Abstract
Myxococcus xanthus cells move on a solid surface by gliding motility. Several genes required for gliding motility have been identified, including those of the A- and S-motility systems as well as the mgl and frz genes. However, the cellular defects in gliding movement in many of these mutants were unknown. We conducted quantitative, high-resolution single-cell motility assays and found that mutants defective in mglAB or in cglB, an A-motility gene, reversed the direction of gliding at frequencies which were more than 1 order of magnitude higher than that of wild type cells (2.9 min−1 for ΔmglAB mutants and 2.7 min−1 for cglB mutants, compared to 0.17 min−1 for wild-type cells). The average gliding speed of ΔmglAB mutant cells was 40% of that of wild-type cells (on average 1.9 μm/min for ΔmglAB mutants, compared to 4.4 μm/min for wild-type cells). The mglA-dependent reversals and gliding speeds were dependent on the level of intracellular MglA protein: mglB mutant cells, which contain only 15 to 20% of the wild-type level of MglA protein, glided with an average reversal frequency of about 1.8 min−1 and an average speed of 2.6 μm/min. These values range between those exhibited by wild-type cells and by ΔmglAB mutant cells. Epistasis analysis of frz mutants, which are defective in aggregation and in single-cell reversals, showed that a frzD mutation, but not a frzE mutation, partially suppressed the mglA phenotype. In contrast to mgl mutants, cglB mutant cells were able to move with wild-type speeds only when in close proximity to each other. However, under those conditions, these mutant cells were found to glide less often with those speeds. By analyzing double mutants, the high reversing movements and gliding speeds of cglB cells were found to be strictly dependent on type IV pili, encoded by S-motility genes, whereas the high-reversal pattern of mglAB cells was only partially reduced by a pilR mutation. These results suggest that the MglA protein is required for both control of reversal frequency and gliding speed and that in the absence of A motility, type IV pilus-dependent cell movement includes reversals at high frequency. Furthermore, mglAB mutants behave as if they were severely defective in A motility but only partially defective in S motility.
Many bacterial species spread over surfaces as expanding swarms (1, 3, 9, 18). The swarming behavior of Myxococcus xanthus is due to gliding motility of individual cells; M. xanthus cells carry no flagella and cannot swim. Gliding movement consists of translocation along the cell’s long axis on a solid surface with one or the other pole leading the way. Swarms of M. xanthus expand by coordinating the gliding movements of individual cells (15, 18). Two multigene systems, the A- and S-motility systems, and the mgl and frz genes have been identified as affecting the ability of vegetative colonies to swarm on agar plates (2, 12, 13, 27).
Among the M. xanthus motility mutants isolated so far, mglA mutants have the strongest defect in that colonies are rendered nonswarming by a mutation in a single gene (11, 12); their colonies are heaped in the center and have a sharp edge that expands outward due only to cell growth and division. Because the colony shape of mgl mutants is indistinguishable from that of A− S− double mutants, the mgl gene is considered to be required for both A motility and S motility (13). These observations had initially suggested that the MglA protein might be part of the gliding motor (22), but the protein was localized to the cytoplasm and the predicted amino acid sequence resembles those of GDP/GTP binding proteins of the p21Ras type (6, 7). Therefore, it appears to be more likely that the MglA protein is involved in regulating the gliding motor. The mglA gene is cotranscribed with an adjacent gene called mglB. Deletion of the mglB gene reduces the level of mglA protein to about 15% of the wild-type level (8). The swarming behavior of mglB mutants is intermediate between those of mglA mutants and the wild type; mglB mutants spread at about 25% of the rate of wild-type swarms and 10 times faster than mglA mutant swarms (8).
Wild-type swarming of M. xanthus requires both A motility and S motility (13, 15). Previous studies showed that mutants defective in the A-motility system were impaired in swarming, and single cells were found to be unable to glide as individual isolated cells (12, 19b). Molecular analysis of one of the A-motility genes, the cglB gene, showed that this gene encodes a lipoprotein with an unusually high cysteine content (19b). All motility retained in this and other A− mutants is considered to be S motility, because A− S− double mutants do not swarm at all (12). Recently, Wu and Kaiser (23–26) discovered that all of the S-motility genes in the sglI region of M. xanthus encode proteins required for the synthesis, export, assembly, and regulation of type IV pili. How type IV pili produce S motility is unknown; however, these pili are also required for twitching motility (3, 10).
The M. xanthus frz genes were first identified in mutants with an altered aggregation behavior during fruiting-body development (27). The predicted amino acid sequences of the Frz proteins exhibit extensive homology to those of proteins of two-component signal transduction systems, specifically those involved in chemotaxis (che genes) in a variety of prokaryotes and in control of twitching motility in Pseudomonas aeruginosa (5, 19). Single isolated cells of frz mutants are defective in reversing the direction of gliding. With the exception of a frzD mutant, frz mutants (frzA, -B, -C, -E, and -F) reverse less frequently than wild-type cells (2). A transposon insertion in the frzD region of the frzCD gene generated a mutant where the cells reverse more often than wild-type cells (2).
During our motility studies of mgl and cglB mutants, we recognized that both motility mutants reverse the direction of movement more often than the wild type. We then investigated systematically the movement of individual cells with high optical and temporal resolutions (21). With the assay employed, displacements of as low as 0.03 μm and translocation speeds of as low as 1 μm/min were detectable. Movements of single cells of mgl, cglB, frz, and various double mutants were quantified to examine the relationship between the A- and S-motility genes and the mgl and frz genes in controlling single-cell motility.
MATERIALS AND METHODS
Bacteria and growth.
