Abstract
In intermediate risk hormone receptor (HR) positive, HER2 negative breast cancer (BC), the decision regarding adjuvant chemotherapy might be facilitated by multigene expression tests. In all, 142 intermediate risk BCs were investigated using the PAM50-based multigene expression test Prosigna® in a prospective multicentric study. In 119/142 cases, Prosigna® molecular subtyping was compared with local and two central (C1 and C6) molecular-like subtypes relying on both immunohistochemistry (IHC; HRs, HER2, Ki-67) and IHC + tumor grade (IHC+G) subtyping. According to local IHC, 35.4% were Luminal A-like and 64.6% Luminal B-like subtypes (local IHC+G subtype: 31.9% Luminal A-like; 68.1% Luminal B-like). In contrast to local and C1 subtyping, C6 classified >2/3 of cases as Luminal A-like. Pairwise agreement between Prosigna® subtyping and molecular-like subtypes was fair to moderate depending on molecular-like subtyping method and center. The best agreement was observed between Prosigna® (53.8% Luminal A; 44.5% Luminal B) and C1 surrogate subtyping (Cohen’s kappa = 0.455). Adjuvant chemotherapy was suggested to 44.2% and 88.6% of Prosigna® Luminal A and Luminal B cases, respectively. Out of all Luminal A-like cases (locally IHC/IHC+G subtyping), adjuvant chemotherapy was recommended if Prosigna® testing classified as Prosigna® Luminal A at high / intermediate risk or upgraded to Prosigna® Luminal B.
Keywords: Prosigna, PAM50, multigene expression analysis, immunohistochemistry, IHC, tumor grade, breast cancer, chemotherapy
1. Introduction
Invasive breast cancer (IBC) is the most common malignant tumor in women regarding morbidity and mortality. In 2016, the annual number of newly diagnosed breast cancer patients in Germany was 69,660 and 18,736 patients died due to the disease [1]. Therapy options include surgical and radiation therapy as well as systemic therapy such as endocrine therapy and chemotherapy. The treatment strategy is determined individually for each IBC patient, based on biology of both tumor and patient but also according to international and national guidelines [2,3,4,5,6]. Deciding whether a patient with IBC should receive systemic therapy depends on both prognostic and predictive factors. Prognostic (and partly predictive) factors in early, non-metastatic IBC that has not metastasized are: age, tumor size, nodal status, tumor grade, proliferation, angioinvasion, hormone receptor (HR) status, HER2 status [7], and molecular subtype [8]. In general, early HR positive, i.e., estrogen receptor (ER) and/or progesterone receptor (PR) positive, IBC frequently responds to endocrine therapy, whereas HER2 positive IBC benefits from anti-HER2 therapy. IBC that (over-)expresses neither HR nor HER2 [triple negative breast cancer (TNBC)] typically responds well to chemotherapy, which is mostly administered in the neoadjuvant setting [8].
In 2000 and 2001, gene expression analysis led to the identification of IBC molecular subtypes, namely Luminal subtypes, HER2-enriched, Basal-like, and normal breast-like [9,10,11]. The identified molecular subtypes were then revised into Luminal A, Luminal B, HER2-enriched and Basal-like, given their prognostic and predictive value used for therapy recommendation [12]. Patients with Luminal A IBC have a very good prognosis and benefit from endocrine therapy alone but not from chemotherapy in a clinically relevant dimension. However, due to endocrine resistance, patients with Luminal B tumors might have a poorer prognosis if treated with endocrine therapy alone [13] but might profit from chemotherapy [9,10,12]. HER2-enriched IBC are highly sensitive to anti-HER2 agents [14]. Finally, patients with Basal-like IBC benefit from chemotherapy [15].
In daily routine diagnostics, surrogate (molecular-like) IBC subtyping is assessed using the immunohistochemical (IHC) expression of ER and PR, HER2 status [IHC and/or in situ hybridization (ISH)], the IHC expression of the proliferation marker Ki-67, and, depending on the classification definition used, tumor grade. Luminal A-like IBCs show expression of HRs, no overexpression of HER2, low Ki-67 expression (and low/intermediate tumor grade). Luminal B-like HER2 negative tumors display HR positivity, HER2 negativity but high Ki-67 (and/or high-grade morphology), whereas Luminal B-like HER2 positive IBCs express both HRs and HER2 independently from the proliferation rate (tumor grade low, intermediate or high). HER2 positive (non-luminal) IBCs show HER2 positivity but HR negativity. As mentioned above, TNBCs display both negative HR and HER2 status [16,17].
In patients with HR+/HER2- IBC of intermediate risk of recurrence, estimated using conventional clinical and pathological risk factors, the decision on adjuvant chemotherapy is very challenging for clinicians. In order to address this issue, several multigene expression tests have been developed to assess the risk of distant recurrence and, in part, to evaluate the molecular subtypes which, however, differ from test to test due to the different technologies and the gene expression profiles used [11]. These tests include Oncotype DX® (Genomic Health), MammaPrint® (Agendia), EndoPredict® (Sividon Diagnostics/Myriad Genetics), Prosigna®, (Veracyte, formerly: NanoString Technologies)] [8]. Formalin-fixed and paraffin-embedded (FFPE) tumor tissue can be used for all assays. All these multigene expression tests provide both risk scores and discrimination into risk groups. They also provide information about late recurrence (EndoPredict® and Prosigna®) and, partly, even information about the molecular subtype (Prosigna®, Blueprint® if added to MammaPrint®). So far, prospective studies have shown that Oncotype DX® and MammaPrint® reveal patients at low risk of recurrence that would be overtreated with chemotherapy [18,19,20]. Currently, there are no available results from prospective trials regarding the predictive value of EndoPredict® and Prosigna® [21]. However, retrospective studies provide strong evidence that the risk scores of these two gene expression tests predict well both late distant recurrence and patients at low risk [22]. Disadvantages of all multigene expression assays are the high costs (~2–3 k Euro in Germany) and the fact that these tests are not comprehensively available in pathological laboratories.
Given the ability of Prosigna® test to provide both a risk of recurrence and a PAM50-based IBC molecular subtype, the present multicenter study aims to investigate whether Prosigna® assay results [testing centers comprised the Institutes of Pathology of Erlangen [coordinating center, C1), München (C2), Viersen (C3), Halle/Saale (C4), and Essen (C5)] correlate with the molecular-like surrogate subtypes, routinely assessed [locally vs. by C1 vs. by the study site Salzburg (C6)] using immunohistochemistry (+/− tumor grade). Furthermore, the impact of Prosigna® test results on treatment decision is investigated.
2. Results
Prosigna® molecular subtyping was compared with local and two central (C1 and C6) molecular-like subtypes relying on both IHC (HRs, HER2, Ki-67) subtyping and IHC + tumor grade (IHC+G) subtyping. Furthermore, the influence of Prosigna® assay results on chemotherapy treatment decision was investigated.
2.1. Characteristics of Cohort
Prognostic clinical and pathological variables including age at diagnosis, tumor size, Ki-67 expression, and tumor grade are summarized in Table S2. Briefly, median patients’ age at diagnosis was 55 years (range: 39–78 years); tumor size ranged between 0.7 and 15.4 cm (median 1.8 cm). There was a significant association between assessment center (local, C1, or C6) and tumor grade (p-value = 1.6 × 10−9) (Figure S1). Notably, center C6 was positively associated with G1 tumors (21/109) and negatively associated with G3 tumors (7/109) (Table S2). Furthermore, there was a significant difference between the three centers also in Ki-67 expression (p-value = 5.8 × 10−6). Indeed, while local and C1 Ki-67 assessments did not show any significant difference in terms of median expression value, the one reported by center C6 was significantly lower compared to both center C1 (post-hoc pairwise adjusted p-value = 1.2 × 10−4) and local institutions (post-hoc pairwise adjusted p-value = 1.4 × 10−5) (Figure S2). Taken together, these results show that in the evaluation of some prognostic clinical and pathological variables, center C6 performed differently from both local institutions and center C1, which instead provided comparable results.
