perturb estrogen signaling are detrimental to normal uterine development and function. A dramatic example was provided when prescription of the potent synthetic estrogen receptor agonist diethylstilbestrol (DES) to pregnant women resulted in profound in utero developmental alterations in uterine structures of female fetuses (6). This treatment was used in efforts to prevent miscarriage and other pregnancy complications from 1940 to 1970 (7). “DES daughters” have an increased risk for infertility, ectopic pregnancy, miscarriage, and clear cell adenocarcinoma (8, 9). Alternatively, women who have mutations in their estrogen receptor causing estrogen insensitivity or who are unable to produce estrogen because of mutations in the enzyme aromatase have an abnormally small uterus with a thin endometrial lining and are infertile (10-14). Chronic exposure to estrogen without the growth-inhibiting effects of progesterone is a major contributing factor to endometrial hyperplasia and subsequent cancer (15, 16). Estrogen also facilitates the progression of endometriosis by increasing growth of lesions and can affect the severity of endometriosis symptoms (17). Finally, we reported previously that thin endometrium, associated with poor fertility treatment outcomes, is associated with defects in estrogen signaling (18). Therefore, understanding mechanisms for optimal estrogen signaling is a key element in maximizing women’s health.
Estrogen affects endometrial cells via nuclear estrogen receptors (ESR1 and ESR2) (19, 20), or the membrane localized GPER (21). ESRs act as estrogen-dependent transcription factors by binding to estrogen and to specific DNA motifs (GGTCAnnnTGACC), called estrogen-responsive enhancers (EREs). ESR can alternatively bind indirectly or “tether” itself to non-ERE sequences, such as AP1 sites, via interaction with other transcription factors, including AP1, to affect transcriptional rates of genes (19). ESR chromatin interaction is facilitated by pioneer factors, including forkhead and GATA family members, that increase chromatin accessibility in regulatory regions (22). Activation of ESR via binding of agonists, including estradiol (E2), to its ligand binding domain leads to interaction between activation functions in the ESR ligand binding domain and amino terminus A/B domains and mediators of chromatin remodeling. This leads to altered histone modifications, which ultimately affects transcriptional rates of estrogen-responsive genes (19). Using chromatin immunoprecipitation and next-generation sequencing (ChIPseq), the locations of all ESR1-chromatin interaction within cells or tissues can be ascertained, defining the ESR1 cistrome (23, 24). These analyses indicate that receptor interactions are frequently in enhancer regions that are far (in linear sequence length) from the transcriptional start of the responding genes, highlighting the importance of the 3-dimensional arrangement of the chromatin structure by which the ESR1 is brought into contact with its target genes.
Much of the molecular detail of estrogen action has been defined using in vitro cell line models or in vivo in experimental animal models. Although there are many similarities between mechanisms in women and these model systems, application to women’s health is best accomplished by determining the key details in women. In vivo and ex vivo studies in women are, of course, limited by ethical and practical considerations, and so studies conducted in women need to be focused on the most important aspects of endometrial function.
Endometrial epithelial cells provide the first barrier to successful embryo implantation (25), and thus optimal function of endometrial epithelium is key to reproduction. However, studies focused on details intrinsic to the epithelial cells are particularly challenging for 2 main reasons: 1) Biochemical characterization of clinical samples is limited because of the invasive process of endometrial biopsy required to obtain samples from healthy volunteers; and 2) although uterine stromal cells can be cultured and studied, epithelial cells have proven difficult to culture in a model that recapitulates their biological environment and function. The recent development of organoid culture has provided a system in which endometrial epithelial cells can be grown and studied in vitro (26). Established protocols describe isolation and culture of epithelial cells from endometrial biopsies, which are stimulated to proliferate by estrogen treatment (27-29). Conversely, use of WNT signaling inhibitors in the culture media induces differentiation into secretory lineage (30). Therefore, the organoid cultures, once removed from the in vivo endocrine environment and cultured in vitro, can be used to assess mechanistic details of the effect of estrogen on endometrial epithelial cells.
Previous work has characterized ESR1 interactions with chromatin in cultured breast cancer (31) and endometrial cancer cells (32), endometrial tumors (33), and endometrial biopsies from infertile women treated with clomiphene (18). Here, we describe the profile of estrogen receptor interaction with chromatin in human endometrial biopsy samples from healthy volunteers at the estrogen-dominant proliferative phase or the progesterone-dominant mid-secretory phase. Further, we cultured epithelial cells from endometrial biopsies in organoids to describe details of estrogen response in an in vitro system. The transcriptional profiles and ESR1 binding sites derived from the isolated epithelial cells are revealed, and then are compared to transcriptomes and ESR1 cistromes derived from whole endometrium, increasing our understanding of the estrogen receptor–mediated processes that are intrinsic to endometrial epithelial cells. Together, our study allows examination of ESR1 interactions with endogenous enhancers and promoters that mediate transcriptional responses within normal endometrial tissue.
Materials and Methods
Ethics Statement
This project was executed in accordance with the federal regulation governing human subject research. The study was reviewed and approved by the Committee for the Protection of Human Subjects at the University of North Carolina at Chapel Hill Institutional Review Board under file number 05-1757 or NIH Institutional Review Board 99CH0103. Informed consent was obtained from all patients before their participation in this study.
Human Endometrial Samples
Endometrial biopsies were obtained for ESR1 ChIPseq and for isolation of epithelial cells for culture as organoids. Healthy women, aged 19 to 34 years, with a regular intermenstrual interval between 25 and 35 days and no history of infertility or pelvic disease, were invited to participate. Exclusion criteria were the following: a) an intermenstrual interval that varied by more than 3 days; b) use of medication known to affect reproductive hormones or fertility within 60 days prior to enrollment; c) chronic disease; d) a body mass index greater than 29.9 or less than 18.5; and e) history of infertility, defined as a failure to conceive for 1 year or longer despite regular intercourse without contraception.
All participants underwent an endometrial biopsy, taken from the mid-fundus, in the office, using a Pipelle suction curette. For isolation of epithelial cells for organoid culture, mid-secretory–phase endometrial biopsies were taken from volunteers and processed immediately. For ESR1 ChIPseq, biopsies were obtained from proliferative or mid-secretory phases and frozen.
Estrogen receptor α chromatin immunoprecipitation and next-generation sequencing
Endometrial biopsies collected and frozen at the mid-secretory or the proliferative phase or fixed, frozen organoid pellets were shipped to Active Motif Inc for Factor Path ESR1 ChIP (ERα antibody 06935, EMD Millipore) (34) and library preparation. Libraries were shipped to National Institute of Environmental Health Sciences (NIEHS) and sequenced in the NIEHS Genomics Core Laboratory. Raw ChIPseq reads were processed and aligned to the human reference genome hg38 using Bowtie (35). The reads were deduplicated, and peaks were called relative to input controls using MACS2 (36). The mergePeaks function of HOMER (37) was used to find shared ESR1 peaks in donor1 and donor2 organoid samples and to make a Venn diagram of ESR1 peaks in proliferative vs mid-secretory endometrium. Peak Annotation and Visualization (PAVIS) (38) was used to compare the locations of ESR1 peaks relative to genes. Known motifs in ESR1 peaks were identified using HOMER findMotifs. Heat map plots of ESR1 signals centered on organoid ESR1 peaks were created using EaSeq (39). EaSeq was also used to locate the nearest gene transcription start site (TSS) and transcription end site to each ESR1 peak as a way to identify peaks within 100 kb of genes. Data are deposited in GEO GSE200807.
Endometrial RNA Sequencing Analysis
RNAseq fragments per kilobase of transcript per million fragments mapped (FPKM) values from our previous study (40) for proliferative or mid-secretory whole endometrium or isolated epithelium RNA were used (GSE132713). Whole endometrium was analyzed using analysis of variance (ANOVA) and List Manager in Partek Genomics Suite software (Partek Inc) to find differentially expressed genes (DEG, 2-fold, false discover rate [FDR] P value < .05) and filter the DEG list to include DEG within 100 kb of ESR1 peaks. This filtered gene list was imported into Ingenuity Pathway Analysis (IPA, Qiagen) for core analysis.
Organoid Culture and Treatment
Organoids were derived from 3 fresh donor endometrial biopsies collected at the mid-secretory phase as described in published studies (27, 29), which confirmed the epithelial characteristics of the resulting organoids. Samples were washed in wash medium (Dulbecco’s modified Eagle’s medium [DMEM]/F12 [Gibco] + 1x Anti-Anti [Gibco]), minced, and digested in 20 mL 0.4 mg/mL Collagenase V (Sigma) + 2.5 mg/mL Dispase II (Sigma) dissolved in wash medium at 37 °C for 50 minutes. Digestion was stopped by adding 10% fetal bovine serum diluted in wash medium. Remaining tissue debris was allowed to settle, and the suspended tissue digest was passed through a 100-µm Cell Strainer (Falcon) and washed with wash media. Epithelial cells were backwashed from the cell strainer, centrifuged at 232 relative centrifugal force for 10 minutes, resuspended, and washed. The final pellet was resuspended in Matrigel (Corning) to make the final concentration 90% Matrigel, 3 to 4 25-µL drops were plated in each well of 12-well, plate and overlayed with 700 µL of expansion media (Advanced DMEM/F12 [Gibco]) containing B27 (minus vitamin A, Gibco), Insulin-Transferrin-Selenium (ITS, Gibco), Primocin (Invitrogen), GlutaMax (Gibco), N2 supplement (Gibco), 1.25 mM N-acetyl-L-cysteine (Sigma), 500 nM A83-01 (Tocris), 50 µg/mL human HGF (Peprotech), 500 µg/mL human EGF (Peprotech), 100 µg/mL human FGF-10 (Peprotech), 500 µg/mL human Rspondin-1 (Peprotech), 100 µg/mL human Noggin (Peprotech), and 10 nM Nicotinamide (Sigma). Media were replaced every 2 to 3 days. Once organoids formed and grew to fill the Matrigel drop (see Supplementary Fig. S1b) (41), they were passaged by resuspension in Advanced DMEM/F12 + B27, ITS, Primocin, and GlutaMax, centrifuged and washed to release organoids from Matrigel and either frozen in 10% dimethyl sulfoxide (Sigma) in fetal bovine serum and stored in liquid nitrogen, or resuspended and plated in fresh Matrigel. For hormone treatments (see graphic in Supplementary Fig. S1a) (41), 4 days after organoids were plated, media were replaced with expansion media that had N2 omitted and contained either 0.1% ethanol vehicle (V) or 10 nM estradiol (E2, Steraloids). For RNA isolation, 2 (d6), 5 (d9), or 8 (d12) days later fresh media containing V or E2 was added, and organoids were isolated 6 hours later. For the ESR antagonist treatment, on d8, and again on d9 fresh media containing V, E2, or 10 nM E2 + 1 µM ICI, 780 (Tocris) was added, and organoids were collected 6 hours after the media change on d9. For ChIPseq, on d9, fresh media containing 10 nM E2 and 1 µM medroxyprogesterone acetate (Sigma) was added, and organoids were isolated 1 hour later. The progesterone was included for planned future PGR ChIPseq analysis of remaining chromatin.