M. xanthus (14) DK1622 and its motility mutant strains were routinely grown on CTT agar plates (11). To assay gliding motility, single colonies were picked, transferred to 5 ml of CTT broth in 50-ml flasks, and grown at 32°C. Cultures were aerated by rotary shaking at 200 rpm. Strains DK9711 (frzE226::Tn5) and DK9713 (ΔmglAB frzE226::Tn5) were constructed by Mx8 transduction of kanamycin resistance from strain DZ3377 (frzE226::Tn5) to DK1622 (frz+) and to DK6204 (frz+ ΔmglAB), respectively. Strains DK9712 (frzCD224::Tn5) and DK9714 (ΔmglAB frzCD224::Tn5) were constructed by Mx8 transduction of kanamycin resistance from strain DZ4033 (frzCD224::Tn5) to DK1622 and to DK6204, respectively. Strain DK9715 was constructed by Mx8 transduction of kanamycin resistance from strain DK3163 [pilR::Tn5(Ω3163)] to DK6204. Strains DK9716 and DK9717 were constructed by Mx8 transduction of tetracycline resistance from strains DK3164 [pilR::Tn5-132(Ω3164)] and DZ4040 [frzE::Tn5(Ω234)], respectively, to strain JZ315. Strains DZ3377, DZ4033, and DZ4040 were kindly provided by D. Zusman, and strain JZ315 was kindly provided by J. Zissler (2, 16, 19). The M. xanthus strains used and their relevant genotypes are summarized in Table 1.
TABLE 1.
M. xanthus strains used
| Strain | Relevant genotype | Motility phenotype or colony morphology | Reference or construction |
|---|---|---|---|
| DK1622 | mgl+ frz+ | A+ S+ | 15 |
| DK3163 | pilR::Tn5(Ω3163) | A+ S− | 19a |
| DK3164 | pilR::Tn5-132(Ω3163) | A+ S− | 19a |
| DK6204 | ΔmglAB | Nonswarming | 7 |
| DK6206 | ΔmglB | Reduced swarming | 8 |
| DK9711 | frzE226::Tn5 | “Frizzy” | Mx8 (DZ3377) × DK1622, selected Kmr |
| DK9712 | frzCD224::Tn5 | Smooth edge | Mx8 (DZ4033) × DK1622, selected Kmr |
| DK9713 | ΔmglAB frzE226::Tn5 | Nonswarming | Mx8 (DZ3377) × DK6204, selected Kmr |
| DK9714 | ΔmglAB frzCD224::Tn5 | Nonswarming | Mx8 (DZ4033) × DK6204, selected Kmr |
| DK9715 | ΔmglAB pilR::Tn5(Ω3163) | Nonswarming | Mx8 (DK3163) × DK6204, selected Kmr |
| DK9716 | cglB::Tn5phoA pilR::Tn5-132(Ω3164) | A− S− | Mx8 (DK3164) × JZ315, selected Tetr |
| DK9717 | cglB::Tn5phoA frzE::Tn5(Ω234) | A− S+ | Mx8 (DZ4040) × JZ315, selected Tetr |
| DZ3377 | frzE226::Tn5 sglA | “Frizzy” | 2 |
| DZ4033 | frzCD224::Tn5 sglA | Smooth edge | 2 |
| DZ4040 | frzE::Tn5(Ω234) sglA | “Frizzy” | 2 |
| JZ315 | cglB::Tn5phoA | A− S+ | 16 |
Recording and quantification of cell movement by video microscopy.
From cultures grown to 80 to 100 Klett density units (4 × 108 to 5 × 108 cells/ml), 10-μl aliquots were withdrawn and spotted on CTT plates (1.5% agar) which had been prepared the day prior to use (21). The droplets were allowed to dry onto the agar for 10 to 15 min and were immediately examined by microscopy. Recording and analyzing of cell movement were conducted by methods described previously (21).
Swarming motility under starvation conditions.
M. xanthus DK1622, DK9712, DK6204, and DK9714 were grown in CTT broth to a density of 100 Klett units (5 × 108 cells/ml) and concentrated 10 times. Ten-microliter aliquots were spotted on TPM plates (17) containing 0.6% agar and incubated for 24 h at 32°C in the dark. Plates were prepared the day before use.
RESULTS
M. xanthus cells move on solid surfaces by gliding (4, 18, 21). Occasionally, cells reverse the direction of movement, so the leading end of the cell becomes the lagging end. During our video microscopic studies of M. xanthus, we noticed that cells of ΔmglAB mutants and of A-motility (cglB) mutants reversed their direction of gliding more often than wild-type cells. We therefore first studied gliding of M. xanthus wild-type cells with respect to reversals and gliding speed in both directions.
Reversals in wild-type cells.
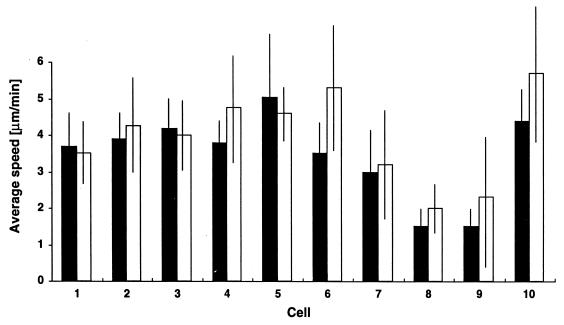
The gliding speed of M. xanthus wild-type cells was measured, using a motility assay with high optical and temporal resolutions (21). M. xanthus wild-type cells (DK1622) that executed at least one reversal during the period of observation were clocked in both their forward and backward directions, and gliding speeds were measured for each. Figure 1 displays the average speed in each direction for 10 cells. No significant difference in gliding speed was evident despite the fourfold variation in average speed among these cells, implying that individual M. xanthus cells have no kinetically preferred gliding direction. Speed variations between M. xanthus cells are common, and the observed speeds fall within the normal range (21).
FIG. 1.
Forward (solid bars) and backward (open bars) translocation velocities of individual DK1622 cells. Ten cells were measured when moving in the forward and backward directions. For each cell all speed values from each direction were pooled, and the average speed and standard deviation were calculated.
A reversal of gliding in a single M. xanthus cell is characterized as the following sequence: movement in the forward direction, stop, and movement in the backward direction. The transition from gliding in one direction to gliding in the reverse direction is observed as a transient decrease in gliding speed to less than 1 μm/min (the minimum speed that is measurable [21]) before a cell resumes movement in the opposite direction. During our studies on the reversal behavior, we noticed that gliding movements which are interrupted by stops can be grouped into three classes: (i) forward movement, stop for less than 9 s, and backward movement (we considered this sequence a reversal); (ii) forward movement, stop for more than 9 s, and forward movement (we called this sequence a pause); and (iii) forward movement, stop for more than 9 s, and backward movement. In this study, we analyzed only the first type of reversals.
