Abstract
Coordinate expression of many virulence genes in the human pathogen Vibrio cholerae is controlled by the ToxR, TcpP, and ToxT proteins. These proteins function in a regulatory cascade in which ToxR and TcpP, two inner membrane proteins, are required to activate toxT and ToxT is the direct activator of virulence gene expression. ToxT-activated genes include those whose products are required for the biogenesis of cholera toxin (CTX) and the toxin-coregulated pilus, the major subunit of which is TcpA. This work examined control of toxT transcription. We tested a model whereby activation of toxT by ToxR and TcpP is required to prime an autoregulatory loop in which ToxT-dependent transcription of the tcpA promoter reads through a proposed terminator between the tcpF and toxT genes to result in continued ToxT production. Primer extension analysis of RNA from wild-type classical strain O395 showed that there are two products encoding toxT, one of which is longer than the other by 105 bp. Deletion of the toxT promoter (toxTΔpro) resulted in the abolishment of toxT transcription, as predicted. Deletion of the tcpA promoter (tcpAΔpro) had no effect on subsequent detection of the smaller toxT primer extension product, but the larger toxT product was not detected, indicating that this product may be the result of transcription from the tcpA promoter and not of initiation directly upstream of toxT. Neither mutant strain produced detectable TcpA, but the CTX levels of the strains were different. The toxTΔpro strain produced little detectable CTX, while the tcpAΔpro strain produced CTX levels intermediate between those of the wild-type and toxTΔpro strains. Dependence of toxT transcription on TcpP and TcpH was confirmed by analyzing RNAs from strains carrying deletions in the genes encoding these regulators. The tcpP defect resulted in undetectable toxT transcription, whereas the tcpH mutation led to a diminishing of toxT RNA but not complete abolishment. Taken together, these results suggest that toxT transcription is dependent on two different promoters; one is directly upstream and is activated in part by TcpP and TcpH, and the other is much further upstream and is activated by ToxT.
Vibrio cholerae is the causative agent of the diarrheal disease cholera, which is usually acquired by oral ingestion of the bacterium with contaminated water or food (10). In response to specific environmental conditions, such as temperature, pH, or osmolarity (11, 23), V. cholerae expresses several virulence determinants, including the cholera toxin (CTX), a toxin-coregulated pilus (TCP), the accessory colonization factor, and a major outer membrane protein (OmpU) (24, 28, 34, 36). CTX is the best-characterized virulence factor and is composed of a single A subunit and five identical B subunits (12, 27). The enzymatically active A subunit is predominantly responsible for fluid loss through an ADP-ribosylation mechanism that results in constitutive cyclic-AMP (cAMP) production in host cells, leading to the opening of normally gated channels (1). Environmental signals optimal for CTX production also stimulate the expression of TcpA (17, 19) and OmpU, a porin that may also function as an adhesin (4, 33, 34). TCP is a pilus in the type IV family that is essential for colonization and virulence. It is made up of a single protein encoded by the tcpA gene, which is part of a pathogenicity island that includes other tcp genes whose products are involved in the biogenesis of the pilus structure, as well as the acf genes (18, 19, 26, 28).
Coordinate expression of ctxAB, tcpA to F, and some acf genes is due to the action of several regulatory proteins. In the current model, these proteins function in a branched regulatory cascade in which two activator proteins, ToxR and TcpP, are required for activation of toxT transcription (13, 32) and ToxT, a member of the AraC family of transcriptional activators (15), activates the expression of other virulence genes, including ctxAB and tcpA to F (9, 32). The ompU gene is in a ToxT-independent branch of this cascade and is activated directly by ToxR (5, 6).
Both ToxR and TcpP are inner membrane proteins with cytoplasmic DNA-binding domains homologous to members of the two-component family of transcriptional activators found in various species of bacteria (13, 24, 25). Each of these proteins is encoded by an operon by which another membrane protein is encoded. For ToxR, this protein is ToxS, and for TcpP, it is TcpH (3, 8, 21). ToxS and TcpH act as effector proteins for ToxR and TcpH, respectively, through a mechanism that likely involves periplasmic interaction between the regulator and the effector (3, 8). The precise mechanism by which ToxRS and TcpPH control toxT transcription is not understood. One observation that may eventually contribute to a better understanding of this system is that overexpression of TcpP suppresses a toxR mutant for toxT expression, while overexpression of ToxR does not suppress a tcpP mutant (13, 14, 19a).
This work examined the transcription of toxT in the context of its location at the end of the tcp gene cluster. Previous genetic analysis demonstrated that insertion mutations in the tcpA gene resulted in downregulation of the production of both TCP and CTX (2). This was interpreted as being due to the polar effects of the insertions on toxT, because RNase protection experiments showed that the tcp gene cluster, including toxT, was transcribed as a long polycistronic message (2). In addition, it was recently shown that the cAMP-cAMP receptor protein complex plays a negative role in regulation of toxin production, potentially through repression of tcpA transcription, which may result in decreased toxT transcription through transcriptional polarity (31, 32). Other work demonstrated that ToxR-dependent activation of toxT occurs at a promoter that is immediately upstream of toxT (14) and that a transcription terminator with 80% efficiency precedes the toxT gene downstream of tcpF.
In one model that would account for these observations, ToxR (in conjunction with TcpP) activates toxT from the proximal promoter and ToxT activation of the tcpA promoter contributes to subsequent expression of the gene through readthrough of the relatively inefficient transcription terminator (compared to a well-characterized λ terminator) between tcpF and toxT (14). This model makes specific predictions about the contributions of different regulatory elements within the tcp gene cluster, and in this work, we tested some of those predictions.
