Abstract
Many bacterial genera, including Bacteroides spp., harbor mobilizable transposons, a class of transfer factors that carry genes for conjugal DNA transfer and, in some cases, antibiotic resistance. Mobilizable transposons are capable of inserting into and mobilizing other, nontransferable plasmids and are implicated in the dissemination of antibiotic resistance. This paper presents the isolation and characterization of Tn5520, a new mobilizable transposon from Bacteroides fragilis LV23. At 4,692 bp, it is the smallest mobilizable transposon reported from any bacterial genus. Tn5520 was captured from B. fragilis LV23 by using the transfer-deficient shuttle vector pGAT400ΔBglII. The termini of Tn5520 contain a 22-bp imperfect inverted repeat, and transposition does not result in a target site repeat. Tn5520 also demonstrates insertion site sequence preferences characterized by A-T-rich nucleotide sequences. Tn5520 has been sequenced in its entirety, and two large open reading frames whose predicted protein products exhibit strong sequence similarity to recombinase-integrase enzymes and mobilization proteins, respectively, have been identified. The transfer, mobilization, and transposition properties of Tn5520 have been studied, revealing that Tn5520 mobilizes plasmids in both B. fragilis and Escherichia coli at high frequency and also transposes in E. coli.
Members of the genus Bacteroides are obligately anaerobic, human colonic bacteria, accounting for about 30% of normal fecal flora. However, they can be significant opportunistic pathogens responsible for a variety of intra-abdominal infections, abscesses, and peritonitis and are an important cause of morbidity and mortality in humans (4–6). Increasing antibiotic resistance in Bacteroides has been reported from around the world (1, 8).
Bacteroides spp. harbor conjugal and mobilizable elements (38, 47). Conjugal elements may be plasmids or chromosomally located tetracycline resistance (TET) elements (37). The plasmids are presumably large, self-transmissible molecules, e.g., pBF4 (41 kb) and pBI136 (80 kb), that transfer via a conjugation-type process (27, 43). TET elements (also called conjugative transposons) range in size from 65 kbp to more than 150 kbp and are divided into three distinct families: the Tcr Emr DOT family, the Tcr Emr 12256 family, and the Tcr Emr 7853 family (35, 37, 39). TET elements manifest unique properties as evidenced by their ability to form circular intermediates, mediate their own transfer from chromosome to chromosome, mobilize coresident plasmids, and mediate excision and circularization of discrete unlinked segments of chromosomal DNA. The last property results in the production of a nonreplicating Bacteroides unit (NBU). Three NBUs have been isolated to date, NBU1, NBU2, and NBU3 (41). NBU1 and NBU2 are mobilized in the presence of the Tcr ERL element in Bacteroides and by the Escherichia coli IncPβ plasmid R751 (21, 50). TET elements are also unique in that most members exhibit tetracycline regulation of self-transfer and mobilization. This has been demonstrated as enhanced frequencies of transfer and mobilization after pretreatment of cells with subinhibitory concentrations of tetracycline in vitro (37).
In contrast to the conjugal elements, Bacteroides mobilizable factors may be plasmids (4 to 15 kb; e.g., pIP417, pIP419, pLV22a, and pBFTM10), transposons (e.g., Tn4399, Tn4555, and Tn4551), or NBUs (NBU1, NBU2, and NBU3) (15, 16, 31, 36, 46, 49). These transfer factors are classified as such since they likely encode functions required for the initiation of the transfer process but not those required for the formation of the conjugation apparatus. In addition, mobilizable transposons (Tn4399, Tn4555, and Tn4451) also encode transposition functions. Tn4399 (9.6 kb) was isolated from Bacteroides fragilis TM4.2321 and mobilizes nonconjugal plasmids in cis in B. fragilis (16, 17). It carries the mocA and mocB genes, which are involved in conjugal DNA transfer (28). Tn4555 (12.5 kb) is a mobilizable cefoxitin resistance transposon, isolated from Bacteroides vulgatus CLA341, that forms circular intermediates during transfer and transposition (44). One transfer protein that has been identified, MobA, exhibits high sequence similarity to the single NBU1 mobilization protein (85% identity at the nucleotide level) (45).
Plasmid or transposon conjugative transfer occurs as a multistep process requiring specific DNA sequences and multiple gene products (20). These include a cis-acting origin of transfer, oriT, and trans-acting proteins (mobilization or Mob proteins), which are involved in the initiation of DNA transfer and replication in the recipient. In addition, other trans-acting proteins that form the conjugation pore or mating apparatus are also required. Conjugal plasmids or transposons encode all of these required proteins and are said to be self-transmissible, since their proteins can perform all initiation and termination functions and also assemble the conjugation apparatus. Unlike the conjugal elements, mobilizable factors harbor an oriT and encode only proteins required for initiation and termination of transfer (Mob proteins). It is believed that the trans-acting proteins required for the formation of the mating apparatus are provided by coresident conjugal plasmids or transposons or by the host chromosome (33, 34).
DNA transfer is initiated when a Mob protein(s) binds at oriT and introduces a single-stranded nick at a specific site (nic). The nicking protein (nickase) then covalently attaches to the 5′ end of nic and leads the nicked DNA strand from donor to recipient through the mating apparatus. DNA replication occurs concomitantly in the donor and recipient to restore the transferred DNA to the double-stranded context. For detailed descriptions of the mechanisms of DNA transfer, see references 11, 13, 20, and 34.
We report here the isolation and partial characterization of a new mobilizable transposon, Tn5520, from B. fragilis LV23. Tn5520 is the smallest mobilizable transposon (4,692 bp) reported to date from any bacterium. Tn5520 exhibited transfer properties in B. fragilis, was mobilizable in E. coli, and also transposed in E. coli. The ends of Tn5520 were determined to contain imperfect inverted repeats, and it was observed that Tn5520 did not modify its target site. DNA sequencing of Tn5520 revealed the presence of two large open reading frames whose predicted protein products exhibited strong sequence similarity to recombinase-integrase enzymes and Bacteroides mobilization proteins, respectively.
