Abstract
The conjugative IncN plasmids pKM101 and pCU1 have previously been shown to contain identical oriT sequences as well as conserved restriction endonuclease cleavage patterns within their tra regions. Complementation analysis and sequence data presented here indicate that these two plasmids encode essentially identical conjugal DNA-processing proteins. This region contains three genes, traI, traJ, and traK, transcribed in the same orientation from a promoter that probably lies within or near the conjugal transfer origin (oriT). Three corresponding proteins were visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and complementation analysis confirmed that this region contains three tra complementation groups. All three proteins resemble proteins of the IncW plasmid R388 and other plasmids thought to have roles in processing of plasmid DNA during conjugation. The hydropathy profile of TraJ suggests a transmembrane topology similar to that of several homologous proteins. Both traK and traI were required for efficient interplasmid site-specific recombination at oriT, while traJ was not required. The leading region of pKM101 contains three genes (stbA, stbB, and stbC), null mutations in which cause elevated levels of plasmid instability. Plasmid instability was observed only in hosts that are proficient in interplasmid recombination, suggesting that this recombination can potentially lead to plasmid loss and that Stb proteins somehow overcome this, possibly via site-specific multimer resolution.
Studies on the conjugal transfer (tra) systems of several plasmids in gram-negative bacteria, including IncF, IncP, IncW, IncQ, IncI, and IncX plasmids, have demonstrated that these systems have functional similarities and extensive sequence similarities at the DNA and protein levels (reviewed in references 32 and 67). This family of DNA transfer systems also includes the virulence (vir) regulon carried by Ti (tumor-inducing) plasmids of Agrobacterium spp., which are responsible for the transfer of tumorigenic DNA to higher plants (75).
Conjugal DNA processing in these plasmids requires plasmid-encoded proteins that interact with a small DNA sequence known as the origin of transfer (oriT) to introduce a strand-specific cleavage at a unique site referred to as the nic site (25, 42, 44, 58, 65). Following cleavage, one protein remains covalently bound to the 5′ end of the cleaved strand, and this strand is unwound in the 5′-to-3′ direction (45). According to a widely held model, complementary-strand synthesis is initiated from the free 3′ end of the cleaved strand in a manner resembling rolling-circle replication (14). In this model, a second cleavage is introduced into the reconstituted oriT after unwinding, and the enzyme ligates the 3′ and 5′ ends of the unwound DNA (46). The mechanism by which the processed strand is transferred into the recipient cell is as yet unknown, but transfer and unwinding are believed to occur simultaneously (32).
In the IncF, IncW, IncP, IncI, and IncQ conjugative systems and the Agrobacterium vir system, cleavage at oriT requires the action of two proteins, the smaller of which binds to the oriT in a plasmid-specific manner (15, 39, 43, 44). It is believed that binding of the smaller protein is required for recognition of the nic site by the second, larger protein, which cleaves and reseals the oriT. Such enzymes are often designated relaxases. Three conserved sequence motifs have been identified within these relaxase domains, suggesting that they may all have a common ancestry (2, 47). In the IncF and IncW proteins, the C-terminal portions of these proteins contain a helicase activity that is believed to unwind the cleaved strand (37, 65), while the IncP, IncI, and IncQ relaxases are thought to lack this helicase activity. Unwinding of plasmid DNA in the latter plasmids is believed to be carried out by a host-encoded helicase (32, 67).
The conjugal transfer systems of two IncN plasmids, pKM101 (51, 52, 70–73) and pCU1 (26, 48, 49, 56), have also been described. The pilus-encoding region of pKM101 was previously described at the sequence level and shown to contain 10 genes that are required both for conjugative transfer of pKM101 and for sensitivity to several donor-specific bacteriophages that bind to the plasmid’s conjugal pilus (8, 51). Earlier complementation experiments indicated that the remaining portion of the tra region contained four complementation groups that are required for conjugation but not required for sensitivity to these phages (70). These complementation groups were therefore thought to direct the processing of plasmid DNA during conjugation. These genes are flanked by the oriT and the fip gene. fip is not required for conjugation but inhibits the fertility of coresident IncP plasmids (72).
Plasmid pCU1 shows striking conservation of restriction sites present in pKM101 over the entire tra region, and the nucleotide sequence of the oriT region is identical to that of pKM101 (9, 48). These nic sites also resemble that of the IncW plasmid R388 (49). The transfer systems of pCU1 and pKM101 are quite similar to that of the IncW plasmid R388 in other respects, including heterologous complementation of tra functions and the pattern of bacteriophage sensitivities imparted by their pili (4, 33, 35).
The experiments presented below characterize the conjugal DNA-processing regions of the IncN plasmids pCU1 and pKM101, showing them to be essentially identical at the DNA sequence level. Although this region was previously thought to contain four tra complementation groups, only three tra genes were identified in the sequence presented in this study. The three genes are transcribed on the same strand, probably from a promoter that lies near or within the oriT. The products of these tra genes rather strongly resemble their counterparts in the IncW plasmid R388 and more weakly resemble Tra proteins of other plasmids. Directly downstream from these genes is the fip gene, whose product is sufficient for the fertility inhibition of coresident IncP plasmids and which may be expressed as part of this putative tra operon. We also present the sequence and functional analysis of the leading region of these plasmids, which is located on the opposite side of oriT. This region contains three genes whose products appear to be required to prevent plasmid instability that can arise as a consequence of interplasmidic homologous recombination.
MATERIALS AND METHODS
Construction of recombinant plasmids.
The bacterial strains, plasmids, and bacteriophages used in this study are listed in Table 1. Standard cloning techniques were employed (57), using buffers and reaction conditions recommended by the enzyme suppliers. pSP34 was constructed by cloning the RK2 oriT as a BamHI fragment from pNH-Kan/oriT (22) into the BamHI site of pCU1 derivative pCU57Δ14 (48). pET-traK was constructed by cloning the traK coding region as a BstYI-HpaI fragment from pSP34 into the BamHI-HincII sites of pET23d. pET-traJ was created by using PCR amplification to create a DNA fragment containing traJ flanked by BamHI and EcoRI sites and cloning this fragment into the BamHI-EcoRI gap of pET23d. pET-traI was constructed by cloning the BglII-HindIII fragment from pSP34 into the BamHI-HindIII sites of pET23d. Each of the resulting junctions between the T7 promoter and the tra gene was checked by DNA sequencing. Plasmid pMIM101 was created by ligating a 3.5-kb BglII fragment containing the traI and fip genes of pKM101 into the BamHI site of pTZ18R such that these genes are transcribed from the Plac promoter of the vector. pSW345 was created by digesting pKM101 Ω155::Tn5 (30) with SmaI and HindIII and inserting the fragment that contains fip, traKJI, stbABC, and orfD into the SmaI-HindIII gap of plasmid pUC12Cm.
TABLE 1.
Strains, plasmids, and bacteriophages used in this study
| Strain, plasmid, or bacteriophage | Description | Reference or source |
|---|---|---|
| Strains | ||
| AB1157 | F−thr-1 leu-6 proA2 his-4 thi-1 argE3 lacY1 galK2 ara-14 xyl-5 mtl-1 tsx-33 rpsL31 supE44 | 68 |
| JC2926 | AB1157 recA13 | 68 |
| JC7623 | AB1157 recB21 recC22 sbcB15 | 68 |
| GW4212 | JC7623 recA::Cmr | 74 |
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 φ80dlacZΔM15 Δ(lacZYA-argF)U169 | 57 |
| HB101 | F−hsdS20 recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 met-1 supE44 λ− | 57 |
| POII1734 | F− MudII1734 lac+ara::(Mu cts) araD139 Δ(lac)X74 galU galK rpsL | 5 |
| CC118 | araD139 Δ(ara leu)7697 ΔlacX74 phoAΔ20 galE galK thi rpsE rpoB argE (Am)recA1 | 38 |
| S17-1 | thi pro hsdR hsdM+recA RP4 tra | 59 |
| BL21(DE3) | F−ompT rB−mB− λDE3 | 61 |
| PN1 | TnphoA in chromosome of CC118 | Iyer lab collection |
| Plasmids | ||
| pTZ18R | rep-ColE1, lacZα Apr | U.S. Biochemical |
| pET23d | rep-ColE1, T7 promoter, Apr | Novagen |
| pGW276 | pKM101 Ω440::Tn5 deleted across SalI sites | 30 |
| pGW277 | pKM101 Ω750::Tn5 deleted across SalI sites | 30 |
| pGW2132 | BglII fragment containing traJ-orfD cloned into BamHI site of pACYC184 | 70 |
| pNH-kan oriT | oriT-RK2 cloned into pUC1318, Kmr Apr | 22 |
| pCU57D14 | traI, traJ, and traK genes of pCU1 cloned into pACYC184, Cmr | 48 |
| pMK2004 | rep-ColE1, Apr Tcr | |
| pSP34 | traI, traJ, and traK genes of pCU1 and oriT of RK2 cloned into pACYC184, Cmr | This study |
| pSP27 | oriT of pCU1 cloned into pMK2004, Apr Tcr | This study |
| pET-traK | traK of pCU1 cloned into pET23d, Apr | This study |
| pET-traJ | traJ of pCU1 cloned into pET23d, Apr | This study |
| pET-traI | traI of pCU1 cloned into pET23d, Apr | This study |
| pSW345 | fip, traKJI, stbABC, and orfD cloned into pUC12Cm | This study |
| pMIM101 | traI and fip of pKM101 cloned into pTZ18R, Apr | This study |
| Bacteriophages | ||
| λ::TnphoA | cI1857 b221 Pam3 rex::TnphoA, Kmr | 69 |
| MudII1734 | lacZYA neo cts, Kmr | 7 |
Transposon mutagenesis.