How often do wild-type cells reverse the direction of gliding? The time interval between two reversals in DK1622 cells was found to be variable. Of 250 cells that all moved during the observation period (Table 2), 13 were found to reverse once or more than once within a period of 20 min. Those cells that reversed performed on average 0.17 reversal per min (Table 2). In an independent experiment, 5 of 90 wild-type cells reversed once or more than once within 10 min. Monitoring cells for longer than 20 min proved to be difficult because cells would often associate with other cells in a group and the cell being tracked could not be identified within the group. Since the longer reversal times were difficult to measure, the true average reversal frequency for wild-type cells may be less than the 0.17 reversal per min recorded.
TABLE 2.
Reversal frequencies of wild-type M. xanthus and mgl and frz motility mutants
| Parameter | Strain:
|
||||||
|---|---|---|---|---|---|---|---|
| DK1622 (wild type) | DK6204 (ΔmglAB) | DK6206 (ΔmglB) | DK9712 (frzD) | DK9711 (frzE) | DK9713 (ΔmglAB frzE) | DK9714 (ΔmglAB frzD) | |
| Average reversal frequency (min−1) (SD) | 0.17 (NDa) | 2.9 (1.4) | 1.8 (1.3) | 1.5 (1.1) | <0.02 (NAb) | 2.7 (1.4) | 3.3 (1.1) |
| No. of reversals counted | 18 | 439 | 393 | 140 | 0 | 222 | 56 |
| Average speed (μm/min) (SD)c | 4.4 (2.2) | 1.9 (1.1) | 2.6 (1.5) | 4.0 (2.3) | 4.3 (2.0) | 2.1 (1.4) | 1.9 (1.0) |
| % of cells exhibiting active movement (no. of cells evaluated) | 100 (250) | 88 (792) | 99 (412) | 100 (265) | 10 (272) | 85 (324) | ND |
ND, not determined.
NA, not applicable.
For frzD mutant cells 3,187 speed values were collected, for frzE mutant cells 1,004 speed values were collected, and for ΔmglAB frzE double-mutant cells 2,927 speed values were collected. See the text for further description and details.
Reversals in mgl mutant cells.
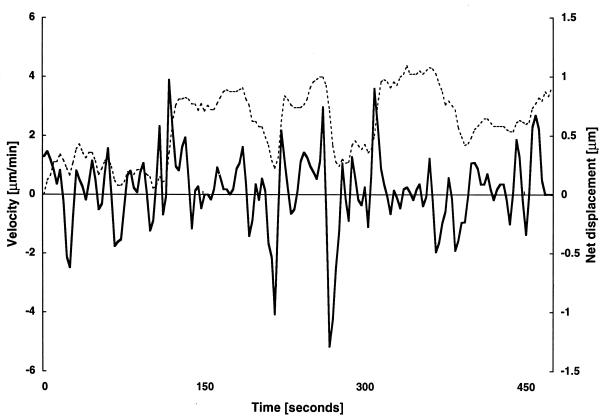
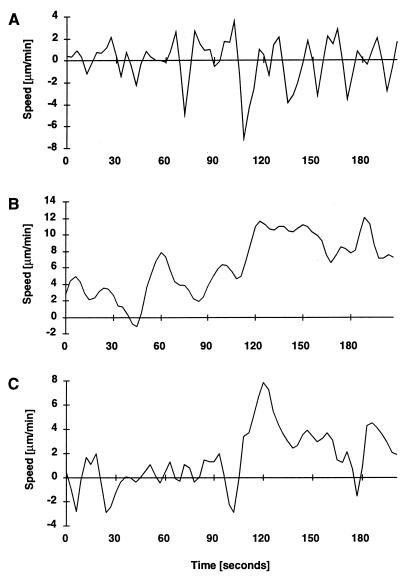
To investigate the function of the MglA protein in gliding, movement of individual ΔmglAB motility mutant cells was quantified. During a 20-min interval, 696 of 792 ΔmglAB cells moved: only 12% failed to move. Figure 2 displays the velocity profile of one cell. As illustrated, the movement pattern of this particular ΔmglAB cell was characterized by frequent reversals and abrupt, often jerky changes in gliding speed (at t = 1 to 140 s and 200 to 310 s) which alternated with intervals where no significant movement was detectable (t = 140 to 160 s and 310 to 400 s). During the total observation time of about 474 s, the cell shown in Fig. 2 completed 14 reversals. The net displacements performed were less than 1 μm in 9 min. Figure 3 summarizes the speed measurements for 50 ΔmglAB cells. The average speed of an actively gliding ΔmglAB mutant cell was 1.9 ± 1.1μm/min. Figure 3 also indicates that 43% of the 2,927 recorded speed values were below 1.0 μm/min, which is the lower limit of reliable measurement (21). Reversal frequencies for 50 ΔmglAB mutant cells were quantified. An average reversal frequency of 2.9 min−1 (standard deviation, 1.4 min−1) was calculated from 439 reversals (Table 2), indicating that the reversal frequency of ΔmglAB mutant cells is at least 1 order of magnitude higher than that of DK1622 cells (Table 2). Cells of ΔmglAB mutants traveled on average 0.3 ± 0.4 μm before reversing their directions. These data are compatible with the time-lapse photographs of mglA mutant cells published by Hodgkin and Kaiser (13), who reported no net movement over a period of 3 h.
FIG. 2.
Velocity profile of an M. xanthus ΔmglAB cell. Strain DK6204 was grown to a density of 5 × 108 cells/ml in CTT broth. Ten microliters was spotted on a 1-day-old CTT plate (1.5% agar). The lagging end of the cell was tracked for 474 s. Velocity (solid line) and net displacement (dashed line) are displayed. The cell reversed the direction of movement 14 times, at t = 15, 30, 63, 75, 96, 102, 192, 222, 261, 288, 363, 399, 444, and 453 s. Reversal frequencies were calculated from intervals t = 15 to 30, 63 to 75, 96 to 102, 222 to 261, and 261 to 288 s.
