Abstract
The function of the extracellular domain (ECD) of Sln1p, a plasma membrane two-transmembrane domain (TMD) sensor of the high-osmolarity glycerol (HOG) response pathway, has been studied in the yeast Saccharomyces cerevisiae. Truncations of SLN1 that retain an intact kinase domain are capable of complementing the lethality of an sln1Δ strain. By observing levels of Hog1p phosphorylation as well as the phosphorylation state of Sln1p, the kinase activities of various SLN1 constructions were determined. In derivatives that do not contain the first TMD, Sln1p activity was no longer dependent on medium osmolarity but appeared to be constitutively active even under conditions of high osmolarity. Removal of the first TMD (ΔTMD1 construct) gave a protein that was strongly phosphorylated whereas Hog1p was largely dephosphorylated, as expected if the active form of Sln1p is phosphorylated. When both TMDs as well as the ECD were deleted, so that the kinase domain is cytosolic, Sln1p was not phosphorylated whereas Hog1p became constitutively hyperphosphorylated. Surprisingly, this hyperactivity of the HOG mitogen-activated protein kinase signaling pathway was not sufficient to result in cell lethality. When the ECD of the ΔTMD1 construct was replaced with a leucine zipper motif, Sln1p was hyperactive, so that Hog1p became mostly unphosphorylated. In contrast, when the Sln1p/leucine zipper construct was crippled by a mutation of one of the internal leucines, the Sln1 kinase was inactive. These experiments are consistent with the hypothesis that the ECD of Sln1p functions as a dimerization and activation domain but that osmotic regulation of activity requires the presence of the first TMD.
The yeast Saccharomyces cerevisiae responds to high external osmolar conditions by increasing the intracellular concentration of glycerol (26). The high-osmolarity glycerol (HOG) pathway in yeast consists of a double two-component phosphorelay cascade coupled to a mitogen-activated protein kinase (MAPK) cascade. The SLN1 gene encodes an enzyme with histidine kinase and aspartate phosphotransferase activities and functions as a plasma membrane sensor. Under normal, low-osmolar conditions, Sln1p actively transfers a phosphate to Ypd1p, which in turn transfers a phosphate to Ssk1p (20). Phosphorylated Ssk1p inhibits the kinase cascade in the HOG MAPK pathway (13). Under high-osmolarity conditions, Sln1p is deactivated. The lack of phosphorelay through the two-component pathway causes inactivation of Ssk1p and activation of the HOG MAPK pathway. The MAPKKKs Ssk2p and Ssk2p are kinases which phosphorylate the MAPKK Pbs2p, which phosphorylates the MAPK Hog1p. Phosphorylated Hog1p has been shown to activate transcription factors that increase the production of enzymes involved in glycerol synthesis and stress response (23).
SLN1 was originally identified as an allele that is synthetically lethal with ubr1Δ, encoding the recognition component of the N-end-rule ubiquitin system (17). The predicted protein sequence is 1,220 amino acids in length and possesses homology to both the sensor histidine autophosphorylation and response regulator proteins of bacterial two-component systems. The histidine and aspartate phosphorylation sites are residues 576 and 1144, respectively (13), encoded by SLN1H576 and SLN1D1144. Sln1p is also predicted to possess two transmembrane domains (TMDs) like some bacterial sensor kinases. The autophosphorylation of histidine residues in the bacterial sensor kinases is believed to occur in trans. That is, bacterial histidine kinases are thought to require dimerization of the protein (29). The homology of Sln1p with the bacterial proteins predicts that Sln1p may also require dimerization for autophosphorylation.
It is known that Sln1p can transfer a phosphate group from the sensor to the receiver domain in trans (13). Neither SLN1H576Q nor SLN1D1144N complements sln1Δ. Complementation is observed, however, when the mutant proteins are coexpressed, demonstrating that an intermolecular transfer of the phosphate group can occur. Likewise, the isolated histidine kinase region (amino acids 450 to 1070) has been observed to transfer a phosphate group to the isolated receiver domain region (amino acids 1059 to 1220) (20). Phosphotransfer was observed between the two protein fragments but not with fragments possessing mutations in either His576 or Asp1144. Although these experiments demonstrate that Sln1p phosphate transfer can occur in trans, they do not show whether Sln1p histidine autophosphorylation and kinase activation normally occur by an intermolecular or intramolecular interaction.
We undertook to address the question of whether Sln1p requires dimerization for histidine autophosphorylation to occur. We also attempted to delimit the structural region of Sln1p that may function as a dimerization domain. These studies also defined regions that are essential for other functions of the protein such as the osmolarity response. Together the results suggest some generalized structure-function relationships for this two-component sensor kinase.
MATERIALS AND METHODS
Strains and plasmids.
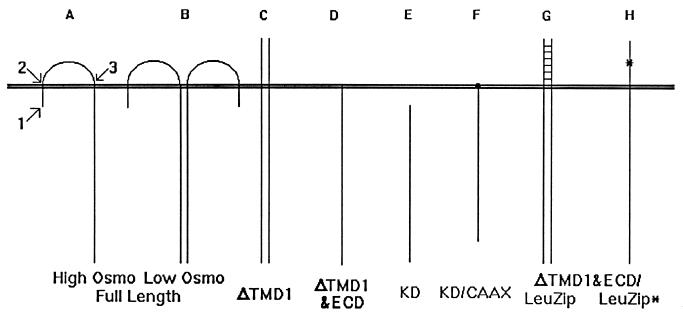
The SLN1/sln1 heterozygous strain (his3-Δ200/his3-Δ200 leu2-3,112/leu2-3,112 lys2-801/lys2-801 SLN1/sln1Δ::HIS3 trp1-1/trp1-1 ura3-52/ura3-52) was provided by I. Ota (17). The SLN1 expression vector was created as follows. The carboxyl-terminal region of SLN1 was isolated from a vector provided by I. Ota (17) and inserted into both pDO105 (high copy) (16) and pDO120 (low copy) (18), using the same restriction sites. The amino-terminal domain was cloned from total genomic DNA of strain Y294 (5) by PCR to add a restriction site just upstream of the initiation codon (Fig. 1A, arrow 1).