2.2. Comparison of Local versus Central Molecular-Like Subtyping
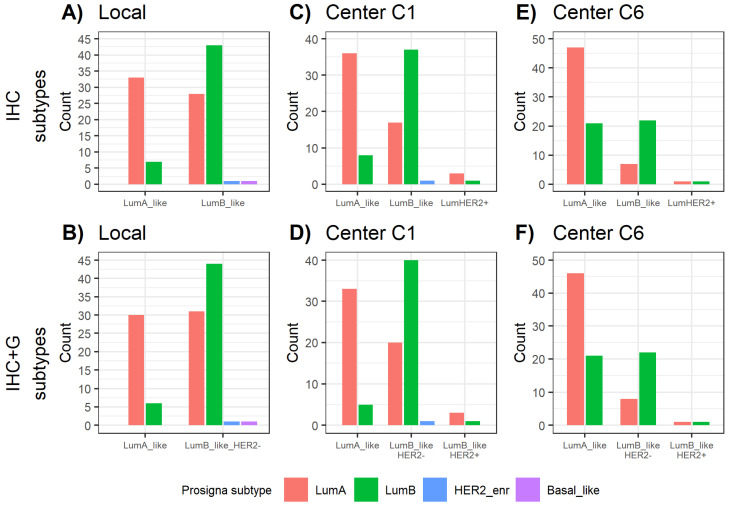
According to local IHC subtyping, 35.4% of cases were Luminal A-like and 64.6% Luminal B-like subtype, whereas according to IHC+G subtyping 31.9% and 68.1% were Luminal A-like and Luminal B-like, respectively. When analyzing both C1 IHC and IHC+G subtyping, the proportion of the different subtypes was comparable to the one reported from local assessments, with Luminal B being the dominant surrogate subtype. In center C6, however, the opposite scenario was observed, with Luminal A-like subtype accounting for more than two thirds of all the cases (Table 1).
Table 1.
Frequency of the different surrogate subtypes locally and centrally (C1, C6) assessed using both immunohistochemistry (IHC) and IHC+G subtyping.
| Surrogate Subtypes | |||
|---|---|---|---|
| Luminal A-like | Luminal B-like HER2 negative | Luminal B-like HER2 positive | |
| Local IHC subtyping | 35.4% | 64.6% | // |
| Local IHC+G subtyping | 31.9% | 68.1% | // |
| C1 IHC subtyping | 42.7% | 53.4% | 3.9% |
| C1 IHC+G subtyping | 36.9% | 59.2% | 3.9% |
| C6 IHC subtyping | 68.7% | 29.3% | 2.0% |
| C6 IHC+G subtyping | 67.7% | 30.3% | 2.0% |
HER2: human epidermal growth factor receptor 2.
The difference between the two centers C1 and C6 in the proportion of surrogate subtypes was further explored. Almost all IHC+G cases declared as Luminal A-like by C6 but not by C1 reported in C1 Ki-67 values ≥ 20% and were thus classified as Luminal B-like (HER2 negative) tumors (Figure S3A). Instead, the IHC+G cases declared as Luminal B-like HER2- by C1, but classified as Luminal A-like by C6, were characterized by Ki-67 values < 20% according to C6 (Figure S3B). Among the IHC+G Luminal A-like cases assessed in center C6, 26.9% (18/67) were graded as G1, whereas the remaining 73.1% (49/67) were of intermediate grade (G2). Only 5.6% (1/18) of the cases assessed as G1 Luminal A-like by C6 were graded as G1 also by C1, while almost all (94.4%, 17/18) were graded as G2. Of these, 76.5% (13/17) were still Luminal A-like cases but 23.5% (4/17) were upgraded to Luminal B-like HER2 negative (Figure S4A) due to Ki-67 expression ≥ 20%. A total of 73.5% (36/49) of G2 Luminal A-like cases detected by C6 were graded as G2 also by C1. Of these, 63.9% (23/36) were still Luminal A-like, 30.5% (11/36) were upgraded to Luminal B-like HER2 negative, and 5.6% (2/36) were Luminal B-like HER2 positive. The remaining 26.5% (13/49) cases were upgraded to G3 by C1 and all classified as Luminal B-like HER2 negative (Figure S4B).
2.3. Distribution of Prosigna® Molecular Subtypes
A total of 53.8% (n = 64/119) of cases were allocated as Prosigna® Luminal A and 44.5% (n = 53/119) as Prosigna® Luminal B. Two cases, however, did not match the local HR positivity (both were locally assessed as Luminal B-like HER2 negative) and were assigned to Basal-like (n = 1/119) and HER2-enriched (n = 1/119) Prosigna® subtypes.
2.4. Comparison between Surrogate Subtyping and Molecular Prosigna® Subtype
In all, 82.5% (33/40) of local IHC Luminal A-like tumors were classified as Prosigna® Luminal A. However, 17.5% (7/40) were upgraded to Prosigna® Luminal B. Within local IHC Luminal B-like IBCs, 58.9% (43/73) matched with Prosigna® Luminal B subtype, whereas 38.4% (28/73) were downgraded to Prosigna® Luminal A (Figure 1A). According to local IHC+G subtyping, 83.3% (30/36) of Luminal A-like cases were classified by Prosigna® as Luminal A and 16.7% (6/36) were upgraded to Prosigna® Luminal B. In regard to local IHC+G Luminal B-like IBCs, 57.1% (44/77) were classified as Prosigna® Luminal B, whereas 40.3% (31/77) were downgraded to Prosigna® Luminal A (Figure 1B).
Figure 1.
Distribution of Prosigna® subtypes across (A) local IHC subtypes, (B) local IHC+G subtypes, (C) C1 IHC subtypes, (D) C1 IHC+G subtypes, (E) C6 IHC subtypes, (F) C6 IHC+G subtypes. HER2_enr = HER2-enriched; IHC = immunohistochemistry; IHC+G = immunohistochemistry + tumor grade; LumA = Luminal A; LumB = Luminal B.
Compared with C6 assessments, local and C1 subtyping showed less cases upgrading from Luminal A-like to Prosigna® Luminal B (Table 2). With respect to center C6, though, they reported more IBC cases downgrading from Luminal B-like to Prosigna® Luminal A (Table 2, Figure 1).
Table 2.
Comparison between local vs. C1 vs. C6 surrogate subtyping and molecular Prosigna® subtype.
| Luminal A-Like | Luminal B-Like | |||
|---|---|---|---|---|
| Match with Prosigna® Luminal A |
Upgrade to Prosigna® Luminal B |
Match with Prosigna® Luminal B |
Downgrade to Prosigna® Luminal A |
|
| Local IHC subtyping | 82.5% (33/40) | 17.5% (7/40) | 58.9% (43/73) | 38.4% (28/73) |
| Local IHC+G subtyping | 83.3% (30/36) | 16.7% (6/36) | 57.1% (44/77) | 40.3% (31/77) |
| C1 IHC subtyping | 81.8% (36/44) | 18.2% (8/44) | 67.3% (37/55) | 30.9% (17/55) |
| C1 IHC+G subtyping | 86.8% (33/38) | 13.2% (5/38) | 65.6% (40/61) | 32.8% (20/61) |
| C6 IHC subtyping | 69.1% (47/68) | 30.9% (21/68) | 75.9% (22/29) | 24.1% (7/29) |
| C6 IHC+G subtyping | 68.7% (46/67) | 31.3% (21/67) | 73.3% (22/30) | 26.7% (8/30) |
IHC: immunohistochemistry.
The best strength of agreement occurred between Prosigna® and center C1 for both IHC and IHC+G subtyping, with a Cohen’s Kappa (κ) of 0.449 and 0.455, respectively. When instead considering the degree of agreement between Prosigna® and center C6, the k value reached 0.379 for IHC subtypes and 0.36 for IHC+G subtypes (Table 3).
Table 3.
Estimates of Cohen’s Kappa (κ) as an index of pairwise agreement between Prosigna® subtypes and local assessments (IHC / IHC+G subtypes), Prosigna® subtypes and C1 assessments (IHC / IHC+G subtypes), Prosigna® subtypes and C6 assessments (IHC / IHC+G subtypes).
| κ (IHC Subtype) |
κ (IHC+G Subtype) |
|
|---|---|---|
| Prosigna vs. local institutes | 0.374 | 0.344 |
| Prosigna vs. center C1 | 0.449 | 0.455 |
| Prosigna vs. center C6 | 0.379 | 0.36 |
IHC: immunohistochemistry.