pSP34 was mutagenized with transposon TnphoA (38) by incubating 1 ml of log-phase CC118(pSP34) with 1 ml of a λ::TnphoA lysate (ca. 109 phage) overnight at 30°C with gentle shaking. The culture was centrifuged and resuspended in 1 ml of Luria-Bertani (LB) medium, and 0.2 ml of this suspension was spread onto LB plates containing kanamycin (50 μg/ml) and chloramphenicol (60 μg/ml). A 0.5-ml portion of a kanamycin solution (10 mg/ml) was added to the center of each plate and allowed to be absorbed into the agar. The plates were incubated overnight at 37°C. The high level of kanamycin caused a zone of clearing containing a small number of isolated colonies. These colonies were selected for further analysis and found to contain TnphoA derivatives of pSP34. pET-traI was mutagenized with TnphoA by transforming it into Escherichia coli PN1, which carries TnphoA on the chromosome of strain CC118. The transformation mixture was incubated in LB medium overnight at 37°C and then plated on LB plates containing chloramphenicol and kanamycin. After overnight incubation at 37°C, the confluent colonies were recovered from the plate and plasmid DNA was extracted. A portion of this DNA was introduced into DH5α by electroporation and plated on LB plates containing chloramphenicol and kanamycin. Transformants were found by restriction digestion to contain TnphoA derivatives of pET-traI. Plasmids pMIM101 and pSW345 were mutagenized with transposon MudII1734 by published procedures (7).
DNA sequencing.
pKM101-derived DNA was sequenced by using derivatives of pMIM101 and pSW345 containing insertions of MudII1734 as template DNA. These plasmids were purified by using SpinBind columns (FMC Bioproducts) and sequenced by using a 373A Stretch DNA sequencer (ABI) and primers that hybridize to the left or right end of MudII1734. Sequencing reactions were carried out on both DNA strands with Taq DNA polymerase and DyeDeoxy Terminator Sequencing kits (ABI) and deoxynucleoside triphosphate substrates. Custom-made primers that hybridize to traI or fip DNA were used as needed to complete the sequence.
pCU1-derived DNA was sequenced either manually, using a modification of the U.S. Biochemical Sequenase protocol described previously (49), or by automated DNA sequencing with a 373A Stretch DNA sequencer (ABI). Sequences was determined from both strands by sequencing outward from pSP34::TnphoA or pET-traI::TnphoA inserts with primers that hybridize to phoA or to IS50 DNA. In the latter case, a restriction fragment containing DNA from only the right IS50, and hence only one primer site, was extracted from an agarose gel via use of GeneClean (Bio/Can Scientific). traI was sequenced by using plasmid pSP60 as a template and custom-made oligonucleotide primers. Inferred protein sequences were used to search public protein sequence databases by using the BLAST algorithm (1).
SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Derivatives of E. coli BL21(DE3) containing a derivative of pET23d containing individual tra genes were cultured to an optical density at 590 nm of 0.4 in M9 medium supplemented with ampicillin (30 μg/ml), thiamine (40 μg/ml), and all 20 amino acids (40 μg/ml) except methionine and cysteine. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to some cultures to a final concentration of 0.4 mM, and incubation was continued at 37°C for 40 min, at which time rifampin (200-μg/ml final concentration) was added. After a further 20 min at 37°C, 10 μCi of [35S]methionine was added to the culture, and incubation was continued at 37°C for 20 min. One milliliter of each culture was centrifuged, resuspended in 50 μl of protein loading buffer, and boiled for 5 min. The lysate was centrifuged for 5 min in a microcentrifuge, and 5 μl of the supernatant was loaded onto a sodium dodecyl sulfate (SDS)–12.5% discontinuous polyacrylamide gel (20). The gel was stained with Coomassie blue and autoradiographed with BioMax X-ray films (Dupont).
Genetic complementation of tra mutations.
Derivatives of strain JC2926 containing a derivative of pKM101 and a derivative of pSW345 were cultured in LB medium supplemented with kanamycin (50 μg/ml) and chloramphenicol (50 μg/ml) to an optical density at 600 nm of approximately 0.5. A single culture of the conjugal recipient (MM294) was cultured to a similar optical density and concentrated 50-fold by centrifugation. Fifty microliters of each donor culture was combined with 50 μl of the concentrated suspension of MM294 and spotted onto Millipore filters that had been placed on prewarmed LB agar medium. These plates were returned to a 37°C incubator for exactly 1 h, at which time the cells were resuspended in 1% NaCl. These cells were serially diluted and plated onto defined medium (AB salts and buffer) containing chloramphenicol (50 μg/ml) to select for transfer of pSW345 derivatives into MM294. Donor cells were enumerated by plating on LB agar supplemented with chloramphenicol (50 μg/ml), kanamycin (50 μg/ml), and streptomycin (250 μg/ml). Transfer efficiency was calculated as the number of recovered transconjugants per recovered donor per hour.
Plasmid cointegration assays.
Filter matings between recipient strain HB101rif and donor strain S17-1 containing pSP27 and a derivative of pSP34 were conducted as previously described (48). pSP34 contains the oriT site of plasmid RK2 and is therefore efficiently mobilized by the tra system of S17-1. pSM34 and pSP27 also contain the oriT of pCU1, and the assay measures the efficiency of cointegration between these sites. Transconjugants were selected on LB plates containing rifampcin (100 μg/ml) and either chloramphenicol (60 μg/ml) to select for transfer of pSP34 or tetracycline (20 μg/ml) to select for cointegrative transfer of pSP34 and pSP27.
Plasmid curing assays.
Strains were cultured from frozen permanent stocks into LB medium containing ampicillin (50 μg/ml) to minimize the accumulation to plasmid-free cells. When these cultures had reached saturation, they were serially diluted 104-fold, and 0.1 ml of the resulting cell suspension was used to inoculate a 10-ml culture of LB medium. Cultures were incubated to the stationary phase (approximately 20 generations) in 125-ml Erlenmeyer flasks with vigorous aeration. They were then serially diluted 106-fold in 100-fold increments. A 0.1-ml portion of the 104-fold dilution was used to inoculate a fresh 10-ml LB culture, while 0.1 ml of the 106 dilution was plated to determine the fraction of plasmid-free colony-forming units. The procedure was repeated two additional times (60 generations total).
Nucleotide sequence accession numbers.
The sequences reported here have been deposited in the GenBank DNA sequence database (accession no. U43676, AF000361, and AF109305).
RESULTS
The conjugal DNA-processing regions of pKM101 and pCU1.
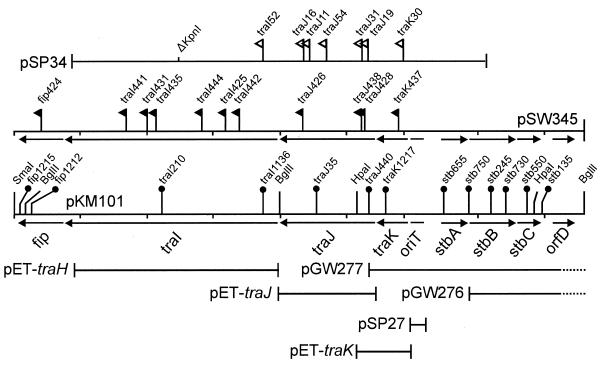
The DNA sequence of an 8.7-kb region between the SmaI-1 site and BglII-3 site of plasmid pKM101 (30) revealed eight open reading frames (ORFs) that are likely to encode proteins (Fig. 1). Four of these ORFs (designated traK, traJ, traI, and fip) are transcribed from right to left, while the remaining four ORFs (stbA, stbB, stbC, and orfD) are transcribed from left to right. traK and stbA are separated by a 513-nucleotide intergenic region that contains the oriT. The intergenic regions between the traK, traJ, traI, and fip genes contain 1, 10, and −1 nucleotides, respectively, suggesting that these genes are probably transcribed as an operon from a promoter that lies within or near oriT. The region containing traK to fip was sequenced independently in the laboratory of one of the authors (R.W.), who obtained an identical sequence (GenBank AF0000361). We also sequenced the traK, traJ, and traI genes of pCU1 (3.4 kb in all), and found that these sequences were identical to those of pKM101 at all but three bases in traI. One of the DNA sequence differences altered TraI residue 206 from alanine to threonine. The two remaining DNA sequence differences did not alter the protein sequence.