MATERIALS AND METHODS
Bacterial strains and plasmids.
V. cholerae and Escherichia coli strains were grown in Luria-Bertani (LB) medium at 30 or 37°C. Strains were maintained at −70°C in LB medium with 20% glycerol. Antibiotics were used at the following concentrations (unless otherwise stated): ampicillin, 100 μg/ml; streptomycin, 100 μg/ml; kanamycin, 30 μg/ml. Plasmids were introduced into E. coli strains by either transformation or electroporation and into V. cholerae strains by conjugation.
DNA manipulation.
Plasmid DNA was purified with Qiagen columns (Qiagen, Inc.). Cloning was performed by using standard protocols (29). PCR was performed with the Expand High Fidelity PCR System (Boehringer Mannheim) by using the manufacturer’s protocols. A recombinant PCR method, gene splicing by overlap extension (16), using primers within and surrounding the toxT promoter region, the tcpA promoter region, tcpP, and tcpH, was performed to create deletions of the corresponding regions (for a description of the construction of toxTΔhth, see reference 5). The DNA templates (15, 36) and primers used for construction of each mutant are listed in Table 1. This procedure requires two PCR rounds, as follows (using the toxT promoter deletion as an example). Outside primer 1 is complementary to a sequence in tcpF (upstream of the toxT promoter), and primer 4 is complementary to a sequence in toxT (downstream of the toxT promoter), and they both have restriction endonuclease sites inserted to mediate cloning. Inside overlapping primers 2 and 3 were designed to delete the toxT promoter region, and they are partly complementary to the sequence upstream and partly complementary to the sequence downstream of the desired deletion. First, two PCRs were carried out, one with primers 1 and 2 and the other with primers 3 and 4. The product of each reaction was purified by agarose gel electrophoresis, followed by gel extraction with the QIAEX II gel extraction system (Qiagen, Inc.), mixed together, and used as the template for a second round of PCR with outermost primers 1 and 4. The amplified PCR fragment was cloned into the corresponding sites of positive-selection suicide plasmid pKAS32, which cannot replicate in V. cholerae in the absence of the Pir protein (30).
TABLE 1.
DNA template and primers used for construction of mutants in this studya
| Strain | DNA template (reference) | Primer 1 | Primer 2 | Primer 3 | Primer 4 |
|---|---|---|---|---|---|
| toxTΔpro mutant | pDH8 (15) | 5′-CCGGAATTCGAAAATGGTCGATATGAT-3′ (EcoRI) | 5′-AGTTATCTTAAAATCGCGAATGTGGCTGTTA-3′ | 5′-TAACAGCCACATTCGCGATTTTAAGATAACT-3′ | 5′-CCGGAATTCTACTTTCGAGAAGAACCC-3′ (EcoRI) |
| tcpAΔpro mutant | ptcpA::phoA2-1 (36) | 5′-GCTCTAGACAGACAGATCCACAAGGT-3′ (XbaI) | 5′-AGCAACACGCACGGTACCGGCCAACTTATTCAATTC-3′ (KpnI) | 5′-AATAAGTTGGCCGGTACCGTGCGTGTTGCTTACGTT-3′ (KpnI) | 5′-TCCCCGCGGTGCAATATATGGGAACAT-3′ (SacII) |
| ΔtcpP mutant | O395 chromosomal DNA | 5′-GGGGTACCGATAACTTTGCAACCGTT-3′ (KpnI) | 5′-TAATTTTTTGTGCATTACTTTACATTTTCT-3′ | 5′-AGAAAATGTAAAGTAATGCACAAAAAATTA-3′ | 5′-TCCCCGCGGGACGATCTCAATACAACT-3′ (SacII) |
| ΔtcpH mutant | O395 chromosomal DNA | 5′-GGGGTACCATAAAAAAATGGGTCGTT-3′ (KpnI) | 5′-ACACTATCTAGGCGGAGCTTTTAATTTTTT-3′ | 5′-AAAAAATTAAAAGCTCCGCCTAGATAGTGT-3′ | 5′-TCCCCGCGGTGTTCTTCTTTTACAAAT-3′ (SacII) |
Restriction sites for each individual primer are underlined and in parentheses if applicable. All primers were designed by using the sequences under GenBank accession no. X64098.
The resulting plasmid was first introduced into E. coli MC4100λpir or DH5αλpir by electroporation, transformed into E. coli SM10λpir, and finally introduced into V. cholerae classical strain O395 by conjugation from SM10λpir. The transconjugants were selected on TCBS agar (Difco) for plasmid-encoded ampicillin (50 μg/ml) resistance. Single recombinants that have the plasmid integrated into the chromosome were grown in the absence of antibiotic selection and then selected again on LB agar containing streptomycin (1 mg/ml) for strains that have undergone resolution of the cointegrate by recombination (30). DNAs from isolates that were both sensitive to ampicillin and resistant to streptomycin were analyzed by PCR using outermost primers 1 and 4. DNA amplified with these primers gave a smaller fragment from DNA of the deletion mutant than from that of the wild type.
RNA analysis.