2.5. Prosigna® Risk Groups and Correlation with Local Surrogate Subtypes
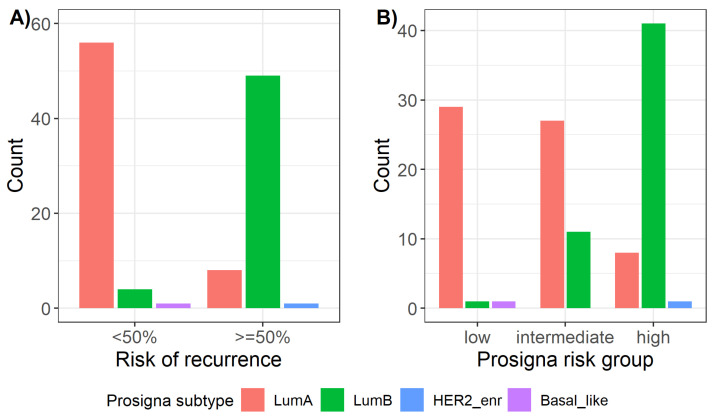
A total of 87.5% (56/64) of all Prosigna® Luminal A tumors showed a risk of recurrence (ROR) < 50%, whereas 92.5% (49/53) of all Prosigna® Luminal B tumors presented with a ROR ≥ 50% (Figure 2A). Furthermore, there was a significant association (p-value = 3.8 × 10−14) between Prosigna® subtypes and Prosigna® risk groups (low, intermediate, and high). Namely, 82% (41/50) of patients at high risk had Prosigna® Luminal B IBC subtype, whereas 71.1% (27/38) of patients at intermediate risk and 93.5% (29/31) at low risk suffered from Prosigna® Luminal A tumors (Figure 2B).
Figure 2.
Distribution of Prosigna® subtypes across (A) Prosigna® risk of recurrence and (B) Prosigna® risk groups. HER2_enr = HER2-enriched; LumA = Luminal A; LumB = Luminal B.
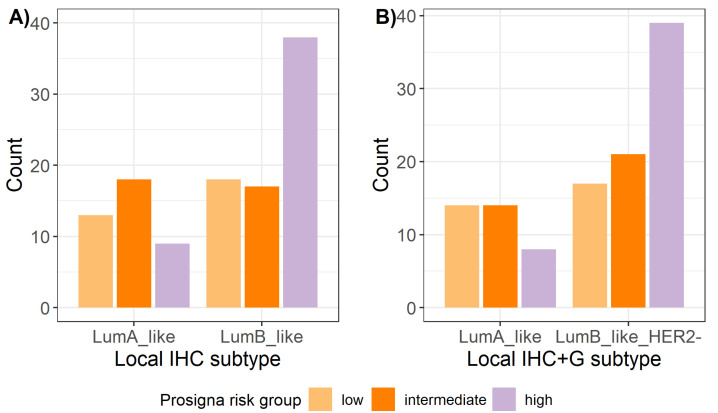
Prosigna® risk group was found to be significantly correlated also with both IHC and IHC+G subtypes (Figure 3), with Prosigna® high risk group being positively associated with Luminal B subtype and negatively associated with Luminal A subtype (p-value = 0.007 and p-value = 0.015, respectively, for IHC and IHC+G subtypes).
Figure 3.
Distribution of Prosigna® risk groups across local surrogate (A) IHC subtypes and (B) IHC+G subtypes. IHC = immunohistochemistry; IHC+G = immunohistochemistry + tumor grade; LumA = Luminal A; LumB = Luminal B.
2.6. Influence of Prosigna® Assay Result on Treatment Recommendation
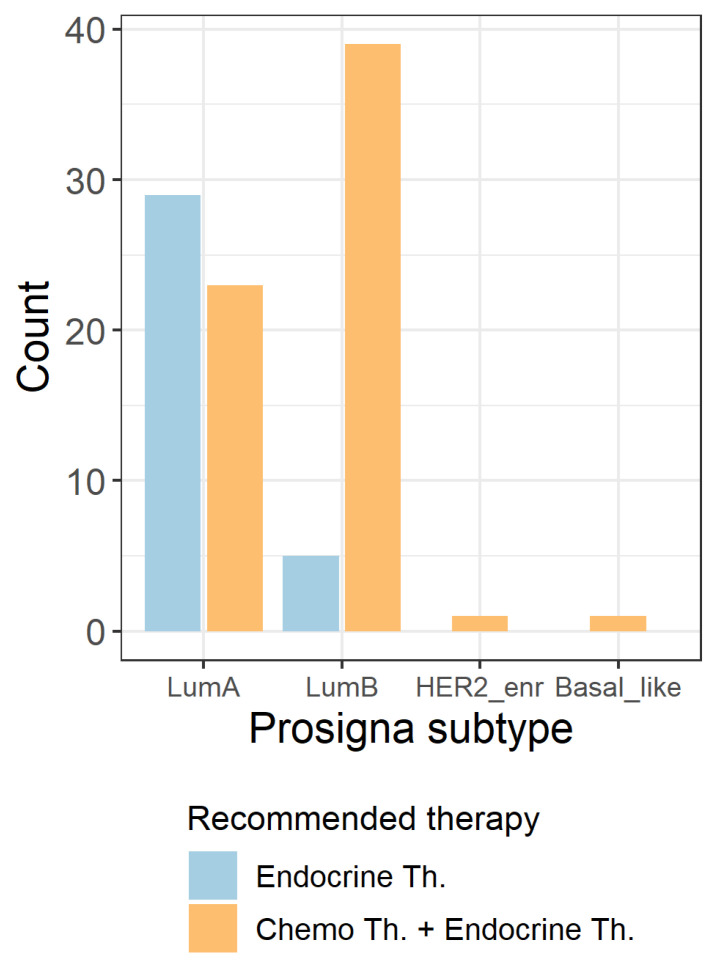
Looking at the distribution of tumor board recommendation within the different Prosigna® subtypes, chemotherapy + endocrine therapy was recommended to 44.2% (23/52) of Prosigna® Luminal A cases and to 88.6% (39/44) of Prosigna® Luminal B cases. The two cases with Prosigna® Basal-like (n = 1) and Prosigna® HER2-enriched (n = 1) IBC were both recommended towards adjuvant chemotherapy + endocrine therapy (Figure 4).
Figure 4.
Distribution of tumor board recommended across the different Prosigna® subtypes. HER2_enr = HER2-enriched; LumA = Luminal A; LumB = Luminal B; Th. = therapy.
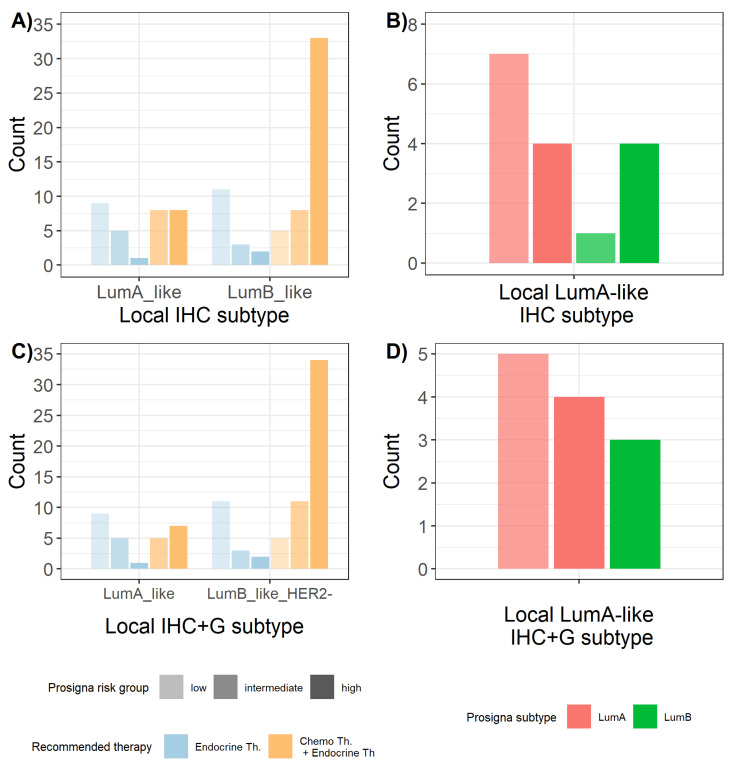
Within local IHC surrogate subtyping, 51.6% (16/31) of Luminal A-like IBCs were recommended towards chemotherapy + endocrine therapy. All these cases were indeed classified by Prosigna® as either at high or intermediate risk (Figure 5A). Notably, looking at the distribution of Prosigna® subtypes within these Luminal A-like IHC cases, 68.8% (11/16) were Prosigna® Luminal A subtypes at high or intermediate risk and the remaining 31.3% (5/16) were upgraded to Prosigna® Luminal B at high or intermediate risk (Figure 5B).
Figure 5.
Influence of Prosigna® results on treatment decision for locally assessed IHC and IHC+G surrogate subtypes. (A) Recommended therapy within locally assessed IHC surrogate subtypes and (B) distribution of Prosigna® subtype within the subset of Luminal A-like IHC subtypes that were recommended towards adjuvant chemotherapy and endocrine therapy. (C) Recommended therapy within locally assessed IHC+G surrogate subtypes and (D) distribution of Prosigna® subtype within the subset of Luminal A-like IHC+G subtypes that were recommended towards adjuvant chemotherapy and endocrine therapy. Colour intensity reflects Prosigna® risk group. IHC = immunohistochemistry; IHC+G = immunohistochemistry + tumor grade; LumA = Luminal A; LumB = Luminal B; Th. = therapy.