FIG. 1.
Genetic map of the conjugal DNA-processing regions of pKM101 and pCU1 and the stb region of pKM101. Open triangles, positions and orientations of TnphoA derivatives of pSP34; filled triangles, insertions of MudII1734 in plasmid pSW345; circles, Tn5 insertion derivatives of pKM101 (70). pGW277 and pGW276 are deletion derivatives of pKM101 (30); solid lines show DNA retained in these plasmids. pSP27 is a derivative of pMK2004 containing oriT of pCU1. Plasmids pET-traK, pET-traJ, and pET-traI are derivatives of pET23d that overexpress the corresponding tra products. Short vertical lines indicate the scale, measured in kilobases.
Visualization of the TraK, TraJ, and TraI proteins.
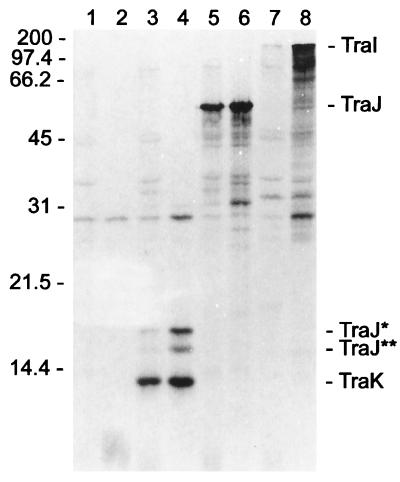
The traK, traJ, and traI genes of pCU1 were each cloned individually into the expression vector pET23d (Fig. 2). These constructs were introduced into E. coli BL21(DE3) to visualize the corresponding Tra proteins by SDS-PAGE. Plasmid pET-traK contains 43 bp upstream of the putative ATG start codon (Fig. 2) and hence should produce a native TraK protein (with 139 amino acids and a molecular mass of 15.3 kDa). pET-traK also contains 113 codons of traJ translationally fused to 33 codons of the pET23d vector (146 codons in all, encoding a peptide of 16.6 kDa). This plasmid directed the synthesis of three detectable proteins (Fig. 3, lane 4). The two most strongly expressed proteins (14.3 and 18 kDa) probably correspond to the native TraK protein and the TraJ fragment, respectively. The fainter protein (16 kDa) could be a truncated version of the TraJ fragment initiated from an internal start codon 20 codons downstream of the ATG used to initiate the 18-kDa TraJ peptide (Fig. 2). The detection of the TraJ peptides strongly suggests that traK and traJ are translationally coupled and therefore are expressed from a single promoter.
FIG. 2.
5′ ends of pCU1 tra genes cloned into the pET23d expression vector. Nucleotide sequences are shown in the upper lines, with corresponding amino acid sequences below. pCU1 nucleotide sequences are shown in lowercase letters, while pET nucleotide sequences are shown in uppercase letters. The amino acid sequences derived from pET23d are shown in roman type, while the amino acid sequences derived from pCU1 genes are italic. Restriction sites, ribosome binding sites (RBS), and putative ATG start codons are underlined.
FIG. 3.
SDS-PAGE of pCU1 Tra proteins. Positions of molecular mass standards (in kilodaltons) are indicated at the left. Lanes 1 and 2, pET23d; lanes 3 and 4, pET-traK; lanes 5 and 6, pET-traJ; lanes 7 and 8, pET-traI. The cultures used to make the cell extracts in lanes 2, 4, 6, and 8 were treated with 0.4 mM IPTG prior to addition of rifampin. Proteins corresponding to predicted Tra proteins are indicated at the right.
pET-traJ contains the native traJ coding region and ribosome binding site. Directly upstream of the start codon are 19 additional codons, most of which are derived from the plasmid vector (Fig. 2). The 58-kDa protein observed on SDS-PAGE (Fig. 3, lane 6) correlates well with the predicted molecular mass (59 kDa) of this TraJ fusion protein.
Plasmid pET-traI was used to visualize the 1,078-amino-acid TraI protein. However, the traI gene of this plasmid contains a 14-codon truncation at its 3′ end. The remainder of traI (codons 1 to 1064) is translationally fused at its 3′ end to 42 codons of an ORF of the pET23d vector, resulting in a fusion protein having a molecular mass of 123 kDa. Although pET23d was designed to provide a ribosome binding site and start codon, this start codon is out of frame with respect to traI, and translation of traI must therefore originate from the predicted traI start codon, which is preceded by a rather weak ribosome binding site (GGA). pET-traI expresses a single protein with a molecular mass of 155 kDa (Fig. 3, lane 8), indicating that this DNA fragment encodes one large protein. This finding confirms the nucleotide sequence analysis but is difficult to reconcile with earlier complementation analysis (70), which predicted that this region contains two complementation groups rather than one (see Discussion).
Amino acid sequence analysis of pCU1 and pKM101 DNA-processing proteins.
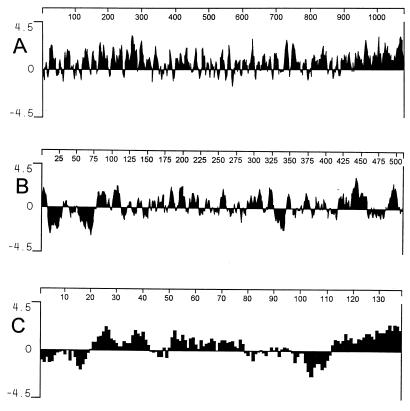
The traK, traJ, and traI genes encode proteins that are predicted to be largely hydrophilic, suggesting that they are soluble in aqueous environments. However, TraJ contains a strongly hydrophobic region between residues 10 and 25 (Fig. 4), which is preceded by arginine residues at residues 4 and 6. This suggests that the amino terminus of TraJ may be exported from the cytoplasm by the general protein export system (41). TraJ contains a second hydrophobic region between residues 54 and 74 followed by positively charged residues at positions 77, 79, 82, 85, 87, 88, and 91. This sequence resembles a stop transfer signal, suggesting that TraJ may have a transmembrane topology, with residues 26 to 53 located in the periplasmic space and the remainder of the protein located in the cytoplasm.
FIG. 4.
Hydropathy profiles of the TraI (A), TraJ (B), and TraK (C) proteins. The algorithm of Kyte and Doolittle (28) was used.
We used the TBLASTN algorithm (1) and the GenBank DNA sequence database to identify proteins having protein sequence similarity to TraK, TraJ, and TraI, and we found extensive sequence similarity to the products of a variety of conjugal transfer genes. The most closely related proteins are TrwA, TrwB, and TrwC of the IncW plasmid R388 (35). TraK and TrwA are 22% identical, with virtually all similarity limited to the carboxyl termini of the proteins (Fig. 5). Similarly, TraJ and TrwB are 37% identical, with similarity distributed along the entire lengths of these proteins. Like TraJ, TrwB is predicted to have a transmembrane topology (35). Finally, TraI and TrwC are 43% identical over their entire lengths, although similarity seems to be strongest at the proteins’ amino termini (Fig. 5). TraI is 108 residues longer than TrwC at its carboxyl terminus, and this nonconserved region contains many acidic amino acid residues. A-Tn9 insertion mutation in this region reduced but did not abolish conjugation (70). TrwA, TrwB, and TrwC are involved in processing of plasmid DNA during conjugation (35). They have been the subject of extensive sequence, genetic, and biochemical analysis, which is summarized in Discussion.
FIG. 5.
Protein sequence similarity between the Tra proteins of pKM101 and the Trw proteins of plasmid R388. The Clustal method (23) was used, with a gap penalty of 20 and a gap length penalty of 20.
Complementation analysis.
Previous complementation studies with pKM101::Tn5 insertion mutants identified five complementation groups: traK, traJ, traH, traI, and fip. Mutations in the region from traH to traK eliminated transfer, while mutations in fip abolished fertility inhibition of the IncP plasmids (70, 72). None of these genes was required for phage sensitivity, suggesting that none is required for synthesis of the conjugal pilus. In those earlier studies, complementation analysis was carried out by using transient heterozygotes that contained two Tn5 derivatives of pKM101, one introduced by transformation. Since the DNA sequence of this region indicates that there are three genes rather than four, we repeated this analysis with stable merodiploids. To do this, a region of pKM101 DNA (Fig. 1) was subcloned into pUC12Cm, creating pSW345. This plasmid was subjected to transposon mutagenesis with MudII1734 (7), and 10 derivatives having insertions in tra genes were isolated. These were introduced into derivatives of E. coli MC4100 containing tra mutants of pKM101, and the resulting merodiploids were tested for the ability to transfer kanamycin resistance to a conjugal recipient. These experiments indicate that this region contains three complementation groups rather than four (Table 2). The mutations previously designated traH alleles have therefore been renamed traI alleles.