For primer extension analysis, an overnight culture of V. cholerae grown in LB medium at 30°C to late log phase (optical density at 600 nm [OD600], >3.0) was subcultured 1:100 into fresh LB medium. After back dilution (t = 0) and at 1-h intervals, aliquots of the cultures were removed and poured over crushed ice prior to centrifugation for cell recovery. Cell pellets were stored at −20°C until ready for RNA isolation. RNA was obtained from cultures of V. cholerae by using TRIzol Reagent (Gibco BRL) by following the manufacturer’s protocols. The RNA samples were then treated with DNase I and quantified by determination of the OD260. The same amount of RNA was used for primer extension as previously described (14), by using a toxT-specific primer (5′-CATTAGTTTGAAAAGATTTTTTCCCAATCAT-3′) or a tcpA-specific primer (5′-TTCTTTTACAAATTTCTTCTTAAAAAGCTGTTTTAA-3′).
Protein analysis.
Total cell lysates were prepared from V. cholerae cells grown to stationary phase overnight (with 1 mM isopropyl-β-d-thiogalactopyranoside [IPTG] if necessary). Samples (1 ml) were removed from the cultures and centrifuged. The harvested pellet was resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer, normalized for OD600, and boiled for 5 min. Aliquots of this lysate were subjected to electrophoresis on a 12% polyacrylamide gel with a 5% stacking gel. For Western blotting, the gels were blotted onto nitrocellulose and probed with anti-TcpA peptide 6 rabbit polyclonal antibodies kindly supplied by Ron Taylor (Dartmouth Medical School).
CTX levels were quantified from supernatants of cultures by the GM1-ganglioside enzyme-linked immunosorbent assay (ELISA) (35) using anti-CtxB rabbit polyclonal antibodies kindly supplied by Michael Bagdasarian (Michigan State University).
RESULTS
Transcription of toxT in the tcp gene cluster.
To analyze transcription of toxT, we used derivatives of classical strain O395 carrying deletions of key elements in the tcp gene cluster (Fig. 1). We had observed that wild-type strain O395 produces little detectable toxT mRNA after overnight growth and exploited this observation to establish a time course experiment as shown in Fig. 2A. When primer extension of toxT RNA was done on samples prepared from wild-type strain O395 after 1:100 back dilution of an overnight culture, we observed that both of the primer extension products previously described (14) became detectable within an hour after back dilution. The smaller primer extension product (99 bp) has been previously shown to initiate from a ToxR-dependent promoter immediately preceding the toxT open reading frame (ORF) (14). The source of the larger product (204 bp) is less clear, but it has been speculated to arise from either an RNA-processing event or a block to reverse transcription due to potential secondary structure in the RNA at that position (14, 15). In this experiment, the smaller product was more abundant within the first hour, but by 2 h, the two products were roughly equivalent. By 5 to 9 h after back dilution, each began to diminish in prevalence (Fig. 2A) and after 9 h, they became undetectable (data not shown).
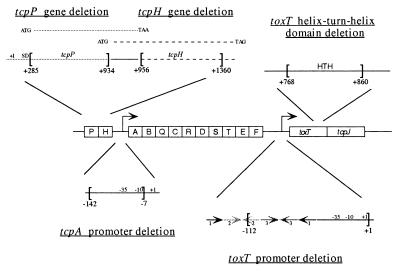
FIG. 1.
Schematic diagram of the V. cholerae tcp gene cluster. Shown in the center is the organization of the tcp gene cluster of wild-type V. cholerae. Each box represents a gene. The right-angle arrows represent promoters. Certain regions are enlarged to show the details of each deletion. Brackets indicate the deleted sequence, and the number below each bracket is the nucleotide position, relative to the +1 start site for transcription, which represents the start or the end of a deletion. Arrows at the toxT promoter region denote three pairs of inverted repeats. tcpP and tcpH are in an operon, and their ORFs overlap. The ORFs are shown above the operon. The leftward-pointing arrows beneath toxT and tcpA represent primers used in the RNA primer extension analyses. SD, Shine-Dalgarno region; HTH, helix-turn-helix motif.
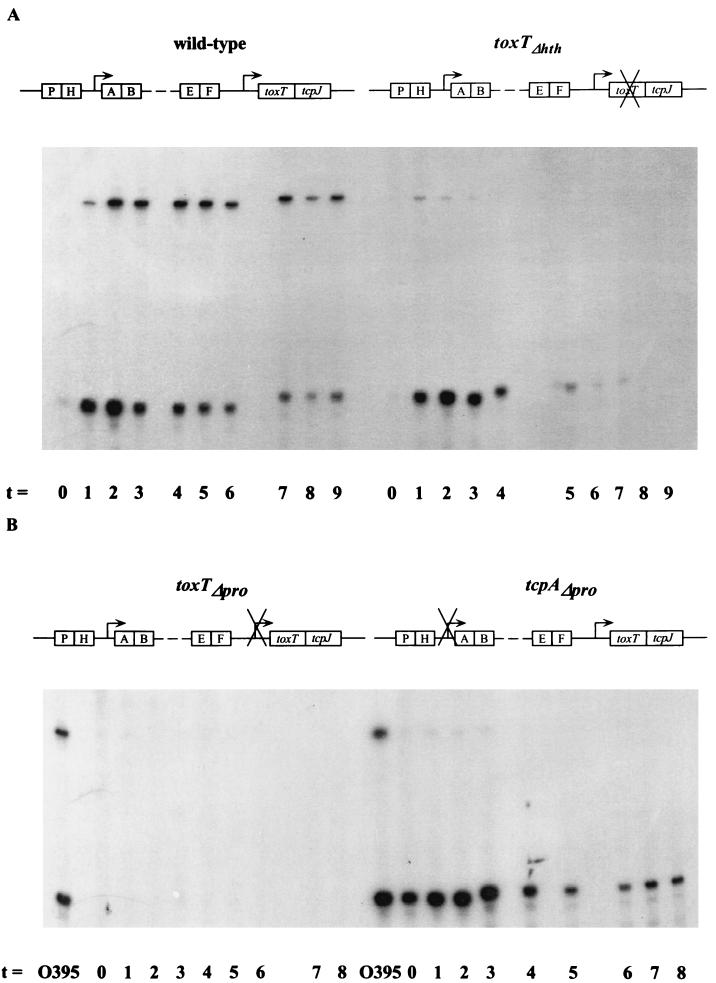
FIG. 2.