For purposes of definition in this paper, the term transferable indicates that a transfer factor is completely autonomous during DNA transfer, whereas mobilizable indicates that the transfer factor is dependent on host cell or other extrachromosomal products. This definition raises no controversy for E. coli, where it has been established that both transfer initiation proteins and a mating apparatus are required for DNA transfer. However, the requirement for a mating apparatus in Bacteroides spp. remains unresolved. Thus, it is difficult to label Bacteroides elements as transferrable or mobilizable by using the E. coli definition. Whether the mating apparatus could be provided by the large TET elements in whose presence the passage of DNA from donor to recipient is enhanced remains to be seen. Also, DNA transfer of small plasmids, like pBFTM10, from B. fragilis strains, like TM4000, that are believed to be devoid of TET elements that enhance DNA transfer has been observed. However, all Bacteroides plasmids and mobilizable elements recovered to date and tested require the presence of the IncPβ plasmid R751 to be mobilized in E. coli. To avoid any confusion, we define the movement of Bacteroides plasmids and transposons from donor to recipient in Bacteroides as transfer and that from E. coli as mobilization.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Media, antibiotics, and growth conditions for Bacteroides spp. and E. coli have been previously described (31). Antibiotics used for the selection of strains and plasmids included the following: ampicillin, 200 μg/ml; chloramphenicol, 40 μg/ml; streptomycin, 50 μg/ml; spectinomycin, 50 μg/ml; and tetracycline, 10 μg/ml (for E. coli) or 5 μg/ml (for Bacteroides spp.). E. coli strains containing R751 were grown on Mueller-Hinton medium (Difco); other E. coli strains were grown in Luria-Bertani (LB) medium supplemented with the appropriate antibiotic when required. Bacteroides spp. were grown in supplemented brain heart infusion medium (BHIS) (3.7%; BBL) supplemented with 0.0005% hemin and 5 g of yeast extract per liter in a Coy anaerobic chamber (5% CO2, 10% H2, and 85% N3).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| B. fragilis | ||
| TM4000 | Rifr | 15 |
| TM4.23 | Tetr Rifr | 15 |
| LV23 | Clinical isolate | |
| E. coli | ||
| HB101 | Smr | 31 |
| DW1030 | Spr | 31 |
| DH5α | 3 | |
| Plasmids | ||
| pBR328 | Mob− Cmr Tetr Apr | 9 |
| pOX38::Kan | Knr, 56-kb HindIII fragment of F plasmid | 18 |
| F′ lac | IncF1, Tra+lac+ | Pasteur Institute |
| pGAT400 | Clnr Apr Tetr (aerobic) Tra+ | 15 |
| pGAT400ΔBglII | Clnr Apr Tetr (aerobic) Tra− | 15 |
| pHB23Δ5 | Clnr Apr Tets (aerobic) Tra+ | This study |
| pKB1 | Apr, 3.7-kb fragment of Tn5520 with bmpH gene in pBR328 | This study |
| pTJ20 | Apr Tra+ | This study |
| pTJ22.3 | Tn1000 insertion in bipH of pTJ20 | This study |
| pTJ22.26 | Tn1000 insertion in bipH of pTJ20 | This study |
| pTJ22.20 | Tn1000 insertion in bipH of pTJ20 | This study |
| pTJ22.25 | Tn1000 insertion outside bipH of pTJ20 | This study |
| pTJ22.10 | Tn1000 insertion outside bipH of pTJ20 | This study |
| pTJ22.36 | Tn1000 insertion outside bipH of pTJ20 | This study |
| pTJ22.24 | Tn1000 insertion outside bipH of pTJ20 | This study |
| pTJ23 | Deletion of B end of Tn5520 | This study |
| pGV2 | Tn1000 insertion in bmpH of pTJ20 | This study |
| pGV3 | Tn1000 insertion in bmpH of pTJ20 | This study |
| pGV4 | Tn1000 insertion outside bmpH (proximal) of pTJ20 | This study |
| pGV5 | Tn1000 insertion outside bmpH (proximal) of pTJ20 | This study |
| pGV6 | Tn1000 insertion outside bmpH (proximal) of pTJ20 | This study |
| pGV7 | Tn1000 insertion outside bmpH (distal) of pTJ20 | This study |
| pGV8 | Tn1000 insertion outside bmpH (distal) of pTJ20 | This study |
| pGV9 | Tn1000 insertion outside bmpH (distal) of pTJ20 | This study |
Rifr, Tetr, Smr, Spr, Cmr, Knr, Clnr, and Apr, resistance to rifampin, tetracycline, streptomycin, spectinomycin, chloramphenicol, kanamycin, clindamycin, and ampicillin, respectively; Tra+ and Tra−, transfer proficiency and deficiency, respectively; Mob−, inability to be mobilized; Tets, tetracycline sensitivity.
Recombinant DNA techniques.
Plasmid DNA was prepared by the miniprep alkaline lysis method or by CsCl equilibrium gradient separation (3) and also by affinity purification (Qiagen Corp., Chatsworth, Calif.). All restriction endonucleases, DNA ligase, and S1 nuclease were purchased from Promega (Madison, Wis.). DNA sequencing from the left end of Tn5520 was performed with a United States Biochemical sequencing kit (Sequenase version 2.0), based on the Sanger dideoxy method.
Primers for Tn5520 sequencing.
Tn5520 sequencing was initiated with primers derived from sequence flanking the insertion site (determined by restriction analysis) of pHB23Δ5 in pGAT400ΔBglII. The 5′ end of Tn5520 (relative to insertion in pGAT400ΔBglII) was sequenced by using the primer 5′-CGGCTAATGGCATCTCACCA-3′ [1037(1)], and the 3′ end was sequenced by using the primer 5′-TAGTTTACACGCCGTAGGGG-3′ [(1026(2)]. As DNA sequence was obtained, new primers were designed to obtain additional Tn5520 sequence. To obtain DNA sequence of the insertion sites of Tn5520, the junctions of pHB23Δ5 in pGAT400ΔBglII were sequenced by using primers reading outward from the left and right ends of Tn5520 {5′-ATTTGACAGCATGGCAACGC-3′ [1056(1)] and 5′-CGTTGGCTCTGCCCTATAGA-3′ [1056(2)]}.
Plasmid mobilization experiments.
Quantitative Bacteroides-to-E. coli filter matings were performed as previously described (31). For these matings, the transfer-deficient shuttle plasmid pGAT400ΔBglII was introduced from E. coli HB101 into B. fragilis LV23, and the resulting transconjugant was used as a donor in experiments involving mobilization into an E. coli HB101 recipient. For E. coli matings, mobilization of plasmids in the presence of R751 was determined by mixing log-phase cultures of E. coli HB101 containing R751 and the plasmid to be assayed for mobilization with E. coli DW1030 (donor-to-recipient ratio, 1:9; total volume, 1.5 ml). After pelleting, the cells were suspended in 100 μl of phosphate-buffered saline (8 mM Na2HPO4, 2 mM NaH2PO4, 145 mM NaCl, pH 6.9) and plated onto sterile 25-mm-pore-size Nalgene GN-6 cellulose nitrate filters (Nalge Co., Rochester, N.Y.) supported on Luria agar plates. The filters were incubated for only 3 h at 37°C to limit secondary mobilization events. The cells were then suspended and serially diluted in phosphate-buffered saline and plated onto the appropriate antibiotic media. The mobilization frequencies in the presence of R751 were calculated by dividing the number of Mob+ plasmid transconjugants by the number of R751 transconjugants in the same experiment.
Analysis of transconjugant DNA.
Transconjugant DNA was prepared by the alkaline lysis method of Birnboim and Doly (3) and analyzed by restriction enzyme analysis with pGAT400ΔBglII digested in a similar manner for comparison.
Cloning of Tn5520 and construction of deletion derivatives.