TABLE 2.
Complementation of mutations in the conjugal DNA-processing region of pKM101
| pSW345 derivative | pKM101 derivativea:
|
|||
|---|---|---|---|---|
| traI210 | traI1136 | traJ35 | traK1217 | |
| traI441 | <1.0 × 10−5 | <1.0 × 10−5 | NDb | ND |
| traI431 | <1.0 × 10−5 | <1.0 × 10−5 | ND | ND |
| traI435 | <1.0 × 10−5 | <1.0 × 10−5 | 2.7 × 10−1 | 8.9 × 10−1 |
| traI444 | <1.0 × 10−5 | <1.0 × 10−5 | 5.0 × 10−1 | ND |
| traI425 | <1.0 × 10−5 | <1.0 × 10−5 | 3.3 × 10−1 | ND |
| traI442 | <1.0 × 10−5 | <1.0 × 10−5 | 5.6 × 10−1 | 5.3 × 10−1 |
| traJ426 | 3.8 × 10−1 | 3.8 × 10−1 | <1.0 × 10−5 | 5.0 × 10−1 |
| traJ438 | ND | 5.7 × 10−1 | <1.0 × 10−5 | 6.7 × 10−1 |
| traJ428 | ND | 6.8 × 10−1 | <1.0 × 10−5 | 3.3 × 10−1 |
| traK437 | 3.9 × 10−1 | 6.9 × 10−1 | 9.1 × 10−1 | <1.0 × 10−5 |
| fip424 | 1.2 | 1.8 | 1.6 | 1.3 |
Values represent numbers of transconjugants per donor per hour.
ND, not determined.
Complementation experiments between Tn5 mutants of pSP34 and Tn5 mutants of pKM101 were also conducted (data not shown). As expected, pSP34 was able to complement each of the pKM101 Tn5 mutants. Complementation experiments between pKM101::Tn5 derivatives and pSP34::Tn5 derivatives produced results similar to those obtained with pKM101::Tn5 and pSW345::MudII1734 derivatives (data not shown).
Intracellular site-specific recombination at oriT.
Intramolecular site-specific recombination between two oriTs on the same plasmid has been observed previously and used as an assay for oriT-processing activity (5, 6). In this study, an assay involving intermolecular site-specific recombination between oriTs carried on two separate plasmids was used to determine which of the tra functions are required for cleavage and religation at oriT during transfer.
In this assay, TnphoA mutants of pSP34 were tested for their ability to mobilize a second plasmid (pSP27) containing the oriT of pCU1 from the E. coli donor strain S17-1 into the E. coli recipient strain HB101rif. pSP34 carries the traK, traJ, and traI genes and oriT of pCU1. This construct also carries the oriT region of IncP plasmid RK2, which allows it to be mobilized efficiently by the IncP tra system carried on the chromosome of strain S17-1. Mobilization of pSP27 from S17-1(pSP34)(pSP27) occurs via a process known as conduction (54), in which pSP27 is integrated into pSP34 at the IncN oriT sites of both plasmids. The resulting cointegrate plasmid is then mobilized by transfer initiated at the IncP oriT. This process requires (i) some or all of the pCU1 tra genes on pSP34 (see below); (ii) the RK2 oriT of pSP34, since pCU57D14, which lacks this site, is not mobilized efficiently from S17-1 (data not shown); and (iii) the pCU1 oriT of pSP27, since pMK2004 lacks this site and is not efficiently mobilized (Table 3).
TABLE 3.
Mobilization of pSP27 by pSP34 tra mutants from S17-1
| Mobilizing plasmid | Mobilized plasmid | Mobilization frequencya | No. of transconjugants with the following plasmid structure:
|
|||
|---|---|---|---|---|---|---|
| Cointegration at oriT | Cointegration in vector | Parental | Other | |||
| pSP34 | pMK2004 | 9.8 × 10−6 | 0 | 5 | 7 | 0 |
| pSP27 | 2.4 × 10−3 | 21 | 0 | 4 | 0 | |
| pSP34 traI52 | pSP27 | 4.6 × 10−6 | 3 | 7 | 6 | 0 |
| pSP34 traIΔKpn | pSP27 | 2.4 × 10−7 | 2 | 0 | 2 | 1 |
| pSP34 traJ16 | pSP27 | 4.7 × 10−4 | 1 | 0 | 9 | 0 |
| pSP34 traJ11 | pSP27 | 5.4 × 10−5 | 6 | 0 | 2 | 0 |
| pSP34 traJ54 | pSP27 | 5.1 × 10−4 | 3 | 0 | 2 | 0 |
| pSP34 traJ31 | pSP27 | 2.9 × 10−4 | 5 | 0 | 0 | 0 |
| pSP34 traJ19 | pSP27 | 1.7 × 10−4 | 7 | 0 | 2 | 1 |
| pSP34 traK30 | pSP27 | 1.0 × 10−5 | 3 | 4 | 4 | 3 |
Mobilization frequencies are given as the number of transconjugants receiving pSP27 (or pMK2004) per transconjugant receiving the pSP34 derivative. In this assay, pSP34 (and derivatives) is efficiently transferred via its RK2 oriT sites and the RK2 tra genes on the chromosome. Transfer of pSP27 requires cointegration with pSP34 via the pCU1 oriT sites found on both plasmids.
The mobilization frequency of pSP27 was decreased to various extents by tra mutations in pSP34 (Table 3). Insertions in traI reduced mobilization of pSP27 by 1,000-fold, while the traK insertion caused a 100-fold reduction and traJ mutations caused only a 10-fold reduction. It seemed possible that not all transconjugants would have plasmids cointegrated at oriT. To test this, we took advantage of the fact that the oriT of pSP34 is contained on a 2.3-kb HpaI fragment, while pSP27 (5.4 kb in length) has no HpaI sites. Cointegration at oriT would therefore create a plasmid containing a diagnostic 7.7-kb HpaI fragment. When S17-1(pSP34)(pSP27) was used as a donor, 84% of the transconjugants (21 of 25) contained a plasmid with a 7.7-kb HpaI fragment. By comparison, when S17-1(pSP34)(pMK2004) was used as a donor, none of the transconjugants had such a fragment, indicating that mobilization of pMK2004 must have occurred by some other mechanism. Integration into the vector portion of the plasmid could also occur via recombination at the oriV sites on either plasmid, as described by Reimmann and Haas (54). The presence of two unaltered parental plasmids would presumably result from the resolution of such a cointegrate following transfer.
As described above, mutations in the tra genes of pSP34 decreased but did not abolish the mobilization of pSP27. However, most of the transconjugants did not have plasmids that had cointegrated via oriT sites. When the donor contained a traK mutation, only 19% of the transconjugants (3 of 16) had plasmids cointegrated at oriT. Similarly, when the donor contained a traI mutation, only 24% of the transconjugants (5 of 21) had this structure. In contrast, when the donor strain contained a traJ mutation, 58% of the transconjugants (22 of 38) had plasmids cointegrated at their oriT sequences. These results provide further support for the idea that both traK and traI are important for cointegration and hence for cleavage and resealing of oriT sites during conjugation, while traJ does not play a central role.
A locus required for stable plasmid inheritance.
Transposon insertions within a 1.5-kb region of pKM101 were found somewhat serendipitously to decrease the stable inheritance of the plasmid, causing plasmid-free bacteria to accumulate during prolonged culturing. This locus was first described as being required for sensitivity to the donor-specific bacteriophages Ike and PRD1, even though it is not required for efficient conjugation (unpublished data). It was subsequently found that strains containing stb mutants of pKM101 formed wild-type plaques on media containing kanamycin or ampicillin, while plaques were not detectable on antibiotic-free media. This was thought to be due to plasmid-free, phage-resistant bacteria overgrowing and obscuring the plaques. Additional alleles of this locus were later identified during two subsequent screens, one for deficiency in the fertility of coresident IncP plasmids (the Fip phenotype) and another for deficiency in entry exclusion (the Eex phenotype). However, when plasmid-free bacteria were eliminated (either by using antibiotics or, as described below, by altering the host genotype), mutants with transposon insertion mutations in this region were found to be Fip+ and Eex+.