FIG. 1.
Schematic representations of Sln1p constructions (see Table 1).
Site-directed mutagenesis (SDM) was performed with the QuikChange mutagenesis system (Stratagene). SDM was accomplished with two oligonucleotides, the first adding a restriction site just after the first TMD (TMD1) (Fig. 1A, arrow 2) and the second adding a restriction site just before TMD2 (Fig. 1A, arrow 3). The latter mutation is silent, while the former creates a conservative amino acid substitution (N49S). The mutagenized amino-terminal region of SLN1 was isolated and added to the vector containing the carboxyl-terminal region to create plasmid pDO108. This construct will be referred to as full length (Table 1).
TABLE 1.
Relative levels of Sln1p and Hog1p phosphorylation with different Sln1p constructs
| Sln1p constructa | Sln1p aaΔb | kDac | sln1Δ compd | Phosphorylatione
|
|
|---|---|---|---|---|---|
| Sln1p | Hog1p | ||||
| Full length/low osmolarity | NAf | 134 | + | + | Low |
| Full length/high osmolarity | NA | 134 | + | − | High |
| ΔTMD1 | 1–49 | 130 | + | + | Low |
| ΔTMD1&ECD | 1–330 | 99 | + | NDg | High |
| KD | 1–438 | 87 | + | − | High |
| ΔKD | 543–1220 | 59 | − | NA | NA |
| KD/CAAX | 1–438 | 88 | + | ND | High |
| ΔECD/LeuZip | 50–329 | 105 | + | ND | High |
| ΔTMD1&ECD/LeuZip | 1–329 | 105 | + | + | Low |
| ΔTMD1&ECD/LeuZip* | 1–329 | 105 | + | − | High |
For details of construction, see Materials and Methods.
Amino acids of wild-type Sln1p deleted in construct.
Predicted molecular mass of construct.
Functional complementation of sln1 null mutant as in Fig. 2.
Relative phosphorylation level determined as in Fig. 5 and 6 (for Hog1p) or Fig. 7 (for Sln1p). Note that results are the same in low- and high-osmolarity media for all constructs with the exception of the full-length construct. Results are also the same with both low- and high-copy-number vectors.
NA, not applicable.
ND, not determined.
To create the ΔTMD1 construct (Table 1), pDO108 was cut with restriction enzymes deleting the amino-terminal intracellular region and TMD1, and annealed oligonucleotides were added. The new amino-terminal coding sequence includes the STE3 signal sequence, MSYKS (7), to direct the protein into the secretory pathway. The ΔTMD1&ECD plasmid was created in the same manner, adding annealed oligonucleotides, which again adds the STE3 signal sequence to the amino terminus. The kinase domain expression plasmid (Table 1, KD) apparently uses Met439 as initiator, as judged by the apparent molecular weight of the protein product (not shown).
To obtain a noncomplementing SLN1 allele, pDO108 was cut with a single restriction enzyme just upstream of the kinase domain, treated with Klenow fragment, and reclosed. The additional four base pairs cause premature termination of the reading frame upstream of the kinase domain (Table 1, ΔKD). To add a CAAX box plasma membrane localization signal to the carboxyl terminus of the SLN1 kinase domain (Table 1, KD/CAAX), the carboxyl terminus of SLN1 was isolated on a plasmid. The vector was then mutagenized with oligonucleotides, adding a silent restriction site near the end of the open reading frame. This vector was cut, and annealed oligonucleotides were inserted. The new sequence adds the amino acids CIIS of the RAS2 gene (21). The modified carboxyl terminus was subsequently isolated and reinserted into pDO108.
To replace the extracellular domain (ECD) of SLN1 with a leucine zipper sequence (Table 1, ΔECD/LeuZip), the relevant sequence from a C/EBP (CCAAT element-binding protein) clone (12) was isolated by PCR and inserted into pDO108. To make the construction single transmembrane (ΔTMD1&ECD/LeuZip), this vector was cut and the same STE3 signal sequence oligonucleotides described above for the ΔTMD1 construct were added. The undimerizable leucine zipper derivative was created by SDM of the leucine zipper fragment described above. This mutagenesis alters the central leucine to a proline. The undimerizable SLN1ΔTMD1&ECD/LeuZip* construct was then created as described above.
To add the FLAG epitope tag to various SLN1 constructions, the mutagenized carboxyl-terminal domain plasmid with the silent restriction site near the end of the open reading frame was used. The plasmid was cut and annealed oligonucleotides were ligated, adding sequence containing the DYKDDDDK FLAG sequence (IBI). The carboxyl-terminal domain of SLN1 in pDO108 was then replaced with the FLAG sequence, and all constructs were recreated as previously described. Immunoblotting and immunoprecipitation used anti-FLAG monoclonal antibodies (Kodak).
To disrupt the SHO1 gene in this strain, we amplified 350- and 500-bp fragments of the SHO1 5′ and 3′ nontranslated regions, respectively, and placed them on either side of a hisG/URA3/hisG cassette (1). This construct was used to disrupt the SHO1 gene in a sln1Δ haploid strain, and selection on 5-fluoorotic acid isolated those cells which had recombined at the hisG repeats and had recovered uracil auxotrophy.
Antisera.
To obtain antisera to the carboxyl-terminal region of Sln1p, a DNA fragment encoding amino acids 494 to 1220 was excised from pDO108. The fragment was ligated into vector pMAL-C2 (New England Biolabs) to create a malE/SLN1 fusion. Isopropyl-β-d-thiogalactopyranoside (IPTG) induction of this plasmid in Escherichia coli produced large amounts of a 124-kDa fusion protein. A cell extract was prepared, and the fusion protein was isolated on an amylose column and eluted with maltose. The Sln1p fusion protein was purified by preparative electrophoresis, electroeluted, dialyzed, and concentrated. The protein was injected into rabbits, and antisera were isolated as described previously (15). Protein gel analysis of various SLN1 constructions used 7.5 to 15% gradient acrylamide gels with a vertical slab gel unit (Hoefer).