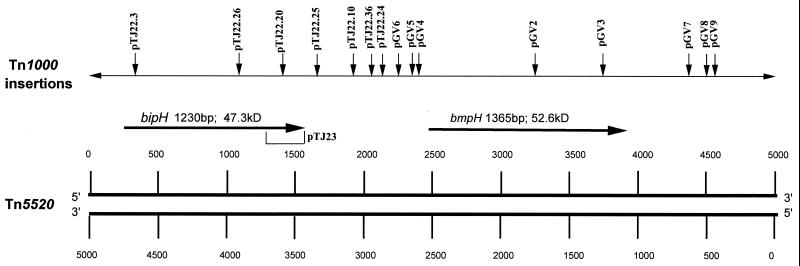
pGAT400ΔBglII is a deletion derivative of pGAT400, a chimeric shuttle vector with Bacteroides and E. coli origins of replication due to the presence of the pBFTM10 plasmid fused to the E. coli plasmid pDG5. pHB23Δ5 is a transfer-proficient plasmid containing an approximately 5-kb insertion in pGAT400ΔBglII (Fig. 1A). A 6.5-kb EcoRI-HindIII fragment of pHB23Δ5 was cloned in pBR328 (9) to give pTJ20 (Fig. 1B). pTJ22.3, pTJ22.26, and pTJ22.20 are derivatives of pTJ20 that harbor Tn1000 insertions within bipH. pTJ22.25, pTJ22.10, pTJ22.36, and pTJ22.24 are derivatives of pTJ20 that harbor Tn1000 insertions downstream of bipH (Fig. 2). pTJ23 is a deletion derivative of pTJ20 that has the right end (1.5 kb) deleted. Similarly, pGV2 to -9 are derivatives of pTJ20 that harbor Tn1000 insertions within and surrounding bmpH (Fig. 2).
FIG. 1.
(A) Map of pGAT400ΔBglII. Vertical arrows denote insertions of Tn5520 and pHB23 plasmids 1 to 15. Numbers above the arrows identify plasmids designated for each cointegrate. ermF, clindamycin resistance marker; Amp, ampicillin resistance tetX*, aerobic tetracycline resistance. AV, AvaI; E, EcoRI; EV, EcoRV; H, HindIII. (B) Restriction analysis of Tn5520 insertions in pGAT400ΔBglII. Except for the 1-kb DNA markers, all DNA was digested with the enzymes AvaI and BglII. Lanes: 1, pGAT400ΔBglII; 2, pHB23Δ1; 3, pHB23Δ2; 4, pHB23Δ7; 5, 1-kb ladder; 6, pHB23Δ10; 7, pHB23Δ11; 8, pHB23Δ14; 9, pHB23Δ16; 10, pHB23Δ17. The arrowhead designating the 5-kb marker indicates the approximate size of the pGAT400ΔBglII insertions. (C) pTJ20 is the 6.5-kb EcoRI-HindIII fragment of pHB23Δ5, containing Tn5520 and flanking sequences cloned into the 3.1-kb EcoRI-HindIII fragment of pBR328. S, SspI.
FIG. 2.
Map of Tn5520, showing open reading frames and predicted protein products. Large, dark arrows indicate open reading frames. The name and length of the open reading frame and the size of the predicted protein product are indicated above each arrow. The schematic above the open reading frames depicts the names and locations of all transposon (Tn1000)-mediated insertions in Tn5520 used in this study. pTJ23 is the only nontransposon disruption of Tn5520 and is a construct that harbors a deletion of the right end of bipH.
Tn1000 transposition insertion mutations within and around bipH and bmpH of Tn5520 were generated by the F′ lac protocol as previously described (31). Each mutant that was determined to contain an insertion in Tn5520 had all internal Tn1000 BglII fragments deleted to prevent further transposition events mediated by the transposon. Plasmid DNA was prepared from the transformants by the boiling method or by CsCl equilibrium gradient separation, and the BglII deletion mutants were checked by restriction analysis. These deletion mutants were then transformed into E. coli DW1030 containing the F-factor derivative pOX38::Kan plasmid, and the resulting transformants were used in transposition assays.
Transposition assays.
Tn5520 was tested for its ability to transpose in E. coli in experiments using the F-factor derivative pOX38::Kan, which can be transferred during conjugation only if mobilized in cis via a pTJ20::pOX38::Kan cointegrate that is transferred to recipients through the pOX38::Kan transfer apparatus. Stationary-phase E. coli DW1030 containing pOX38::Kan and a plasmid to be assayed for transposition was mixed with E. coli HB101 in a 1:2 ratio, diluted 1:4 in LB medium, and incubated for 1 h at 37°C with vigorous agitation and then for 4 h with very slow agitation. The mating broth was then serially diluted, plated to LB agar containing ampicillin and streptomycin to select for pTJ20::pOX38::Kan cointegrates that had been formed by transposition events, and then transferred to HB101 by the pOX38 transfer apparatus. The diluted mating broth was also plated to LB agar containing kanamycin and streptomycin to select for pOX38::Kan transfer. The transposition frequency was calculated as (CFU of ampicillin-resistant transconjugants per milliliter)/(CFU of kanamycin-resistant transconjugants per milliliter). Transconjugant plasmid DNA was analyzed by restriction analysis for cointegrate formation.
Digital imaging.
Ethidium bromide-stained agarose gels were photographed under UV light. Polaroid prints were scanned at high resolution (1,000 dpi) with a ScanJet 4c flatbed scanner (Hewlett-Packard, Louisville, Ky.). Scanned graphics files were imported into the application Microsoft Powerpoint, and text and labels were added. Images were printed at high resolution (1,440 dots/in.) on a Stylus Color 1520 printer (Epson America, Inc., Torrance, Calif.), using heavy satin-gloss photographic paper (Hewlett-Packard).
Nucleotide accession number.
The Tn5520 DNA sequence (4,960 bp) was deposited in the GenBank database with the accession number AF038866.
RESULTS
Isolation of Tn5520.
We used an interspecies mobilization assay to recover elements from B. fragilis that possessed transfer properties. The transfer-deficient shuttle plasmid pGAT400ΔBglII was introduced by conjugation from E. coli HB101 into B. fragilis LV23. The resulting transconjugant was used as a donor in experiments involving transfer into an E. coli HB101 recipient. pGAT400ΔBglII contains a deletion of the pBFTM10 transfer genes btgA and btgB and is normally incapable of transfer (31). Following the mating of B. fragilis LV23 and E. coli HB101, 88 transconjugants were obtained, with a transfer frequency of (4.5 ± 3.1) × 10−8. Plasmid DNA from 15 transconjugants was analyzed by restriction analysis, with all transconjugants containing approximately 5 kbp of new DNA. Additional endonuclease restriction analysis demonstrated that the new DNA was present in different locations in pGAT400ΔBglII. These multiple insertion sites suggested the presence of a transposable element (Fig. 1A). Thirteen of 15 insertions of the new DNA were clustered in a region downstream of the Tn4400 transposon contained in the pBFTM10 portion of pGAT400ΔBglII. Figure 1B shows the restriction patterns of eight insertions compared with that of pGAT400ΔBglII. One plasmid, pHB23Δ5, was chosen for further analysis because of the insertion of the new DNA in a region of known sequence. A 6.5-kb EcoRI-HindIII fragment from pHB23Δ5 containing the insertion and flanking sequences was cloned into the 3.1-kb EcoRI-HindIII fragment of pBR328, to give pTJ20 (Fig. 1C).