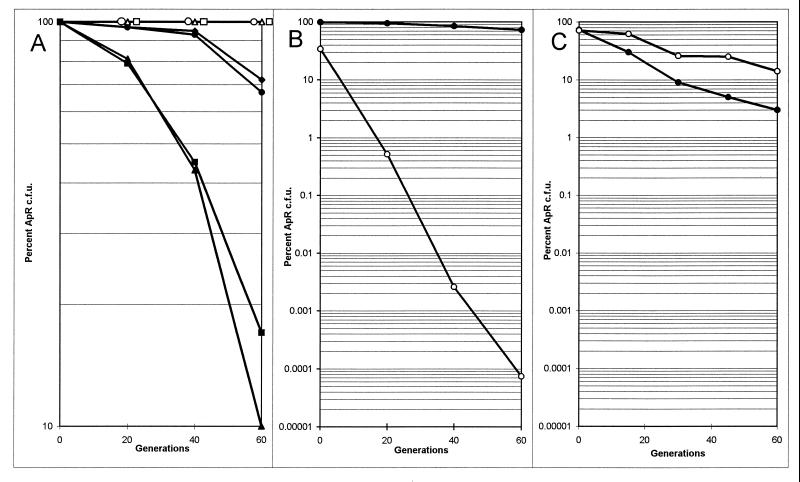
We quantitated the rate of loss of Apr from cells containing pKM101 and its stb derivatives. Two strains containing stb::Tn5 derivatives of pKM101 lost Apr at a 10-fold-higher rate than pKM101 itself (Fig. 6A). Both curves tend to arc downwards, suggesting that plasmid-free cells may also have a slightly higher division rate than plasmid-containing cells. These data indicate that about 4% of the cells of a culture containing an stb mutant plasmid became Aps per generation, while cells containing pKM101 became Aps at a rate of about 0.3% per generation.
FIG. 6.
Curing of pKM101 and its derivatives during prolonged culturing in the absence of antibiotic selection. (A) AB1157(pKM101) (●), AB1157(pKM101 stbA655::Tn5) (■), AB1157(pKM101 stbC135::Tn5) (▴), AB1157(pKM101 Ω155::Tn5) (⧫), JC2926(pKM101) (○), JC2926(pKM101 stbA655::Tn5) (□), and JC2926(pKM101 stbC135::Tn5) (▵). (B) JC7623(pGW277) (●) and JC7623(pGW276) (○). (C) GW4212(pGW277) (●) and GW4212(pGW276) (○).
We attempted to complement this deficiency by using pGW2132 (a derivative of pACYC184 containing the traJ, traK, stbABC, and orfD genes of pKM101, [70]). The complementation assays described above were originally carried out within a recA strain of E. coli. To our surprise, stb pKM101 derivatives were fully stable in this genetic background, even in the absence of a second plasmid (see below). These assays were therefore repeated with a recombination-proficient host, and it was found that pGW2132 did not increase the stability of coresident stb derivatives of pKM101 (data not shown). However, interpretation of this result is difficult because both pGW2132 and its parent pACYC184 appeared to destabilize pKM101. Therefore, nonspecific competition between these two plasmids or other interactions may have obscured any complementation.
We subsequently compared plasmid stability in a RecA+ host (AB1157) and an isogenic RecA− host (JC2926). In the recA strain, we did not detect loss of either pKM101 or an stb insertion derivative even after 60 generations (Fig. 6A, open symbols). We conclude that plasmid instability requires homologous recombination and that proteins encoded by the stb locus prevent this recombination-dependent instability.
Several multicopy plasmids have been observed to be particularly unstable in a recB recC sbcB strain (27). The stabilities of several pKM101 derivatives were therefore tested in this background. These tests were done with a set of deletion derivatives of pKM101 rather than transposon insertion derivatives. When we attempted to introduce these plasmids into strain JC7623 by transformation, we found that pKM101 itself and deletion derivatives that retained stb could be readily introduced but that all deletion derivatives lacking stb yielded either no transformants or a small number of slow-growing colonies (data not shown). We quantitated the stabilities of pGW276 (stb) and pGW277 (stb+) (Fig. 1). The former plasmid was lost at a rate of 20% per generation, while the latter plasmid was lost at a rate of only 0.5% per generation (Fig. 6B). To determine whether this instability was dependent upon homologous recombination, we introduced the same two plasmids into GW4212, which contains mutations in the recB, recC, sbcB, and recA genes (69). Although both strains lost Apr at detectable rates, the two plasmids seem to be about equally stable (Fig. 6C). If anything, pGW276 (which lacks stb) appeared to be slightly more stable in this host than pGW277, although this difference was very slight. We conclude that the plasmid instability observed in JC7623(pGW276) is dependent upon a recombination-proficient genotype.
Sequence analysis of this region revealed four ORFs, designated stbA, stbB, stbC, and orfD, encoding products having predicted molecular masses of 15.7, 26.4, 13.5, and 13.2 kDa, respectively. These ORFs are identical to the orfA, orfB, orfC, and orfD described by Delver and Belogurov (10). Insertions in stbA, stbB, or stbC confer the unstable plasmid segregation phenotype, while an insertion in orfD did not affect stability (data not shown). The stbA and stbB genes overlap by 17 nucleotides, and stbB and stbC are separated by 1 nucleotide, suggesting that these three genes are transcribed as an operon from a promoter that lies near oriT. In contrast, orfD is separated from stbC by 181 nucleotides, suggesting that expression of this gene would require a promoter located within this intergenic region. All four proteins contain predominantly hydrophilic amino acid residues, although the carboxyl terminus of StbC is hydrophobic. None of the products of these four ORFs showed any significant sequence similarity to other known proteins.
The DNA sequence just upstream of stbA contains a striking pattern of repeated DNA sequences (Fig. 7A). Thirteen such direct repeats were found, each of which showed a remarkable similarity to a consensus sequence (Fig. 7B). Directly upstream of these repeats is an extensive dyad symmetry (inverted arrows in Fig. 7A) and the oriT sequence (shaded residues in Fig. 7A). Directly downstream of these repeats is the putative ribosome binding site for stbA. Within the region containing these repeats is a possible promoter for the stb operon (underlined). Therefore, if these repeats provide a binding site for one or more Stb proteins, binding of these proteins might plausibly repress transcription of the stbA promoter.
FIG. 7.
DNA sequence of the intergenic region between traK and stbA. (A) Putative promoters, ribosome binding sites (RBS), and translation start sites for traK and stbA are underlined. The oriT sequence is shaded, while the nucleotides that flank the proposed nic site are double underlined. A dyad symmetry is indicated by inverted arrows, and repeated sequences are indicated by square brackets. (B) Alignment of the repeated sequences.
DISCUSSION
Restriction maps of pCU1 and pKM101 suggested that they contain very similar conjugal transfer regions. The data presented in this paper demonstrate that their conjugal DNA-processing regions are identical at all but 3 nucleotides over a 5,879-bp region encompassing oriT, traK, traJ, and traI. pCU1 and pKM101 also have strong sequence similarity at the 2,366-bp kik region, which has been sequenced for both plasmids in previous studies (51, 55). The variation observed over both regions represents a substitution rate of less than 0.2%. This high degree of sequence identity at both regions combined with the lack of any restriction site polymorphisms over the entire tra region strongly suggests that the tra regions of pCU1 and pKM101 are extremely similar and diverged from a common ancestor relatively recently. Furthermore, it is possible that this identity extends past the tra region. pCU1 and pKM101 have identical restriction maps except for the regions that encode antibiotic resistances. pKM101 is a deletion derivative of a larger plasmid, R46, which contains resistance determinants against ampicillin, streptomycin-spectinomycin, sulfanomides, tetracycline, and arsenate. Some of these genes are found within an integron (18). In contrast, pCU1 confers resistance to ampicillin and streptomycin-spectinomycin. It is possible that the only significant differences between pCU1, pKM101, and R46 are found in these clustered antibiotic resistance determinants. The deletion that created pKM101 appears to have been mediated by insertion sequence IS26 (17, 31). It is possible that pCU1 was also derived from an R46-like plasmid by a similar in vivo deletion.
The nucleotide sequences of the conjugal DNA-processing genes of pKM101 and pCU1 identify three genes in this region that are required for conjugation. TraI is homologous to TrwC of the IncW plasmid R388 (Fig. 5), and both proteins are also related to the TraI protein of plasmid F. Both the R388 and F plasmid proteins are known to contain an amino-terminal oriT-specific nucleolytic function and a carboxyl-terminal helicase function (36, 37, 65). The intermolecular-recombination results presented in this study strongly suggest that the IncN TraI has a comparable nucleolytic activity. The corresponding relaxases of the TraI protein of plasmid RK2 and the VirD2 protein of Agrobacterium tumefaciens have tyrosine residues (at residues 22 and 29, respectively) which are involved in covalent binding of the protein to the 5′ end of the cleaved strand and are considered to be part of the catalytic centers of these enzymes (46, 47, 66). The IncN TraI protein has four tyrosine residues at positions 18, 19, 26, and 27. This arrangement is reminiscent of the two tyrosine residues separated by three amino acids in the A protein of phage φX174, which is required for rolling-circle replication (21). For the φX174 system, it was postulated that the two tyrosine residues alternate in cleaving within the replication origin. This situation could assist in the intermolecular recombination event observed in this study. It is conceivable that each tyrosine residue (or one from each pair) cleaves the DNA strand and binds to separate pSP27 and pSP34 oriTs, bringing them into close proximity to each other to allow the transesterification step between the free 3′ OH of one plasmid and the 5′ phosphate of the other, resulting in the formation of a cointegrate plasmid.