RNA primer extension analyses of V. cholerae wild-type O395 and a toxTΔhth mutant strain (A) and toxTΔpro and tcpAΔpro mutant strains (B). Samples were collected at 1-h intervals after back dilution of an overnight culture grown in LB medium at 30°C 1:100 into fresh LB medium. A radiolabeled toxT primer was used. RNA collected from wild-type strain O395 after 2 to 3 h of growth was used as a positive control for the mutant strains in panel B.
In order to determine whether the transcripts shown in Fig. 2A require de novo transcription or accumulated over the time of the experiment, rifampin was added to the cultures 2 h after back dilution. When this was done, RNA diminished in abundance very rapidly with a half-life of less than 5 min for each, as judged by primer extension (data not shown), indicating that new transcription is continuously required to generate the pattern shown in Fig. 2A.
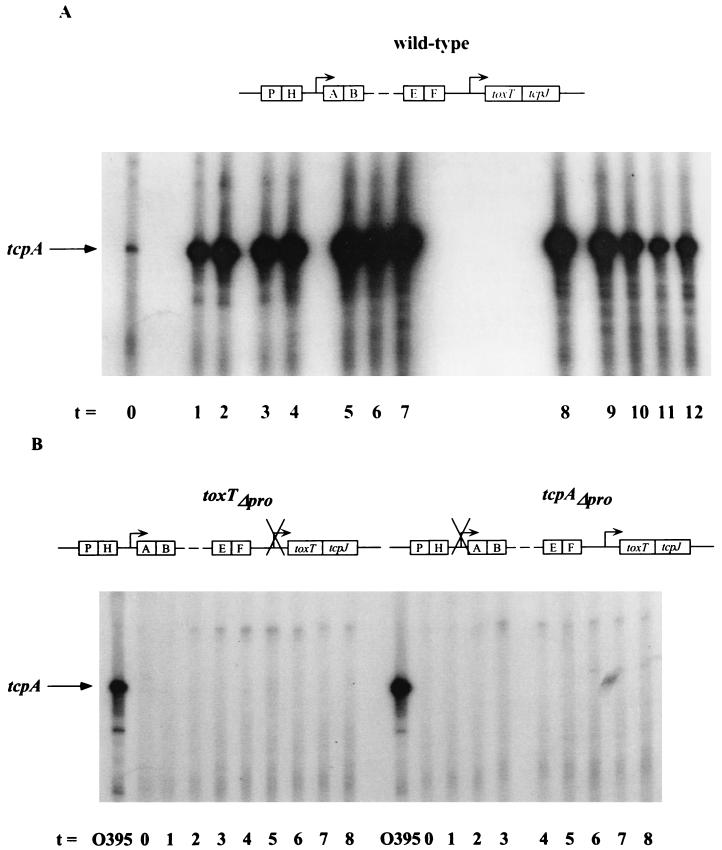
Analysis of tcpA transcription in the wild-type strain by primer extension showed that immediately after the back dilution, tcpA mRNA (120 bp) was minimal but increased steadily after that until 7 h, when its prevalence as a percentage of the total mRNA began to decrease (Fig. 3A). This is similar to the transient nature of toxT mRNA production shown in Fig. 2A.
FIG. 3.
RNA primer extension analyses of V. cholerae strain O395 wild-type (A) and mutant strains toxTΔpro and tcpAΔpro (B). Samples were collected at 1-h intervals after back dilution of an overnight culture grown in LB medium at 30°C 1:100 into fresh LB medium. A radiolabeled tcpA primer was used. RNA collected from wild-type strain O395 after 2 to 3 h of growth was used as a positive control for the mutant strains in panel B. The position of the tcpA transcript is indicated.
In order to ascertain whether functional ToxT is required for the wild-type pattern of toxT transcription, a similar time course experiment was performed by using RNA from a derivative of strain O395 that expresses a null allele of toxT, called toxTΔhth, that lacks the predicted helix-turn-helix DNA-binding domain of ToxT (5). In contrast to what we observed in the wild type, in the toxTΔhth strain, the larger primer extension product was severely diminished in quantity, while the smaller product was produced at levels similar to the wild-type level (Fig. 2A). Thus, transcription of toxT from a near promoter is not dependent on functional ToxT. This result also shows that the larger product requires a promoter that is ToxT dependent.
We next addressed specifically the consequence of ToxR-dependent activation of the toxT promoter. To do this, we analyzed a strain in which the previously mapped ToxR binding site in the toxT promoter (14) had been deleted. This strain, toxTΔpro, was predicted not to synthesize any toxT from the near promoter due to the lesion in the ToxR binding site and the basal promoter element, and that is what we observed in the primer extension experiment shown in the left side of Fig. 2B. No larger primer extension product was detected in this experiment.