Sequence analysis of Tn5520.
An initial DNA sequence of Tn5520 was obtained by using primers from the junctions of Tn5520 in pGAT400ΔBglII (with the pHB23Δ5 plasmid). As DNA sequence of Tn5520 was obtained, new primers were designed so that the fragment could be further sequenced. Tn5520 was sequenced in its entirety, and the sequence was deposited in the GenBank database.
DNA sequence analysis revealed the presence of two large (>500-bp) open reading frames (Fig. 2) designated orf1 (renamed bipH for Bacteroides integration protein) and orf2 (renamed bmpH for Bacteroides mobilization protein). bipH (1,230 bp; predicted protein BipH, 47.3 kDa) was found to exhibit sequence identity at both the nucleotide and predicted protein levels to a variety of integrase-recombinase proteins in a BLAST search of the Swiss-Prot database. These included YqkM (skin element) from Bacillus subtilis (26), integrase-recombinase XerD from E. coli K-12 (25), and integrase-recombinase XerD from Haemophilus influenzae Rd (10) (P values of 1.4e−6 [41%], 1.2e−4 [42%], and 2.5e−4 [39%], respectively).
bmpH (1,365 bp; predicted protein BmpH, 58.2 kDa) was found to be highly similar to the Bacteroides uniformis mobilization proteins NBU1 and NBU2 (P values of 6.0e−43 [56%] and 1.0e−42 [56%], respectively) (22, 23, 45) and also to the B. vulgatus Tn4555 mobilization protein MobA (P value of 4.0e−40 [42%]) (46).
DNA sequence analysis further revealed that the region preceding bmpH harbored multiple repeat sequences (two palindromic repeats, three inverted repeats, and four direct repeats). In addition, this region also contained the E. coli RP4 plasmid consensus nick site sequence (32), 2396CTTGCCC2403, located immediately adjacent to the largest inverted repeat (17 bp). An amino acid alignment of the predicted BmpH protein with Mob (NBU1) and MobA (Tn4555) revealed that nickase protein motif III residues were present (amino acids 137 to 151), including highly conserved aspartate (D139) and histidine (H144) residues (alignment not shown).
Characterization of Tn5520 ends.
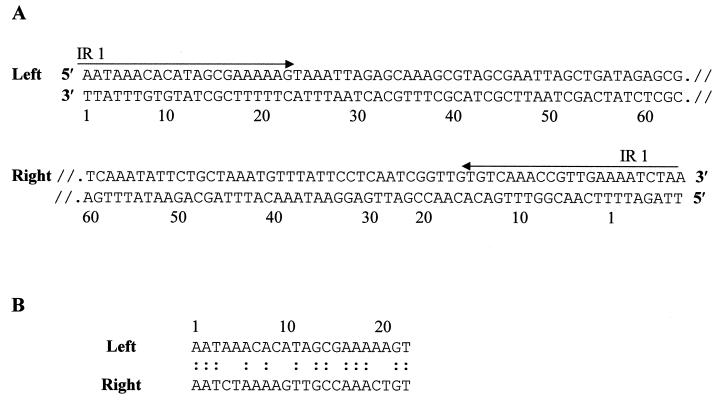
To determine the nature of the 5′ and 3′ termini of Tn5520, DNA sequence analysis of the termini of Tn5520 in pGAT400ΔBglII was performed with the software package DNAsis version 2.0. It was observed that the ends were delimited by the presence of an imperfect inverted repeat 22 bp in length. Figure 3A depicts the DNA sequences of the ends of the element, and Fig. 3B shows the structure of the imperfect inverted repeat.
FIG. 3.
(A) DNA sequence analysis of the ends of Tn5520. (B) Sequence of IR1, the 22-bp imperfect inverted repeat.
Insertion sites preferred by Tn5520.
All 15 insertions of similar size in pGAT400ΔBglII were determined to be of Tn5520 by sequencing the junctions of the element. In this study, we analyzed these junction sequences at both the 5′ and 3′ termini to determine any patterns or sequence preferences for insertion. We observed that the pGAT400ΔBglII sequence AATAA was present distal to the right end of the Tn5520 element in 12 of 15 insertions analyzed (Fig. 4). In the remaining three cases, the sequence ATAA (two cases) or TTAA (one case) was located proximal to the left end of the element.
FIG. 4.
Insertion sites preferred by Tn5520. Preferred sited are underlined; insertions of 15 transconjugants are shown. The locations of these sites in pGAT400ΔBglII are shown in Fig. 1A. Uppercase letters are Tn5520 sequence. Lowercase letters are pGAT400ΔBglII sequence.
Transfer of Tn5520 in B. fragilis.
The transfer and mobilization properties of Tn5520 were tested in B. fragilis and E. coli, respectively (Table 2). pHB23D5 was mobilized into B. fragilis TM4000 and TM4.23 from E. coli HB101 containing RK231. TM4000 is a B. fragilis donor strain that does not contain any known TET elements. TM4.23 harbors a TET element responsive to tetracycline that promotes plasmid and transposon transfer, and it is otherwise isogenic to TM4000. TM4.23 is the product of a mating between B. fragilis TMP230 and B. fragilis TM4000 that selected for the transfer of a TET element (15). pGAT400 and pGAT400ΔBglII were used as transfer-proficient and negative controls, respectively. When TM4000 was used as a donor and the constructs were assayed for transfer into E. coli HB101, it was observed that pGAT400 and pHB23D5 transferred at similar (but very low) frequencies [(7.9 ± 3.8) × 10−9 and (4.5 ± 3.1) × 10−8], while transfer of pGAT400ΔBglII was not detected (≤10−9). When TM4.23 was used as a donor with tetracycline induction, pHB23D5 and pGAT400 were transferred at frequencies that were approximately the same but 4 orders of magnitude greater than that from TM4000 [(3.6 ± 1.6) × 10−5 and (1.5 ± 0.83) × 10−4, respectively]. Transfer of pGAT400ΔBglII was not detected (≤10−9).
TABLE 2.