Intermolecular recombination at the pCU1 oriT required both the traI and traK genes, suggesting that TraK may be a functional homologue of the IncF TraY protein. The TraY proteins of both the F plasmid and R100 are required for DNA cleavage both in vivo and in vitro (25, 42) and for oriT-mediated recombination (6). Alignment of the TraK protein with the F TraY protein shows little sequence similarity. On the other hand, there is limited sequence identity, primarily in their carboxyl termini, between TraK and the TrwA protein of IncW plasmid R388. TrwA is not required for intramolecular recombination between two R388 oriT sites in vivo (33) but has been shown to enhance in vitro cleavage by TrwC (39). The fact that R388 TrwA is not required for recombination at oriT while pKM101 TraK is required may be related to the sensitivities of these two recombination assays. In the IncW study, recombination was identified by the loss of an antibiotic marker situated between two copies of the oriT (33). Detection of a recombination event required that all copies of the plasmid within the cell carry the deletion. As a result, individual recombination events within the cell could be masked by the presence of plasmids that had not undergone recombination. The assay described in this study selects for plasmids which have undergone recombination, since only those pSP27 plasmids which cointegrate into the larger pSP34 plasmid are transferred.
TraJ is the only Tra protein in this region whose hydropathy profile suggests a transmembrane topology. Homologous proteins are found in virtually all conjugation systems. Members of this family of proteins have similar hydropathy profiles and in some cases have been shown to be associated with the inner membrane (43). It has been postulated that such a protein could be used to bind both the relaxosome and the mating pore and, by doing so, to bring the DNA in juxtaposition to the pore. This could explain why traJ mutations caused such a slight reduction in conduction of pSP27. Binding of pSP27 and pSP34 DNA to the membrane via TraJ prior to cleavage could enhance intermolecular recombination by bringing the two plasmids into closer proximity. Involvement of the traJ function in the site-specific recombination event has not been observed for the TraJ homologue of any other transfer system studied to date (33, 42, 46). Alternatively, traJ::Tn5 mutations could exert polar effects on expression of the downstream traI gene, although these same mutations did not appear to be polar in complementation assays.
Downstream of traI is a gene designated fip, which abolishes the conjugation of coresident IncP plasmids (72). fip appears to be part of the traKJI operon, although the significance of this coexpression is not understood. Immediately downstream from fip is the nuc gene, which is transcribed convergently to fip (53). Like fip, nuc is also the last gene in a tra operon and has no direct role in conjugation.
It was initially surprising that three tra genes were found in this region rather than four, since this region was previously thought to have four complementation groups (70). We therefore carried out additional complementation studies, using stable heterodiploid strains rather than the transient heterodiploids used previously. We identified three complementation groups, in agreement with the sequence data. Mutations previously designated as being in the traH complementation group were therefore renamed traI mutations. It is far from clear why the earlier study indicated that traI mutations fell into two complementation groups. However, the sequence of traI suggests an internal translation start site at codon 482 (AAGAAGG-N5-ATG). This region is immediately upstream of the putative helicase domain. This would suggest that TraI could be made in two forms, a full-length form and a truncated form containing only a helicase domain. The TraI protein of F has extremely similar properties, since a protein designated TraI* is translated by using an internal initiation codon. TraI* contains the helicase domain but not the relaxase domain (65). However, if this explanation is correct, is does not explain why only three complementation groups were obtained in the present study. We therefore cannot at this time fully understand the discrepancy between the older analysis and the present one.
We have also described a locus of pKM101 that appears to play a role in preventing recombination-mediated plasmid instability. While the cause of this instability is unclear, several other plasmids have been reported to have functionally similar genes, and in some cases these genes have been shown to encode site-specific recombination systems. These include the parA gene of RP4 (12), the D protein of mini-F (29), the per gene of R46 (11), and the cre gene of bacteriophage P1 (59), among others. The ParA and Per proteins are homologous to the resolvase protein of Tn3, while the D and Cre proteins are not homologous to other known proteins, and none is similar to StbA, StbB, or StbC or pKM101. The hallmark of most of these systems is that they are needed only in recombination-proficient hosts. It is believed that two identical copies of any multicopy plasmid can undergo homologous recombination, resulting in a head-to-tail dimer, which would effectively decrease plasmid copy number and could lead to inefficient partitioning to daughter cells during cell division (61). These site-specific recombination systems are thought to convert dimeric plasmids to monomers, thereby enhancing plasmid stability. We postulate that the stb locus of pKM101 may play a similar role. If so, it is not clear why three proteins would be required, since all the site-specific recombination systems listed above require just one protein. Perhaps only stbC is required for stability, and insertion mutations in stbA or stbB prevent expression of stbC by transcriptional polarity.
Each of these recombinases act at a particular DNA sequence, denoted the par site in RP4, the rfsF site in mini-F, the per site in R46, and the lox site in P1. Each of these sites is composed of direct or inverted repeats that provide binding sites for the resolvase proteins. In all cases except the lox-cre system, these sites are located directly upstream of the respective recombinase genes, and each recombinase serves as a transcriptional autorepressor as well as a recombinase. This provides a simple mechanism for the synthesis of sufficient amounts of protein to saturate the binding site. We hypothesize that if in fact the stb operon encodes a site-specific recombinase, the direct repeats found directly upstream of stbA could provide a cognate resolution site. If so, binding of one or more of these proteins could cause negative autoregulation.
As described above, one of these recombinase systems (per) is found on plasmid R46. Interestingly, R46 is the direct parent of pKM101, which was derived by an in vivo deletion of 15 kb of R46 DNA (17, 31). This deleted DNA includes the per gene. Therefore, if our model for stb function is accurate, it would appear that R46 has two such systems, one encoded by stb and the other encoded by per. In agreement with previous studies, we find that R46 is not detectably lost from a population of bacteria even after 60 generations (unpublished data). In constrast, pKM101 is lost at a detectable rate, which is consistent with published data that per mutants of R46 are detectably unstable (11). In all cases, instability occurred only in a recombination-proficient host.
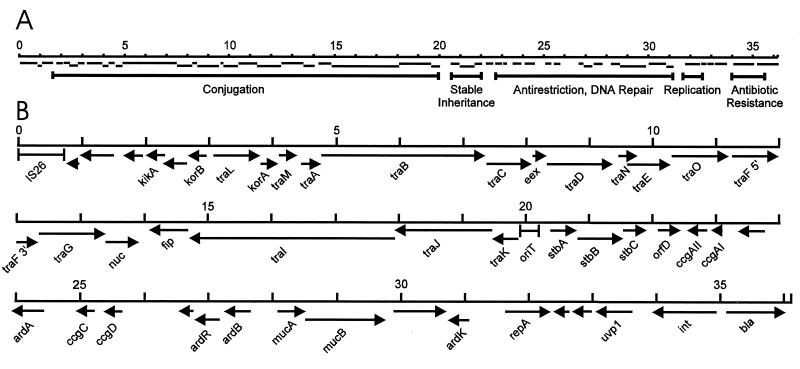
Plasmid pKM101 (36,255 bp) has now been sequenced in its entirety. Figure 8 shows the positions and transcriptional orientations of all previously characterized genes of this plasmid as well as 10 uncharacterized ORFs. Approximately half of the DNA of this plasmid encodes proteins that play some role in conjugation. pKM101 also contains genes that direct vegetative replication, stable plasmid inheritance, inhibition of host restriction systems, and error-prone repair of damaged DNA (see the legend to Fig. 8 for more detail). A more detailed description of pKM101 and its parent R46 is in preparation and will be the subject of a future study.
FIG. 8.
Functional map (A) and physical map (B) of pKM101. Labelled arrows denote characterized genes, while unlabelled arrows denote uncharacterized ORFs. Previous maps of pKM101 started at the unique EcoRI site (31). To avoid confusion between the locations of genes in pKM101 and its parental plasmid R46, we have numbered the sequence from the first nucleotide of IS26 (which is common to both plasmids). Gene designations: bla, oxa2 β-lactamase (19); IS26, insertion sequence IS26, (formerly denoted IS46); kikA, required for killing of Klebsiella strains during conjugation (24); traM to -G, required for conjugation and for sensitivity to donor-specific bacteriophages (51); traI to -K, conjugal DNA processing (this study); korA and korB, corepressors of the korB, traL, and traN promoters (40); eex, entry exclusion (52); nuc, periplasmic endonucleolytic DNase (53); fip, fertility inhibition of coresident IncP plasmids (70); stbA to -C: stable plasmid inheritance (this study); ardA and ardB, inhibition of host DNA restriction enzymes (3); ardR and ardK, regulators of ardA, ardB, ccgAI, ccgAII, ccgC, ccgD, and repA (10); ccgAI, ccgAII, ccgC, and ccgD, genes of unknown function that are regulated by ArdK and ArdR (10); mucA and mucB, error-prone DNA repair (50); mpr, unknown function, possible metalloprotease (10); repA, plasmid vegetative replication (10); uvp1, site-specific recombinase (64); int, conserved gene within integron (19). The complete DNA sequence of pKM101 was compiled by using sequence data contained in the GenBank DNA sequence database (accession no. AF000360, U09868, U43676, U72482, U00430, U00434, L09114, M81860, Y00358, and X06046).