RNA Isolation and Analysis
RNA was isolated from organoids using Trizol Reagent (Invitrogen) according to the manufacturer’s instructions. For reverse-transcription polymerase chain reaction (RT-PCR), complementary DNA was prepared using Superscript II (Invitrogen) with Random Hexamers (Invitrogen), as previously described (42, 43). Sequences of primers (Sigma) used are: ESR1 F-CTGCAGGGAGAGGAGTTTGTGT R-TCCAGAGACTTCAGGGTGCT, IHH F-GACCGCGAC CGCAATAAGTA R-TGGGCCTTTGACTCGTAATAC, GAPDH F-ATGGGGAAGGTGAAGGTCG R-GGGGTCATT GATGGCAACAATA, GREB1 F-ATGGGAAATTCTTTACGC TGGAC R-CACTCGGCTACCACCTTCT, PGR F-GACG TGGAGGGCGCATAT R-AGCAGTCCGCTGTCCTTTTCT. PCR was performed using SsoAdvanced Universal SYBR green Supermix (BioRad) with a CFX instrument (BioRad).
For RNAseq, RNA (3 replicates each of V or E2-treated samples from donor 1 and donor 2) was DNAse treated and cleaned up using the RNeasy Mini kit (Qiagen) or the RNA Clean and Concentrator 5 kit (Zymo). RNA was submitted to the NIEHS sequencing core for library preparation and paired-end sequencing using Illumina’s Ribo-Zero Gold kit for donor 1 RNA and Illumina Stranded messenger RNA Prep for donor 2. Raw data were filtered to remove low-quality reads, mapped to hg38 using TopHat (44), and deduplicated using Picard tools (2.18.15; “Picard Toolkit.” 2019. Broad Institute, GitHub Repository. https://broadinstitute.github.io/picard/; Broad Institute). BAM files were imported into Partek Genomics Suite for analysis. Reads per kilobase per million reads (RPKM) for RefSeq genes were determined, and the maximum RPKM value from any sample for each gene (MAX) was listed. We noted differences between donor 1 and donor 2 data sets that reflect differences in RNA and library quality. For this reason, we analyzed the data sets from each donor separately, and then compared the DEG, so that each would be relative to its own V control. Genes for which the MAX was less than 1% of the mean RPKM for the whole data set were filtered out (< 0.02 for donor 1, < 0.002 for donor 2). Values of 0 for V samples were replaced with a value of 0.05% of MAX to allow calculation of E2/V. After principal component analysis, it was noted that donor 1 V-1 and donor 1 E2-1 samples were outliers, potentially due to low reads, and were excluded from further analysis. Partek ANOVA was used to determine E2 vs V fold changes, and the List Manager function was used to find DEGs (2-fold, FDR P value < .05). Donor 1 or donor 2 DEG data were analyzed individually using IPA, and analyses were compared using the comparison function. Partek Venn diagrams were used to construct a list of all DEGs in either donor to combine E2 vs V fold changes of the data sets for visualization. Data are deposited in GEO (GSE200807).
Results
Endometrial Estrogen Receptor α Cistrome
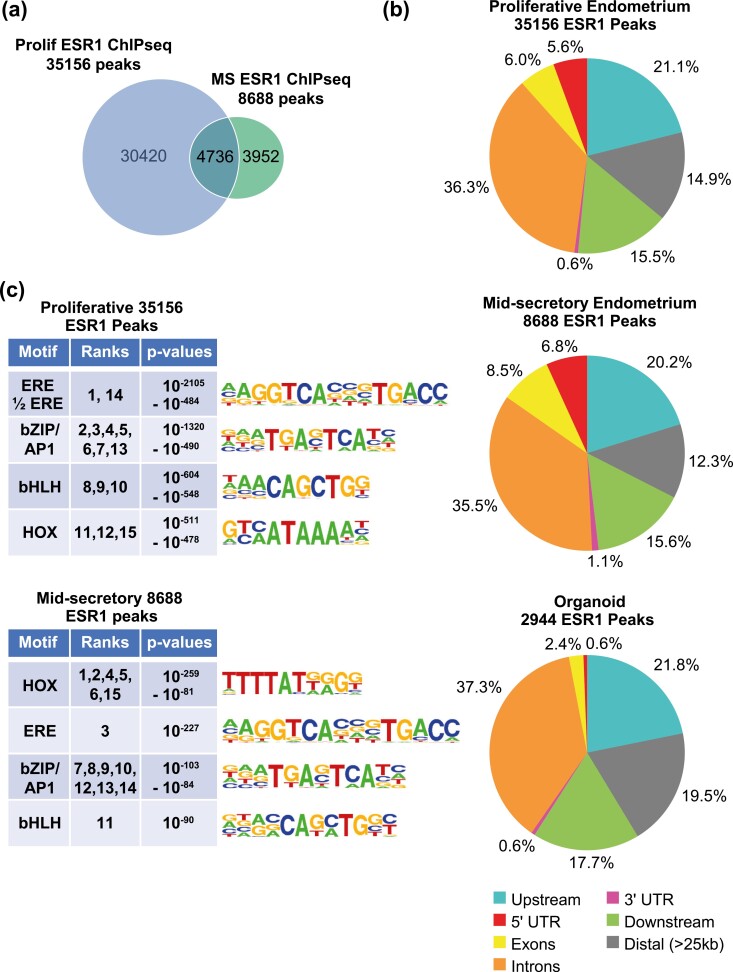
We previously described PGR interaction with chromatin (ChIPseq) together with transcriptomic (RNAseq) analysis in proliferative- or mid-secretory–stage endometrial biopsies from healthy volunteers (40). Here, we have extended those findings by evaluating ESR1 binding using the same techniques in additional endometrial biopsy samples. In the whole endometrial samples, there are 4-fold more ESR1 peaks in the proliferative sample (35 156 peaks) than the mid-secretory sample (8688 peaks), consistent with the estrogen-dominant proliferative phase (Fig. 1A) in the figure panels, the letters are lower case, so this is inconsistent. We examined the locations of the ESR1 peaks relative to annotated genes. ESR1 peaks were distributed comparably in the proliferative and mid-secretory endometrium (Fig. 1B), with about half of peaks located at genes (49% of proliferative and 52% of mid-secretory ESR1 peaks at 5′ untranslated region [UTR], exons, introns, and 3′ UTR). We also compared the ESR1 peaks identified here to PGR peaks in proliferative and mid-secretory endometrial biopsies that were previously described (40). A total of 3459 of the 35156 ESR1 peaks in proliferative samples overlapped with PGR peaks (Supplementary Fig. S2c) (41). We recently reported that 2616 of the 8688 ESR1 peaks in mid-secretory samples overlapped with PGR peaks (45). Motif analysis of the ESR1 peaks indicates that estrogen-responsive enhancer (ERE) and homeobox factor (HOX) motifs are enriched both in proliferative and mid-secretory samples (Fig. 1C). ERE is the most significantly enriched motif in proliferative samples (see Fig. 1C), and HOX motifs are the top enriched motifs in mid-secretory ESR1 peaks (see Fig. 1C). In addition, bZIP/AP1 and bHLH motifs were seen both in proliferative and mid-secretory ESR1 binding peaks.
Figure 1.
ESR1 ChIPseq of proliferative and mid-secretory endometrial biopsies. A, Venn diagram comparing number of ESR1 peaks in one proliferative (Prolif) endometrial biopsy to ESR1 peaks in one mid-secretory (MS) endometrial biopsy. B, Locations of ESR1 peaks from proliferative or mid-secretory endometrium or from organoids, relative to annotated genes (RefSeq). C, Summaries of the top 15 HOMER known motifs enriched in ESR1 peaks from proliferative or mid-secretory endometrial biopsies. The ranks of each motif, as determined by P value, along with the range of P values covered by the ranks are indicated. Motif logos are shown.
To further evaluate the effect of ESR1 chromatin interactions on uterine functions, we determined the ESR1 binding sites that are within less than 100 kb of an annotated Refseq gene in the proliferative (31 814 peaks) or mid-secretory (7931 peaks) samples (Supplementary Fig. S2) (41). Next, we analyzed the endometrial transcriptome, which we previously described (40), by identifying genes that were expressed in proliferative and mid-secretory RNA (FPKM ≥ 1 in at least one sample) and determining those that were differentially expressed between proliferative and mid-secretory phases (1628 DEG 2-fold, FDR P-value < .05; Supplementary Table S1a) (41). Genes that differ between the estrogen-dominant proliferative and progesterone-dominant mid-secretory phases include ESR1 targets that are either increased or decreased by estrogen. We then filtered the DEG to include genes that are less than 100 kb from the proliferative (902 genes; Supplementary Table S1b) (41) or mid-secretory (394 genes) ESR1 peaks, respectively (Supplementary Fig. S2a) (41) as genes that are candidate ESR1 downstream targets.
We focused on 902 proliferative endometrium DEGs that are candidate ESR1/estrogen targets and used IPA to assess their effect on biological functions and signaling (Tables 1-3 and Supplementary Table S1c-e) (41). Our analysis indicates effects on multiple overlapping processes including estrogen and progesterone signaling, proliferation, chromatin modification, transcription, endometrial physiology–associated signals, lipids, growth factor/receptor tyrosine kinase signaling, signaling via other receptors, and hematological system development.
Table 1.
Biological functions (overlap P < 10–5) enriched in 902 proliferative vs mid-secretory differentially expressed genes that are less than 100 kb from a proliferative estrogen receptor α chromatin immunoprecipitation and next-generation sequencing peak
| Function | Activation z score | No. of genes |
|---|---|---|
| Hematological system development and function | 2.18 | 43 |
| DNA replication | 1.99 | 32 |
| Cell survival | 1.96 | 162 |
| Colony formation | –1.73 | 61 |
| Morbidity or mortality | –1.77 | 267 |
| Transport | –1.77 | 157 |
| Quantity of carbohydrate | –1.83 | 61 |
| Aggregation of cells | –2.06 | 42 |
| Cellular homeostasis | –2.22 | 163 |
| Synthesis of lipid | –2.50 | 87 |
Table 2.