We observed no detectable tcpA mRNA in the toxTΔpro strain (Fig. 3B), which clearly demonstrates the cascade of regulation in the ToxR regulon: activation of tcpA transcription is ultimately dependent on activation of toxT by ToxR. That the phenotype of the toxTΔpro strain produces undetectable levels of toxin is worth noting (Table 2), as it strongly implies that there is no source of ToxT in the cell in the absence of ToxR-dependent transcription. Lack of toxin production by the toxTΔpro strain also confirms an earlier observation that functional ToxT must be produced in V. cholerae in order for the ctxAB promoter to be activated (5), irrespective of the fact that ToxR alone can activate ctxAB transcription when tested in E. coli (22).
TABLE 2.
Toxin production in V. cholerae wild-type and mutant strainsa
| Strain | Relevant genetic map | Avg toxin production (ng/ml/OD600) ± SDb |
|---|---|---|
| O395 (wild type) | 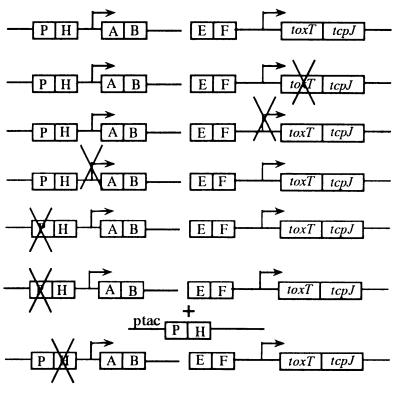 |
1,370 ± 130 |
| toxTΔhth mutant | 0.038 ± 0.015 | |
| toxTΔpro mutant | 0.027 ± 0.0021 | |
| tcpAΔpro mutant | 760 ± 150 | |
| ΔtcpP/pMMB66EH mutantc | 0.043 ± 0.031 | |
| ΔtcpP/ptcpPH mutantc | 5,670 ± 720 | |
| ΔtcpH mutant | 19 ± 3 | |
Strains were grown overnight in LB medium, and supernatants were assayed by GM1-ganglioside ELISA.
Toxin levels represent three individual experiments.
Plasmid experiments included 1 mM IPTG for induction of genes under tac promoter control.
The data presented above suggest that the larger toxT primer extension product arises from a ToxT-dependent transcript. The logical site for initiation of this transcript is at the tcpA promoter. To test this hypothesis, we performed toxT primer extension on RNA from a strain in which the tcpA promoter had been deleted (tcpAΔpro). The smaller, ToxR-dependent product was observed (Fig. 2B), whereas the larger product was nearly undetectable. To confirm that the deletion had, in fact, abolished tcpA transcription, we performed primer extension on the same RNA samples by using a tcpA-specific primer and observed no tcpA primer extension product (Fig. 3B).
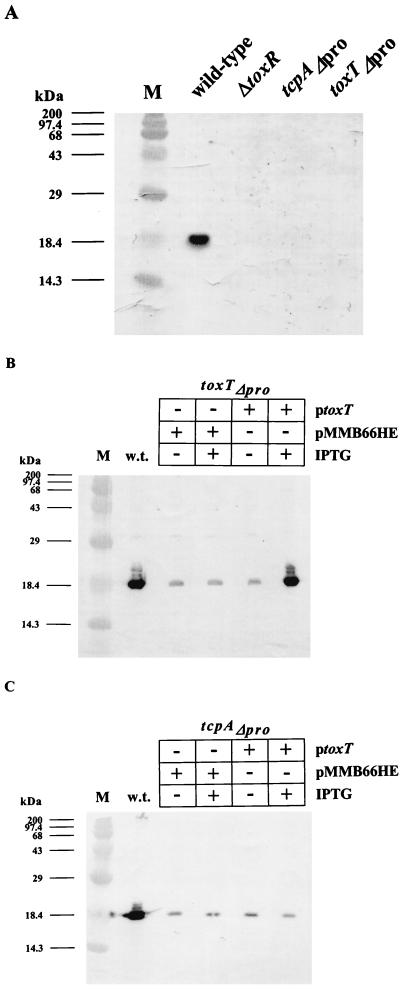
Figure 4 shows TcpA immunoblot data confirming the phenotypes of the mutant strains described above. The blot in panel A demonstrates the dependence of TcpA production on ToxR, as well as on both the tcpA and toxT promoters. As shown in Fig. 4B, the toxTΔpro strain could be complemented for TcpA production by a plasmid encoding an IPTG-inducible toxT gene, indicating that the tcpA promoter in this background remains responsive to ToxT. In contrast, expression of ToxT in the tcpAΔpro strain did not restore TcpA production (Fig. 4C), thereby confirming that the deletion removed sites critical for ToxT-dependent promoter activation.
FIG. 4.
Western blot analysis using anti-TcpA antibody. V. cholerae wild-type O395 and mutant strains ΔtoxR, toxTΔpro, and tcpAΔpro (A); wild-type O395 and mutant strain toxTΔpro/pMMB66HE or toxTΔpro/ptoxT (B); and wild-type O395 and mutant strain tcpAΔpro/pMMB66HE or tcpAΔpro/ptoxT (C) were grown overnight in LB medium at 30°C. IPTG (1 mM) was added to induce toxT transcription in strains carrying plasmids. ptoxT harbors the toxT ORF on low-copy-number plasmid pMMB66HE under the control of the IPTG-inducible tac promoter. Protein molecular size standards (lane M) are indicated on the left. w.t., wild type.
TcpP and TcpH are required for toxT transcription and coordinate regulation.