ACKNOWLEDGMENTS
We thank Peter Diamandis for help with pKM101 stability assays.
This study was supported by Public Health Service grants GM42893 to S.C.W. and CA21615 to G.C.W.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Balzer B, Pansegrau W, Lanka E. Essential motifs of relaxase (TraI) and TraG proteins involved in conjugative transfer of plasmid RP4. J Bacteriol. 1994;176:4285–4295. doi: 10.1128/jb.176.14.4285-4295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belogurov A A, Delver E P, Rodzevich O V. Plasmid pKM101 encodes two nonhomologous antirestriction proteins (ArdA and ArdB) whose expression is controlled by homologous regulatory sequences. J Bacteriol. 1993;175:4843–4850. doi: 10.1128/jb.175.15.4843-4850.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolland S, Llosa M, Avila P, de la Cruz F. General organization of the conjugal transfer genes of the IncW plasmid R388 and interactions between R388 and IncN and IncP plasmids. J Bacteriol. 1990;172:5795–5802. doi: 10.1128/jb.172.10.5795-5802.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brasch M A, Meyer R J. A 38 base-pair segment of DNA is required in cis for conjugative mobilization of broad host-range plasmid R1162. J Mol Biol. 1987;198:361–369. doi: 10.1016/0022-2836(87)90286-5. [DOI] [PubMed] [Google Scholar]
- 6.Carter J R, Porter R D. traY and traI are required for oriT-dependent enhanced recombination between lac-containing plasmids and lambda plac5. J Bacteriol. 1991;173:1027–1034. doi: 10.1128/jb.173.3.1027-1034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castilho B A, Olfson P, Casadaban M J. Plasmid insertion mutagenesis and lac gene fusion with mini-Mu bacteriophage transposon. J Bacteriol. 1984;158:488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cellini C, Kalogeraki V S, Winans S C. The hydrophobic TraM protein of pKM101 is required for conjugal transfer and sensitivity to donor-specific bacteriophage. Plasmid. 1997;37:181–187. doi: 10.1006/plas.1997.1291. [DOI] [PubMed] [Google Scholar]
- 9.Coupland G M, Brown A M C, Willetts N S. The origin of transfer (oriT) of the conjugative plasmid R46: characterization by deletion analysis and DNA sequencing. Mol Gen Genet. 1987;208:219–225. doi: 10.1007/BF00330445. [DOI] [PubMed] [Google Scholar]
- 10.Delver E P, Belogurov A A. Organization of the leading region of IncN plasmid pKM101 (R46): a regulation controlled by CUP sequence elements. J Mol Biol. 1997;271:13–30. doi: 10.1006/jmbi.1997.1124. [DOI] [PubMed] [Google Scholar]
- 11.Dodd H M, Bennett P M. Location of the site-specific recombination system of R46: a function necessary for plasmid maintenance. J Gen Microbiol. 1986;132:1009–1020. doi: 10.1099/00221287-132-4-1009. [DOI] [PubMed] [Google Scholar]
- 12.Eberl L, Kristensen C S, Givskov M, Grohmann E, Gerlitz M, Schwab H. Analysis of the multimer resolution system encoded by the parCBA operon of broad-host-range plasmid RP4. Mol Microbiol. 1994;12:131–141. doi: 10.1111/j.1365-2958.1994.tb01002.x. [DOI] [PubMed] [Google Scholar]
- 13.Eberl L, Givskov M, Schwab H. The divergent promoters mediating transcription of the par locus of plasmid RP4 are subject to autoregulation. Mol Microbiol. 1992;6:1969–1979. doi: 10.1111/j.1365-2958.1992.tb01370.x. [DOI] [PubMed] [Google Scholar]
- 14.Erikson M J, Meyer R J. The origin of greater-than-unit-length plasmids generated during bacterial conjugation. Mol Microbiol. 1993;7:289–298. doi: 10.1111/j.1365-2958.1993.tb01120.x. [DOI] [PubMed] [Google Scholar]
- 15.Furuya N, Komano T. Specific binding of the NikA protein to one arm of 17-base-pair inverted repeat sequences within the oriT region of plasmid R64. J Bacteriol. 1995;177:46–51. doi: 10.1128/jb.177.1.46-51.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grandoso G, Llosa M, Zabala J C, de la Cruz F. Purification and biochemical characterization of TrwC, the helicase involved in plasmid R388 conjugal DNA transfer. Eur J Biochem. 1994;226:403–412. doi: 10.1111/j.1432-1033.1994.tb20065.x. [DOI] [PubMed] [Google Scholar]
- 17.Hall R M. pKM101 is an IS46-promoted deletion of R46. Nucleic Acids Res. 1987;15:5479. doi: 10.1093/nar/15.13.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall R M, Collis C M. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 19.Hall R M, Vockler C. The region of the IncN plasmid R46 coding for resistance to beta-lactam antibiotics, streptomycin/spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn21-like transposons. Nucleic Acids Res. 1987;15:7491–7501. doi: 10.1093/nar/15.18.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hames B D. An introduction to polyacrylamide gel electrophoresis. In: Hames B D, Rickwood D, editors. Gel electrophoresis of proteins. A practical approach. Washington, D.C: IRL Press Ltd.; 1981. pp. 1–89. [Google Scholar]
- 21.Hanai R, Wang J C. The mechanism of sequence specific DNA cleavage and strand transfer by X174 gene A* protein. J Biol Chem. 1993;268:23830–23838. [PubMed] [Google Scholar]
- 22.Hengen P N, Iyer V N. DNA cassettes containing the origin of transfer (oriT) of two broad-host-range transfer systems. BioTechniques. 1992;13:58–62. [PubMed] [Google Scholar]
- 23.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. Cabios. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 24.Holcik M, Iyer V N. Structure and mode of action of kikA, a genetic region lethal to Klebsiella oxytoca and associated with conjugative antibiotic-resistance plasmids of the IncN group. Plasmid. 1996;35:189–203. doi: 10.1006/plas.1996.0021. [DOI] [PubMed] [Google Scholar]
- 25.Inamoto S, Yoshioka Y, Ohtsubo E. Site- and strand-specific nicking in vitro at oriT by the TraY-TraI endonuclease of plasmid R100. J Biol Chem. 1991;266:10086–10092. [PubMed] [Google Scholar]
- 26.Iyer V N. IncN group plasmids and their genetic systems. In: Thomas C M, editor. Promiscuous plasmids of Gram-negative bacteria. London, United Kingdom: Academic Press; 1989. pp. 165–183. [Google Scholar]
- 27.Kusano K, Nakayama K, Nakayama H. Plasmid-mediated lethality and plasmid multimer formation in an Escherichia coli recBC sbcBC mutant. Involvement of RecF recombination pathway genes. J Mol Biol. 1989;209:623–634. doi: 10.1016/0022-2836(89)90000-4. [DOI] [PubMed] [Google Scholar]
- 28.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 29.Lane D, de Feyter R, Kennedy M, Phua S H, Semon D. D protein of mini-F plasmid acts as a repressor of transcription and as a site-specific resolvase. Nucleic Acids Res. 1986;14:9713–9728. [PMC free article] [PubMed] [Google Scholar]
- 30.Langer P J, Shanabruch W G, Walker G C. Functional organization of plasmid pKM101. J Bacteriol. 1981;145:1310–1316. doi: 10.1128/jb.145.3.1310-1316.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langer P J, Walker G C. Restriction endonuclease cleavage map of pKM101: relationship to parental plasmid R46. Mol Gen Genet. 1981;182:268–272. doi: 10.1007/BF00269669. [DOI] [PubMed] [Google Scholar]
- 32.Lanka E, Wilkins B M. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 33.Llosa M, Bolland S, de la Cruz F. Structural and functional analysis of the origin of conjugal transfer of the broad-host-range IncW plasmid R388 and comparison with the related IncN plasmid R46. Mol Gen Genet. 1991;226:467–472. doi: 10.1007/BF00260661. [DOI] [PubMed] [Google Scholar]
- 34.Llosa M, Bolland S, de la Cruz F. Conjugation-independent, site-specific recombination at the oriT of the IncW plasmid R388 mediated by TrwC. J Bacteriol. 1994;176:3210–3217. doi: 10.1128/jb.176.11.3210-3217.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Llosa M, Bolland S, de la Cruz F. Genetic organization of the conjugal DNA processing region of the IncW plasmid R388. J Mol Biol. 1994;235:448–464. doi: 10.1006/jmbi.1994.1005. [DOI] [PubMed] [Google Scholar]
- 36.Llosa M, Grandoso G, de la Cruz F. Nicking activity of TrwC directed against the origin of transfer of the IncW plasmid R388. J Mol Biol. 1995;246:54–62. doi: 10.1006/jmbi.1994.0065. [DOI] [PubMed] [Google Scholar]