Upstream regulators (overlap P < .0003; activation z score > 2 or < –2) Enriched in 902 proliferative vs mid-secretory differentially expressed genes that are less than 100 kb from a proliferative estrogen receptor α chromatin immunoprecipitation and next-generation sequencing peak
| Regulators | ||
|---|---|---|
| Process | Increased activity | Decreased activity |
| Chromatin | EP400 | NUPR1, SMARCB1, let-7 |
| Cytokine signaling | IL10RA | IFNG, IL1B, IL6 |
| Estrogen | ESR1, E2 | |
| Growth factor | FGF1, VEGFB, PDGF BB | |
| Growth factor/RTK | ERBB2 | |
| RTK signaling | Insulin, MAPK1 | |
| Progesterone | P4, PGR | |
| Proliferation | FOXM1, CKAP2L, RABL6, CCND1, CCND1 | CDKN2A, TP53, Rb |
| Receptor signaling | RARA, AGT | DEX, calcitriol, NR3C1 |
| Transcription | E2f, TBX2, MYC, CEBPB, SOX4 | HNF4A, EHMT1 |
Abbreviations: AGT, angiotensin; CEBPB, CCAAT enhancer binding protein β; CDKN2A, cyclin-dependent kinase inhibitor 2A; CKAP2L, cytoskeleton associated protein 2 like; DEX, dexamethasone; E2, estradiol; EHMT1, euchromatic histone lysine methyltransferase 1; EP400, E1A binding protein p400; ERBB2, Erb-b2 receptor tyrosine kinase 2; ESR1, estrogen receptor α; FGF1, fibroblast growth factor 1; FOXM1, forkhead box M1; HNF4A, hepatocyte nuclear factor 4 α; IFNG, interferon γ; IL1B, interleukin 1B; IL6, interleukin 6; MAPK1, mitogen-activated protein kinase 1; NUPR1, nuclear protein 1, transcriptional regulator; PDGF BB, platelet-derived growth factor BB; PGR, progesterone receptor; RABL6, RAS oncogene family like 6; RARA, retinoic acid receptor a; RTK, receptor tyrosine kinase; SMARCB1, SWI/SNF component SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily b, member 1; SOX4, SRY-box transcription factor 4; TBX2, T-box transcription factor 2; TP53, tumor protein 53; VEGFB, vascular endothelial growth factor B.
Table 3.
Pathways (overlap P < .03); activation z score > 1.4 or < –1.9 enriched in 902 proliferative vs mid-secretory differentially expressed genes that are less than 100 kb from a proliferative estrogen receptor α chromatin immunoprecipitation and next-generation sequencing peak
| Pathway | ||
|---|---|---|
| Process | Increased activity | Decreased activity |
| Cancer | Basal cell carcinoma, tumor microenvironment | |
| Chromatin | HOTAIR regulatory pathway | |
| Cytokine signaling | Acute-phase response | |
| Endometrial physiology | Inhibition of MMPs | |
| Lipid | LXR/RXR | |
| Proliferation | Cell cycle/chromosomal replication, estrogen-mediated S-phase entry, cyclins/cell cycle, PTEN | |
| Receptor signaling | AHR | |
| Transcription | BEX2 signaling | |
Abbreviations: AHR, aryl hydrocarbon receptor; BEX2, brain expressed X-linked 2; HOTAIR, HOX transcript antisense RNA; LXR/RXR, liver X receptor/retinoid X receptor; MMP, matrix metalloproteinase; PTEN, phosphatase and tensin homolog.
We noted increased estrogen/ESR1 activity and decreased progesterone/PGR activity (see Table 2), as expected of genes that differ between estrogen-dominant proliferative phase vs the progesterone-dominant mid-secretory phase and validating that our approach identifies potential ESR1-regulated endometrial pathways.
Proliferation-associated functions, signaling pathways, and regulators are enriched, including DNA replication (see Table 1), chromosome replication, E2-mediated S-phase entry, cyclins, and cell cycle (see Table 3) together with the regulators mitotic spindle component cytoskeleton-associated protein 2 like (CKAP2L), cell growth factor RAB, member RAS oncogene family like 6 (RABL6), cyclin CCND1, mitosis factor forkhead box M1 (FOXM1), and inhibition of cell-cycle inhibitors Rb tumor suppressor and tumor protein 53 (TP53) and TP53 stabilizer cyclin-dependent kinase inhibitor 2A (CDKN2A; see Table 2). Altogether, these indicate increased proliferation, which corresponds to estrogen-stimulated endometrial growth that is characteristic of the proliferative phase.
The gene set includes chromatin modifiers including activation of HOX transcript antisense RNA (HOTAIR; see Table 3), a long noncoding RNA associated with multiple cancers that acts as a guide and scaffold to deliver Polycomb repressive complex 2 (PRC2) chromatin modifiers to specific gene loci (46), histone acetyl transferase (HAT) complex factor E1A binding protein p400 (EP400), and inhibition of micro RNA let-7, which targets HOTAIR. Additional chromatin regulators affected include SWI/SNF component SWI/SNF related, matrix-associated, actin-dependent regulator of chromatin, subfamily b, member 1 (SMARCB1), which removes repressive chromatin marks, and HAT activity factor nuclear protein 1, transcriptional regulator (NUPR1; see Table 2).
Candidate estrogen/ESR1 gene responses were consistent with effects on activities of multiple transcription factors (increased activity of transcription factor brain expressed X-linked 2; BEX2) pathway (see Table 3), increased activity of E2f, T-box transcription factor 2 (TBX2), MYC proto-oncogene, CCAAT enhancer binding protein β (CEBPB), and SRY-box transcription factor 4 (SOX4), inhibition of transcription factor hepatocyte nuclear factor 4 α (HNF4A), and transcription inhibitor euchromatic histone lysine methyltransferase 1 (EHMT1; see Table 2). Effects on chromatin and transcription align with estrogen/ESR1 having roles in chromatin modifications that influence accessibility of transcriptional mediators.
Several signaling pathways known to be important in endometrial physiology and pathophysiology were enriched. For example, the pattern of gene expression suggests increased activity of the protein kinase B inhibitor and tumor suppressor phosphatase and tensin homolog (PTEN) was increased. PTEN has an essential role as a cell-cycle checkpoint regulator to prevent premature mitosis and resulting gene mutations (47) and is frequently inactivated in endometrial tumors (48). Further, estrogen inhibition of endometrial cytokine signals are reflected by decreased acute-phase response (see Table 3) and cytokine signaling (interleukin 1B [IL-1B], IL-6, interferon γ; see Table 3) and increased interleukin 10 receptor subunit A (IL-10RA) signaling (see Table 3) in the proliferative-phase ESR1-associated genes, aligning with suppression of cytokine signaling during the proliferative phase and later activation associated with implantation (49, 50). There are indications that ESR1 signaling affects lipids, as reflected by decreased activity of lipid synthesis (see Table 1) and liver X receptor/retinoid X receptor (LXR/RXR) activation (see Table 3).
Several mediators of growth factor/receptor tyrosine kinase (RTK) pathways are affected as well. The RTK Erb-b2 receptor tyrosine kinase 2 (ERBB2) is activated, as is the downstream modulator mitogen-activated protein kinase 1 (MAPK1; see Table 2). The activity of growth factors platelet-derived growth factor BB (PDGFBB), vascular endothelial growth factor B (VEGFB), and fibroblast growth factor 1 (FGF1) are decreased (see Table 2). Signaling via other receptors is also evident, including activation of aryl hydrocarbon receptor (AHR; see Table 3) and retinoic acid receptor a (RARA; see Table 2), and inhibition of calcitriol (vitamin D agonist) and dexamethasone (glucocorticoid receptor agonist; see Tables 2 and 3).
Hematological system development and function was enriched (see Table 1), which includes enhanced signaling from angiotensin (AGT; see Table 2). This observation aligns with studies indicating angiogenesis, vascular remodeling, and regulation of blood flow in the endometrium and placenta are essential processes regulated by the renin-angiotensin system (51-55). Overall, our analysis of ESR1-associated candidate estrogen-regulated processes aligns with estrogen’s multiple roles in the proliferative-phase endometrium, both previously described and novel.
Estrogen Responses of Organoids
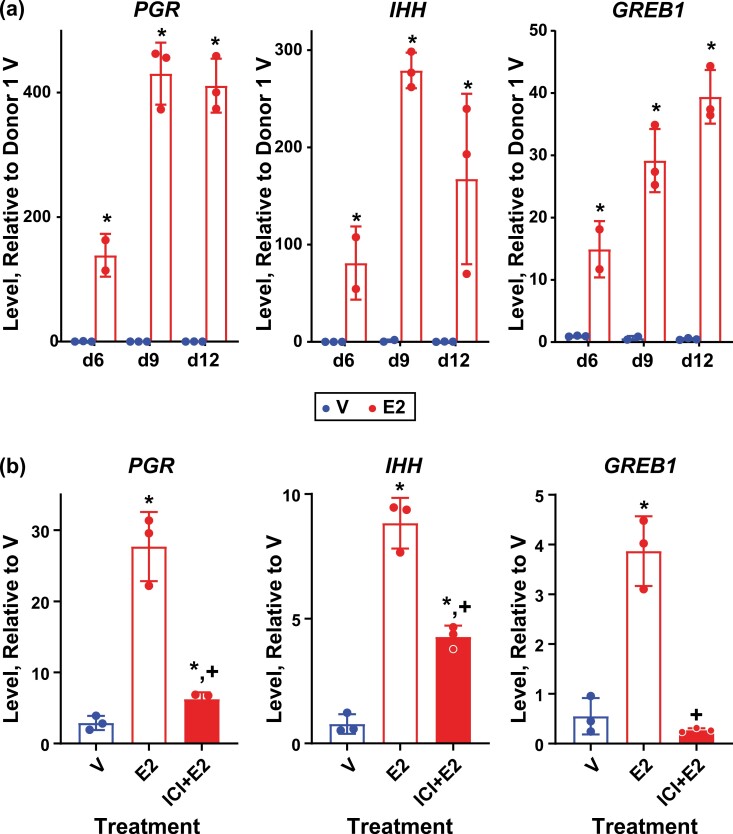
The development of organoid models derived from epithelial cells isolated from endometrial biopsies has provided an in vitro system in which to manipulate and study hormone-signaling mechanisms (27, 29, 56). E2 responses were evaluated in organoids cultured as previously described by Fitzgerald et al (29) with some modification. Organoids were plated and allowed to form for 4 days, then treated with E2 or V for 2, 5, or 8 more days (d6, d9, d12; see Fig S1a) (41). Samples were collected 6 hours following a change to fresh media containing V or E2 (see “Materials and Methods” for details) to capture acute responses. Organoids derived from endometrial samples obtained from 3 different donors were compared to assess individual donor variability. The levels of transcript for 3 estrogen-regulated endometrial genes (PGR, IHH, and GREB1) were measured. All 3 genes were robustly induced by E2 in all 3 donors as early as the first day examined (6; see Fig. 2A and Supplementary S3a and S3b) (41). In the donor 1– and donor 3–derived organoids, notable E2-induced increases in PGR and IHH were observed on d9 compared to d6, while the level of PGR induced by E2 on d9 in the donor 2–derived organoids decreased, and the level of IHH induced by E2 did not change relative to the levels observed on d6 (see Fig. 2A and Supplementary Fig. S3a and S3b)(41)). The transcripts were all calculated relative to levels observed in donor 1, revealing that, in general, estrogen induced all 3 transcripts more robustly in donor 1–derived organoids than in the other 2. The expression of ESR1 transcript was similar in all 3 sets of organoids (see Supplementary Fig. S3c) (41) but was lower in donor 2, which may contribute to the decreased response of this donor, although we have not quantified ESR1 protein levels in any of the donors. In donor 2 samples, on d9, induction of PGR, IHH, or GREB1 with E2 could be inhibited using the ESR1 antagonist, ICI 182 780, indicating an ESR-mediated response (see Fig. 2B). Although the media the organoids are grown in contains phenol red, which has weak estrogenic activity (57), the antagonist does not decrease activity below baseline, indicating any estrogenic activity from the phenol red is minimal. Based on the responses observed, for comprehensive analysis of organoid transcriptomes, and ESR1 cistrome, we focused on samples from 2 donor-derived organoids (donors 1 and 2) on d9.