The HOG1 open reading frame was cloned from total genomic Y294 (5) DNA by PCR. The product was ligated into pET23B (Novagen) to create a His6-tagged Hog1p. IPTG induction of this plasmid in E. coli produced a 45-kDa product. The protein was isolated on a Ni-nitrilotriacetic acid agarose column (Qiagen) and used to create antisera as described above.
Hog1p phosphorylation assay.
Strains were grown in YPD (rich) or SD (defined) medium (10) at 28°C. For high-osmolarity conditions, sorbitol or NaCl was added to the cultures to a final concentration of 1 or 0.3 M, respectively. The cultures were incubated in high-osmolarity medium for 10 min for low-copy-number constructs or 12 h for high-copy-number constructs unless otherwise indicated. Subsequent steps were performed at 5°C. Mid-log-phase cultures (2 × 107 to 3 × 107 cells/ml) were harvested by centrifugation. Cells were broken in protein isolation buffer (25 mM HEPES [pH 7.4], 10% glycerol, 1 mM Na2EDTA) with protease inhibitors (87 μg of phenylmethylsulfonyl fluoride, 2.5 μg of Nα-p-tosyl-l-lysine chloromethyl ketone, 0.75 μg of pepstatin A, 0.5 μg of leupeptin, and 0.5 μg of aprotinin per ml) with 500 μm-diameter acid-washed glass beads in a Mini-Bead Beater (BioSpec Products). In some cases, a phosphatase inhibitor cocktail (10 mM NaF, 5 mM β-glycerol phosphate, 1 mM sodium vanadate) was added to the breakage buffer. The extracts were subjected to centrifugation for 1 h at 100,000 × g in a TL-100 ultracentrifuge (Beckman), and the supernatant was collected in order to isolate the cytosolic fraction. Aliquots containing 100 μg of cytosolic protein were used for immunoprecipitation (22) with 10 μl of anti-Hog1p antisera and 5 mg of protein A-Sepharose CL-4B (Pharmacia). To remove the immunoglobulin G from the immunoprecipitate, the final washed pellets were resuspended in 1% sodium dodecyl sulfate (SDS) and centrifuged (8). Supernatants were collected and placed in SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer.
Hog1p immunoblot analyses used 10% Ready Gels in the Mini-Protean cell system (Bio-Rad) which were blotted to polyvinylidene difluoride membranes in a Mini-V8 apparatus (Life Technologies). Immunoblot analyses used the PhotoBlot chemiluminescence detection system (Gibco-BRL). Rabbit polyclonal antiphosphotyrosine antisera was obtained from Transduction Laboratories. The relative levels of Hog1p phosphorylation were normalized to amounts of Hog1p immunoprecipitated by scanning densitometry of a duplicate immunoblot that used anti-Hog1p antisera. A comparison of extracts prepared with or without addition of phosphatase inhibitors demonstrated that there was no significant difference in the levels of Hog1p phosphorylation found.
Cell fractionation and membrane localization.
Cells were grown and harvested as described above. Cells were broken with glass beads in membrane isolation buffer (300 mM sucrose, 50 mM Tris-HCl [pH 7.5], 10 mM β-mercaptoethanol, 1 mM Na2EDTA) with protease inhibitors at 5°C. Unbroken cells and glass beads were removed by centrifugation at 1,000 × g for 5 min. Membrane and cytosolic fractions were separated as before. Pellets (membrane fractions) were directly resuspended in SDS-PAGE loading buffer or were used for additional fractionation as described below, while supernatants (cytosol) were first concentrated with a Centricon-10 concentrator (Amicon) at 4,500 × g for 4 h at 5°C.
Plasma membranes were isolated as described previously (24), with modifications (14). Measurement of plasma membrane H+-ATPase activity (11) was used to assess the purity of this fraction. Vanadate-sensitive ATPase activity accounted for greater than 95% of the total ATPase activity in the plasma membrane fraction. Mitochondrial fractions were isolated from cell extracts by differential centrifugation (1,000 to 20,000 × g) (2). Measurement of mitochondrial ATPase activity was used to assess the purity of this fraction. Sodium azide-sensitive ATPase activity accounted for greater than 98% of the total ATPase activity in the mitochondrial membrane fraction. A crude microsomal membrane fraction was also isolated from cell extracts by differential centrifugation as previously described (27,000 to 100,000 × g) (6). All membrane fractions were suspended in membrane isolation buffer to a volume equivalent to that of the cytosolic fraction.
Sln1p phosphorylation assay.
Strains containing different FLAG-tagged SLN1 plasmids were labeled with 32Pi (375 μCi/ml) in low-phosphate medium (27) with or without 1 M sorbitol. Cells were harvested, broken in radioimmunoprecipitation assay buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris [pH 8.0]) (8) with protease inhibitors (see above) and phosphatase inhibitors (10 mM NaF, 5 mM β-glycerophosphate, and 1 mM sodium vanadate), and total membrane and cytosolic fractions were separated as described above. Sln1p was isolated from both fractions by immunoprecipitation with FLAG monoclonal antibodies (Kodak). The immunoprecipitates were separated on polyacrylamide gels in duplicate. One gel was transferred to membranes for immunoblot analysis with anti-Sln1p antisera to assess total immunoprecipitated protein levels, while the second was exposed to X-ray film at −80°C for 2 days with an intensifying screen to assess phosphorylation levels.
RESULTS AND DISCUSSION
SLN1 mutagenesis.