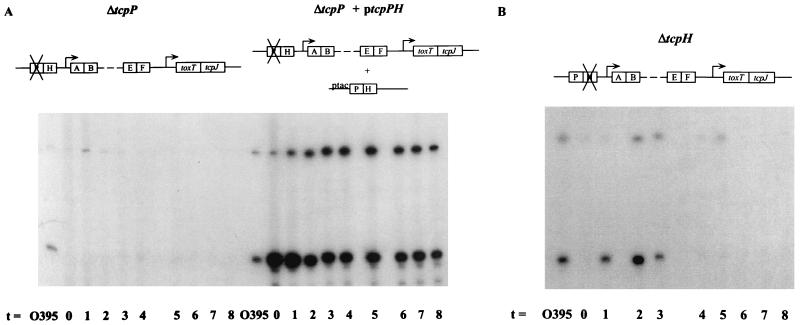
Two other gene products encoded in the tcp gene cluster, TcpP and TcpH, have been implicated in coordinate regulation in V. cholerae through their effects on toxT transcription (3, 13, 20). To characterize the role of TcpP and TcpH in the context of the regulatory loop model analyzed above, mutant strains carrying deletions in these genes (ΔtcpP and ΔtcpH) were constructed and their RNAs were used in primer extension reactions with a radiolabeled toxT primer. Deletion of the tcpP gene resulted in complete abolishment of both toxT-specific primer extension products (Fig. 5A) and reduction of TcpA to the background level (data not shown). Both toxT transcription and TcpA production were restored upon introduction of a tcpPH-encoding plasmid into the tcpP mutant.
FIG. 5.
RNA primer extension analyses of V. cholerae mutant strains ΔtcpP/pMMB66EH and ΔtcpP/ptcpPH (A) and mutant strain ΔtcpH (B). Samples were collected at 1-h intervals after back dilution of a culture grown overnight in LB medium at 30°C 1:100 into fresh LB medium. IPTG (1 mM) was used to induce transcription in strains carrying plasmids. A radiolabeled toxT primer was used. RNA collected from wild-type strain O395 after 2 to 3 h of growth was used as a positive control for all of the mutant strains. ptcpPH harbors the tcpP and tcpH ORFs on low-copy-number plasmid pMMB66EH under the control of the IPTG-inducible tac promoter.
In contrast to the abolishment of toxT transcription in the tcpP mutant, the tcpH mutant strain synthesized detectable toxT mRNA, although the pattern of transcription was altered relative to the wild-type pattern (Fig. 5B). This intermediate phenotype for toxT transcription caused by the tcpH lesion was also seen in toxin production measured by GM1-ganglioside ELISA. As seen in Table 2, the ΔtcpP mutant strain synthesized a barely detectable level of CTX, whereas the ΔtcpH mutant strain made a low but easily detectable level of CTX. These data suggest that TcpP is an absolute requirement for toxT transcription and subsequent expression of CTX and TCP, while TcpH is required for production and/or maintenance of wild-type levels of toxT transcription without being strictly required for transcription activation.
DISCUSSION
Previous studies showed that virulence gene expression in V. cholerae is controlled through a regulatory cascade by the ToxR-ToxT system (7, 9, 15). According to this model, activation of toxT expression requires ToxR, and gene fusion studies and electrophoretic mobility shift assays showed that there is a ToxR binding site between −73 and −114 relative to the toxT transcript initiation site that is required for activation of toxT transcription (14). Virulence gene expression is activated by ToxT, whose carboxyl terminus is similar to the DNA-binding domain of the AraC transcription activator of E. coli and Salmonella typhimurium (15); thus, it likely acts in a manner similar to that of AraC.
Previous work suggested that the ToxR-dependent promoter is not the only source of toxT mRNA (2, 14). This conclusion rests on the observation that insertions in the tcpA gene have a negative effect on toxin production, which suggests that a long tcp transcript initiating at the tcpA promoter also encodes toxT. RNase protection analysis supported this possibility (2). The presence of a transcription terminator between tcpF and toxT, albeit a relatively weak one compared to a well-characterized λ terminator, indicates that not all transcripts initiating at the tcpA promoter would read through to toxT (14). While the results presented in this report generally support the autoregulatory loop of transcription in the tcp operon proposed by Brown and Taylor (2), there is a slight discrepancy between our results and theirs in the levels of CTX produced by strains with mutations in tcpA. The tcpAΔpro strain we constructed for this study synthesizes nearly wild-type levels of CTX, with, at most, a twofold decrease compared with the wild type. In contrast, tcpA transposon insertion mutants in the study of Brown and Taylor (such as CS2-1 and RT110.21) synthesize approximately 10-fold less CTX than the wild type (2). The basis of this discrepancy is not clear.
The observation that a toxT-lacZ fusion is activated to high levels in wild-type V. cholerae but that ToxR does not activate toxT-lacZ expression in E. coli led to the suggestion that ToxR is necessary but not sufficient for activation of toxT (14). The TcpP gene product had been implicated in coordinate regulation in the ToxR system by others (20, 37), including Häse and Mekalanos, who most recently demonstrated that a tcpP mutant V. cholerae strain does not activate expression of a toxT-lacZ gene fusion (13). These investigators also showed that overexpressed TcpP activates toxT transcription in the absence of ToxR, although wild-type expression of TcpP does not (13). We also note that overexpression of TcpPH in the ΔtcpP strain led to toxT expression that was sustained and stronger than that observed in the wild type (cf. Fig. 2A and 5A) and a concomitant increase in CTX compared to the wild type (Table 2). This implies that TcpP and TcpH are limiting in this system and, if this is so, may provide evidence for another level of control in the ToxR regulatory system. One hypothesis that might account for the elevated CTX levels when TcpP and TcpH are overexpressed is that TcpP activates ctxAB transcription directly. However, overexpression of TcpP and TcpH in the toxTΔpro strain did not result in detectable CTX production (data not shown), confirming that TcpP and TcpH mediate their effect on CTX expression solely through the toxT promoter. Combined with the fact that ToxR is normally required for toxT transcription, we conclude that ToxR and TcpP work together in some way to activate the toxT promoter.