Within local IHC+G subtyping, 44.4% (12/27) Luminal A-like tumors were recommended towards chemotherapy + endocrine therapy (Figure 5C). In all, 75% (9/12) of these were classified as Luminal A subtypes at high or intermediate risk group according to Prosigna® assay, whereas the remaining 25% (3/12) were upgraded to Prosigna® Luminal B subtype at high risk (Figure 5D).
3. Discussion
In the present study, we correlated the results of a PAM50-based multigene expression test (Prosigna®) with surrogate subtyping of 119 IBC patients and investigated the influence of Prosigna® results on therapy decision. We showed that the agreement between Prosigna® molecular subtypes and molecular-like subtyping using IHC +/− tumor grade was fair to moderate depending on surrogate subtyping method and center. For Luminal A-like cases locally assessed either with IHC or IHC+G subtyping, results showed that adjuvant chemotherapy + endocrine therapy was recommended by the interdisciplinary tumor board to those cases that Prosigna® testing classified as Luminal A at high/intermediate risk or upgraded to Luminal B.
Commercially available breast cancer multigene expression tests (e.g., Oncotype DX®, MammaPrint®, EndoPredict®, Prosigna®) have been developed to help oncologists in the decision for or against adjuvant chemotherapy in patients with early HR+ IBC at intermediate risk. In prospective–retrospective (Oncotype DX®, EndoPredict®, Prosigna®) and prospective (Oncotype DX®, MammaPrint®) trials, respectively, it has been shown that multigene expression tests can identify IBC patients with low risk of recurrence that can be treated with adjuvant endocrine therapy but do not need additional chemotherapy [13,18,19,20,23,24,25,26,27,28,29,30,31,32,33,34]. However, application of these tests should be restricted to a narrow IBC patient cohort in which the use of multigene expression tests is reasonable [35,36].
In detail, the Prosigna® assay was validated to predict the outcome in (a) post-menopausal women with HR+, lymph node-negative, Stage I or II IBC and (b) post-menopausal women with hormone receptor-positive (HR+), lymph node-positive (1–3 positive nodes), Stage II IBC after standard of care loco-regional treatment and adjuvant endocrine therapy alone. If used in conjunction with other clinical and pathological risk factors, the Prosigna® assay may provide additional information about the probable distant recurrence-free survival at 10 years [37]. Indeed, besides the PAM50-based molecular subtype, Prosigna® assay predicts the risk of distant recurrence within 10 years and the risk group [38]. In our study of HR+ (locally assessed) IBC cases, 53.8% were classified as Prosigna® Luminal A subtype and 44.5% as Prosigna® Luminal B subtype. Two outlier cases were found, one Basal-like and one HER2-enriched. Both were locally classified as Luminal B-like HER2 negative. Within the Prosigna® high risk group, most cases were of the Prosigna® Luminal B subtype; vice versa, almost the entire low risk group was characterized by Prosigna® Luminal A IBCs. This is in line with both the biological understanding of molecular subtyping of IBC and its prognostic and predictive value. Indeed, Luminal A tumors are associated with a better prognosis compared to Luminal B IBCs; hence, therapy stratification can be modified accordingly [8]. As mentioned above, surrogate IBC subtyping using IHC +/- tumor grade has been used in daily routine diagnostics instead of mRNA-based molecular subtyping for the last decades. Indeed, compared to multigene expression assays, surrogate subtyping benefits from several advantages: it is cheaper, of low turn around time, and available even in small pathological laboratories. However, as highlighted by our study, the expression of single biomarkers (e.g., Ki-67) can differ from site to site due to pre- and post-analytical reasons [39,40]. Hence, oncologists have to consider the (post-) analytical standards of their local pathologists for therapy stratification. Notably, when comparing local and two central (C1, C6) assessments of surrogate subtypes, differences emerged between center C6 and both the local and C1 institutions. Indeed, while both local and C1 laboratories detected a higher proportion of Luminal B-like cases compared to Luminal A-like cases, in center C6 the majority of samples were classified as Luminal A-like. This is of great importance, since therapy stratification may vary. Discrepancies in surrogate subtyping were due to significant differences in both Ki-67 expression values and tumor grade assessments. These findings confirm that, to date, distinction between Luminal A-like and Luminal B-like tumors by IHC is still problematic and controversial. Some people suggest emphasizing tumor grade and PR expression in regard to luminal subtype distinction [17,41,42], while others advise the use of Ki-67 IHC [16]. Although big standardization efforts have been made [39,43,44], there is still a lack of common internationally accepted guidelines for the use, standardized scoring method, and optimal cut off for Ki-67 expression [16,39,45,46]. One should note that the use of Ki-67 IHC for therapy decision depends on (inter-)national guidelines: It is recommended by the St. Gallen International Expert Consensus but not by the American Society of Clinical Oncology [16,46,47,48].
When comparing molecular-like subtypes with Prosigna® subtyping, we found a match up to 86.8% for Luminal A(-like) cases and up to 75.9% for Luminal B(-like) cases depending on center and surrogate subtyping method. Accordingly, a fair-to-moderate agreement was found between Prosigna® molecular subtypes and molecular-like subtyping. This is in line with the findings of Bastien et al., who reported some inconsistency between IHC surrogate subtyping and PAM50 gene expression subtypes. In their study, ESR1 and ERBB2 gene expression showed more prognostic impact than the corresponding IHC markers [49]. In another study that correlated IHC-based surrogate subtyping with PAM50 molecular subtypes, 38.4% of IBCs were discrepantly subtyped [50]. Furthermore, agreement between PAM50 HER2 enriched tumors and HER2 positive subtype defined by standard IHC/ISH is not always given [51,52], which might lead to confusion and fundamental changes in regard to therapy recommendation. In a TNBC cohort, 84.3% of cases matched with PAM50 Basal-like subtype, 16.7% were HER2-enriched, and 5.2% showed a luminal gene signature (4.2% Luminal A, 1.0% Luminal B) [53].
In our study, the distribution of Prosigna® risk groups within local surrogate subtypes (both IHC and IHC+G) highlighted a significant association between Prosigna® high risk tumors and local Luminal B-like subtyping, whereas local Luminal A-like IBCs positively associated with Prosigna® low risk cases, which is in line with the expected biological behaviour of surrogate subtypes.
When comparing IHC / IHC+G molecular-like subtypes with Prosigna® molecular subtypes, 13.2% to 31.3% of Luminal A-like tumors were upgraded to Prosigna® Luminal B, whereas 24.1% to 40.3% of Luminal B-like IBCs were downgraded to molecular Luminal A. Especially for Luminal A-like cases that are classified as Prosigna® Luminal B, this upgrade may influence oncologists’ decision on additional chemotherapy to adjuvant endocrine therapy. In our study, Luminal A-like tumors that were recommended to be treated with chemotherapy were either of higher risk and/or upgraded to Prosigna® Luminal B. Hence, the combination of Prosigna® molecular subtyping and risk estimation may have an impact on therapy stratification, especially for HR+ HER2- IBC patients with intermediate Ki-67 level / tumor grade.
A notable limitation of this study is the relatively small number of cases. A total of 18 samples had indeed to be excluded due to missing surrogate subtyping. The lack of clinical and pathological parameters was mainly attributable to those cases for which routine diagnostic was performed in laboratories not offering Prosigna® testing assay and that turned to central institutes (i.e., C1-C5) for gene expression testing.
Regarding the decision for or against chemotherapy, only the interdisciplinary tumor board decision after multigene expression assay performance was available. Hence, we could describe the (surrogate) subtypes and risk groups that were recommended to be treated with chemotherapy + endocrine therapy. However, we were not able to track any changes, either in gynecological oncologists-recommended therapy or patient’s attitude, in favor of adding adjuvant chemotherapy. To address this issue, an ongoing single-center study (C1) is currently evaluating prospective questionnaires to assess patients’ decision about adding adjuvant chemotherapy before and after molecular testing. Furthermore, due to missing survival information and the small number of cases, we could not correlate Prosigna® risk estimation with patients’ outcome and menopausal status. Therefore, we could analyze which surrogate subtyping matches best with Prosigna® subtyping but not which subtyping method (IHC vs. IHC+G, local vs. central, surrogate vs. molecular subtyping) has the greatest impact on prognosis.