To test the in vivo effects of different structural alterations in Sln1p, three restriction enzyme sites were introduced into SLN1 by SDM (Fig. 1A): one immediately upstream of the initiator ATG (arrow 1), a second immediately downstream of TMD1 (arrow 2), and a third immediately upstream of TMD2 (arrow 3). This mutated copy of SLN1 was cloned behind the ADH1 promoter on both low- and high-copy-number shuttle plasmids. We used the ADH1 promoter to obviate any potential expression regulatory effects that might be caused by the nascent SLN1 promoter in these constructs.
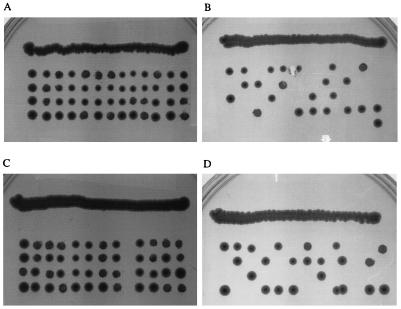
A SLN1/sln1Δ heterozygous strain was used to test the ability of the mutagenized SLN1 plasmid to complement sln1Δ. The strain containing this plasmid was sporulated, and tetrads were dissected. Because an SLN1 null mutation is lethal in this strain, complementation could be tested by growth of the spores. Four viable colonies were obtained with the mutagenized SLN1 plasmid with every tetrad (Fig. 2A), while only two were found with the parent vector (Fig. 2B). The restriction site mutations incorporated into SLN1 therefore do not affect complementation of sln1Δ (Table 1). No difference was detected in the growth rates of strains containing mutagenized or wild-type SLN1 in either low- or high-osmolarity medium. These data were observed with both low- and high-copy-number SLN1-containing vectors.
FIG. 2.
SLN1/sln1Δ tetrad dissections with various SLN1 expression constructs. (A) Full-length SLN1 with mutations incorporating restriction enzyme sites; (B) parental vector control; (C) SLN1KD (Δ1-438); (D) SLN1ΔKD (Δ543-1220). These results were obtained with both low- and high-copy-number SLN1-containing vectors.
SLN1 deletion studies.
As a first step to understanding the relationship between the structure of Sln1p and its function in cells, various structural deletion constructs were tested for the ability to complement the lethal sln1Δ phenotype. The restriction sites that were introduced into the gene were used to remove sequence corresponding to the short amino-terminal intracellular region and TMD1. This construct, ΔTMD1, possesses a deletion of the first 49 amino acids of Sln1p (Table 1). In hopes of correctly orienting the truncated protein in membranes, the STE3 signal sequence (7) was added to the amino terminus of the construction. The amino terminus should therefore be extracellular, with only a single TMD (Fig. 1C). This plasmid was used to transform the SLN1/sln1Δ strain, which was subsequently sporulated and subjected to tetrad dissection to test the ability of the deletion to complement sln1Δ. The construct was able to complement the mutation (Table 1). These data were observed with both low- and high-copy-number SLN1-containing vectors.
Continuing the deletion of the structural domains of Sln1p, the ECD was deleted from the ΔTMD1 construct (Fig. 1D). This construct, ΔTMD1&ECD, possesses a deletion of the first 330 amino acids of Sln1p (Table 1). Subsequently, the second TMD was deleted to yield a kinase domain construct (KD) possessing a deletion of the first 438 amino acids of Sln1p (Fig. 1E). The KD construct did not contain the secretion signal sequence used with the other truncation mutants. Surprisingly, both of these deletion protein constructions complemented sln1Δ (Fig. 2C; Table 1).
As a negative control, a mutation that retained all the membrane structural elements but deleted the kinase domain (ΔKD; amino acids 543 to 1120) (Fig. 2D) was used. Complementation was not observed (Table 1).
Subcellular localization.
To observe the localization of the different Sln1p truncations, rabbit polyclonal antibodies were prepared against the carboxyl-terminal region of Sln1p that contains the kinase domain. The antisera were used to show that the constitutively expressed full-length mutant SLN1p was present in approximately fivefold excess over wild-type levels (not shown). No difference in expression level was observed between the full-length construct and any of the deletion constructs. This result suggests that the deletion constructs are overexpressed to the same extent and are as stable as the full-length construct in cells. The sizes of the different truncated proteins as determined by gel migration were as predicted (not shown).
Crude membrane preparations were separated from cytosol by centrifugation and analyzed for the presence of the SLN1 constructs by immunoblot analysis (not shown). The full-length, ΔTMD1, ΔTMD1&ECD, and ΔKD constructs were all found by immunoblot analysis to be exclusively associated with the membrane fraction. The KD construct, which possesses no membrane structural components or localization signals, was found to be completely cytosolic.
Membrane localization experiments were used to determine the organelles in which the truncated proteins were present. Crude membrane preparations were separated by differential centrifugation and analyzed for purity by ATPase activity specific for each organellar membrane (see Materials and Methods). With the exception of the KD construct mentioned above, each showed a similar pattern of localization in that between 30 and 50% of the protein was specifically associated with the plasma membrane, with the remainder associated with microsomes (endoplasmic reticulum, Golgi complex, and vesicles). Wild-type cells and strains expressing the truncation constructs from low-copy-number vectors show a higher percentage (∼80%) of Sln1p in the plasma membrane fraction (Fig. 3). No localization to mitochondrial membranes was observed (not shown).
FIG. 3.
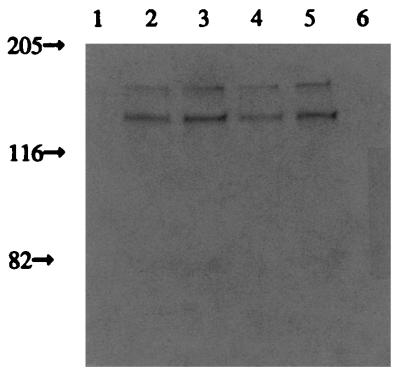
Sln1ΔTMD1p subcellular localization. The SLN1ΔTMD1-expressing strain was examined for Sln1p subcellular distribution by immunoblot analysis using anti-Sln1p antisera. Cells were grown to the mid-logarithmic phase of growth, harvested, and lysed, and membrane-associated and cytosolic proteins separated. The total membrane fraction was used to make a crude plasma membrane preparation by differential centrifugation. The purity of the plasma membrane fraction was tested by the ability of plasma membrane H+-ATPase activity to be specifically inhibited by vanadate (see Materials and Methods). The remaining membranes were termed the microsomal fraction. Lanes 1 to 3, high-copy-number vector; lanes 4 to 6, low-copy-number vector. Lanes 1 and 6, total cytosolic protein; lanes 2 and 5, plasma membrane fraction; lanes 3 and 4, microsomal fraction. Sizes here and in Fig. 5 to 7 are in kilodaltons.