Transfer of Tn5520 in B. fragilis and mobilization in E. coli
| Process | Donor strain | Plasmid | Frequency of transfer (mean ± SE)a |
|---|---|---|---|
| Plasmid transfer (B. fragilis to E. coli) | TM4000 | pGAT400 | (7.9 ± 3.8) × 10−9 |
| pGAT400ΔBglII | NDb | ||
| pHB23Δ5 | (4.5 ± 3.1) × 10−8 | ||
| TM4.23 | pGAT400 | (3.6 ± 1.6) × 10−5 | |
| pGAT400ΔBglII | ND | ||
| pHB23Δ5 | (1.5 ± 0.83) × 10−4 | ||
| R751 mobiliza-tion (E. coli to E. coli) | HB101(R751)c | pBR328 | ND |
| pGAT400 | (0.4 ± 0.17) × 10−2 | ||
| pTJ20 | (3.2 ± 0.05) × 10−1 | ||
| pKB1 | (0.3 ± 0.1) × 10−2 | ||
| pGV2d | ND | ||
| pGV3d | ND | ||
| pGV4e | (3.7 ± 2.9) × 10−7 | ||
| pGV5e | ND | ||
| pGV6e | (3.3 ± 1.5) × 10−6 | ||
| pGV7f | (1.6 ± 0.09) × 10−2 | ||
| pGV8f | (1.02 ± 0.095) × 10−2 | ||
| pGV9f | (1.4 ± 0.05) × 10−2 |
Experiments were performed at least in triplicate.
ND, transfer or mobilization not detectable (≤10−9 [Bacteroides] or no transconjugants on selective plates).
The IncPβ plasmid R751 was coresident in the donor during E. coli-to-E. coli mobilization experiments.
Tn1000 insertion in bmpH.
Tn1000 insertion proximal to bmpH.
Tn1000 insertion distal to bmpH.
Mobilization of Tn5520 in E. coli.
The 4.6-kb insertion from pHB23D5 was subcloned as a 6.5-kb EcoRI-HindIII fragment in pBR328, to form pTJ20, and tested for mobilization in E. coli. These experiments were performed with donor strain HB101, containing the IncPβ plasmid R751 and either pTJ20 or a positive control, mated with the recipient E. coli DW1030. We and others have demonstrated that other Bacteroides transfer factors can be mobilized when R751 is present (15, 40). R751 presumably provides the conjugation apparatus necessary for mobilization of similar Bacteroides transfer factors. Results of matings revealed that pTJ20 was mobilized efficiently in E. coli [(3.2 ± 0.05) × 10−1] compared with the control cloning vector, pBR328 (≤10−5, the limit of detectability) (Table 2). The frequencies of mobilization of pTJ20 were comparable to those of the transfer-proficient pGAT400 control [(0.4 ± 0.17) × 10−2]. This indicated that Tn5520 was capable of transferring from Bacteroides in an interspecies mating and was mobilized in E. coli in an intraspecies manner.
Since the bmpH product exhibited sequence identity to mobilization proteins, a smaller region containing only bmpH was cloned into the mobilization-deficient vector pBR328 to give pKB1. pKB1 was also tested for mobilization in E. coli when coresident with R751 and was found to be mobilized at high frequencies [(0.3 ± 0.1) × 10−2]. Transposon Tn1000-mediated disruptions in and surrounding bmpH were then generated and tested in similar mobilization experiments. Disruptions of bmpH completely abolished mobilization, while those distal to bmpH did not affect mobilization (Fig. 2; Table 2). Insertions proximal to bmpH reduced mobilization but did not abolish it completely [(3.7 ± 2.9) × 10−7 and (3.3 ± 1.5) × 10−6]. DNA sequence analysis indicated that a region suggestive of the Tn5520 oriT (containing multiple direct, palindromic, and inverted repeats and a consensus nick site sequence) may be localized in this bmpH-proximal sequence.
Transposition of Tn5520 in E. coli.
Table 3 summarizes the results of transposition assays in which pTJ20 (which harbors he entire Tn5520) and transposon insertion derivatives of pTJ20 (mapping inside or outside bipH) were tested for transposition in E. coli. It was observed that pTJ20 formed cointegrates with pOX38::Kan and was transferred efficiently in E. coli [frequency of (1.1 ± 0.2) × 10−5] compared with the negative control pBR328 [(7.7 ± 1.9) × 10−8]. Insertions disrupting bipH (pTJ22.23, pTJ22.26, and pTJ22.20) resulted in almost complete loss of cointegrate formation [frequencies of (1.9 ± 1.2) × 10−7, (1.2 ± 0.05) × 10−7, and (1.2 ± 0.32) × 10−7, respectively]. Insertions upstream and downstream of bipH (pTJ22.25, pTJ22.10, pTJ22.36, and pTJ22.24) had no effect on cointegrate formation [frequencies of (7.5 ± 2.3) × 10−5, (2.9 ± 0.9) × 10−5, (1.9 ± 0.6) × 10−5, and (5.1 ± 2.9) × 10−5, respectively]. Transconjugants obtained from the transposition assays were analyzed by restriction analysis, which confirmed that cointegrate plasmids were present.
TABLE 3.
Transposition of Tn5520 in E. coli
| Donor | Transposition frequency (mean ± SE)a |
|---|---|
| DW1030(pOX38::Kan, pTJ20)b | (1.1 ± 0.2) × 10−5 |
| DW1030(pOX38::Kan, pBR322)c | (7.7 ± 1.9) × 10−8 |
| pTJ22.23d | (1.9 ± 1.2) × 10−7 |
| pTJ22.26d | (1.2 ± 0.05) × 10−7 |
| pTJ22.20d | (1.2 ± 0.3) × 10−7 |
| pTJ22.25e | (7.5 ± 2.3) × 10−5 |
| pTJ22.10e | (2.9 ± 0.9) × 10−5 |
| pTJ22.36e | (1.9 ± 0.6) × 10−5 |
| pTJ22.24e | (5.1 ± 2.9) × 10−5 |
| pTJ23f | (7.7 ± 1.9) × 10−8 |
Experiments were performed at least in triplicate.
Positive control.
Negative control.
Mutant with Tn1000 insertion in bipH.
Mutant with Tn1000 insertion outside bipH.
Deletion of Tn5520 B end.
Deletion of the right end of Tn5520 resulted in a loss of transposition (assayed as described above), with the frequency being reduced to the level exhibited by the negative control pBR328 [(7.7 ± 1.9) × 10−8]. The left end of Tn5520 is only 268 bp away from the beginning of the transposase gene and was not tested in similar experiments.
DISCUSSION
We have isolated and characterized a new 4.69-kb mobilizable transposon from B. fragilis LV23, designated Tn5520. DNA sequence analysis revealed the presence of two open reading frames (later named bipH and bmpH) whose predicted protein products were 47.3 and 58.2 kDa, respectively. Computer homology searches of the DNA and predicted protein sequences against available databases confirmed that bipH exhibited high sequence similarity to a variety of integrase-recombinase proteins and that bmpH was highly similar to Bacteroides mobilization proteins. Sequence analysis also revealed that Tn5520 did not modify its insertion site and that the termini of Tn5520 were characterized by a 22-bp imperfect inverted repeat. It was also evident from sequence analysis that Tn5520 demonstrated sequence preferences for insertion characterized by A-T-rich sequences. Analyses to assess the nature and requirements for transfer and mobilization of the element were performed. We demonstrated that bmpH was required for transfer of Tn5520 and that disruptions of bmpH completely abolished mobilization of Tn5520 in E. coli. In addition, we demonstrated that bmpH alone could confer mobilization properties on a nonmobilizable plasmid in E. coli. We also showed that, unlike other mobilizable transposons, Tn5520 was capable of transposition in E. coli. We determined that disruptions of bipH completely abolished transposition in E. coli, whereas disruptions elsewhere in Tn5520 had no effect on transposition.