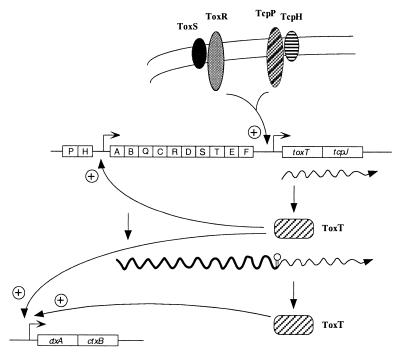
These data support the following model for transcription control of toxT and coordinate regulation of virulence in V. cholerae (Fig. 6). ToxR, ToxS, TcpP, and TcpH in the inner membrane activate toxT transcription from the toxT promoter, generating the transcript represented by the smaller extension product seen in our experiments. ToxT made from this transcript activates the tcpA promoter, producing a long transcript that reads through a weak terminator between tcpF and toxT, thereby generating another source of toxT mRNA. The readthrough transcript may be processed at the site to which the longer primer extension product maps. ToxT from both transcripts is necessary for maximal expression of CTX.
FIG. 6.
Model for control of toxT transcription and coordinate regulation of virulence in V. cholerae. ToxR, ToxS, TcpP, and TcpH in the inner membrane activate toxT transcription from the toxT promoter. ToxT protein then activates transcription of the tcpA promoter, producing more ToxT from the readthrough transcript. ToxT also activates transcription of other virulence genes, including ctx, as shown here. The tcp gene cluster and the ctx operon are shown with each box representing a gene. Symbols: , promoter; ∧∧∧→, transcript of the relevant genes; ∥○ stem-loop structure which is a putative RNA-processing site for the readthrough transcript initiating from the tcpA promoter.
There are alternative hypotheses that would account for our data regarding the two RNA species observed in the primer extension experiments. One is that the larger one originates from a promoter activated by a product of one of the genes coexpressed with tcpA. We do not favor this possibility because none of the ORFs downstream of tcpA encodes a protein with homology to transcription activators. Another hypothesis is that readthrough transcription could result in the utilization of another upstream toxT promoter. Although this has not been ruled out, there are no putative −35 and −10 sequences preceding the site to which the larger toxT primer extension product maps.
The fact that two different activators, ToxR and TcpP, are required for priming of the autoregulatory loop leading to toxT expression raises questions about how this system evolved. One way to view this issue is to assume that prior to acquisition of the V. cholerae pathogenicity island (18) by virulent V. cholerae, ToxR controlled only ompU expression (as well as that of the ToxR-repressed ompT gene). While the island encodes an activator of toxT—TcpP—the level of toxT activation by TcpP alone in V. cholerae may not have been sufficient for a competitive advantage. However, under conditions appropriate for activation of ompU by ToxR, perhaps there was a competitive advantage for strains that also expressed CTX and TCP. This would have been a driving force allowing ToxR to take control of the TcpP-dependent activation of toxT with subsequent CTX and TCP expression. That TcpP remains a requirement in this system (i.e., that ToxR did not gain complete control over toxT transcription) may indicate that its role in toxT expression is not limited to transcription activation. One possibility, for example, is that TcpP is an important component of the signaling pathway leading to toxT expression.
ACKNOWLEDGMENTS
This work was supported by grants AI 31645 (to V.J.D.) and RR 00200 (to the Unit for Laboratory Animal Medicine, University of Michigan) from the National Institutes of Health. R.R.Y. is a trainee of the University of Michigan Genetics Training Grant (T32 GM 07544). DNA sequence analysis was supported in part by grant MO1-RR 00042 to the University of Michigan General Clinical Research Center.
We thank Ana Coelho, Eric Krukonis, and Adam Crawford for insightful comments on the manuscript.