Hog1p phosphorylation assay.
To observe the effect of the different Sln1p truncation mutants on activation of the HOG pathway, the relative levels of Hog1p phosphorylation were determined. To accomplish this, cytosolic protein extracts were used for immunoprecipitation of the Hog1 protein with anti-Hog1p antisera, separated by SDS-PAGE, and subjected to immunoblot analysis with antiphosphotyrosine antisera.
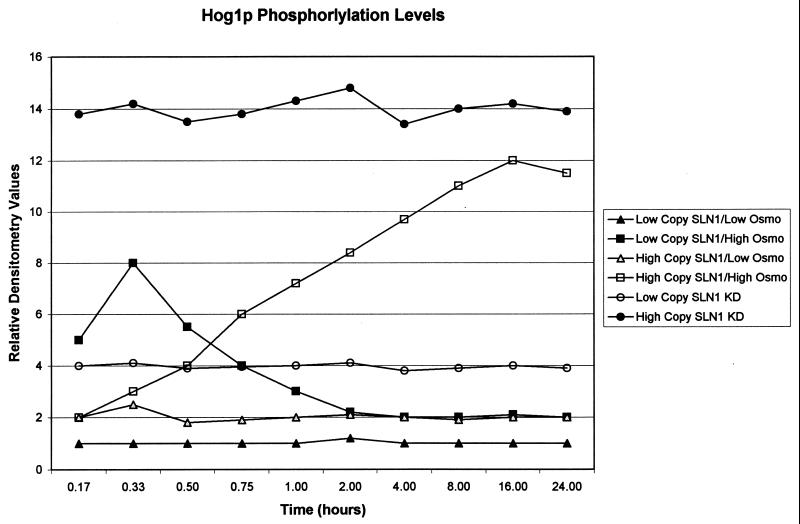
It was found that the maximal induction of the pathway with high-copy-number SLN1-containing plasmids was obtained when the cells were grown in high-osmolarity medium for approximately eight generations (Fig. 4). These data for the high-copy-number constructs contrast with reports that HOG pathway signaling peaks within 5 min and returns to undetectable phosphorylation levels within 20 min (3, 4, 19, 28). The overexpression of SLN1 in our system lengthened the time of HOG pathway induction, as evidenced by Hog1p phosphorylation levels. Results in agreement with the literature were obtained when a wild-type, nonoverexpressed SLN1 strain was used or when SLN1 was expressed from a low-copy-number plasmid in an sln1Δ strain. Under these conditions, Hog1p phosphorylation peaked within 30 min and fell to basal levels within 75 min (Fig. 4). Interestingly, Maeda and coworkers found that overexpression of SSK1, the second aspartate phosphoacceptor of the pathway, leads to an increased duration of Hog1p phosphorylation in response to high-osmolarity growth (13).
FIG. 4.
Hog1p phosphorylation time course. The relative levels of Hog1p phosphorylation were determined at different times of incubation with high-osmolarity medium with different SLN1 constructs. Total cytosolic protein was subjected to immunoprecipitation with anti-Hog1p antisera, separated by SDS-PAGE, and immunoblotted with antiphosphotyrosine antisera, and the intensities of the bands corresponding to Hog1p were quantified by scanning densitometry. All values were corrected for the amount of Hog1p loaded (as judged with duplicate anti-Hog1p immunoblots) and normalized to the average value observed with the low-copy-number wild-type SLN1 construct grown in low-osmolarity medium. Data represent averages of three independent experiments; standard deviations are less than 5%. Data for wild-type SLN1 strains are not substantially different from those for sln1Δ strains containing SLN1 on a low-copy-number plasmid; data for the SLN1 KD-containing strains are not substantially different when grown in high- or low-osmolarity medium.
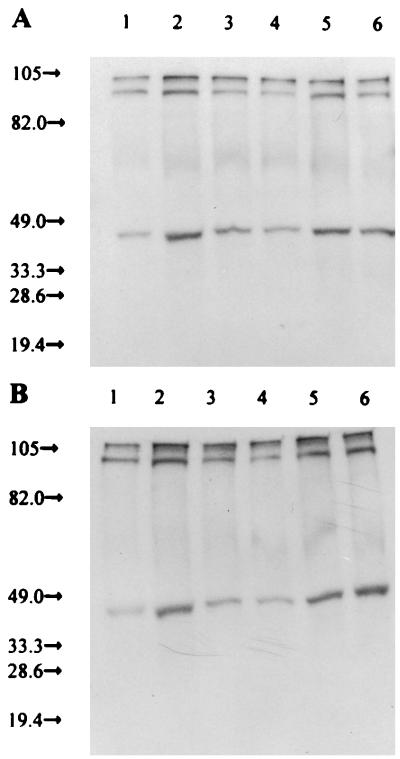
With full-length SLN1, Hog1p phosphorylation increases when cells are grown in high- as opposed to low-osmolarity medium (Fig. 5, lanes 1 and 2). (The identities of the 100- and 105-kDa bands are unknown). Therefore, as in wild-type cells, Sln1p kinase activity appears to be high in low-osmolarity medium and the HOG pathway is inactive (Table 1). This situation is reversed in high-osmolarity medium.
FIG. 5.