The ends of Tn5520 were characterized by 22-bp imperfect inverted repeats. Compared with other transposons (19), the inverted-repeat-harboring termini of Tn5520 were A-T rich (15 of 22 nucleotides; 68.1% compared to an overall GC content of 46.9% for Tn5520) and exhibited ∼57% identity of the terminal nucleotides. This percentage is low; however, some other integrases with imperfect inverted repeats also exhibit low identity (<70%) at their termini (19). Tn5520 did not duplicate its target site upon insertion, as evidenced by sequence analysis. This is in contrast to Tn4399, which creates a 3-bp duplication of the target site upon insertion, with an additional 5 bp at the right end (17). We also observed that the sequence AATAA was found distal to the right end of Tn5520 in 12 of 15 insertion sites analyzed, which indicated that Tn5520 exhibits orientation and sequence preferences for insertion. Classic transposons of different families, like IS1 and Tn3, as well as conjugative transposons, like Tn916 from Enterococcus faecalis, target A-T-rich sequences (7, 12, 19). A preference for A-T-rich targets is also a feature of phage lambda integration, which is catalyzed by the Int protein (19).
The sequence specificity described above may be different from target site specificity, which may be random or nonrandom. Independent insertions of the 65-kb cryptic conjugative transposon XBU4422 occur at a specific site upstream of the tetX gene (42). The termini of XBU4422 are characterized by 23-bp imperfect inverted repeats that have identity with the tetX target site. (tetX is a cryptic Bacteroides tetracycline resistance determinant that is expressed in aerobically growing E. coli [37]. Of the 15 Tn5520 insertions that we isolated, the one used for DNA sequencing was located within the tetX gene in pGAT400ΔBglII.) The only identity to the target exhibited by the termini of Tn5520 is at the right end, where the last three nucleotides, TAA, are also present in the A-T-rich target, AATAA, found in 12 of 15 insertions.
DNA sequencing of Tn5520 revealed the presence of two large (≥500-bp) open reading frames. bipH (1,230 bp; predicted protein, 47.3 kDa) exhibited strong similarity upon translation to recombinase-integrase proteins from a variety of genera, including those of NBU1 and NBU2 (2, 10, 24–26, 30, 51). Most of the proteins to which BipH exhibits sequence similarity belong to the phage-integrase family of proteins. Members of this family perform integration reactions analogous to that catalyzed by phage lambda Int protein and are characterized by having no requirement for high-energy cofactors, having A-T-rich target sites, and being stimulated by supercoiled substrates (19). Unlike lambda integration, which results in a 7-bp 5′ extension, Tn5520 integration does not alter its target site, and as yet any requirement for host-encoded factors is unknown. Thus, the integrase of Tn5520 may belong to the phage-integrase class of cutting-and-rejoining enzymes, although its properties of insertion may be unique. Other mobilizable and conjugative transposons, like Tn4555 and Tn916, and NBU1 have also been found to integrate in a lambdoid phage-like process (7, 48).
bmpH (1,365 bp; predicted protein, 52.6 kDa) exhibited strong sequence similarity upon translation to mobilization proteins from NBU1, NBU2, and Tn4555 (22, 23, 46). As with Tn5520, NBU1, NBU2, and Tn4555 harbor single mobilization proteins (22, 46). The recently discovered Tn4551 from C. perfringens also contains a single mobilization protein, TnpZ. We presume that the single mobilization protein of Tn5520 is multifunctional, performing most, if not all, of the reactions required for the initiation of DNA transfer (recognition, binding, and specific cutting at the nick site). Of the other mobilizable transposons examined to date, Tn4399 harbors two proteins involved in mobilization (MocA and MocB), as do Bacteroides mobilizable plasmids (pIP417, pLV22a, and pBFTM10) (14, 28, 29, 49). Unlike BmpH, the latter mobilization proteins are smaller, averaging ∼235 amino acids in length (∼50% smaller than Tn4555 MobA and BmpH); thus, the larger size of BmpH may reflect its ability to be multifunctional. Secondary-structure predictions revealed that BmpH may be a stable, basic protein with no significant hydrophobic regions or transmembrane domains. However, a motif search predicted four myristoylation sites, one of which (amino acids 267GSLGSN272) also corresponded to a very weak transmembrane domain. This may indicate that BmpH is targeted or localized to the membrane surface but does not span the membrane. If BmpH is multifunctional, this prediction would be consistent with the nicking protein recruiting the nicked DNA strand to the cell membrane for transfer to a recipient.
Following isolation, we were able to study the transfer of Tn5520 from B. fragilis and its mobilization in E. coli. It was observed that transfer of Tn5520 in B. fragilis occurred without the requirement of coresident TET elements, as demonstrated by transfer from the TET element-devoid B. fragilis TM4000. In fact, the transfer of pHB23Δ5 from B. fragilis TM4000 was consistently 10-fold higher than that of the positive control pGAT400. Transfer of pHB23Δ5 and pGAT400 also occurred from TM4.23 at a frequency approximately 3 orders of magnitude greater than that from TM4000. The presence of TET elements in TM4.23 likely contributes to this increased level of transfer from TM4.23, as seen with plasmid (pBFTM10 and pLV22a) transfer, although the mechanism is unclear.
We also observed that Tn5520 was efficiently mobilized in E. coli in the presence of the IncPβ plasmid R751. Transposon insertions in bmpH completely abolished transfer, while those outside and distal to bmpH had no effect, indicating that BmpH was required and sufficient for mobilization. From DNA sequence analysis, we identified multiple repeat sequences (four direct, three inverted, and two palindromic) immediately proximal to bmpH, and we presume that the Tn5520 oriT is located in this region. A consensus nick site sequence based on the E. coli RP4 plasmid model (2396CTTGCCC2403) was also identified, indicating that the Tn5520 oriT may be present in this bmpH-proximal region (32). Transposon insertions in this region (pGV4, pGV5, and pGV6) reduced mobilization of Tn5520, indicating that the region was required and that the insertions may have altered the oriT such that transfer initiation processes were less efficient.