Although the multigene expression assays mentioned above provide independent, and partly similar predictions on prognosis, the various tests accurately described by Sinn et al. cannot be compared directly with each other. Indeed, there is a considerable “inter-assay heterogeneity” which includes variability in test development, different test measures, only partly overlapping gene signatures, and variable risk scores [11,54,55]. Furthermore, classifying special histological types of IBC (e.g., adenoid-cystic breast carcinoma, secretory breast carcinoma) using gene expression tests without closer examination of underlying mechanisms may not reflect the distinct biology and outcome, and may thus require additional investigation [56].
In intermediate risk hormone receptor (HR) positive, HER2 negative breast cancer (BC), the decision regarding adjuvant chemotherapy might be also supported by using the PAM50-based genomic signature, the chemoendocrine score (CES), which is predictive of poorer relapse-free survival in patients with ROR-intermediate IBC treated with either adjuvant endocrine therapy only or no adjuvant systemic therapy, but not in patients treated with adjuvant chemotherapy [57]. Unfortunately, since PAM50-based molecular subtype in our cohort was performed by using the commercial Prosigna® assay, we were not able to calculate the CES values which might have added further valuable information on prognosis in our cases with the Prosigna® intermediate risk group.
McVeigh reported that the use of Oncotype DX influenced the choice of therapy, leading to 57% of the patients being spared from chemotherapy [58]. Similarly, we demonstrated that 48.4% of patients with surrogate Luminal A-like tumors at intermediate risk were recommended to receive endocrine therapy alone. Several decision impact and cost-effectiveness analyses have been conducted, showing multigene expression testing being cost-effective or one being more effective than another test [59,60,61,62]. One of the main disadvantages of these impact studies, however, is mostly the lack of integration with patients’ outcome [54].
4. Materials and Methods
4.1. Study Design
A total number of n = 142 IBC cases were prospectively included into the study between 2015 and 2016 and analyzed for further risk stratification within diagnostic setting using multigene expression testing (Prosigna® assay, to date of testing: NanoString Technologies, Seattle, WA, U.S.A.) at the Institutes of Pathology of Erlangen (coordinating center, C1), München (C2), Viersen (C3), Halle/Saale (C4), and Essen (C5). Histopathological diagnoses of invasive breast cancer (IBC) were made either in one of the aforementioned centers or decentralised in a peripheral pathological laboratory according to the German guidelines and recommendations relevant during those years [63]. Clinical and pathological data (e.g., age, histological subtype, tumor size, nodal status, tumor grade, IHC expression of ER, PR, and Ki-67, and HER2 status) were collected and, whenever available, also local tumor board’s therapy recommendations from original patients’ records and pathological reports. For all decentral pathological laboratories not offering Prosigna® testing, gene expression analysis was performed by one of the five study centers. All cases with sufficient formalin-fixed and paraffin-embedded (FFPE) tissue availability underwent further testing in center C1 and study site Salzburg (C6). These two centers retrospectively performed a second “central” assessment of tumor grade and IHC / ISH (ER, PR, Ki-67, HER2). Within each of the two centers C1 and C6, two pathologists, blinded to local data and experienced in breast cancer pathology, evaluated the surrogate subtypes. In case of disagreement between the two pathologists of the same center, consensus was reached after having reviewed together the respective assessments. Details in tissue processing for routine diagnostics, immunohistochemistry, and HER2 chromogenic in situ hybridization (CISH) can be found in the Supplement. Approval of the local academic ethics committee of the Friedrich-Alexander-Universität Erlangen-Nürnberg was obtained.
4.2. Molecular-Like Surrogate Subtyping
Based on local and central (C1 and C6) pathology, respectively, which involves the assessment of IHC expression of ER, PR, Ki-67 as well as HER2 status, and tumor grade, IBC samples were classified in surrogate subtypes according to:
(1) IHC (+/− ISH) alone (“IHC subtyping”; local vs. C1 vs. C6) (Table 4);
Table 4.
Criteria for surrogate IHC subtyping according to [16].
| Surrogate Subtype | Subgroup | ER | PR | HER2 | Ki-67 (%) | |
|---|---|---|---|---|---|---|
| Luminal A-like | + | +/− | − | and | Low (<20%) | |
| Luminal B-like | HER2 negative | + | +/− | − | and | High (≥20%) |
| HER2 positive | + | +/− | + | Any value | ||
| HER2 positive (non-luminal) | − | − | + | Any value | ||
| Triple negative | − | − | − | Any value |
ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; IHC: immunohistochemistry; Ki-67: proliferation marker; PR: progesterone receptor.
(2) IHC (+/− ISH) and tumor grade (”IHC+G subtyping”; local vs. C1 vs. C6) (Table 5).
Table 5.
| Surrogate Subtype | Subgroup | ER | PR | HER2 | Tumor Grade | Ki-67 (%) | ||
|---|---|---|---|---|---|---|---|---|
| Luminal A-like | + | +/− | − | and | G1, G2 | or | Low (<20%) | |
| Luminal B-like | HER2 negative | + | +/− | − | and | G3 | or | High (≥20%) |
| HER2 positive | + | +/− | + | G1, G2, G3 | Any value | |||
| HER2 positive (non-luminal) | − | − | + | G1, G2, G3 | Any value | |||
| Triple negative | − | − | − | G1, G2, G3 | Any value |
ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; IHC: immunohistochemistry; Ki-67: proliferation marker; PR: progesterone receptor.
4.3. Prosigna® Assay
The Prosigna® test (Veracyte, South San Francisco, CA, USA; formerly: NanoString Technologies, Seattle, WA, USA) was established in our laboratories between 2014 and 2016. For establishment, inter-laboratory agreement on n = 15 IBC cases (Institutes of Pathology Erlangen, München, and Heidelberg) was evaluated. Both tumor molecular subtype estimation and risk group assessment correlated in 100% of samples. Before RNA isolation, experienced pathologists reviewed the IBC cases regarding adequate tumor tissue on H&E slides, marked tumor region, and assessed tumor content on each tumor slide. RNA extraction and the multigene expression assay were performed by trained technicians according to the manufacturer’s manual. Assay measurements were done fully automatically by nCounter® Prep Station and nCounter Dx Digital Analyzer. The report file in output provided information about the molecular subtype, risk of recurrence (ROR), risk group (low, intermediate, or high), and probability of distant recurrence for each patient [65].
4.4. Statistical Analysis
All statistical analyses were performed within the R environment v.4.0.3 [66]. p-values (or, where applicable, adjusted p-values) < 0.05 were considered statistically significant. Details can be found in the Supplement.
5. Conclusions
In conclusion, we could demonstrate that IHC/IHC+G surrogate subtyping and molecular Prosigna® subtyping correlate moderately. The best pairwise agreement occurred, for both IHC and IHC+G, between Prosigna® and central C1 surrogate subtyping. Local Luminal A-like cases (IHC/IHC+G) were recommended towards chemotherapy + endocrine therapy if they were classified as Prosigna® Luminal A at high/intermediate risk or upgraded to Prosigna® Luminal B. Hence, the additional use of molecular subtyping and risk profiling might influence therapy recommendation or confirm oncologists’ choice of therapy in HR+ HER2- IBCs with intermediate prognostic risk assessed using conventional clinical and pathological risk factors.
Acknowledgments
Our special thanks to all patients, all oncologists, and interdisciplinary tumor board members, involved. Furthermore, we thank all technicians and pathologists in our laboratories for performance of Prosigna® assay.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23158716/s1 [67,68,69,70,71,72].
Author Contributions
Conceptualization, R.E., M.P.L., E.P., K.W.S., A.B., M.V., C.T., K.S.(Karl Sotlar), D.M., M.W.B., A.H.; methodology, R.E., R.S., D.U.-G., A.B., R.F.H.W., M.V., K.S. (Karl Sotlar), A.S.; formal analysis, M.A., F.F.; investigation, R.E., R.S., D.U.-G., A.B., R.F.H.W., M.V., K.S.(Karl Sotlar), A.S., K.S.(Katharina Stering), A.H.; resources, R.E., A.H., M.P.L., E.P., A.B., K.W.S., C.T., D.M., F.K., P.A.F., M.W.B.; data curation, R.E.; writing—original draft preparation, R.E., A.H.; writing—review and editing, all authors; visualization, R.E., M.A.; supervision, R.E., F.F., R.S., D.U.-G., R.F.H.W., M.V., K.S. (Karl Sotlar), D.M.; project administration, R.E.; All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
All procedures performed in studies involving human participants were in accordance with the ethical standards of the local national research committee and with the 1964 Helsinki Declaration and its later amendments, or comparable ethical standards. Approval for the study was obtained from the local ethics committee of the Friedrich-Alexander-Universität Erlangen-Nürnberg. This article did not involve any studies with animals performed by any of the authors.