Hog1p phosphorylation with various SLN1 truncation mutations. Antiphosphotyrosine immunoblot analysis was performed on immunoprecipitated Hog1p separated by SDS-PAGE. (A) Rich medium; (B) defined medium. Odd-numbered lanes, low-osmolarity medium; even-numbered lanes, high-osmolarity medium. Lanes 1 and 2, full-length SLN1 with restriction sites (Fig. 1A); lanes 3 and 4, ΔTMD1 construction (Fig. 1C); lanes 5 and 6, KD construction (Fig. 1E). Duplicate anti-Hog1p immunoblots demonstrated that the same amounts of Hog1p were immunoprecipitated in all cases.
Sln1p with a single TMD (ΔTMD1) fails to show any response to the osmolarity of the medium, and Hog1p appears to be primarily unphosphorylated (Fig. 5, lanes 3 and 4). Therefore, without TMD1, Sln1p appears to be constitutively active, and the HOG pathway is inactive as in low-osmolarity medium (Table 1). When the SLN1 kinase domain alone (KD) is expressed, again no response to changes in osmolarity is observed. However, Hog1p appears to possess a relatively high level of phosphorylation (Fig. 5, lanes 5 and 6). Therefore, the soluble kinase domain peptide appears to lack activity, as the HOG pathway is as active as it is with the wild-type SLN1 construct in high-osmolarity medium (Table 1). These experiments were conducted in both rich (Fig. 5A) and defined (Fig. 5B) media. The ΔTMD1&ECD construct also resulted in constitutive activation of the HOG pathway (Table 1), showing no change when grown in media of different osmolarities (not shown). The use of low-copy-number constructs produced results that are substantially the same (not shown).
To control for the effect of Sho1p in these experiments, the gene encoding the protein was disrupted in these strains. Sho1p is another plasma membrane osmosensor that directly activates the MAPKK Pbs2p, which in turn phosphorylates Hog1p (20). No substantive differences in the relative levels of Hog1p were observed with these constructs in an sln1Δ sho1Δ strain (not shown).
These results demonstrate that only full-length Sln1p is capable of altering kinase activity in response to differences in medium osmolarity. Further, the results suggest that Sln1p anchoring to the membrane is insufficient for activation of kinase activity.
Plasma membrane localization.
The observation that the ΔTMD1&ECD construct appeared to be mostly inactive suggested that plasma membrane localization alone is insufficient to fully activate Sln1p kinase. As further evidence, a CAAX farnesylation sequence was added to the carboxyl terminus of the KD construct (Fig. 1F, KD/CAAX) to tether it to the plasma membrane (25). (This effort was assisted by the fact that the carboxyl-terminal region of Sln1p has numerous basic amino acids, a necessary feature of the CAAX motif [21].) This construct was shown to complement sln1Δ lethality (Table 1). By immunoblot analysis, approximately 80% of this protein was found associated with the membrane fraction. However, 100% of the membrane-associated protein was found specifically associated with the plasma membrane (not shown). As with ΔTMD1&ECD, this construct caused constitutive Hog1p phosphorylation (Table 1). These experiments demonstrate that membrane localization alone is not responsible for the full activation of the kinase activity of Sln1p.
Sln1p ECD dimerization.
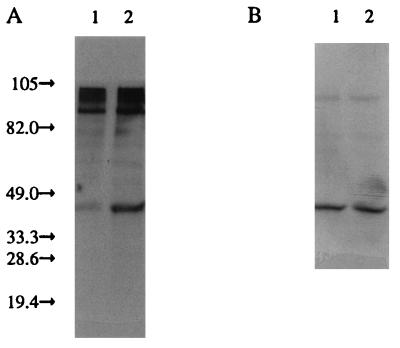
Sln1p seems to be more active with an untethered ECD, while it is nearly inactive in the absence of the ECD. Therefore, it was hypothesized that the ECD of Sln1p is involved in dimerization of the protein and that this is essential for kinase activity. To test this hypothesis, the restriction enzyme sites incorporated into SLN1 were used to replace the Sln1p ECD with the rat C/EBP leucine zipper region (Table 1, ΔECD/LeuZip). This construction retains the first intracellular and transmembrane domains. Immunoblot analysis revealed that this construct is, like the previous Sln1p truncation constructions, predominantly associated with the plasma membrane. This construct complemented sln1Δ but resulted in high levels of Hog1p phosphorylation (Table 1), which suggests that the Sln1p kinase activity is minimal with this construct. The leucine zipper motif is much shorter than the nascent Sln1p ECD (50 as opposed to 280 amino acids) and is known to adopt an alpha-helical motif in its native conformation. Therefore, it was hypothesized that the physical constraint of having both sides of the zipper tethered to the membrane might prevent dimerization, resulting in a less active form of Sln1p kinase. To test this hypothesis, the first intracellular and transmembrane domains of SLN1 were removed in this construct, allowing the amino-terminal leucine zipper motif to freely adopt its native conformation (Fig. 1G; Table 1). The resulting protein was again observed to be membrane associated, primarily with the plasma membrane. This construct resulted in only basal levels of Hog1p phosphorylation (Fig. 6A, lane 1; Table 1), suggesting that the unconstrained zipper motif was free to dimerize and activate the Sln1p kinase domains.
FIG. 6.
Hog1p phosphorylation with Sln1p/leucine zipper constructions. (A) Antiphosphotyrosine immunoblot analysis of immunoprecipitated Hog1p. Lane 1, cells containing the ΔTMD1&ECD/LeuZip construct; lane 2, cells containing the ΔTMD1&ECD/LeuZip* (mutated, undimerizable leucine zipper) construct. (B) Duplicate immunoblot analysis of panel A with anti-Hog1p antisera showing similar amounts of Hog1p immunoprecipitated in all cases. The cells were grown in low-osmolarity medium.