Tn5520 also transposes in E. coli, as evidenced by cointegrate formation with the pOX38::Kan plasmid. This is the first report of a mobilizable transposon exhibiting transposition activity in a genus other than that from which it was isolated. Cointegrates generated by the transposition event appeared to be stable, and transconjugant plasmid DNA recovered after transfer showed no resolution (data not shown). We reasoned that this may indicate a broad-host-range characteristic of Tn5520, and consequently we tested 100 anaerobes from different genera for the presence of Tn5520. We observed that 35% of the isolates tested positive by Southern hybridization (under conditions of high stringency) for the presence of Tn5520 (data not shown). This indicates that Tn5520 or Tn5520-like elements are widespread in anaerobes. The nature of the transposition event (in E. coli and Bacteroides spp.) still needs to be characterized, which will involve determination of whether Tn5520 is duplicated during cointegrate formation, determination of the nature of the junctions, and detection of any resolving activity.
In summary, Tn5520 is the smallest mobilizable transposon from any bacterium and presents an interesting case for speculation on the minimal size of a factor that can possess both transposition and transfer properties. Such a minimalistic design might be exploited as a template for the integration or addition of extraneous DNA (like antibiotic resistance cassettes), singly or in combination, to yield larger transferable drug resistance elements.
ACKNOWLEDGMENTS
This work was supported by VA Merit Review no. 001 to D.W.H.
We acknowledge helpful discussions with V. K. Viswanathan, Leonid Sitailo, and Kathleen Bass.
REFERENCES
- 1.Aldridge K E. The occurrence, virulence, and antimicrobial resistance of anaerobes in polymicrobial infections. Am J Surg. 1995;169:S2–S7. [PubMed] [Google Scholar]
- 2.Arendt E K, Daly C, Fitzgerald G F, van de Guchte M. Molecular characterization of lactococcal bacteriophage Tuc2009 and their use for site-specific plasmid integration in the chromosome of Tuc2009-resistant Lactococcus lactis MG1363. Appl Environ Microbiol. 1994;60:2324–2329. doi: 10.1128/aem.60.7.2324-2329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1998. [Google Scholar]
- 4.Beiluch V M, Tally F P. Pathophysiology of abscess formation. Clin Obstet Gynecol. 1983;10:93–103. [PubMed] [Google Scholar]
- 5.Bodner S J, Koenig M G, Goodman J S. Bacteremic Bacteroides infections. Ann Intern Med. 1970;73:537–544. doi: 10.7326/0003-4819-73-4-537. [DOI] [PubMed] [Google Scholar]
- 6.Brook I. Aerobic and anaerobic bacteriology of intracranial abscesses. Pediatr Neurol. 1992;8:210–214. doi: 10.1016/0887-8994(92)90070-f. [DOI] [PubMed] [Google Scholar]
- 7.Clewell D B, Flannagan S E, Jaworski D D. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 1995;3:230–236. doi: 10.1016/s0966-842x(00)88930-1. [DOI] [PubMed] [Google Scholar]
- 8.Cornick N A, Cuchural G J, Snydman D R, Jacobus N V, Iannini P B, Hills G B, Cleary T J, O’Keefe J P, Pierson C L, Finegold S M. The antimicrobial susceptibility patterns of the Bacteroides fragilis group in the United States. J Antimicrob Chemother. 1990;25:1011–1019. doi: 10.1093/jac/25.6.1011. [DOI] [PubMed] [Google Scholar]
- 9.Covarrubias L, Cervantes L, Covarrubias A, Soberon X, Vichido I, Blanco A, Kupersztoch-Portnoy Y, Bolivar F. Construction and characterization of new cloning vehicles. V. Mobilization and coding properties of pBR322 and several deletion derivatives including pBR327 and pBR328. Gene. 1981;13:25–35. doi: 10.1016/0378-1119(81)90040-8. [DOI] [PubMed] [Google Scholar]
- 10.Fleischmann R D, et al. Whole genome random sequencing and assembly of Haemophilus influenzae. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 11.Forste J P, Pansegrau W, Ziegelin G, Kroger M, Lanka E. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc Natl Acad Sci USA. 1989;86:1771–1775. doi: 10.1073/pnas.86.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grindley N D, Reed R R. Transpositional recombination in prokaryotes. Annu Rev Biochem. 1985;54:863–896. doi: 10.1146/annurev.bi.54.070185.004243. [DOI] [PubMed] [Google Scholar]
- 13.Guiney D G, Lanka E. Conjugative transfer of IncP plasmids. In: Thomas C M, editor. Promiscuous plasmids of gram negative bacteria. London, United Kingdom: Academic Press Ltd.; 1989. pp. 27–56. [Google Scholar]
- 14.Haggoud A, Reyssett G, Azeddoug H, Sebald M. Nucleotide sequence analysis of two 5-nitroimidazole resistance determinants from Bacteroides strains and of a new insertion sequence upstream of the two genes. Antimicrob Agents Chemother. 1994;38:1047–1051. doi: 10.1128/aac.38.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecht D W, Jagielo T J, Malamy M H. Conjugal transfer of antibiotic resistance factors in Bacteroides fragilis: the btgA and btgB genes of plasmid pBFTM10 are required for its transfer from Bacteroides fragilis and for its mobilization by IncPβ plasmid R751 in Escherichia coli. J Bacteriol. 1991;173:7471–7480. doi: 10.1128/jb.173.23.7471-7480.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hecht D W, Malamy M H. Tn4399, a conjugable transposable mobilizing element of Bacteroides fragilis. J Bacteriol. 1989;171:3603–3608. doi: 10.1128/jb.171.7.3603-3608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hecht D W, Thompson J S, Malamy M H. Characterization of the termini and transposition products of Tn4399, a conjugal mobilization transposon of Bacteroides fragilis. Proc Natl Acad Sci USA. 1989;86:5340–5344. doi: 10.1073/pnas.86.14.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ippen-Ihlen K A, Minkley E G. The conjugation system of F, the fertility factor of Escherichia coli. Annu Rev Genet. 1986;20:593–624. doi: 10.1146/annurev.ge.20.120186.003113. [DOI] [PubMed] [Google Scholar]
- 19.Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- 20.Lanka E, Wilkens B. DNA processing reactions in bacterial conjugation. Ann Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Shoemaker N B, Salyers A A. Excision, transfer, and integration of NBU1, a mobilizable site-selective insertion element. J Bacteriol. 1993;175:6588–6598. doi: 10.1128/jb.175.20.6578-6587.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L Y, Shoemaker N B, Wang G, Cole S P, Hashimoto M K, Wang J, Salyers A A. The mobilization regions of two Bacteroides integrated elements, NBU1 and NBU2, have only a single mobilization protein and may be on a cassette. J Bacteriol. 1995;177:3940–3945. doi: 10.1128/jb.177.14.3940-3945.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L Y, Shoemaker N B, Salyers A A. Characterization of the mobilization region of a Bacteroides insertion element (NBU1) that is excised and transferred by Bacteroides conjugative transposons. J Bacteriol. 1993;175:6588–6598. doi: 10.1128/jb.175.20.6588-6598.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lillehauq D, Birkeland N K. Characterization of genetic elements required for site-specific integration of the temperate lactococcal bacteriophage phi LC3 and construction of integration-negative phi LC3 mutants. J Bacteriol. 1993;175:1745–1755. doi: 10.1128/jb.175.6.1745-1755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lovett S T, Kolodner R D. Nucleotide sequence of the Escherichia coli recJ chromosomal region and construction of recJ overexpression plasmids. J Bacteriol. 1991;173:353–364. doi: 10.1128/jb.173.1.353-364.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizuno M, Masuda S, Takemaru K, Hosono S, Sato T, Takeuchi M, Kobayashi Y. Systematic sequencing of the 283kb 210 degrees-232 degrees region of the Bacillus subtilis genome containing the skin element and many sporulation genes. Microbiology. 1996;142:3103–3111. doi: 10.1099/13500872-142-11-3103. [DOI] [PubMed] [Google Scholar]