Informed Consent Statement
Informed consent was obtained from all patients for Prosigna® testing as part of diagnostics. For central surrogate subtyping, the study got an informed consent waiver by the ethics committee of the Friedrich-Alexander-Universität Erlangen-Nürnberg due to the use of archival tissues with retrospective character.
Data Availability Statement
Detailed data not shown in the manuscript are available on request.
Conflicts of Interest
P.A.F. has received honoraria from Roche, Pfizer, Novartis, and Celgene. His institution conducts research for Novartis, Cepheid, and BioNTech. A.H. has received honoraria for lectures or consulting/advisory boards for Abbvie, AstraZeneca, Biontech, BMS, Boehringer Ingelheim, Cepheid, Diaceutics, Illumina, Ipsen, Janssen, Lilly, MSD, NanoString, Novartis, Qiagen, QUIP GmbH, Roche, 3DHistotech. R.E. has received honoraria from Roche, Eisai, Pfizer, Mindpeak, AstraZeneca, and Novartis and travel grants from BioNTech. The institution of A.H. and R.E. conducts research for AstraZeneca, Roche, Janssen-Cilag, Zytomed Systems, NanoString Technologies, Veracyte, Biocartis, Novartis, Cepheid, and BioNTech. N.H. reports honoraria for lectures and/or consulting from Amgen, AstraZeneca, Daiichi-Sankyo, Exact Sciences, Lilly, MSD, Novartis, Pierre Fabre, Pfizer, Roche, Sandoz/Hexal, and Seattle Genetics. R.W. served as advisor/consultant/speaker for and/or received travel grants by Agendia, Amgen, Aristo, AstraZeneca, Boeringer Ingelheim, Carl Zeiss, Celgene, Daiichi-Sankyo, Eisai, Exact Sciences, Genomic Health, Gilead, Glaxo Smith Kline, Hexal, Lilly, Medstrom Medical, MSD, Mundipharma, Mylan, Nanostring, Novartis, Odonate, Paxman, Palleos, Pfizer, Pierre Fabre, PumaBiotechnolgogy, Riemser, Roche, Sandoz/Hexal, Sanofi Genzyme, Seattle Genetics/Seagen, Tesaro Bio, Teva, Veracyte, and Viatris. O.H. received honoraria for lectures or consulting/advisory boards for Riemser, Roche, AstraZeneca, Amgen, Pfizer, Eisai, Hexal, MSD, Novartis, SeaGen, and Daiichi-Sankyo. M.P.L. reports honoraria for advisory boards for Lilly, AstraZeneca, MSD, Novartis, Pfizer, Gilead, Eisai, Exact Sciences, Daiichi-Sankyo, Grünenthal, Pierre Fabre, PharmaMar, and Roche; honoraria for lectures for Lilly, Roche, MSD, Novartis, Pfizer, Exact Sciences, Daiichi-Sankyo, Grünenthal, AstraZeneca, and Eisai; editorial board for medac; travel grants for Roche, and Pfizer. The other authors have nothing to declare.
Funding Statement
This research received no external funding (intramural funding). Furthermore, this work was supported by the Interdisciplinary Center for Clinical Research (IZKF, Clinician Scientist Program) of the Medical Faculty of Friedrich-Alexander-Universtität Erlangen-Nürnberg.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1. [(accessed on 27 April 2020)]. Available online: https://www.krebsdaten.de/Krebs/DE/Content/Krebsarten/Brustkrebs/brustkrebs_node.html.
- 2.Interdisziplinäre S3-Leitlinie für die Früherkennung, Diagnostik, Therapie und Nachsorge des Mammakarzinoms. Langversion 4. 3 February 2020 AWMF-Registernummer: 032-045OL. [(accessed on 2 August 2022)]. Available online: https://www.leitlinienprogramm-onkologie.de/leitlinien/mammakarzinom/
- 3.Arbeitsgemeinschaft Gynäkologische Onkologie e.V. Guidelines Breast Version 2020.1 (Download Gesamtdateien) [(accessed on 26 April 2020)]. Available online: www.ago-online.de.
- 4.Allison K.H., Hammond M.E.H., Dowsett M., McKernin S.E., Carey L.A., Fitzgibbons P.L., Hayes D.F., Lakhani S.R., Chavez-MacGregor M., Perlmutter J., et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J. Clin. Oncol. 2020;38:1346–1366. doi: 10.1200/JCO.19.02309. [DOI] [PubMed] [Google Scholar]
- 5.Wolff A.C., Hammond M.E.H., Allison K.H., Harvey B.E., Mangu P.B., Bartlett J.M.S., Bilous M., Ellis I.O., Fitzgibbons P., Hanna W., et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer. Am. Soc. Clin. Oncol./Coll. Am. Pathol. Clin. Pract. Guidel. Focused Update. 2018;142:1364–1382. doi: 10.5858/arpa.2018-0902-SA. [DOI] [PubMed] [Google Scholar]
- 6.Lokuhetty D., White V.A., Watanabe R., Cree I.A. WHO Classification of Tumours Series. 5th ed. Volume 2. International Agency for Research on Cancer; Lyon, France: 2019. WHO Classification of Tumours Editorial Board. Breast Tumours; pp. 88–97. [Google Scholar]
- 7.Lux M.P., Önügören O., Wunderle M., Hein A., Rauh C., Schrauder M.G., Fasching P.A., Beckmann M.W. (Neo-)Adjuvanten und First-Line-Therapie—Prognostische und prädiktive Faktoren. Gynäkologe. 2015;48:876–884. doi: 10.1007/s00129-015-3806-9. [DOI] [Google Scholar]
- 8.Cardoso F., Kyriakides S., Ohno S., Penault-Llorca F., Poortmans P., Rubio I.T., Zackrisson S., Senkus E. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. 2019;30:1194–1220. doi: 10.1093/annonc/mdz173. [DOI] [PubMed] [Google Scholar]
- 9.Perou C.M., Sorlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 10.Sorlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.B., van de Rijn M., Jeffrey S.S., et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schettini F., Brasó-Maristany F., Kuderer N.M., Prat A. A perspective on the development and lack of interchangeability of the breast cancer intrinsic subtypes. npj Breast Cancer. 2022;8:85. doi: 10.1038/s41523-022-00451-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parker J.S., Mullins M., Cheang M.C., Leung S., Voduc D., Vickery T., Davies S., Fauron C., He X., Hu Z., et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paik S., Shak S., Tang G., Kim C., Baker J., Cronin M., Baehner F.L., Walker M.G., Watson D., Park T., et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 14.Schettini F., Prat A. Dissecting the biological heterogeneity of HER2-positive breast cancer. Breast. 2021;59:339–350. doi: 10.1016/j.breast.2021.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Early Breast Cancer Trialists’ Collaborative Group Adjuvant chemotherapy in oestrogen-receptor-poor breast cancer: Patient-level meta-analysis of randomised trials. Lancet. 2008;371:29–40. doi: 10.1016/S0140-6736(08)60069-0. [DOI] [PubMed] [Google Scholar]
- 16.Goldhirsch A., Winer E.P., Coates A.S., Gelber R.D., Piccart-Gebhart M., Thurlimann B., Senn H.J., Panel M. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Von Minckwitz G., Untch M., Blohmer J.-U., Costa S.D., Eidtmann H., Fasching P.A., Gerber B., Eiermann W., Hilfrich J., Huober J., et al. Definition and Impact of Pathologic Complete Response on Prognosis after Neoadjuvant Chemotherapy in Various Intrinsic Breast Cancer Subtypes. J. Clin. Oncol. 2012;30:1796–1804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 18.Sparano J.A., Gray R.J., Makower D.F., Pritchard K.I., Albain K.S., Hayes D.F., Geyer C.E.J., Dees E.C., Perez E.A., Olson J.A.J., et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2015;373:2005–2014. doi: 10.1056/NEJMoa1510764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gluz O., Nitz U.A., Christgen M., Kates R.E., Shak S., Clemens M., Kraemer S., Aktas B., Kuemmel S., Reimer T., et al. West German Study Group Phase III PlanB Trial: First Prospective Outcome Data for the 21-Gene Recurrence Score Assay and Concordance of Prognostic Markers by Central and Local Pathology Assessment. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2016;34:2341–2349. doi: 10.1200/JCO.2015.63.5383. [DOI] [PubMed] [Google Scholar]
- 20.Cardoso F., van’t Veer L.J., Bogaerts J., Slaets L., Viale G., Delaloge S., Pierga J.-Y., Brain E., Causeret S., DeLorenzi M., et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N. Engl. J. Med. 2016;375:717–729. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 21. [(accessed on 10 April 2021)]. Available online: https://www.ago-online.de/fileadmin/ago-online/downloads/_leitlinien/kommission_mamma/2021/Einzeldateien_Literatur/2021D_05_Prognostische_und_praediktive_Faktoren_MASTER_final_20210302_inklRef.pdf.