- 37.Llosa M, Grandoso G, Hernando M, de la Cruz F. Functional domains in protein TrwC of plasmid R388: dissected DNA strand transferase and DNA helicase activities reconstitute protein function. J Mol Biol. 1996;264:56–67. doi: 10.1006/jmbi.1996.0623. [DOI] [PubMed] [Google Scholar]
- 38.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moncalian G, Grandoso G, Llosa M, de la Cruz F. OriT-processing and regulatory roles of TrwA protein in plasmid R388 conjugation. J Mol Biol. 1987;270:188–200. doi: 10.1006/jmbi.1997.1082. [DOI] [PubMed] [Google Scholar]
- 40.Moré M I, Pohlman R F, Winans S C. Genes encoding the pKM101 conjugal mating pore are negatively regulated by the plasmid-encoded KorA and KorB proteins. J Bacteriol. 1996;178:4392–4399. doi: 10.1128/jb.178.15.4392-4399.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy C K, Beckwith J. Export of proteins to the cell envelope in Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 967–978. [Google Scholar]
- 42.Nelson W C, Howard M T, Sherman J A, Matson S W. The traY gene product and integration host factor stimulate Escherichia coli DNA helicase I-catalyzed nicking at the F plasmid oriT. J Biol Chem. 1995;270:28374–28380. [PubMed] [Google Scholar]
- 43.Panicker M M, Minkley E G., Jr Purification and properties of the F sex factor TraD protein, an inner membrane conjugal transfer protein. J Biol Chem. 1992;267:12761–12766. [PubMed] [Google Scholar]
- 44.Pansegrau W, Balzer D, Kruft V, Lurz R, Lanka E. In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc Natl Acad Sci USA. 1990;87:6555–6559. doi: 10.1073/pnas.87.17.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pansegrau W, Ziegelin G, Lanka E. Covalent association of the traI gene product of plasmid RP4 with the 5′-terminal nucleotide at the relaxation nick site. J Biol Chem. 1990;265:10637–10644. [PubMed] [Google Scholar]
- 46.Pansegrau W, Schröder W, Lanka E. Relaxase (TraI) of IncP plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc Natl Acad Sci USA. 1993;90:2925–2929. doi: 10.1073/pnas.90.7.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pansegrau W, Schröder W, Lanka E. Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J Biol Chem. 1994;269:2782–2789. [PubMed] [Google Scholar]
- 48.Paterson E S, Iyer V N. The oriT region of the conjugative transfer system of plasmid pCU1 and specificity between it and the mob region of other N tra plasmids. J Bacteriol. 1992;174:499–507. doi: 10.1128/jb.174.2.499-507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paterson E S, Iyer V N. Localization of the nic site of IncN plasmid pCU1 through the formation of a hybrid oriT. J Bacteriol. 1997;179:5768–5776. doi: 10.1128/jb.179.18.5768-5776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perry K L, Elledge S J, Mitchell B B, Marsh L, Walker G C. umuDC and mucAB operons whose products are required for UV light- and chemical-induced mutagenesis: UmuD, MucA, and LexA proteins share homology. Proc Natl Acad Sci USA. 1985;82:4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pohlman R F, Genetti H D, Winans S C. Common ancestry between IncN conjugal transfer genes and macromolecular export systems of plant and animal pathogens. Mol Microbiol. 1994;14:655–668. doi: 10.1111/j.1365-2958.1994.tb01304.x. [DOI] [PubMed] [Google Scholar]
- 52.Pohlman R F, Genetti H D, Winans S C. Entry exclusion of the IncN plasmid pKM101 is mediated by a small hydrophilic protein containing a lipid attachment motif. Plasmid. 1994;31:158–165. doi: 10.1006/plas.1994.1017. [DOI] [PubMed] [Google Scholar]
- 53.Pohlman R F, Liu F, Wang L, Moré M I, Winans S C. Genetic and biochemical analysis of an endonuclease encoded by the IncN plasmid pKM101. Nucleic Acids Res. 1993;21:4867–4872. doi: 10.1093/nar/21.21.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reimmann C, Haas D. Mobilization of chromosomes and non-conjugative plasmids by cointegrative mechanisms. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 137–188. [Google Scholar]
- 55.Rivas S, Bolland S, Cabezon E, Goni F M, de la Cruz F. TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J Biol Chem. 1997;272:25583–25590. doi: 10.1074/jbc.272.41.25583. [DOI] [PubMed] [Google Scholar]
- 56.Rodriguez M, Holcik M, Iyer V N. Lethality and survival of Klebsiella oxytoca evoked by conjugative IncN group plasmids. J Bacteriol. 1995;177:6352–6361. doi: 10.1128/jb.177.22.6352-6361.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 58.Scherzinger E R, Lurz R, Otto S, Dobrinski B. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 1992;20:41–48. doi: 10.1093/nar/20.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- 60.Sternberg N, Sauer B, Hoess R, Abremski K. Bacteriophage P1 cre gene and its regulatory region. Evidence for multiple promoters and for regulation by DNA methylation. J Mol Biol. 1986;187:197–212. doi: 10.1016/0022-2836(86)90228-7. [DOI] [PubMed] [Google Scholar]
- 61.Studier F W, Rosenberg A H, Dunn J J, Dunbendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 62.Summers D K, Beton C W, Withers H L. Multicopy plasmid instability: the dimer catastrophe hypothesis. Mol Microbiol. 1993;8:1031–1038. doi: 10.1111/j.1365-2958.1993.tb01648.x. [DOI] [PubMed] [Google Scholar]
- 63.Thatte V, Bradley D E, Iyer V N. N conjugative transfer system of plasmid pCU1. J Bacteriol. 1985;163:1229–1236. doi: 10.1128/jb.163.3.1229-1236.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tosini F, Venanzi S, Boschi A, Battaglia P A. The uvp1 gene on the R46 plasmid encodes a resolvase that catalyzes site-specific resolution involving the 5′-conserved segment of the adjacent integron In1. Mol Gen Genet. 1998;258:404–411. doi: 10.1007/s004380050748. [DOI] [PubMed] [Google Scholar]
- 65.Traxler B A, Minkley E G., Jr Evidence that DNA helicase I and oriT site-specific nicking are both functions of the F TraI protein. J Mol Biol. 1988;204:205–209. doi: 10.1016/0022-2836(88)90609-2. [DOI] [PubMed] [Google Scholar]
- 66.Vogel A M, Das A. Mutational analysis of Agrobacterium tumefaciens VirD2: tyrosine 29 is essential for endonuclease activity. J Bacteriol. 1992;174:303–308. doi: 10.1128/jb.174.1.303-308.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilkins B M, Lanka E. DNA processing and replication during plasmid transfer between Gram-negative bacteria. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 105–136. [Google Scholar]
- 68.Willetts N S, Clark A J, Low B. Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J Bacteriol. 1969;97:244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilmes-Riesenberg M R, Wanner B. TnphoA and TnphoA′ elements for making and switching fusions for study of transcription, translation, and cell surface localization. J Bacteriol. 1992;174:4558–4575. doi: 10.1128/jb.174.14.4558-4575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winans S C, Walker G C. Conjugal transfer system of the IncN plasmid pKM101. J Bacteriol. 1985;161:402–410. doi: 10.1128/jb.161.1.402-410.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Winans S C, Walker G C. Entry exclusion determinant(s) of the IncN plasmid pKM101. J Bacteriol. 1985;161:411–416. doi: 10.1128/jb.161.1.411-416.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winans S C, Walker G C. Fertility inhibition of RP1 by IncN plasmid pKM101. J Bacteriol. 1985;161:425–427. doi: 10.1128/jb.161.1.425-427.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winans S C, Walker G C. Identification of pKM101-encoded loci specifying potentially lethal gene products. J Bacteriol. 1985;161:417–424. doi: 10.1128/jb.161.1.417-424.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Winans S C, Elledge S J, Krueger J H, Walker G C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J Bacteriol. 1985;161:1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Winans S C. Two-way chemical signalling in Agrobacterium-plant interactions. Microbiol Rev. 1992;56:12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]