- 27.Morgan R M, Macrina F L. bctA: a novel pBF4 gene necessary for conjugal transfer in Bacteroides spp. Microbiology. 1997;143:2155–2165. doi: 10.1099/00221287-143-7-2155. [DOI] [PubMed] [Google Scholar]
- 28.Murphy C G, Malamy M H. Characterization of a “mobilization cassette” in transposon Tn4399 from Bacteroides fragilis. J Bacteriol. 1993;175:5814–5823. doi: 10.1128/jb.175.18.5814-5823.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy C G, Malamy M H. Requirements for strand- and site-specific cleavage within the oriT region of Tn4399, a mobilizing transposon from Bacteroides fragilis. J Bacteriol. 1995;177:3158–3165. doi: 10.1128/jb.177.11.3158-3165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nauta A, van Sinderen D, Karsens H, Smit E, Venema G, Kok J. Inducible gene expression mediated by a repressor-operator system isolated from Lactococcus lactis bacteriophage rlt. Mol Microbiol. 1996;19:1331–1341. doi: 10.1111/j.1365-2958.1996.tb02477.x. [DOI] [PubMed] [Google Scholar]
- 31.Novicki T J, Hecht D W. Characterization and DNA sequence of the mobilization region of pLV22a from Bacteroides fragilis. J Bacteriol. 1995;177:4466–4473. doi: 10.1128/jb.177.15.4466-4473.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pansegrau W, Lanka E. Common sequence motifs in DNA relaxases and nick regions from a variety of DNA transfer systems. Nucleic Acids Res. 1991;19:3455. doi: 10.1093/nar/19.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pansegrau W, Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Prog Nucleic Acid Res Mol Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 34.Pansegrau W, Lanka E. Mechanisms of initiation and termination reactions in conjugative DNA processing. J Biol Chem. 1996;271:13068–13076. doi: 10.1074/jbc.271.22.13068. [DOI] [PubMed] [Google Scholar]
- 35.Rasmussen J, Odelson D, Macrina F. Complete nucleotide and transcription of ermF, a macrolide-lincosamide-sterptogramin B resistance determinant from Bacteroides fragilis. J Bacteriol. 1986;168:523–533. doi: 10.1128/jb.168.2.523-533.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reyssett G, Haggoud A, Su W, Sebald M. Genetic and molecular analysis of pIP417 and pIP419: Bacteroides plasmids encoding 5-nitroimidazole resistance. Plasmid. 1992;27:181–190. doi: 10.1016/0147-619x(92)90020-b. [DOI] [PubMed] [Google Scholar]
- 37.Salyers A A. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin Microbiol Rev. 1992;5:387–399. doi: 10.1128/cmr.5.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salyers A A, Shoemaker N B. Resistance gene transfer in anaerobes: new insights, new problems. Clin Infect Dis. 1997;23:S36–S43. doi: 10.1093/clinids/23.supplement_1.s36. [DOI] [PubMed] [Google Scholar]
- 39.Salyers A A, Shoemaker N B, Li L Y, Stevens A M. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shoemaker N B, Getty C, Guthrie E P, Salyers A A. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J Bacteriol. 1986;166:959–965. doi: 10.1128/jb.166.3.959-965.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shoemaker N B, Salyers A A. Tetracycline-dependent appearance of plasmid-like forms in Bacteroides uniformis 0061 mediated by Bacteroides tetracycline resistance elements. J Bacteriol. 1988;170:1651–1657. doi: 10.1128/jb.170.4.1651-1657.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoemaker N B, Salyers A A. A cryptic 65-kilobase-pair transposon-like element isolated from Bacteroides uniformis has homology with Bacteroides conjugal tetracycline resistance elements. J Bacteriol. 1990;172:1694–1702. doi: 10.1128/jb.172.4.1694-1702.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith C J. Characterization of Bacteroides ovatus plasmid pBI136 and structure of its clindamycin resistance region. J Bacteriol. 1985;161:1069–1073. doi: 10.1128/jb.161.3.1069-1073.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith C J, Parker A C. Identification of a circular intermediate in the transfer and transposition of Tn4555, a mobilizable transposon from Bacteroides spp. J Bacteriol. 1993;175:2682–2691. doi: 10.1128/jb.175.9.2682-2691.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith C J, Parker A C. A gene product related to TraI is required for the mobilization of Bacteroides mobilizable transposons and plasmids. Mol Microbiol. 1996;20:741–750. doi: 10.1111/j.1365-2958.1996.tb02513.x. [DOI] [PubMed] [Google Scholar]
- 46.Smith C J, Parker A C. The transfer origin for Bacteroides mobilizable transposon Tn4555 is related to a plasmid family from gram-positive bacteria. J Bacteriol. 1998;180:435–439. doi: 10.1128/jb.180.2.435-439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith C J, Tribble G D, Bayley D P. Genetic elements of Bacteroides species: a moving story. Plasmid. 1998;40:12–29. doi: 10.1006/plas.1998.1347. [DOI] [PubMed] [Google Scholar]
- 48.Tribble G D, Parker A C, Smith C J. The Bacteroides mobilization transposon Tn4555 integrates by a site-specific recombination mechanism similar to that of the gram-positive bacterial element Tn916. J Bacteriol. 1997;179:2731–2739. doi: 10.1128/jb.179.8.2731-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trinh S, Haggoud A, Reyssett G. Conjugal transfer of the 5-nitroimidazole resistance plasmid pIP417 from Bacteroides vulgatus BV-17: characterization and nucleotide sequence analysis of the mobilization region. J Bacteriol. 1996;178:6671–6676. doi: 10.1128/jb.178.23.6671-6676.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valentine P J, Shoemaker N B, Salyers A A. Mobilization of Bacteroides plasmids by Bacteroides conjugal elements. J Bacteriol. 1988;170:1319–1324. doi: 10.1128/jb.170.3.1319-1324.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu A, Bertani L E, Haggard-Ljungquist E. Control of prophage integration and excision in bacteriophage P2: nucleotide sequences of the int gene and att sites. Gene. 1989;80:1–11. doi: 10.1016/0378-1119(89)90244-8. [DOI] [PubMed] [Google Scholar]