Figure 2.
Estrogen-responsive genes are induced in organoid cultures. A, RT-PCR of RNA isolated from donor 1 derived organoid cultures treated as described in methods and in Supplementary Fig. S1 (41) with 0.1% ethanol vehicle (V) or with 10 nM estradiol (E2) 6, 9, or 12 days after initial plating (d6, d9, d12). Progesterone receptor (PGR), Indian hedgehog (IHH), and growth-regulating estrogen receptor binding 1 (GREB1). Bar indicates mean, error bars indicate SD; * indicates P < .05 vs V based on 2-way ANOVA with Fisher LSD multiple comparisons test. N = 3 for all but d6 E2 (n = 2). B, RT-PCR of RNA isolated from donor 2 derived organoids 9 days after plating treated with 0.1% ethanol V, 10 nM E2, or with 1 µM ICI 182780 + 10 nM E2 (ICI + E2). Bars indicates mean, error bars indicate SD; * indicates P < .05 vs V; +indicates P < .05 vs E2 based on 2-way ANOVA with Fisher LSD multiple comparisons test. N = 3 for all.
Estrogen Regulates the Organoid Transcriptome
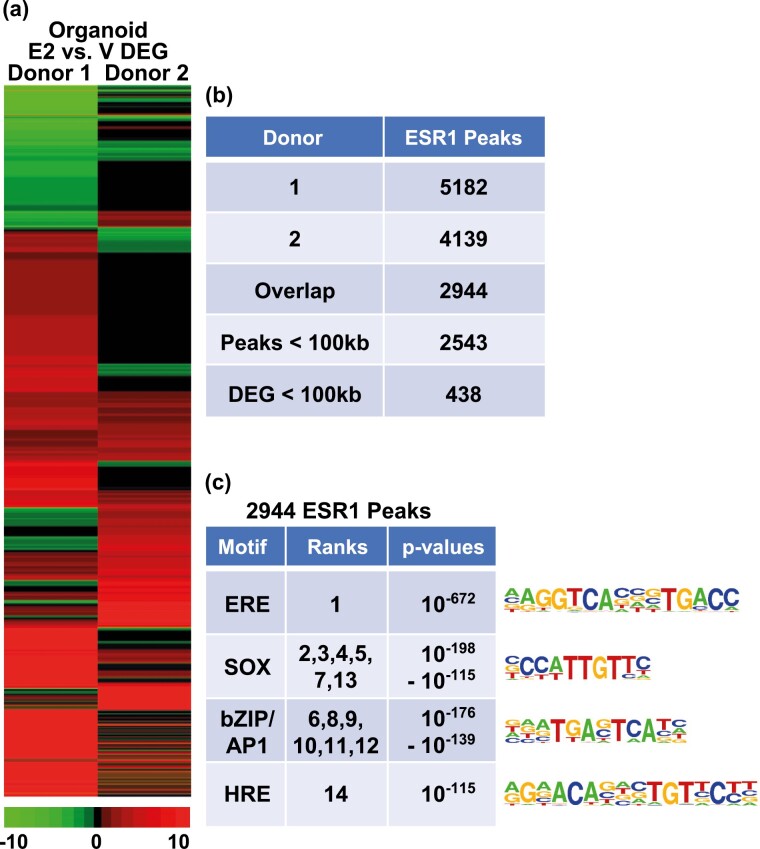
We used RNAseq to compare the transcriptomes of donor 1– and donor 2–derived organoid sets after treatment with V or estrogen. Transcripts expressed in each donor were compared, indicating 10 652 genes were detected in both donors (Supplementary Fig. S4a) (41). We then compared these genes to the 15 127 and 15 337 genes detected in proliferative and mid-secretory isolated endometrial epithelial cells (40), respectively (Supplementary Fig. S4b) (41), confirming that most of the organoid transcripts (76%) represent genes expressed in intact epithelium. Further analysis of the organoid transcripts revealed that, as was observed with the estrogen-responsive genes evaluated by RT-PCR (see Fig. 2 and Supplementary Fig. S3) (41), donor 1–derived organoids exhibited more robust estrogen responses than donor 2 (donor 1 E2/V 1907 DEG; donor 2 E2/V 695 DEG; 2-fold, FDR < 0.05; Fig. 3A and Supplementary Table S2a and S2b) (41). We also compared the organoid and endometrial responses (Supplementary Fig. S4b) (41), which indicates the estrogen-regulated organoid genes are similar to the DEGs between proliferative and mid-secretory endometrium. The estrogen-regulated organoid transcripts were analyzed using IPA, revealing, as previously reported (28), that E2 treatment of organoids promotes formation of cellular protrusions/ciliagenesis (Table 4 and Supplementary Table S2c) (41). As in the whole endometrium, signals and regulators affected overlapping processes, including chromatin modification, RTK-mediated signals, and transcription (Tables 5 and 6 and Supplementary Table S2c-S2e) (41). Evaluation of the effect of gene regulations on signaling or regulatory pathways revealed activation of estrogen signaling, as well as signal transducer and activator of transcription 3 (STAT3) and transforming growth factor β (TGFβ)-Smad family member (SMAD)-mediated signals (see Tables 5 and 6), all of which have demonstrated importance in endometrial growth and function (58, 59). The pattern of gene regulation of donor 2 is consistent with decreased activity of Wnt family member 5A (WNT5A), which is involved in polarity of epithelial cells during embryo implantation (60) (see Table 5). Activity of some functions or signals are selectively enriched in one donor, for example, γ-aminobutyric acid (GABA) and TP53 in donor 2, and AGT and SMAD in donor 1.
Figure 3.
Human epithelial organoid ESR1 transcriptome and cistrome. A, Hierarchical cluster comparing E2 vs V fold-changes of significantly regulated genes (2-fold, FDR P < .05) in either donor. B, ESR1 ChIPseq of chromatin isolated from organoids. The number of peaks of each donor sample is indicated, as well as those shared by both donors. A total of 2543 of the 2944 overlapping ESR1 peaks are within 100 kb of a gene. C, Summaries of the top 14 HOMER known motifs enriched in the 2944 shared ESR1 peaks in organoids. The motif’s ranks, as determined by P value, along with the range of P values covered by the motif, are indicated.
Table 4.
Biological functions (overlap P value < .05) enriched in estrogen-regulated organoid genes
| Activation z score | ||
|---|---|---|
| Function | Donor 1 | Donor 2 |
| Organization of cytoskeleton | 4.50 | 4.88 |
| Organization of cytoplasm | 4.50 | 4.88 |
| Microtubule dynamics | 4.62 | 4.72 |
| Formation of cellular protrusions | 3.42 | 4.10 |
| Formation of cilia | 4.16 | N/A |
| Transport of ion | 3.44 | N/A |
| Outgrowth of cells | –0.33 | 2.18 |
Abbreviation: N/A, not available.
Table 5.
Upstream regulators (overlap P value < .05; activation z score < –1.5 or > 1.5) enriched in estrogen-regulated organoid genes
| Donor 1 regulators | Donor 2 regulators | |||
|---|---|---|---|---|
| Process | Increased activity | Decreased activity | Increased activity | Decreased activity |
| Chromatin | KDM1A | IKZF1 | ||
| Estrogen | E2, ESR1 | E2 | ||
| Proliferation | TP53 | mir-1 | ||
| Receptor signaling | DEX, AGT, RXRA, PI3K | PI3K, DEX | GABA | |
| TGFβ signaling | Smad2/3-Smad4 | Tgf β | ||
| Cytokine signaling | STAT3 | STAT3 | ||
| Transcription | EHMT1, ERG | LDB1, LMO2 | ERG | |
| WNT signaling | WNT5a | |||
Abbreviations: AGT, angiotensin; E2, estradiol; EHMT1, euchromatic histone lysine methyltransferase 1; ESR1, estrogen receptor α; DEX, dexamethasone; GABA, γ-aminobutyric acid; STAT3, signal transducer and activator of transcription 3; TGFβ, transforming growth factor β; WNT5a, Wnt family member 5A.
Table 6.
Signaling pathways (overlap P value < .05; activation z score > 1.5) enriched in estrogen-regulated organoid genes
| Process | Donor 1 | Donor 2 |
|---|---|---|
| Endometrial physiology | Autophagy, relaxin | |
| Estrogen | ESR | |
| Receptor signaling | Adrenomedullin | Ephrin receptor, oxytocin, androgen, melatonin, opioid, eNOS, VEGF-VEGFR |
| Receptor tyrosine kinase signaling | G-β-γ, CRH |
Abbreviations: eNOS, endothelial nitric oxide synthase; ESR, estrogen receptor α; VEGF, vascular endothelial growth factor.
Organoid Estrogen Receptor α Cistrome
The gene-regulation patterns revealed using RNAseq analysis indicated a robust response to E2. To better evaluate the direct effects of ESR1, sites of interaction with chromatin in organoids were evaluated by ESR1 ChIPseq. Comparable numbers of ESR1 peaks were identified in hormone treated organoids from either donor (Fig. 3B). For subsequent analysis, we focused on 2944 ESR1 peaks shared by both donors. Examination of the locations of ESR1 peaks relative to transcripts shows that organoid ESR1 is more distal than the whole endometrial ESR1 (see Fig. 1B), with a smaller proportion of binding in 5′ UTR and exons and a larger proportion located in distal regions. As was observed in the proliferative endometrium, ERE was the most significantly enriched motif in the ESR1 peaks (Fig. 3c). However, unlike in the whole endometrium, SOX motifs were significantly enriched. This likely reflects the role of the SOX transcription factor activity specific to epithelial cells. Like the whole endometrium, bZIP/AP1 motifs were enriched. The HRE motif, which interacts with PGR, Androgen Receptor (AR), and Glucocorticoid Receptor (GR), was enriched as well. A total of 2543 of the 2944 ESR1 peaks are within 100 kb of an annotated gene (Fig. 3B and Supplementary Fig. S4c) (41); 438 genes that are estrogen regulated in either donor are less than 100 kb from one of the 2543 ESR1 peaks (see Fig. 3B and Supplementary Table S3a) (41). Pathway analysis of these 438 genes that are candidate ESR1 downstream targets indicates some of the signals and functions noted previously (Tables 7 and 8 and Supplementary Table S3b and S3c) (41), including formation of cellular protrusions, estrogen, STAT3 and TGFβ signaling, were enriched (see Tables 3, 7, and 8). The pattern of gene regulation by these ESR1 targets revealed functions and regulators that were not apparent in the analysis of all the DEGs (see Tables 4-6) including progesterone activation, and lipid concentration (see Tables 7 and 8).
Table 7.
Functions (overlap P value < .05) enriched in estrogen-regulated organoid genes that are less than 100 kb from proliferative estrogen receptor α chromatin immunoprecipitation and next-generation sequencing peaks
| Activation z score | ||
|---|---|---|
| Function | Donor 1 | Donor 2 |
| Chemotaxis | 2.34 | 0.6 |
| Formation of cellular protrusions | 2.26 | 1.28 |
| Invasion of cells | 1.75 | 1.46 |
| Quantity of Ca2+ | 1.37 | 1.57 |
| Concentration of lipid | –1.92 | –1.22 |
Table 8.