FIG. 3.
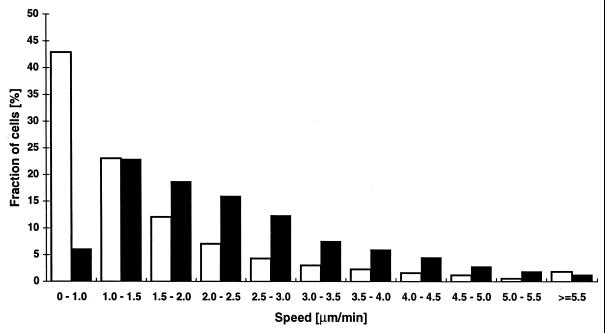
Distribution of gliding speeds of ΔmglAB (DK6204) and ΔmglB (DK6206) cells. Gliding speeds from 50 ΔmglAB cells (open bars) and 50 ΔmglB cells (solid bars) were measured. The speed values for each strain were grouped, and the number of speed values qualifying for each category was determined. For ΔmglAB cells 2,927 values were collected, and for ΔmglB cells 11,169 values were collected.
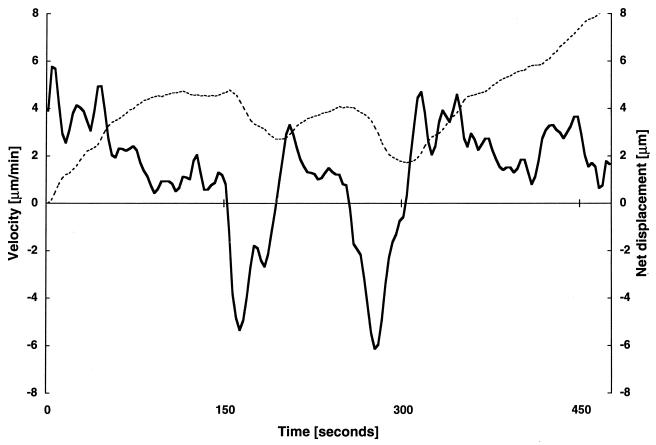
The mglA gene is cotranscribed with another gene, called mglB. Cells of a ΔmglB mutant strain contain only 15% of the wild-type level of the MglA protein (8). This reduction in MglA protein level is not due to transcriptional control. In contrast to mglA point mutants and mglAB deletion mutants, some swarming of mglB colonies is observed on a 1.5% agar surface. However, the swarm expansion rate is one-quarter of that of the wild type (8). We examined 412 individual ΔmglB mutant cells by video microscopy. More than 98% of these cells moved during a 20-min time interval. The gliding profile of a single ΔmglB mutant cell is displayed in Fig. 4. In contrast to the ΔmglAB mutant cell (Fig. 2), this ΔmglB cell moved during the entire period of observation. The cell maintained movement in one direction for up to 160 s before reversing. Gliding speeds for 50 ΔmglB mutant cells were determined and are represented in Fig. 3. Their average speed is 2.6 ± 1.5 μm/min. Of the 11,169 speed values obtained, 6% were below the minimal detectable speed, indicating that a ΔmglB cell is actively gliding for 94% of the time. A particular cell (Fig. 4) accomplished four reversals in 474 s. Reversals for 50 ΔmglB mutant cells were quantified, and of 393 reversals counted, the average value was calculated to be 1.8 reversals per min with a standard deviation of 1.3 (Table 2). To test whether the difference in the average reversal frequencies of mglAB and mglB mutant cells is statistically significant, the two distributions were subjected to a paired Student t test. Taking each measurement as being independent, the probability that the two distributions were derived from one and the same distribution and appeared to be different by chance was found to be less than 10−17. Although the mean values are close (2.9 and 1.8 min−1, respectively), the average reversal frequencies are significantly different. Thus, a decrease in the level of mglA protein is reflected by an increase in the frequency of reversal.
FIG. 4.
Velocity profile of an M. xanthus ΔmglB cell. Strain DK6206 was grown to a density of 5 × 108 cells/ml in CTT broth. Ten microliters was spotted on a 1-day-old CTT plate (1.5% agar). The lagging end of the cell was followed for 474 s. The solid line indicates velocity, and the dashed line indicates net displacement. The cell conducted four reversals (at t = 150, 195, 255, and 303 s).
Reversals in A− mutant cells.
Colonies of mutants defective in the A-motility system exhibit sharp edges, and no single cells are detectable at the perimeter (12, 19b). Because no agl null mutant is available, single-cell analyses were conducted with strain JZ315, which carries a Tn5phoA insertion in the cglB gene (16). Cells of this strain and agl mutant strains are nonmotile when separated from each other by more than 2 μm (12, 19b). However, when in close proximity to each other (<2 μm), cglB mutant cells showed active movements, presumably reflecting their S motility. These movements included an increased and highly variable pattern of cell reversals. The patterns are illustrated in the velocity profiles of three individual cells (Fig. 5). During the approximately 200-s observation period, the cell whose pattern is shown in Fig. 5A performed abrupt movements that included 15 reversals. If movement of cglB cells included such frequent reversals and if most of the runs in one direction lasted for 30 s or less, then we considered these cells to be moving in a high-reversal mode. More than half (54%) of 50 cells that were analyzed in detail moved in the high-reversal mode as illustrated in Fig. 5A. Considering reversals of 50 cells in a high-reversal mode, the average number of reversals was 2.7 (±1.4) min−1 (Table 3). Another one-third of the cglB cells investigated moved in a pattern that is characterized by the absence of reversals, such as is shown in Fig. 5B. This cell translocated at high speed for a period of up to 150 s without a reversal. This fast forward movement was not smooth but was subject to abrupt changes in translocation speed; after the first reversal, the speed fluctuated between the following approximated values (micrometers per minute): 8, 2, 6, 4, 11, 8, 12, and 8. The apparent bimodal movement pattern (high reversal [Fig. 5A] or low reversal [Fig. 5B]) of cglB cells is not due to two subpopulations in the culture, because the same cell can move in both modes, as shown in Fig. 5C. About 16% of the cells investigated were able to move in a pattern where periods of high reversals (Fig. 5C, 30 to 100 s) alternated with intervals of unidirectional, relatively fast movement (Fig. 5C, 100 to 170 s). Thus, for cglB mutant cells in close proximity to each other, each cell is able to move in either a high-reversal or a low-reversal mode, with a higher probability of moving in the former mode. A correlation between a particular reversal mode and the cell-to-cell distance or the cell orientation could not be addressed, because these movements were observed for cells in contact with each other.