The same construct was mutagenized to convert the central leucine codon of the zipper motif to a proline, a mutation that is reported to abolish dimerization of a related leucine zipper (Fig. 1H) (9). This construct demonstrated constitutive high-level Hog1p phosphorylation (Table 1; Fig. 6A, lane 2). These observations support the hypothesis that Sln1p must be dimerized in order to achieve full kinase activity.
Sln1p phosphorylation.
The model for osmotically induced signal transduction implies that Hog1p phosphorylation levels are opposite those of Sln1p (13). To directly confirm this, the phosphorylation state of Sln1p was observed. A restriction enzyme site was introduced into the carboxyl-terminal domain of SLN1 by SDM and used to introduce DNA sequence encoding the FLAG epitope. This tag was inserted into the full-length, ΔTMD1, and KD, as well as both wild-type and undimerizable mutant forms, of ΔTMD1&ECD/LeuZip expression constructs. Immunoblot analysis both with anti-FLAG and anti-Sln1p antibodies demonstrated all three SLN1 constructs were expressed in similar amounts (not shown).
The FLAG-tagged constructs were used to test the phosphorylation state of Sln1p under different growth conditions. Cells were labeled with 32Pi in both high- and low-osmolarity media. The cells were harvested and broken, and cytosol was separated from the membrane fraction. The Sln1p in each fraction was immunoprecipitated with anti-FLAG antibodies, separated by SDS-PAGE, and visualized by autoradiography. Duplicate immunoblots with anti-Sln1p antisera demonstrated approximately equal amounts of Sln1p immunoprecipitated in each case (Fig. 7B and D).
FIG. 7.
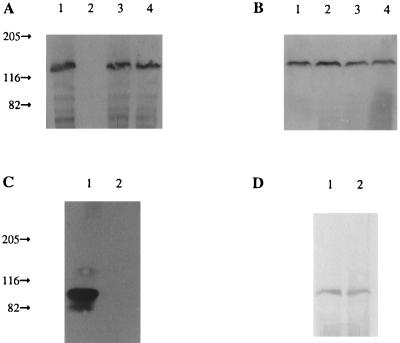
Phosphorylation of various Sln1p constructions. (A) Autoradiogram of a polyacrylamide gel of FLAG-tagged Sln1p proteins immunoprecipitated from the membrane fraction of 32Pi-labeled cells. Lanes 1 and 3, low-osmolarity medium; lanes 2 and 4, high-osmolarity medium; lanes 1 and 2, full-length Sln1p; lanes 3 and 4, ΔTMD1. (B) Duplicate immunoblot analysis of panel A with anti-Sln1p antisera. (C) Autoradiogram of a polyacrylamide gel of FLAG-tagged Sln1p immunoprecipitated from the membrane fraction of 32Pi-labeled cells. Lane 1, ΔTMD1/LeuZip; lane 2, ΔTMD1/LeuZip* (mutated, undimerizable leucine zipper). (D) Duplicate immunoblot analysis of panel C with anti-Sln1p antisera.
Full-length Sln1p was associated only with the membrane fraction and was highly labeled when cells were grown in low-osmolarity medium (Fig. 7A, lane 1). When the cells were grown in high-osmolarity medium, full-length Sln1p was unlabeled (Fig. 7A, lane 2). This result is consistent with the Hog1p phosphorylation experiments described above (Table 1). ΔTMD1 was also associated exclusively with the membrane fraction but was highly labeled in both low- and high-osmolarity media (Fig. 7A, lanes 3 and 4), consistent with the hypothesis that the ECD of this construct results in a high percentage of dimerization and kinase activity (Table 1). The KD construct was present only in the cytosolic fraction (not shown) and was found to be unlabeled under all conditions tested (Table 1). The protein was present in cells, however, as observed with a duplicate anti-Sln1p immunoblot (not shown).
The ΔTMD1&ECD/LeuZip construct showed high levels of Sln1p phosphorylation, while the undimerizable mutant did not (Fig. 7C). The interpretation of the data assumes that the phosphorylation being observed with these SLN1 alleles is due to autophosphorylation of the protein through the function of its histidine kinase activity. However, the possibility that the observed phosphorylation is due to the activity of another kinase cannot be completely ruled out. Nevertheless, these results are consistent with the hypothesis that dimerization of Sln1p is essential for autophosphorylation.
Summary.
The results suggest that the function of Sln1p is dependent on several structural aspects of the protein. First, regulation of kinase activity in response to osmolarity requires the presence of TMD1. All truncated forms of the protein that lacked this domain failed to respond to changes in the osmolarity of the growth medium, regardless of the apparent level of histidine kinase activity. No changes in either Sln1p autophosphorylation or Hog1p phosphorylation levels were detected with any of the TMD1 truncations upon addition of high concentrations of sorbitol or salt to the growth medium.
Second, the ECD appears to play an important role in regulating the level of activity of the kinase domain. Deletion of the ECD resulted in a marked reduction of the amount of phosphorylated Sln1p and, as judged by the high level of phosphorylated Hog1p, an attenuation of kinase activity. Substitution of the Sln1 ECD with a known dimerization motif led to a high level of Sln1p autophosphorylation, reflected by inactivation of the HOG MAPK pathway. Mutations shown to prevent dimerization led to the opposite condition, high-level constitutive signaling. Although we have not explicitly shown that Sln1p kinase activation is dependent on dimerization of the protein, these results are consistent with the hypothesis that dimerization is essential for activity of the kinase and that dimerization is effected by the ECD.
Third, in agreement with previous studies (20), it is clear that the cytoplasmic histidine kinase and receiver domains of Sln1p are essential for viability. Mutant proteins in which the kinase domain is disrupted or eliminated are unable to support growth of the sln1Δ strain. The toxicity of the sln1 null allele has been attributed to hyperactivation of the HOG MAPK cascade (13). Therefore, growth of the sln1Δ strain should reflect the level of activity of the SLN1p kinase. In several of our constructs (sln1Δ strains expressing ΔTMD1&ECD, KD, KD/CAAX, and ΔTMD1&ECD/LeuZip* [Table 1]), the level of Hog1 phosphorylation appeared to be high regardless of the osmolarity. In these strains, phosphorylation of Sln1p was undetectable. While this suggests a lack of activity, sln1Δ strains expressing these mutant proteins grew normally. As we also observed this complementation with low-copy-number derivatives of the high-copy-number constructs, it would appear that the truncated Sln1p derivatives possess sufficient kinase activity to relieve the toxicity associated with constitutive HOG pathway signaling.