Upstream regulator (overlap P value < .05; activation z scores < –1.7 or > 1.3) enriched in estrogen-regulated organoid genes that are less than 100 kb from estrogen receptor α chromatin immunoprecipitation and next-generation sequencing peaks
| Donor 1 regulators | Donor 2 regulators | |||
|---|---|---|---|---|
| Process | Increased activity | Decreased activity | Increased activity | Decreased activity |
| Chromatin | let-7 | let-7 | ||
| Cytokine signaling | STAT3, IL6 | IL22 | STAT3, IL6 | IL22 |
| Estrogen | E2, ESR1 | E2, ESR1 | ||
| Growth factor/receptor tyrosine kinase signaling | ERBB2 | |||
| Lipid | ACOX1 | ACOX1 | ||
| NR signaling | NCOA2 | |||
| Progesterone | P4 | P4 | ||
| TGFβ signaling | SMAD4 | SMAD4 | ||
| Transcription | PPARG, BCL6 | EOMES | ||
| WNT signaling | TCFL7, LGR4 | |||
Abbreviations: ACOX1, acyl-CoA oxidase 1; E2, estradiol; ESR1, estrogen receptor α; IL6, interleukin 6; IL22, interleukin 22; NR, nuclear receptor; PPARG, peroxisome proliferator activated receptor γ.
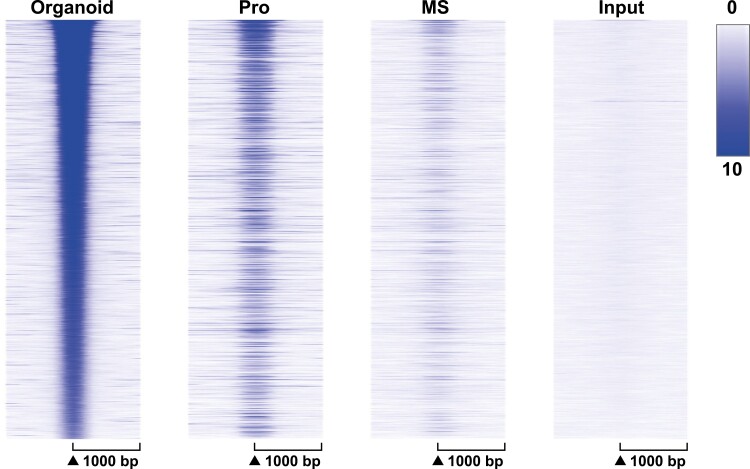
To compare the effect of estrogen signaling in organoids and endometrium, ESR1 peak signal was compared in heat maps centered on organoid ESR1 peaks (Fig. 4). Locations of organoid ESR1 are more like proliferative than mid-secretory ESR1 peaks (see Fig. 4 and Supplementary Fig S4c) (41), with more proliferative-phase than mid-secretory–phase ESR1 signal at locations of organoid ESR1 peaks in the ranked heat map (see Fig. 4) as well as more overlap of organoid ESR1 peaks with proliferative than mid-secretory ESR1 peaks indicated on the Venn diagrams (see Supplementary Fig. S4c) (41). To evaluate the effect of estrogen in whole endometrium and organoids, we compared the signaling and functions regulated by genes within 100 kb of ESR1 peaks in each. As with separate analyses (see Tables 1-3, 7, and 8), estrogen affects multiple processes. Estrogen-E2 signaling is activated in both (Table 9), whereas progesterone signaling is inactivated in proliferative endometrium and activated in organoids (see Table 9 and Supplementary Table S4) (41). Some processes/functions are selectively enriched in organoids, such as formation of cellular protrusions, associated with cilia formation (Table 10). Some are shared, such as ERBB2 activation, activation of lipid metabolic enzyme acyl-CoA oxidase 1 (ACOX1), dopamine, and AGT signaling, whereas others, including forkhead box O1 (FOXO1) and peroxisome proliferator activated receptor γ (PPARG) signaling, are inhibited in the whole endometrium and activated in the organoids (see Table 9). Altogether this suggests that some processes intrinsic to the epithelial cells may be masked by whole endometrial signals and that culturing organoids reveals estrogen signaling processes that are not otherwise observable. Alternatively, the differences could reflect alteration of epithelial cell signals by their isolation and culture separate from the other cell types and other factors found in whole endometrium.
Figure 4.
Organoid ESR1 cistrome resembles proliferative endometrium. ESR1 ChIPseq signal centered on locations of ESR1 peaks (± 1000 bp) in organoid samples. MS, mid-secretory whole endometrium; Pro, proliferative whole endometrium.
Table 9.
Upstream regulators (overlap P value < .05; activation z scores < –1.7 or > 1.3) enriched in proliferative vs mid-secretory endometrial and estrogen-regulated organoid genes that are less than 100 kb from estrogen receptor α chromatin immunoprecipitation and next-generation sequencing peaks
| Pro/MS regulators | Donor 1 regulators | Donor 2 regulators | ||||
|---|---|---|---|---|---|---|
| Process | Increased activity | Decreased activity | Increased activity | Decreased activity | Increased activity | Decreased activity |
| Chromatin | let-7 | let-7 | let-7 | |||
| Cytokine signaling | IL10RA | IL6, IFNG, | IL10RA, STAT3, IL6 | STAT3 | ||
| Endometrial physiology | FOXO1 | FOXO1 | ||||
| Estrogen | E2, ESR1 | E2, ESR1 | E2, ESR1 | |||
| Growth factor | FGF1 | FGF1 | ||||
| Growth factor/receptor tyrosine kinase signaling | ERBB2 | ERBB2 | ||||
| Lipid | ACOX1 | ACOX1 | ACOX1 | |||
| Progesterone | P4 | P4 | P4 | |||
| Proliferation | CDKN2A | |||||
| Receptor signaling | Dopamine, AGT | NR3C1, RXR | Dopamine, AGT, NR3C1 | Dopamine | NR3C1, RXR | |
| Receptor tyrosine kinase signaling | p38 MAPK | |||||
| Transcription | GLI2, LHX1 | PPARG, EHMT1, GATA1 | LHX1, PPARG, EHMT1, GATA1 | GLI2 | ||
Abbreviations: ACOX1, acyl-CoA oxidase 1; AGT, angiotensin; CDKN2A, cyclin-dependent kinase inhibitor 2A; E2, estradiol; EHMT1, euchromatic histone lysine methyltransferase 1; ERBB2, Erb-b2 receptor tyrosine kinase 2; ESR1, estrogen receptor α; FGF1, fibroblast growth factor 1; FOXO1, forkhead box O1; IL6, interleukin 6; MS, mid-secretory; NR, nuclear receptor; PPARG, peroxisome proliferator activated receptor γ; Pro, proliferative.
Table 10.
Biological functions (overlap P value < .05) enriched in proliferative vs mid-secretory endometrial and estrogen-regulated organoid genes that are less than 100 kb from estrogen receptor α chromatin immunoprecipitation and next-generation sequencing peaks
| Activation z score | |||
|---|---|---|---|
| Function | Pro/MS | Donor 1 | Donor 2 |
| Formation of cellular protrusions | –1 | 2.26 | 1.28 |
| Concentration of lipid | –1.15 | –1.92 | –1.22 |
| Cellular homeostasis | –2.22 | 1.94 | –0.05 |
Abbreviations: MS, mid-secretory; Pro, proliferative.
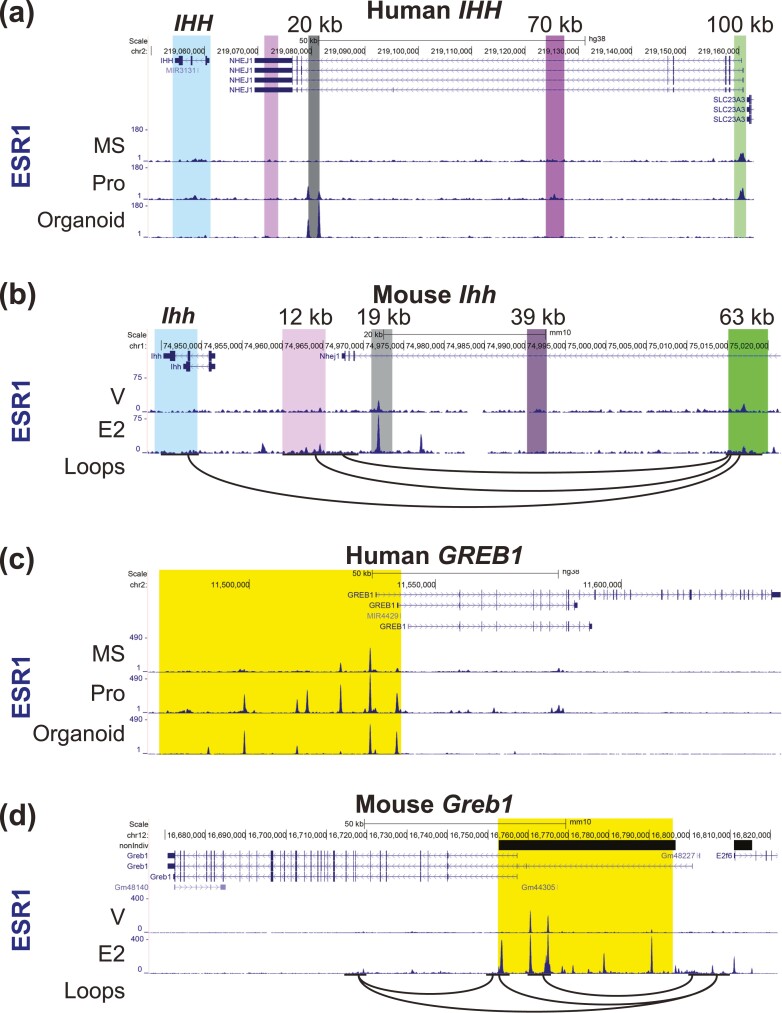
Endometrial and organoid ESR1 peaks relative to the IHH and GREB1 genomic regions are shown in Fig. 5. Two ESR1 peaks are located 20 kb 5′ of the IHH TSS in organoids and in proliferative-phase whole endometrium (Fig. 5A). A previous study described a similar region 19 kb 5′ of the mouse uterus Ihh gene (Fig. 5B), with demonstrated hormone-dependent enhancer activity (61). ESR1 binds 70 kb 5′ of the IHH TSS in proliferative endometrium, and 100 kb 5′ of IHH 5′ both in proliferative and mid-secretory endometrium (see Fig. 5A). Chromosome-conformation capture sequencing (HiC) from the ovariectomized mouse uterus (62) indicates that distal regions interact with the Ihh transcript (see Fig. 5B). Multiple ESR1 peaks are present in a region beginning 50 kb 5′ of the GREB1 gene (highlighted in yellow in Fig. 5C) in organoids and endometrium. Several of the peaks in this region are more prominent in the proliferative sample as compared to the mid-secretory sample. The comparable region of mouse uterine Greb1 was identified as a super-enhancer with multiple ESR1 binding peaks in a previous study (62). In the mouse uterus, HiC indicates interactions between the ESR1 binding super-enhancer region and the Greb1 gene (Fig. 5D). The pattern of ESR1 binding near these estrogen-regulated genes suggest potential enhancer regions involved in gene regulation by estrogen.