FIG. 5.
Movement patterns of cglB cells (JZ315) [A− S+]) when in close proximity to each other. Panels A, B, and C show results for three different cells. For details, see the text.
TABLE 3.
Reversal frequencies of A− and S− motility mutants
| Parameter | Strain:
|
|||
|---|---|---|---|---|
| JZ315 (cglB) (high reversing)a | DK9716 (cglB pilR) | DK9717 (cglB frzE) (high reversing)a | DK9715 (ΔmglAB pilR) | |
| Average reversal frequency (min−1) (SD) | 2.7 (1.4) | 0 (NAc) | 2.6 (1.4) | 0 [2.0]b (1.4) |
| No. of reversals counted | 446 | 0 | 296 | 73 |
| Average speed (μm/min) (SD) | 2.5 (1.7) | 2.0 (1.0) | 2.1 (1.1) | 1.9 (1.0) |
| % of cells exhibiting active movement (no. of cells evaluated)d | ≥98 (373) | ∼10 (164) | ≥98 (240) | 30 (310) |
Only those cells for which most of the runs between two reversals lasted for 30 s or less were considered. The switching of a single cglB cell between the high-reversal and the low-reversal mode is also indicated by the high standard deviation of the average reversal frequency.
Only 30% of the cells moved, and the average reversal frequency of those cells was 2.0 min−1. See the text for more details.
NA, not applicable.
See the text for further description and details.
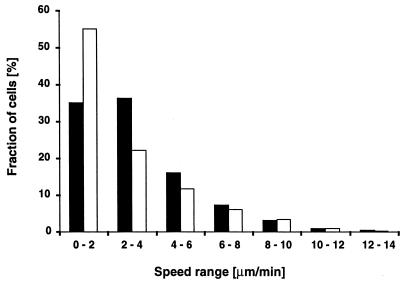
We examined the average translocation speed of cglB cells when in close proximity. For 2,621 speed measurements obtained from 50 cells, the average speed of high-reversal cells was determined to be 2.5 (±1.7) μm/min (Table 3). This value is below the average speed of 5.0 μm/min for wild-type cells when close to each other (21). However, for low-reversal cells the average velocity was 4.7 (±3.0) μm/min, and those cglB cells were able to glide with the same high velocities as wild-type cells (Fig. 6). Both wild-type and cglB cells utilize the same high speeds at approximately the same frequency (Fig. 6, speed ranges of >4 μm/min). One noticeable difference is that cglB cells moved significantly more often at speeds of 1 to 2 μm/min. Because only a small fraction of these cells were conducting unidirectional, high-speed movements and because the average speed value integrates all speed measurements, this average value does not reflect adequately the ability of A− cells to move with wild-type speeds in close proximity.
FIG. 6.
Distribution of gliding speeds of wild-type (solid bars) and cglB (open bars) cells when in close proximity (cell-cell distance of ≤1 μm). The gliding speeds of 50 cells were measured for each strain. Distributions were calculated from 3,446 speed measurements for DK1622 cells and 2,621 measurements for JZ315 cells.
Reversals in double-mutant cells containing frz mutations.
Individual cells of M. xanthus mutants defective in the frz genes had previously been described to be altered in control of reversal frequency (2). However, the phenotype of a frz deletion mutant is opposite from that of a Δmgl mutant; most frz mutants have a decreased rather than increased reversal frequency (2) (Table 2). Under our experimental conditions, a frzE mutant (DK9711) moved with an average speed of 4.3 ± 2.0 μm/min and an average reversal frequency of <0.02 reversal per min (Table 2).
One exception to the pattern of decreased reversal frequency of frz mutants is a Tn5 insertion in the 3′ end of the frzCD gene. Such a frzD mutant cell (strain DK9712) glides on average with a speed of 4.0 ± 2.3 μm/min and performs reversals at a rate of 1.5 ± 1.1 per min. This average reversal frequency is comparable to the value determined earlier by Blackhart and Zusman (2). To investigate the possibility of a connection between the frz-dependent and the mgl-dependent control of reversal frequency, two double-mutant strains, a ΔmglAB frzE mutant (DK9713) and a ΔmglAB frzD mutant (DK9714), were constructed, and the behavior of individual double-mutant cells was examined by high-resolution video microscopy. As shown in Table 2, cells of the ΔmglAB frzE double mutant reversed the direction of movement with an average frequency of about 2.7 ± 1.4 min−1. This frequency was very similar to the 2.9 ± 1.4 min−1 which was measured for ΔmglAB mutants and was clearly different from the frequency found for frzE mutant cells (<0.02 min−1) (Table 2). In addition, the average gliding speed of 2.1 ± 1.4 μm/min for ΔmglAB frzE double mutants was similar to that for ΔmglAB mutants (1.9 ± 1.1 μm/min) but markedly different from that for frzE mutant cells (4.3 ± 2.0 μm/min) (Table 2). Single cells of the ΔmglAB frzD strain (DK9714) also showed a reversal frequency and average gliding speed that are very similar to those of ΔmglAB (Table 2).
We also examined the effect of a frzE mutation on the high-reversal phenotype of cglB cells by analyzing the single-cell movement of a cglB frzE double mutant (Table 3). About 98% of the 240 cells examined were actively moving. Detailed analysis of 50 cglB frzE cells revealed that the high-reversal phenotype of cglB cells was retained (Table 3). The average number of reversals for the double-mutant cells moving in a high-reversal mode was 2.6 ± 1.4 min−1. Similar to the cglB strain, cells were observed to move also in a low-reversal mode with increased speed (3.4 ± 2.8 μm/min [data not shown]). Thus, a frzE mutation does not affect the high-reversal behavior of cglB cells.