- 22.Sestak I., Buus R., Cuzick J., Dubsky P., Kronenwett R., Ferree S., Sgroi D., Schnabel C., Baehner R., Mallon E., et al. Abstract S6-05: Comprehensive comparison of prognostic signatures for breast cancer in TransATAC. Cancer Res. 2017;77((Suppl. S4)):S06-05. doi: 10.1158/1538-7445.SABCS16-S6-05. [DOI] [Google Scholar]
- 23.Dubsky P., Brase J.C., Jakesz R., Rudas M., Singer C.F., Greil R., Dietze O., Luisser I., Klug E., Sedivy R., et al. The EndoPredict score provides prognostic information on late distant metastases in ER+/HER2- breast cancer patients. Br. J. Cancer. 2013;109:2959–2964. doi: 10.1038/bjc.2013.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubsky P., Filipits M., Jakesz R., Rudas M., Singer C.F., Greil R., Dietze O., Luisser I., Klug E., Sedivy R., et al. EndoPredict improves the prognostic classification derived from common clinical guidelines in ER-positive, HER2-negative early breast cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2013;24:640–647. doi: 10.1093/annonc/mds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Filipits M., Rudas M., Jakesz R., Dubsky P., Fitzal F., Singer C.F., Dietze O., Greil R., Jelen A., Sevelda P., et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011;17:6012–6020. doi: 10.1158/1078-0432.CCR-11-0926. [DOI] [PubMed] [Google Scholar]
- 26.Martin M., Brase J.C., Calvo L., Krappmann K., Ruiz-Borrego M., Fisch K., Ruiz A., Weber K.E., Munarriz B., Petry C., et al. Clinical validation of the EndoPredict test in node-positive, chemotherapy-treated ER+/HER2− breast cancer patients: Results from the GEICAM 9906 trial. Breast Cancer Res. 2014;16:R38. doi: 10.1186/bcr3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paik S., Tang G., Shak S., Kim C., Baker J., Kim W., Cronin M., Baehner F.L., Watson D., Bryant J., et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 28.Wolmark N., Mamounas E.P., Baehner F.L., Butler S.M., Tang G., Jamshidian F., Sing A.P., Shak S., Paik S. Prognostic Impact of the Combination of Recurrence Score and Quantitative Estrogen Receptor Expression (ESR1) on Predicting Late Distant Recurrence Risk in Estrogen Receptor-Positive Breast Cancer after 5 Years of Tamoxifen: Results from NRG Oncology/National Surgical Adjuvant Breast and Bowel Project B-28 and B-14. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2016;34:2350–2358. doi: 10.1200/jco.2015.62.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albain K.S., Barlow W.E., Shak S., Hortobagyi G.N., Livingston R.B., Yeh I.T., Ravdin P., Bugarini R., Baehner F.L., Davidson N.E., et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: A retrospective analysis of a randomised trial. Lancet Oncol. 2010;11:55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sgroi D.C., Sestak I., Cuzick J., Zhang Y., Schnabel C.A., Schroeder B., Erlander M.G., Dunbier A., Sidhu K., Lopez-Knowles E., et al. Prediction of late distant recurrence in patients with oestrogen-receptor-positive breast cancer: A prospective comparison of the breast-cancer index (BCI) assay, 21-gene recurrence score, and IHC4 in the TransATAC study population. Lancet Oncol. 2013;14:1067–1076. doi: 10.1016/S1470-2045(13)70387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filipits M., Nielsen T.O., Rudas M., Greil R., Stöger H., Jakesz R., Bago-Horvath Z., Dietze O., Regitnig P., Gruber-Rossipal C., et al. The PAM50 risk-of-recurrence score predicts risk for late distant recurrence after endocrine therapy in postmenopausal women with endocrine-responsive early breast cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014;20:1298–1305. doi: 10.1158/1078-0432.CCR-13-1845. [DOI] [PubMed] [Google Scholar]
- 32.Martín M., Prat A., Rodríguez-Lescure A., Caballero R., Ebbert M.T., Munárriz B., Ruiz-Borrego M., Bastien R.R., Crespo C., Davis C., et al. PAM50 proliferation score as a predictor of weekly paclitaxel benefit in breast cancer. Breast Cancer Res. Treat. 2013;138:457–466. doi: 10.1007/s10549-013-2416-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gnant M., Filipits M., Greil R., Stoeger H., Rudas M., Bago-Horvath Z., Mlineritsch B., Kwasny W., Knauer M., Singer C., et al. Predicting distant recurrence in receptor-positive breast cancer patients with limited clinicopathological risk: Using the PAM50 Risk of Recurrence score in 1478 postmenopausal patients of the ABCSG-8 trial treated with adjuvant endocrine therapy alone. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2014;25:339–345. doi: 10.1093/annonc/mdt494. [DOI] [PubMed] [Google Scholar]
- 34.Gnant M., Sestak I., Filipits M., Dowsett M., Balic M., Lopez-Knowles E., Greil R., Dubsky P., Stoeger H., Rudas M., et al. Identifying clinically relevant prognostic subgroups of postmenopausal women with node-positive hormone receptor-positive early-stage breast cancer treated with endocrine therapy: A combined analysis of ABCSG-8 and ATAC using the PAM50 risk of recurrence score and intrinsic subtype. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015;26:1685–1691. doi: 10.1093/annonc/mdv215. [DOI] [PubMed] [Google Scholar]
- 35.Andre F., Ismaila N., Henry N.L., Somerfield M.R., Bast R.C., Barlow W., Collyar D.E., Hammond M.E., Kuderer N.M., Liu M.C., et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: ASCO Clinical Practice Guideline Update—Integration of Results From TAILORx. Journal of Clinical Oncology. 2019;37:1956–1964. doi: 10.1200/JCO.19.00945. [DOI] [PubMed] [Google Scholar]
- 36.Krop I., Ismaila N., Andre F., Bast R.C., Barlow W., Collyar D.E., Hammond M.E., Kuderer N.M., Liu M.C., Mennel R.G., et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women with Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline Focused Update. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017;35:2838–2847. doi: 10.1200/JCO.2017.74.0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. [(accessed on 9 April 2021)]; Available online: https://www.accessdata.fda.gov/cdrh_docs/reviews/K130010.pdf.
- 38. [(accessed on 9 April 2021)]. Available online: https://www.prosigna.com/de/overview/
- 39.Dowsett M., Nielsen T.O., A’Hern R., Bartlett J., Coombes R.C., Cuzick J., Ellis M., Henry N.L., Hugh J.C., Lively T., et al. Assessment of Ki67 in breast cancer: Recommendations from the International Ki67 in Breast Cancer working group. J. Natl. Cancer Inst. 2011;103:1656–1664. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris L., Fritsche H., Mennel R., Norton L., Ravdin P., Taube S., Somerfield M.R., Hayes D.F., Bast R.C., Jr. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 41.Prat A., Cheang M.C., Martin M., Parker J.S., Carrasco E., Caballero R., Tyldesley S., Gelmon K., Bernard P.S., Nielsen T.O., et al. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013;31:203–209. doi: 10.1200/JCO.2012.43.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braun L., Mietzsch F., Seibold P., Schneeweiss A., Schirmacher P., Chang-Claude J., Peter Sinn H., Aulmann S. Intrinsic breast cancer subtypes defined by estrogen receptor signalling—Prognostic relevance of progesterone receptor loss. Mod. Pathol. Off. J. United States Can. Acad. Pathol. Inc. 2013;26:1161–1171. doi: 10.1038/modpathol.2013.60. [DOI] [PubMed] [Google Scholar]
- 43.Rimm D.L., Leung S.C.Y., McShane L.M., Bai Y., Bane A.L., Bartlett J.M.S., Bayani J., Chang M.C., Dean M., Denkert C., et al. An international multicenter study to evaluate reproducibility of automated scoring for assessment of Ki67 in breast cancer. Mod. Pathol. Off. J. United States Can. Acad. Pathol. Inc. 2019;32:59–69. doi: 10.1038/s41379-018-0109-4. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen T.O., Leung S.C.Y., Rimm D.L., Dodson A., Acs B., Badve S., Denkert C., Ellis M.J., Fineberg S., Flowers M., et al. Assessment of Ki67 in Breast Cancer: Updated Recommendations from the International Ki67 in Breast Cancer Working Group. J. Natl. Cancer Inst. 2020 doi: 10.1093/jnci/djaa201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polley M.Y., Leung S.C., McShane L.M., Gao D., Hugh J.C., Mastropasqua M.G., Viale G., Zabaglo L.A., Penault-Llorca F., Bartlett J.M., et al. An international Ki67 reproducibility study. J. Natl. Cancer Inst. 2013;105:1897–1906. doi: 10.1093/jnci/djt306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gnant M., Thomssen C., Harbeck N. St. Gallen/Vienna 2015: A Brief Summary of the Consensus Discussion. Breast Care. 2015;10:124–130. doi: 10.1159/000430488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. [(accessed on 10 April 2021)]. Available online: https://documents.cap.org/protocols/cp-breast-biomarker-20-1400.pdf.