Figure 5.
ESR1 peaks near human and mouse estrogen-responsive transcripts IHH, and GREB1. For the human genes, ESR1 ChIPseq from mid-secretory (MS) and proliferative (Pro) whole endometrium and from organoids is shown. For the mouse genes, ESR1 ChIPseq from V or E2 treated whole uterus is shown. Interacting loops from mouse uterus HiC are also shown as black arcs. A, Human IHH. The transcript is highlighted, regions with ESR1 peaks at 20 kb 5′ of the IHH transcription start site (TSS), at 70 kb 5′ of IHH, and at 100 kb 5′ of IHH are also highlighted. B, Mouse Ihh. Regions comparable to the human gene are highlighted as described in A. C, Human GREB1. The ESR1-binding super-enhancer region is highlighted. D, Mouse Greb1. ESR1-binding super-enhancer is highlighted. Orientation on the genome browser view is the opposite of the human GREB1.
Discussion
By combining cistromic and transcriptional data derived from endometrial biopsies or cultured epithelial organoids, we have shown details of estrogen-mediated response in human uterine tissue. Here, we provide the first description of the ESR1 cistrome of endometrial biopsies from healthy women. Our previous study described PGR ChIPseq analysis of endometrial biopsies and integrated it with RNAseq (40). Other published ESR1 ChIPseq studies used cultured endometrial stromal cells (63), uterine adenocarcinoma epithelial cell lines (32, 64, 65), endometrial tumors (33), or endometrial biopsies from infertile women (18). In these and other ESR1 ChIPseq analyses, most binding is observed either at distal enhancers or within genes, and motif analysis reveals that ERE is a highly enriched motif in ESR1 ChIPseq peaks from either stage ( (23, 24) see Fig. 1B and 1C). One novel finding we report is preferential enrichment of HOX motifs in the mid-secretory ESR1 peaks (see Fig. 1C). HOX proteins are transcription factors and chromatin modifiers involved in developmental patterning (66) that also facilitate stromal decidualization (67). Enrichment of HOX motifs was previously noted in mouse uterine ESR1 peaks that lacked ERE motifs (68). An ESR1-binding super-enhancer was described at the mouse Hoxd cluster (62) and is present in the proliferative human endometrium (not shown). Mouse models with disruption of Hoxa 9, 10, 11; Hoxc 9, 10, 11; and Hoxd 9, 10, 11 exhibit impaired uterine development and function (69, 70), and a mutation in HOXA11 was identified in a patient with a septate uterus (71). Together these observations reflect the importance of HOX factors to uterine biology.
bHLH motifs are enriched in proliferative and mid-secretory ESR1 peaks as well. HAND2 is a member of the bHLH family that modulates uterine stromal-epithelial signaling in mice (72). A recent study reported a single-nucleotide variation that interacts with HAND2 as a potential causal factor for preterm birth (73). Enrichment of motifs that potentially bind the bHLH factor HAND2 within ESR1 peaks, together with its essential role in uterine biology, hint at an ability to affect overlapping genes and processes.
The “tethering” (indirect interaction) mode of ESR1-mediated transcriptional responses was derived from studies indicating ESR1 can interact with AP1 transcription factors to drive an AP1-luciferase reporter gene (74). Findings based on in vitro model gene systems did not account for chromatin architecture; major advances in our general understanding of cell-specific gene regulation have highlighted the importance of modulation of chromatin accessibility, status of histone modifications, interaction between enhancers and promoters, and relative location within the nucleus (75). In this broader context, what appears as “tethering” in the simplified model likely indicates protein-protein interactions between ESR1 and other transcription factors (76) including bZIP (AP1), bHLH, and HOX motif-binding proteins, resulting in the enrichment of these motifs in the ESR1-binding sequences. Analysis of pathways affected by estrogen in organoids or in proliferative endometrium included multiple chromatin modifiers (see Tables 1-8), pointing to the importance of chromatin dynamics in estrogen response. Transcripts for the factors that could bind DNA motifs enriched in endometrial ESR1 peaks (HOX, bZIP/AP1, and bHLH) are detected in the endometrial RNAseq data (Supplementary Fig. S5a) (41), supporting their potential roles in estrogen signaling.
In organoids, ESR1 binding is more distal relative to genes than in the whole endometrium (see Fig. 1B. Whether this is a characteristic of epithelial cells or is a result of culturing isolated cells will be important to investigate. bZIP/AP1 motifs were enriched in organoid ESR1 ChIPseq peaks, and organoid RNAseq data sets indicate expression of several AP1 family members (Supplementary Fig. S5b) (41). Unlike the whole-endometrium ESR1 peaks, SOX motifs were enriched in organoids. Multiple SOX transcripts are expressed, as reflected by the RNAseq (Supplementary Fig. S5b) (41). A role for SOX in uterine epithelial cell function is supported by findings showing that SOX17 is detected in uterine epithelial cells both in mice and humans (61). In mice SOX17 binds to an enhancer 19 kb 5′ of Ihh that additionally binds multiple transcriptional regulators including ESR1 and PGR; this 19 kb enhancer is required for uterine Ihh expression (see Fig. 5B and [61]). bHLH motifs were highly enriched in proliferative and mid-secretory endometrium, but not in organoids (see Figs. 1C and 3C), consistent with the expression of HAND2 in uterine stromal cells (77). HOX motifs were not enriched in organoid ESR1 peaks and were the most significantly enriched motifs in mid-secretory endometrium, also consistent with the elevated expression of HOXA10 in mid-secretory stromal cells (78, 79).
The organoid ESR1 cistrome is more like proliferative than the mid-secretory endometrium ESR1 cistrome (see Fig. 4), and while estrogen-stimulated transcriptional profiles of organoids compared to proliferative endometrium indicate some signaling processes are conserved, others differ. One limitation of the analysis is that the whole-endometrium data rely on biological samples from women at proliferative and mid-secretory phases (estrogen dominant or progesterone dominant), which is not directly comparable to organoids treated with vehicle or estrogen. Genes that differ between the endometrial phases include ESR1 targets, but will also be influenced by progesterone, which inhibits estrogen activity in the mid-secretory phase. Enrichment of estrogen signaling activation both in the endometrial analysis and in organoids (see Tables 9 and 11) indicates that the approach does capture estrogen targets in both systems and supports the use of the organoid model to reflect endometrial epithelial-cell estrogen response.
Table 11.
Signaling pathways (overlap P value < .05) enriched in proliferative vs mid-secretory endometrial and estrogen regulated organoid genes that are less than 100 kb from estrogen receptor α chromatin immunoprecipitation and next-generation sequencing peaks
| Activation z score | ||||
|---|---|---|---|---|
| Signaling pathway | Pro/MS | Donor 1 | Donor 2 | Process |
| HOTAIR | 2.24 | 2.45 | 1 | Chromatin |
| Tumor microenvironment | 1.81 | 2.12 | 0.45 | Cancer |
| ESR | 0.63 | 1.27 | 2.12 | Estrogen |
Abbreviations: ESR, estrogen receptor α; HOTAIR, HOX transcript antisense RNA; MS, mid-secretory; Pro, proliferative.
In both whole endometrium and in organoids, candidate ESR1 target signaling affects chromatin remodeling–associated processes (see Tables 1-11), including HOTAIR, a long noncoding RNA encoded in the noncoding strand of the HOXC cluster. HOTAIR mediates epigenetic changes that repress genes by shuttling PRC2 and LSD1 to their targets (80). In multiple cancers HOTAIR promotes tumor growth, metastasis, migration, and epithelial mesenchymal transition (80-82). HOTAIR is increased by estrogen in breast cancer cells (83), is associated with poor prognosis in endometrial cancer (84), and mediates endometrial cancer cell proliferation and invasion (85). Further investigation of estrogen regulation of this pathway in normal endometrium will be the focus of future work.
FOX01 signaling has been studied in mouse models, demonstrating expression in uterine epithelial cells and importance in uterine function (61, 86, 87), and shows differences in enrichment between the epithelial cell organoids and the whole endometrium (see Table 9), highlighting the ability of the organoid model to reveal epithelial processes that are masked by the responses occurring in other endometrial cell types of whole-endometrial samples. Our analysis also indicates estrogen affects lipid and fatty acid metabolism, as ACOX1, a fatty acid oxidase, is activated and concentration of lipid is inhibited (see Tables 9 and 10). Lipid levels are vital for uterine receptivity and fetal development, and as sources for steroid hormone synthesis (88). Hormones affect lipid metabolism in mouse uterine epithelial cells (89); however, the mechanisms and biological importance of estrogen/ESR1 regulation of endometrial lipid has not yet been well characterized.
Overall, our study provides novel details of ESR1 chromatin interactions underlying estrogen responses in the endometrium that can be further studied using epithelial cells cultured in organoids. We detect processes in the organoids that mirror those of the proliferative endometrium, supporting their usefulness as a model to study estrogen responses intrinsic to epithelial cells.
Acknowledgments
We are grateful to Jana Philips for coordinating endometrial sample donations for the studies.
Glossary
Abbreviations
- AGT
angiotensin
- ANOVA
analysis of variance
- CEBPB
CCAAT enhancer binding protein β
- CDKN2A
cyclin-dependent kinase inhibitor 2A
- ChIPseq
chromatin immunoprecipitation and next-generation sequencing
- CKAP2L
, cytoskeleton-associated protein 2 like
- DEG
differentially expressed gene
- DES
diethylstilbestrol
- DMEM
Dulbecco’s modified Eagle’s medium
- E2
estradiol
- EHMT1
euchromatic histone lysine methyltransferase 1
- EP400
E1A binding protein p400
- ERBB2
Erb-b2 receptor tyrosine kinase 2
- ERE
estrogen-responsive enhancer
- ESR1
estrogen receptor α
- FDR
false discovery rate
- FGF1
fibroblast growth factor 1
- FOXM1
forkhead box M1
- HNF4A
hepatocyte nuclear factor 4 α
- HOTAIR
HOX transcript antisense RNA
- HOX
homeobox factor
- IFNG
interferon γ
- IL1B
interleukin 1B
- IL6
interleukin 6
- IPA
Ingenuity Pathway Analysis
- MAPK1
mitogen-activated protein kinase 1
- MAX
maximum RPKM value from any sample for each gene
- NIEHS
National Institute of Environmental Health Sciences
- NUPR1
nuclear protein 1, transcriptional regulator
- PDGF BB
platelet-derived growth factor BB
- PGR
progesterone receptor
- PTEN
phosphatase and tensin homolog
- RABL6
RAS oncogene family like 6
- RARA
retinoic acid receptor a
- RNAseq
RNA sequencing
- RPKM
reads per kilobase per million reads
- RTK
receptor tyrosine kinase
- RT-PCR
reverse-transcription polymerase chain reaction
- SMARCB1
SWI/SNF component SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily b, member 1
- SOX4
SRY-box transcription factor 4
- TBX2
T-box transcription factor 2
- TGFβ
transforming growth factor β
- TP53
tumor protein 53
- TSS
transcription start site
- UTR
untranslated region
- V
vehicle
- VEGFB
vascular endothelial growth factor B
Contributor Information
Sylvia C Hewitt, Pregnancy & Female Reproduction, RDBL, NIEHS, Research Triangle Park, North Carolina 27709, USA.