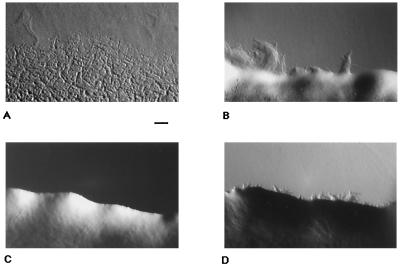
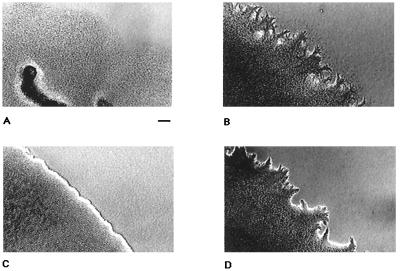
mglA (17) and frz (27) are also important for cell movement under starvation conditions that induce fruiting-body development. Swarming motility integrates the gliding movements of individual cells and can be easily detected with the unaided eye. To further investigate the connection between the frz-dependent and mglA-dependent control of cell movement, the macroscopic swarming behaviors of the mglA frz double mutants under growth and starvation conditions were compared. To examine swarming edges under vegetative conditions, CTT plates (1.5% agar) were inoculated with single cells of strains DK1622, DK6204, DK9712, and DK9714 and incubated at 32°C. Swarming edges of single colonies were recorded after 5 days (Fig. 7). To study swarming edges under starvation conditions, cells of these strains were grown in CTT medium, concentrated 10 times, and spotted on petri plates containing TPM starvation medium with 0.6% agar. Edges of swarms were examined after incubation at 32°C for 24 h (Fig. 8). As expected, ΔmglAB mutants exhibited a sharp swarming edge (Fig. 7C and 8C) compared to the wild type (Fig. 7A and 8A). Swarms of frzD mutants showed an edge containing numerous pronounced flares and projections (Fig. 7B and 8B). ΔmglAB frzD double-mutant cells had projections (Fig. 7D and 8D) like those of frzD mutant cells (Fig. 7B and 8B), which was qualitatively different from the ΔmglAB mutant (Fig. 7C and 8C). The wave-shaped swarming edge of strain DK9714 (ΔmglAB frzCD224::Tn5), which is clearly different from that of DK6204 (ΔmglAB) cells, implies active net cell movement. The swarming pattern of ΔmglAB frzE mutant cells under the same conditions was indistinguishable from that of ΔmglAB mutant cells; i.e., there was a sharp edge like that in Fig. 7C and 8B (data not shown). The swarming behavior of the wild type and the ΔmglAB and ΔmglAB frz mutants on TPM starvation plates containing 1.5% (data not shown) or 0.6% (Fig. 8) agar was observed. The swarming effect of ΔmglAB frzD was most pronounced on the support with the lower percentage of agar.
FIG. 7.
Swarming patterns of M. xanthus motility strains under vegetative conditions on CTT plates (1.5% agar). Cells were streaked on CTT agar plates, which were prepared 1 day before use. The plates were incubated at 32°C for 5 days in the dark. (A) DK1622; (B) DK9712; (C) DK6204; (D) DK9714. Bar, 100 μm.
FIG. 8.
Swarming patterns of M. xanthus motility strains on TPM starvation agar. The agar plates contained 10 mM Tris-HCl (pH 7.6), 1 mM potassium phosphate, 8 mM magnesium sulfate, and 0.6% agar. The plates were prepared on the day prior to use. Cells were grown in CTT broth to a density of 5 × 108 cells/ml and concentrated 10 times, and 10-μl drops were spotted on the agar plates. The plates were incubated at 32°C for 24 h in the dark. (A) DK1622; (B) DK9712; (C) DK6204; (D) DK9714. Bar, 100 μm.
Dependence of cell reversals on S motility.
In M. xanthus, movement of cells in close proximity is controlled by the S-motility system (12, 15). Recently, Wu and Kaiser (23–26) showed that the S-motility genes in the sglI region encode type IV pili. To examine the effect of S motility on the high-reversal phenotype of mglAB mutants, we analyzed 310 cells of an mglAB pilR double mutant. In general, the motility of this strain was strongly impaired. Of those cells, only about 30% showed visible movement. In contrast to DK6204, the movement consisted of short translocations, and only 11 of the 20 actively moving cells that were analyzed in detail retained a mode of frequent reversals. In those cells, the average number of reversals was reduced to 2.0 (±1.4) per min (Table 3). This observation indicates that the S-motility defect reduced the extent of motility significantly but only partially affected the high-reversal mode of mglAB mutant cells. Interestingly, the average speed of mglAB pilR double-mutant cells was 1.9 (±1.0) μm/min, as determined from 1,694 measurements, and thus was similar to speeds of ΔmglAB cells.
In order to examine the effect of S motility on the high-reversal pattern of cglB mutant cells, we constructed a cglB pilR double mutant and analyzed the movement of individual cells by video microscopy. The high-reversal phenotype in these A− S− double-mutant cells was completely abolished (Table 3). During the 20-min observation period, 18 of the 164 cells conducted only a short, one-stroke displacement of about 1.2 (±0.9) μm and then stopped. During these short movements, the average speed was 2.0 (±1.0) μm/min (determined from 904 speed measurements). In contrast to the mglAB pilR double-mutant phenotype, no reversals were observed. Therefore, type IV pili are required for cglB mutant cells to exhibit both the high-reversal mode and the fast movements when in close proximity.
DISCUSSION
A high-resolution motility assay (21) was used to examined the motility behavior of mglAB and cglB mutant cells. Quantification of the gliding movement of both strains revealed that individual cells moved in a jerky fashion with a high reversal frequency and a reduced speed compared to DK1622 wild-type cells (Fig. 2 and 5; Table 2). Thus, a decrease in continuous gliding activity, a high reversal frequency, and a reduced speed can explain the absence of net movement observed as the nonswarming phenotype of growing mgl colonies (11), as well as the absence of net movement after 3 h (12). We will first elaborate on the phenotype of a cglB strain (JZ315) in order to discuss the behavior of mgl strains.
S motility in single cells.