ACKNOWLEDGMENTS
We thank David Lach, Pramathesh Patel, and Ramakrishna Seethala for assistance with E. coli protein expression, Beverly Remsburg and Michael Kornacker for assistance with the C/EBP leucine zipper, and Irene Ota for providing the SLN1/sln1 heterozygous strain and the SLN1 plasmid.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belendiuk G, Mangnall D, Tung B, Westley J, Getz G S. CTP-phosphatidic acid cytidyltransferase from Saccharomyces cerevisiae: partial purification, characterization, and kinetic behavior. J Biol Chem. 1978;253:4555–4565. [PubMed] [Google Scholar]
- 3.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 4.Fassler J S, Gray W M, Malone C L, Tao W, Lin H, Deschenes R J. Activated alleles of yeast SLN1 increase Mcm1-dependent reporter gene expression and diminish signaling through the Hog1 osmosensing pathway. J Biol Chem. 1997;272:13365–13371. doi: 10.1074/jbc.272.20.13365. [DOI] [PubMed] [Google Scholar]
- 5.Fedor-Chaiken M, Deschenes R J, Broach J R. SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell. 1990;61:329–340. doi: 10.1016/0092-8674(90)90813-t. [DOI] [PubMed] [Google Scholar]
- 6.Fischl A S, Carman G M. Phosphatidylinositol biosynthesis in Saccharomyces cerevisiae: purification and properties of microsome-associated phosphatidylinositol synthase. J Bacteriol. 1983;154:304–311. doi: 10.1128/jb.154.1.304-311.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hagen D C, McCaffrey G, Sprague G F., Jr Evidence the yeast STE3 gene encodes a receptor for the peptide pheromone a factor: gene sequence and implications for the structure of the presumed receptor. Proc Natl Acad Sci USA. 1986;83:1418–1422. doi: 10.1073/pnas.83.5.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 9.Hu J C, O’Shea E K, Kim P S, Sauer R T. Sequence requirements for coiled-coils: analysis with λ repressor-GCN4 leucine zipper fusions. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 10.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 11.Koland J G, Hammes G G. Steady state kinetic studies of purified yeast plasma membrane proton-translocating ATPase. J Biol Chem. 1986;261:5936–5942. [PubMed] [Google Scholar]
- 12.Landschulz W H, Johnson P F, Adashi E Y, Graves B J, McKnight S L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988;2:786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- 13.Maeda T, Wurgler-Murphy S M, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Science. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 14.Monk B C, Kurtz M B, Marrinan J A, Perlin D S. Cloning and characterization of the plasma membrane H+-ATPase from Candida albicans. J Bacteriol. 1991;173:6826–6836. doi: 10.1128/jb.173.21.6826-6836.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostrander D B, Gorman J A, Carman G M. Regulation of profilin localization in Saccharomyces cerevisiae by phosphoinositide metabolism. J Biol Chem. 1995;270:27045–27050. doi: 10.1074/jbc.270.45.27045. [DOI] [PubMed] [Google Scholar]
- 16.Ostrander D B, O’Brien D J, Gorman J A, Carman G M. Effect of CTP synthetase regulation by CTP on phospholipid synthesis in Saccharomyces cerevisiae. J Biol Chem. 1998;273:18992–19001. doi: 10.1074/jbc.273.30.18992. [DOI] [PubMed] [Google Scholar]
- 17.Ota I M, Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 18.Park, T.-S., D. B. Ostrander, A. Pappas, and G. M. Carman. Identification of Ser424 as the protein kinase A phosphorylation site in CTP synthetase from Saccharomyces cerevisiae: phosphorylation plays a role in the regulation of phospholipid metabolism. J. Biol. Chem., in press. [DOI] [PubMed]
- 19.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 20.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 21.Powers S, Kataoka T, Fasano O, Goldfarb M, Strathern J, Broach J R, Wigler M. Genes in S. cerevisiae encoding proteins with domains homologous to the mammalian RAS proteins. Cell. 1984;36:607–612. doi: 10.1016/0092-8674(84)90340-4. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Schüller C, Brewster J L, Alexander M R, Gustin M C, Ruis H. The HOG pathway controls osmotic regulation of transcription via the stress response element STRE of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serrano R. H+-ATPase from plasma membranes of Saccharomyces cerevisiae and Avena sativa roots: purification and reconstitution. Methods Enzymol. 1988;157:533–544. doi: 10.1016/0076-6879(88)57102-1. [DOI] [PubMed] [Google Scholar]
- 25.Stokoe D, MacDonald S G, Cadwallader K, Symons M, Hancock J F. Activation of Raf as a result of recruitment to the plasma membrane. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 26.Varela J C S, Mager W H. Response of Saccharomyces cerevisiae to changes in external osmolarity. Microbiology. 1996;142:721–731. doi: 10.1099/00221287-142-4-721. [DOI] [PubMed] [Google Scholar]
- 27.Warner J R. Labeling of RNA and phosphoproteins in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:423–428. doi: 10.1016/0076-6879(91)94033-9. [DOI] [PubMed] [Google Scholar]
- 28.Wurgler-Murphy S M, Maeda T, Witten E A, Saito H. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol. 1997;17:1289–1297. doi: 10.1128/mcb.17.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wurgler-Murphy S M, Saito H. Two-component signal transducers and MAPK cascades. Trends Biochem Sci. 1997;22:172–176. doi: 10.1016/s0968-0004(97)01036-0. [DOI] [PubMed] [Google Scholar]