San-pin Wu, Pregnancy & Female Reproduction, RDBL, NIEHS, Research Triangle Park, North Carolina 27709, USA.
Tianyuan Wang, Integrative Bioinformatics Support Group, NIEHS, Research Triangle Park, North Carolina 27709, USA.
Madhumita Ray, Pregnancy & Female Reproduction, RDBL, NIEHS, Research Triangle Park, North Carolina 27709, USA.
Marja Brolinson, Program in Reproductive and Adult Endocrinology, NICHD, Bethesda, Maryland 20847, USA.
Steven L Young, Department of Obstetrics and Gynecology, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.
Thomas E Spencer, Department of Obstetrics, Gynecology and Women’s Health, University of Missouri, Columbia, Missouri 65211, USA.
Alan DeCherney, Program in Reproductive and Adult Endocrinology, NICHD, Bethesda, Maryland 20847, USA.
Francesco J DeMayo, Pregnancy & Female Reproduction, RDBL, NIEHS, Research Triangle Park, North Carolina 27709, USA.
Financial Support
This work was supported by the Intramural Research Program of the National Institutes of Health (Project Z1AES103311-01 to F.J.D. and Z1AHD008926 to A.D.) as well as the National Institutes of Health (P01 HD106485 to S.L.Y. and 1R01HD096266-04 to T.E.S.).
Disclosures
The authors have nothing to disclose.
Data Availability
Original data generated and analyzed during this study are included in this published article or are available from GEO, as detailed in “Materials and Methods” and “Results.”
References
- 1. Rodriguez AC, Blanchard Z, Maurer KA, Gertz J. Estrogen signaling in endometrial cancer: a key oncogenic pathway with several open questions. Horm Cancer. 2019;10(2-3):51-63. doi: 10.1007/s12672-019-0358-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chantalat E, Valera MC, Vaysse C, et al. . Estrogen receptors and endometriosis. Int J Mol Sci. 2020;21(8):2815. doi: 10.3390/ijms21082815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mazur EC, Large MJ, DeMayo FJ. Chapter 24 - Human oviduct and endometrium: changes over the menstrual cycle. In: Plant TM, Zeleznik AJ, eds. Knobil and Neill’s Physiology of Reproduction. 4th ed. Academic Press; 2015:1077-1097. doi: 10.3390/ijms21082815 [DOI] [Google Scholar]
- 4. Roy A, Matzuk MM. Reproductive tract function and dysfunction in women. Nat Rev Endocrinol. 2011;7(9):517-525. doi: 10.1038/nrendo.2011.79 [DOI] [PubMed] [Google Scholar]
- 5. Hawkins SM, Matzuk MM. The menstrual cycle: basic biology. Ann N Y Acad Sci. 2008;1135:10-18. doi: 10.1196/annals.1429.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ben-Baruch G, Menczer J, Mashiach S, Serr DM. Uterine anomalies in diethylstilbestrol-exposed women with fertility disorders. Acta Obstet Gynecol Scand. 1981;60(4):395-397. doi: 10.3109/00016348109154132 [DOI] [PubMed] [Google Scholar]
- 7. Office of Cancer Communications, National Cancer Institute, National Institutes of Health, Department of Health, Education, and Welfare. Exposure in utero to diethylstilbestrol and related synthetic hormones. Association with vaginal and cervical cancers and other abnormalities. JAMA. 1976;236(10):1107-1109. doi: 10.1001/jama.1976.03270110011002 [DOI] [PubMed] [Google Scholar]
- 8. Huo D, Anderson D, Palmer JR, Herbst AL. Incidence rates and risks of diethylstilbestrol-related clear-cell adenocarcinoma of the vagina and cervix: update after 40-year follow-up. Gynecol Oncol. 2017;146(3):566-571. doi: 10.1016/j.ygyno.2017.06.028 [DOI] [PubMed] [Google Scholar]
- 9. Hoover RN, Hyer M, Pfeiffer RM, et al. . Adverse health outcomes in women exposed in utero to diethylstilbestrol. N Engl J Med. 2011;365(14):1304-1314. doi: 10.1056/nejmoa1013961 [DOI] [PubMed] [Google Scholar]
- 10. Blakemore J, Naftolin F. Aromatase: contributions to physiology and disease in women and men. Physiology. 2016;31(4):258-269. doi: 10.1152/physiol.00054.2015 [DOI] [PubMed] [Google Scholar]
- 11. Quaynor SD, Stradtman EW Jr, Kim HG, et al. . Delayed puberty and estrogen resistance in a woman with estrogen receptor alpha variant. N Engl J Med. 2013;369(2):164-171. doi: 10.1056/NEJMoa1303611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bernard V, Kherra S, Francou B, et al. . Familial multiplicity of estrogen insensitivity associated with a loss-of-function ESR1 mutation. J Clin Endocrinol Metab. 2016;102(1):93-99. doi: 10.1210/jc.2016-2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iguchi T, Sato T, Nakajima T, Miyagawa S, Takasugi N. New frontiers of developmental endocrinology opened by researchers connecting irreversible effects of sex hormones on developing organs. Differentiation. 2021;118:4-23. doi: 10.1016/j.diff.2020.10.003 [DOI] [PubMed] [Google Scholar]
- 14. Delcour C, Khawaja N, Gonzalez-Duque S, et al. . Estrogen receptor alpha inactivation in 2 sisters: different phenotypic severities for the same pathogenic variant. J Clin Endocrinol Metab. 2022;107(6):e2553-e2562. doi: 10.1210/clinem/dgac065 [DOI] [PubMed] [Google Scholar]
- 15. Mills AM, Longacre TA. Endometrial hyperplasia. Semin Diagn Pathol. 2010;27(4):199-214. doi: 10.1053/j.semdp.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 16. Emons G, Mustea A, Tempfer C. Tamoxifen and endometrial cancer: a Janus-headed drug. Cancers (Basel). 2020;12(9):2535. doi: 10.3390/cancers12092535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liang Y, Xie H, Wu J, Liu D, Yao S. Villainous role of estrogen in macrophage-nerve interaction in endometriosis. Reprod Biol Endocrinol. 2018;16(1):122. doi: 10.1186/s12958-018-0441-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hawkins Bressler L, Fritz MA, Wu SP, et al. . Poor endometrial proliferation after clomiphene is associated with altered estrogen action. J Clin Endocrinol Metab. 2021;106(9):2547-2565. doi: 10.1210/clinem/dgab381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hewitt SC, Korach KS. Estrogen receptors: new directions in the new millennium. Endocr Rev. 2018;39(5):664-675. doi: 10.1210/er.2018-00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Binder AK, Winuthayanon W, Hewitt SC, Couse JF, Korach KS. Steroid receptors in the uterus and ovary. In: Plant TM, Zeleznik AJ, eds. Knobil and Neill’s Physiology of Reproduction. Academic Press; 2015:1099-1193. doi: 10.1016/b978-0-12-397175-3.00025-9 [DOI] [Google Scholar]
- 21. Luo J, Liu D. Does GPER really function as a G protein-coupled estrogen receptor in vivo? Front Endocrinol (Lausanne). 2020;11:148. doi: 10.3389/fendo.2020.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25(21):2227-2241. doi: 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mayayo-Peralta I, Prekovic S, Zwart W. Estrogen receptor on the move: cistromic plasticity and its implications in breast cancer. Mol Aspects Med. 2021;78:100939. doi: 10.1016/j.mam.2020.100939 [DOI] [PubMed] [Google Scholar]
- 24. Droog M, Mensink M, Zwart W. The estrogen receptor α-cistrome beyond breast cancer. Mol Endocrinol. 2016;30(10):1046-1058. doi: 10.1210/me.2016-1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nat Med. 2012;18(12):1754-1767. doi: 10.1038/nm.3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heremans R, Jan Z, Timmerman D, Vankelecom H. Organoids of the female reproductive tract: innovative tools to study desired to unwelcome processes. Front Cell Dev Biol. 2021;9:867. doi: 10.3389/fcell.2021.661472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Turco MY, Gardner L, Hughes J, et al. . Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat Cell Biol. 2017;19(5):568-577. doi: 10.1038/ncb3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haider S, Gamperl M, Burkard TR, et al. . Estrogen signaling drives ciliogenesis in human endometrial organoids. Endocrinology. 2019;160(10):2282-2297. doi: 10.1210/en.2019-00314 [DOI] [PubMed] [Google Scholar]
- 29. Fitzgerald HC, Dhakal P, Behura SK, Schust DJ, Spencer TE. Self-renewing endometrial epithelial organoids of the human uterus. Proc Natl Acad Sci U S A. 2019;116(46):23132-23142. doi: 10.1073/pnas.1915389116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garcia-Alonso L, Handfield LF, Roberts K, et al. . Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat Genet. 2021;53(12):1698-1711. doi: 10.1038/s41588-021-00972-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Welboren WJ, van Driel MA, Janssen-Megens EM, et al. . ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28(10):1418-1428. doi: 10.1038/emboj.2009.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carleton JB, Berrett KC, Gertz J. Multiplex enhancer interference reveals collaborative control of gene regulation by estrogen receptor α-bound enhancers. Cell Syst. 2017;5(4):333-344.e5. doi: 10.1016/j.cels.2017.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Droog M, Nevedomskaya E, Dackus GM, et al. . Estrogen receptor α wields treatment-specific enhancers between morphologically similar endometrial tumors. Proc Natl Acad Sci U S A. 2017;114(8):E1316-E1325. doi: 10.1073/pnas.1615233114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Data from: RRID:AB_310305. https://scicrunch.org/resources/data/record/nif-0000-07730-1/AB_310305/resolver?q=AB_310305&l=AB_310305&i=rrid:ab_310305-1987644.