Recently, it was shown that the sglI locus in M. xanthus encodes genes necessary for the structure, export, assembly, function, and regulation of type IV pili (15, 23–26). These extracellular appendages are known to be involved in two types of bacterial surface translocation mechanisms: (i) twitching motility, where cells translocate for a few micrometers by sudden, jerky movements (3, 10), and (ii) S-motility gliding. Currently, details of twitching movement are unknown, and the mechanistic basis of twitching is obscure. Wild-type cells in close proximity to each other move with high speeds (5.0 ± 2.6 μm/min) compared to speeds of isolated cells (3.8 ± 1.4 μm/min) (21). In the cglB strain JZ315 (A− S+), movement is detectable only when cells are in close proximity to each other. Cells were observed to translocate either by jerky movement in a high-reversal mode or by unidirectional movement at high, variable velocities. The latter feature is also observed in wild-type cells (Fig. 6; Table 3). Since the cglB cells are defective in A motility, both movement patterns are solely due to S motility. Single-cell studies on other A− S+ strains, including agl mutants, should reveal whether these motility patterns are representative of all A-motility mutants. As determined in preliminary experiments, cglC mutant cells (DK1219) exhibited a phenotype similar to the one observed for cglB mutants (data not shown).
Considering that movement by S motility depends on type IV pili, it seems plausible that S-motility in M. xanthus is related to twitching; both motility forms require type IV pili and are comprised of jerky movements. However, in contrast to twitching, movement by S motility in M. xanthus is mostly in the direction of a cell’s long axis. S motility is observed only when cells are in close proximity to each other, i.e., when other cells provide part or all of the surface to move on (e.g., during aggregation and fruiting-body formation). It was previously shown that the A- and the S-motility systems provide advantages for M. xanthus cells to translocate on different surfaces (20).
Motility defect of mgl mutants.
It was previously shown that the cellular level of the MglA protein in an mglB deletion mutant is reduced to about 15% of the wild-type level (8). Here, we show that the average reversal frequency and the gliding speed in mgl mutant cells also correlate with the level of MglA protein; on average, wild-type cells glide with a speed of 4.4 ± 2.2 μm/min and 0.17 reversal per min, ΔmglB mutant cells (containing 15% of the wild-type MglA level) glide at 2.6 ± 1.5 μm/min and with 1.8 reversals per min, and ΔmglAB mutant cells (containing no MglA) glide at 1.9 ± 1.1 μm/min and with 2.9 reversals/min (Table 2). An alteration in reversal frequency is not necessarily coupled to a change in gliding speed, as is evident from the motility phenotype of frzD mutant cells (Table 2). These cells move as isolated cells with about wild-type speed (4.0 ± 2.3 μm/min) but have an increased reversal frequency (1.5 reversals/min). In contrast, ΔmglB mutant cells reverse at a frequency (1.8 reversals/min) which is similar to that of frzD mutants (1.5 reversals/min) but have a decreased gliding speed (2.6 ± 1.5 μm/min). Thus, mgl mutant cells carry two motility defects: one in single-cell gliding speed and one in control of reversal frequency.
In M. xanthus, the reversal frequency is controlled not only by mglA but also by the frz genes. Null mutations in frz and mglAB have opposite effects; single cells of frzE::Tn5 insertion mutants showed a low number of reversals (<0.02 min−1), whereas cells of ΔmglAB mutants exhibited a high number of reversals (2.9 min−1) (Table 2). The single-cell motility assay (Fig. 2, 3, and 4; Table 2) and a swarming assay (Fig. 7 and 8) were used to examine the possible connection between mgl-dependent and frz-dependent reversals of gliding movements by analyzing mglAB, frz, and mgl frz mutant cells. The observation that mglAB frzE double mutants exhibit a high reversal frequency rules out the possibility that reversals in M. xanthus are generated independently by MglA and FrzE.
One important question is how a swarming pattern relates to the gliding speed and reversal frequency of single cells. Wild-type cells glide with an average speed of 4.4 μm/min and an average reversal frequency of 0.17 reversal per min (21) (Table 2). The edge of a swarming wild-type colony shows many single cells and cells in loose groups (Fig. 7A). Single cells of frzD mutants glide with an average speed of 4.0 μm/min and an average of 1.5 reversals per min (Table 2) (2). This reversal frequency is about 10-fold higher than that of wild-type cells. Edges of growing colonies of frzD mutant cells are sharp and coherent, and no single cells are visible (Fig. 7B). Finger-like projections or flares can be observed (Fig. 7B). Colonies of ΔmglAB mutants have a similar morphology except that they are smaller and do not have finger-like projections (Fig. 7C). Single-cell analysis of ΔmglAB mutant cells revealed a reversal frequency of 2.9 reversals per min, which is about 20-fold above the wild-type reversal frequency. Additionally, the gliding speed of ΔmglAB cells was dramatically reduced, to 1.9 μm/min from the 4.4 μm/min for wild-type cells (Table 2). Thus, it seems that a 10-fold increase in reversal frequency to above that found for wild-type cells is sufficient for the formation of a sharp edge of an M. xanthus colony. It is also important to notice that reversals in frz mutants have been observed for isolated cells, whereas with cglB and most mglAB mutant cells, reversals were detected for cells in close proximity to each other.
Studies of single-cell movement and of swarming patterns under vegetative and developmental conditions for mgl, frz, and mgl frz mutants revealed an interesting connection between the two sets of genes: the average gliding speed and reversal frequency observed for mglAB mutant cells are epistatic in a frzE mutant background (Table 2). The motility behavior of mglAB frzD double-mutant cells seems to be quite different from that of mglAB and mglAB frzE mutant cells. Under vegetative (Fig. 7D) and developmental (Fig. 8D) conditions, the flares at the edges of mglAB frzD double-mutant swarms resemble those of frzD mutants but not those of mglAB mutants. Flare formation suggests that movement occurred in mglAB frzD cells to an extent that is different from that in mglAB cells. Analysis of the average gliding speed and reversal frequency of single mglAB and mglAB frzD cells did not reveal a significant difference between the two mutant strains (Table 2). Small differences in gliding speed and reversal frequency between different mutant strains may not be detectable by quantitative video microscopy under the conditions employed and may be recognized as a difference in swarming behavior only after prolonged incubation. Flares observed in mglAB frzD swarms were noticeable only after incubation for more than 4 days on CTT agar (Fig. 7A). Thus, a frzD mutation can partially suppress the S-motility (swarming) defect of mglAB mutant cells. Since mglAB colonies do not swarm, the suppression of this defect by frzD seems to be a gain of function in S motility.