REFERENCES
- 1.Betley M J, Miller V L, Mekalanos J J. Genetics of bacterial enterotoxins. Annu Rev Microbiol. 1986;40:577–605. doi: 10.1146/annurev.mi.40.100186.003045. [DOI] [PubMed] [Google Scholar]
- 2.Brown R C, Taylor R K. Organization of the tcp, acf, and toxT genes within a ToxT-dependent operon. Mol Microbiol. 1995;16:425–439. doi: 10.1111/j.1365-2958.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 3.Carroll P A, Tashima K T, Rogers M B, DiRita V J, Calderwood S B. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol Microbiol. 1997;25:1099–1111. doi: 10.1046/j.1365-2958.1997.5371901.x. [DOI] [PubMed] [Google Scholar]
- 4.Chakrabarti S R, Chaudhuri K, Sen K, Das J. Porins of Vibrio cholerae: purification and characterization of OmpU. J Bacteriol. 1996;178:524–530. doi: 10.1128/jb.178.2.524-530.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champion G A, Neely M N, Brennan M A, DiRita V J. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol Microbiol. 1997;23:323–331. doi: 10.1046/j.1365-2958.1997.2191585.x. [DOI] [PubMed] [Google Scholar]
- 6.Crawford J A, Kaper J B, DiRita V J. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol Microbiol. 1998;29:235–246. doi: 10.1046/j.1365-2958.1998.00925.x. [DOI] [PubMed] [Google Scholar]
- 7.DiRita V J. Co-ordinate expression of virulence genes by ToxR in Vibrio cholerae. Mol Microbiol. 1992;6:451–458. doi: 10.1111/j.1365-2958.1992.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 8.DiRita V J, Mekalanos J J. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell. 1991;64:29–37. doi: 10.1016/0092-8674(91)90206-e. [DOI] [PubMed] [Google Scholar]
- 9.DiRita V J, Parsot C, Jander G, Mekalanos J J. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkelstein R A. Cholera. Crit Rev Microbiol. 1973;2:553–623. [Google Scholar]
- 11.Gardel C L, Mekalanos J J. Regulation of cholera toxin by temperature, pH, and osmolarity. Methods Enzymol. 1994;235:517–526. doi: 10.1016/0076-6879(94)35167-8. [DOI] [PubMed] [Google Scholar]
- 12.Gill D M. The arrangement of subunits in cholera toxin. Infect Immun. 1976;52:1242–1248. doi: 10.1021/bi00651a011. [DOI] [PubMed] [Google Scholar]
- 13.Häse C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins D E, DiRita V J. Transcriptional control of toxT, a regulatory gene in the ToxR regulon of Vibrio cholerae. Mol Microbiol. 1994;14:17–29. doi: 10.1111/j.1365-2958.1994.tb01263.x. [DOI] [PubMed] [Google Scholar]
- 15.Higgins D E, Nazareno E, DiRita V J. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J Bacteriol. 1992;174:6974–6980. doi: 10.1128/jb.174.21.6974-6980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 177–183. [Google Scholar]
- 17.Iredell J R, Manning P A. Biotype-specific tcpA genes in Vibrio cholerae. FEMS Microbiol Lett. 1994;121:47–54. doi: 10.1111/j.1574-6968.1994.tb07074.x. [DOI] [PubMed] [Google Scholar]
- 18.Karaolis D K R, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufman M R, Shaw C E, Jones I D, Taylor R K. Biogenesis and regulation of the Vibrio cholerae toxin-coregulated pilus: analogies to other virulence factor secretory systems. Gene. 1993;126:43–49. doi: 10.1016/0378-1119(93)90588-t. [DOI] [PubMed] [Google Scholar]
- 19a.Krukonis, E. S., and V. J. DiRita. Unpublished data.
- 20.Manning P A. The tcp gene cluster of Vibrio cholerae. Gene. 1997;192:63–70. doi: 10.1016/s0378-1119(97)00036-x. [DOI] [PubMed] [Google Scholar]
- 21.Miller V L, DiRita V J, Mekalanos J J. Identification of toxS, a regulatory gene whose product enhances ToxR-mediated activation of the cholera toxin promoter. J Bacteriol. 1989;171:1288–1293. doi: 10.1128/jb.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by ToxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 25.Ogierman M A, Voss E, Meaney C, Faast R, Attridge S R, Manning P A. Comparison of the promoter proximal regions of the toxin-co-regulated tcp gene cluster in classical and El Tor strains of Vibrio cholerae O1. Gene. 1996;170:9–16. doi: 10.1016/0378-1119(95)00744-x. [DOI] [PubMed] [Google Scholar]
- 26.Ogierman M A, Zabihi S, Mourtzios L, Manning P A. Genetic organization and sequence of the promoter-distal region of the tcp gene cluster of Vibrio cholerae. Gene. 1993;126:51–60. doi: 10.1016/0378-1119(93)90589-u. [DOI] [PubMed] [Google Scholar]
- 27.Pearson G D, Mekalanos J J. Molecular cloning of Vibrio cholerae enterotoxin genes in Escherichia coli K-12. Proc Natl Acad Sci USA. 1982;79:2976–2980. doi: 10.1073/pnas.79.9.2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson K M, Mekalanos J J. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun. 1988;56:2822–2829. doi: 10.1128/iai.56.11.2822-2829.1988. . (Erratum, 57:660, 1989.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Skorupski K, Taylor R K. Broad-host-range positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 31.Skorupski K, Taylor R K. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc Natl Acad Sci USA. 1997;94:265–270. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skorupski K, Taylor R K. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 33.Sperandio V, Bailey C, Girón J A, DiRita V J, Silveira W D, Vettore A L, Kaper J B. Cloning and characterization of the gene encoding the OmpU outer membrane protein of Vibrio cholerae. Infect Immun. 1996;64:5406–5409. doi: 10.1128/iai.64.12.5406-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sperandio V, Giron J A, Silveira W D, Kaper J B. The OmpU outer membrane protein, a potential adherence factor of Vibrio cholerae. Infect Immun. 1995;63:4433–4438. doi: 10.1128/iai.63.11.4433-4438.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svennerholm A M, Holmgren J. Identification of the Escherichia coli heat-labile enterotoxin by means of a ganglioside immunosorbent assay (GM1-ELISA) procedure. Curr Microbiol. 1978;1:19–23. [Google Scholar]
- 36.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas S, Williams S G, Manning P A. Regulation of tcp genes in classical and El Tor strains of Vibrio cholerae O1. Gene. 1995;166:43–48. doi: 10.1016/0378-1119(95)00610-x. [DOI] [PubMed] [Google Scholar]