- 48.Harris L.N., Ismaila N., McShane L.M., Andre F., Collyar D.E., Gonzalez-Angulo A.M., Hammond E.H., Kuderer N.M., Liu M.C., Mennel R.G., et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women with Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2016;34:1134–1150. doi: 10.1200/JCO.2015.65.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bastien R.R.L., Rodríguez-Lescure Á., Ebbert M.T.W., Prat A., Munárriz B., Rowe L., Miller P., Ruiz-Borrego M., Anderson D., Lyons B., et al. PAM50 Breast Cancer Subtyping by RT-qPCR and Concordance with Standard Clinical Molecular Markers. BMC Med. Genom. 2012;5:44. doi: 10.1186/1755-8794-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim H.K., Park K.H., Kim Y., Park S.E., Lee H.S., Lim S.W., Cho J.H., Kim J.Y., Lee J.E., Ahn J.S., et al. Discordance of the PAM50 Intrinsic Subtypes Compared with Immunohistochemistry-Based Surrogate in Breast Cancer Patients: Potential Implication of Genomic Alterations of Discordance. Cancer Res. Treat. Off. J. Korean Cancer Assoc. 2019;51:737–747. doi: 10.4143/crt.2018.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sweeney C., Bernard P.S., Factor R.E., Kwan M.L., Habel L.A., Quesenberry C.P., Shakespear K., Weltzien E.K., Stijleman I.J., Davis C.A., et al. Intrinsic subtypes from PAM50 gene expression assay in a population-based breast cancer cohort: Differences by age, race, and tumor characteristics. Cancer Epidemiol. Biomark. Prev. A Publ. Am. Assoc. Cancer Res. Cosponsored By Am. Soc. Prev. Oncol. 2014;23:714–724. doi: 10.1158/1055-9965.EPI-13-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Picornell A.C., Echavarria I., Alvarez E., López-Tarruella S., Jerez Y., Hoadley K., Parker J.S., del Monte-Millán M., Ramos-Medina R., Gayarre J., et al. Breast cancer PAM50 signature: Correlation and concordance between RNA-Seq and digital multiplexed gene expression technologies in a triple negative breast cancer series. BMC Genom. 2019;20:452. doi: 10.1186/s12864-019-5849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinn P., Aulmann S., Wirtz R., Schott S., Marmé F., Varga Z., Lebeau A., Kreipe H., Schneeweiss A. Multigene Assays for Classification, Prognosis, and Prediction in Breast Cancer: A Critical Review on the Background and Clinical Utility. Geburtshilfe Und Frauenheilkd. 2013;73:932–940. doi: 10.1055/s-0033-1350831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prat A., Parker J.S., Fan C., Cheang M.C.U., Miller L.D., Bergh J., Chia S.K.L., Bernard P.S., Nielsen T.O., Ellis M.J., et al. Concordance among gene expression-based predictors for ER-positive breast cancer treated with adjuvant tamoxifen. Ann. Oncol. 2012;23:2866–2873. doi: 10.1093/annonc/mds080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weigelt B., Reis-Filho J.S. Histological and molecular types of breast cancer: Is there a unifying taxonomy? Nat. Rev. Clin. Oncol. 2009;6:718–730. doi: 10.1038/nrclinonc.2009.166. [DOI] [PubMed] [Google Scholar]
- 57.Prat A., Lluch A., Turnbull A.K., Dunbier A.K., Calvo L., Albanell J., de la Haba-Rodríguez J., Arcusa A., Chacón J.I., Sánchez-Rovira P., et al. A PAM50-Based Chemoendocrine Score for Hormone Receptor-Positive Breast Cancer with an Intermediate Risk of Relapse. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017;23:3035–3044. doi: 10.1158/1078-0432.CCR-16-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McVeigh T.P., Hughes L.M., Miller N., Sheehan M., Keane M., Sweeney K.J., Kerin M.J. The impact of Oncotype DX testing on breast cancer management and chemotherapy prescribing patterns in a tertiary referral centre. Eur. J. Cancer. 2014;50:2763–2770. doi: 10.1016/j.ejca.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ward S., Scope A., Rafia R., Pandor A., Harnan S., Evans P., Wyld L. Gene expression profiling and expanded immunohistochemistry tests to guide the use of adjuvant chemotherapy in breast cancer management: A systematic review and cost-effectiveness analysis. Health Technol. Assess. 2013;17:1–302. doi: 10.3310/hta17440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seguí M.Á., Crespo C., Cortés J., Lluch A., Brosa M., Becerra V., Chiavenna S.M., Gracia A. Genomic profile of breast cancer: Cost–effectiveness analysis from the Spanish National Healthcare System perspective. Expert Rev. Pharm. Outcomes Res. 2014;14:889–899. doi: 10.1586/14737167.2014.957185. [DOI] [PubMed] [Google Scholar]
- 61.Blohmer J.U., Rezai M., Kümmel S., Kühn T., Warm M., Friedrichs K., Benkow A., Valentine W.J., Eiermann W. Using the 21-gene assay to guide adjuvant chemotherapy decision-making in early-stage breast cancer: A cost-effectiveness evaluation in the German setting. J. Med. Econ. 2013;16:30–40. doi: 10.3111/13696998.2012.722572. [DOI] [PubMed] [Google Scholar]
- 62.Hall P.S., Smith A., Hulme C., Vargas-Palacios A., Makris A., Hughes-Davies L., Dunn J.A., Bartlett J.M.S., Cameron D.A., Marshall A., et al. Value of Information Analysis of Multiparameter Tests for Chemotherapy in Early Breast Cancer: The OPTIMA Prelim Trial. Value Health. 2017;20:1311–1318. doi: 10.1016/j.jval.2017.04.021. [DOI] [PubMed] [Google Scholar]
- 63.Arbeitsgemeinschaft Gynäkologische Onkologie e.V. Guidelines Breast Versions of 2015/2016 (Chapter Pathologie) [(accessed on 7 April 2021)]. Available online: www.ago-online.de.
- 64.Von Minckwitz G., Blohmer J.U., Costa S.D., Denkert C., Eidtmann H., Eiermann W., Gerber B., Hanusch C., Hilfrich J., Huober J., et al. Response-guided neoadjuvant chemotherapy for breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013;31:3623–3630. doi: 10.1200/JCO.2012.45.0940. [DOI] [PubMed] [Google Scholar]
- 65. [(accessed on 7 April 2021)]. Available online: https://www.prosigna.com/en-gb/downloads/
- 66.R Core Team R: A Language and Environment for Statistical Computing. 2020. [(accessed on 19 December 2021)]. Available online: https://www.R-project.org/
- 67.Elston C.W., Ellis I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 68.Hammond M.E., Hayes D.F., Dowsett M., Allred D.C., Hagerty K.L., Badve S., Fitzgibbons P.L., Francis G., Goldstein N.S., Hayes M., et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 2010;28:2784–2795. doi: 10.1200/jco.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolff A.C., Hammond D.E., Hicks D.G., Dowsett M., McShane L.M., Allison K.H., Allred D.C., Bartlett J.M., Bilous M., Fitzgibbons P., et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 2013;31:3997–4013. doi: 10.1200/jco.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 70.Gamer M., Lemon J., Fellows I., Singh P. irr: Various Coefficients of Interrater Reliability and Agreement. [(accessed on 26 January 2019)]. Available online: https://CRAN.R-project.org/package=irr.
- 71.Landis J.R., Koch G.G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 72.Bewick V., Cheek L., Ball J. Statistics review 8: Qualitative data–tests of association. Crit. Care. 2003;8:1–8. doi: 10.1186/cc2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Detailed data not shown in the manuscript are available on request.