Although colonies of both mglAB and A− S− mutants exhibit round, sharp edges, the motility patterns of single cells are clearly different (Fig. 2, 5, and 6; Tables 2 and 3). In contrast to A− S− cells, ΔmglAB cells show a residual movement pattern of high reversals that requires the presence of type IV pili (encoded by S-system genes [Table 3]). Furthermore, a frzD mutation partially suppresses the group swarming defect of ΔmglAB cells. This effect is more visible under conditions of low percent agar concentration, where M. xanthus moves mostly by S motility (20). Comparing the gliding movements of ΔmglAB and A− S− cells, it seems that the ΔmglAB mutant behaves as if it is a strong A-motility system mutant with only partially defective S motility.
ACKNOWLEDGMENTS
We thank Hans Warrick for many helpful discussions and advice on using the optical and electronic equipment.
This work was supported by National Science Foundation grant MCB 9423182 to D.K. A.M.S. was a recipient of a postdoctoral fellowship from the Max-Planck-Gesellschaft.
REFERENCES
- 1.Alberti L, Harshey R M. Differentiation of Serratia marcescens 274 into swimmer and swarmer cells. J Bacteriol. 1990;172:4322–4328. doi: 10.1128/jb.172.8.4322-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackhart B D, Zusman D R. “Frizzy” genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci USA. 1985;82:8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradley D E. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol. 1980;26:146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 4.Burchard R P. Studies on gliding motility in Myxococcus xanthus. Arch Microbiol. 1974;99:271–280. doi: 10.1007/BF00696242. [DOI] [PubMed] [Google Scholar]
- 5.Darzins A. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol. 1994;11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 6.Hartzell P. Complementation of sporulation and motility defects in a prokaryote by a eukaryotic GTPase. Proc Natl Acad Sci USA. 1997;94:9881–9886. doi: 10.1073/pnas.94.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartzell P, Kaiser D. Function of MglA, a 22-kilodalton protein essential for gliding in Myxococcus xanthus. J Bacteriol. 1991;173:7615–7624. doi: 10.1128/jb.173.23.7615-7624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartzell P, Kaiser D. Upstream gene of the mgl operon controls the level of MglA protein in Myxococcus xanthus. J Bacteriol. 1991;173:7625–7635. doi: 10.1128/jb.173.23.7625-7635.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henrichsen J. Bacterial surface translocation: survey and a classification. Bacteriol Rev. 1972;36:478–503. doi: 10.1128/br.36.4.478-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henrichsen J. Twitching motility. Annu Rev Microbiol. 1983;37:81–93. doi: 10.1146/annurev.mi.37.100183.000501. [DOI] [PubMed] [Google Scholar]
- 11.Hodgkin J, Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci USA. 1977;74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol Gen Genet. 1979;171:167–176. [Google Scholar]
- 13.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 14.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser D, Crosby C. Cell movement and its coordination in swarms of Myxococcus xanthus. Cell Motil. 1983;3:227–245. [Google Scholar]
- 16.Kalos M, Zissler J F. Defects in contact-stimulated gliding during aggregation by Myxococcus xanthus. J Bacteriol. 1990;172:6476–6493. doi: 10.1128/jb.172.11.6476-6493.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kroos L, Hartzell P, Stephens K, Kaiser D. A link between cell movement and gene expression argues that motility is required for cell-cell signaling during fruiting body development. Genes Dev. 1988;2:1677–1685. doi: 10.1101/gad.2.12a.1677. [DOI] [PubMed] [Google Scholar]
- 18.Kühlwein H, Reichenbach H. Schwarmentwicklung und Morphogenese bei Myxobakterien: Archangium-Myxococcus-Chondromyces. Göttingen, Germany: Institut für den wissenschaftlichen Film; 1968. [Google Scholar]
- 19.McBride M J, Weinberg R A, Zusman D R. “Frizzy” aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc Natl Acad Sci USA. 1989;86:424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Morandi, D. Unpublished data.
- 19b.Rodriguez, A. M., and A. M. Spormann. Genetic and molecular analysis of cglB, a gene essential for single cell gliding in Myxococcus xanthus. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 20.Shi W, Zusman D R. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc Natl Acad Sci USA. 1993;90:3378–3382. doi: 10.1073/pnas.90.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spormann A M, Kaiser D. Gliding movements in Myxococcus xanthus. J Bacteriol. 1995;177:5846–5852. doi: 10.1128/jb.177.20.5846-5852.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephens K, Kaiser D. Genetics of gliding motility in Myxococcus xanthus: molecular cloning of the mgl locus. Mol Gen Genet. 1987;207:256–266. [Google Scholar]
- 23.Wu S S, Kaiser D. Genetic and functional evidence that type IV pili are required for social motility in Myxococcus xanthus. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 24.Wu S S, Kaiser D. Markerless deletion of pil genes in Myxococcus xanthus generated by counterselection with the Bacillus subtilis sacB gene. J Bacteriol. 1996;178:5817–5821. doi: 10.1128/jb.178.19.5817-5821.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu S S, Kaiser D. The Myxococcus xanthus pilT locus is required for social gliding although pili are still produced. Mol Microbiol. 1997;23:109–121. doi: 10.1046/j.1365-2958.1997.1791550.x. [DOI] [PubMed] [Google Scholar]
- 26.Wu S S, Kaiser D. Regulation of expression of the pilA gene of Myxococcus xanthus. J Bacteriol. 1997;179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zusman D R. “Frizzy” mutants: a new class of aggregation-defective developmental mutants of Myxococcus xanthus. J Bacteriol. 1982;150:1430–1437. doi: 10.1128/jb.150.3.1430-1437.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]