- 35. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Y, Liu T, Meyer CA, et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heinz S, Benner C, Spann N, et al. . Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576-589. doi: 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang W, Loganantharaj R, Schroeder B, Fargo D, Li L. PAVIS: a tool for peak annotation and visualization. Bioinformatics. 2013;29(23):3097-3099. doi: 10.1093/bioinformatics/btt520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat Struct Mol Biol. 2016;23(4):349-357. doi: 10.1038/nsmb.3180 [DOI] [PubMed] [Google Scholar]
- 40. Chi RPA, Wang TY, Adams N, et al. . Human endometrial transcriptome and progesterone receptor cistrome reveal important pathways and epithelial regulators. J Clin Endocrinol Metab. 2020;105(4):21. doi: 10.1210/clinem/dgz117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hewitt SC. Supplemental data for “The estrogen receptor α cistrome in human endometrium and epithelial organoids.” 2022. Uploaded on July 20, 2022. https://datadryad.org/stash/share/-2qdP20K9P8sZ3Bt_ IV587oByNJeMQ4OrpOAXl_Yp5Y
- 42. Hewitt SC, Deroo BJ, Hansen K, et al. . Estrogen receptor-dependent genomic responses in the uterus mirror the biphasic physiological response to estrogen. Mol Endocrinol. 2003;17(10):2070-2083. doi: 10.1210/me.2003-0146 [DOI] [PubMed] [Google Scholar]
- 43. Hewitt SC, Lierz SL, Garcia M, et al. . A distal super enhancer mediates estrogen-dependent mouse uterine-specific gene transcription of Igf1 (insulin-like growth factor 1). J Biol Chem. 2019;294(25):9746-9759. doi: 10.1074/jbc.RA119.008759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105-1111. doi: 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hewitt SC, Wu SP, Wang T, Young SL, Spencer TE, DeMayo FJ. Progesterone signaling in endometrial epithelial organoids. Cells. 2022;11(11):1760. doi: 10.3390/cells11111760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Balas MM, Johnson AM. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res. 2018;3(3):108-117. doi: 10.1016/j.ncrna.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brandmaier A, Hou SQ, Shen WH. Cell cycle control by PTEN. J Mol Biol. 2017;429(15):2265-2277. doi: 10.1016/j.jmb.2017.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tashiro H, Blazes MS, Wu R, et al. . Mutations in PTEN are frequent in endometrial carcinoma but rare in other common gynecological malignancies. Cancer Res. 1997;57(18):3935-3940. [PubMed] [Google Scholar]
- 49. Dimitriadis E, White CA, Jones RL, Salamonsen LA. Cytokines, chemokines and growth factors in endometrium related to implantation. Hum Reprod Update. 2005;11(6):613-630. doi: 10.1093/humupd/dmi023 [DOI] [PubMed] [Google Scholar]
- 50. Guzeloglu-Kayisli O, Kayisli UA, Taylor HS. The role of growth factors and cytokines during implantation: endocrine and paracrine interactions. Semin Reprod Med. 2009;27(1):62-79. doi: 10.1055/s-0028-1108011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Buharalioglu CK, Song CY, Yaghini FA, et al. . Angiotensin II-induced process of angiogenesis is mediated by spleen tyrosine kinase via VEGF receptor-1 phosphorylation. Am J Physiol Heart Circ Physiol. 2011;301(3):H1043-H1055. doi: 10.1152/ajpheart.01018.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Anton L, Merrill DC, Neves LAA, et al. . The uterine placental bed renin-angiotensin system in normal and preeclamptic pregnancy. Endocrinology. 2009;150(9):4316-4325. doi: 10.1210/en.2009-0076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Herr D, Bekes I, Wulff C. Local renin-angiotensin system in the reproductive system. Front Endocrinol (Lausanne). 2013;4:150. doi: 10.3389/fendo.2013.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lumbers ER, Delforce SJ, Arthurs AL, Pringle KG. Causes and consequences of the dysregulated maternal renin-angiotensin system in preeclampsia. Front Endocrinol (Lausanne). 2019;10:563. doi: 10.3389/fendo.2019.00563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang Y, Lumbers ER, Sykes SD, Pringle KG. Regulation of the renin-angiotensin system pathways in the human decidua. Reprod Sci. 2015;22(7):865-872. doi: 10.1177/1933719114565029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Boretto M, Maenhoudt N, Luo X, et al. . Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat Cell Biol. 2019;21(8):1041-1051. doi: 10.1038/s41556-019-0360-z [DOI] [PubMed] [Google Scholar]
- 57. Berthois Y, Katzenellenbogen JA, Katzenellenbogen BS. Phenol red in tissue culture media is a weak estrogen: implications concerning the study of estrogen-responsive cells in culture. Proc Natl Acad Sci U S A. 1986;83(8):2496-2500. doi: 10.1073/pnas.83.8.2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pawar S, Starosvetsky E, Orvis GD, Behringer RR, Bagchi IC, Bagchi MK. STAT3 regulates uterine epithelial remodeling and epithelial-stromal crosstalk during implantation. Mol Endocrinol. 2013;27(12):1996-2012. doi: 10.1210/me.2013-1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Eritja N, Felip I, Dosil MA, et al. . A Smad3-PTEN regulatory loop controls proliferation and apoptotic responses to TGF-β in mouse endometrium. Cell Death Differ. 2017;24(8):1443-1458. doi: 10.1038/cdd.2017.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cha J, Bartos A, Park C, et al. . Appropriate crypt formation in the uterus for embryo homing and implantation requires Wnt5a-ROR signaling. Cell Rep. 2014;8(2):382-392. doi: 10.1016/j.celrep.2014.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang X, Li X, Wang T, et al. . SOX17 regulates uterine epithelial-stromal cross-talk acting via a distal enhancer upstream of Ihh. Nat Commun. 2018;9(1):4421. doi: 10.1038/s41467-018-06652-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hewitt SC, Grimm SA, Wu SP, DeMayo FJ, Korach KS. Estrogen receptor α (ERα)-binding super-enhancers drive key mediators that control uterine estrogen responses in mice. J Biol Chem. 2020;295(25):8387-8400. doi: 10.1074/jbc.RA120.013666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yilmaz BD, Sison CAM, Yildiz S, et al. . Genome-wide estrogen receptor-α binding and action in human endometrial stromal cells. F S Sci. 2020;1(1):59-66. doi: 10.1093/humupd/dmz005 [DOI] [PubMed] [Google Scholar]
- 64. Gertz J, Savic D, Varley KE, et al. . Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell. 2013;52(1):25-36. doi: 10.1016/j.molcel.2013.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. La Greca A, Bellora N, Le Dily F, et al. . Chromatin topology defines estradiol-primed progesterone receptor and PAX2 binding in endometrial cancer cells. Elife. 2022;11:e66034. doi: 10.7554/eLife.66034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Carnesecchi J, Pinto PB, Lohmann I. Hox transcription factors: an overview of multi-step regulators of gene expression. Int J Dev Biol. 2018;62(11-12):723-732. doi: 10.1387/ijdb.180294il. [DOI] [PubMed] [Google Scholar]
- 67. Du H, Taylor HS. The role of Hox genes in female reproductive tract development, adult function, and fertility. Cold Spring Harb Perspect Med. 2015;6(1):a023002. doi: 10.1101/cshperspect.a023002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hewitt SC, Li L, Grimm SA, et al. . Research resource: whole-genome estrogen receptor α binding in mouse uterine tissue revealed by ChIP-seq. Mol Endocrinol. 2012;26(5):887-898. doi: 10.1210/me.2011-1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mucenski ML, Mahoney R, Adam M, Potter AS, Potter SS. Single cell RNA-seq study of wild type and Hox9,10,11 mutant developing uterus. Sci Rep. 2019;9(1):4557. doi: 10.1038/s41598-019-40923-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Raines AM, Adam M, Magella B, et al. . Recombineering-based dissection of flanking and paralogous Hox gene functions in mouse reproductive tracts. Development. 2013;140(14):2942-2952. doi: 10.1242/dev.092569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhu Y, Cheng Z, Wang J, et al. . A novel mutation of HOXA11 in a patient with septate uterus. Orphanet J Rare Dis. 2017;12(1):178. doi: 10.1186/s13023-017-0727-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li Q, Kannan A, DeMayo FJ, et al. . The antiproliferative action of progesterone in uterine epithelium is mediated by Hand2. Science. 2011;331(6019):912-916. doi: 10.1126/science.1197454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sakabe NJ, Aneas I, Knoblauch N, et al. . Transcriptome and regulatory maps of decidua-derived stromal cells inform gene discovery in preterm birth. Sci Adv. 2020;6(49):eabc8696. doi: 10.1126/sciadv.abc8696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kushner PJ, Agard DA, Greene GL, et al. . Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74(5):311-317. doi: 10.1016/s0960-0760(00)00108-4 [DOI] [PubMed] [Google Scholar]
- 75. Misteli T. The self-organizing genome: principles of genome architecture and function. Cell. 2020;183(1):28-45. doi: 10.1016/j.cell.2020.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vockley CM, McDowell IC, D’Ippolito AM, Reddy TE. A long-range flexible billboard model of gene activation. Transcription. 2017;8(4):261-267. doi: 10.1080/21541264.2017.1317694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Murata H, Tanaka S, Tsuzuki-Nakao T, et al. . The transcription factor HAND2 up-regulates transcription of the IL15 gene in human endometrial stromal cells. J Biol Chem. 2020;295(28):9596-9605. doi: 10.1074/jbc.ra120.012753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Daftary GS, Taylor HS. Endocrine regulation of HOX genes. Endocr Rev. 2006;27(4):331-355. doi: 10.1210/er.2005-0018 [DOI] [PubMed] [Google Scholar]
- 79. Modi D, Godbole G. HOXA10 signals on the highway through pregnancy. J Reprod Immunol. 2009;83(1-2):72-78. doi: 10.1016/j.jri.2009.07.009 [DOI] [PubMed] [Google Scholar]
- 80. Abba MC, Fabre ML, Lee J, Tatineni P, Kil H, Aldaz CM. HOTAIR modulated pathways in early-stage breast cancer progression. Front Oncol. 2021;11:783211. doi: 10.3389/fonc.2021.783211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xin X, Li Q, Fang J, Zhao T. LncRNA HOTAIR: a potential prognostic factor and therapeutic target in human cancers. Front Oncol. 2021;11:679244. doi: 10.3389/fonc.2021.679244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gupta RA, Shah N, Wang KC, et al. . Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071-1076. doi: 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol Biol. 2013;425(19):3707-3722. doi: 10.1016/j.jmb.2013.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. He X, Bao W, Li X, et al. . The long non-coding RNA HOTAIR is upregulated in endometrial carcinoma and correlates with poor prognosis. Int J Mol Med. 2014;33(2):325-332. doi: 10.3892/ijmm.2013.1570 [DOI] [PubMed] [Google Scholar]
- 85. Huang J, Ke P, Guo L, et al. . Lentivirus-mediated RNA interference targeting the long noncoding RNA HOTAIR inhibits proliferation and invasion of endometrial carcinoma cells in vitro and in vivo. Int J Gynecol Cancer. 2014;24(4):635-642. doi: 10.1097/IGC.0000000000000121 [DOI] [PubMed] [Google Scholar]
- 86. Adiguzel D, Sahin P, Kuscu N, Ozkavukcu S, Bektas NI, Celik-Ozenci C. Spatiotemporal expression and regulation of FoxO1 in mouse uterus during peri-implantation period. PLoS One. 2019;14(5):e0216814. doi: 10.1371/journal.pone.0216814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vasquez YM, Wang X, Wetendorf M, et al. . FOXO1 regulates uterine epithelial integrity and progesterone receptor expression critical for embryo implantation. PLoS Genet. 2018;14(11):e1007787. doi: 10.1371/journal.pgen.1007787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ye Q, Zeng X, Cai S, Qiao S, Zeng X. Mechanisms of lipid metabolism in uterine receptivity and embryo development. Trends Endocrinol Metab. 2021;32(12):1015-1030. doi: 10.1016/j.tem.2021.09.002 [DOI] [PubMed] [Google Scholar]
- 89. Stacey K, Beasley B, Wilce PA, Martin L. Effects of female sex hormones on lipid metabolism in the uterine epithelium of the mouse. Int J Biochem. 1991;23(3):371-376. doi: 10.1016/0020-711x(91)90121-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Original data generated and analyzed during this study are included in this published article or are available from GEO, as detailed in “Materials and Methods” and “Results.”